Abstract
Transmembrane protease, serine 6 (TMPRSS6), is likely to be involved in iron metabolism through its pleiotropic effect on hepcidin concentrations. Recently, genome-wide association studies have identified common variants in the TMPRSS6 gene to be linked to anaemia and low iron status. To get a more precise evaluation of identified TMPRSS6 single nucleotide polymorphism associations with iron status in cohorts of differing continental ancestry, we conducted a systematic review with meta-analyses. We searched the literature using HuGE Navigator, Pubmed and Scopus databases for primarily genome-wide association studies using TMPRSS6 as a free term. Fixed-effects meta-analysis was used to obtain summary estimates of associations. Eleven studies comprised Caucasian populations, four included an Asian population and one study included an African-American population. Differences in minor allele frequencies of 8 TMPRSS6 SNPs (rs855791, rs4820268, rs2111833, rs1421312, rs228921, rs228918, rs228919 and rs575620) across ethnic groups were observed, with the MAF of rs855791 significantly higher in Asian populations than in Caucasians (0.55 vs 0.42, P < 0.0001). In the meta-analysis, the A allele of rs855791 was associated with lower Hb and ferritin concentrations in all populations. This allele was also associated with increased serum transferrin receptor and transferrin concentrations. We observed similar associations for the G allele in rs4820268. Clear disparities in associations were found for the African-American population, although not statistically significant. Associations between TMPRSS6 SNPs and anaemia are consistent across Caucasian and Asian populations. This study highlights the need to conduct studies in African populations where iron deficiency is of utmost public health significance.
Keywords: Systematic review, Meta-analyses, TMPRSS6, Iron status
Introduction
Iron deficiency has previously been hypothesized to originate entirely from dietary and/or other environmental factors. However, several discoveries regarding disorders of iron metabolism have indicated that there is a genetic contribution to the development of iron deficiency (Leboeuf et al. 1995; Morse et al. 1999; Finberg 2009). In particular, polymorphisms in the Trans Membrane Protease Serine 6 (TMPRSS6) gene have been implicated as influencing iron metabolism in both animal and human studies. In the mask mutant mouse, loss of the catalytic domain of matriptase-2 (protein produced due to TMPRSS6 expression) results in an increase in hepcidin expression in the liver, lowered dietary iron absorption and severe microcytic anaemia (Du et al. 2008). Similar findings have been observed in TMPRSS6 knockout mice (Tmprss6 2/2; Folgueras et al. 2008) and in zebrafish overexpressing mutant matriptase-2 (Silvestri et al. 2008). Contrariwise, overexpression of wild-type matriptase-2 in human hepatoblastoma (HepG2) cells has been observed to result in suppression of hepcidin gene promoter activation (Du et al. 2008). Mutations in the TMPRSS6 gene have also been implicated in iron deficiency anaemia refractory to oral iron therapy within Caucasian populations (Finberg et al. 2008; Guillem et al. 2008; Melis et al. 2008). Further evidence of the association of TMPRSS6 polymorphisms with iron status in persons not affected by overt genetic disorders of iron metabolism have been observed in several GWAS though not all findings are consistent across studies (Benyamin et al. 2009; Chambers et al. 2009; Ganesh et al. 2009; Tanaka et al. 2010).
At the population level, various studies have observed a geographic disparity in iron status leading to the hypothesis that dissimilarities in genetic alterations across ethnicities may indeed contribute to differences in iron status (Gordeuk et al. 1992; Barton et al. 1995; Wurapa et al. 1996; Moyo et al. 1998). Specifically, studies have shown that Asia and Africa have the highest proportion of individuals affected by anaemia according to the WHO regional prevalence estimates (WHO 2008). Furthermore, in studies conducted in the US, African-Americans appear to have lower haemoglobin concentrations and serum transferrin saturation (TS) as compared to Caucasians (Williams 1981; Jackson et al. 1983; Perry et al. 1992; McLaren et al. 2001; Beutler et al. 2003). Disparities in iron status have also been observed between Mexican-American and non-Hispanic white women of childbearing age (Looker et al. 1997). It is not known, however, if these differences contain a genetic component.
In order to shed light on the contradictory findings from GWAS and to evaluate the role of ethnicity as an explanatory factor, we performed a systematic review with meta-analysis on TMPRSS6 loci identified in cohorts of differing continental ancestry.
Methods
Search strategy
An electronic literature search was conducted using HuGE Navigator which is a database of published population-based epidemiological studies of human genes extracted and curated from PubMed since 2001. The search term ‘tmprss6 × [Text MesH]’ was used. The latest search was conducted on 31st August 2013. In addition, the PubMed and Scopus databases were searched to include any articles that may have been published on the topic prior to 2001. Searches in both databases were conducted with TMPRSS6 as a free term. Manual searching of reference lists of original articles was also conducted.
Eligibility criteria
The first step in the study selection was exclusion of duplicates followed by examination of titles and abstracts obtained. Articles were included when they were: original research articles; conducted in humans; and when testing for TMPRSS6 SNP associations with iron status measures i.e. haemoglobin (Hb), serum or plasma ferritin (SF/PF) and/or serum transferrin receptor (sTfR), was undertaken. In addition, we included studies on associations of dichotomous outcomes (anaemia, iron deficiency, or iron deficiency anaemia) with TMPRSS6 SNPs. Animal, single patient studies and studies on disorders of iron metabolism [iron-refractory iron deficiency anaemia (IRIDA), hemojuvelin, hereditary hemochromatosis] were excluded. The full text of each remaining study was reviewed to establish eligibility, and all relevant information and data were extracted.
Data extraction
Data extraction was conducted by one author (WGW) and repeated by two other authors (AMB and EJF) for 30 % of the papers that met the inclusion criteria. The latter was done for quality control purposes. For each article, information on authors, publication year, sample size, ethnicity, health status of the population (e.g. type 2 diabetes type patients), study design, mean age, gender distribution, minor allele frequencies, genotyping platform, call rate, beta values, standard errors, confidence intervals, reported measure of variance, agreement with Hardy–Weinberg equilibrium and model adjustments were extracted. In the case of GWA studies, information on the main and replication study was reported separately. Additionally, where several cohorts were included in a study, information on each cohort was reported separately. In the case that information provided was missing, insufficient or unclear, authors were contacted for further information. Any recalculation of values required prior to meta-analyses was performed by one author (WGW).
Statistical methods
Meta-analyses were performed on genetic variants with information from more than two studies or cohorts and results ordered by ethnicity. Results were presented at the sub-group and overall levels. The sub-groups were designated as Caucasian, Asian and Mixed, with Mixed implying that the study results presented were from more than one ethnicity.
All data for specific iron status measures i.e. Hb, SF/PF and sTfR were transformed into identical units before meta-analyses. In the case where all extracted values had different transformations, values were back transformed to comparable units to enable comparison. New standard error values were obtained by calculating the ratio between the untransformed beta values and their standard errors and applying this ratio to the recalculated beta value.
Our goal in conducting the meta-analyses was to compute the common effect size for the identified populations, and not to generalize the findings to other populations. Additionally, the studies with complete information were few. For these reasons, we used the fixed-effects model to assign study weights as well as combine summary statistics. We also conducted a random-effect analyses to check for heterogeneity between studies.
