Abstract
SUMMARY
We performed massively parallel sequencing of paired tumor/normal samples from 203 multiple myeloma (MM) patients and identified significantly mutated genes and copy number alterations, and discovered putative tumor suppressor genes by determining homozygous deletions and loss-of-heterozygosity. We observed frequent mutations in KRAS (particularly in previously treated patients), NRAS, BRAF, FAM46C, TP53 and DIS3 (particularly in non-hyperdiploid MM). Mutations were often present in subclonal populations, and multiple mutations within the same pathway (e.g. KRAS, NRAS and BRAF) were observed in the same patient. In vitro modeling predicts only partial treatment efficacy of targeting subclonal mutations, and even growth promotion of non-mutated subclones in some cases. These results emphasize the importance of heterogeneity analysis for treatment decisions.
INTRODUCTION
We previously reported the sequencing of 38 matched tumor/normal MM pairs, and that report of the genomic landscape of MM pointed to a number of recurrently mutated genes (e.g. FAM46C, DIS3) that are likely causal drivers of the disease (Chapman et al., 2011). However, that study design was only powered to detect commonly mutated genes, but not less commonly mutated genes, due to the weak statistical power provided by the small sample size. It also did not examine copy number alterations, leading to homozygous deletions or loss of heterozygosity (LOH), or clonal heterogeneity due to the modest sequence coverage (~ 30X) of those whole genome sequences.
The identification of driver mutations in MM holds great promise for personalized medicine, whereby patients with particular mutations would be treated with the appropriate targeted therapy (Fonseca et al., 2009; Mahindra et al., 2012; Palumbo and Anderson, 2011). However, if the mutation is present in only a fraction of the cells, one might doubt whether such targeted therapy would be clinically efficacious. Recent studies have documented the existence of clonal heterogeneity in solid tumors and acute myeloid leukemia, albeit in small numbers of patients (Campbell et al., 2010; Carter et al., 2012; Ding et al., 2012; Gerlinger et al., 2012; Nik-Zainal et al., 2012; Shah et al., 2012; Walter et al., 2012). These studies demonstrated how acquisition of genetic alterations over time leads to clonal evolution. Systemic treatment with chemotherapy may affect the fitness of some subclones more than others, and thus may alter the tumor composition by promoting particular subclones (Landau et al., 2013b). Consequently, the full breadth of tumor heterogeneity, particularly in solid malignancies, may not be captured in a single biopsy, which represents a challenge for cancer therapy (Gerlinger et al., 2012). Clonal heterogeneity and clonal evolution have also been observed in MM by either whole exome sequencing or array CGH, albeit in a modest number of patients (Egan et al., 2012; Keats et al., 2012; Walker et al., 2012).
We therefore sought to estimate the extent of clonal heterogeneity in MM in a large-scale MM genome sequencing dataset capturing a breadth of untreated and previously treated patients, and to infer the timing of genetic events in MM. In the work presented here, we address several important questions: 1) Can we identify significantly mutated genes by integrating evidence from both point mutations and copy number analysis? 2) How do the mutation profile and the clonal and subclonal composition of MM differ between hyperdiploid and non-hyperdiploid and between treated and untreated MM? 3) Can the contribution of subclones in a patient be reconstructed from a single biopsy to inform targeted therapy?
RESULTS
We first set out to create a MM genome dataset that would be sufficiently powered to comprehensively assess the genetic diversity of the disease and the extent to which subclonal heterogeneity is observed within patients. A total of 203 tumor-normal pairs were analyzed; 177 by whole exome sequencing and 26 by whole genome sequencing (16 and 23, respectively, have been previously reported (Chapman et al., 2011)). The average depth of coverage for the whole exomes and whole genomes was 89X and 30X, respectively. To estimate the statistical significance of mutation frequency (as a measure of positive selection), we used a new version of the MutSig algorithm (MutSigCV) that compares observed mutation frequencies against sequence context-specific, tumor-specific and gene-specific background mutation frequencies (Lawrence et al., 2013). Additionally, we developed analytical tools to further prioritize homozygous somatic single nucleotide variants (SSNVs) or genes, which harbor mutations that are positionally clustered or preferentially affecting highly conserved amino acids (Supplemental Experimental Procedures). Analysis of the 203 tumor-normal pairs showed that 11 genes were recurrently mutated using a standard significance threshold of q < 0.1 (Figure 1 and S1). The individual and combined p and q values for these prioritization procedures are shown in Tables S1 and S2. Mutation validation studies were performed on 140 mutations, with a rate of 90.4%, in line with other large-scale cancer genome sequencing studies (Table S2).
