Abstract
The ability of microalbuminuria to predict early progressive renal function decline in type-1 diabetic patients has been questioned. To resolve this, we determined the plasma proteome differences between microalbuminuric patients with type-1 diabetes and stable renal function (controls) and patients at risk for early progressive renal function decline (cases) and asked whether these differences have value as surrogate biomarkers. Mass spectrometry was used to analyze small (less than 3 kDa) plasma peptides isolated from well-matched case and control plasma obtained at the beginning of an 8-12 year follow-up period. Spearman analysis of plasma peptide abundance and the rate of renal function decline during follow-up identified seven masses with a significant negative correlation with early progressive renal function decline. Tandem mass spectrometry identified three fragments of high molecular weight kininogen. Increased plasma high molecular weight kininogen in the cases was confirmed by immunoblot. One peptide, des-Arg9-BK(1-8), induced Erk1/2 phosphorylation when added apically to two proximal tubular cell lines grown on permeable inserts. Thus, we have identified plasma protein fragments, some of which have biological activity with moderate to strong correlation, with early progressive renal function decline in microalbuminuric patients with type-1 diabetes. Other peptides are candidates for validation as candidate biomarkers of diabetes-associated renal dysfunction.
Introduction
Microalbuminuria (MA) has been considered the primary diagnostic tool to identify type 1 diabetes mellitus (T1D) patients at risk for progressive renal dysfunction1,2. However, the correlation of MA with future renal dysfunction in diabetics has now been called into question. Several findings indicate that MA may not reliably herald the beginning of renal dysfunction. First, only approximately 20% of patients with MA will progress to proteinuria3; second, many patients with MA can revert to normoalbuminuria4-6; and third, many individuals with T1D have already experienced early progressive renal function decline (ERFD) before or coincidental with MA onset7,8. These findings have called into question the model of diabetic nephropathy in which MA conveyed a high risk of progressive renal dysfunction and support a new model in which only a subset of those with MA develop progressive ERFD. This change in our understanding of diabetic renal disease also is indicative of our incomplete understanding of the mechanisms of ERFD, a process that takes place while measured renal function is still in the normal or even elevated range. These findings emphasize the need for further studies to understand the pathophysiology of ERFD in patients with MA and to identify those T1D patients at risk for early renal damage.
We addressed the hypothesis that qualitative differences in plasma proteins might provide insight into ERFD pathophysiology and serve as candidate biomarkers of the risk of progressive ERFD and progressive renal function loss. To address this hypothesis we have analyzed plasma samples obtained during the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes using LC-MALDI-TOF MS to compare the low molecular weight protein (less than 3,000 Daltons) or peptidomic plasma fraction. We analyzed the plasma peptidome of patients matched for cystatin C estimated glomerular filtration rate (eGFR), MA, and medications (among other clinical parameters) comparing those who retained stable renal function to those that developed ERFD during subsequent 8-12 years of follow-up. We hypothesized that qualitative differences in the low molecular weight plasma proteome (the peptidome) might provide insight into the etiology of early progressive RFD and serve as putative biomarkers of future progression. We observed a striking correlation between the rate of future renal function decline and components of the kallikrein-kininogen system. These protein fragments should now be considered as candidates for confirmation in larger studies as candidate biomarkers of ERFD and predictors of renal dysfunction in T1D.
Results
Characteristics of the Study Population
The study population was comprised of the patients whose onset of MA was documented in the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes. Additional eligibility criteria included follow-up examinations spanning at least 8-12 years after MA onset for estimating the rate of GFR decline and availability of a 6 ml aliquot of stored urine for peptide analysis9. Thirty-three patients (16 cases and 17 controls) selected from a previous urinary biomarker study were who met all eligibility criteria (cases with renal function decline defined as a decline of 3.3% or more per year (range: −3.3 to −16.1% per year), and controls with lesser rates of renal function decline (range: +1.9 to −3.2% per year) had contemporaneous plasma samples avaiable for the current study.
Correlation of Discriminating Peptides with the Rate of Future Renal Function Decline
To detect peptides whose abundance strictly correlated with the linear estimate of renal function and not simply a discrete clinical group, a Spearman rank order correlation analysis was performed comparing peptide abundance with the rate of renal function decline. A total of seven peptides were identified with Spearman correlation value ranked between an absolute value of −0.45 and −0.51 (p<0.001) (Table 1). As such if validated, they may have a value to identify patients with an increased risk of the development of ERFD.
Table 1. Characterization of plasma peptides whose abundance strongly correlates with the rate of future renal function decline.
The integrated area under the curve for plasma peptide data was extracted from aligned LC-MALDI-TOF MS data sets and analyzed by Spearman correlation analysis to the estimated rate of renal function decline (estimated using serum concentration of cystatin C and changes in renal function were estimated by slopes). Peptides with strong correlation (between −0.45 and −1.0) to progressive renal function decline are listed. The difference in the observed abundance of plasma peptides selected by the Spearman correlation was estimated using unpaired t-test with Welch’s correction for unequal variation in case and control peptide abundance data sets.
