Abstract
Rhipicephalus sanguineus, the common brown dog tick, produces several chemokine-binding proteins which are secreted into the host in its saliva to modulate the host response during feeding. Two of these demonstrate very restricted selectivity profiles. Here, we describe the characterization of the third, which we named Evasin-4. Evasin-4 was difficult to produce recombinantly using its native signal peptide in HEK cells, but expressed very well using the urokinase-type plasminogen activator signal peptide. Using SPR, Evasin-4 was shown to bind most CC chemokines. Investigation of the neutralization properties by inhibition of chemokine-induced chemotaxis showed that binding and neutralization did not correlate in all cases. Two major anomalies were observed: no binding was observed to CCL2 and CCL13, yet Evasin-4 was able to inhibit chemotaxis induced by these chemokines. Conversely, binding to CCL25 was observed, but Evasin-4 did not inhibit CCL25-induced chemotaxis. Size-exclusion chromatography confirmed that Evasin-4 forms a complex with CCL2 and CCL18. In accordance with the standard properties of unmodified small proteins, Evasin-4 was rapidly cleared following in vivo administration. To enhance the in vivo half-life and optimize its potential as a therapeutic agent, Fc fusions of Evasin-4 were created. Both the N- and C-terminal fusions were shown to retain binding activity, with the C-terminal fusion showing a modest reduction in potency.
Keywords: anti-inflammatory, binding protein, Fc fusion, hemokine, tick
Introduction
Vertebrates have developed a sophisticated immune system that, under healthy circumstances, balances the ability to defend the host against pathogens while maintaining tolerance to self proteins. Among the array of proteins associated with the immune response, chemokines play a key role in the control of leukocyte migration towards the site of infection. Despite the limited number of successes to date in drug-discovery programs targeting chemokines, understanding the complexity of the chemokine system and the effects of its inhibition remain an intense area of research activity.
Because pathogens try to evade the host immune system, evolution has led them to develop chemokine and cytokine mediators with the ability to interfere with the host immune response 1. For example, because viruses depend on living cells for their replication, they have developed many strategies to remain undetected in their hosts. They express chemokine and receptor homologs such as vMIP-II, a viral chemokine 2, and US28, an HCMV-encoded chemokine receptor homolog 3,4. Chemokine-binding proteins (CKBPs) have also been identified in viral genomes. These molecules are able to sequester chemokines and either block their interaction with their receptor, with glycosaminoglycans or both 5, with both of these interactions being necessary for chemokine activity in vivo 6. The first eukaryotic CKBP was isolated in 2005 from the eggs of the Schistosoma mansoni worm and was shown to bind members of all families, notably CXCL8, CCL2, CCL3, CCL5 and CX3CL1 7.
Five years ago, our laboratory identified a family of CKBPs produced by the salivary gland of the tick Rhipicephalus sanguineus, which we termed Evasins 8,9. Ticks are blood-sucking ectoparasites which can feed on their host for several days without being detected by its immune system. The Evasins are small proteins secreted in the tick saliva and are probably required by the tick to inhibit the chemokine-mediated recruitment of leukocytes to the bite site. We have previously reported the recombinant expression of Evasin-1 and Evasin-3, where sufficient quantities of these CKBPs were produced in SF9 insect cells, and Escherichia coli respectively, to allow the resolution of their 3D structures 8–10. Both CKBPs exhibit a restricted binding profile and are in this aspect very different from viral CKBPs, which tend to have very broad binding specificities 5. Evasin-1 binds and neutralizes CCL3, CCL4 and CCL18, whereas Evasin-3 only binds ELR+ CXC-chemokines. In vivo studies have shown that both these Evasins inhibit cellular recruitment in different murine models of inflammation and lead to a reduction of the symptoms consistent with an anti-inflammatory activity of these proteins 8.
Evasin-4 was identified by cross-linking to iodinated CCL5 and CCL11 with the same expression cloning strategy that was used for Evasin-1 and Evasin-3 8,9. However, production of the recombinant protein proved difficult using standard procedures. Here, we report the strategies we used to produce Evasin-4, which included the use of a signal peptide of an unrelated secreted protein. Extensive characterization of the protein revealed that, in contrast to Evasin-1 9, it has a broad selectivity profile even though both proteins show a conserved pattern of disulfides, suggesting that they share a common structural fold. With the aim of testing its properties as an anti-inflammatory agent, we created Fc fusions with Evasin-4 to provide a half-life that would be suitable for use in vivo in disease settings.
Results
Expression and purification
The expression of Evasin-4 with its natural signal peptide was very poor in HEK293 cells (Fig. 1) or insect cells (SF9 cells, data not shown). The signal peptide was therefore replaced by that of Evasin-1 or urokinase-type plasminogen activator (uPA) (Fig. S1) which had previously been shown to induce strong expression in our laboratory. Although Evasin-1 is expressed at high levels in both insect and mammalian expression systems 9, its signal peptide did not improve expression of Evasin-4. However, as shown in Fig. 1, Evasin-4 was expressed at very high levels using the uPA signal peptide, as shown by the broad band observed by the western blot. Despite the fact that expression of carboxy-tagged Evasin-4–6His was good, surprisingly the majority of the protein did not bind to the nickel-affinity column and the yields of protein obtained after elution were negligible, and moreover were highly contaminated (data not shown). The 6His tag was then placed on the N-terminus of the protein with an intervening caspase 8 cleavage site. The N-terminally tagged protein expressed well and was purified at a yield of 20 mg·L−1 by a nickel-affinity chromatography followed by size-exclusion chromatography (SEC) to remove high molecular mass contaminants. None of the conditions used for the caspase 8 cleavage removed the tag from the Evasin-4 protein (data not shown).
Figure 1.
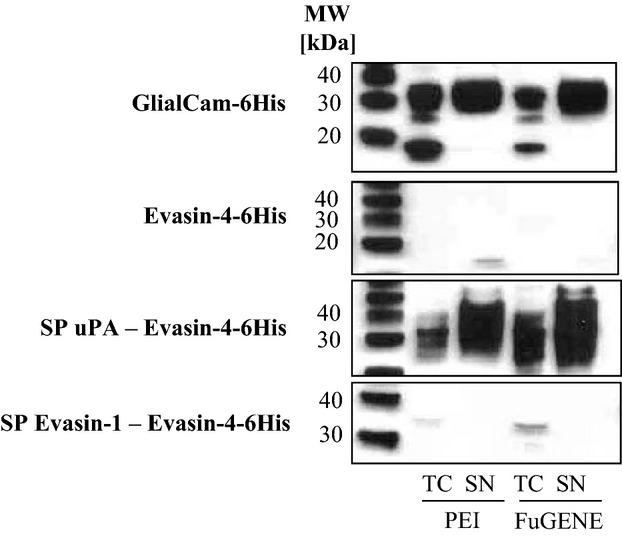
Impact of signal peptide on Evasin-4–6His expression. Anti-His western blot analysis of Evasin-4–6His expression in HEK293 cells after transfection using PEI or FuGENE transfection reagent (1 min exposure). Samples were analyzed 6 days post transfection. From the top, expression of: recombinant GlialCam–6His used as positive control, recombinant Evasin-4–6His, recombinant Evasin-4–6His with the uPA signal peptide and recombinant Evasin-4–6His with the Evasin-1 signal peptide. TC, total cells; SN, supernatant; SP, signal peptide.