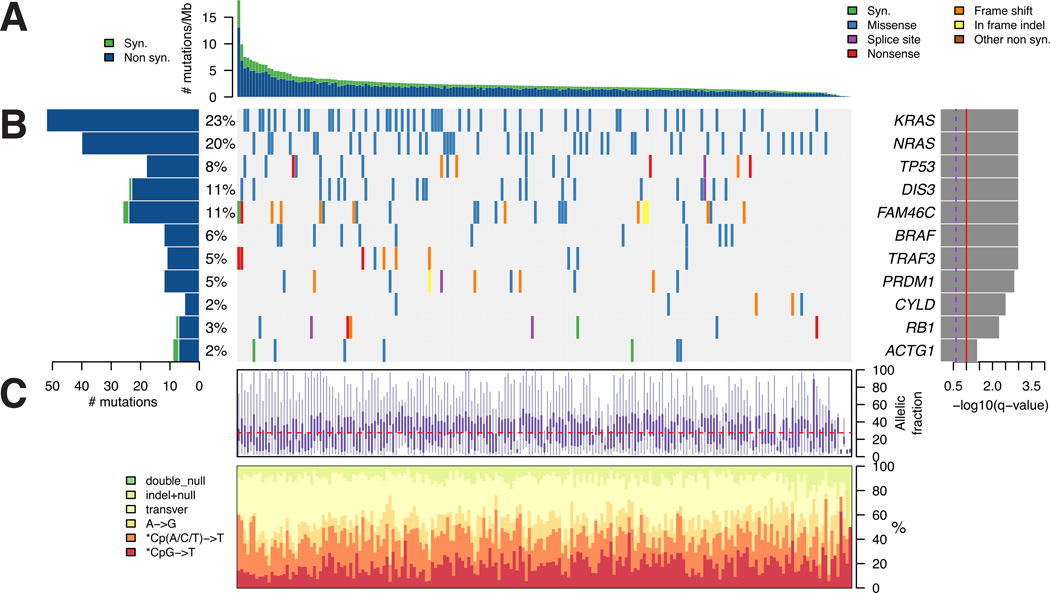
Figure 1. Determining significantly mutated genes in 203 patients with MM.
(A) The rate of synonymous and nonsynonymous mutations is displayed as mutations per megabase (of exome), with individual MM samples ranked by total number of mutations. (B) The heat map represents individual mutations in 203 patient samples, color-coded by type of mutation. Only one mutation per gene is shown if multiple mutations were found in a sample. Left: Histogram shows the number of mutations in each gene. Percentages represent the fraction of tumors with at least one mutation in the specified gene. Right: The 11 genes with the lowest q value (q-combined in Table S1), ranked by level of significance. (C) Base substitution and allelic fraction distribution of individual samples, ranked in the same order as in A. See also Figure S1 and Tables S1–S3.
Among the 11 significantly mutated genes were 5 genes (KRAS, NRAS, FAM46C, DIS3 and TP53) previously identified as the most commonly mutated genes in our 38-patient pilot MM genome study (Chapman et al., 2011). An additional 4 genes (BRAF, TRAF3, CYLD, RB1) have been implicated in the pathogenesis of MM (Annunziata et al., 2007; Chapman et al., 2011; Demchenko et al., 2010; Keats et al., 2007; Walker et al., 2012). PRDM1 is a transcriptional repressor that is involved in plasmacytic differentiation, and it acts as a tumor suppressor gene in activated B cell-like diffuse large B cell lymphoma (DLBCL). Mutations that disrupt its function have been described in DLBCL (Mandelbaum et al., 2010), but are not known to play a role in MM. PRDM1 has been shown to promote survival of transformed plasma cells (Lin et al., 2007), and transgenic mice prone to plasmacytoma development show reduced plasmacytoma incidence if one or two PRDM1 alleles are knocked out (D'Costa et al., 2009). We find a recurrent missense mutation (S552C) in two patients, with two additional patients harboring closely clustered missense mutations (S605R, S606I), and an additional 5 patients with truncating frame shift or splice site mutations, supporting a role of PRDM1 as a tumor suppressor (Figure 1 and S1; Table S1 and S2).
Additionally, several recurrently mutated and biologically relevant genes fall just below the significance threshold (Table S1). For example, EGR1 was previously shown to abrogate JUN-induced MM growth inhibition and cell death when knocked down in MM cells, and has been reported as a mechanism of resistance to MM therapy (Chen et al., 2010). We found that EGR1 mutations were clustered toward the 5’ end of the gene (Table S1 and S2; Figure S1), a pattern of mutation often associated with somatic hypermutation (Pasqualucci et al., 2001). To further explore this possibility, we asked whether the observed mutations occurred within WRCY motifs known to be the targets of activation-induced cytidine deaminase (AID), a key enzyme that catalyzes somatic hypermutation. This analysis revealed that EGR1 indeed had significant enrichment of mutations in WRCY motifs (q < 0.1; Table S3), consistent with a somatic hypermutation mechanism. Whether these mutations act as “drivers”, and are positively selected, or merely constitute “passengers”, remains to be seen.