| Peptide Mass (m/z) |
Spearman’s Rank Correlation Coefficient (rs) |
P-value* For comparison of cases and controls |
Mascot MOWSE Score** |
Peptide or protein fragment |
|---|---|---|---|---|
| Identified | ||||
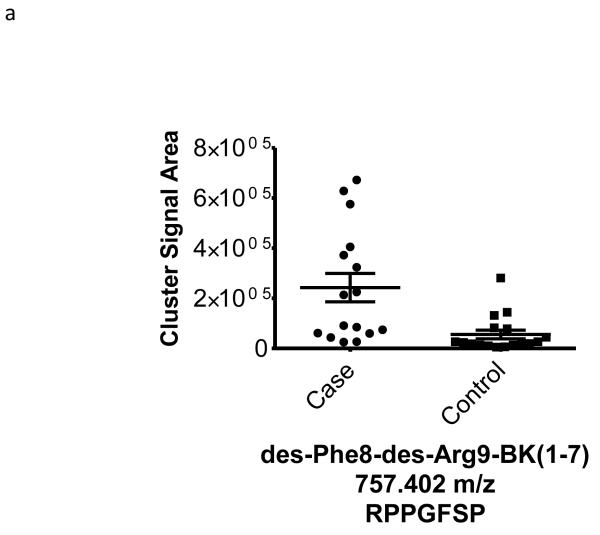
| 757.402 | −0.51 | 0.006 | 36 | des-Phe8-des-Arg9-Bradykinin (1-7) (K)-RPPGFSP-(F) |
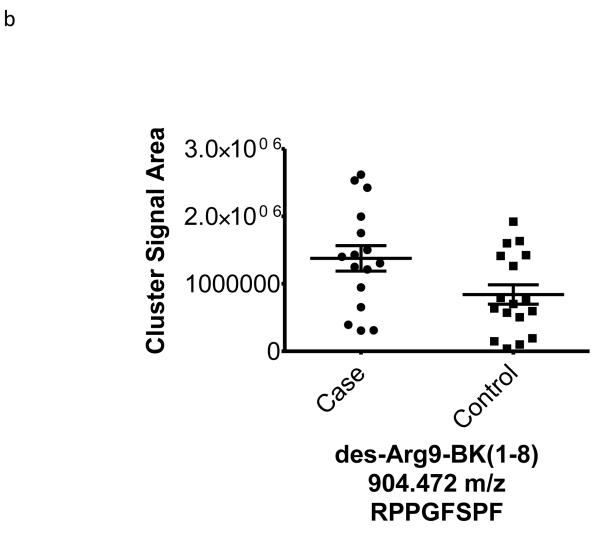
| 904.472 | −0.46 | 0.03 | 44 | des-Arg9-Bradykinin (1-8) (K)-RPPGFSPF-(R) |
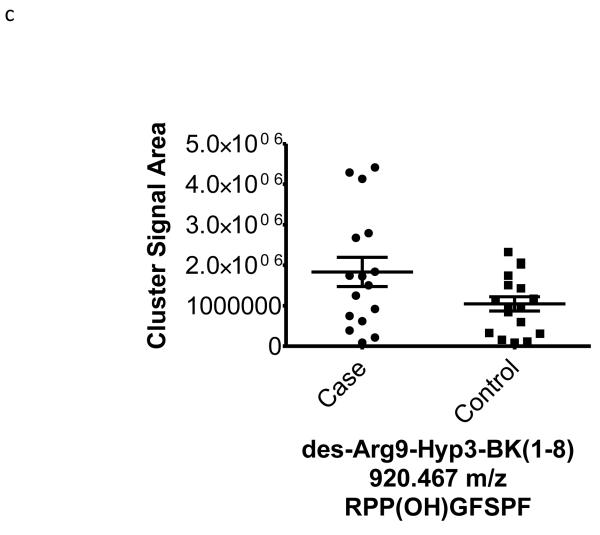
| 920.467 | −0.45 | 0.06 | 44 | Hyp3-des-Arg9-Bradykinin (1-8) (K)-RP-Hyp-GFSPF-(R) |
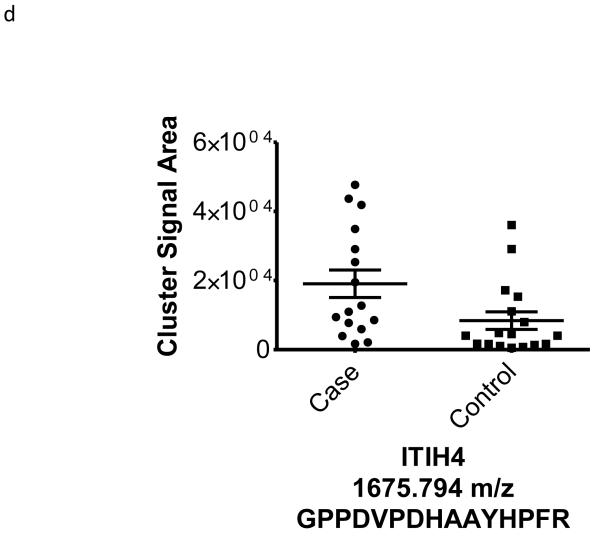
| 1675.794 | −0.50 | 0.03 | 96 | Kallikrein sensitive glycoprotein (ITIH4) (P)-GPPDVPDHAAYHPFR-(R) |
| Unidentified | ||||
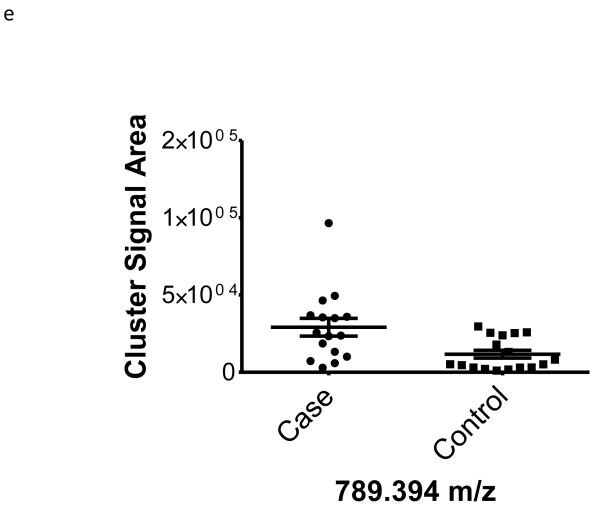
| 789.394 | −0.48 | 0.01 | na | |
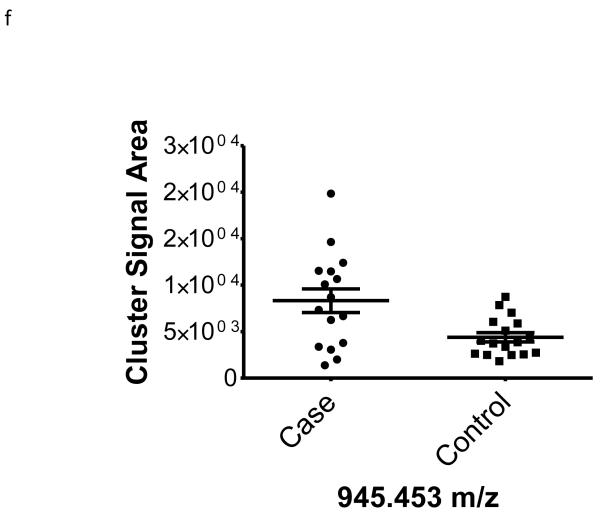
| 945.453 | −0.46 | 0.01 | na | |
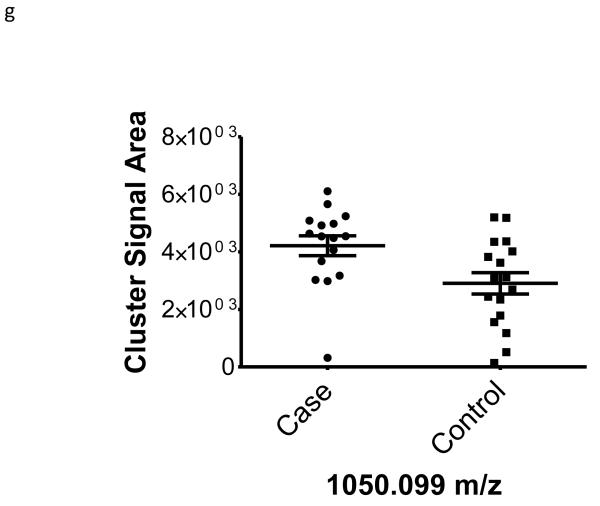
| 1050.099 | −0.46 | 0.02 | na | |
Analysis by T-test (with Welch’s correction for unequal variances) of differences in abundance of peptides in case (n= 16) and control (n= 17) samples.