The untagged protein was expressed and purified following a three-step chromatography protocol with a final purity > 95%, as estimated by the analytical methods used, at a yield of 15 mg·L−1. The predicted mass of recombinant Evasin-4 is 11.3 kDa, but all the Evasin-4 constructs migrated as a broad band at a molecular mass of 40–60 kDa on SDS/PAGE. Therefore, the protein appears to be highly glycosylated when produced in HEK293 cells. Primary sequence analysis of Evasin-4 with netngly software (http://www.cbs.dtu.dk/services/NetNGlyc/) highlighted six putative N-glycosylation sites that support this assumption. In addition, it should be noted that the protein rapidly lost its stain after Coomassie Brilliant Blue staining, a characteristic of highly glycosylated proteins, so the gels necessitated scanning immediately.
The Fc constructs fused either to the N- or the C-terminus of the Evasin-4 expressed well in mammalian cells and a yield of 30 mg·L−1 (Evasin-4–Fc) or 115 mg·L−1 (Fc–Evasin-4) and a purity higher than 96% were reached by purifying the proteins using protein A chromatography followed by SEC.
Analysis of selectivity
Immobilization of recombinant, native Evasin-4 on CM4 chips at pH 4 for SPR analysis was unsuccessful, presumably because of its acidic nature, with a theoretical isoelectric point of 3.82, which could be further modified by its extensive glycosylation. Therefore, the initial screening was performed by immobilization of the chemokines (Table 1). It was immediately apparent that Evasin-4 was selective for CC chemokines because it bound all those tested, with the exception of CCL2 and CCL13, but did not bind CXCL8, CXCL12 or XCL1. However, as shown in Table 1, the affinities were rather poor, which was surprising in view of the fact that Evasin-1 binds CC chemokines with subnanomolar affinity 9 and Evasin-3 demonstrates single digit nanomolar affinity for its ligands 8. The affinities for the chemokines were similar for the various N-terminal Evasin-4 fusions produced, demonstrating that extensions at the N-terminus did not affect activity (data not shown).
Table 1.
Comparison of the kinetic characteristics of Evasin-4 binding to CC chemokines by SPR immobilizing either the chemokine or 6His–Evasin-4
| Chemokine | Immobilized chemokine | Immobilized 6His–Evasin-4 |
|---|---|---|
| KD Evasin-4 (nm) | KD 6His–Evasin-4 (nm) | |
| CCL1 | 112.1 | 0.20 |
| CCL3 | 33.1 | 0.06 |
| CCL5 E26A | 193.6 | 0.09 |
| CCL11 | 127.7 | 0.34 |
| CCL15 | 132.5 | 2.31 |
| CCL17 | 126.2 | 0.61 |
| CCL23 | 14.1 | 4.43 |
We therefore took advantage of the N-terminally tagged construct to immobilize the protein in order to examine binding using the chemokines as analytes. The addition of the tag including the 6His moiety resulted in a higher pI, allowing coating on CM4 chips. Direct coating of small proteins such as chemokines is likely to mask epitopes or deform the protein, therefore as expected the affinities were two orders of magnitude higher (Tables1 and 2). Moreover, analysis of most of the CC subclass was facilitated, demonstrating that the affinities measured ranged from picomolar to single digit nanomolar. No binding was observed for CCL2, -4, -13, -20, -27, CXCL1, -8, -10, -11, -12, XCL1 and CX3CL1 (Fig. 2A and Table 2). Using wild-type CCL5, binding was observed but kinetic parameters could not be calculated because complete dissociation did not occur, probably due to the oligomerization characteristics of this chemokine (results not shown). Thus using the obligate tetrameric variant, E26A, a KD of 0.09 nm was obtained. However, the ability of Evasin-4 to bind and inhibit wild-type CCL5 was confirmed by chemotaxis assays as described below.
Table 2.
Kinetic characteristics of Evasin-4 binding to CC chemokines by SPR
| Immobilized 6His–Evasin-4 | |||
|---|---|---|---|
| Chemokine | ka × 106 (m−1·s−1) | kd × 10−3(s−1) | KD (nm) |
| CCL1 | 16.6 | 3.26 | 0.20 |
| CCL2 | No binding | ||
| CCL3 | 13.8 | 0.89 | 0.06 |
| CCL4 | No binding | ||
| CCL5 E26A | 26.0 | 2.23 | 0.09 |
| CCL7 | 14.1 | 9.88 | 0.70 |
| CCL8 | 7.11 | 1.76 | 0.25 |
| CCL11 | 12.9 | 4.35 | 0.34 |
| CCL13 | No binding | ||
| CCL14 | 10.4 | 1.48 | 0.14 |
| CCL15 | 40.5 | 93.6 | 2.31 |
| CCL16 | 12.0 | 3.06 | 0.26 |
| CCL17 | 17.6 | 10.7 | 0.61 |
| CCL18 | 19.6 | 0.53 | 0.03 |
| CCL19 | 2.57 | 0.35 | 0.14 |
| CCL20 | No binding | ||
| CCL21 | 11.0 | 0.08 | 0.01 |
| CCL22 | 8.9 | 3.80 | 0.43 |
| CCL23 | 0.006 | 0.03 | 4.43 |
| CCL24 | 30.5 | 12.5 | 0.41 |
| CCL25 | 0.008 | 0.54 | 69.6 |
| CCL26 | 0.17 | 0.15 | 0.88 |
| CCL27 | No binding | ||
Figure 2.
