Abstract
The transdifferentiation of epithelial cells into motile mesenchymal cells, a process known as epithelial–mesenchymal transition (EMT), is integral in development, wound healing and stem cell behaviour, and contributes pathologically to fibrosis and cancer progression. This switch in cell differentiation and behaviour is mediated by key transcription factors, including SNAIL, zinc-finger E-box-binding (ZEB) and basic helix-loop-helix transcription factors, the functions of which are finely regulated at the transcriptional, translational and post-translational levels. The reprogramming of gene expression during EMT, as well as non-transcriptional changes, are initiated and controlled by signalling pathways that respond to extracellular cues. Among these, transforming growth factor-β (TGFβ) family signalling has a predominant role; however, the convergence of signalling pathways is essential for EMT.
The idea that epithelial cells can downregulate epithelial characteristics and acquire mesenchymal characteristics arose in the early 1980s from observations made by Elizabeth Hay1, who described epithelial to mesenchymal phenotype changes in the primitive streak of chick embryos. Initially described as ‘epithelial to mesenchymal transformation’, this differentiation process is now commonly known as epithelial–mesenchymal transition (EMT) to emphasize its transient nature; mesenchymal–epithelial transition (MET) describes the reverse process. The ability of epithelial cells to transition into mesenchymal cells and back, either partially or fully, illustrates an inherent plasticity of the epithelial phenotype. During EMT, epithelial cells lose their junctions and apical–basal polarity, reorganize their cytoskeleton, undergo a change in the signalling programmes that define cell shape and reprogramme gene expression; this increases the motility of individual cells and enables the development of an invasive phenotype2,3. EMT is integral to development, and the processes underlying it are reactivated in wound healing, fibrosis and cancer progression3–5. EMT and MET also regulate embryonic stem cell differentiation, induced pluripotency and cancer stem cell behaviour (BOX 1). Depending on the tissue and signalling contexts, epithelial cells may lose only some characteristics or may show some epithelial and mesenchymal properties; this can be considered as partial EMT. Epithelial plasticity, therefore, covers a range of changes in differentiation and cell behaviour, with full epithelial integrity at one end and EMT as the full realization of a plasticity response at the other end6. Cell culture models of EMT enable the complex molecular mechanisms that underlie it to be characterized, and they have revealed cooperation and crosstalk between signalling pathways and transcriptional, translational and post-translational regulation.
Box 1 | EMT, MET and stem cells.
Epithelial–mesenchymal transition (EMT) and mesenchymal– epithelial transition (MET) have been closely linked to ‘stemness’ in development and cancer. The pluripotent embryonic stem (ES) cells in the inner mass of the blastocyst have epithelial characteristics229. In gastrulation, pluripotent epithelial epiblast cells ingress to form, through EMT, the primary mesoderm123. EMT thus represents an initial differentiation event in the generation of the three germ layers from pluripotent cells. Illustrating the importance of EMT in early differentiation, ES cell or epiblast cell colonies in culture give rise to peripheral cells with a mesenchymal phenotype, as judged by loss of epithelial cadherin (E-cadherin) expression and the expression of vimentin and neural cadherin (N-cadherin)229,230.
Conversely, the reprogramming of fibroblasts into induced pluripotent stem (iPS) cells requires the transition from a mesenchymal phenotype to an epithelial phenotype. This reprogramming recapitulates an MET process as it involves the repression of mesenchymal genes, including some that encode transcription factors with a role in EMT, and the activation of epithelial genes encoding epithelial cell junction proteins231,232. Consistent with the role of transforming growth factor-β (TGFβ) family proteins in EMT, TGFβ receptor inhibitors increase, whereas TGFβ decreases, the efficiency of iPS cell generation233. Furthermore, MET represents an initiation step that is required for progression towards pluripotency and which depends on, and is promoted by, bone morphogenetic protein (BMP) signalling234. As is the case in EMT, microRNAs (miRNAs) regulate MET, which is required for iPS cell reprogramming. Indeed, MET is promoted by the BMP-induced expression of the epithelial miR-200 family of miRNAs and miR-205 that repress zinc-finger E-box-binding 1 (ZEB1) and ZEB2 expression100,234. Additionally, downregulation of TGFβ receptor type II (TβRII) and RHOC expression by miR-302 or miR-372 increases reprogramming efficiency235.
EMT has also been associated with epithelial and carcinoma stem cell properties. Expression of SNAIL or TWIST in mammary epithelial cells induces a mesenchymal cell population marked with a CD44hiCD24low phenotype, which is similar to that observed in epithelial stem cells236. The correlation of EMT with stemness extends to carcinomas. These contain a subpopulation of self-renewing tumour-initiating cells, known as cancer stem cells (CSCs), which efficiently generate new tumours. In mammary carcinomas, induction of EMT promotes the generation of CD44hiCD24low CSCs that are able to form mammospheres, and similarly defined CSCs isolated from tumours express EMT markers236. Consistent with the reversible nature of EMT, differentiated cancer cells can transition into CSCs, and vice versa, enabling oncogenic mutations that arose in differentiated cancer cells to integrate through EMT into CSCs. As EMT promotes cell invasion that leads to tumour cell dissemination, this scenario enables CSCs with new oncogenic mutations to clonally expand, following invasion, dissemination and MET in secondary tumours237,238.
In cancer, both EMT and CSC generation have been associated with TGFβ signalling. For example, breast cancer CSCs show higher levels of TGFβ1 and TβRII expression than the more differentiated cells, and inhibition of TGFβ signalling in CSCs reestablishes an epithelial phenotype239. Also, WNT and Notch signalling are associated with CSCs. Colon CSCs show a high level of WNT signalling, with nuclear β-catenin at the invasive cancer front and in scattered tumour cells240,241. Notch signalling contributes to the generation of CSCs in other cancers242, including pancreatic adenocarcinomas243, and the inhibition of Notch signalling suppresses EMT and CSCs in a xenograft model244. As is the case in EMT- and MET-based cell reprogramming, miRNAs contribute to the generation and maintenance of CSCs. For example, the miR-106b-25 cluster induces EMT and tumour-initiating characteristics in breast cancer by repressing SMAD7 to increase TGFβ signalling245. However, it also promotes MET and iPS cell reprogramming by targeting TβRII, possibly reflecting context-dependent differences in its functions246.
This Review describes the molecular processes that lead to EMT. It first outlines the main changes that occur in cells undergoing EMT, before focusing on mechanisms that direct changes in gene expression and the signalling pathways that control the initiation and progression of EMT.
EMT in development and disease
Epithelia are established as single cell layers or multilayer tissues with various functions. Epithelial cells show apical–basal polarity, adhere and communicate with each other through specialized intercellular junctions and are positioned on a basement membrane that helps to define their physiology; for example, through the interaction of basement membrane proteins with integrins. In this way, epithelia function as permeability barriers that delineate tissues and organs6. The transition of epithelial cells into mesenchymal cells, in development or pathologically, follows a common and conserved programme with hall-marks. However, it also has an inherent flexibility and some variation, which depends on the cell type, tissue context and signals that activate the EMT programme. Indeed, EMT has been assigned three distinct subtypes, which are dependent on the physiological context4. Furthermore, the plasticity of the epithelial phenotype enables cells to transition through multiple rounds of EMT and MET (FIG. 1).
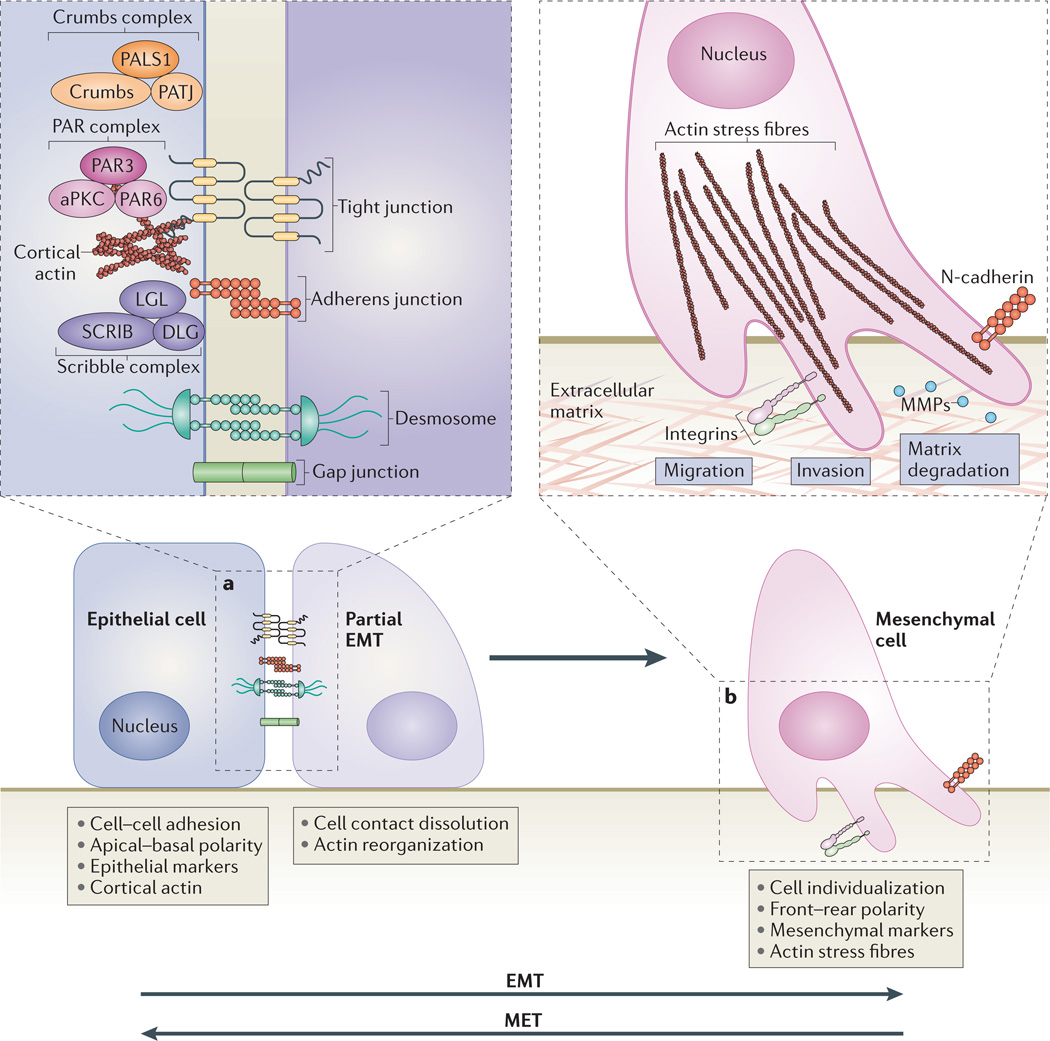
Figure 1. Cellular events during EMT.
a | The first steps of epithelial–mesenchymal transition (EMT) are the disassembly of epithelial cell–cell contacts — that is, tight junctions, adherens junctions, desmosomes and gap junctions — and the loss of cell polarity through the disruption of the Crumbs, partitioning defective (PAR) and Scribble (SCRIB) polarity complexes. The expression of epithelial genes is repressed, concomitantly with the activation of mesenchymal gene expression. b | Next, the epithelial actin architecture reorganizes, and cells acquire motility and invasive capacities by forming lamellipodia, filopodia and invadopodia, and by expressing matrix metalloproteinases (MMPs) that can degrade extracellular matrix (ECM) proteins. The process of mesenchymal–epithelial transition (MET) enables the cells that have undergone EMT to revert to the epithelial state. Three types of EMT can be discerned depending on the physiological tissue context4. Type 1 EMT occurs in embryogenesis and organ development262, type 2 EMT is important for tissue regeneration and organ fibrosis4,263, and type 3 EMT is associated with cancer progression4 and cancer stem cell properties237. MET also contributes to development (for example, kidney development264) and the generation of metastatic carcinomas238,265. Consecutive rounds of EMT and MET occur during development262,264. In primary EMT, ectodermal or epithelial cells without prior history of EMT differentiate into mesenchymal cells. In secondary EMT, cells that have already undergone EMT and reverted to the epithelial state initiate a new EMT process3,262. Similarly, following dissemination, cancer cells can revert through MET to an epithelial state, leading to the formation of secondary carcinomas with phenotypes similar to the primary tumour265. aPKC, atypical protein kinase C; DLG, discs large; LGL, lethal giant larvae; N-cadherin, neural cadherin; PALS1, protein associated with Lin-7 1; PATJ, PALS1-associated tight-junction protein.
In all tissue contexts, key events in EMT are the dissolution of the epithelial cell–cell junctions; loss of a pical–basal polarity and acquisition of a front–rear polarity; reorganization of the cytoskeletal architecture and changes in cell shape; downregulation of an epithelial gene expression signature and activation of genes that help to define the mesenchymal phenotype; increased cell protrusions and motility; and, in many cases, an ability to degrade extracellular matrix (ECM) proteins to enable invasive behaviour (FIG. 1). Additionally, cells that have undergone EMT acquire resistance to senescence and apoptosis3. Similarly to epithelial cells, endothelial cells can transition into a mesenchymal phenotype, and the mesenchymal transdifferentiation of epithelial or endothelial cells can result in the expression of myofibroblast genes (BOX 2).
Box 2 | EndMT and the expression of smooth muscle actin.
Similarly to epithelial cells, endothelial cells can transition to a mesenchymal phenotype, a process known as endothelial-mesenchymal transition (EndMT)247,248. EndMT is developmentally required in endocardial cushion formation, and contributes to fibroblast accumulation in mouse models of renal, pulmonary and cardiac fibrosis249. EndMT also provides a major source of cancer-associated fibroblasts that contribute to carcinoma progression250. Additionally, endothelial cells generate chondrocytes and osteoblasts in heterotypic bone formation, which characterizes fibrodysplasia ossificans progressiva251.
During EndMT, as in epithelial–mesenchymal transition (EMT), endothelial cells downregulate the expression of cell junction proteins, specifically vascular endothelial (VE)-cadherin and cluster of differentiation 31 (CD31; also known as PECAM1) (components of adherens junctions)120, and the endothelial tight junction protein claudin 5 (REFS 248,252). The transcription programmes and signalling pathways that drive EndMT share extensive similarities with EMT. SNAIL transcription factors are induced in EndMT and directly repress the expression of the genes encoding VE-cadherin and CD31, while activating the expression of invasion-associated genes and the migratory phenotype213,248. In heart development, SNAIL1, SNAIL2 and TWIST1 cooperate with the transcription factors hairy/enhancer-of-split related with YRPW motif protein 1 (HEY1) and HEY2 downstream of Notch signalling, to drive EndMT in a signalling crosstalk that is balanced by bone morphogenetic protein 2 (BMP2) and NOTCH1 (REF. 253). Disturbing this balance leads to septal or valve malformation253.
As in EMT, transforming growth factor-β (TGFβ) family signalling through SMADs induces EndMT, which further emphasizes the similarities between EMT and EndMT. TGFβ activates the expression of SNAIL, which drives EMT and EndMT, and represses the expression of VE-cadherin, CD31 and claudin 5 (REF. 248). A dominant-negative form of SMAD4 blocks TGFβ-induced EndMT, which correlates with decreased SNAIL expression, attenuated decrease in VE-cadherin and CD31 expression, and increased fibroblast-specific protein 1 (FSP1) and α-smooth muscle actin (αSMA) expression254. As TGFβ2 and BMP4 can induce EndMT through the activin receptor-like kinase 2 (ALK2) receptor129, SMAD1 and/or SMAD5 may have important roles in controlling endothelial plasticity. The epidermal growth factor (EGF) receptor human EGF receptor 2 (HER2) also participates in EndMT during heart valve formation, in an ERK MAPK signalling-dependent manner255.