Mascot MOWSE scores ≥ 20 are considered significant (p-value < 0.05) and provide for identity.
Identification of Peptide Amino Acid Sequence by Mass Spectrometry
To better understand the possible role of the selected peptide masses in the etiology of ERFD we sought to identify partial amino acid sequence tagging information using tandem mass spectrometry. Two of the seven masses were not fragmented due to low intensity and proximity of the monoisotopic peak to larger more intense ions. Using the parent peptide mass, the peptide fragmentation data (see supplemental data for peak list information and fragmentation spectra) gained from the tandem MS experiments, and the data analysis software, Matrix Science Mascot, amino acid sequences were assigned to a total of four of the five peptides submitted for MS/MS analysis (Table 1). The results of these analyses identified fragments of two plasma proteins including kininogen-1 (three fragments) and a fragment of plasma kallikrein-sensitive glycoprotein (inter-alpha-trypsin inhibitor heavy chain H4, ITIH4). The comparison of the fragmentation spectra for synthetic peptides with experimental data supported these peptide assignments (see supplemental data). The kinin peptides were modified by C-terminal proteolysis and/or prolyl-specific hydroxylation. While the strength of the assignments for these peptides as candidate biomarkers for ERFD in T1D is based on the Spearman Correlation analysis, we provide a figure (Figure1) to illustrate the comparison of the means for these plasma peptide abundances. These data suggest that in these patients the abundances of specific plasma peptides can begin to predict significant declines in GFR. In general, a comparison between cases and controls for these peptides identified a 30-50% increased mean abundance in patients who were at risk of ERFD (Figure1, Table 1).
Figure 1. Graphical illustration for the abundances of plasma peptides that correlate with future severe progressive renal function decline demonstrating increased abundance in decliner (case) over non-decliner (control) plasma samples.
Aligned MS data sets were constructed from peptide mass and retention time, and estimated peptide abundance was calculated from the MS ion cluster signal area or inferred from baseline when peptides were not detected.
Differential Abundance of High Molecular Weight Kininogen in Plasma
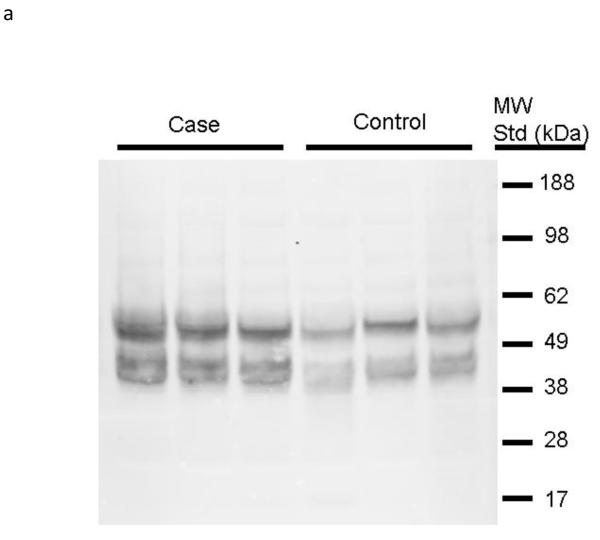
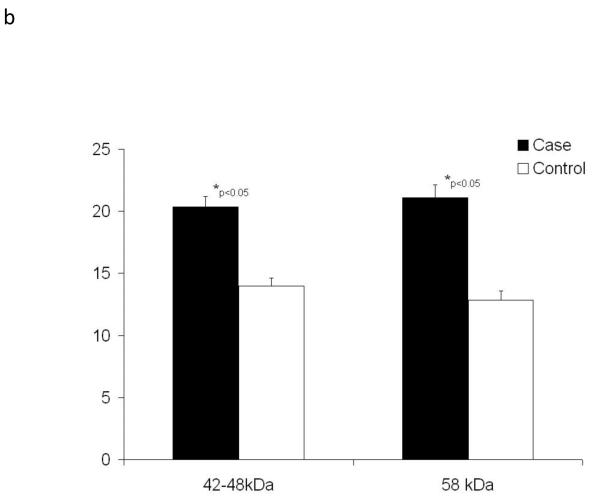
We then examined whether the increased abundance of bradykinin (BK) forms in case plasma might result from increased amounts of its precursor kininogen. Using an antibody developed against recombinant high molecular weight kininogen (rhKininogen: aa 19-644), we analyzed by immunoblot the expression of high molecular weight kininogen within the plasma sample set used for the peptidomic analysis. The products of high molecular weight kininogen precursor is proteolysis by plasma kallikrein are BK and a high molecular weight kininogen α1ß1 heterodimer held together by a single disulfide chain. Because the α1 and ß1 monomers have masses similar to albumin, we first depleted the plasma samples of albumin prior to immunoblot analysis. Additionally, for this comparison samples were selected from the patient populations with the most severe loss of renal function and most stable renal function. These samples were then resolved by 10% SDS-PAGE and putative kininogen heavy (α) and light (ß) chains were identified by immunoblot. Shown in Figure 2 are immunoblot data for the comparison of three cases and three control samples. Immunoblot densitometry measurements of a total of six patient samples were used to estimate differential abundance of kininogen immune-positive bands. Two sets of immune-positive bands were detectable in plasma and by t-test were significantly (p<0.05) increased approximately 30-40% in case plasma.