An estimate of potential publication bias was carried out by generating funnel plots. The symmetry of the funnel plot was assessed both visually and formally by using Egger’s test (Egger et al. 1997). The Chi square test was conducted to test for heterogeneity and I2values reported. We conducted an independent samples T test on summary estimates per SNP for each outcome measure to determine whether differences observed between ethnicities were statistically significant.
The R program for statistical computing version 2.15.2 (R Core Team 2013) was used to perform all analyses.
Results
We found 20 articles through HuGE Navigator, 84 articles through PubMed and 86 articles through Scopus and four articles through manual search of the references.1 In total we obtained 14 articles that contained information from various cohorts (Fig. 1; Table 1). Articles identified as eligible for meta-analyses (n = 14) contained complete information on two TMPRSS6 variants. In addition, 11 articles on MAF comparisons and 3 articles on 4 TMPRSS6 variants were included in the systematic review (Fig. 1). Only one study corrected for iron intake in association analyses. Additional data on covariates considered in association analyses in the various studies can be found in Table 1. The majority of the articles were based on studies conducted entirely in subjects of Caucasian ethnicity (n = 11; 73 %). Three studies had multiple ethnicities as part of the cohorts investigated (Chambers et al. 2009; Lee 2009; McLaren et al. 2012). We did not find any studies conducted among individuals from the African population (Tables 1 and 2). No publication bias was present in the meta-analyses performed except for the association between rs855791 and transferrin in the Caucasian population.
Fig. 1.
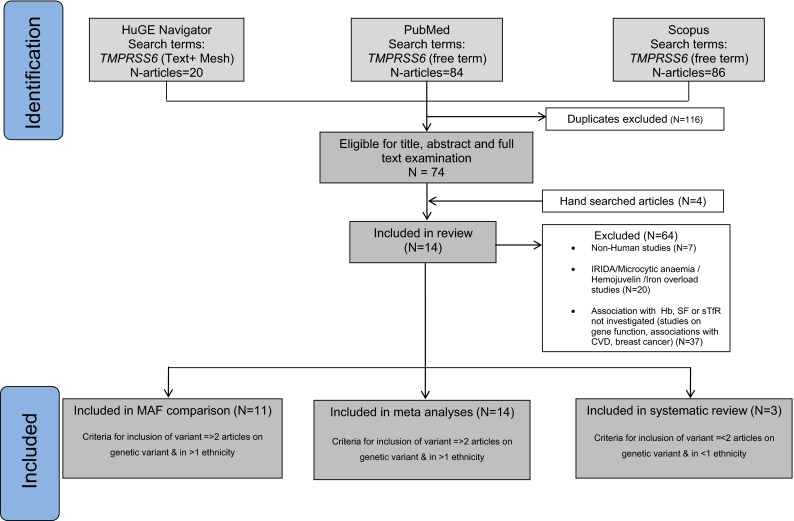
Study selection process
Table 1.
Characteristics of cohorts included in the review
| Cohort | Location | Ethnicity | Mean age years (SD) | n | Gender ratio male/female | Iron status marker/erythrocyte trait studied | Confounders considered in association analyses | References |
|---|---|---|---|---|---|---|---|---|
| – | China | Han (East) | 62.2 (7.7) | 1,141 | 0/100 | Hb, SI, SF, sTfRN, FEP, TIBC | Age | An et al. (2012) |
| – | China | Han (North) | 58.2 (7.5) | 354 | 0/100 | Hb, SI, SF, sTfRN, FEP, TIBC | ||
| – | China | Zhuang | 59.4 (7.9) | 630 | 0/100 | Hb, SI, SF, sTfRN, FEP, TIBC | ||
| Health professional follow-up study (HPFS) and Nurses health study (NHS) | USA | Caucasian | 43.3 (6.7) | 2,422 | – | Ferritin, TfR, TRFN | None | He et al. (2012) |
| KORA F3, KORA F4, Micors, InCHIANTI, Croat-Vis | Europe | Caucasian | 58.9 (12.8) | ~6,600 | – | sTfR, SI, TRFN, TRFN saturation | Age, sex | Oexle et al. (2011) |
| – | China | Han | 58.3(5.9) | 1,574 | 45.2/45.8 | Hb, PF | Age, sex, dietary iron and BMI | Gan et al. (2012) |
| – | USA | Mixed | 56.5(13.5) | 48 | 0/100 | Hb, SI, SF, MCV, TIBC | Age, presence or absence of pica | Lee (2009) |
| Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium | Europe | Caucasian | 58.5 (8.3) | 24,731 | 43/57 | Hb, Hct, MCH, MCHC, MCV, Erythrocyte count | None | Ganesh et al. (2009) |
| Haematology genetics (HAEMGEN) study | Italy | Caucasian | – | 9,456 | – | Hb, Hct, MCH, MCHC, MCV, Erythrocyte count | None | Ganesh et al. (2009) |
| InCHIANTI | USA | Caucasian | 68.3 (15.5) | 1,206 | 44/56 | SI, SF, Hb, Hct, MCV, RBC count, RBC width, platelets | Age and gender | Tanaka et al. (2010) |
| Women’s Healthy Aging Study (WHAS) I | USA | Caucasian | 78.3 (8.0) | 375 | 0/100 | SI, SF, Hb, Hct, MCV, RBC count, RBC width, platelets | None | Tanaka et al. (2010) |
| WHAS II | USA | Caucasian | 74 (2.7) | 194 | 0/100 | SI, SF, Hb, Hct, MCV, RBC count, RBC width, platelets | None | Tanaka et al. (2010) |
| InCHIANTI, BLSA, SardiNIA stage 2 | Italy and USA | Caucasian | 59.5 (15.8) | 3,200 | 46/54 | Fe, TRFN, Ferritin, sTfR | Sex, age | Pichler et al. (2011) |
| – | Spain | Caucasian | 24.3 (4.8) | 270 | 0/100 | SI, SF, Hb, Hct, MCV, RBC count, TRFN, Transferrin saturation, TIBC | None | Blanco-Rojo et al. (2011) |
| Australian Twin families | Australia | Caucasian | Adolescents | ~2,512 | 40/60 | Fe, TRFN, Transferrin saturation, Hb, MCV | Sex, age, sex * age | Benyamin et al. (2009) |
| Nijmegen biomedical study (NBS) | NL | Caucasian | – | 1,832 | – | Fe, TRFN, Transferrin saturation, Hb, MCV | None | Benyamin et al. (2009) |
| London Life Sciences Population (LOLLIPOP) | UK | Caucasian | 54.5 (9.8) | 1,599 | 87.3/12.7 | Hb, RBC, MCV, MCH, MCHC | None | Chambers et al. (2009) |
| LOLLIPOP | UK | Indian Asians | 53.6 (10.8) | 9,685 | 88.8/12.2 | Hb, RBC, MCV, MCH, MCHC | None | Chambers et al. (2009) |
| Salzburg Atherosclerosis Prevention Program in Subjects at High Individual Risk (SAPHIR) | Austria | Caucasian | Range Male 39–66 years Females 39–67 years |
1,726 | 63/37 | Hb, RBC count, SI | Age and gender | Kloss-Brandstatter et al. (2012) |
| ValBorbera (VB) study | Italy | Caucasian | 55.4 (17.8) | ~1,650 | 44/56 | Hb, Hct, MCV, MCH, MCHC, SI, TRFN, ferritin, TRFN saturation | None | Traglia et al. (2011) |
| Hemochromatosis and Iron Overload Screening (HEIRSa) study | USA | Caucasian | 60.1 (10.4) | 1,084 | 24/76 | SI, UIBC, SF, sTfR, TIBC | Age, sex, ln(CEA), ln(CRP), ln(GGT), C282Y/H63D | McLaren et al. (2012) |
| HEIRSa | USA | African-American | 57.5 (12.4) | 221 | 36/64 | SI, UIBC, SF, sTfR, TIBC | ln(CRP), ln(GGT), CagA | McLaren et al. (2012) |
| HEIRSa | USA | Hispanic | 56.3 (10.1) | 239 | 25/75 | SI, UIBC, SF, sTfR, TIBC | ln(CEA) | McLaren et al. (2012) |
| HEIRSa | USA | Asian | 55.7 (10.9) | 153 | 36/64 | SI, UIBC, SF, sTfR, TIBC | ln(GGT) | McLaren et al. (2012) |
Ethnicities with similar vowels next to them were analysed and compared in the same study
C282Y/H63D genotype, CagA strain infection status (infected vs non-infected)
ln natural log transformation, CEA carcinoembryonic antigen, CRP c-reactive protein , GGT gamma-glutamyltransferase, HFE gene, Hb Haemoglobin, SI serum iron, SF serum ferritin, PF plasma ferritin, TRFN transferrin, sTfRN serum transferrin, sTfR serum transferrin receptor, FEP free erythrocyte porphyrins, TIBC total iron-binding capacity, UIBC unsaturated iron-binding capacity, Hct hematocrit, MCH mean corpuscular haemoglobin, MCHC mean corpuscular haemoglobin concentration, MCV mean corpuscular volume, RBC red blood cell
aAdjustments considered in association testing with serum ferritin
– Information not available
Table 2.