EMT and EndMT can generate mesenchymal cells that express αSMA, as seen in myofibroblasts, and these processes have been named EMyT and EndMyT248,256,257. It is, however, unclear whether functional myofibroblasts seen in fibrosis or cancer derive from epithelial or endothelial cells. In EMyT, the transcription factor serum response factor (SRF) and myocardin family transcription factors, including myocardin and myocardin-related transcription factors (MRTFs; also known as MKLs), drive expression of the myofibroblast markers αSMA and smooth muscle protein 22α (SM22α; also known as transgelin). Zinc-finger E-box-binding 1 (ZEB1) and SMAD3 interact with SRF to activate αSMA expression142, and SMAD3 interacts with myocardin to activate SM22α expression258. In response to TGFβ, SMAD3 also induces expression and nuclear import of MRTFs, and cooperates with MRTFs to induce myofibroblast differentiation, partly by inducing the expression of SNAIL2 (REFS 141,259). In kidney epithelial cells, SMAD3 has a biphasic role in TGFβ-induced EMyT; it drives EMT at an early stage of differentiation, but it is degraded at a later stage. Persistent SMAD3 expression interferes with SRF–MRTF complex formation, thus repressing αSMA expression260. In addition, β-catenin, which is released from junctions, antagonizes the inhibitory effect of SMAD3 on the MRTF–SRF complex, which in turns increases αSMA expression261.
Deconstructing cell junctions and polarity
Specialized cell surface protein complexes form epithelial cell–cell junctions that are essential for epithelial integrity. Vertebrate cells contact each other through subapical tight junctions, adherens junctions and desmosomes at lateral surfaces, and scattered gap junctions at lateral surfaces6 (FIG. 1). Upon the initiation of EMT, these junctions are deconstructed and the junction proteins are relocalized and/or degraded. The dissolution of tight junctions during EMT is accompanied by decreased claudin and occludin expression, and the diffusion of zonula occludens 1 (ZO1; also known as TJP1) from cell–cell contacts6. During the destabilization of adherens junctions, epithelial cadherin (E-cadherin) is cleaved at the plasma membrane and subsequently degraded7. Consequently, β-catenin can no longer interact with E-cadherin and it is either degraded or protected from degradation (for example, in response to WNT signalling), so that it can act in transcription8. p120 catenin (also known as catenin δ1) also accumulates in the nucleus and participates in transcription following a decrease in E-cadherin levels9. EMT initiation also disrupts desmosomes6,7, and the integrity of gap junctions is compromised by decreased connexin levels10. As EMT progresses, the expression of junction proteins is transcriptionally repressed, which stabilizes the loss of epithelial junctions11,12.
Instructed through contacts with the basement membrane, epithelial cells display apical–basal polarity, which is organized by polarity complexes that are physically and functionally integrated with the cell junction architecture. In vertebrate cells, partitioning-defective (PAR) complexes (comprising PAR6, PAR3 and atypical protein kinase C (aPKC)) and Crumbs complexes (comprising Crumbs (CRB), protein associated with Lin-7 1 (PALS1) and PALS1-associated tight-junction protein (PATJ)) localize apically in association with tight junctions and define the apical compartment; Scribble complexes (comprising Scribble (SCRIB), Discs large (DLG) and lethal giant larvae (LGL)) define the basolateral compartment13 (FIG. 1). Consequently, the dissolution of epithelial junctions during EMT confers a loss of apical–basal polarity6. In support of this association, decreased expression of E-cadherin in tumour cells prevents SCRIB from interacting with the lateral plasma membrane14, and decreasing SCRIB or E-cadherin expression reduces adhesion and increases cell motility15. After EMT is initiated, the expression of polarity complex proteins, such as CRB3 and LGL2, is repressed16, which further destabilizes the polarized phenotype.
Cytoskeletal changes and motility
Cells that undergo EMT reorganize their cortical actin cytoskeleton into one that enables dynamic cell elongation and directional motility2,7,17 (FIG. 1). New actin-rich membrane projections facilitate cell movement and act as sensory extensions of the cytoskeleton. These projections include sheet-like membrane protrusions called lamellipodia and spike-like extensions called filopodia at the edge of lamellipodia18. Actin-rich invadopodia exert a proteolytic function in ECM degradation, thus facilitating cell invasion18,19. Finally, EMT is characterized by increased cell contractility and actin stress fibre formation. These dynamic changes in actin organization are probably mediated by regulatory proteins such as moesin20, but the molecular mechanisms controlling F-actin dynamics during EMT remain to be elucidated.
RHO GTPases regulate actin dynamics and control actin rearrangement during EMT. Among these, RHOA promotes actin stress fibre formation, whereas RAC1 and CDC42 mainly promote the formation of lamellipodia and filopodia. The activation of RHO GTPases is tightly regulated by guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs) and guanine nucleotide dissociation inhibitors (GDIs)7,18. Following RHO activation at the onset of EMT, the RHO-associated kinase (ROCK) cooperates with the formin diaphanous 1 (DIA1) to promote actin polymerization. ROCK also induces myosin light chain phosphorylation, which enhances acto myosin contractility and activates LIM kinase (LIMK) to in activate the actin severing factor cofilin21. Following activation by RAC1 or CDC42, the kinase p21 activated kinase 1 (PAK1) activates targets that are involved in cell spreading and motility22. RHO GTPases also regulate the formation of cell–cell junction complexes and the stability of adherens junctions. Cytoplasmic p120 catenin, which is generated during EMT, represses RHO activity. This may facilitate the dissociation of cell junctions and activates RAC and CDC42 to induce the formation of membrane protrusions and cell motility23.
The conversion from apical–basal polarity to front– rear polarity involves interplay between RHO GTPases and proteins that define apical–basal polarity 24,25. T o achieve directional polarity, the PAR and Scribble complexes together with PATJ of the Crumbs complex relocalize to the leading edge of the cell, where RAC1 and CDC42 activate actin polymerization and membrane protrusion formation. By contrast, RHOA localizes at the rear of the cell and promotes the disassembly of adhesion complexes and cell retraction24. PI3K plays a central part in initiating front–rear polarity and participates in the recruitment of CDC42 and RAC GEFs to the leading edge24. The front–rear polarity is maintained through feedback mechanisms that involve polarity proteins and small GTPases. For example, RAC1 activation at the leading edge stimulates PI3K, which recruits RAC GEFs. RHO GTPases also control the reorganization of the microtubule cytoskeleton. For instance, RAC1 promotes microtubule polymerization and is itself activated by it at the leading edge of cells, which sets up a positive feedback loop that participates in the reorganization of the microtubule cytoskeleton26. Additionally, RAC1 promotes the clustering of integrins towards the front of the cell, and the localization of PAR3 and aPKC at the front of the cell requires PATJ, PALS1 and PAR3 (REF. 24). At the rear of the cell, RHOA signalling inhibits RAC1 activation and prevents formation of PAR complexes24,25. The reorganization of the cytoskeletal architecture and polarity complexes, which result in cell shape changes, cell elongation, membrane protrusions and front–rear polarity, are essential in EMT and enable directional migration.
Overview of changes in gene expression
During EMT, cells downregulate the expression of epithelial proteins, including those that are part of cell junction complexes6,11 (FIGs 1,2). They also redirect their gene expression programme to promote changes in cytoskeletal architecture, to promote adhesion to mesenchymal cells and to alter the interaction of cells with the ECM7,12,17. Variations in EMT-associated gene expression profiles are apparent and depend on cell and tissue type and on the degree of progression towards mesenchymal differentiation.
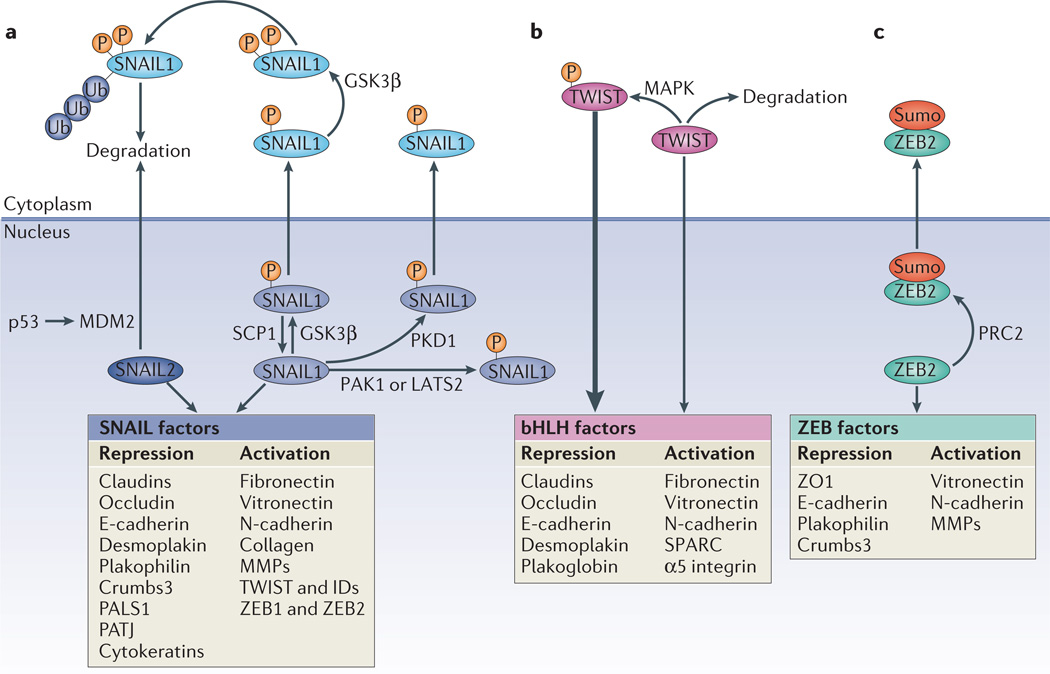
Figure 2. Roles and regulation of major EMT transcription factors.
Epithelial–mesenchymal transition (EMT) is driven by SNAIL, zinc-finger E-box-binding (ZEB) and basic helix–loop–helix (bHLH) transcription factors that repress epithelial marker genes and activate genes associated with the mesenchymal phenotype. Post-translational modifications regulate their activities, subcellular localization and stability. a | Glycogen synthase kinase-3β (GSK3β) phosphorylates (P) SNAIL1 at two motifs; phosphorylation of the first motif facilitates the nuclear export of SNAIL1, and phosphorylation of the second motif enables the ubiquitin (Ub)-mediated degradation of SNAIL1. Phosphorylation of SNAIL1 by protein kinase D1 (PKD1) also leads to its nuclear export. Conversely, phosphorylation of SNAIL1 by p21 activated kinase 1 (PAK1) or large tumour suppressor 2 (LATS2), or dephosphorylation of SNAIL1 by small C-terminal domain phosphatase1 (SCP1) promotes the nuclear retention of SNAIL1 and enhances its activity. SNAIL2 is degraded as a result of its p53-mediated recruitment to the p53–mouse double minute 2 (MDM2) complex. b | TWIST is phosphorylated by the MAPK p38, JUN N-terminal kinase (JNK) and ERK, which protects it from degradation, and thus promotes its nuclear import and functions. c | ZEB2 is sumoylated (Sumo) by Polycomb repressive complex 2 (PRC2) and subsequently exported from the nucleus, which reduces its activity as a transcription factor. E-cadherin, epithelial cadherin; ID, inhibitor of differentiation; MMP, matrix metalloproteinase; N-cadherin, neural cadherin; PALS1, protein associated with Lin-7 1; PATJ, PALS1-associated tight-junction protein; SPARC, secreted protein acidic and rich in Cys; ZO1, zonula occludens 1.
A hallmark of EMT is the downregulation of E-cadherin to reinforce the destabilization of adherens junctions. Additionally, the repression of genes encoding claudins and occludin, and desmoplakin and plakophilin, stabilizes the dissolution of apical tight junctions and desmosomes, respectively6. These changes in gene expression prevent the de novo formation of epithelial cell–cell junctions and result in the loss of the epithelial barrier function11. The repression of genes encoding epithelial (or endothelial (BOX 1) cell junction proteins is accompanied by the activation of genes, the protein products of which promote mesenchymal adhesion. Specifically, the downregulation of E-cadherin is balanced by the increased expression of mesenchymal neural cadherin (N-cadherin), which results in a ‘cadherin switch’ that alters cell adhesion7,27. Through this switch, the transitioning cells lose their association with epithelial cells and acquire an affinity for mesenchymal cells through homotypic N-cadherin interactions; these interactions are weaker than homotypic E-cadherin interactions and facilitate cell migration and invasion28. N-cadherin connects to the cytoskeleton through α-catenin and β-catenin and also interacts with p120 catenin, signalling mediators29 and receptor tyrosine kinases (RTKs) such as platelet-derived growth factor (PDGF) and fibroblast growth factor receptors (FGFRs)7,30. EMT also activates the expression of neural cell adhesion molecule (NCAM), another adhesion molecule that interacts with N-cadherin to modulate the activity of RTKs that are associated with it7,31. NCAM interacts with the SRC family tyrosine kinase FYN to facilitate the assembly of focal adhesions, migration and invasion32.
Alterations in the expression of genes encoding cytoskeletal and polarity complex proteins also contribute to EMT. The intermediate filament composition changes with the repression of cytokeratin and the activation of vimentin expression6. Keratin and vimentin filaments regulate the trafficking of organelles and membrane-associated proteins, but they show differences in the proteins that they target to the membrane. For example, keratin, but not vimentin, directs E-cadherin to the membrane33. Changes in intermediate filament composition also enable cell motility, possibly owing to the interaction of vimentin with motor proteins34. To enable directional motility, cells undergoing EMT repress the expression of the apical Crumbs complex proteins Crumbs3 and PATJ, and of basolateral Scribble complex proteins, such as LGL2 (REFS 7,16,24).
Remodelling of the ECM and changes to cell interactions with the ECM are essential in the initiation and progression of EMT. Integrin complexes enable cells to receive signals from ECM proteins through interactions with signalling mediators, such as integrin-linked kinase (ILK), particularly interesting new Cys–His protein 1 (PINCH; also known as LIMS1) and parvin7,30. As epithelial cells differentiate into mesenchymal cells, they do not interact with a basement membrane and communicate with a different ECM. Therefore, the cells downregulate some epithelial integrins, but activate the expression of others; some of these newly expressed integrins have key roles in EMT progression7. For example, EMT down-regulates the expression of the epithelial α6β4 integrins that mediate contacts with the basement membrane through epigenetically silencing the gene encoding β4 (REF. 35). Additionally, epithelial α3β1 integrin, which binds laminin but also associates with E-cadherin, is required for progression through EMT and integrates β-catenin and transforming growth factor-β (TGFβ)– SMAD signalling36. Increased α5β1 integrin expression during EMT increases cell adhesion to fibronectin, the expression of which is also activated during EMT and promotes cell migration37,38. The increased expression of α1β1 or α2β1 integrins and their interactions with type I collagen facilitate the disruption of E-cadherin complexes and the nuclear translocation of β-catenin39.
Changes to the integrin repertoire during EMT correlate with the increased expression of proteases, such as the matrix metalloproteinases MMP2 and MMP9, thus enhancing ECM protein degradation and enabling invasion40. MMPs additionally target some transmembrane proteins, which results in, for example, the release of the extracellular domain of E-cadherin, hence contributing to the loss of adherens junctions40. Furthermore, MMP3 can induce EMT through increased expression and activity of RAC1B, which results in increased levels of cellular reactive oxygen species, leading to increased SNAIL1 expression41. Increased αvβ6 integrin expression during EMT correlates with an increase in the expression of proteases, and the colocalization of these proteins in invadopodia mediates invasive cell behaviour7,19. Also, αvβ3 integrin, the expression of which is increased with EMT, contributes to pro-invasive functions at the leading edge of cancer cells42. Finally, localized ECM degradation by invading cells may release stored growth factors that then act on cells. Some proteases that mediate invasion and some integrins, such as αvβ6, activate the differentiation factor TGFβ that is stored in a latent form in the ECM43. This exposes the cells to increased TGFβ signalling, which promotes EMT (see below) and stimulates the expression of some ECM proteins, such as collagens and fibronectin, enhancing the remodelling of the ECM into a matrix with a different composition and properties2,3.
Transcription factors driving EMT
The changes in gene expression that contribute to the repression of the epithelial phenotype and activation of the mesenchymal phenotype involve master regulators, including SNAIL, TWIST and zinc-finger E-box-binding (ZEB) transcription factors. Their expression is activated early in EMT, and they thus have central roles in development, fibrosis and cancer (FIG. 2). As these transcription factors have distinct expression profiles, their contributions to EMT depend on the cell or tissue type involved and the signalling pathways that initiate EMT. They often control the expression of each other and functionally cooperate at target genes11, and additional transcription factors further define the EMT transcription programme and drive EMT progression. Together, the EMT transcription factors coordinate the repression of epithelial genes and the induction of mesenchymal genes, and often the same transcription factors direct both repression and activation12.