Figure 2. High molecular weight kininogen is increased in plasma of patients with future renal dysfunction.
The plasma samples of cases (n=6) and controls (n=6) were depleted of albumin, divided between two 10% gels, separated and transferred onto nitrocellulose membranes and probed with mono-specific antibodies to high molecular weight kininogen (one of two blots shown, Panel 2a). Pooled data for all samples were analyzed by densitometry and are shown in Panel 2b). Statistical differences were estimated using a Student’s t-test.
Biological Activity of the des-Arg9-BK(1-8) Fragment in a Renal Tubular Cell Line
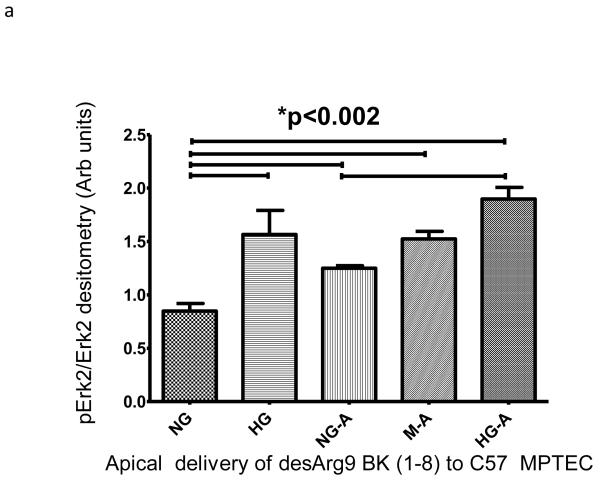
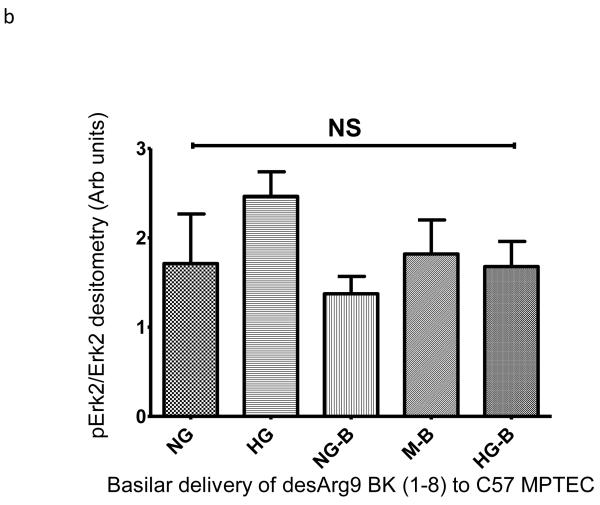
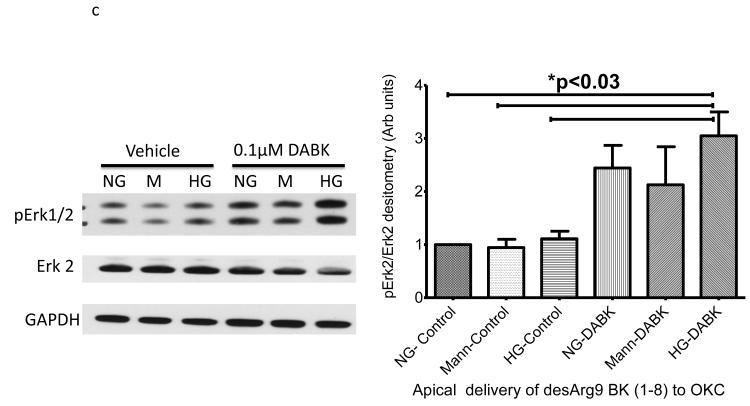
We wished to examine if one of the BK peptides prevalent in patients with ERFD exhibited biological activity in renal cells. We chose the des-Arg9-BK(1-8) peptide because it is reported to exhibit activity only when it binds to the bradykinin-1 receptor (B1R); whose expression is increased in diabetic mice 10. We examined the endpoint of ERK phosphorylation as it has been shown to be a downstream signaling molecule in the B1R pathway11. Initially, we incubated immortalized mouse proximal tubule cells grown on plastic with 5mM glucose, or 30 mM glucose, or 5 mM glucose + mannitol 25mM (as an osmotic control) in the presence and absence of 100 nM des-Arg9 BK (1-8) and observed des-Arg9 BK (1-8) effects in normal glucose (5 mM) or in the osmotic control to have comparable ERK phosphorylation to that induced by high glucose alone (30 mM) (supplemental figure S3a). The most substantial increase in ERK phosphorylation was seen when cells were exposed to des-Arg9 BK (1-8) in the presence of high glucose (30 mM). As epithelial cells grown on plastic may not recapitulate a true polarized phenotype, we repeated these experiments with the mouse proximal tubule cell line grown on permeable inserts and evaluated the effects on Erk1/2 phosphorylation following either apical or basilar delivery of the peptide. In comparison with vehicle, apical but not basilar delivery of the des-Arg9 BK (1-8) resulted in a statistically significant difference by ANOVA analysis in Erk1/2 phosphorylation with comparing 5mM glucose control samples with 30mM glucose treated cells or des-Arg9 BK (1-8) treated cells (Figure 3a-b). Finally, to determine if this observation was isolated to the mouse renal proximal tubular cell line these apical delivery experiments were repeated using an opossum kidney cell (OKC) proximal tubular cell line. While the OKC cell line did not respond robustly for induction of Erk1/2 phosphorylation by media alone (30mM glucose), apical treatment of the OKC in 5mM glucose, 30mM glucose, and 5mM glucose plus 25mM mannitol with des-Arg9 BK (1-8) induced a statistically significant increase in Erk1/2 phosphorylation compared to normal control (Figure 3c).
Figure 3. Induction of extracellular-signal-regulated kinases (ERK1/2) phosphorylation by glucose and des-Arg9-BK (1-8) (DABK) in proximal tubule cell lines.