Minor allele frequencies of TMPRSS6 SNPs in various populations
| SNP | Cohort | Ethnicity | MAF | References |
|---|---|---|---|---|
| rs855791 (A) | – | Han (East) | 0.53 | An et al. (2012) |
| – | Han (North) | 0.55 | An et al. (2012) | |
| – | Zhuang | 0.63 | An et al. (2012) | |
| – | Han | 0.53 | Gan et al. (2012) | |
| CHARGE + HAEMGEN | Caucasian | 0.39 | Ganesh et al. (2009) | |
| InCHIANTI + BLSA + WHAS I + WHAS II | Caucasian | 0.41 | Tanaka et al. (2010) | |
| Australian Twin families | Caucasian | 0.42 | Benyamin et al. (2009) | |
| LOLLIPOP | Caucasian | 0.34 | Chambers et al. (2009) | |
| LOLLIPOP | Indian Asians | 0.53 | Chambers et al. (2009) | |
| SAPPHIR | Caucasian | 0.43 | Kloss-Brandstatter et al. (2012) | |
| NBS | Caucasian | 0.46 | Kloss-Brandstatter et al. (2012) | |
| – | Mixed | 0.37 | Lee (2009) | |
| ValBorbera study | Caucasian | 0.45 | Traglia et al. (2011) | |
| NBS | Caucasian | 0.46 | Galesloot et al. (2013) | |
| rs4820268 (G) | – | Han (East) | 0.51 | An et al. (2012) |
| Han (North) | 0.53 | An et al. (2012) | ||
| Zhuang | 0.61 | An et al. (2012) | ||
| – | Han | 0.50 | Gan et al. (2012) | |
| InCHIANTI + BLSA + WHAS I + WHAS II | Caucasian | 0.46 | Tanaka et al. (2010) | |
| Caucasian | 0.47 | Benyamin et al. (2009) | ||
| LOLLIPOP | Caucasian | 0.43 | Chambers et al. (2009) | |
| LOLLIPOP | Indian Asians | 0.55 | Chambers et al. (2009) | |
| SAPPHIR | Caucasian | 0.45 | Kloss-Brandstatter et al. (2012) | |
| NBS | Caucasian | 0.47 | Kloss-Brandstatter et al. (2012) | |
| rs2111833 (T) | HEIRS | Caucasian | 0.34 | McLaren et al. (2012) |
| HEIRS | African-American | 0.39 | McLaren et al. (2012) | |
| HEIRS | Hispanic | 0.20 | McLaren et al. (2012) | |
| HEIRS | Asian | 0.31 | McLaren et al. (2012) | |
| rs1421312 (G) | HEIRS | Caucasian | 0.40 | McLaren et al. (2012) |
| HEIRS | African-American | 0.61 | McLaren et al. (2012) | |
| HEIRS | Hispanic | 0.34 | McLaren et al. (2012) | |
| HEIRS | Asian | 0.31 | McLaren et al. (2012) | |
| InCHIANTI + BLSA + WHAS I + WHAS II | Caucasian | 0.39 | Tanaka et al. (2010) | |
| LOLLIPOP | Caucasian | 0.46 | Chambers et al. (2009) | |
| LOLLIPOP | Indian Asians | 0.54 | Chambers et al. (2009) | |
| rs228921 (G) | LOLLIPOP | Caucasian | 0.41 | Chambers et al. (2009) |
| LOLLIPOP | Indian Asians | 0.48 | Chambers et al. (2009) | |
| rs228918 (C) | LOLLIPOP | Caucasian | 0.47 | Chambers et al. (2009) |
| LOLLIPOP | Indian Asians | 0.48 | Chambers et al. (2009) | |
| rs228919 (T) | LOLLIPOP | Caucasian | 0.40 | Chambers et al. (2009) |
| LOLLIPOP | Indian Asians | 0.48 | Chambers et al. (2009) | |
| rs5756520 (A) | LOLLIPOP | Caucasian | 0.41 | Chambers et al. (2009) |
| LOLLIPOP | Indian Asians | 0.48 | Chambers et al. (2009) |
In rs855791, MAF was higher in the Asian than in Caucasian populations (0.55 vs 0.42, P < 0.0001). The MAF of the rs2111833 SNP ranged from 0.20 in the Hispanic population to 0.47 in the Caucasian population, while the MAF of rs1421312 was 0.31 in the Asian population and 0.47 in the Caucasian population. MAF for rs228921, rs228918, rs228919 and rs575620 were comparable in Caucasian and Indian Asians (Table 1).