We also found 4 missense mutations in the interferon regulatory factor IRF4, with 3 of the mutations being identical (K123R) (Chapman et al., 2011), establishing K123R as a recurrent, “hotspot” mutation in IRF4 (Figure S1; Table S2). IRF4 has previously been reported as a MM survival factor, wherein a loss-of-function, RNA-interference screen showed that IRF4 inhibition results in loss of viability of MM cell lines (Shaffer et al., 2008). SP140 is the lymphoid-restricted homolog of SP100, expressed in plasma cells, and a genome-wide association study identified SP140 as a susceptibility locus for chronic lymphocytic leukemia (Di Bernardo et al., 2008), with risk alleles being associated with reduced levels of SP140 mRNA. We identified missense, frame shift and splice site alterations in 8 patients, with LOH observed for two of these alterations, consistent with its possible role as a tumor suppressor in MM.
The available clinical characteristics of the patients in the study are shown in Figure 2 and Table S4. Identifying significantly mutated genes exclusively in patients with t(4;14) and t(11;14), as obtained for 50 patients subjected to routine clinical FISH testing, did not reveal additional significantly mutated genes (Table S1). In general, there was no strong statistically significant association between particular mutations and clinical features, tumor ploidy, or history of prior treatment, but hypothesis-generating trends could be observed (Figure 2; Table S1 and S5). The sample size may need to be larger to definitively address such associations.
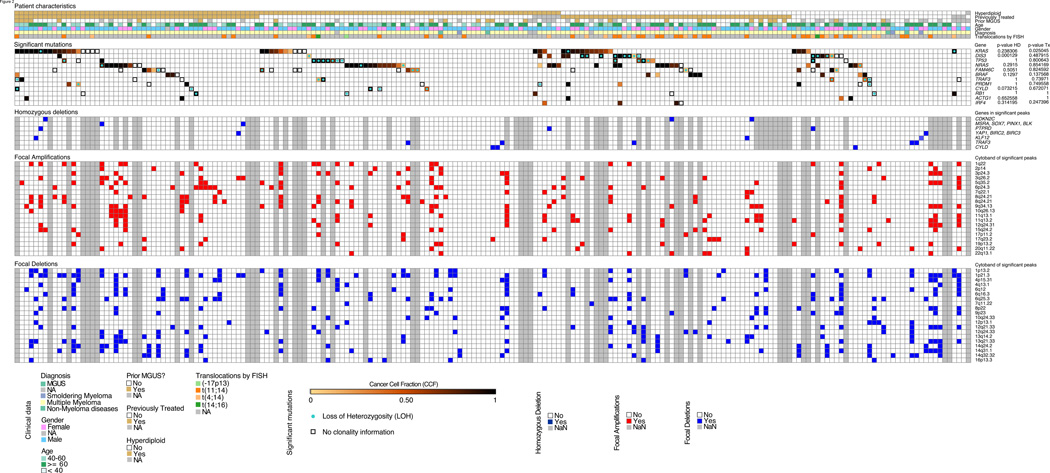
Figure 2. Mutational profile, LOH, and copy number profile in subtypes of MM.
Data for all 203 patient samples for which whole genome or whole exome sequencing was performed are displayed in columns. The first panel from the top displays patient characteristics (“NA”, if information on a characteristic was unavailable). Classification into hyperdiploid versus non-hyperdiploid samples was performed as described in Experimental Procedures. The second panel displays the 11 significantly mutated genes and IRF4 (which harbors K123R mutations in 3 patients), color-coded by the cancer cell fraction in which these mutations occur, and circles within symbols representing LOH. p value HD and p value Tx represent differences in the prevalence of mutations in the indicated gene between hyperdiploid and non-hyperdiploid, or between previously treated and untreated samples, respectively. The third panel highlights samples harboring homozygous deletions at the most significant loci. Only selected genes within those loci are displayed. Grey symbols denote samples with unavailable high density copy number array or ABSOLUTE data. The lower two panels display focal deletions and amplifications across 153 patients with high density copy number arrays, as determined by GISTIC analysis. Grey symbols denote samples without high density copy number array. See also Figure S2 and Tables S4–S9.