Proximal tubule cell lines were used to determine the effects of 10 minute des-Arg9- BK treatment on Erk1/2 phosphorylation. Cells were seeded onto permeable inserts, grown to 70% confluence, serum starved and then cultured in medium containing 0.5% FBS plus normal glucose (5mM glucose, NG), osmotic control (5mM glucose + 25mM mannitol, M) or high glucose (30mM glucose, HG). After 24h the cells were treated for 10 min with agonist vehicle (PBS) or agonist (DABK). A C57 mouse proximal tubular cell line (compliments of Jeffery Schelling, MD, Case Western Reserve University) was used to ascertain peptide effects as a function of apical (A) or basilar (B) treatment. An opossum kidney (OK) cell line was used to confirm apical delivery (C) findings of C57 cell line experiment. Cell lysates from treated cells were immunoblotted for phospho-Erk1/2 (p-Erk1/2) then stripped and re-probed for Erk-2. The percentage of Erk-2 phosphorylation induced by culture or treatment was estimated from band densitometry ratios for (p-Erk-2 band of p-Erk 1/2 IB densitometry) to (Erk-2). Statistical differences indicated in plots are estimated by ANOVA analysis (Kruskal-Wallis) with Dunn’s post hoc comparison of means. Data are plotted as mean and standard error of the mean.
DISCUSSION
The goal of this study was to identify plasma proteome components associated with the risk of progressive ERFD, a phase of diabetic nephropathy associated with the initiation of progressive renal dysfunction while GFR is still in the normal range7. To achieve this goal we studied plasma samples obtained soon after enrollment in the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes. The samples studied were well-matched for microalbuminuria, estimated GFR and other clinical factors. These patients had been followed for 8 to 12 years, thereby allowing us to classify patients into groups with either long-term stable renal function or progressive renal function deterioration. As such, the samples could be used to either correlate the presence of a peptide detected in the enrollment plasma sample with the rate of future renal function decline or to detect peptides that identify patients that retain stable renal function as compared to those diabetics with ERFDs demonstrated here. MS analysis was used to select and characterize changes in plasma abundance of high molecular weight kininogen derived peptides and kinin cleavage products. These peptides included two forms of des-Arg9-BK (1-8), des-Phe8-des-Arg9-BK (1-7) form, as well as a fragment of the plasma kallikrein-sensitive glycoprotein (inter-alpha trypsin inhibitor H4, ITIH4). The novel findings in these studies are that quantifiable changes in plasma low and high molecular weight protein expression occur very early in DN12. We examined at least one mechanism by which case samples could have increased expression of BK variants by measuring high molecular weight kininogen protein from which BK is derived. We observed increased expression of high molecular weight kininogen heavy chain and light chains in the plasma of cases compared controls, a finding that may explain the increased amount of BK available for post-translational modification.
The plasma kallikrein-kininogen system (KKS) plays a substantial role in regulation of coagulation and inflammation. Kallikrein is a highly abundant plasma protein that is activated by contact with negatively charged surfaces. When active, kallikrein cleaves one of its substrates, high molecular weight kininogen, resulting in release of BK. BK exerts its actions promoting vasodilation, vascular permeability and edema by stimulating cognate cell surface receptors, B1R and B2R. B2R are activated by binding of BK and are constitutively expressed in vascular tissues, while B1R are activated by binding of desArg9 BK(1-8) and their expression is induced by cellular ischemia, inflammation and in some instances diabetes10,12. In humans and rodents, BK is degraded C-terminally by three proteases including angiotensin-converting enzyme-1 (ACE1), aminopeptidase P (APP) and carboxypeptidase N (CPN)13,14. These enzymes convert BK to desArg9 BK (1-8), desArg1-Pro2 BK (3-9) and desPhe8-Arg9-BK (1-7) respectively. Recently a second angiotensin-converting enzyme (ACE2) has been identified. ACE2 is the principle route of desArg9-BK (1-8) degradation and as has been recently reviewed is insensitive to standard ACE inhibitors15.
The association of the KKS in diabetic nephropathy has been examined extensively. In human studies, Jaffa et al studied plasma samples from patients enrolled in the Diabetes Control and Complications Trial and observed that plasma pre-kallikrein levels correlated with hypertension, albumin excretion rates, and the development of MA16. The same group also demonstrated that diabetic patients with glomerular hyper-filtration had increased urinary excretion of active kallikrein. Animal studies of the role of the KKS product BK in diabetic nephropathy have produced contradictory findings. In Akita diabetic mice, B2R deletion exacerbated albuminuria, interstitial fibrosis and glomerulosclerosis17,18. The same investigators also demonstrated that B2R antagonists in several murine diabetic nephropathy models attenuated the beneficial effect of ACE inhibitors. In contrast, Tan et al observed an attenuation of diabetic nephropathy in B2R knockout mice made diabetic with streptozotocin19. These conflicting data may result from disparities in the animal strains or diabetic models used. Our data do not bring clarity to the conflicting results in these animal studies. However, our data do underscore the finding that the KKS system is strongly regulated in type -1 diabetics with ERFD.
One of the goals of these studies was to discover plasma peptides that correlate with ERFD and that could then serve as candidate biomarkers of progressive renal dysfunction in MAT1D. We observed seven peptides that had a moderate to strong correlation with the rate of future renal function decline having Spearman correlation coefficients from −0.45 to −0.51. In contrast to our previous peptide biomarker studies, these seven peptides had a high prevalence in both case and control samples and thus are good biomarker candidates9. The confirmation of these prevalent peptides as biomarkers of ERFD and future renal function in T1D will require validation studies using large patient populations and their predictive value may also be strengthened by inclusion in multiple marker panels20.