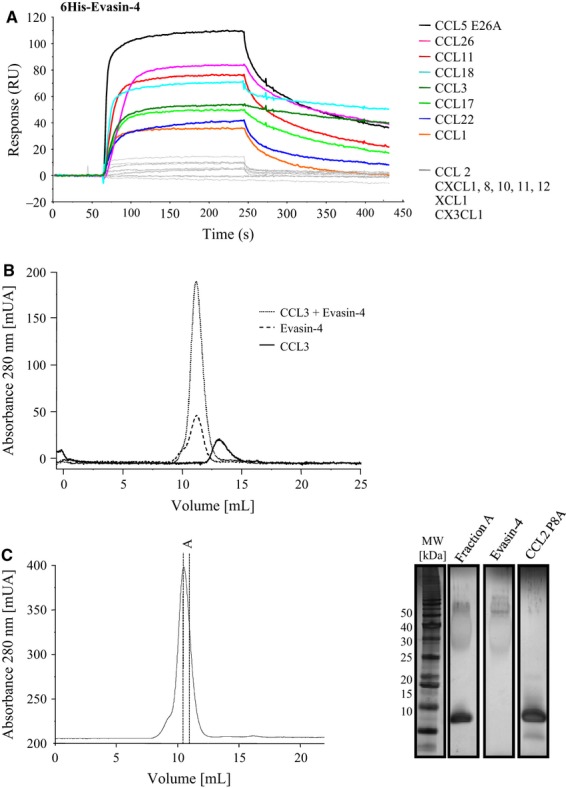
Selectivity of recombinant Evasin-4. (A) Chemokines suspended at 0.1 μg·mL−1 in running buffer were injected for 3 min on coated 6His–Evasin-4 followed by 2.5 min of dissociation. CCL5 was replaced by the mutant CCL5 E26A to avoid oligomerization 27. CCL1, -3, -5, -11, -17, -18, -22 and -26 showed strong binding to Evasin-4, whereas sensograms of CCL2, CXCL1, -8, -10, -11, -12, XCL1 and CX3CL1 indicated no binding of these chemokines to Evasin-4. (B) Complex formation between Evasin-4 and human CCL3. Evasin-4, CCL3 and an equimolar mixture of the two proteins were subjected one by one to SEC. Observation of a single peak when analyzing the mixture confirmed the formation of a complex between the two proteins. (C) SEC analysis of the complex between Evasin-4 and the obligate monomer CCL2 P8A. Equimolar amounts of Evasin-4 and CCL2 P8A were incubated and loaded onto a SEC column (left). A fraction of the peak analyzed by SDS/PAGE followed by silver staining confirmed the presence of both Evasin-4 and CCL2 (right).
We were intrigued by the fact that no binding was observed for certain chemokines, particularly the closely related chemokines CCL2 and CCL13. As discussed below, Evasin-4 was able to inhibit the chemotactic activity of these chemokines, albeit with low potency. The other interesting observation is that both Evasin-1 and Evasin-4 bind CCL18, a chemokine for which the human cognate receptor is unknown, and for which no species ortholog is known in the other hosts that R. sanguineus feeds on. We were able to demonstrate that the Evasins formed 1 : 1 complexes with their ligands by using SDS/PAGE analysis following SEC analysis for Evasin-3 and CXCL8, and Evasin-4 for human and murine CCL3 and CCL5 (for an example, see Fig. 2B), and to demonstrate that there was no complex formed by Evasin-4 and CXCL8 (results not shown). However, a complex was observed between Evasin-4 and CCL2 (Fig. 2C). The binding to CCL18 was also confirmed because it eluted as a complex with both Evasin-1 and Evasin-4, but as expected, not with Evasin-3 11.
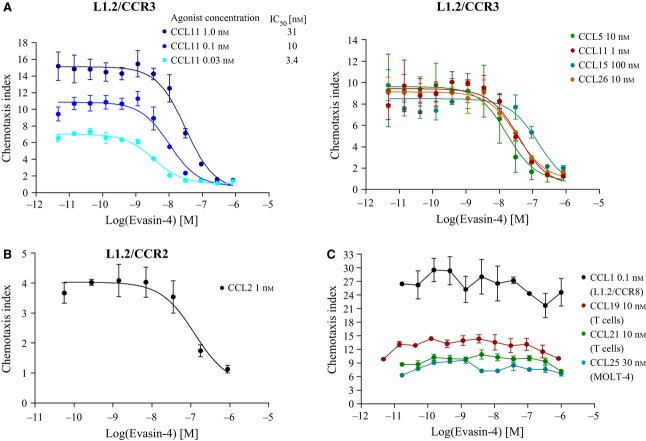
The ability of Evasin-4 to inhibit the chemotactic activity of chemokines was then assessed either using transfected cell lines or freshly isolated primary cells, and in some cases both, as summarized in Table 3 and exemplified for some CC chemokines in Fig. 3A. Because the half-maximal inhibitory concentration (IC50) value is dependent on the concentration of the agonist used for the measurement of inhibition as shown in Fig. 3A, the experiments were conducted using the agonist concentration that induces 80% of the maximal response (EC80) and are given in Table 3. Mostly there was good concordance between the affinities determined by SPR and the ability to inhibit chemotaxis, however, several disparities were evident. Despite repeated experiments with different preparations of CCL2, including the obligate monomer P8A–CCL2, no binding was observed to Evasin-4, whereas binding was observed for the other CCR2 ligands, CCL7 and CCL8. However, inhibition of L1.2/CCR2 chemotaxis in response to CCL2 was repeatedly observed, although the potency was close to micromolar (Fig. 3B and Table 2). This result was corroborated by the formation of a complex as assessed by SEC (Fig. 2C).
Table 3.
Inhibition of chemotaxis by Evasin-4. Data are presented as the mean of at least three experiments ± SD. –, no inhibition observed
| Chemokine | Cells | EC80 (nm) | IC50 Evasin-4 (nm) |
|---|---|---|---|
| CCL1 | L1.2/CCR8 | 0.1 | – |
| CCL2 | L1.2/CCR2 | 1 | 615 ± 276 |
| CCL3 | L1.2/CCR5 | 40 | 4.4 ± 0.9 |
| 300.19CCR1 | 1 | 2.0 ± 0.5 | |
| CCL3L1 | L1.2/CCR5 | 1 | 3.5 ± 2.0 |
| CCL5 | Monocytes | 10 | 1.5 ± 1.1 |
| 300.19/CCR1 | 1 | 2.2 ± 1.2 | |
| L1.2/CCR5 | 1 | 4.8 ± 1.5 | |
| CCL7 | Eosinophils | 10 | 5.0 ± 2.8 |
| L1.2/CCR2 | 5 | 5.0 ± 3.2 | |
| CCL8 | Eosinophils | 10 | 3.1 ± 1.6 |
| L1.2/CCR3 | 5 | 3.6 ± 2.4 | |
| CCL11 | Eosinophils | 1 | 4.9 ± 1.5 |
| L1.2/CCR3 | 1 | 14.0 ± 4.2 | |
| CCL13 | L1.2/CCR2 | 10 | 167 ± 70 |
| CCL14 | 300.19/CCR1 | 1010 | 2.4 ± 1.3 |
| CCL15 | L1.2/CCR3 | 100 | 88 ± 51 |
| CCL16 | 300.19/CCR1 | 250 | 304 ± 20 |
| CCL17 | L1.2/CCR4 | 1 | 69 ± 24 |
| CCL19 | T lymphocytes | 10 | – |
| CCL21 | T lymphocytes | 10 | – |
| CCL22 | L1.2/CCR4 | 1 | 38 ± 25 |
| CCL23 | 300.19/CCR1 | 10 | 2.2 ± 0.4 |
| CCL24 | L1.2/CCR3 | 10 | 95 ± 31 |
| CCL25 | MOLT-4 | 100 | – |
| CCL26 | Eosinophils | 50 | 101 ± 63 |
| L1.2/CCR3 | 10 | 68 ± 30 |
Figure 3.