Tumor suppressor genes can be inactivated not only by point mutation, but also by biallelic deletion. We therefore searched for genes with a statistically significant excess of homozygous deletion (using a modification of the GISTIC algorithm (Beroukhim et al., 2007; Mermel et al., 2011)) across the 153 patients in our study for whom high density copy number array data were available. Deletions were identified as being homozygous using the ABSOLUTE algorithm (Carter et al., 2012). We identified 7 significant regions, containing 32 genes, including known tumor suppressor genes such as CDKN2C, as well as genes associated with the regulation of the NF-κB signaling pathway including TRAF3, BIRC2, BIRC3, and CYLD (Figure 2; Table S6). CYLD was also found significantly mutated in 5 patients (Figure 1 and Figure 2) and its inactivation through deletions and mutations has been described in MM (Demchenko et al., 2010; Keats et al., 2007). Similarly, the exonuclease-encoding DIS3 gene is subject to point mutations with LOH (as determined by ABSOLUTE (Carter et al., 2012)), strongly implicating DIS3 as a tumor suppressor in 11% of MM patients (Figure 2). In order to designate samples as either hyperdiploid or non-hyperdiploid with high resolution, we developed and validated a classification method using WES and WGS (Figure S2A, Supplemental Experimental Procedures). Interestingly, DIS3 aberration was more commonly seen among the 86 non-hyperdiploid MM cases compared to the 116 hyperdiploid cases (Fisher’s Exact test, p = 0.00013; Table S5), with a non-significant trend towards a greater fraction of LOH in DIS3 mutated non-hyperdiploid patients, compared to hyperdiploid samples (Fisher’s Exact test p = 0.13). We also found an excess of homozygous deletions in the gene encoding the tyrosine phosphatase PTPRD, which has recently been implicated as a tumor suppressor in MM, glioblastoma, and other cancers (Kamada et al., 2012). PTPRD dephosphorylates STAT3, which promotes signaling from IL6, a well-recognized MM survival factor. Whether IL6-signaling is indeed the mechanistic target of PTPRD deletion remains to be established. We also found homozygous deletions with a peak at 8p23.1 (with 18 genes), containing several candidate tumor suppressor genes (BLK, MSRA, PINX1, SOX7), for which a connection to MM has not been previously established.
We next asked whether there was evidence to support pathway-level patterns of mutation, whereby mutations in individual genes may lack statistical significance but when multiple members of a pathway or functionally-related gene set are mutated (albeit rarely), significance is observed. We first tested the 3 gene set hypotheses that emerged from a pilot analysis of the MM genome, namely the NF-κB pathway, histone modifying enzymes and the coagulation cascade (Chapman et al., 2011). We find that indeed all 3 gene sets retain statistical significance across our collection of 203 patients (p < 0.05), if tested as individual hypotheses (Table S7). Next, we tested a collection of 612 curated gene sets (taken from MSigDB (Subramanian et al., 2005)), and found that 6 gene sets reached statistical significance after correction for multiple hypothesis testing (Table S7). These gene sets primarily include mutated components of the cell cycle machinery (including CDKN1B and CCND1), and serve to highlight genes that are only borderline significant when analyzed individually, but reach significance as part of these gene sets. For example, we observed 3 coding mutations (accompanied by LOH) in the transcription factor MAX, as part of a significantly mutated gene set, which functions as a heterodimerization partner for MYC, which is well known to be dysregulated in MM (Shou et al., 2000). Interestingly, MAX has been implicated as a tumor suppressor susceptibility gene in pheochromocytoma (Comino-Mendez et al., 2011), and a small molecule inhibitor of MYC-MAX heterodimerization has been reported to result in myeloma cell death (Holien et al., 2012).
Of the 203 patients in the study, 131 (65%) had evidence of mutations in one or more of the 11 recurrently mutated genes, and 119 (59%) had mutations of a gene within a statistically significant gene set (new and previously published in Table S8), accounting for a total of 154 patients (76%). Of the remaining 24% of tumors lacking such obviously functionally important mutations, some are likely to be driven by rare mutations in bona fide driver genes (e.g. the tumors with mutations in MYD88 or CARD11, previously reported to be recurrently mutated in the B-cell malignancy DLBCL (Lohr et al., 2012; Morin et al., 2011) (Table S1 and S2)). Tumors lacking such mutations might alternatively be driven by focal copy number alterations or chromosomal rearrangements. Of the 153 patients from whom copy number array data were available, 119 patients (including 40 of the 60 patients lacking SSNV in the most significantly mutated genes), had evidence of at least one focal gene copy number gain or loss within a significant peak (Figure 2 and S2B; Table S8). 21 patients (of 139 patients with high density copy number array and ABSOLUTE data) harbored homozygous deletions in significant peaks (Figure 2). Similarly, structural variants were found in all of the 26 patients from whom whole genome sequencing was available (Chapman et al., 2011), including 3 previously unpublished patients (Table S9). Whether such gene rearrangements are indeed causal of MM in these patients remains to be proven.