In many instances, disease biomarkers are simply indicators of disease activity and are not reflections of disease mechanisms. However, as BK agonists have been shown to alter renal cell behavior21,22 we examined if one of the modified BK forms of higher abundance in patients with ERFD had biological activity in renal proximal tubular cells. We chose the des-Arg9-BK(1-8) for several reasons including a Spearman rank correlation analysis that showed correlation values of −0.37 in angiotensin converting enzyme inhibitor (ACEi)-naïve patients and −0.74 in ACEi-treated patients (data not shown); suggesting patients on ACEi might experience greater exposure to the effects of circulating des-Arg9-BK(1-8). Des-Arg9-BK(1-8) is the ligand for the inducible B1R whose expression can be induced in diabetic proximal tubule cells. Our data demonstrated that des-Arg9-BK(1-8) stimulated ERK phosphorylation in a mouse proximal tubule cell line grown on plastic as well as when delivered apically to these cells and to second OKC proximal tubular cell line. The des-Arg9 BK (1-8) augmented the ERK phosphorylation induced by high glucose concentrations for the mouse proximal tubular cell line was not observed with cells grown on permeable inserts and may be the results of loss of polarization or simultaneous stimulation of both apical and basolateral populations of B1R receptors. These data are consistent with the literature that B1R agonists activate ERK and indicate that at least one of the prevalent BK forms in cases can exhibit biological activity in the kidney. The consequences of such activity are currently under investigation.
In summary, we have identified peptides in the low molecular fraction of plasma that correlate with ERFD and progressive nephropathy in type-1 diabetes. These peptides reflect changes in the abundance and post-translationally modified forms of bioactive peptides. These differences in concentration and modification may define a new mechanism by which diabetic microvascular is initiated or progresses. The usefulness of these discriminating peptides as biomarkers of diabetes associated renal function decline should be determined in additional rigorous studies in a larger patient population.
Materials and Methods
Study Subjects and Plasma Sample Collection
Patients who were enrolled in the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes were eligible for this pilot study7. The study protocol and patient consent procedures were approved by the Committee on Human Studies of the Joslin Diabetes Center. Urinary MA (defined as an excretion rate of 30 to 299μg/min) was established by repeat urine measurements during the first two years of patient follow-up. Patients observed to develop new-onset MA during the first four years following initial evaluation were invited to provide blood samples for further study. Serial measurements of cystatin C were examined to determine trends in renal function changes. Cystatin C is a low molecular weight, cationic, non-glycosylated, protease inhibitor that is constitutively expressed by nucleated cells. Cystatin C has been demonstrated to perform equal to or more superior to serum creatinine as an estimation of glomerular filtration rates in adults with GFR’s greater than 60mL/min8.
Peptidomic Analysis
The low and high molecular weight plasma proteomes were separated and isolated peptides were analyzed as described previously9. Briefly proteins were precipitated using organic solvent, the soluble peptide fractions following lyophilization and quantification using a modified Lowery/BCA method, were separated with reversed phase (RP) capillary-high performance liquid chromatography, robotically spotted onto archival MALDI-TOF MS plates (Opti-TOF plates) and MALDI-TOF MS data acquired using an Applied Biosystems (Foster City, CA) AB4700 Proteomics Analyzer operating in reflectron mode. LC MALDI-TOF MS ion chromatograms were constructed using Data Explorer software (Applied Biosystems) and then exported as peak list text files and used for determination of differential peptide abundance. Additionally, individual LC-MS spectra were concatenated to produce plasmapeptide LC-MALDI-TOF MS ion chromatogram per each sample analyzed.
Statistical Analysis of Plasma Peptide Abundance
LC-MALDI-TOF MS ion chromatograms were extracted in the form of integrated signal area for the peptide isotopic series (cluster area under the curve, AUC) from the ABI4700 .t2d files using Data Explorer (Applied Biosystems) software and exported to MarkerView (Applied Biosystems) software to chromatographically align and array the tabulated peptide peak list data using a signal-to-noise filter of 10. Three LC fractions collected prior to initiating the reversed phase elution gradient and following re-equilibration of the column from 100% solvent B to 100 solvent A were used to establish a null peptide mass list. These masses were subtracted from the aggregate data set.
The data were analyzed using a Spearman rank order correlation to correlate peptide abundance with renal function decline (estimated from -slope of linear regressions for annual measurements of serum cystatin C; whose individual measurements are considered useful for estimating the glomerular filtration rate, GFR)7,9. Prior to the calculation of the Spearman rank order correlation, the data were preprocessed to filter-out and eliminate the peptides that were detected in less than 80% of all samples. In each case of a missing peptide abundance value (cluster AUC), MALDI TOF MS spectra were manually reviewed and background signal for 1 m/z centroided around the reported mass were estimated and recorded. The requirement for a given peptide’s presence in 80%+ of all samples was used to provide for a robust peptide candidate biomarker panel. Spearman rank order correlation values between −1.0 and −0.45 or between +1.0 and +0.45 were considered meaningful. In addition abundance of peptides falling within these two ranges were compared between cases and controls by un-paired t-test with Welch’s correction to address for un-equal variances.
Computer Assisted Tandem-MS Data Analysis
Candidate peptide m/z values selected from Spearman rank order correlation analyses were analyzed in tandem MS experiments to better understand their importance in diabetic ERFD. The selected peptide masses were used to establish a MALDI ion inclusion list and were fragmented using the AB4700 Proteomics Analyzer as previously described9. Final fragmentation data were collected as averaged data from 1500 laser shots. The peptide fragmentation information was searched against the Swiss-Prot database using Matrix Science Mascot software (v1.9) to identify a peptide with the highest correlative amino acid sequence. Criteria used for analysis were a) unconstrained proteolytic search (no enzyme fragmentation criteria stipulated) b) partial peptide modification of sodiation at C-termini, glutamic and aspartic acid side chains, c) Swiss Protein database (20051115), Homo sapiens taxonomy, d) 0.15Da mass accuracy for precursor peptides and e) 0.3Da mass accuracy for peptide fragment ion mass measurement. The resulting search yielded the likelihood of peptide homology or identity by a given total ion MOWSE (Molecular Weight Search) score. Given the above stipulations, peptides with a total ion score ≥ 20 provide for assignment of the peptide amino acid sequence with absolute identification or with near absolute homology. To confirm peptide assignments for the four candidate peptides, synthetic peptides were purchased and MALDI-TOF/TOF spectra acquired and compared to experimental spectra.