SNAIL transcription factors
Of the three vertebrate SNAIL proteins, SNAIL1 (also known as SNAIL) and SNAIL2 (also known as SLUG) activate the EMT programme during development, fibrosis and cancer44 (FIG. 2). They repress epithelial genes by binding to E-box DNA sequences through their carboxy-terminal zinc-finger domains11,45 (FIG. 2; TABLE 1). How SNAIL represses gene expression is well illustrated by the activity of SNAIL1 at the E-cadherin promoter46–52. Upon binding of E-box sequences in the proximal promoter region of the E-cadherin gene, SNAIL1 recruits the Polycomb repressive complex 2 (PRC2), which contains the methyltransferases enhancer of zeste homologue 2 (EZH2), G9a and suppressor of variegation 3–9 homologue 1 (SUV39H1), the co-repressor SIN3A, histone deacetylases 1, 2 and/or 3, and the Lys-specific demethylase 1 (LSD1); all of these components coordinate histone modifications, specifically methylation and acetylation at histone H3 Lys 4 (H3K4), H3K9 and H3K27 (REFS 48–53). H3K9 methylation and H3K27 methylation mark repressive chromatin, and H3K4 methylation and H3K9 acetylation mark active chromatin. Repressive and active marks in ‘bivalent domains’, as seen in the E-cadherin promoter, cooccur at many promoters in embryonic stem cells54 and create a poised state for the promoter that enables timely activation while maintaining repression in the absence of differentiation signals. The bivalent control of the E-cadherin promoter may contribute to the reversible nature of EMT. In addition to repressing epithelial genes, SNAIL1 activates genes that contribute to the mesenchymal phenotype (TABLE 1). This may also involve bivalent domains with, for example, repressive H3K9 trimethylation and activating H3K18 acetylation, which enables mesodermal goosecoid expression in response to the TGFβ-related Nodal55.
Table 1.
EMT transcription factors, their direct targets and the signalling pathways that induce their activity
| Transcription factor |
Downregulated expression during EMT |
Upregulated expression during EMT |
Regulatory signalling pathways | Refs |
|---|---|---|---|---|
| SNAIL1 and SNAIL2 |
E-cadherin, claudins, occludin, Crumbs3, PALS1, PATJ, cytokeratins, desmoplakin and plakophilin |
Fibronectin, N-cadherin, collagen, MMP15, MMP2, MMP9, TWIST, ID1, ID2, ZEB1 and ZEB2 |
TGFβ–SMAD3, WNT–β-catenin, Notch, PI3K–AKT, NF-κB, EGF and FGF |
11,45,59,60,61, 66,159,185 |
| TWIST1 | E-cadherin, claudins, occludin, desmoplakin and plakoglobin |
Fibronectin, N-cadherin and α5 integrin |
MAPK | 11,45,67,68,72 |
| ZEB1 and ZEB2 | E-cadherin, ZO1, Crumbs3 and plakophilin |
N-cadherin and MMPs | TGFβ–SMAD3, WNT–β-catenin and RAS–MAPK |
11,45,73–75, 77,142,143 |
| FOXD3 | Unknown | Unknown | β1 integrin and laminin | 266 |
| FOXC2 | E-cadherin | Fibronectin, vimentin, N-cadherin and αSMA |
TGFβ–SMAD3 | 267 |
| FOXF1 | E-cadherin, claudin1, occludin, desmoglein1β, desmoglein2, desmocollin2 and desmoplakin |
Fibronectin and N-cadherin | Unknown | 268 |
| FOXQ1 | E-cadherin | Fibronectin, N-cadherin and vimentin |
Unknown | 269 |
| FOXO3A | E-cadherin | SNAIL1 | AKT | 270 |
| FOXA1 | E-cadherin | Fibronectin, vimentin and SNAIL1 | TGFβ, HGF and AKT | 271 |
| FOXA2 | E-cadherin and ZO1 | Fibronectin, vimentin, N-cadherin, SNAIL1 and SNAIL2 |
TGFβ, HGF and AKT | 271 |
| Serpent, GATA4 and GATA6 |
E-cadherin, Crumbs and claudins |
N-cadherin and MMP1 | Unknown | 81 |
| HMGA2 | E-cadherin | SNAIL1, SNAIL2 and TWIST | TGFβ–SMAD3 | 103 |
| SOX9 | Unknown | SNAIL2 | BMPs and PKA | 83 |
| KLF8 | E-cadherin | MMP9 | Unknown | 272 |
| CBFA–KAP1 | Unknown | FSP1 | Unknown | 273 |
| ZNF703 (also known as Zeppo1 in mice) |
E-cadherin | Vimentin, N-cadherin, SNAIL1 and cytokeratin |
RHO-GTPase | 274 |
| PRX1 | E-cadherin | Vimentin and laminin | BMP2 and TGFβ | 275 |
αSMA, α-smooth muscle actin; BMPs, bone morphogenetic proteins; CBFA, CArG-binding factor-A; E-cadherin, epithelial cadherin; EGF, epidermal growth factor; EMT, epithelial-mesenchymal transition; FGF, fibroblast growth factor; FOX, forkhead box; FSP1, fibroblast-specific protein 1; HGF, hepatocyte growth factor; HMGA2, high mobility group A2; ID, inhibitor of differentiation; KAP1, KRAB-associated protein 1 (also known as TIF1β); KLF8, Krueppel-like factor 8; MMP, matrix metalloproteinase; N-cadherin, neural cadherin; NF-κB, nuclear factor-κB; TGFβ, transforming growth factor-β; PALS1, protein associated with Lin-7 1; PATJ, PALS1-associated tight-junction protein; PKA, protein kinase A; PRX1, paired-related homeobox 1; SOX, SRY box; ZNF703, zinc-finger 703; ZO1, zonula occludens 1; ZEB, zinc-finger E-box-binding.
Multiple signalling pathways cooperate in the initiation and progression of EMT (see below), and they often activate SNAIL1 expression. TGFβ and WNT family proteins, Notch, and growth factors that act through RTKs, all activate SNAIL1 expression depending on the physiological context11. SNAIL1 and SNAIL2 co operate with other transcription regulators to control gene expression. For example, SNAIL1 cooperates with ETS1, which is activated by MAPK, to activate MMP expression56. It also cooperates with the SMAD3–SMAD4 complex to cause the TGFβ-mediated repression of E-cadherin and occludin expression57.
Post-translational modifications that are initiated by signalling pathways also control the localization and degradation, and thus the activity, of SNAIL1 (REF. 11). Glycogen synthase kinase-3β (GSK3β)-mediated SNAIL1 phosphorylation at two Ser-rich motifs in activates its transcriptional activity. Phosphorylation of Ser97 and Ser101 in the first motif facilitates the nuclear export of SNAIL1, and the subsequent phosphorylation of Ser108, Ser112, Ser116 and Ser120 in the second motif labels SNAIL1 for ubiquitin-mediated degradation58 (FIG. 2). Several pathways increase SNAIL1 activity by influencing GSK3β-mediated phosphorylation. The WNT and PI3K–AKT pathways inhibit SNAIL1 phosphorylation by GSK3β59, and Notch and nuclear factor-κB (NF-κB) signalling disrupt the GSK3β–SNAIL1 interactions60,61; both mechanisms increase SNAIL1 stability. Conversely, small C-terminal domain phosphatase 1 (SCP1; also known as CTDSP1) antagonizes phosphorylation by GSK3β and retains SNAIL1 in the nucleus where it represses transcription62. Additionally, PKD1 phosphorylates SNAIL1 at Ser11 to facilitate nuclear export63, whereas SNAIL1 phosphorylation by PAK1 at Ser246 or large tumour suppressor 2 (LATS2) at Thr203 increases its nuclear retention and promotes EMT64,65. The tumour suppressor p53 recruits SNAIL2 for degradation following its interaction with the ubiquitin ligase mouse double minute 2 (MDM2), consequently suppressing cancer invasion66 (FIG. 2).
bHLH transcription factors
Homodimeric and heterodimeric basic helix–loop–helix (bHLH) transcription factors function as master regulators of lineage specification and differentiation. Among these, E12 and E47, TWIST1 and TWIST2, and inhibitor of differentiation (ID) proteins have key roles in EMT progression11,12. As with SNAIL, TWIST expression downregulates epithelial gene expression and activates mesenchymal gene expression45 (FIG. 2; TABLE 1). In cancer cells, TWIST1 represses E-cadherin and induces N-cadherin expression independently of SNAIL and probably through the association with other proteins45,67–69. TWIST recruits the methyltransferase SET8 (also known as SETD8 in humans), which mediates H4K20 monomethylation, a histone mark that is associated with repression at E-cadherin promoters and with activation at N-cadherin promoters69. In head and neck cancer cells, TWIST1 activates the expression of B lymphoma Mo-MLV insertion region 1 homologue (BMI1), a component of PRC1, and subsequently cooperates with BMI1 at E-box sequences to repress the expression of E-cadherin and the cell cycle inhibitor p16 (also known as INK4A; encoded by CDKN2A)67. This cooperation involves recruitment of PRC2, which trimethylates H3K27 at the E-cadherin and CDKN2A gene promoters67.
Diverse signalling pathways activate TWIST expression during development and tumorigenesis11,45. Most notably, the transcription factor hypoxia-inducible factor 1α (HIF1α) induces TWIST expression under hypoxic conditions, thus promoting EMT and tumour cell dissemination68. Additionally, mechanical stress induces Twist expression in Drosophila melanogaster epithelia, in a manner dependent on β-catenin70. The activities of TWIST greatly depend on its dimer composition. TWIST1 and TWIST2 form homodimers, as well as heterodimers, with E12 or E47 to regulate E-box DNA binding and transcription regulation. ID proteins that cannot bind DNA associate with TWIST, E12 or E47, and hence inhibit TWIST function. Consequently, repression of ID gene expression by TGFβ results in the derepression of TWIST (or other bHLH proteins) and increases its activity in EMT71. As with SNAIL, the stability of TWIST1 is regulated by phosphorylation; MAPKs phosphorylate TWIST1 at Ser68, protecting it from ubiquitin-mediated degradation and increasing its activity72.
ZEB transcription factors
The two vertebrate ZEB transcription factors, ZEB1 and ZEB2, bind regulatory gene sequences at E-boxes and can repress or activate transcription11,45. ZEB-mediated transcriptional repression often involves the recruitment of a C-terminal-binding protein (CTBP) co-repressor; although in some cancer cells ZEB1 represses E-cadherin expression independently of CTBP by recruiting the Switch/sucrose nonfermentable (SWI/SNF) chromatin remodelling protein BRG1 (REF. 73). ZEB1 can also interact with the transcriptional coactivators p300/CBP-associated factor (PCAF; also known as KAT2B) and p300, which switches it from a transcriptional repressor to a transcriptional activator74. Additionally, ZEB1 can recruit the Lys-specific demethylase 1 (LSD1), possibly linking it to histone demethylation in EMT75. Therefore, like SNAIL and TWIST, ZEBs bind E-boxes and function as transcriptional repressors and activators, thereby repressing some epithelial junction and polarity genes and activating mesenchymal genes that define the EMT phenotype11,45 (FIG. 2).
ZEB expression often follows activation of SNAIL expression, consistent with SNAIL1 directly targeting the ZEB1 gene. Additionally, TWIST1 cooperates with SNAIL1 in the induction of ZEB1 expression76. ZEB expression is induced in response to TGFβ and WNT proteins, and growth factors that activate RAS– MAPK signalling45. The induction of ZEB expression by TGFβ signalling involves ETS1, which is activated by MAPK signalling77. ZEB expression is post-transcriptionally repressed by microRNAs (miRNAs) (see below), and the post-translational sumoylation of ZEB2 by PRC2 prevents its association with CTBP and promotes its cytoplasmic localization, which attenuates ZEB2-mediated gene repression78.
Novel transcription factors regulating EMT
In addition to the established EMT transcription factors, other transcription factors were recently shown to induce or regulate EMT, either in development or in cancer (TABLE 1). Several of these are forkhead box (FOX) transcription factors, which are defined by a DNA-binding forkhead domain79. Others belong to the GATA family, which is characterized by a DNA-binding dual zinc-finger module and controls the differentiation of diverse cell lineages80; they promote or are required for EMT by regulating the expression of genes that are required for epithelial cell junctions or polarity complexes81. Other inducers of EMT are SRY box (SOX) transcription factors82 that synergize with SNAIL1 or SNAIL2 in driving EMT and/or cell invasion83,84. The regulation of these more recently identified EMT transcription factors, their roles in EMT and functional relationships with SNAIL, TWIST or ZEB transcription factors are not yet well defined. Regardless, it is clear that diverse transcription factors coordinate gene expression reprogramming during EMT, and that some of these are master regulators and others may have more restricted functions depending on the tissue context.
Regulating EMT at the RNA level
In addition to the direct effects of EMT transcription factors on gene expression, changes at the RNA level regulate EMT progression. The differential splicing of nascent RNAs into mRNAs that generate proteins with structural and functional differences and the miRNA-mediated degradation of gene transcripts define the activities of key proteins in EMT.
Alternative splicing in EMT
EMT is marked by extensive changes in the splicing of many mRNAs, which generates diverse protein isoforms in mesenchymal cells as compared to epithelial cells. The functional consequences of differential splicing in EMT are well illustrated by p120 catenin, the adhesion protein cluster of differentiation 44 (CD44), the RTK FGFR2 (REFS 85–88) and extensive isoform changes, as a result of alternative splicing, in various additional proteins that regulate EMT89. Many of the changes in splicing result from the rapid downregulation, during EMT, of the expression of epithelial splicing regulatory protein 1 (ESRP1) and ESRP2, two RNA binding proteins that control the splicing of many gene transcripts. Their downregulation results in mesenchymal protein isoforms that help to define alterations in adhesion, motility and signalling pathways90. Other changes in splicing result from the increased expression, during EMT, of RNA binding protein FOX1 homologue 2 (RBFOX2), another RNA binding protein that contributes to EMT and cell invasion89,91. EMT is also frequently marked by increased expression and/or activity of the splicing factor Ser-Arg-rich splicing factor 1 (SRSF1), which promotes EMT by splicing the mRNA encoding the RTK recepteur d’origine nantais 1 (RON1) and the GTPase RAC1 — further highlighting the key contributions of cell type-dependent splicing to this transdifferentiation92,93. These changes in splicing impose another layer of complexity on the gene expression changes that occur during EMT.
miRNA-mediated control of EMT
Non-coding miRNAs that selectively bind mRNAs, thus inhibiting their translation or promoting their degradation, also regulate the epithelial phenotype and EMT94. Some of these control the expression of EMT master transcription factors. For example, miR-29b and miR-30a repress SNAIL1 expression95,96; hence, increased miR-29b expression can reverse EMT and decrease cell invasion95. Additionally, miR-1 and miR-200b can repress SNAIL2 expression, and SNAIL2 represses the expression of miR-1 and miR-200b, resulting in a double-negative feedback mechanism97. A similar feedback loop also occurs between miR-34 and SNAIL1 (REF. 98), and miR-203 and SNAIL1 (REF. 99).
Members of the miR-200 family and miR-205 repress the translation of ZEB1 and ZEB2 mRNAs100, and double-negative feedback controls ZEB and miR-200 expression, with ZEB proteins repressing the expression of miR-200 miRNAs, and miR-200 suppressing ZEB expression101. During EMT, decreased miR-200 expression results in increased ZEB1 and ZEB2 levels and EMT progression101. Additionally, p53 represses liver carcinoma cell EMT by increasing the expression of miR-200 and miR-192 (a miR-215 homologue), which target and reduce ZEB1 and ZEB2 expression102. Finally, the expression of high mobility group A2 (HMGA2), which activates SNAIL1 and TWIST expression103, is downregulated by the miRNA let-7 in a pancreas carcinoma model104, and by miR-365 in lung adenocarcinoma cells105.