We next addressed the extent to which clonal heterogeneity exists in MM. To do this, we computed the allelic fraction of each somatic single nucleotide variant (SSNV). The allelic fraction estimation alone (Figure 1), however, cannot be used to assess the fraction of cancer cells harboring the mutation because it does not take into account i) the copy number at that locus, or ii) tumor purity (whereby normal cell contamination can lead to the spurious impression of mutation subclonality). We therefore used the ABSOLUTE algorithm (Carter et al., 2012) to estimate the cancer cell fraction (CCF) of each SSNV by modeling the observed wild-type and mutated allele counts, taking into account local somatic copy number and sample purity, calculated from high density SNP array or sequencing data (Figure S3) (Landau et al., 2013a). The CCF estimates for all SSNVs in a given sample were then analyzed with a Bayesian clustering algorithm (Landau et al., 2013b) to estimate the number of subclonal cell populations present in each tumor (Figure S3, Experimental Procedures).
Looking across the entire coding region, we found that of 153 patient samples with a purity greater than 0.7, nearly all had evidence of clonal heterogeneity (Figure 3A). Most patients harbored at least 3 detectable subclones (beyond the major clone), with some patients having as many as 7. For comparison, the same analysis applied to ovarian cancer (The Cancer Genome Atlas Research Network, 2011) (sequenced to similar depth and with similar purities) showed that a lower proportion of MM patients (8%) had one or no subclones compared to ovarian cancer (19%) (Fisher’s exact test, p = 0.0024) (Figure 3A).
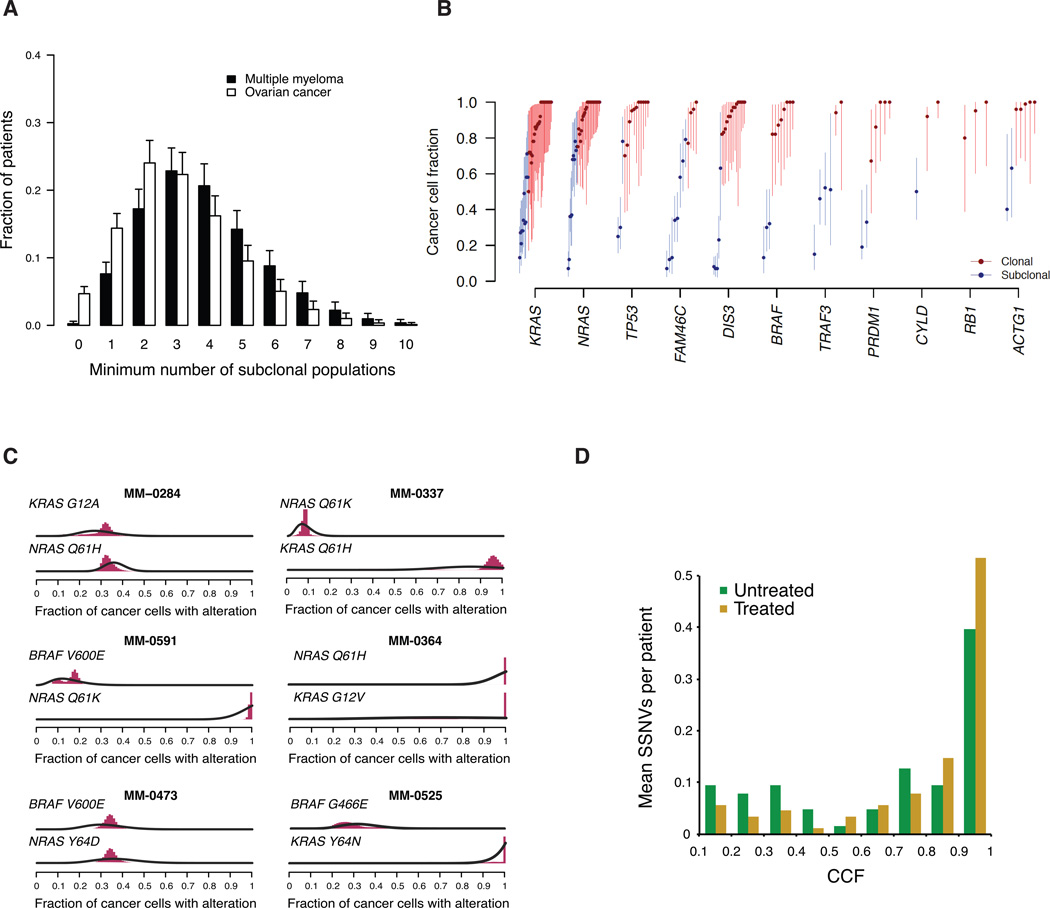
Figure 3. Clonal heterogeneity of significantly mutated genes in MM.