Immunoblot Analysis for Plasma Protein Abundance
In order to examine if the increased plasma abundance of kininogen fragments was the result of increased kininogen protein expression and abundance of protein degradation products, immunoblot experiments using reduced and denatured plasma protein samples were conducted as described previously23. In brief, plasma proteins were isolated following albumin depletion. Albumin was removed from 200 μg aliquots with the VivaPure anti-HSA kit for human albumin depletion (Vivascience, Sartorius Group). Albumin depleted plasma samples were concentrated to dryness with a vacuum concentrator (Thermo Savant SC210A Speedvac concentrator), re-dissolved in 100 μl 10mM Tris HCl, pH 7.8 and protein assayed. Samples (10 μg) were mixed with NuPAGE LDS Sample Buffer (Invitrogen, Inc.) and DTT, then heated to 70C for 10 min. Samples were applied to NuPAGE 4-12% Bis-Tris gels (Invitrogen, Inc.) and electrophoresed for 40 minutes at 200V. The proteins were transferred to 0.45 μm nitrocellulose membrane, 90 min at 30V using NuPAGE transfer buffer (Invitrogen, Inc.) with 20% methanol. After transfer, membranes were blocked with Odyssey™ Blocking buffer (LiCor Biosciences, Inc.) for 1 hr and incubated with Goat anti-Kininogen antibody (R&D Systems cat# AF1569) (1:500 in Odyssey™ blocking buffer/0.1%Tween 20) at 4C. Membranes were washed with TTBS (Tris-buffered saline/0.1% Tween 20). Following 1 hour incubation with IRdye800-conjugated Donkey anti-Goat (1:5000 in Odyssey™/Tween) membranes were scanned using the Odyssey™ infrared imaging system (LiCorBioSciences, Inc.).
Immunoblot Analysis of extracellular-signal-regulated kinase (ERK) phosphorylation by glucose and des-Arg9 BK (1-8)
An immortalized C57 black mouse proximal tubule cell line (compliments of Jeffery Schelling, MD, Case Western Reserve University) were cultured in T-25 flasks at 33 C in DMEM:F-12 (1:1) medium supplemented with 10% heat inactivated FBS to 60% confluence. The cells were then serum starved overnight at 33 C in normal glucose, low serum medium (0.5% serum) and then exchanged into low serum treatment medium (5mM glucose, NG; 5mM glucose + 25mM mannitol, M; 30mM glucose, HG) plus 0.5% serum. After 24h at 33 C the cells were treated for 10 min with agonist vehicle (PBS) or agonist (des-Arg9 BK) followed by collecting lysates using RIPA buffer. Total lysate (5 mg) was loaded under reducing/denaturing conditions, separated using gradient PAGE gels and immunoblotted as previously described onto 0.45um nitrocellulose.24,25 The blotted membrane was blocked in 1% milk overnight, probed with primary antibody at 1:1000 (sc-7383, Santa Cruz Biotechnology, Inc, Santa Cruz, CA), secondary antibody at 1:5000 (sc-93, Santa Cruz Biotechnology, Inc, Santa Cruz, CA), and bands imaged using luminol and film. The phospho-Erk2 densitometry was normalized to total-Erk1/2 and then normalized to the mannitol-PBS control experiment. Differences in immunoblot densitometry were determined by an analysis of variance (ANOVA) of immunoblot experiment data and Newman-Kuels post hoc analysis of treatment pairs by t-test.
To determine the effects of polarization on Erk1/2 phosphorylation, immortalized C57 black mouse proximal tubule cell line were seeded onto permeable inserts in 6-well at approximately 10-20 percent density, grown at 33 C in DMEM:F-12 (1:1) medium supplemented with 10% heat inactivated FBS to 70-80% confluence. The cells were then serum starved overnight at 33 C in normal glucose, low serum medium (0.5% serum) and then exchanged into low serum treatment medium (5mM glucose, NG; 5mM glucose + 25mM mannitol, M; 30mM glucose, HG) plus 0.5% serum. After 24h at 33 C the cells were treated apical or basilar administration of the vehicle or peptide for 10 min followed by collecting lysates using RIPA buffer.
To determine the effects of cell type on Erk1/2 phosphorylation by glucose and apical peptide delivery, an opossum kidney proximal tubule cell line (OKC) were seeded onto permeable inserts in 6-well at approximately 10-20 percent density, grown at 37 C in DMEM medium supplemented with 10% heat inactivated FBS to 70-80% confluence. The cells were then serum starved overnight at 37 C in normal glucose, low serum medium (0.5% serum) and then exchanged into low serum treatment medium (5mM glucose, NG; 5mM glucose + 25mM mannitol, M; 30mM glucose, HG) plus 0.5% serum. After 24h at 37 C the cells were treated apical administration of the vehicle or peptide for 10 min followed by collecting lysates using RIPA buffer.
Supplementary Material
Acknowledgements
We would like to thank Nina Lesousky for Technical Support regarding OK cells. This work was supported by National Institutes of Health grants R01 DK067638 (ASK and JBK), R01 DK041526 (ASK), ADA Grant 7-03-MN628 (MAN), U01 DK085673-01 (MLM and JBK), R01 DK091584-01A1 (MLM), DOE DE-FG02-05ER6406 (JBK and MLM), and the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.B.K.).
Footnotes
Disclosure. All the authors declared no competing interests.