In addition to controlling the expression of EMT transcription factors, miRNAs also target genes that help to define the epithelial or mesenchymal phenotype, such as those encoding adhesion junction and polarity complex proteins, and signalling mediators. For example, increased miR-9 expression, which occurs in mammary carcinomas, represses E-cadherin expression and promotes a mesenchymal phenotype with increased cell migration and invasion106. N-cadherin expression is repressed by miR-194 (REFS 107,108), the expression of which is attenuated in advanced stage gastric cancer cells107. Additionaly, increased miR-194 expression in mesenchymal liver cancer cells results in decreased N-cadherin levels, cell migration and invasion108. In TGFβ-induced EMT, miR-491-5p represses PAR3 expression, which helps to destabilize tight junctions109. miR-661 represses the expression of Nectin 1 (also known as poliovirus receptor-related protein 1), a protein that is involved in cell–cell adhesion, hence contributing to epithelial cell junction disassembly in SNAIL-expressing cancer cells110. Several miRNAs control the expression and/or activities of RHOA, and thus affect actin organization and tight junction stability. Among these, miR-155, which is expressed in response to TGFβ, targets the mRNA encoding RHOA, resulting in the dissolution of tight junctions111. TGFβ also induces the expression of miR-24, which targets neuroepithelial cell-transforming 1A (NET1A), a RHO-GEF that activates RHOA, therefore promoting EMT through the disruption of adherens and tight junctions112. Additionally, miR-31 reduces breast cancer cell dissemination by targeting RHOA expression113, and miR-124 decreases EMT, cell invasion and metastasis by targeting ROCK2, an effector of RHOA signalling114. Clearly, the regulated activities of miRNAs represent an extensive regulatory network that the cells impose on the gene expression programme to control EMT.
TGFβ family proteins: inducers of EMT
The TGFβ family comprises three TGFβs, two activins, many bone morphogenetic proteins (BMPs) and other homodimers and heterodimers of ligands that all act through binary combinations of transmembrane dual specificity kinase receptors (that is, receptors that act as Ser/Thr kinases, as well as Tyr kinases). In development, TGFβ1 and TGFβ2 expression is associated with EMT-like events in the formation of endocardial cushions115, whereas TGFβ3 drives EMT that mediates palate fusion116. Postnatally, TGFβ1 induces EMT in wound healing, fibrosis and cancer. For example, increased levels of TGFβ1 have been linked to EMT in mesangial cells before kidney fibrosis117, biliary epithelial cells before liver disease progression to hepatic fibrosis118 and fibroblasts from patients with pulmonary fibrosis119. Cardiac fibrosis through endothelial–mesenchymal transition (EndMT) also results from increased TGFβ signalling120. In carcinomas, increased expression and activation of TGFβ1 promotes an epithelial plasticity response that may progress to EMT, a prerequisite for cancer cell invasion and dissemination3,4,121. For example, squamous carcinomas convert into invasive spindle cell carcinomas in response to increased expression of activated TGFβ1 (REF. 122).
Other TGFβ family proteins also regulate EMT, primarily in development. Nodal and vegetal 1 (VG1) participate in EMT during cell ingression to form mesoderm in mouse and Xenopus laevis embryos, respectively123. BMP signalling cooperates with WNT and FGF signalling to induce neural crest delamination124, and it also participates in EMT that is required in heart cushion formation125. Additionally, anti-Müllerian hormone mediates EMT in coelomic epithelium during Müllerian duct regression126. In pathological contexts, BMP signalling is involved in both EMT and MET127,128. BMP2, BMP4 and BMP7 promote EMT and invasiveness in pancreatic cancer cells, which is apparent by the loss of E-cadherin expression and an increase in MMP2 expression127. In renal fibrosis, BMP7 induces MET of renal fibroblasts with decreased type I collagen secretion and restored E-cadherin expression128. Finally, TGFβ and BMP signalling induce endothelial cells to acquire mesenchymal stem cell-like properties that contribute to fibrodysplasia ossificans progressiva, in which heterotypic ossification occurs as a result of activating mutations in the gene encoding activin receptor-like kinase 2 (ALK2; also known as ACTR1), a type I TGFβ and BMP receptor129.
TGFβ signalling through SMADs in EMT
Binding of TGFβ family proteins to tetrameric cell surface receptor complexes enables the ‘type II’ TGFβ family receptors to phosphorylate and thus to activate the ‘type I’ transmembrane kinases, which then phosphorylate the C termini of the intracellular signalling effectors — that is, SMADs. In response to TGFβ, which generally acts through complexes of TβRII (type II; also known as TGFR2) and TβRI (type I; also known as TGFR1) receptors, the receptor-activated SMAD2 and/or SMAD3 combine with SMAD4 to form trimeric SMAD complexes; SMAD1 and/or SMAD5 interact with SMAD4 in response to BMPs. Following their translocation into the nucleus, the SMAD complexes combine with DNA-binding transcription factors at regulatory gene sequences and activate or repress transcription by interacting with transcriptional coactivators and/or transcriptional co-repressors130,131 (FIG. 3). SMAD activation is negatively regulated by inhibitory SMADs (SMAD6 and SMAD7), which compete with SMAD2 and SMAD3, or SMAD1 and SMAD5, for binding to the type I receptors132.
Figure 3. Molecular mechanisms of TGFβ-induced EMT.
The initiation of, and progression through, epithelial-mesenchymal transition (EMT) are regulated at the transcriptional, post-transcriptional, translational and post-translational levels. Transforming growth factor-β (TGFβ) induces EMT by acting at several of these levels and through SMAD-mediated and non-SMAD signalling. TGFβ signals through a tetrameric complex of type I and type II receptors (TβRI and TβRII) to activate SMAD2 and SMAD3, which then combine with SMAD4. The trimeric SMAD complex translocates into the nucleus and cooperates with transcription regulators in the repression or activation of target genes. TGFβ –SMAD signalling activates the expression of EMT transcription factors, and SMAD complexes cooperate with these transcription factors to increase their transcriptional activities. TGFβ also induces the expression of microRNAs (miRNAs) that repress the expression of epithelial proteins, and EMT transcription factors can repress the expression of miRNAs that target mesenchymal components, thus promoting EMT. TGFβ signalling also decreases the expression of the epithelial splicing regulatory proteins (ESRPs), which leads to a differential splicing programme following EMT. TGFβ can also induce non-SMAD signalling pathways that contribute to EMT. It activates PI3K–AKT-mammalian TOR complex 1 (mTORC1) signalling, which increases translation and cell size; active AKT also derepresses the translation of specific mRNAs by phosphorylating heterogeneous nuclear ribonucleoprotein E1 (hnRNPE1). TGFβ also increases cell junction dissolution and induces cytoskeletal changes by regulating RHO-GTPases. TGFβ induces TβRII association with a TβRI-partitioning defective 6 (PAR6) complex at tight junctions, which enables TβRII to phosphorylate PAR6; this results in the recruitment of the E3 ubiquitin ligase SMAD ubiquitylation regulatory factor 1 (SMURF1), RHOA ubiquitylation and degradation, and the loss of tight junctions. TGFβ also induces RHOA activity; this promotes actin reorganization by leading to the activation of diaphanous (DIA1) and also RHO-associated kinase (ROCK), which phosphorylates myosin light chain (MLC) to activate LIM kinase (LIMK) and thus inhibit cofilin. RAC and CDC42 also participate in cytoskeletal changes through p21 activated kinase 1 (PAK1) and direct the formation of lamellipodia and filopodia. 4E–BP1, eukaryotic translation initiation factor 4E–binding protein 1; S6K1, ribosomal S6 kinase 1.
Dominant-negative forms of TβRII or TβRI, and pharmacological inhibition of the kinase activity of TβRI, block TGFβ-induced EMT in many cell types122,133,134, whereas a constitutively activated TβRI protein initiates EMT135. Expression of inactive SMAD2, SMAD3 or SMAD4, as well as decreased SMAD4 or increased SMAD7 expression, prevent TGFβ-induced EMT133,136,137. Interestingly, although SMAD3 drives gene expression that initiates EMT, SMAD2 may antagonize TGFβ-induced EMT138–140; accordingly, SMAD2-deficient keratinocytes undergo EMT and promote tumour formation owing to the increased activity of SMAD3–SMAD4 (REF. 138).
In response to TGFβ, SMAD complexes not only activate the expression, but also increase the activity of EMT transcription factors. TGFβ induces SNAIL1 expression through SMAD3-dependent transcription138, whereas SNAIL2 expression is induced indirectly by the SMAD3-induced expression of myocardin-related transcription factor (MRTF)141. SMAD3–SMAD4 also cooperates with SNAIL1 in response to TGFβ, and thus relays the TGFβ-activated repression of the genes encoding E-cadherin and occludin57. TGFβ also induces the expression of ZEB1, which is further controlled by MAPK signalling77, and SMAD3–SMAD4 complexes interact with ZEB1 and ZEB2 to mediate TGFβ-regulated gene expression77,142,143. SMAD3– SMAD4 complexes also interact with activating transcription factor 3 (ATF3) to repress ID1 expression, hence enhancing TWIST expression and activity71. Finally, SMAD3 and SMAD4 also activate the expression of HMGA2 in TGFβ-induced EMT144.
Other changes in gene expression during EMT occur without directly requiring EMT transcription factors and are controlled by TGFβ-activated SMADs140,145. SMADs directly activate the expression of some mesenchymal genes; for example, those encoding fibronectin, vimentin and collagen αI146,147. Moreover, the association of Nodal-induced SMAD complexes with the PHD-Bromo domain protein tripartite interaction motif 33 (TRIM33) mediates its interactions with histone marks, which leads to the activation of mesodermal goosecoid expression55.
Non-SMAD signalling in EMT
Complementing its signalling through SMADs, TGFβ also induces signalling through RHO-like GTPases, PI3K and MAPK pathways148,149, which also contribute to EMT45,137,150 (FIG. 3). Activation of RHO, RAC and CDC42 GTPases drives actin reorganization, lamellipodia and filopodia formation18 (see above). Early in EMT, PAR6 interacts with TGFβ receptors at tight junctions. Following TGFβ stimulation, PAR6 is phosphorylated by TβRII and recruits the E3 ubiquitin ligase SMAD ubiquitylation regulatory factor 1 (SMURF1), which locally mediates RHOA ubiquitylation and degradation as the tight junctions dissolve151. TGFβ also induces RHOA activation152, partly by promoting the expression of GEFs153, and the activation of ROCK and LIMK152,154. Consequently, decreased RHOA expression or the chemical inhibition of ROCK prevents the actin reorganization that is required for TGFβ-induced EMT152,154,155. Finally, RHOA activation also contributes to changes in gene expression, and mammalian epithelial cells that are unable to express a RHOA GEF show reduced α-smooth muscle actin (αSMA) expression in response to TGFβ156.
In epithelial cells undergoing EMT, TGFβ activates AKT through PI3K, which results in the activation of mammalian TOR complex 1 (mTORC1) and mTORC2 (REFS 134,157,158). Pharmacological inhibition of PI3K prevents TGFβ-induced EMT157, illustrating the essential role of the PI3K–AKT pathway in this process. In TGFβ-induced EMT, mTORC1 contributes to the increased cell size, protein synthesis, motility and invasion that take place134, whereas mTORC2 is required for the transition from an epithelial to a mesenchymal phenotype158. Inhibition of AKT decreases the level of SNAIL1 expression158,159, attenuating the repression of E-cadherin and the activation of MMP9 expression, and mTORC2 inhibition impedes invasive behaviour and metastatic potential158. Phosphorylation of GSK3β by AKT inhibits its activity, stabilizing SNAIL1 to repress the expression of several genes, including the gene encoding E-cadherin58,160. Finally, TGFβ induces the AKT-mediated phosphorylation of heterogeneous nuclear ribonucleoprotein E1 (hnRNPE1; also known as PCBP1), which results in the release of hnRNPE1 from the 3′ untranslated regions of disabled 2 (DAB2) and interleukin (IL)-like EMT inducer (ILEI; also known as FAM3C) mRNA, thus derepressing their translation and enabling EMT161 (FIG. 3).
TGFβ proteins also activate the ERK, p38 and JUN N-terminal kinase (JNK) MAPK pathways148,149, probably as a result of the dual specificity kinase activities of TGFβ receptors162. As dual specificity kinases generally have much lower Tyr phosphorylation activity than Ser/Thr phosphorylation activity, the TGFβ receptors induce a much lower level of MAPK activation than growth factor-activated RTKs162. TGFβ induces ERK MAPK signalling through the adaptor protein SRC homology 2 domain-containing-transforming A (SHCA; also known as SHC1), which associates with the TβRI receptor and is phosphorylated at Tyr and Ser by TβRI in response to TGFβ (FIG. 4). The phosphorylation of SHCA on Tyr provides a docking site for growth factor receptor-bound protein 2 (GRB2) and son of sevenless (SOS), and initiates the RAS–RAF–MEK–ERK (also known as MAPKK) MAPK pathway163. TGFβ-induced p38 MAPK and JNK activation results from the association of the ubiquitin ligase TNF receptor-associated factor 6 (TRAF6) with the TGFβ receptor complex, which activates TAK1, a kinase upstream of p38 MAPK and JNK164,165. Although the contributions of these MAPK pathways to TGFβ-induced EMT remain to be defined, inhibition of ERK or p38 MAPK kinase activity represses TGFβ-induced EMT166,167. ERK MAPK signalling increases TGFβ-induced transcription, leading to increased repression of E-cadherin and the activation of N-cadherin and MMP expression168,169. Additionally TGFβ-induced activation of ERK5 MAPK stabilizes SNAIL1 by inhibiting GSK3β, thus increasing SNAIL1 activity170. Activation of TGFβ-activated kinase 1 (TAK1; also known as MAP3K7) and p38 MAPK are required for TGFβ-induced expression of the transcription factor ATF2 and consequently for its cooperation with SMAD3–SMAD4 in TGFβ-induced gene expression171. Finally, JNK activates c-JUN, a component of the activator protein 1 (AP1) complex, hence mediating TGFβ-induced transcription in cooperation with SMAD3–SMAD4 (REF. 172) (FIG. 4).
Figure 4. Signalling pathways involved in EMT.
Epithelial-mesenchymal transition (EMT) progression is regulated by signalling pathways that can cooperate to induce full EMT responses. In addition to promoting EMT through SMAD proteins, transforming growth factor-β (TGFβ) can activate the PI3K–AKT, ERK MAPK, p38 MAPK and JUN N-terminal kinase (JNK) pathways. TβRI phosphorylates the adaptor protein SRC homology 2 domain-containing-transforming A (SHCA), which creates a docking site for growth factor receptor-bound protein 2 (GRB2) and son of sevenless (SOS) and initiates the RAS-RAF-MEK-ERK MAPK pathway. TGFβ-induced p38 MAPK and JNK activation results from the association of TNF receptor-associated factor 6 (TRAF6) with the TGFβ receptor complex, which activates TGFβ-activated kinase 1 (TAK1) and p38 MAPK and JNK as a result. Several growth factors that act through receptor tyrosine kinases (RTKs), including epidermal growth factor (EGF), fibroblast growth factor (FGF), hepatocyte growth factor (HGF) and vascular endothelial growth factor (VEGF), can induce EMT. The RAS-RAF-MEK-ERK MAPK signalling cascade represents a major pathway that is activated by RTKs in response to growth factors. Once activated ERK1 and ERK2 MAPK can facilitate EMT by increasing the expression of EMT transcription factors and regulators of cell motility and invasion, such as RHO GTPases and the 90 kDa ribosomal protein S6 kinase. Other signalling pathways, such as the WNT, Notch and Hedgehog (HH) pathways, also participate in EMT. WNT signalling promotes EMT by inhibiting glycogen synthase kinase-3β (GSK3β) to stabilize β-catenin, which translocates to the nucleus to engage the transcription factors lymphoid enhancer-binding factor 1 (LEF) and T cell factor (TCF) and promote a gene expression programme that favours EMT. In HH signalling, glioma 1 (GLI1) can induce SNAIL1 expression, and the intracellular domain of Notch can activate SNAIL2 expression; thus HH and Notch signalling promote a decrease in epithelial cadherin (E-cadherin) levels. The cell microenvironment also regulates EMT. During inflammation and in cancer, interleukin-6 (IL-6) can promote EMT through Janus kinase (JAK)-signal transducer and activator of transcription 3 (STAT3)-induced SNAIL1 expression. Hypoxia in the tumour environment can promote EMT through hypoxia-inducible factor 1α (HIF1α), which activates the expression of TWIST. EMT responses can be increased through crosstalk and cooperation between distinct pathways. For example, RTK- or integrin-induced AKT activation can induce SNAIL expression through nuclear factor-κB (NF-κB) and stabilize SNAIL and β-catenin by inhibiting GSK3β , thus cooperating with WNT signalling. TGFβ signalling can also increase EMT responses initiated by growth factors such as FGF or EGF. ECM, extracellular matrix; FZD, frizzled; ILK, integrin-linked kinase; MKK, MAPK kinase; mTORC2, mammalian TOR complex 2; Notch-IC , intracellular fragment of Notch; PTCH1, patched 1; SHH, sonic HH; SMO, smoothened.