Associations of rs855791 with Hb and iron status
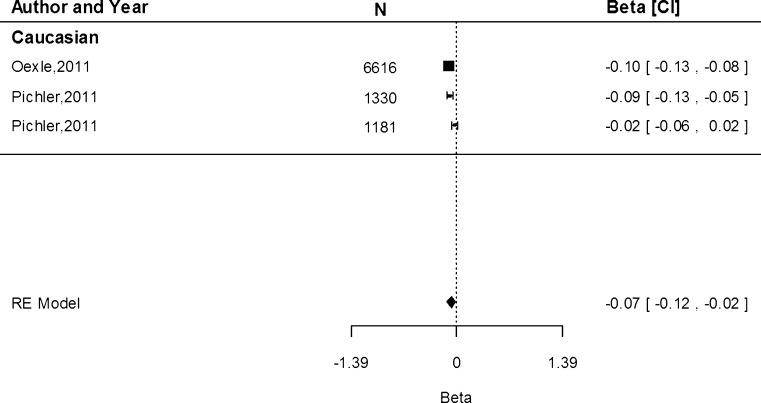
The rs855791 (c.2207T>C) SNP causes the matriptase-2 valine to alanine amino acid substitution (p.Val736Ala; Nai et al. 2011). In our association analyses we considered the A allele which is the minor allele on the reverse strand (similar to the T allele on the forward strand). The meta-analysis of the association of rs855791 with Hb indicates that each A allele (encoding for Valine [Val]) was associated with 0.11 g/dL (95 % CI −0.11, −0.10) lower Hb concentrations overall, with values of −0.08 (95 % CI −0.11, −0.04), −0.15 (95 % CI −0.18, −0.12) and −0.11 (95 % CI −0.11, −0.10) g/dL in the Caucasian, Asian and mixed populations, respectively (Fig. 2). The difference in the effect estimates between Caucasian and Asian populations, based on 11 and 5 separate study populations, respectively, was not significant (P = 0.85). Heterogeneity was high among the studies within the Caucasian population (I2 = 96.2 %, P = <0.0001, n = 11) and in the mixed populations (I2 = 96.3 %, P < 0.0001, n = 2), but was absent for the Asian cohorts (I2 = 0.00 %, P = 0.47, n = 5).
Fig. 2.
Fixed-effects meta-analysis of observational studies evaluating association of rs855791 with haemoglobin concentration (g/dL)
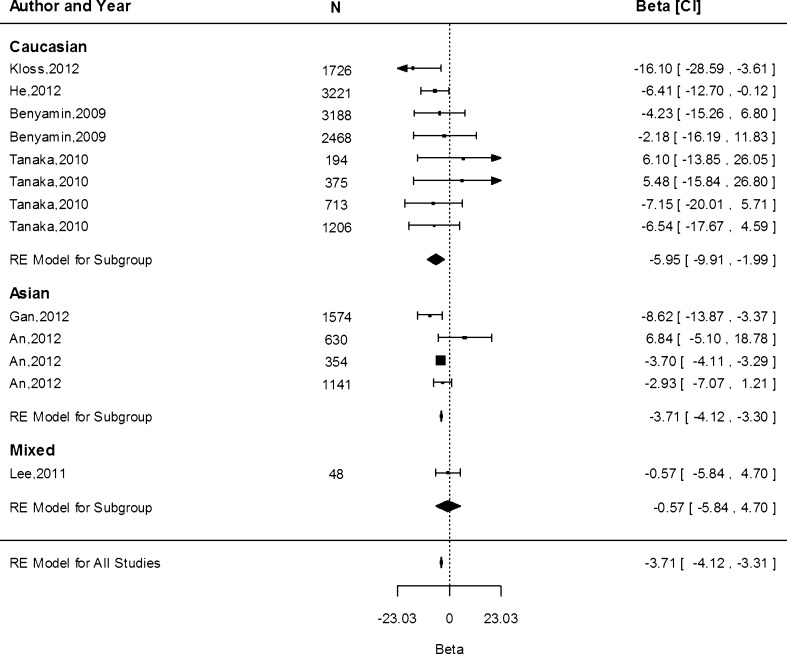
Data from 13 study populations were available to study the associations of the A allele and serum ferritin concentration. Overall, a reduction of 3.71 μg/L (95 % CI −4.12, −3.31) was observed, without significant differences between ethnicities (P > 0.05; Fig. 3). As for Hb, heterogeneity was high in the Caucasian population (I2 = 65.2 %, P = 0.02) but not in the Asian populations (I2 = 0.00 %).
Fig. 3.
Fixed-effects meta-analysis of observational studies evaluating association of rs855791 with ferritin (µg/L)
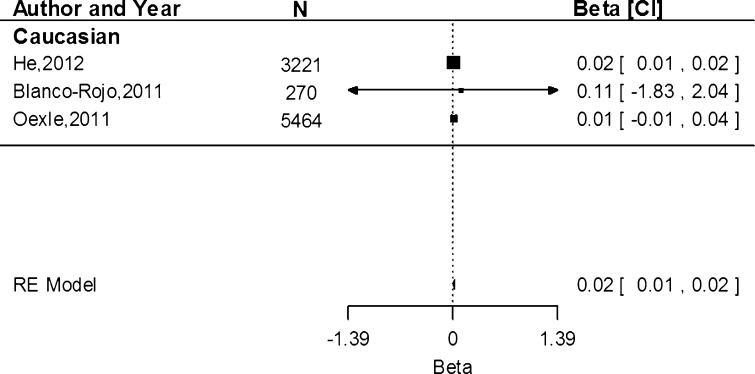
Associations of rs855791 with serum transferrin receptor concentration were only conducted within the Caucasian population. Based on three study populations, the estimate shows that the A allele was associated with an increase of 0.02 mg/dL (95 % CI 0.01, 0.02) in serum transferrin receptor concentration (Fig. 4). Heterogeneity between the studies was low (I2 = 0.00 %, P = 0.93).
Fig. 4.
Fixed-effects meta-analysis of observational studies evaluating association of rs855791 with serum transferrin receptor (mg/dL)
In determining the association with transferrin, overall meta-analysis of five study populations indicated that the A allele had no effect 0.00 mg/dL (95 % CI −0.05, 0.06; Fig. 5). Heterogeneity between the studies was high (I2 = 99.7 %, P = 0.01). Publication bias was detected in the meta-analyses performed (P = 0.01).
Fig. 5.
Fixed-effects meta-analysis of observational studies evaluating association of rs855791 with rs855791 with transferrin (mg/dL)
Associations of rs4820268 with Hb and iron status
The minor allele at the rs4820268 (c.1563C>T) leads to a synonymous change at nucleotide position 521 (p.Asp521Asp). In our association analyses, we considered the G allele which is the minor allele on the forward strand (similar to the C allele on the reverse strand). The meta-analysis of the association of rs4820268 with Hb and ferritin within the Caucasian and Asian ethnicities indicated that the G allele results in lower concentrations of Hb by 0.12 g/dL (95 % CI −0.16, −0.01) and 0.16 g/dL (95 % CI −0.22, −0.10), respectively (Fig. 6). The difference in the effect estimates between four Caucasian and Asian populations was not significant (P > 0.05). We observed a non-significant amount of heterogeneity within studies in both the Caucasian (I2 = 6.20 %, P = 0.34) and Asian population (I2 = 0.61 %, P = 0.23).
Fig. 6.