(A) The numbers of predicted subclones by clustering of cancer cell fractions are shown, as described in Supplemental Experimental Procedures. As a comparison the predicted distribution of the number of subclones is also shown for a cohort of patients with ovarian cancer. Error bars represent standard deviation. (B) The CCF, i.e. the expected fraction of MM cells that harbor a coding mutation in the indicated gene, is shown. Each symbol represents a somatic mutation in an individual patient. The most significantly mutated genes are shown. Based on the probability distribution, mutations were determined to be either clonal (red circles, upper bound of CCF confidence interval ≥ 0.95) or subclonal (blue circles, upper bound of CCF confidence interval < 0.95). Error bars represent the 95% confidence interval. (C) Co-occurrence of significant mutations in the same patient is depicted. Results of the Bayesian clustering procedure applied to SSNV CCF distributions for KRAS, NRAS, and BRAF in samples which harbor mutations in at least two of these 3 oncogenes. Probability distributions over CCF for the co-occurring SSNV in the indicated oncogenes before clustering (black curves), and after clustering (filled red bars). (D) The fraction of somatic mutations that are present at the indicated CCF are shown for the 11 most significantly mutated genes. Mutations in significantly mutated genes occur at significantly higher CCFs in previously treated patients, compared to untreated patients (p = 0.007, Wilcoxon rank sum test). See also Figure S3.
We next asked whether certain types of mutations tended to be clonal (consistent with early events) whereas others might tend to be subclonal (consistent with later events). It was conceivable that true driver mutations (e.g. those reaching statistical significance based on mutation frequency, or those with strong connections to MM biology) might be exclusively clonal events. This, however, was not the case. Mutations in most of these genes were found to be clonal in some patients and subclonal in others, including some cases with subclonal coding mutations occurring on segments with subclonal copy number change (Figure 3B, Table S2). For example, of the 44 patients with coding KRAS mutations that were analyzed for clonality, 32 (73%) had clonal KRAS mutations, whereas for 12 patients (27%), the KRAS mutations were subclonal, detectable in as few as 13% of cells in some patients.
In some cases, we observed multiple significant mutations in the same tumor sample, including mutations in oncogenes whose function might be expected to be redundant. For example, some patients had mutations in two of three oncogenes (NRAS, BRAF, and KRAS) (Figure 3C), or two mutations in KRAS (Table S2), despite the fact that these mutations similarly activate the MAP kinase pathway, and therefore seemed unlikely to occur in the same tumor. We therefore asked whether there is evidence to support these mutations being present in the same clones, or rather in different subclones within the tumor. We reasoned that if they occurred in the same cell, we should find some cases in which both mutations were clonal. This analysis indicated that consistent with their biological function, KRAS, NRAS and BRAF mutations were rarely simultaneously clonal in our patient samples (1 sample); instead they were mostly either both subclonal within the tumor, or they occurred in a nested fashion (i.e. one clone being the subclone of another), (9 samples). While these data indicate that mutations in KRAS, NRAS, and BRAF can coexist in the same cell, such subclones often did not appear to have sufficient selective advantage to grow to clonality. In contrast, we found that DIS3 and KRAS mutations were often simultaneously clonal (Table S2). DIS3 is known to encode a ribonuclease involved in RNA processing, but how loss-of-function DIS3 mutations are oncogenic, and how they interact with KRAS in cellular transformation remains to be determined. Interestingly, we found that in general, significantly recurrent mutations were more often clonal in previously treated compared to untreated patients (p = 0.007, Wilcoxon Rank Sum Test) (Figure 3D). This suggests that treatment may accelerate the fixation of certain subclones by eliminating less fit clones.