Tyrosine kinase receptors induce EMT
In contrast to TGFβ family proteins, growth factors that act through transmembrane RTKs often stimulate cell proliferation. Ligand binding induces the autophosphorylation of these receptors on Tyr; this enables them to activate the PI3K–AKT pathway, and the ERK MAPK, p38 MAPK and JNK pathways (the roles of which in EMT were discussed above) and SRC signalling. Growth factors that act through RTKs can induce partial or full EMT. In fact, the induction of EMT by RTKs was uncovered before TGFβ was found to induce EMT2,173.
PI3K–AKT and ERK MAPK activation by growth factors
The activation of EMT by RTKs highlights the roles of the PI3K–AKT and ERK MAPK pathways in this transdifferentiation process. As with TGFβ-induced EMT, signalling through AKT and mTORC2 is essential for RTK-induced EMT158,174. ERK MAPK pathway activation, not only by growth factors but also by mutations in the genes encoding RAS or RAF that occur in cancer cells, also contributes to EMT. RAS and RAF signalling activate the expression of SNAIL1 and/or SNAIL2, and induce the activation of RHO-GTPases, therefore promoting cell motility and invasive behaviour in cancer-associated EMT175. Activation of the 90 kDa ribosomal protein S6 kinase by ERK MAPK contributes to the invasive properties of epithelial and carcinoma cells by inducing the expression of the transcription factor FOS-related antigen 1 (FRA1)176. Of note, cells in culture exhibit autocrine TGFβ signalling, and RTK signalling induces TGFβ1 expression, thus enhancing autocrine TGFβ signalling. These results position growth factor activity in the context of the cell response to TGFβ family signalling.
FGF signalling and EMT
FGFs induce mesenchymal characteristics in epithelial cells. FGF1-induced EMT of bladder carcinoma cells is accompanied by SNAIL2 expression, the destabilization of desmosomes, and the expression of α2β1 integrin and MMP13 (REFS 177–179). FGFs act as EMT inducers in development, for example, in gastrulation. Mouse embryos lacking FGFR1 show aberrant mesoderm formation in the primitive streak and maintain E-cadherin and β-catenin at the cell surface180. In FGF8-deficient mouse embryos, cells in the primitive streak fail to migrate away to generate mesoderm181. Later in development, FGF signalling is required for neural crest cell migration, and studies in X. laevis have revealed roles for fgf2 and fgf8 in the expression of neural crest cell markers124.
Hepatocyte growth factor signalling and EMT
Hepatocyte growth factor (HGF), which acts through the RTK c-MET (also known as HGFR), was identified as ‘scatter factor’, as it was seen to convert kidney epithelial cells into migratory fibroblast-like cells173. HGF induces the expression of SNAIL1 or SNAIL2, which depends on cell type, to drive EMT178,182. The induction of SNAIL expression by HGF requires the ERK MAPK pathway and involves binding of the transcription factor early growth response 1 (EGR1) to the SNAIL gene promoter182. HGF also induces desmosome destabilization and the repression of the gene encoding desmoplakin178. The c-MET receptor was found to associate with E-cadherin183, but the role of this interaction in EMT remains to be clarified. Finally, in development, HGF participates in EMT in the formation of somites and the endocardial cushion184.
Insulin-like growth factor 1 and EMT
Insulin-like growth factor 1 (IGF1) induces EMT in some cell culture models. IGF1 receptor activation causes EMT in mammary epithelial cells, resulting in loss of E-cadherin, and gain of N-cadherin, vimentin and fibronectin185. In these cells, increased SNAIL1 expression in response to IGF1 depends on NF-κB185. In other cells, however, IGF1-induced EMT was accompanied by ZEB1 expression that was dependent on activation of the ERK MAPK pathway186. Also, the PI3K–AKT pathway is required for IGF1-induced EMT187. Interestingly, in one cell system, AKT1 and AKT2 act differently, with downregulation of AKT1 expression stimulating EMT, and this induction is blocked following downregulation of AKT2 expression187. Finally, the IGF1 receptor forms complexes with E-cadherin and αv integrin188, as was also seen with c-MET183, and IGF1-induced disruption of these complexes promotes cell migration188.
Epidermal growth factors and EMT
In cell culture, epidermal growth factor (EGF) induces the endocytosis of E-cadherin, as well as SNAIL1 or TWIST expression, which leads to a reduction in E-cadherin levels189,190. EGF also induces EMT in epithelial explants, resulting in increased cell motility and MMP2- and MMP9-mediated proteolysis that is dependent on ILK kinase and the ERK MAPK pathway191. Activation of human EGF receptor 2 (HER2; also known as ERBB2) in mammary epithelial cells induces tumours with properties of EMT that are able to escape immune surveillance192,193. The EGF-related Cripto1 (also known as TDGF1) may also have a role in EMT during development. Its expression correlates with primitive streak formation, and mesoderm and endoderm specification, whereas a decrease in its expression confers defects in mesoderm formation194. Cripto1 may also have a role in mammary gland morphogenesis194. Not only does its expression increase with mammary development, but adding Cripto1 to, or overexpressing it in, cultured mammary epithelial cells triggers a fibroblast morphology, decreases the level of E-cadherin and increases the levels of N-cadherin, vimentin and SNAIL expression195. Increased Cripto1 expression has also been associated with the increased motility and invasion of tumour cells, and tumour progression194. Rather than acting as a ligand, the activity of Cripto1 in EMT may relate to its role as Nodal co-receptor and its functions in WNT signalling196,197.
Platelet-derived growth factor in EMT
In colon adenocarcinoma cells, platelet-derived growth factor (PDGF) induces the dissolution of adherens junctions, the nuclear localization of β-catenin and the repression of E-cadherin expression198. Ablation of the PDGF receptor α-subunit results in craniofacial and heart valve anomalies owing to defective mesenchymal cell migration and decreased MMP2 activity199. The PDGF-related growth factor vascular endothelial growth factor (VEGF), an inducer of angiogenesis, also induces EMT. VEGF induces SNAIL expression in breast cancer cells, partly by inhibiting GSK3β activity200, as well as EMT that correlates with SNAIL1, SNAIL2 and Twist expression in pancreas carcinoma cells201. Additionally, SNAIL1 increases VEGF expression, possibly conferring feedback amplification of the EMT response202. VEGF-induced EMT may link EMT or EndMT with angiogenesis, which could greatly benefit cancer progression and invasion.
Other extracellular signals regulate EMT
Other differentiation factors activate or regulate EMT, although the underlying mechanisms are less well-defined (FIG. 4). In canonical WNT signalling, the binding of WNT ligands to Frizzled receptors results in the inhibition of GSK3β, hence preventing β-catenin phosphorylation, ubiquitylation and degradation, and enabling β-catenin to regulate gene expression8. Inhibition of the GSK3β kinase increases SNAIL stability, thus promoting EMT58. WNT signalling has roles in developmental EMT. WNT3-deficient mouse embryos do not form a primitive streak and mesoderm203, while ectopic WNT8C expression induces the formation of multiple primitive streaks204. Canonical WNT signalling also enables the generation and proliferation of neural crest precursors205, whereas GSK3β-independent WNT signalling is required for neural crest cell migration206. WNT signalling also promotes EMT associated with cancer progression, and β-catenin-mediated gene expression is increased in cells at the invasive front of colorectal tumours where EMT occurs207.
In Hedgehog (HH) signalling, ligand binding to patched (PTC) receptors activates glioma (GLI) family transcription factors208. During development, cells in the ventral somite region transition into mesenchyme to form the sclerotome, where sonic HH (SHH) induces the expression of sclerotomal cell markers209. Ectopic expression of GLI1 in rat kidney epithelial cells induces SNAIL1 expression, the loss of E-cadherin210, increased SHH expression and signalling associated with increased SNAIL1 expression in epithelial cancers, including in the invasive front of neuroendocrine tumours211.
Notch signalling also participates in developmental EMT and possibly in EMT during cancer progression. Interactions of Delta-like or Jagged ligands with Notch receptors initiate signalling through the proteolytic release of the Notch intracellular domain that then regulates target gene expression212. In development, Notch signalling is required for EMT during endocardial cushion formation, and reduced SNAIL1 expression together with defective cushion formation occurs in Notch1-deficient mice213. Additionally, the Notch intracellular domain directly activates SNAIL2 expression214. Notch1 inhibition partially reverts EMT in lung adenocarcinoma cells and decreases their invasive behaviour215.
Additional factors in the tissue or tumour microenvironment facilitate the initiation of EMT. For example, the hypoxic conditions in growing tumours facilitate EMT by inducing expression of the transcription factor HIF1α, which activates TWIST expression and thus initiates EMT that leads to an invasive phenotype68. HIF1α also induces SNAIL1 expression in endothelial cells and ovarian carcinoma cells, resulting in the loss of vascular endothelial cadherin (VE-cadherin) or E-cadherin, respectively216,217. The release of inflammatory cytokines, besides TGFβ1, by immune cells, endothelial cells and cancer-associated fibroblasts contributes to EMT. For example, IL-6 promotes EMT of breast cancer cells in culture, which correlates with a decrease in E-cadherin levels, an increase in N-cadherin, vimentin, SNAIL1 and TWIST levels, and cell invasion218. Ectopic TWIST expression in these cells induces IL-6 expression and activates the transcription factor signal transducer and activator of transcription 3 (STAT3)218, which then activates SNAIL1 expression219. The transcription factor brachyury promotes EMT in tumour cells, partly by increasing the secretion of IL-8, which in turn maintains the mesenchymal phenotype of tumour cells by increasing brachyury expression220. The mechanisms through which inflammatory cytokines other than TGFβ contribute to EMT remain to be defined, although some cytokines may induce NF-κB signalling to directly activate SNAIL1 or ZEB expression221. Considering the link between inflammation and fibrosis, these inflammatory cytokines may also contribute to EMT in fibrosis.
Crosstalk of signalling pathways in EMT
As is apparent from this overview, growth and differentiation factors, and other extracellular cues, can activate a differentiation response in epithelial cells that leads to EMT. Distinct pathways have predominant roles in the initiation and progression of EMT and regulate changes in morphology and gene expression that convert epithelial cells into motile mesenchymal cells. EMT-inducing extracellular factors often activate several pathways, and the expression of EMT transcription factors can be activated by different pathways, strongly suggesting the presence of signalling cooperation and the convergence of these pathways on common targets during EMT (FIG. 4). It should be noted that the studies on signalling in EMT use cell culture systems in which key pathways show basal autocrine activation, which is in part defined by the culture conditions.
Cooperation between signalling pathways in EMT is seen in vivo as well as in cell culture. For example, WNT signalling cooperates with FGF and TGFβ family members to regulate EMT during gastrulation222 and neural crest cell delamination124, whereas Notch and TGFβ signalling regulate endocardial cushion formation213. Synergy between TGFβ signalling and RTK signalling, activated by, for example, the EGF-related TGFα or FGF, is apparent in EMT of cancer cells. Indeed, TGFβ facilitates EGF- or FGF-induced EMT in cancer cell models, thus increasing the epithelial to mesenchymal gene expression switch86,169,223, and ERK MAPK pathway activation in response to growth factors or mutant RAS increases TGFβ-induced EMT137. Cooperation between signalling pathways can also inhibit EMT. For example, HGF, which can induce EMT173, can inhibit TGFβ-induced EMT and myofibroblast differentiation in renal fibrosis by promoting the expression of the SMAD transcriptional co-repressor ski-related novel protein N (SNON)224. Additionally phosphorylation of SMADs by ERK MAPK can attenuate their nuclear translocation and transcriptional activities in mink lung epithelial cells131.
WNT and TGFβ signalling seem to cooperate primarily to regulate gene expression changes during EMT. Destabilization of adherens junctions in response to TGFβ enables β-catenin to accumulate in the nucleus and feed into canonical WNT signalling2. In palate medial-edge epithelial cells, TGFβ3 induces the formation of a complex between SMAD2, SMAD4 and lymphoid enhancer-binding factor 1 (LEF1), which binds E-cadherin gene sequences and represses E-cadherin gene expression to promote EMT146. In this way, the interactions of LEF or T cell factor (TCF) transcription factors with TGFβ-or BMP-activated SMAD complexes may have key roles in the regulation of gene expression during EMT. Additionally, GSK3β, which is inhibited by WNT signalling, integrates signalling pathways and controls EMT. GSK3β-mediated SNAIL1 phosphorylation increases the cytoplasmic retention and degradation of SNAIL1, and phosphorylation of GSK3β by AKT promotes GSK3β ubiquitylation and degradation, hence stabilizing SNAIL1 and enhancing the repression of E-cadherin expression58. GSK3β-mediated phosphorylation also decreases the stability of TGFβ-and BMP-activated SMADs and, thus, attenuates SMAD-mediated responses225,226. Also, PDGF induces the phosphorylation of a nuclear p68 (also known as DDX5) RNA helicase, which interacts with β-catenin to promote its nuclear accumulation independently of WNT signalling198.
Finally, functional crosstalk between TGFβ and Notch signalling has been reported. Inactivation of the genes encoding Notch1 or Jκ-recombination signal-binding protein (RBPjk), a target of Notch signalling, impedes endocardial EMT in mouse embryos, which is accompanied by reduced SNAIL1 expression, maintenance of VE-cadherin expression in endocardial cells, and impaired TGFβ2 and TβRI expression213,227. Conversely, TGFβ can induce, through SMAD3, the expression of the Notch signalling target hairy and enhancer of split-related with YRPW motif 1 (HEY1) in several EMT cell models228.
Concluding remarks
Epithelial plasticity responses need to be seen as a spectrum of changes and transitions, depending on the tissue and signalling context. The process of MET, although less well characterized than EMT, illustrates the reversible nature of this transition. The initiation and progression of EMT involve distinct signalling pathways and signalling crosstalk. TGFβ has emerged as a potent inducer of EMT, not only through SMAD-mediated changes in gene expression and the activation of EMT transcription factors but also through signalling pathways that are not mediated by SMADs. Quantifying the extent of EMT and the degree of its progression may be challenging, as the expression of EMT markers often depends on the cell type and initiating signalling pathway in question. Furthermore, characterizing EMT in vivo is impeded by its transient and reversible nature. Recent advances in imaging, including intravital techniques, will increase our knowledge of how the cellular environment controls EMT. Our understanding of EMT gained from cell culture systems, and from development and cancer, may enable us to target EMT associated with fibrosis and cancer progression.
Acknowledgements
The authors apologize to their colleagues whose work could not be cited owing to space limitations. Relevant research in the laboratory of R.D. is sponsored by US National Institutes of Health grants from the National Cancer Institute. S.L. and J.X. were recipients of scientist development awards by the US American Heart Association.
Glossary
- Apical–basal polarity
The unequal distribution of proteins and other materials between the apical side (facing the exterior) and the basal side (facing the interior) in epithelial cells.
- CD44hiCD24low phenotype
The profile of the cell surface expression of two cell surface proteins, cluster of differentiation 44 (CD44) and CD24, which correlates with epithelial and carcinoma stem cell properties and behaviour.
- Front–rear polarity
Unequal distribution of proteins between the front of a migrating cell, defined by the leading edge of the membrane, and the rear, where adhesion complexes disassemble. Front–rear polarity is required for directional migration.
- Tight junctions
Adhesion protein complexes between adjacent epithelial or endothelial cells. Composed of integral membrane proteins, including claudins and occludin, and the cytoplasmic proteins zonula occludens 1 (ZO1), ZO2 and ZO3, which link the transmembrane proteins to the actin cytoskeleton and signalling proteins.