Fixed-effects meta-analysis of observational studies evaluating association of rs4820268 with haemoglobin concentration (g/dL)
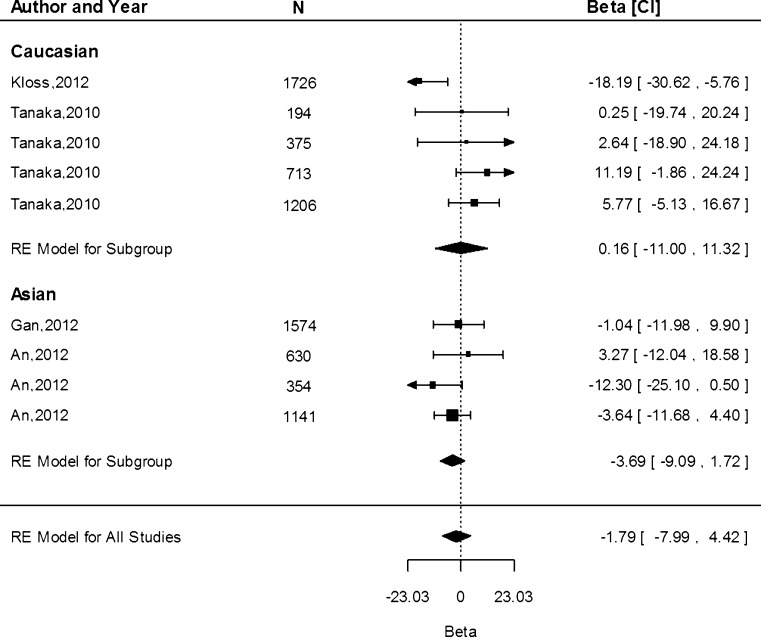
We obtained data from 13 study populations on the associations of the G allele and serum ferritin concentration. The G allele resulted in an increased ferritin concentration of 0.12 µg/L (95 % CI −6.16, 6.39) in the Caucasian population and a decrease of 3.69 µg/L (95 % CI −9.09, 1.72) in the Asian population (Fig. 7). There was no significant difference in the effect estimates between Caucasian and Asian populations (P > 0.05). We observed a significant amount of heterogeneity in the studies within the Caucasian population (I2 = 65.15 %, P = 0.02), but not in the Asian studies (I2 = 0.00 %, P = 0.43).
Fig. 7.
Fixed-effects meta-analysis of observational studies evaluating association of rs4820268 with ferritin (µg/L)
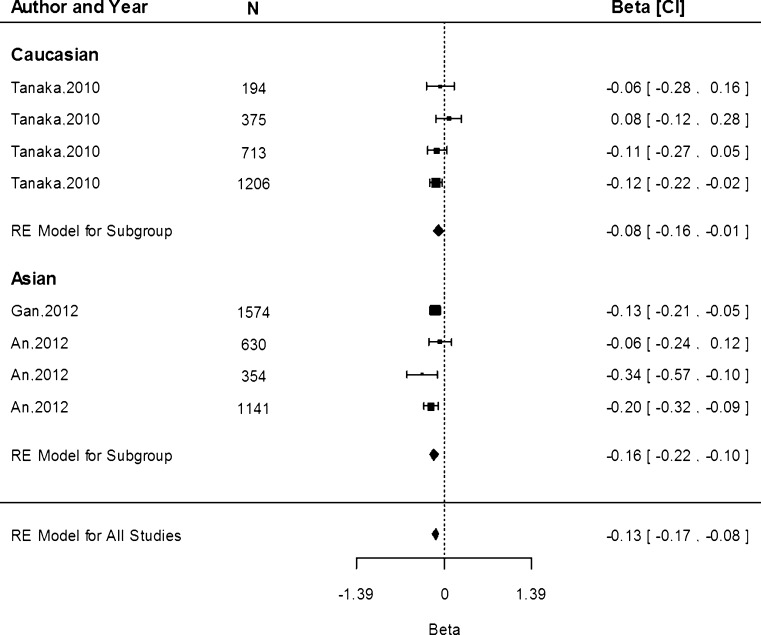
Associations of rs4820268 with sTfR concentration were only conducted in three studies within the Caucasian population. The G allele was associated with a reduction by 0.08 mg/dL (95 % CI −0.10, −0.06) in the Caucasian population (Fig. 8). There was a significant amount of heterogeneity detected (I2 = 65.15 %, P = 0.02).
Fig. 8.
Fixed-effects meta-analysis of observational studies evaluating association of rs4820268 with serum transferrin receptor (mg/dL)
TMPRRS6 variants not eligible for meta-analysis
Since the associations of rs2111833 (c.1083G>A) and rs1421312 (c.659-1988T>C) with iron status among Caucasians, Asians, Hispanics and African-Americans was described in only one study by McLaren and McLachlan (McLaren et al. 2012), these variants were not eligible for the meta-analysis. In this study, MAF across the populations for rs211833 indicated that the T allele has the lowest frequency among the Hispanic population (20 %) while the Caucasian, Asian and African-American populations had a MAF of approximately 30 % (Table 2). When investigating the association of rs2111833 with serum ferritin, the T allele was associated with an increase in serum ferritin concentrations across all ethnicities. For rs2111833 and sTfR, the estimates in each ethnicity indicated an association of the T allele with a decrease in concentrations (McLaren et al. 2012). However, none of the observations were significant (P > 0.05).
The SNP rs1421312 was also studied in four ethnicities. The MAF of this SNP in the African-American population was 0.61, higher than that in the Caucasian population 0.40, Asian population 0.41, and Hispanic population, 0.34. Overall, the G allele of rs1421312 was associated with a non-significant decrease in serum ferritin concentration. However, each additional G allele resulted in a non-significant decrease of 30.75 µg/L (95 % CI −159.62, 98.12) in SF concentrations within the African-American population, but in increase in the other ethnic populations. For sTfR, this SNP was associated with non-significant negative associations in Caucasians (−0.23 (95 % CI −0.45, −0.01) and Asians (−0.25 (95 % CI −0.78, 0.28) and a positive association in African-Americans (0.88 (95 % CI −0.04, 1.80) and Hispanics [0.19 (95 % CI −0.48, 0.86; McLaren et al. 2012].
We also identified studies in the Chinese population that have observed rs855791 and rs4820268 polymorphisms as genetic risk factors for developing anaemia, iron deficiency and iron deficiency anaemia (An et al. 2012; Gan et al. 2012). We additionally identified another study conducted among Danish blood donors that has observed that the T and G alleles of rs855791 and rs4820268, respectively, are not associated with low serum ferritin (Sorensen et al. 2012).
Discussion
To our knowledge, this is the first review to focus on the association of TMPRSS6 genetic variants with iron-related parameters in different ethnic groups. Sample sizes included in the meta-analyses were sufficient to investigate the research question. We observed that the A allele of rs855791 is associated with lower concentrations of Hb and ferritin across all populations. Additionally, this allele is associated with increased sTfR and transferrin concentrations. Based on our meta-analyses, the overall effect estimate of the Asian studies indicates that for each A and G allele of rs855791 and rs4820268, respectively, an additional decrease in Hb values of 0.07 and 0.12 g/dL is observed, as compared to the Caucasian population. Similarly, ferritin concentration is lowered by an additional 2.24 and 3.85 µg/L, respectively, in the Asian populations. The directions and magnitudes of associations were identical in both fixed-effects and random-effects model.
Although the observed differences in iron status may not be impressive from a clinical point of view, it may be of significance on the population level especially in populations that already have marginal iron status due to low dietary iron intake.