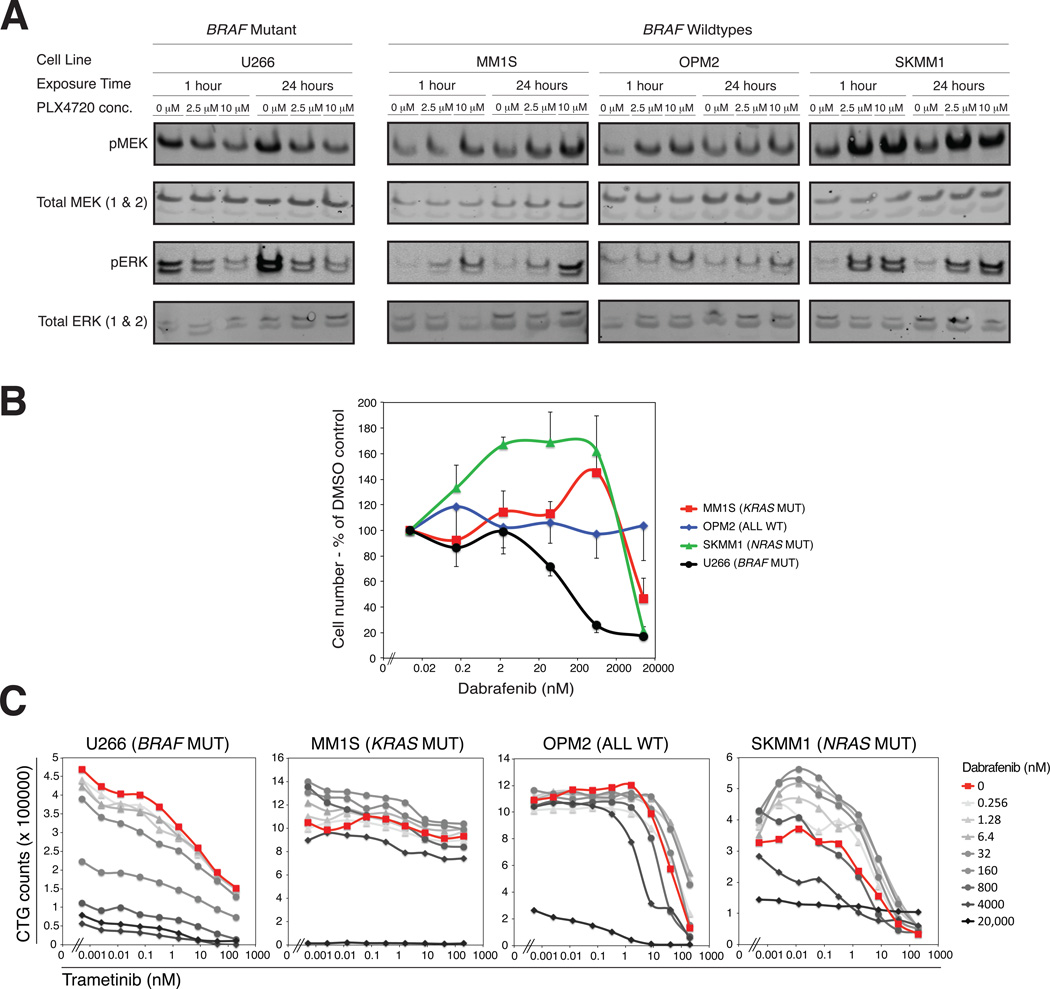
We next explored the therapeutic implications of the observed clonal heterogeneity. Specifically, we focused on BRAF, because the observation of BRAF activating mutations in MM has stimulated clinical exploration of BRAF inhibitors in this disease. Indeed, a recent report of a single BRAF-mutant MM patient showing durable response to a BRAF inhibitor is encouraging (Andrulis et al., 2013). Consistent with that clinical response, we found that U266 cells, which express the BRAF-K601N mutant (COSMIC database http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) that has been shown to cause elevated phospho-MEK and phospho-ERK levels in other malignancies (Dahlman et al., 2012), were more sensitive to treatment with the BRAF inhibitor PLX4720 compared to BRAF-wildtype cell lines (Figure 4A and 4B). In addition, BRAF inhibition downregulated MAP kinase pathway activity only in BRAF-mutant MM cells, whereas in BRAF-wildtype cells, the pathway was paradoxically upregulated (Figure 4A), similar to reports in melanoma (Poulikakos et al., 2010). Strikingly, paradoxical pathway activation was even more pronounced in the presence of KRAS or NRAS mutations, and this increased MAP kinase activity was associated with BRAF inhibitor-induced growth stimulation of KRAS- or NRAS-mutant MM cell lines (Figure 4B). Taken together, these results suggest that treatment of patients harboring subclonal BRAF mutations may at best have only partial responses when treated with BRAF inhibitors.
Figure 4. Heterogeneity composition determines the response to targeted therapy.
(A) The BRAF WT MM cell lines OPM2 (NRAS and KRAS WT, FGFR3 K650E), MM1S (KRAS G12A), SKMM1 (NRAS G12D) and the BRAF-mutant MM cell line U266 (BRAF K601N) were treated with the BRAF-inhibitor PLX4720 at the indicated concentrations. Phosphorylated and total MEK and ERK were detected by western blot at the indicated timepoints. (B) The indicated cell lines were cultured for 5 days in the absence or presence of increasing concentrations of the BRAF-inhibitor dabrafenib. Cell numbers were determined by flow cytometry on day 5 of culture and normalized to the cell number at a dabrafenib concentration of 0 µM (=100%). Error bars represent standard deviation. (C) The indicated MM cell lines were cultured in the presence of the MEK-inhibitor trametinib with or without dabrafenib at varying doses. The cell number on day 5 of culture was determined by cell titer glo. Curves with darker shades of grey represent higher concentrations of dabrafenib.