- Adherens junctions
Form an adhesion belt between cells and provide homophilic interactions between epithelial cadherin (E-cadherin) molecules on opposing cell surfaces. The cytoplasmic proteins β-catenin and p120 catenin associate with E-cadherin and with the actin cytoskeleton through actin-associated proteins such as α-catenin.
- Desmosomes
Characterized by a ‘patch’ that holds cells on opposing lateral cell surfaces together, using similar protein complexes to adherens junctions. Desmosomal cadherin, desmogleins and desmocollins associate with plakoglobins and plakophilins underneath the plasma membrane, and connect to intermediate filaments through desmoplakin.
- Gap junctions
Protein complexes that connect neighbouring cells and enable the passage of ions and small molecules between them through hemi-channels formed by hexamers of connexin proteins. They also provide adherence.
- Apical compartment
The membrane-associated compartment at the apical side of a polarized epithelial cell.
- Basolateral compartment
The membrane-associated compartment of an epithelial cell combining both the lateral sides that mediate cell–cell interactions and the basal side that enables the cell to interact with the underlying basement membrane.
- Cortical actin cytoskeleton
The organization of the actin bundles and associated proteins underneath the membranes of epithelial cells that mediate cell–cell and cell– extracellular matrix interactions.
- Actin stress fibre
Dynamic structures of actin filaments and associated proteins that have important roles in cell motility and contractility.
- Endocardial cushion
An accumulation of cells, mostly arising from endothelial cells, in the primordial heart that will give rise to the valves and septa of the heart.
- Fibrodysplasia ossificans progressiva
Very rare progressive connective tissue disease resulting in the gradual ossification of fibrous tissue, either spontaneously or in response to damage. It is caused by an activating mutation in the activin receptor type 1 (ACVR1) gene that encodes the transforming growth factor-β family type I receptor activin receptor-like kinase 2 (ALK2).
- Guanine nucleotide exchange factors
(GEFs). Proteins that facilitate the exchange of GDP for GTP in the nucleotide-binding pocket of a GTP-binding protein.
- GTPase-activating proteins
(GAPs). Proteins that inactivate small GTPases by stimulating them to hydrolyse GTP into GDP.
- Guanine nucleotide dissociation inhibitors
(GDIs). Cytosolic proteins that bind to prenylated RAB proteins in their inactive, GDP-bound state. They are involved in RAB GTPase delivery to, and removal from, membranes.
- Coelomic epithelium
The epithelium that lines the surface of the body wall and the abdominal organs.
- Sclerotome
Part of the somite in vertebrate development that gives rise to the vertebrae and much of the skull.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 2.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nature Rev. Mol. Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 3.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman HA. Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu. Rev. Physiol. 2011;73:413–435. doi: 10.1146/annurev-physiol-012110-142225. [DOI] [PubMed] [Google Scholar]
- 6.Huang RY, Guilford P, Thiery JP. Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J. Cell Sci. 2012;125:4417–4422. doi: 10.1242/jcs.099697. [DOI] [PubMed] [Google Scholar]
- 7.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 8.Niehrs C. The complex world of WNT receptor signalling. Nature Rev. Mol. Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 9.Kourtidis A, Ngok SP, Anastasiadis PZ. p120 catenin: an essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog. Mol. Biol. Transl. Sci. 2013;116:409–432. doi: 10.1016/B978-0-12-394311-8.00018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bax NA, et al. Epithelial-to-mesenchymal transformation alters electrical conductivity of human epicardial cells. J. Cell. Mol. Med. 2011;15:2675–2683. doi: 10.1111/j.1582-4934.2011.01266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature Rev. Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 12.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nature Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 13.St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Navarro C, et al. Junctional recruitment of mammalian Scribble relies on E-cadherin engagement. Oncogene. 2005;24:4330–4339. doi: 10.1038/sj.onc.1208632. [DOI] [PubMed] [Google Scholar]
- 15. Qin Y, Capaldo C, Gumbiner BM, Macara IG. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J. Cell Biol. 2005;171:1061–1071. doi: 10.1083/jcb.200506094.. Silencing the expression of the polarity complex protein SCRIB results in loss of cell–cell adhesion, EMT and migration, similarly to silencing E-cadherin expression. These results illustrate the intimate functional linkage of epithelial cell adhesion and apical–basal polarity, and their roles in EMT.
- 16.Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27:6958–6969. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- 17.Yilmaz M, Christofori G. Mechanisms of motility in metastasizing cells. Mol. Cancer Res. 2010;8:629–642. doi: 10.1158/1541-7786.MCR-10-0139. [DOI] [PubMed] [Google Scholar]
- 18.Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 19.McNiven MA. Breaking away: matrix remodeling from the leading edge. Trends Cell Biol. 2013;23:16–21. doi: 10.1016/j.tcb.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haynes J, Srivastava J, Madson N, Wittmann T, Barber DL. Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol. Biol. Cell. 2011;22:4750–4764. doi: 10.1091/mbc.E11-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 22.Whale A, Hashim FN, Fram S, Jones GE, Wells CM. Signalling to cancer cell invasion through PAK family kinases. Front. Biosci. 2011;16:849–864. doi: 10.2741/3724. [DOI] [PubMed] [Google Scholar]
- 23.Anastasiadis PZ, Reynolds AB. Regulation of Rho GTPases by p120-catenin. Curr. Opin. Cell Biol. 2001;13:604–610. doi: 10.1016/s0955-0674(00)00258-1. [DOI] [PubMed] [Google Scholar]
- 24.Nelson WJ. Remodeling epithelial cell organization: transitions between front-rear and apical-basal polarity. Cold Spring Harb. Perspect Biol. 2009;1:a000513. doi: 10.1101/cshperspect.a000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godde NJ, Galea RC, Elsum IA, Humbert PO. Cell polarity in motion: redefining mammary tissue organization through EMT and cell polarity transitions. J. Mammary Gland Biol. Neoplasia. 2010;15:149–168. doi: 10.1007/s10911-010-9180-2. [DOI] [PubMed] [Google Scholar]
- 26.Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J. Cell Biol. 2003;161:845–851. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J. Cell Sci. 2008;121:727–735. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 28.Theveneau E, Mayor R. Cadherins in collective cell migration of mesenchymal cells. Curr. Opin. Cell Biol. 2012;24:677–684. doi: 10.1016/j.ceb.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brieher WM, Yap AS. Cadherin junctions and their cytoskeleton(s) Curr. Opin. Cell Biol. 2013;25:39–46. doi: 10.1016/j.ceb.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Hansen SM, Berezin V, Bock E. Signaling mechanisms of neurite outgrowth induced by the cell adhesion molecules NCAM and N-cadherin. Cell. Mol. Life Sci. 2008;65:3809–3821. doi: 10.1007/s00018-008-8290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nature Rev. Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 32.Lehembre F, et al. NCAM-induced focal adhesion assembly: a functional switch upon loss of E-cadherin. EMBO J. 2008;27:2603–2615. doi: 10.1038/emboj.2008.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toivola DM, Tao GZ, Habtezion A, Liao J, Omary MB. Cellular integrity plus: organelle-related and protein-targeting functions of intermediate filaments. Trends Cell Biol. 2005;15:608–617. doi: 10.1016/j.tcb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Pursell B, Lu S, Chang TK, Mercurio AM. Regulation of β4-integrin expression by epigenetic modifications in the mammary gland and during the epithelial-to-mesenchymal transition. J. Cell Sci. 2009;122:2473–2480. doi: 10.1242/jcs.049148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim Y, et al. Integrin α3β1-dependent β-catenin phosphorylation links epithelial Smad signaling to cell contacts. J. Cell Biol. 2009;184:309–322. doi: 10.1083/jcb.200806067.. Illustrates the crucial role of an integrin, and the remarkable crosstalk and interdependence of signalling systems, in the control of EMT. Loss of a specific integrin impairs E-cadherin turnover and results in dysregulation of TGFβ signalling and β-catenin–SMAD cooperation.
- 37.Maschler S, et al. Tumor cell invasiveness correlates with changes in integrin expression and localization. Oncogene. 2005;24:2032–2041. doi: 10.1038/sj.onc.1208423. [DOI] [PubMed] [Google Scholar]
- 38.Mise N, et al. Zyxin is a transforming growth factor-β (TGF-β)/Smad3 target gene that regulates lung cancer cell motility via integrin α5β1. J. Biol. Chem. 2012;287:31393–31405. doi: 10.1074/jbc.M112.357624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koenig A, Mueller C, Hasel C, Adler G, Menke A. Collagen type I induces disruption of E-cadherin-mediated cell-cell contacts and promotes proliferation of pancreatic carcinoma cells. Cancer Res. 2006;66:4662–4671. doi: 10.1158/0008-5472.CAN-05-2804. [DOI] [PubMed] [Google Scholar]
- 40.Nistico P, Bissell MJ, Radisky DC. Epithelial-mesenchymal transition: general principles and pathological relevance with special emphasis on the role of matrix metalloproteinases. Cold Spring Harb Perspect Biol. 2012;4:a011908. doi: 10.1101/cshperspect.a011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Radisky DC, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688.. Provides a remarkable example of how the tumour microenvironment can induce EMT. Exposure of breast epithelial cells to the stromal matrix metalloprotease MMP3 induces the expression of an isoform of the RAC1 GTPase, and, in turn, an increase in cellular reactive oxygen species. This novel pathway activates the expression of SNAIL and the EMT programme.
- 42.Shah PP, Fong MY, Kakar SS. PTTG induces EMT through integrin αvβ3-focal adhesion kinase signaling in lung cancer cells. Oncogene. 2012;31:3124–3135. doi: 10.1038/onc.2011.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheppard D. Integrin-mediated activation of latent transforming growth factor β. Cancer Metastasis Rev. 2005;24:395–402. doi: 10.1007/s10555-005-5131-6. [DOI] [PubMed] [Google Scholar]
- 44.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 45.Xu J, Lamouille S, Derynck R. TGF-β-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Batlle E, et al. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 47. Cano A, et al. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nature Cell Biol. 2000;2:76–83. doi: 10.1038/35000025.. References 46 and 47 identify SNAIL as a repressor of E-cadherin expression and demonstrate that it promotes EMT in development and cancer by repressing the E-cadherin promoter.
- 48.Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong ZT, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2012;31:583–594. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- 51.Herranz N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong C, et al. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene. 2013;32:1351–1362. doi: 10.1038/onc.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dong C, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest. 2012;122:1469–1486. doi: 10.1172/JCI57349.. Histone methylation is shown to be crucial in the repression of E-cadherin expression, and in the initiation of EMT. The cooperation of SNAIL with a methyltransferase is shown to be essential in the control of this differentiation programme.
- 54.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 55.Xi Q, et al. A poised chromatin platform for TGF-β access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorda M, et al. Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J. Cell Sci. 2005;118:3371–3385. doi: 10.1242/jcs.02465. [DOI] [PubMed] [Google Scholar]
- 57. Vincent T, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-β mediated epithelial-mesenchymal transition. Nature Cell Biol. 2009;11:943–950. doi: 10.1038/ncb1905.. This study provides the first evidence at the molecular level for direct cooperation of TGFβ– SMAD signalling with a crucial EMT transcription factor, SNAIL1, in the initiation of EMT.
- 58.Zhou BP, et al. Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial-mesenchymal transition. Nature Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 59. Yook JI, et al. A Wnt-Axin2-GSK3β cascade regulates Snail1 activity in breast cancer cells. Nature Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508.. Reference 58 and 59 demonstrate, in molecular detail, the regulation of SNAIL1 activity by GSK3β-mediated phosphorylation, and provide insight into the control of EMT by WNT signalling.
- 60.Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl Acad. Sci. USA. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Y, et al. Stabilization of Snail by NF-κB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009;15:416–428. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y, Evers BM, Zhou BP. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J. Biol. Chem. 2009;284:640–648. doi: 10.1074/jbc.M806916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Du C, Zhang C, Hassan S, Biswas MH, Balaji KC. Protein kinase D1 suppresses epithelial-to-mesenchymal transition through phosphorylation of snail. Cancer Res. 2010;70:7810–7819. doi: 10.1158/0008-5472.CAN-09-4481. [DOI] [PubMed] [Google Scholar]
- 64.Yang Z, et al. Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail’s subcellular localization and functions. Cancer Res. 2005;65:3179–3184. doi: 10.1158/0008-5472.CAN-04-3480. [DOI] [PubMed] [Google Scholar]
- 65.Zhang K, et al. Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. EMBO J. 2012;31:29–43. doi: 10.1038/emboj.2011.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang SP, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nature Cell Biol. 2009;11:694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 67. Yang MH, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nature Cell Biol. 2010;12:982–992. doi: 10.1038/ncb2099.. The Polycomb complex protein BMI1, which is required for stem cell self-renewal, is shown to functionally cooperate with the EMT transcription factor TWIST1 in the repression of E-cadherin expression and in EMT. These results illustrate a role for chromatin remodelling in EMT.
- 68.Yang MH, et al. Direct regulation of TWIST by HIF-1α promotes metastasis. Nature Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 69.Yang F, et al. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012;31:110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farge E. Mechanical induction of Twist in the Drosophila foregut/stomodeal primordium. Curr. Biol. 2003;13:1365–1377. doi: 10.1016/s0960-9822(03)00576-1. [DOI] [PubMed] [Google Scholar]
- 71.Kang Y, Chen CR, Massagué J. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 72.Hong J, et al. Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Res. 2011;71:3980–3990. doi: 10.1158/0008-5472.CAN-10-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanchez-Tillo E, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 74.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 76.Dave N, et al. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J. Biol. Chem. 2011;286:12024–12032. doi: 10.1074/jbc.M110.168625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by 5EF1 proteins in epithelial mesenchymal transition induced by TGF-β. Mol. Biol. Cell. 2007;18:3533–3544. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Long J, Zuo D, Park M. Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J. Biol. Chem. 2005;280:35477–35489. doi: 10.1074/jbc.M504477200. [DOI] [PubMed] [Google Scholar]
- 79.Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nature Rev. Mol. Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 80.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J. Biol. Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Campbell K, Whissell G, Franch-Marro X, Batlle E, Casanova J. Specific GATA factors act as conserved inducers of an endodermal-EMT. Dev. Cell. 2011;21:1051–1061. doi: 10.1016/j.devcel.2011.10.005.. A GATA transcription factor is found to have a crucial role in the induction of EMT, in Drosophila melanogaster and mammalian cells, through the downregulation of junctional E-cadherin, and repression of expression of the apical-basal polarity complex protein Crumbs.
- 82.Kondoh H, Kamachi Y. SOX-partner code for cell specification: Regulatory target selection and underlying molecular mechanisms. Int. J. Biochem. Cell Biol. 2010;42:391–399. doi: 10.1016/j.biocel.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 83.Sakai D, Suzuki T, Osumi N, Wakamatsu Y. Cooperative action of Sox9, Snail2 and PKA signaling in early neural crest development. Development. 2006;133:1323–1333. doi: 10.1242/dev.02297. [DOI] [PubMed] [Google Scholar]
- 84.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown RL, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shirakihara T, et al. TGF-β regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J. 2011;30:783–795. doi: 10.1038/emboj.2010.351.. TGFβ is shown to induce a switch in splicing patterns, specifically of FGF receptors, illustrating a role of differential splicing in the control of EMT. This study also highlights the functional crosstalk of TGFβ signalling with RTK signalling through the ERK MAPK pathway in defining the EMT phenotype.
- 87.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yanagisawa M, et al. A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J. Biol. Chem. 2008;283:18344–18354. doi: 10.1074/jbc.M801192200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shapiro IM, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218.. Documents the extensive changes in splicing patterns that accompany EMT and the key roles of splicing mediators in the control of EMT. Analyses of the alternative splicing signature that defines EMT enable subtype classification of mammary carcinoma cells and tissues.