The exact mechanism through which TMPRSS6 action is exerted is still under investigation. It had generally been thought that TMPRSS6 polymorphisms affect hepcidin transcription, thereby altering hepcidin concentrations in response to systemic iron concentrations (Du et al. 2008; Finberg et al. 2008; De Falco et al. 2013). However, two recent studies (Traglia et al. 2011; Galesloot et al. 2013) did not confirm an intermediate role for hepcidin in the SNP–iron status parameters associations. These studies instead indicate pleiotropic SNP effects on hepcidin and iron status parameters. Currently, it is proposed that matriptase-2 could regulate hepcidin expression by cleaving HJV to decrease BMP–SMAD signalling (Meynard et al. 2014). Further studies are required to elucidate the role of hepcidin in TMPRSS6–iron status parameter associations.
We have observed similar directions of the effect estimates between rs855791 and rs4820268 in associations with iron status parameters. This is probably because these two SNPs are in linkage disequilibrium (Nai et al. 2011). However, we also observed differences in the direction of association of the risk allele in rs1421312 with body iron, serum ferritin and serum iron in African-Americans. Similar alleles in different ethnic groups could result in variations in the expression of genes (Spielman et al. 2007). This further strengthens the need to conduct ethnicity-specific studies when considering the role of genetics in disease outcome.
We observed that the risk allele frequencies of rs855791 and rs4820268 were as high as 63 % in the Asian populations, while in the Caucasian populations, it was generally less than 45 %. This difference in MAF across ethnicities has also been observed in the SNPs rs211833 and rs1421312. Indeed, allele frequencies for SNPs can vary greatly across ethnic groups (Ioannidis et al. 2004), and this may impact the prevalence and incidence of disease across ethnic groups in case of associations (Lan et al. 2007; Myles et al. 2008). The high frequency of the risk alleles of the rs855791 and rs4820268 SNPs in Asian populations and the stronger negative associations with Hb and ferritin predispose this population to iron deficiency. Environmental factors such as a diets low in bioavailable iron and high inflammation burden further aggravate this predisposition. In this regard, it would have been interesting to include in this review studies conducted within the African population as well. However, at the time the search was completed, there were no studies conducted within the African population other than the one performed in the African-American population. However, this is an admixed population (Parra et al. 1998), and therefore, observations are not readily transferrable to the African population. Besides this, it may be that the phenotypic expression for the same alleles investigated are different within the genetically diverse African population (Tishkoff et al. 1996; Tishkoff and Williams 2002; Tishkoff and Verrelli 2003; Tishkoff and Kidd 2004; Frazer et al. 2007; Garrigan et al. 2007; Jakobsson et al. 2008; Li et al. 2008; Tishkoff et al. 2009).
Several limitations of our study need mentioning. We focussed on SNP–iron status measure associations that were investigated in at least two studies and in more than one ethnicity. Consequently, significant SNP–iron status measure associations that have only been researched in one ethnicity were not described. Additionally, since we only included SNPs described in greater than two cohorts and more than one ethnicity, we were only able to include two genetic variants from the selected SNPs in the meta-analyses. For this reason, there may be ethnic differences in other genetic variations that have not been included in this review.
Secondly, high heterogeneity was observed among the Caucasian studies. We attribute this to the variation in population characteristics and study designs. Most studies that were conducted within the Caucasian populations have previously been designed to study other outcomes, whereas the studies within the Asian population were specifically designed to study genetic influences of iron metabolism and are fewer in number.
Thirdly, in conducting association analyses, only one study (Gan et al. 2012) included dietary iron as a predictor of iron status within the model. This investigation on the interactions between SNPs and the environment is crucial in order to quantify the contribution of genetic alterations to the development of iron deficiency. Further to this, only two studies considered inflammation as a potential confounder in association analyses. These studies either excluded subjects with CRP levels above a pre-defined cut-off (Traglia et al. 2011; An et al. 2012; Gan et al. 2012) or corrected for inflammation (McLaren et al. 2012). Correction for inflammation or exclusion of values of inflamed subjects would ensure that what is observed is a true association. Ferritin and Hb are influenced by inflammation and sTfR concentrations might also be influenced negatively by inflammatory cytokines (Seiser et al. 1993).
In conclusion, we observe that the risk alleles A and G in rs855791 and rs4820268 are associated with a reduction in Hb and serum ferritin concentrations and an increase in transferrin and serum transferrin receptor concentrations in most populations investigated. Our study highlights key information gaps in several areas regarding the associations of SNPs in TMPRSS6 and iron status. First of all, more information on the (factors involved in) regulation of TMPRSS6 is required as well as how exactly TMPRSS6 exerts its effects on body iron. Moreover, our study also highlights the need to conduct further studies in ethnicities or populations where iron deficiency is of public health significance. The question is whether populations with high prevalence of risk alleles may require higher recommendations for iron intake in order to maintain normal erythropoiesis. This is especially relevant in combination with unfavourable environmental conditions such as diets with low bioavailable iron and high inflammation burden. Knowledge of genetic factors influencing iron status could improve care and advice given to populations at risk for iron deficiency.
Acknowledgments
We thank Toshiko Tanaka, National Institute on Aging, National Institutes of Health, USA, for in-depth advice. The authors’ responsibilities were as follows—WNGW: literature search, design of study, data extraction, statistical analyses, interpretation of data, and writing of the manuscript; AMB and EJF: design of study, data extraction, statistical analyses, interpretation of data; MBZ, GWT and DS: interpretation of data and provision of advice; and all authors: contributed to subsequent drafts of the manuscript and approved the submitted version of the manuscript.
Conflict of interest
Wanjiku N Gichohi-Wainaina, G Wayne Towers, Dorine W Swinkels, Michael B Zimmermann, Edith J Feskens and Alida Melse-Boonstra declare that they have no conflict of interest.
Footnotes
For Figs. 1, 2, 3, 4, 5, 6, 7 and 8, black squares are point estimates for each study, horizontal lines are 95 % CIs, black diamonds are summary estimates with the lateral tips of the open diamond indicating the standard errors for the summary estimates. Subtitle Mixed refers to a cohort consisting of more than one ethnicity. Effect allele is A for all associations involving rs855791.