Evasin-4 inhibits cell recruitment in vitro. Inhibition of the chemotactic activity of the CC chemokines by increasing concentrations of Evasin-4. (A) Evasin-4 inhibits a broad range of chemokines with IC50 values dependent on the concentration of chemokine used. (B) Evasin-4 is able to inhibit CCL2-mediated chemotaxis, whereas no binding was observed by SPR. (C) By contrast, Evasin-4 does not show any inhibition of CCL1, CCL19, CCL21 and CCL25 but binds to these chemokines by SPR. Data are shown as mean ± SD and are representative of two to seven experiments.
Also, although SPR had shown convincing binding data for CCL1, CCL19, CCL21 and CCL25, Evasin-4 was unable to demonstrate potent inhibition of chemotaxis (Fig. 3C).
Evasin-4–Fc constructs
The Fc constructs were analyzed using the same methods as for Evasin-4. Using SPR, we observed that the constructs retain binding to CCL3, -5, -7, -8, -11, -14, -15, -18 and -26, with profiles similar to that of Evasin-4 (Fig. 4A and Table 4). Furthermore, inhibition of cell migration in vitro allowed us to confirm that the Fc proteins are still active. Fc–Evasin-4 has IC50 values similar to those of Evasin-4, for example, with inhibition of CCL8 in the nanomolar range, whereas Evasin-4–Fc appears to be a less potent inhibitor of the eotaxins. Its IC50 values for the inhibition of CCL11 and CCL24 were, respectively, 100 and 7 times higher than those of Evasin-4 (Fig. 4B and Table 5).
Figure 4.
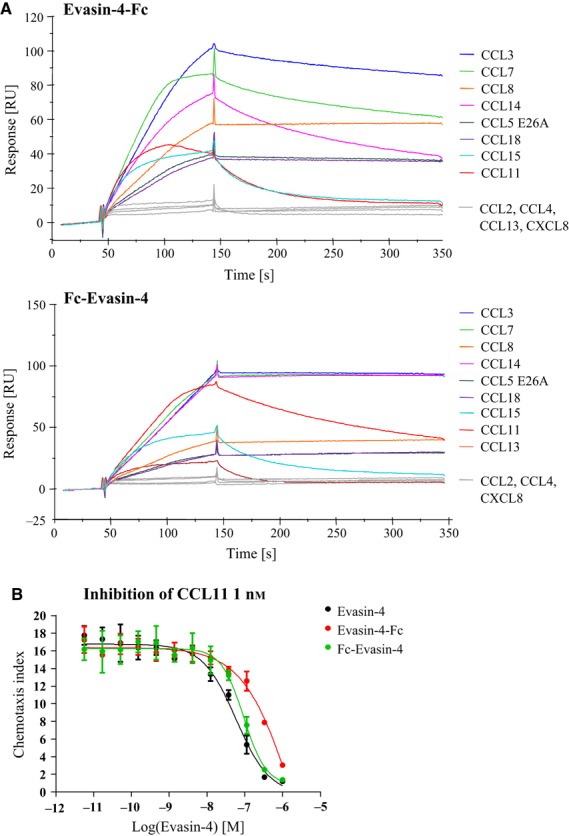
Characterization of Fc constructs. (A) SPR analysis of coated Fc constructs with the indicated chemokines as analytes. Similar to Evasin-4, the Fc constructs showed good binding to CCL3, -5, -7, -11, -14, -15, -18 and -26. No binding between the Fc constructs and CCL2, CCL4 or CXCL8 was observed. CCL13 appears to be bound by Fc–Evasin-4 but not by Evasin-4–Fc. (B) Inhibition of CCL11-mediated chemotaxis using L1.2/CCR3 cells with increasing concentrations of the Fc constructs. Data are presented as mean of three experiments ± SD.
Table 4.
Kinetic characteristics of Fc constructs binding to CC chemokines
| Chemokine | Immobilized Evasin-4 | ||
|---|---|---|---|
| KD 6His–Evasin-4 (nm) | KD Evasin-4–Fc (nm) | KD Fc–Evasin-4 (nm) | |
| CCL1 | 1.81 | 0.097 | 0.276 |
| CCL3 | 0.393 | 6.79 | 0.277 |
| CCL5 E26A | 0.002 | 1.72 | 0.020 |
| CCL7 | 0.045 | 0.053 | 0.134 |
| CCL8 | 0.220 | 0.015 | 0.088 |
| CCL11 | 0.069 | 3.6 | 0.099 |
| CCL15 | 0.199 | 0.016 | 0.04 |
| CCL18 | 0.198 | 3.63 | 0.083 |
| CCL23 | 10.76 | 0.232 | 0.124 |
| CCL24 | 142.4 | 707.9 | 4.836 |
| CCL26 | 11.4 | 15.56 | 17.26 |
Table 5.
Inhibition of chemotaxis by Fc constructs. IC50 values. Data are presented as mean of at least two experiments ± SD. n.d., not determined
| Chemokine | Cell line | EC80 (nm) | IC50 Evasin-4 (nm) | IC50 Evasin-4–Fc (nm) | IC50 Fc–Evasin-4 (nm) |
|---|---|---|---|---|---|
| CCL5 | L1.2 CCR5 | 1 | 4.8 ± 1.5 | 9.6 ± 5.1 | n.d. |
| CCL8 | L1.2 CCR2 | 5 | 3.6 ± 2.4 | 6.0 ± 4.1 | 6.4 ± 3.2 |
| CCL11 | L1.2 CCR3 | 1 | 14.0 ± 4.2 | 1539 ± 332 | 88 ± 8 |
| CCL13 | L1.2 CCR2 | 10 | 167 ± 70 | 83 ± 23 | 213 ± 172 |
| CCL22 | L1.2 CCR4 | 1 | 39 ± 18 | 38 ± 7 | n.d. |
| CCL24 | L1.2 CCR3 | 10 | 95 ± 31 | 642 ± 431 | 113 ± 23 |
We then investigated the properties of the molecules in vivo in order to assess the half-life extension effect achieved through the fusion to the Fc domain. Following a single subcutaneous injection of 10 mg·kg−1 of the protein and subsequent sampling, analysis of blood protein levels over a 14-day period led us to estimate the plasma half-life of Evasin-4–Fc and Fc–Evasin-4 to be 62.7 and 90.6 h, respectively, versus 8.7 h for Evasin-4. Performing the same experiment using an intravenous injection gave a half-life of 149 h for Evasin-4–Fc and 110 h for Fc–Evasin-4 (Fig. 5). Therefore, the fusion of the Fc domain confers an approximate 8- to 18-fold increase in half-life compared with Evasin-4 and results in a molecule that has pharmacokinetic properties suitable for testing in in vivo experiments.