- 90.Warzecha CC, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Braeutigam C, et al. The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene. 2013 doi: 10.1038/onc.2013.50. http://dx.doi.org/10.1038/onc.2013.50. [DOI] [PubMed] [Google Scholar]
- 92.Goncalves V, Matos P, Jordan P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum. Mol. Genet. 2009;18:3696–3707. doi: 10.1093/hmg/ddp317. [DOI] [PubMed] [Google Scholar]
- 93.Valacca C, et al. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J. Cell Biol. 2010;191:87–99. doi: 10.1083/jcb.201001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr. Opin. Cell Biol. 2013;25:200–207. doi: 10.1016/j.ceb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ru P, et al. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol. Cancer Ther. 2012;11:1166–1173. doi: 10.1158/1535-7163.MCT-12-0100. [DOI] [PubMed] [Google Scholar]
- 96.Zhang J, et al. miR-30 inhibits TGF-β1-induced epithelial-to-mesenchymal transition in hepatocyte by targeting Snail1. Biochem. Biophys. Res. Commun. 2012;417:1100–1105. doi: 10.1016/j.bbrc.2011.12.121. [DOI] [PubMed] [Google Scholar]
- 97. Liu YN, et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2012;32:296–306. doi: 10.1038/onc.2012.58.. A mutually inhibitory loop, with miR-1 and miR-200 microRNAs repressing SNAIL2 expression, and SNAIL2 repressing miR-1 and miR-200 expression, is described. This double feedback mechanism enables for the gradual amplification of cancer-associated EMT.
- 98.Siemens H, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 99.Moes M, et al. A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition. PLoS ONE. 2012;7:e35440. doi: 10.1371/journal.pone.0035440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722.. miR-200 and miR-205 microRNAs are shown to repress ZEB1 and ZEB2 expression. They have a crucial role in the progression of EMT, and are able to induce MET.
- 101.Bracken CP, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 102.Kim T, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thuault S, et al. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J. Biol. Chem. 2008;283:33437–33446. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Watanabe S, et al. HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am. J. Pathol. 2009;174:854–868. doi: 10.2353/ajpath.2009.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qi J, et al. MiR-365 regulates lung cancer and developmental gene thyroid transcription factor 1. Cell Cycle. 2012;11:177–186. doi: 10.4161/cc.11.1.18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ma L, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024.. The miRNA miR-9 represses E-cadherin expression, leading to activation of β-catenin signalling and, consequently, to VEGF expression, thus promoting angiogenesis and metastasis. The expression of this metastasis-promoting miRNA is activated by MYC, and correlates with MYCN amplification, in breast cancers.
- 107.Song Y, et al. Inverse association between miR-194 expression and tumor invasion in gastric cancer. Ann. Surg. Oncol. 2012;19(Suppl. 3):S509–S517. doi: 10.1245/s10434-011-1999-2. [DOI] [PubMed] [Google Scholar]
- 108.Meng Z, et al. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148–2157. doi: 10.1002/hep.23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou Q, et al. TGF-β-induced MiR-491-5p expression promotes Par-3 degradation in rat proximal tubular epithelial cells. J. Biol. Chem. 2010;285:40019–40027. doi: 10.1074/jbc.M110.141341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vetter G, et al. miR-661 expression in SNAI1-induced epithelial to mesenchymal transition contributes to breast cancer cell invasion by targeting Nectin-1 and StarD10 messengers. Oncogene. 2010;29:4436–4448. doi: 10.1038/onc.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 111.Kong W, et al. MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Papadimitriou E, et al. Differential regulation of the two RhoA-specific GEF isoforms Net1/Net1A by TGF-β and miR-24: role in epithelial-to-mesenchymal transition. Oncogene. 2012;31:2862–2875. doi: 10.1038/onc.2011.457. [DOI] [PubMed] [Google Scholar]
- 113.Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Concurrent suppression of integrin α5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 2010;70:5147–5154. doi: 10.1158/0008-5472.CAN-10-0410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Zheng F, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 115.Mercado-Pimentel ME, Runyan RB. Multiple transforming growth factor-β isoforms and receptors function during epithelial-mesenchymal cell transformation in the embryonic heart. Cells Tissues Organs. 2007;185:146–156. doi: 10.1159/000101315. [DOI] [PubMed] [Google Scholar]
- 116.Nawshad A, LaGamba D, Hay ED. Transforming growth factor β (TGFβ) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT) Arch. Oral Biol. 2004;49:675–689. doi: 10.1016/j.archoralbio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 117.Schnaper HW, Hayashida T, Hubchak SC, Poncelet AC. TGF-β signal transduction and mesangial cell fibrogenesis. Am. J. Physiol. Renal Physiol. 2003;284:F243–F252. doi: 10.1152/ajprenal.00300.2002. [DOI] [PubMed] [Google Scholar]
- 118.Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-β in hepatic fibrosis. Front Biosci. 2002;7:d793–d807. doi: 10.2741/A812. [DOI] [PubMed] [Google Scholar]
- 119.Willis BC, Borok Z. TGF-β-induced EMT: mechanisms and implications for fibrotic lung disease. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;293:L525–L534. doi: 10.1152/ajplung.00163.2007. [DOI] [PubMed] [Google Scholar]
- 120. Zeisberg EM, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nature Med. 2007;13:952–961. doi: 10.1038/nm1613.. This study illustrates the important pathological contribution of EndMT to fibrosis, and its control by TG F β family proteins, with TGFβ promoting and BMP7 antagonizing EndMT.
- 121.Katsuno Y, Lamouille S, Derynck R. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr. Opin. Oncol. 2013;25:76–84. doi: 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- 122.Portella G, et al. Transforming growth factor β is essential for spindle cell conversion of mouse skin carcinoma in vivo: implications for tumor invasion. Cell Growth Differ. 1998;9:393–404. [PubMed] [Google Scholar]
- 123.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J. Clin. Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nature Rev. Mol. Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 125.Kruithof BP, Duim SN, Moerkamp AT, Goumans MJ. TGFβ and BMP signaling in cardiac cushion formation: lessons from mice and chicken. Differentiation. 2012;84:89–102. doi: 10.1016/j.diff.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 126.Klattig J, Englert C. The Müllerian duct: recent insights into its development and regression. Sex. Dev. 2007;1:271–278. doi: 10.1159/000108929. [DOI] [PubMed] [Google Scholar]
- 127.Gordon KJ, Kirkbride KC, How T, Blobe GC. Bone morphogenetic proteins induce pancreatic cancer cell invasiveness through a Smad1-dependent mechanism that involves matrix metalloproteinase-2. Carcinogenesis. 2009;30:238–248. doi: 10.1093/carcin/bgn274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zeisberg M, Shah AA, Kalluri R. Bone morphogenic protein-7 induces mesenchymal to epithelial transition in adult renal fibroblasts and facilitates regeneration of injured kidney. J. Biol. Chem. 2005;280:8094–8100. doi: 10.1074/jbc.M413102200. [DOI] [PubMed] [Google Scholar]
- 129. Medici D, et al. Conversion of vascular endothelial cells into multipotent stem-like cells. Nature Med. 2010;16:1400–1406. doi: 10.1038/nm.2252.. Endothelial cells are shown to convert into mesenchymal cells and acquire stem cell properties, dependent on the TGFβ family receptor ALK2. Lineage tracing suggests an endothelial cell origin of chondrocytes and osteoblasts in heterotypic ossification associated with fibrodysplasia ossificans progressiva.
- 130.Feng XH, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 131.Massagué J. TGFβ signalling in context. Nature Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miyazono K, ten Dijke P, Heldin CH. TGF-β signaling by Smad proteins. Adv. Immunol. 2000;75:115–157. doi: 10.1016/s0065-2776(00)75003-6. [DOI] [PubMed] [Google Scholar]
- 133.Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-β and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol. Biol. Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lamouille S, Derynck R. Cell size and invasion in TGF-β-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J. Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146.. This study illustrates the crucial role of AKT–mTOR signalling in EMT. mTORC1 is required for the increased cell size, cell motility and invasion that accompany EMT.
- 135.Piek E, Moustakas A, Kurisaki A, Heldin CH, ten Dijke P. TGF-β type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 1999;112:4557–4568. doi: 10.1242/jcs.112.24.4557. [DOI] [PubMed] [Google Scholar]
- 136.Deckers M, et al. The tumor suppressor Smad4 is required for transforming growth factor β-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006;66:2202–2209. doi: 10.1158/0008-5472.CAN-05-3560. [DOI] [PubMed] [Google Scholar]
- 137.Zavadil J, Böttinger EP. TGF-β and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 138.Hoot KE, et al. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J. Clin. Invest. 2008;118:2722–2732. doi: 10.1172/JCI33713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ju W, et al. Deletion of Smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. Mol. Cell. Biol. 2006;26:654–667. doi: 10.1128/MCB.26.2.654-667.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang YC, et al. Hierarchical model of gene regulation by transforming growth factor β. Proc. Natl Acad. Sci. USA. 2003;100:10269–10274. doi: 10.1073/pnas.1834070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Morita T, Mayanagi T, Sobue K. Dual roles of myocardin-related transcription factors in epithelial mesenchymal transition via slug induction and actin remodeling. J. Cell Biol. 2007;179:1027–1042. doi: 10.1083/jcb.200708174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nishimura G, et al. δEF1 mediates TGF-β signaling in vascular smooth muscle cell differentiation. Dev. Cell. 2006;11:93–104. doi: 10.1016/j.devcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 143.Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFβ/BMP signaling pathway. EMBO J. 2003;22:2443–2452. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Thuault S, et al. Transforming growth factor-β employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zavadil J, et al. Genetic programs of epithelial cell plasticity directed by transforming growth factor-β. Proc. Natl Acad. Sci. USA. 2001;98:6686–6691. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nawshad A, Medici D, Liu CC, Hay ED. TGFβ3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J. Cell Sci. 2007;120:1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kaimori A, et al. Transforming growth factor-β1 induces an epithelial-to-mesenchymal transition state in mouse hepatocytes in vitro . J. Biol. Chem. 2007;282:22089–22101. doi: 10.1074/jbc.M700998200. [DOI] [PubMed] [Google Scholar]
- 148.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 149.Moustakas A, Heldin CH. Non-Smad TGF-β signals. J. Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 150.Lamouille S, Derynck R. Emergence of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin axis in transforming growth factor-β-induced epithelial-mesenchymal transition. Cells Tissues Organs. 2011;193:8–22. doi: 10.1159/000320172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ozdamar B, et al. Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718.. TGFβ signalling is shown to confer a highly localized effect in epithelial cells, inducing RHOA degradation at tight junctions, thus contributing to epithelial disassembly and EMT.
- 152.Bhowmick NA, et al. Transforming growth factor-β 1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shen X, et al. The activity of guanine exchange factor NET1 is essential for transforming growth factor-β-mediated stress fiber formation. J. Biol. Chem. 2001;276:15362–15368. doi: 10.1074/jbc.M009534200. [DOI] [PubMed] [Google Scholar]
- 154.Vardouli L, Moustakas A, Stournaras C. LIM-kinase 2 and cofilin phosphorylation mediate actin cytoskeleton reorganization induced by transforming growth factor-β. J. Biol. Chem. 2005;280:11448–11457. doi: 10.1074/jbc.M402651200. [DOI] [PubMed] [Google Scholar]
- 155.Tavares AL, Mercado-Pimentel ME, Runyan RB, Kitten GT. TGF β-mediated RhoA expression is necessary for epithelial-mesenchymal transition in the embryonic chick heart. Dev. Dyn. 2006;235:1589–1598. doi: 10.1002/dvdy.20771. [DOI] [PubMed] [Google Scholar]
- 156.Tsapara A, et al. The RhoA activator GEF-H1/Lfc is a transforming growth factor-β target gene and effector that regulates α-smooth muscle actin expression and cell migration. Mol. Biol. Cell. 2010;21:860–870. doi: 10.1091/mbc.E09-07-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor β-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 2000;275:36803–36810. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 158. Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R. TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J. Cell Sci. 2012;125:1259–1273. doi: 10.1242/jcs.095299.. This study, together with reference 134, illustrates the crucial role of AKT-mTOR signalling in EMT. mTORC2 is required for the transition from the epithelial to the mesenchymal phenotype.
- 159.Julien S, et al. Activation of NF-κB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene. 2007;26:7445–7456. doi: 10.1038/sj.onc.1210546. [DOI] [PubMed] [Google Scholar]
- 160.Bachelder RE, Yoon SO, Franci C, de Herreros AG, Mercurio AM. Glycogen synthase kinase-3 is an endogenous inhibitor of Snail transcription: implications for the epithelial-mesenchymal transition. J. Cell Biol. 2005;168:29–33. doi: 10.1083/jcb.200409067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Chaudhury A, et al. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nature Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029.. Uncovers a TGFβ-induced translational control mechanism that contributes to EMT. Phosphorylation of hnRNPE1 by AKT derepresses select target mRNAs, resulting in translational activation of DAB2 and ILEI, which are required for EMT.
- 162.Kang JS, Liu C, Derynck R. New regulatory mechanisms of TGF-β receptor function. Trends Cell Biol. 2009;19:385–394. doi: 10.1016/j.tcb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 163.Lee MK, et al. TGF-β activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007;26:3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yamashita M, et al. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-β. Mol. Cell. 2008;31:918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sorrentino A, et al. The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nature Cell Biol. 2008;10:1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- 166.Xie L, et al. Activation of the Erk pathway is required for TGF-β1-induced EMT in vitro. Neoplasia. 2004;6:603–610. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yu L, Hébert MC, Zhang YE. TGF-β receptor-activated p38 MAP kinase mediates Smad-independent TGF-β responses. EMBO J. 2002;21:3749–3759. doi: 10.1093/emboj/cdf366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Grande M, et al. Transforming growth factor-β and epidermal growth factor synergistically stimulate epithelial to mesenchymal transition (EMT) through a MEK-dependent mechanism in primary cultured pig thyrocytes. J. Cell Sci. 2002;115:4227–4236. doi: 10.1242/jcs.00091. [DOI] [PubMed] [Google Scholar]
- 169.Uttamsingh S, et al. Synergistic effect between EGF and TGF-β 1 in inducing oncogenic properties of intestinal epithelial cells. Oncogene. 2008;27:2626–2634. doi: 10.1038/sj.onc.1210915. [DOI] [PubMed] [Google Scholar]
- 170.Marchetti A, et al. ERK5/MAPK is activated by TGFβ in hepatocytes and required for the GSK-3p–mediated Snail protein stabilization. Cell Signal. 2008;20:2113–2118. doi: 10.1016/j.cellsig.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 171.Sano Y, et al. ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-β signaling. J. Biol. Chem. 1999;274:8949–8957. doi: 10.1074/jbc.274.13.8949. [DOI] [PubMed] [Google Scholar]
- 172.Santibanez JF. JNK mediates TGF-β 1-induced epithelial mesenchymal transdifferentiation of mouse transformed keratinocytes. FEBS Lett. 2006;580:5385–5391. doi: 10.1016/j.febslet.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 173.Stoker M, Perryman M. An epithelial scatter factor released by embryo fibroblasts. J. Cell Sci. 1985;77:209–223. doi: 10.1242/jcs.77.1.209. [DOI] [PubMed] [Google Scholar]
- 174.Gulhati P, et al. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011;71:3246–3256. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Makrodouli E, et al. BRAF and RAS oncogenes regulate Rho GTPase pathways to mediate migration and invasion properties in human colon cancer cells: a comparative study. Mol. Cancer. 2011;10:118. doi: 10.1186/1476-4598-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Doehn U, et al. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol. Cell. 2009;35:511–522. doi: 10.1016/j.molcel.2009.08.002.. Highlights a key contribution of the RAS–ERK MAPK pathway to EMT. The ERK MAPK-activated protein 90 kDa ribosomal protein S6 kinase is required for the induction of motile and invasive behaviour of epithelial and carcinoma cells following mesenchymal transition, through the induction of defined genes.