References
- An P, Wu Q, Wang H, Guan Y, Mu M, Liao Y, Zhou D, Song P, Wang C, Meng L, Man Q, Li L, Zhang J, Wang F. TMPRSS6, but not TF, TFR2 or BMP2 variants are associated with increased risk of iron-deficiency anemia. Hum Mol Genet. 2012;21:2124–2131. doi: 10.1093/hmg/dds028. [DOI] [PubMed] [Google Scholar]
- Barton JC, Edwards CQ, Bertoli LF, Shroyer TW, Hudson SL. Iron overload in African Americans. Am J Med. 1995;99:616–623. doi: 10.1016/S0002-9343(99)80248-4. [DOI] [PubMed] [Google Scholar]
- Benyamin B, Ferreira MA, Willemsen G, Gordon S, Middelberg RP, McEvoy BP, Hottenga JJ, Henders AK, Campbell MJ, Wallace L, Frazer IH, Heath AC, de Geus EJ, Nyholt DR, Visscher PM, Penninx BW, Boomsma DI, Martin NG, Montgomery GW, Whitfield JB. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat Genet. 2009;41:1173–1175. doi: 10.1038/ng.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E, Barton JC, Felitti VJ, Gelbart T, West C, Lee PL, Waalen J, Vulpe C. Ferroportin 1 (SCL40A1) variant associated with iron overload in African-Americans. Blood Cells Mol Dis. 2003;31:305–309. doi: 10.1016/S1079-9796(03)00165-7. [DOI] [PubMed] [Google Scholar]
- Blanco-Rojo R, Baeza-Richer C, Lopez-Parra AM, Perez-Granados AM, Brichs A, Bertoncini S, Buil A, Arroyo-Pardo E, Soria JM, Vaquero MP. Four variants in transferrin and HFE genes as potential markers of iron deficiency anaemia risk: an association study in menstruating women. Nutr Metab (Lond) 2011;8:69. doi: 10.1186/1743-7075-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Zhang W, Li Y, Sehmi J, Wass MN, Zabaneh D, Hoggart C, Bayele H, McCarthy MI, Peltonen L, Freimer NB, Srai SK, Maxwell PH, Sternberg MJ, Ruokonen A, Abecasis G, Jarvelin MR, Scott J, Elliott P, Kooner JS. Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nat Genet. 2009;41:1170–1172. doi: 10.1038/ng.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco L, Sanchez M, Silvestri L, Kannengiesser C, Muckenthaler MU, Iolascon A, Gouya L, Camaschella C, Beaumont C. Iron refractory iron deficiency anemia. Haematologica. 2013;98:845–853. doi: 10.3324/haematol.2012.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM, Beutler E, Beutler B. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320:1088–1092. doi: 10.1126/science.1157121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finberg KE. Iron-refractory iron deficiency anemia. Semin Hematol. 2009;46:378–386. doi: 10.1053/j.seminhematol.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos K, Andrews NC, Fleming MD. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA) Nat Genet. 2008;40:569–571. doi: 10.1038/ng.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueras AR, de Lara FM, Pendas AM, Garabaya C, Rodriguez F, Astudillo A, Bernal T, Cabanillas R, Lopez-Otin C, Velasco G. Membrane-bound serine protease matriptase-2 (Tmprss6) is an essential regulator of iron homeostasis. Blood. 2008;112:2539–2545. doi: 10.1182/blood-2008-04-149773. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Sun W, Wang H, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galesloot TE, Geurts-Moespot AJ, den Heijer M, Sweep FC, Fleming RE, Kiemeney LA, Vermeulen SH, Swinkels DW. Associations of common variants in HFE and TMPRSS6 with iron parameters are independent of serum hepcidin in a general population: a replication study. J Med Genet. 2013;50:593–598. doi: 10.1136/jmedgenet-2013-101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Guan Y, Wu Q, An P, Zhu J, Lu L, Jing L, Yu Y, Ruan S, Xie D, Makrides M, Gibson RA, Anderson GJ, Li H, Lin X, Wang F. Association of TMPRSS6 polymorphisms with ferritin, hemoglobin, and type 2 diabetes risk in a Chinese Han population. Am J Clin Nutr. 2012;95:626–632. doi: 10.3945/ajcn.111.025684. [DOI] [PubMed] [Google Scholar]
- Ganesh SK, Zakai NA, van Rooij FJ, Soranzo N, Smith AV, Nalls MA, Chen MH, Kottgen A, Glazer NL, Dehghan A, Kuhnel B, Aspelund T, Yang Q, Tanaka T, Jaffe A, Bis JC, Verwoert GC, Teumer A, Fox CS, Guralnik JM, Ehret GB, Rice K, Felix JF, Rendon A, Eiriksdottir G, Levy D, Patel KV, Boerwinkle E, Rotter JI, Hofman A, Sambrook JG, Hernandez DG, Zheng G, Bandinelli S, Singleton AB, Coresh J, Lumley T, Uitterlinden AG, Vangils JM, Launer LJ, Cupples LA, Oostra BA, Zwaginga JJ, Ouwehand WH, Thein SL, Meisinger C, Deloukas P, Nauck M, Spector TD, Gieger C, Gudnason V, van Duijn CM, Psaty BM, Ferrucci L, Chakravarti A, Greinacher A, O’Donnell CJ, Witteman JC, Furth S, Cushman M, Harris TB, Lin JP. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB, Pilkington MM, Wilder JA, Cox MP, Soodyall H, Strassmann B, Destro-Bisol G, de Knijff P, Novelletto A, Friedlaender J, Hammer MF. Inferring human population sizes, divergence times and rates of gene flow from mitochondrial, X and Y chromosome resequencing data. Genetics. 2007;177:2195–2207. doi: 10.1534/genetics.107.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordeuk V, Mukiibi J, Hasstedt SJ, Samowitz W, Edwards CQ, West G, Ndambire S, Emmanual J, Nkanza N, Chapanduka Z, et al. Iron overload in Africa. Interaction between a gene and dietary iron content. N Engl J Med. 1992;326:95–100. doi: 10.1056/NEJM199201093260204. [DOI] [PubMed] [Google Scholar]
- Guillem F, Lawson S, Kannengiesser C, Westerman M, Beaumont C, Grandchamp B. Two nonsense mutations in the TMPRSS6 gene in a patient with microcytic anemia and iron deficiency. Blood. 2008;112:2089–2091. doi: 10.1182/blood-2008-05-154740. [DOI] [PubMed] [Google Scholar]
- He M, Workalemahu T, Manson JE, Hu FB, Qi L. Genetic determinants for body iron store and type 2 diabetes risk in US men and women. PLoS ONE. 2012;7:e40919. doi: 10.1371/journal.pone.0040919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- Jackson RT, Sauberlich HE, Skala JH, Kretsch MJ, Nelson RA. Comparison of hemoglobin values in black and white male U.S. military personnel. J Nutr. 1983;113:165–171. doi: 10.1093/jn/113.1.165. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, Bras JM, Schymick JC, Hernandez DG, Traynor BJ, Simon-Sanchez J, Matarin M, Britton A, van de Leemput J, Rafferty I, Bucan M, Cann HM, Hardy JA, Rosenberg NA, Singleton AB. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Kloss-Brandstatter A, Erhart G, Lamina C, Meister B, Haun M, Coassin S, Seifert M, Klein-Franke A, Paulweber B, Kedenko L, Kollerits B, Swinkels DW, Vermeulen SH, Galesloot TE, Kronenberg F, Weiss G. Candidate gene sequencing of SLC11A2 and TMPRSS6 in a family with severe anaemia: common SNPs, rare haplotypes, no causative mutation. PLoS ONE. 2012;7:e35015. doi: 10.1371/journal.pone.0035015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, Shen M, Garcia-Rossi D, Chanock S, Zheng T, Berndt SI, Puri V, Li G, He X, Welch R, Zahm SH, Zhang L, Zhang Y, Smith M, Wang SS, Chiu BC, Linet M, Hayes R, Rothman N, Yeager M. Genotype frequency and F ST analysis of polymorphisms in immunoregulatory genes in Chinese and Caucasian populations. Immunogenetics. 2007;59:839–852. doi: 10.1007/s00251-007-0253-3. [DOI] [PubMed] [Google Scholar]