Figure 5.
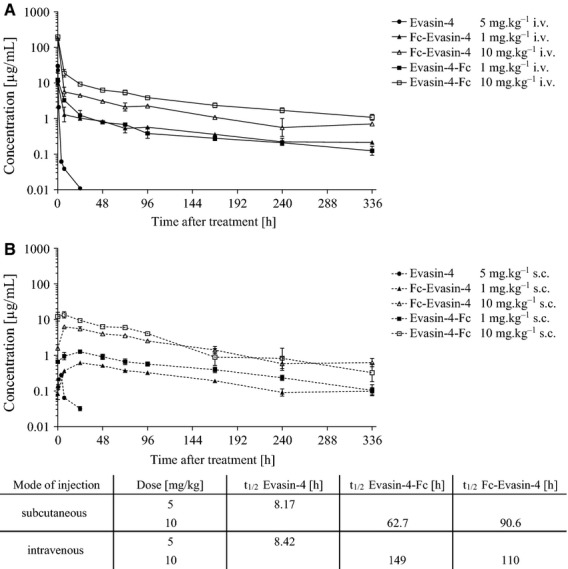
Pharmacokinetic profiles of Evasin-4 and Fc constructs. The indicated amount of proteins was injected intravenously (A) or subcutaneously (B) into C57BL/6 mice and the concentration of the compound in plasma was determined at different time points (n = 3 for each measurement).
Discussion
Blood is the only source of food for ticks. They have therefore developed sophisticated ways, including the subversion of the host immune system, to remain on their host as long as is necessary for them to be sated. We have previously reported the identification and isolation of three selective CKBPs named Evasins and the characterization of Evasin-1 and -3 8,9. Evasin-4, the CKBP described in this study, has a broader selectivity pattern than Evasin-1 and -3 and SPR analysis has shown that it is able to bind at least 18 CC chemokines.
The three CKBP are, by definition, secreted proteins, and although presumably secreted into the saliva by the ticks in a similar manner, had very different behavior when produced recombinantly. Evasin-1 expressed well in both mammalian and insect cells, and Evasin-3, although it expressed satisfactorily in mammalian expression systems, was produced in large quantities in E. coli in a soluble form. By contrast, Evasin-4 was difficult to express recombinantly and was only achieved after replacement of its signal peptide with that from an unrelated protein, uPA. It is interesting to note that replacement of its signal peptide with that of the closely related protein Evasin-1 did not confer a significant improvement in recombinant protein production.
R. sanguineus, the tick species from which the Evasins have been identified, has a preference for the dog as its host, therefore often referred to as the common brown dog tick, but will also feed on rodents, cats, humans, cattle and deer 12. Because the majority of chemokines which are commercially available are human and murine, our studies have used the chemokines from these two species. Therefore, it is possible that the selectivity we have described could be slightly different for chemokines from the other species that this tick feeds on, although chemokines amongst species are generally very similar.
For the majority of the CC chemokines tested, the binding observed by SPR was supported by inhibition of the chemotactic activity in vitro. However, this was not the case for CCL1, -19, -21 and -25. By contrast, no binding between Evasin-4 and CCL2 or CCL13 was observed by SPR even though it was able to inhibit chemotaxis mediated by these chemokines, albeit only with potencies in the 100–1000 nm range. This disparity may be due to the differences in the time course of the experiments: using SPR, the association phase was performed for 3 min under flow conditions, whereas in chemotaxis assays, the Evasin remains in presence of the chemokine for the duration of the experiment, i.e. for 4 h. If the binding of Evasin-4 to CCL2 or CCL13 has a slow on-rate, it may be that the SPR experiment is not long enough to allow the detection of the interaction between the two molecules.
The two Evasins previously characterized both inhibit neutrophil recruitment, because Evasin-3 inhibits ELR chemokines from all species tested, and Evasin-1 inhibits CCL3, a CCR1 ligand that plays a role in neutrophil recruitment in mice. Neutrophils are an essential component in the early immune or innate response to infection by pathogens. Looking more closely at the results obtained in chemotaxis, we noticed that all CC chemokines inducing the migration of eosinophils (CCL3, -5, -7, -8, -11, -13, -14, -15, -16, -23, -24 and -26) and/or of mast cells (CCL5, -7, -11, -13, -14, -15, -16, -24 and -26) were inhibited by Evasin-4 13. Eosinophils are important for the defense against parasites 14. Thus it is seems reasonable that ticks need to inhibit the migration of these cells to the bite site in order to decrease rejection of the arthropod by the host. Mast cells are tissue-resident rather than circulating leukocytes, and their degranulation and activation lead to the immediate release of inflammatory mediators, stimulating inflammation and the host response against pathogens 15.
CCL2 and CCL13 are closely related agonists of CCR2 and are responsible for the recruitment of monocytes towards sites of inflammation 16,17. Because monocytes are part of the early response occurring during the innate immune response where their influx follows the initial neutrophil influx, it would be surprising if ticks did not inhibit the recruitment of these cells. Previous studies have consistently highlighted the presence of an anti-CCL2 activity in saliva from various tick species 18,19 although in preliminary experiments performed in our laboratory we were unable to observe it with R. sanguineus saliva 8. We tested the hypothesis that perhaps Evasin-4 would be more potent against the murine form of CCL2, JE, but the IC50 was also close to micromolar (data not shown). Therefore, the amount of Evasin-4 needed to inhibit CCL2 and CCL13 appears to be too high to be relevant under physiological conditions. It is possible that other salivary proteins or as yet unidentified Evasins might be responsible for the inhibition of CCL2 or that R. sanguineus produces an anti-CCL2 binding protein at a later stage during its feeding period.