- 177.Valles AM, Boyer B, Tarone G, Thiery JP. a2p1 integrin is required for the collagen and FGF-1 induced cell dispersion in a rat bladder carcinoma cell line. Cell Adhes. Commun. 1996;4:187–199. doi: 10.3109/15419069609014222. [DOI] [PubMed] [Google Scholar]
- 178.Savagner P, Yamada KM, Thiery JP. The zinc-finger protein Slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J. Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Billottet C, et al. Modulation of several waves of gene expression during FGF-1 induced epithelial-mesenchymal transition of carcinoma cells. J. Cell Biochem. 2008;104:826–839. doi: 10.1002/jcb.21667. [DOI] [PubMed] [Google Scholar]
- 180.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 181.Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1 -mediated upregulation of Snail. EMBO J. 2006;25:3534–3545. doi: 10.1038/sj.emboj.7601213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hiscox S, Jiang WG. Association of the HGF/SF receptor, c-met, with the cell-surface adhesion molecule, E-cadherin, and catenins in human tumor cells. Biochem. Biophys. Res. Commun. 1999;261:406–411. doi: 10.1006/bbrc.1999.1002. [DOI] [PubMed] [Google Scholar]
- 184.Romano LA, Runyan RB. Slug is an essential target of TGFβ2 signaling in the developing chicken heart. Dev. Biol. 2000;223:91–102. doi: 10.1006/dbio.2000.9750. [DOI] [PubMed] [Google Scholar]
- 185.Kim HJ et al. Constitutively active type I insulin-like growth factor receptor causes transformation and xenograft growth of immortalized mammary epithelial cells and is accompanied by an epithelial-to-mesenchymal transition mediated by NF-κB and Snail. Mol. Cell. Biol. 2007;27:3165–3175. doi: 10.1128/MCB.01315-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Graham TR, et al. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2008;68:2479–2488. doi: 10.1158/0008-5472.CAN-07-2559. [DOI] [PubMed] [Google Scholar]
- 187.Irie HY, et al. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J. Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Canonici A, et al. Insulin-like growth factor-I receptor, E-cadherin and αv integrin form a dynamic complex under the control of α-catenin. Int. J. Cancer. 2008;122:572–582. doi: 10.1002/ijc.23164. [DOI] [PubMed] [Google Scholar]
- 189.Lu Z, Ghosh S, Wang Z, Hunter T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of β-catenin, and enhanced tumor cell invasion. Cancer Cell. 2003;4:499–515. doi: 10.1016/s1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- 190.Lo HW, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–9076. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ahmed N, et al. Molecular pathways regulating EGF-induced epithelio-mesenchymal transition in human ovarian surface epithelium. Am. J. Physiol. Cell Physiol. 2006;290:C1532–C1542. doi: 10.1152/ajpcell.00478.2005. [DOI] [PubMed] [Google Scholar]
- 192.Moody SE, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 193.Knutson KL, et al. Immunoediting of cancers may lead to epithelial to mesenchymal transition. J. Immunol. 2006;177:1526–1533. doi: 10.4049/jimmunol.177.3.1526. [DOI] [PubMed] [Google Scholar]
- 194.Rangel MC, et al. Role of Cripto-1 during epithelial-to-mesenchymal transition in development and cancer. Am. J. Pathol. 2012;180:2188–2200. doi: 10.1016/j.ajpath.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Strizzi L, et al. Epithelial mesenchymal transition is a characteristic of hyperplasias and tumors in mammary gland from MMTV-Cripto-1 transgenic mice. J. Cell. Physiol. 2004;201:266–276. doi: 10.1002/jcp.20062. [DOI] [PubMed] [Google Scholar]
- 196.Morkel M, et al. β-catenin regulates Cripto- and Wnt3-dependent gene expression programs in mouse axis and mesoderm formation. Development. 2003;130:6283–6294. doi: 10.1242/dev.00859. [DOI] [PubMed] [Google Scholar]
- 197.Tao Q, et al. Maternal Wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120:857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 198. Yang L, Lin C, Liu ZR. p68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036.. In PDGF-induced EMT, tyrosine phosphorylation of the RNA helicase p68 is shown to promote the nuclear translocation of β-catenin through a WNT-independent pathway. Nuclear import of β-catenin and its cooperation with TCF/LEF transcription factors are required for EMT.
- 199.Robbins JR, McGuire PG, Wehrle-Haller B, Rogers SL. Diminished matrix metalloproteinase 2 (MMP-2) in ectomesenchyme-derived tissues of the Patch mutant mouse: regulation of MMP-2 by PDGF and effects on mesenchymal cell migration. Dev. Biol. 1999;212:255–263. doi: 10.1006/dbio.1999.9373. [DOI] [PubMed] [Google Scholar]
- 200.Wanami LS, Chen HY, Peiro S, Garcia de Herreros A, Bachelder RE. Vascular endothelial growth factor-A stimulates Snail expression in breast tumor cells: implications for tumor progression. Exp. Cell Res. 2008;314:2448–2453. doi: 10.1016/j.yexcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yang AD, et al. Vascular endothelial growth factor receptor-1 activation mediates epithelial to mesenchymal transition in human pancreatic carcinoma cells. Cancer Res. 2006;66:46–51. doi: 10.1158/0008-5472.CAN-05-3086. [DOI] [PubMed] [Google Scholar]
- 202.Peinado H, et al. Snail and E47 repressors of E-cadherin induce distinct invasive and angiogenic properties in vivo . J. Cell Sci. 2004;117:2827–2839. doi: 10.1242/jcs.01145. [DOI] [PubMed] [Google Scholar]
- 203.Liu P, et al. Requirement for Wnt3 in vertebrate axis formation. Nature Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 204.Popperl H, et al. Misexpression of Wnt8C in the mouse induces an ectopic embryonic axis and causes a truncation of the anterior neuroectoderm. Development. 1997;124:2997–3005. doi: 10.1242/dev.124.15.2997. [DOI] [PubMed] [Google Scholar]
- 205.Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- 206.Carmona-Fontaine C, et al. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Brabletz T, et al. Variable β-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl Acad. Sci. USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature Rev Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 209.Monsoro-Burq AH. Sclerotome development and morphogenesis: when experimental embryology meets genetics. Int. J. Dev. Biol. 2005;49:301–308. doi: 10.1387/ijdb.041953am. [DOI] [PubMed] [Google Scholar]
- 210.Li X, et al. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene. 2006;25:609–621. doi: 10.1038/sj.onc.1209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fendrich V, et al. Snail and Sonic Hedgehog activation in neuroendocrine tumors of the ileum. Endocr. Relat. Cancer. 2007;14:865–874. doi: 10.1677/ERC-07-0108. [DOI] [PubMed] [Google Scholar]
- 212.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J. Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Timmerman LA, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Niessen K, et al. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 2008;182:315–325. doi: 10.1083/jcb.200710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Xie M, et al. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J. Cell Biochem. 2012;113:1501–1513. doi: 10.1002/jcb.24019. [DOI] [PubMed] [Google Scholar]
- 216.Imai T, et al. Hypoxia attenuates the expression of E-cadherin via up-regulation of SNAIL in ovarian carcinoma cells. Am. J. Pathol. 2003;163:1437–1447. doi: 10.1016/S0002-9440(10)63501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Luo D, Wang J, Li J, Post M. Mouse Snail is a target gene for HIF. Mol. Cancer Res. 2011;9:234–245. doi: 10.1158/1541-7786.MCR-10-0214. [DOI] [PubMed] [Google Scholar]
- 218.Sullivan NJ, et al. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene. 2009;28:2940–2947. doi: 10.1038/onc.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yadav A, Kumar B, Datta J, Teknos TN, Kumar P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol. Cancer Res. 2011;9:1658–1667. doi: 10.1158/1541-7786.MCR-11-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fernando RI, Castillo MD, Litzinger M, Hamilton DH, Palena C. IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res. 2011;71:5296–5306. doi: 10.1158/0008-5472.CAN-11-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Jing Y, Han Z, Zhang S, Liu Y, Wei L. Epithelial-mesenchymal transition in tumor microenvironment. Cell Biosci. 2011;1:29. doi: 10.1186/2045-3701-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Heisenberg CP, Solnica-Krezel L. Back and forth between cell fate specification and movement during vertebrate gastrulation. Curr. Opin. Genet. Dev. 2008;18:311–316. doi: 10.1016/j.gde.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wendt MK, Smith JA, Schiemann WP. Transforming growth factor-β-induced epithelial-mesenchymal transition facilitates epidermal growth factor-dependent breast cancer progression. Oncogene. 2010;29:6485–6498. doi: 10.1038/onc.2010.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yang J, Dai C, Liu Y. A novel mechanism by which hepatocyte growth factor blocks tubular epithelial to mesenchymal transition. J. Am. Soc. Nephrol. 2005;16:68–78. doi: 10.1681/ASN.2003090795. [DOI] [PubMed] [Google Scholar]
- 225.Aragon E, et al. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Fuentealba LC, et al. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Grego-Bessa J, Diez J, Timmerman L, de la Pompa JL. Notch and epithelial-mesenchyme transition in development and tumor progression: another turn of the screw. Cell Cycle. 2004;3:718–721. [PubMed] [Google Scholar]
- 228.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-β/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Eastham AM, et al. Epithelial-mesenchymal transition events during human embryonic stem cell differentiation. Cancer Res. 2007;67:11254–11262. doi: 10.1158/0008-5472.CAN-07-2253. [DOI] [PubMed] [Google Scholar]
- 230.Ullmann U, et al. Epithelial-mesenchymal transition process in human embryonic stem cells cultured in feeder-free conditions. Mol. Hum. Reprod. 2007;13:21–32. doi: 10.1093/molehr/gal091. [DOI] [PubMed] [Google Scholar]
- 231.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Esteban MA, et al. The mesenchymal-to-epithelial transition in somatic cell reprogramming. Curr. Opin. Genet. Dev. 2012;22:423–428. doi: 10.1016/j.gde.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 233.Maherali N, Hochedlinger K. TGFβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015.. Illustrates the key roles of MET and BMP signalling in the reprogramming of somatic cells towards induced pluripotency, and identifies BMP-dependent induction of select miRNAs as a key regulatory axis in the initiation of MET.
- 235. Subramanyam D, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature Biotech. 2011;29:443–448. doi: 10.1038/nbt.1862.. The miR-302 and miR-372 miRNAs are shown to promote MET during reprogramming of human fibroblasts into pluripotent cells, and to inhibit TG F β-induced EMT of human keratinocytes, partly through inhibition of targets that promote EMT.
- 236. Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027.. Provides the first direct link between EMT and gain of epithelial stem cell properties. Induction of EMT in normal and malignant mammary epithelial cells results in the acquisition of stem cell-like characteristics.
- 237. Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin. Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001.. This thoughtful review conceptualizes the roles of the phenotypic plasticity associated with EMT in cancer cell behaviour and cancer progression.
- 238.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nature Rev. Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 239.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 240.Vermeulen L, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nature Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 241.Brabletz T, et al. Nuclear overexpression of the oncoprotein β-catenin in colorectal cancer is localized predominantly at the invasion front. Pathol. Res. Pract. 1998;194:701–704. doi: 10.1016/s0344-0338(98)80129-5. [DOI] [PubMed] [Google Scholar]
- 242.Espinoza I, Pochampally R, Xing F, Watabe K, Miele L. Notch signaling: targeting cancer stem cells and epithelial-to-mesenchymal transition. Onco Targets Ther. 2013;6:1249–1259. doi: 10.2147/OTT.S36162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ma J, Xia J, Miele L, Sarkar FH, Wang Z. Notch signaling pathway in pancreatic cancer progression. Pancreat. Disord. Ther. 2013;3 [PMC free article] [PubMed] [Google Scholar]
- 244.Palagani V, et al. Epithelial mesenchymal transition and pancreatic tumor initiating CD44+/EpCAM+ cells are inhibited by γ-secretase inhibitor IX. PLoS ONE. 2012;7:e46514. doi: 10.1371/journal.pone.0046514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Smith AL, et al. The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31:5162–5171. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Medici D, Kalluri R. Endothelial-mesenchymal transition and its contribution to the emergence of stem cell phenotype. Semin. Cancer Biol. 2012;22:379–384. doi: 10.1016/j.semcancer.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.van Meeteren LA, ten Dijke P. Regulation of endothelial cell plasticity by TGF-β. Cell Tissue Res. 2012;347:177–186. doi: 10.1007/s00441-011-1222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.von Gise A, Pu WT. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ. Res. 2012;110:1628–1645. doi: 10.1161/CIRCRESAHA.111.259960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127.. Provides evidence that endothelial conversion into mesenchymal cells substantially contributes to the generation of carcinoma-associated stromal fibroblasts.
- 251.Medici D, Olsen BR. The role of endothelial-mesenchymal transition in heterotopic ossification. J. Bone Miner. Res. 2012;27:1619–1622. doi: 10.1002/jbmr.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kokudo T, et al. Snail is required for TGFβ-induced endothelial-mesenchymal transition of embryonic stem cell-derived endothelial cells. J. Cell Sci. 2008;121:3317–3324. doi: 10.1242/jcs.028282. [DOI] [PubMed] [Google Scholar]
- 253.Luna-Zurita L, et al. Integration of a Notch-dependent mesenchymal gene program and BMP2-driven cell invasiveness regulates murine cardiac valve formation. J. Clin. Invest. 2010;120:3493–3507. doi: 10.1172/JCI42666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Medici D, Potenta S, Kalluri R. Transforming growth factor-β2 promotes Snail-mediated endothelial-mesenchymal transition through convergence of Smad-dependent and Smad-independent signalling. Biochem. J. 2011;437:515–520. doi: 10.1042/BJ20101500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rivera-Feliciano J, et al. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133:3607–3618. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Quaggin SE, Kapus A. Scar wars: mapping the fate of epithelial-mesenchymal-myofibroblast transition. Kidney Int. 2011;80:41–50. doi: 10.1038/ki.2011.77. [DOI] [PubMed] [Google Scholar]
- 257.Radisky DC, Kenny PA, Bissell MJ. Fibrosis and cancer: do myofibroblasts come also from epithelial cells via EMT? Cell Biochem. 2007;101:830–839. doi: 10.1002/jcb.21186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Qiu P, et al. Myocardin enhances Smad3-mediated transforming growth factor-β1 signaling in a CArG box-independent manner: Smad-binding element is an important cis element for SM22alpha transcription in vivo . Circ. Res. 2005;97:983–991. doi: 10.1161/01.RES.0000190604.90049.71. [DOI] [PubMed] [Google Scholar]
- 259.Mihira H, et al. TGF-β-induced mesenchymal transition of MS-1 endothelial cells requires Smad-dependent cooperative activation of Rho signals and MRTF-A. J. Biochem. 2012;151:145–156. doi: 10.1093/jb/mvr121. [DOI] [PubMed] [Google Scholar]
- 260.Masszi A, et al. Fate-determining mechanisms in epithelial-myofibroblast transition: major inhibitory role for Smad3. J. Cell Biol. 2010;188:383–399. doi: 10.1083/jcb.200906155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Charbonney E, Speight p, Masszi A, Nakano H, Kapus A. β-catenin and Smad3 regulate the activity and stability of myocardin-related transcription factor during epithelial-myofibroblast transition. Mol. Biol. Cell. 2011;22:4472–4485. doi: 10.1091/mbc.E11-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 263.Zeisberg M, Kalluri R. Cellular mechanisms of tissue fibrosis. 1. Common and organ-specific mechanisms associated with tissue fibrosis. Am. J. Physiol. Cell Physiol. 2013;304:C216–C225. doi: 10.1152/ajpcell.00328.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Chaffer CL, Thompson EW, Williams ED. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs. 2007;185:7–19. doi: 10.1159/000101298. [DOI] [PubMed] [Google Scholar]
- 265.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 266.Cheung M, et al. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 267.Mani SA, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc. Natl Acad. Sci. USA. 2007;104:10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nilsson J, et al. Nuclear Janus-activated kinase 2/nuclear factor 1-C2 suppresses tumorigenesis and epithelial-to-mesenchymal transition by repressing Forkhead box F1. Cancer Res. 2010;70:2020–2029. doi: 10.1158/0008-5472.CAN-09-1677. [DOI] [PubMed] [Google Scholar]
- 269.Qiao Y, et al. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 2011;71:3076–3086. doi: 10.1158/0008-5472.CAN-10-2787. [DOI] [PubMed] [Google Scholar]
- 270.Belguise K, Guo S, Sonenshein GE. Activation of FOXO3a by the green tea polyphenol epigallocatechin-3-gallate induces estrogen receptor α expression reversing invasive phenotype of breast cancer cells. Cancer Res. 2007;67:5763–5770. doi: 10.1158/0008-5472.CAN-06-4327. [DOI] [PubMed] [Google Scholar]
- 271.Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70:2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nature Struct. Mol. Biol. 2011;18:867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Venkov CD, et al. A proximal activator of transcription in epithelial-mesenchymal transition. J. Clin. Invest. 2007;117:482–491. doi: 10.1172/JCI29544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Slorach EM, Chou J, Werb Z. Zeppo1 is a novel metastasis promoter that represses E-cadherin expression and regulates p120-catenin isoform expression and localization. Genes Dev. 2011;25:471–484. doi: 10.1101/gad.1998111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ocana OH, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]