- Leboeuf RC, Tolson D, Heinecke JW. Dissociation between tissue iron concentrations and transferrin saturation among inbred mouse strains. J Lab Clin Med. 1995;126:128–136. [PubMed] [Google Scholar]
- Lee P. Role of matriptase-2 (TMPRSS6) in iron metabolism. Acta Haematol. 2009;122:87–96. doi: 10.1159/000243792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Looker AC, Dallman PR, Carroll MD, Gunter EW, Johnson CL. Prevalence of iron deficiency in the United States. JAMA. 1997;277:973–976. doi: 10.1001/jama.1997.03540360041028. [DOI] [PubMed] [Google Scholar]
- McLaren CE, Li KT, Gordeuk VR, Hasselblad V, McLaren GD. Relationship between transferrin saturation and iron stores in the African American and US Caucasian populations: analysis of data from the third National Health and Nutrition Examination Survey. Blood. 2001;98:2345–2351. doi: 10.1182/blood.V98.8.2345. [DOI] [PubMed] [Google Scholar]
- McLaren CE, McLachlan S, Garner CP, Vulpe CD, Gordeuk VR, Eckfeldt JH, Adams PC, Acton RT, Murray JA, Leiendecker-Foster C, Snively BM, Barcellos LF, Cook JD, McLaren GD. Associations between single nucleotide polymorphisms in iron-related genes and iron status in multiethnic populations. PLoS ONE. 2012;7:e38339. doi: 10.1371/journal.pone.0038339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis MA, Cau M, Congiu R, Sole G, Barella S, Cao A, Westerman M, Cazzola M, Galanello R. A mutation in the TMPRSS6 gene, encoding a transmembrane serine protease that suppresses hepcidin production, in familial iron deficiency anemia refractory to oral iron. Haematologica. 2008;93:1473–1479. doi: 10.3324/haematol.13342. [DOI] [PubMed] [Google Scholar]
- Meynard D, Babitt JL, Lin HY. The liver: conductor of systemic iron balance. Blood. 2014;123:168–176. doi: 10.1182/blood-2013-06-427757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse AC, Beard JL, Jones BC. A genetic developmental model of iron deficiency: biological aspects. Proc Soc Exp Biol Med. 1999;220:147–152. doi: 10.3181/00379727-220-44357. [DOI] [PubMed] [Google Scholar]
- Moyo VM, Mandishona E, Hasstedt SJ, Gangaidzo IT, Gomo ZA, Khumalo H, Saungweme T, Kiire CF, Paterson AC, Bloom P, MacPhail AP, Rouault T, Gordeuk VR. Evidence of genetic transmission in African iron overload. Blood. 1998;91:1076–1082. [PubMed] [Google Scholar]
- Myles S, Davison D, Barrett J, Stoneking M, Timpson N. Worldwide population differentiation at disease-associated SNPs. BMC Med Genomics. 2008;1:22. doi: 10.1186/1755-8794-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nai A, Pagani A, Silvestri L, Campostrini N, Corbella M, Girelli D, Traglia M, Toniolo D, Camaschella C. TMPRSS6 rs855791 modulates hepcidin transcription in vitro and serum hepcidin levels in normal individuals. Blood. 2011;118:4459–4462. doi: 10.1182/blood-2011-06-364034. [DOI] [PubMed] [Google Scholar]
- Oexle K, Ried JS, Hicks AA, Tanaka T, Hayward C, Bruegel M, Gogele M, Lichtner P, Muller-Myhsok B, Doring A, Illig T, Schwienbacher C, Minelli C, Pichler I, Fiedler GM, Thiery J, Rudan I, Wright AF, Campbell H, Ferrucci L, Bandinelli S, Pramstaller PP, Wichmann HE, Gieger C, Winkelmann J, Meitinger T. Novel association to the proprotein convertase PCSK7 gene locus revealed by analysing soluble transferrin receptor (sTfR) levels. Hum Mol Genet. 2011;20:1042–1047. doi: 10.1093/hmg/ddq538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GS, Byers T, Yip R, Margen S. Iron nutrition does not account for the hemoglobin differences between blacks and whites. J Nutr. 1992;122:1417–1424. doi: 10.1093/jn/122.7.1417. [DOI] [PubMed] [Google Scholar]
- Pichler I, Minelli C, Sanna S, Tanaka T, Schwienbacher C, Naitza S, Porcu E, Pattaro C, Busonero F, Zanon A, Maschio A, Melville SA, Grazia Piras M, Longo DL, Guralnik J, Hernandez D, Bandinelli S, Aigner E, Murphy AT, Wroblewski V, Marroni F, Theurl I, Gnewuch C, Schadt E, Mitterer M, Schlessinger D, Ferrucci L, Witcher DR, Hicks AA, Weiss G, Uda M, Pramstaller PP. Identification of a common variant in the TFR2 gene implicated in the physiological regulation of serum iron levels. Hum Mol Genet. 2011;20:1232–1240. doi: 10.1093/hmg/ddq552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- Seiser C, Teixeira S, Kuhn LC. Interleukin-2-dependent transcriptional and post-transcriptional regulation of transferrin receptor mRNA. J Biol Chem. 1993;268:13074–13080. [PubMed] [Google Scholar]
- Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8:502–511. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen E, Grau K, Berg T, Simonsen AC, Magnussen K, Erikstrup C, Hansen MB, Ullum H. A genetic risk factor for low serum ferritin levels in Danish blood donors. Transfusion. 2012;52:2585–2589. doi: 10.1111/j.1537-2995.2012.03629.x. [DOI] [PubMed] [Google Scholar]
- Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Roy CN, Yao W, Matteini A, Semba RD, Arking D, Walston JD, Fried LP, Singleton A, Guralnik J, Abecasis GR, Bandinelli S, Longo DL, Ferrucci L. A genome-wide association analysis of serum iron concentrations. Blood. 2010;115:94–96. doi: 10.1182/blood-2009-07-232496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Kidd KK. Implications of biogeography of human populations for ‘race’ and medicine. Nat Genet. 2004;36:S21–S27. doi: 10.1038/ng1438. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 2002;3:611–621. doi: 10.1038/ni0702-611. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traglia M, Girelli D, Biino G, Campostrini N, Corbella M, Sala C, Masciullo C, Vigano F, Buetti I, Pistis G, Cocca M, Camaschella C, Toniolo D. Association of HFE and TMPRSS6 genetic variants with iron and erythrocyte parameters is only in part dependent on serum hepcidin concentrations. J Med Genet. 2011;48:629–634. doi: 10.1136/jmedgenet-2011-100061. [DOI] [PubMed] [Google Scholar]
- WHO (2008) Worldwide prevalence of anaemia 1993–2005
- Williams DM. Racial differences of hemoglobin concentration: measurements of iron, copper, and zinc. Am J Clin Nutr. 1981;34:1694–1700. doi: 10.1093/ajcn/34.9.1694. [DOI] [PubMed] [Google Scholar]
- Wurapa RK, Gordeuk VR, Brittenham GM, Khiyami A, Schechter GP, Edwards CQ. Primary iron overload in African Americans. Am J Med. 1996;101:9–18. doi: 10.1016/S0002-9343(96)00053-8. [DOI] [PubMed] [Google Scholar]