One intriguing feature of Evasin-4 is that this CKBP is both selective in binding to only some CC chemokines and yet has a broad specificity by recognizing almost 20 different proteins. In this sense, it is very different from Evasin-1, the other CC chemokine-binding protein, which shows an exquisite selectivity for CCL3, -4 and -18. We therefore tried to identify the amino acid(s) targeted by Evasin-4 by aligning the sequences of the CC chemokines but no obvious motif was found (Fig. S2). We are currently trying to identify the pharmacophore to understand the binding mode of Evasin-4 and hope to pinpoint the residues involved. Understanding how selectivity is achieved in both Evasin-1 and Evasin-4 could allow the design of CKBPs with an engineered selectivity profile which would block specific chemokines of interest but not the others.
Today it is well-established that chemokines and chemokine receptors are associated with chronic inflammation in many pathologies, including autoimmune diseases and tumors. Despite intensive efforts and positive outcomes in animal models, clinical trials with chemokine inhibitors have often been unsuccessful 20,21. Because the chemokine system is highly promiscuous: many chemokines bind to several receptors; certain receptors recognize multiple chemokines; cells often express more than one chemokine receptor; it has been suggested that blocking one single ligand or one single receptor is not enough to observe significant improvement in vivo 22. Because Evasin-4 binds to almost all inflammatory CC chemokines, it would be an excellent tool to study the simultaneous inhibition of multiple chemokines in animal models. However, recombinant, native Evasin-4 is unsuitable to use in its current format due to its short half-life compared with the long plasma exposure obtained by proteins such as antibodies (Fig. 5). We therefore investigated the ability to improve its pharmacokinetic properties by a classical fusion to an Fc domain. Whereas Evasin-4 fused either to the N- or C-terminus of the Fc moiety retained biological activity, the C-terminal format Fc–Evasin-4 fusion protein appears to best conserve the binding and inhibition characteristics of Evasin-4, and now has an extended half-life.
Poxviruses express a class of 35 kDa CKBPs which only bind CC chemokines, being in this way similar to Evasin-4. Previous studies were made in which the CC chemokine inhibitor from vaccinia virus, vCCI, was fused to an Fc moiety and it was found to reduce monocyte recruitment both in acute (zymosan-induced peritonitis) or sustained (collagen-induced arthritis) models of inflammation 23,24. In the arthritis model, prophylactic treatment with vCCI–Fc led to a delayed onset of the disease and a significant reduction of the clinical score 23, confirming the benefit of simultaneously inhibiting several CC chemokines. However, Evasin-4 and vCCI do not share the same specificity profile and Evasin-4 might therefore further elucidate the involvement of specific chemokines in inflammation. For example, vCCI blocks the chemotaxis mediated by the homeostatic chemokines CCL19 and CCL21, whereas Evasin-4 has no effect on the migration of cells towards these chemokines. Moreover, vCCI inhibits CCL2 and its murine equivalent JE, which is not the case of Evasin-4. In addition, vCCI–Fc was shown to be strongly immunogenic in mouse 23, while bioinformatics predictions and first results with Evasins are more promising 8. We are currently developing in vivo models to test the therapeutic properties of the Fc fusions of Evasin-4 and we hope to soon determine if concurrent inhibition of CC chemokines improves the clinical evolution in models of inflammatory diseases.
Material and methods
Material
Human chemokines were produced as previously described 25 or obtained from Peprotech (Rocky Hill, NJ, USA). Chemicals reagents were purchased from Sigma-Aldricht (St. Louis, MO, USA).
Expression and purification
The sequences of the constructs are shown in Fig. S1. Recombinant Evasin-4 was initially expressed as a C-terminally His-tagged protein in insect and mammalian expression systems, as previously described 9. Because it was expressed very poorly, the natural signal peptide was replaced with alternative signal peptides, derived from Evasin-1 and uPA. An N-terminal His-tagged protein was generated by the construction of a pEAK12d expression vector in which the signal peptide sequence of uPA was fused to a five amino acid flexible linker sequence followed by a 6His tag sequence, which was separated from the sequence encoding amino acids 24–127 of Evasin-4 by a second flexible linker and a caspase 8 cleavage site. Untagged Evasin-4 was expressed by direct fusion of the sequence encoding amino acids 24–127 of Evasin-4 to the uPA signal peptide sequence. Lastly, two constructs linking an Fc moiety to the N- or C-terminus of Evasin-4, again using the uPA signal peptide, were cloned into Gateway™ expression vectors (Life Technologies, Grand Island, NY, USA) as follows. PCR fragments containing the coding sequences of Evasin-4, Fc domain and the uPA signal peptide were amplified in separate PCRs. PCR primers used to generate the C-terminal Evasin-4–Fc fusion protein contained sequence overlaps at the 5′-end of Evasin-4 to the 3′ signal peptide of uPA, and at the 3′-end of Evasin-4 to the Fc domain. For PCR primers used to generate the N-terminal Evasin-4 Fc fusion protein, the signal peptide sequence contained a 3′ sequence which overlapped with the 5′ sequence of the Fc domain, and the 5′-end of Evasin-4 contained an overlap with 3′-end of the Fc domain. Full-length cDNAs were generated by assembly PCR. The Evasin-4 and the Fc portions in both of the formats were separated by a GSGSGGG linker sequence.
Expression of the recombinant proteins was monitored by western blots with an anti-His IgG (Santa Cruz, Dallas, TX, USA) for the His-tagged proteins and by autoradiography SDS/PAGE gels following cross-linking of the supernatants with iodinated CCL11 or CCL5, as described previously 8, or by SDS gel electrophoresis for the Fc fusion constructs. The 6His-tagged proteins were purified by nickel-affinity chromatography followed by a SEC to remove any remaining contaminants, and the Fc fusions by protein A chromatography, using an Akta Purifier system with standard procedures (GE Healthcare Life Sciences, Little Chalfont, UK). The untagged protein was produced in serum-free conditions and purified by applying the supernatant from 500- or 3000-mL cultures of HEK293–EBNA cells, harvested 6 days after transfection, onto a Q Sepharose Fast Flow (GE Healthcare) column of 38 mL, previously equilibrated with 50 mm Tris/HCl pH 7.5, and proteins were eluted with a linear gradient of 0-350 mm NaCl. The fractions containing the recombinant proteins were concentrated, supplemented with 2 m (NH4)2SO4 and applied onto a HiScreen Phenyl HP column (4.7 mL) (GE Healthcare) previously equilibrated with 50 mm Tris/HCL pH 7.5 buffer containing 2 m (NH4)2SO4. Proteins were eluted with a linear gradient of 2 to 0 m (NH4)2SO4 in Tris/HCl pH 7.5 and then dialysed against 5 L of 50 mm ammonium bicarbonate pH 8.0, quantified by UV absorption at 280 nm, lyophilized and stored at −20 °C. If necessary, a third step of purification was performed, consisting of reverse-phase chromatography on a PLRP-S column (7 mL) previously equilibrated in 0.2% trifluoroacetic acid (TFA). Bound proteins were eluted with 0–15% CH3CN in H2O containing 0.2% TFA over 5 CV, 15-90% CH3CN in H2O containing 0.2% TFA over 25 CV at 2 mL·min−1, 90% CH3CN in H2O containing 0.2% TFA over 2 CV and finally 0% CH3CN in H2O containing 0.2% TFA over 3 CV. The eluted protein was lyophilized and stored at −20 °C.