References
- 1.Mogensen CE, et al. Prevention of diabetic renal disease with special reference to microalbuminuria. Lancet. 1995;346:1080–1084. doi: 10.1016/s0140-6736(95)91747-0. [DOI] [PubMed] [Google Scholar]
- 2.Viberti GC, et al. Microalbuminuria as a predictor of clinical nephropathy in insulin-dependent diabetes mellitus. Lancet. 1982;1:1430–1432. doi: 10.1016/s0140-6736(82)92450-3. [DOI] [PubMed] [Google Scholar]
- 3.Perkins BA, Krolewski AS. Early nephropathy in type 1 diabetes: a new perspective on who will and who will not progress. Curr Diab Rep. 2005;5:455–463. doi: 10.1007/s11892-005-0055-7. [DOI] [PubMed] [Google Scholar]
- 4.Giorgino F, et al. Factors associated with progression to macroalbuminuria in microalbuminuric Type 1 diabetic patients: the EURODIAB Prospective Complications Study. Diabetologia. 2004;47:1020–1028. doi: 10.1007/s00125-004-1413-8. [DOI] [PubMed] [Google Scholar]
- 5.Perkins BA, et al. Regression of microalbuminuria in type 1 diabetes. N.Engl.J.Med. 2003;348:2285–2293. doi: 10.1056/NEJMoa021835. [DOI] [PubMed] [Google Scholar]
- 6.Araki S, et al. Factors associated with frequent remission of microalbuminuria in patients with type 2 diabetes. Diabetes. 2005;54:2983–2987. doi: 10.2337/diabetes.54.10.2983. [DOI] [PubMed] [Google Scholar]
- 7.Perkins BA, et al. Microalbuminuria and the risk for early progressive renal function decline in type 1 diabetes. J Am Soc Nephrol. 2007;18:1353–1361. doi: 10.1681/ASN.2006080872. [DOI] [PubMed] [Google Scholar]
- 8.Perkins BA, et al. Detection of renal function decline in patients with diabetes and normal or elevated GFR by serial measurements of serum cystatin C concentration: results of a 4-year follow-up study. J.Am.Soc.Nephrol. 2005;16:1404–1412. doi: 10.1681/ASN.2004100854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merchant ML, et al. Urinary peptidome may predict renal function decline in type 1 diabetes and microalbuminuria. J Am Soc Nephrol. 2009;20:2065–2074. doi: 10.1681/ASN.2008121233. doi:ASN.2008121233 [pii]1681/ASN.2008121233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakoki M, Takahashi N, Jennette JC, Smithies O. Diabetic nephropathy is markedly enhanced in mice lacking the bradykinin B2 receptor. Proc Natl Acad Sci U S A. 2004;101:13302–13305. doi: 10.1073/pnas.0405449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Dynamic receptor-dependent activation of inducible nitric-oxide synthase by ERK-mediated phosphorylation of Ser745. J Biol Chem. 2007;282:32453–32461. doi: 10.1074/jbc.M706242200. doi:M706242200[pii]10.1074/jbc.M706242200. [DOI] [PubMed] [Google Scholar]
- 12.Leeb-Lundberg LM, Marceau F, Muller-Esterl W, Pettibone DJ, Zuraw BL. International union of pharmacology. XLV. Classification of the kinin receptor family: from molecular mechanisms to pathophysiological consequences. Pharmacol Rev. 2005;57:27–77. doi: 10.1124/pr.57.1.2. doi:57/1/27 [pii]10.1124/pr.57.1.2. [DOI] [PubMed] [Google Scholar]
- 13.Dendorfer A, Wolfrum S, Wagemann M, Qadri F, Dominiak P. Pathways of bradykinin degradation in blood and plasma of normotensive and hypertensive rats. Am J Physiol Heart Circ Physiol. 2001;280:H2182–2188. doi: 10.1152/ajpheart.2001.280.5.H2182. [DOI] [PubMed] [Google Scholar]
- 14.Kuoppala A, Lindstedt KA, Saarinen J, Kovanen PT, Kokkonen JO. Inactivation of bradykinin by angiotensin-converting enzyme and by carboxypeptidase N in human plasma. Am J Physiol Heart Circ Physiol. 2000;278:H1069–1074. doi: 10.1152/ajpheart.2000.278.4.H1069. [DOI] [PubMed] [Google Scholar]
- 15.Fleming I, Kohlstedt K, Busse R. New fACEs to the renin-angiotensin system. Physiology (Bethesda) 2005;20:91–95. doi: 10.1152/physiol.00003.2005. doi:20/2/91 [pii]10.1152/physiol.00003.2005. [DOI] [PubMed] [Google Scholar]
- 16.Jaffa AA, et al. Plasma prekallikrein: a risk marker for hypertension and nephropathy in type 1 diabetes. Diabetes. 2003;52:1215–1221. doi: 10.2337/diabetes.52.5.1215. [DOI] [PubMed] [Google Scholar]
- 17.Kakoki M, et al. Senescence-associated phenotypes in Akita diabetic mice are enhanced by absence of bradykinin B2 receptors. J Clin Invest. 2006;116:1302–1309. doi: 10.1172/JCI26958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakoki M, McGarrah RW, Kim HS, Smithies O. Bradykinin B1 and B2 receptors both have protective roles in renal ischemia/reperfusion injury. Proc Natl Acad Sci U S A. 2007;104:7576–7581. doi: 10.1073/pnas.0701617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Y, et al. Targeted deletion of B2-kinin receptors protects against the development of diabetic nephropathy. Am J Physiol Renal Physiol. 2007;293:F1026–1035. doi: 10.1152/ajprenal.00203.2007. [DOI] [PubMed] [Google Scholar]
- 20.Hewitt SM, Dear J, Star RA. Discovery of protein biomarkers for renal diseases. Journal of the American Society of Nephrology. 2004;15:1677–1689. doi: 10.1097/01.asn.0000129114.92265.32. [DOI] [PubMed] [Google Scholar]
- 21.Tan Y, Wang B, Keum JS, Jaffa AA. Mechanisms through which bradykinin promotes glomerular injury in diabetes. Am J Physiol Renal Physiol. 2005;288:F483–492. doi: 10.1152/ajprenal.00165.2004. [DOI] [PubMed] [Google Scholar]
- 22.Tang SC, et al. Bradykinin and high glucose promote renal tubular inflammation. Nephrol Dial Transplant. 2010;25:698–710. doi: 10.1093/ndt/gfp599. doi:gfp599 [pii]10.1093/ndt/gfp599. [DOI] [PubMed] [Google Scholar]
- 23.Barati MT, et al. Proteomic analysis defines altered cellular redox pathways and advanced glycation end-product metabolism in glomeruli of db/db diabetic mice. Am J Physiol Renal Physiol. 2007;293:F1157–1165. doi: 10.1152/ajprenal.00411.2006. doi:00411.2006 [pii]10.1152/ajprenal.00411.2006. [DOI] [PubMed] [Google Scholar]
- 24.Holthouser KA, et al. Ouabain stimulates Na-K-ATPase through a sodium/hydrogen exchanger-1 (NHE-1)-dependent mechanism in human kidney proximal tubule cells. Am J Physiol Renal Physiol. 2010;299:F77–90. doi: 10.1152/ajprenal.00581.2009. doi:ajprenal.00581.2009 [pii]10.1152/ajprenal.00581.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rood IM, et al. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010 doi: 10.1038/ki.2010.262. doi:ki2010262 [pii]10.1038/ki.2010.262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.