The 6His–Evasin-4 fusion protein was subjected to caspase 8 cleavage as previously described 26 with varying incubation times up to 24 h. The purity of the recombinant proteins was assessed by SDS/PAGE, reverse-phase HPLC, SEC and MALDI-TOF analyses, and the authenticity assessed by Edman degradation.
Analysis of selectivity
The selectivity profile was analysed using SPR on BIAcore 3000 or A100 system (GE Healthcare). The analyses were performed either by immobilizing the chemokines or by immobilizing the 6His–Evasin-4 protein on a CM4 chip. For binding analysis with immobilized chemokines using the BIAcore A100 system, the chemokines (CCL1, -2, -3, -5 E26A, -11, -15, -16, -17, -18, -22, -23, -26, CXCL1, -8, XCL1 and CX3CL1) were suspended at 25 μg·mL−1 in 10 mm sodium acetate buffer pH 5 or in 10 mm sodium acetate buffer pH 4 for CCL3 and CCL4. Chemokines were immobilized to reach a level of 700–1000 response units (RU) and different concentrations of Evasin-4 (12.5 to 200 nm) in running buffer were injected for 3 min at 30 μL·min−1. For analyses with immobilized Evasin-4 using the BIAcore 3000, 6His–Evasin-4 was suspended at 50 μg·mL−1 in 10 mm sodium acetate buffer pH 4.5 and immobilized by amine coupling chemistry to obtain a response of 800–1000 RU. Chemokines were suspended at 0.1 μg·mL−1 in HBS-P+ running buffer (0.01 m Hepes pH 7.4, 0.15 M NaCl, 0.05% surfactant P20) and injected for 3 min at 30 μL·min−1. To determine the kinetic characteristics, five dilutions of the analyte (either Evasin-4 or CC chemokines) were prepared and injected for 3 min followed by a dissociation time of 15 min. In each case, the CM4 chip was regenerated using 50 mm glycine buffer pH 2 for 30 s. To characterize the Fc constructs, the same method was used except that Evasin-4 Fc constructs were suspended at 20 μg·mL−1 in 10 mm sodium acetate buffer pH 4.5 and immobilized to reach a response of 1200 RU.
Neutralization assays
The ability to inhibit chemokine-induced in vitro chemotaxis was assessed using ChemoTx System chemotaxis plates (NeuroProbe Inc., Gaithersburg, MD, USA) at the EC80 concentration of agonist, determined prior to the inhibition assays. Inhibition was tested using L1.2/chemokine receptor transfectants or primary cell lines or both. Human primary cells were isolated from human blood by Ficoll gradient centrifugation (GE Healthcare) as previously described 11 followed by selection using MACS isolation kits (Miltenyi Biotec, Bergisch Gladbach, Germany). Lastly the ability of Evasin-4 to form complexes with certain chemokines was assessed by SEC. A 500-μg sample of Evasin-4 and an equimolar amount of the chemokine were incubated for 45 min at room temperature in NaCl/Pi and then applied to a Superdex75 column (GE Healthcare), previously calibrated with the following proteins: coalbumin, 75 kDa; ovalbumin, 44 kDa; carbonic anhydrase, 29 kDa; ribonuclease A, 13.7 kDa; aprotinin, 6.5 kDa. Fractions collected were subsequently analyzed on silver stained SDS/PAGE gels.
Analysis of pharmacokinetic profiles
C57BL/6 female mice were injected intravenously (i.v.) or subcutaneously (s.c.) with 5 mg·kg−1 of Evasin-4 or 1 or 10 mg·kg−1 of each of the Fc constructs on day 0 (n = 9 for each group) and monitored for signs of discomfort, pain or distress throughout the study period. Repeated submandibular blood samples were taken from groups of three mice at 0.083, 0.25, 0.5, 1, 4, 7 and 24 h for Evasin-4 and at 0.166, 7 h and 1, 2, 3 and 4 days for Evasin-4 Fc fusions. Intracardiac samples were taken under terminal anesthesia using inhaled isofluorane prior to sacrifice by cervical dislocation on days 7, 10 and 14. Plasma was prepared for each blood sample. A bioanalytical ELISA method was developed using a polyclonal anti-Evasin antibody raised in rabbits (Eurogentec, Seraing, Belgium) and was used to quantify the concentration of Evasin-4 at each time point. Pharmacokinetic parameters were calculated from the measured plasma concentrations using winnonlin v. 6.1. All animal studies were conducted in accordance with the Swiss Animal Welfare Act under supervision of the Geneva Cantonal Office for Veterinary Affairs.
Acknowledgments
The research leading to these results has received funding from the European Union Seventh Framework Programme [FP7-2007-2013] under grant agreement n° HEALTH-F4-2011-281608 (TIMER). We thank Prof. A. Mantovani for L1.2 receptor transfectants, A. Garin and A. Hermant for help with chemokine experiments and biology, C. Arod, N. Barillat, F. Bollin, F. Borlat, V. Dechavanne, L. Dupin, J.-P. Gaudry, L. Glez, C. Losberger, M. de Tiani and L. Chevalet for advice in protein expression and purification, P. Graber for advice in SPR, D. Bertschy-Meier, C. Martin and S. Carboni for help with in vivo experiments and P. Rueckert and A. Bernhardt for the PK analysis.
Glossary
- CKBP
chemokine-binding protein
- SEC
size-exclusion chromatography
- uPA
urokinase-type plasminogen activator
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s web site
Fig S1 Sequences of Evasin-4 constructs.
Fig S2 CC chemokines recognized by Evasin-4.
References
- 1.Fallon PG, Alcami A. Pathogen-derived immunomodulatory molecules: future immunotherapeutics? Trends Immunol. 2006;27:470–476. doi: 10.1016/j.it.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Singh UP, Singh S, Ravichandran P, Taub DD, Lillard JW., Jr Viral macrophage-inflammatory protein-II: a viral chemokine that differentially affects adaptive mucosal immunity compared with its mammalian counterparts. J Immunol. 2004;173:5509–5516. doi: 10.4049/jimmunol.173.9.5509. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn DE, Beall CJ, Kolattukudy PE. The cytomegalovirus US28 protein binds multiple CC chemokines with high affinity. Biochem Biophys Res Commun. 1995;211:325–330. doi: 10.1006/bbrc.1995.1814. [DOI] [PubMed] [Google Scholar]
- 4.Chee MS, Satchwell SC, Preddie E, Weston KM, Barrell BG. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature. 1990;344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- 5.Alcami A, Saraiva M. Chemokine binding proteins encoded by pathogens. Adv Exp Med Biol. 2009;666:167–179. doi: 10.1007/978-1-4419-1601-3_13. [DOI] [PubMed] [Google Scholar]
- 6.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci USA. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith P, Fallon RE, Mangan NE, Walsh CM, Saraiva M, Sayers JR, McKenzie AN, Alcami A, Fallon PG. Schistosoma mansoni secretes a chemokine binding protein with antiinflammatory activity. J Exp Med. 2005;202:1319–1325. doi: 10.1084/jem.20050955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deruaz M, Frauenschuh A, Alessandri AL, Dias JM, Coelho FM, Russo RC, Ferreira BR, Graham GJ, Shaw JP, Wells TN. Ticks produce highly selective chemokine binding proteins with antiinflammatory activity. J Exp Med. 2008;205:2019–2031. doi: 10.1084/jem.20072689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frauenschuh A, Power CA, Deruaz M, Ferreira BR, Silva JS, Teixeira MM, Dias JM, Martin T, Wells TN, Proudfoot AE. Molecular cloning and characterization of a highly selective chemokine-binding protein from the tick Rhipicephalus sanguineus. J Biol Chem. 2007;282:27250–27258. doi: 10.1074/jbc.M704706200. [DOI] [PubMed] [Google Scholar]
- 10.Dias JM, Losberger C, Deruaz M, Power CA, Proudfoot AE, Shaw JP. Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3. PLoS ONE. 2009;4:e8514. doi: 10.1371/journal.pone.0008514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krohn S, Garin A, Gabay C, Proudfoot AE. The activity of CCL18 is principally mediated through interaction with glycosaminoglycans. Front Immunol. 2013;4:193. doi: 10.3389/fimmu.2013.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dantas-Torres F, Figueredo LA, Brandao-Filho SP. Rhipicephalus sanguineus (Acari: Ixodidae), the brown dog tick, parasitizing humans in Brazil. Rev Soc Bras Med Trop. 2006;39:64–67. doi: 10.1590/s0037-86822006000100012. [DOI] [PubMed] [Google Scholar]
- 13.Palmqvist C, Wardlaw AJ, Bradding P. Chemokines and their receptors as potential targets for the treatment of asthma. Br J Pharmacol. 2007;151:725–736. doi: 10.1038/sj.bjp.0707263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janeway CA, Travers P, Walport M, Shlomchik M. The humoral immune response. In: Austin P, Lawrence E, editors. Immunobiology: the Immune System in Health and Disease. Garland Publishing, New York; 2001. pp. 341–380. [Google Scholar]
- 15.Stone KD, Prussin C, Metcalfe DD. IgE, mast cells, basophils, and eosinophils. J Allergy Clin Immunol. 2010;125:S73–S80. doi: 10.1016/j.jaci.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurihara T, Warr G, Loy J, Bravo R. Defects in macrophage recruitment and host defense in mice lacking the CCR2 chemokine receptor. J Exp Med. 1997;186:1757–1762. doi: 10.1084/jem.186.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boring L, Gosling J, Chensue SW, Kunkel SL, Farese RV, Jr, Broxmeyer HE, Charo IF. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J Clin Invest. 1997;100:2552–2561. doi: 10.1172/JCI119798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajnicka V, Vancova I, Kocakova P, Slovak M, Gasperik J, Slavikova M, Hails RS, Labuda M, Nuttall PA. Manipulation of host cytokine network by ticks: a potential gateway for pathogen transmission. Parasitology. 2005;130:333–342. doi: 10.1017/s0031182004006535. [DOI] [PubMed] [Google Scholar]
- 19.Peterkova K, Vancova I, Hajnicka V, Slovak M, Simo L, Nuttall PA. Immunomodulatory arsenal of nymphal ticks. Med Vet Entomol. 2008;22:167–171. doi: 10.1111/j.1365-2915.2008.00726.x. [DOI] [PubMed] [Google Scholar]
- 20.Allegretti M, Cesta MC, Garin A, Proudfoot AE. Current status of chemokine receptor inhibitors in development. Immunol Lett. 2012;145:68–78. doi: 10.1016/j.imlet.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Schall TJ, Proudfoot AE. Overcoming hurdles in developing successful drugs targeting chemokine receptors. Nat Rev Immunol. 2011;11:355–363. doi: 10.1038/nri2972. [DOI] [PubMed] [Google Scholar]
- 22.Koelink PJ, Overbeek SA, Braber S, de Kruijf P, Folkerts G, Smit MJ, Kraneveld AD. Targeting chemokine receptors in chronic inflammatory diseases: an extensive review. Pharmacol Ther. 2012;133:1–18. doi: 10.1016/j.pharmthera.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Buatois V, Fagete S, Magistrelli G, Chatel L, Fischer N, Kosco-Vilbois MH, Ferlin WG. Pan-CC chemokine neutralization restricts splenocyte egress and reduces inflammation in a model of arthritis. J Immunol. 2010;185:2544–2554. doi: 10.4049/jimmunol.1000182. [DOI] [PubMed] [Google Scholar]
- 24.White GE, McNeill E, Christou I, Channon KM, Greaves DR. Site-directed mutagenesis of the CC chemokine binding protein 35K-Fc reveals residues essential for activity and mutations that increase the potency of CC chemokine blockade. Mol Pharmacol. 2011;80:328–336. doi: 10.1124/mol.111.071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Proudfoot AE, Borlat F. Purification of recombinant chemokines from E. coli. Methods Mol Biol. 2000;138:75–87. doi: 10.1385/1-59259-058-6:75. [DOI] [PubMed] [Google Scholar]
- 26.Severin IC, Gaudry JP, Johnson Z, Kungl A, Jansma A, Gesslbauer B, Mulloy B, Power C, Proudfoot AE, Handel T. Characterization of the chemokine CXCL11–heparin interaction suggests two different affinities for glycosaminoglycans. J Biol Chem. 2010;285:17713–17724. doi: 10.1074/jbc.M109.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Appay V, Brown A, Cribbes S, Randle E, Czaplewski LG. Aggregation of RANTES is responsible for its inflammatory properties. Characterization of nonaggregating, noninflammatory RANTES mutants. J Biol Chem. 1999;274:27505–27512. doi: 10.1074/jbc.274.39.27505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1 Sequences of Evasin-4 constructs.
Fig S2 CC chemokines recognized by Evasin-4.