The presence of MAP kinase pathway activation in MM has similarly increased interest in the clinical testing of MEK inhibitors in MM (Annunziata et al., 2011; Tai et al., 2007). In melanoma, the combination of MEK and BRAF inhibitors appears efficacious with a favorable toxicity profile, and the combination appears to abrogate the paradoxical activation of the MAP kinase pathway in BRAF-wildtype cells (Flaherty et al., 2012). In MM cell lines, we found that combination treatment resulted in increased killing of BRAF-mutant cells, whereas a combination benefit was not observed in BRAF-wildtype cell lines (Figure 4C). These results support the clinical exploration of combination BRAF/MEK inhibitors in clonal, BRAF-mutant MM.
DISCUSSION
The modern oncology paradigm holds that the characterization of tumor genomes will reveal a coherent view of the pathogenesis of cancer, and this in turn will lead to the development of targeted therapies. Our characterization of 203 MM genomes represents by far the most comprehensive effort reported to date, elucidating with statistical confidence the recurrent point mutations and copy number alterations associated with the disease. In particular, the integration of copy number and mutation analysis led to the identification of genes whose recurrent mutation is also accompanied by loss of heterozygosity (LOH), a hallmark of loss-of-function of tumor suppressor genes. Based on these methods, biologically important driver genes may be prioritized, even though they occur at a low frequency. Also of interest were mutations in EGR1 (seen in 7 of 203 patients). The 5’ bias of the mutations and their occurring within a WRCY motif all suggest that they occur as a result of somatic hypermutation as a consequence of AID activity that is most commonly associated with the normal process of immunoglobulin gene rearrangement in B-cells (Lenz and Staudt, 2010). We recently reported in the B-cell malignancy DLBCL that somatic hypermutation of the BCL2 gene occurred as a result of chromosomal translocations that bring the immunoglobulin heavy chain enhancer in proximity to the BCL2 locus (Lohr et al., 2012). Similarly, somatic hypermutation may occur when genes are dysregulated by IGH translocations in MM. For example, we identified 39 coding and non-coding mutations in the CCND1 locus, with some samples harboring multiple mutations. In at least 4 of these patients t(11;14) translocations were also detected, suggesting the possibility that these mutations might also result from somatic hypermutation.
Interestingly, several genes, while not reaching statistical significance on their own, were part of frequently mutated pathways or processes that have been causally implicated in MM (including the NF-κB pathway, chromatin-modifying enzymes, and RNA processing molecules). Mutations in the RNA-binding proteins DIS3 and FAM46C were collectively observed in 21% of patients. It remains unknown why these mutations occur at such high frequency in MM, and yet are seen only rarely, if ever, in other types of cancer. DIS3 mutations were often accompanied by LOH, and were most commonly seen in non-hyperdiploid MM.
Additionally, we observed an accumulation of mutations in components of the cell cycle machinery, as well as in members of the MAPK pathway. A key opportunity for the future will be to relate these mutations to the promising preclinical and clinical results that have recently been reported in MM using cyclin-dependent kinase inhibitors and RAF kinase inhibitors (Andrulis et al., 2013; Cirstea et al., 2013).
Perhaps the most striking finding of our study was that MM tumors are highly heterogenous. Lower resolution genetic analyses (e.g. cytogenetics, FISH and array CGH) have pointed to the existence of clonal heterogeneity in MM, and recent studies using exome sequencing and copy number analyses in a small number of samples similarly documented clonal diversity (Egan et al., 2012; Keats et al., 2012; Walker et al., 2012). The present study of 203 patients is unprecedented in its comprehensiveness, and the analytical approach allowed us to i) identify subclonal mutations, ii) estimate the cancer cell fraction in which these mutations occur, iii) estimate the minimum number of subclones. Our method is statistically powered to detect subclones representing at least 10% of the overall tumor sample. It is therefore likely that our finding that MM tumor samples contain on average at least ~ 5 subclones underestimates the clonal diversity of the disease. It is conceivable that a much larger number of additional subclones may also exist, either below our detection sensitivity or in non-sampled MM tissues. More comprehensive characterization of MM tissue will likely be required to resolve these questions.
Interestingly, point mutations in the most significantly mutated genes were found to be clonal in some patients but subclonal in others. That is, these mutated genes appear to be able to function both as initiators of MM and also as potentiators of the disease. For example, BRAF mutations were often subclonal, and in some cases, co-existent with NRAS mutations. In our cohort these mutations did not co-occur clonally. Rather, at least one of them was always subclonal. With increasing numbers of samples and greater depth of sequencing, many more such cases, including many different genes, may be identified. Other examples of such evolution in cancer have been reported (Campbell et al., 2010; Ding et al., 2012; Gerlinger et al., 2012; Jovanovic et al., 2010; Nik-Zainal et al., 2012; Shah et al., 2012; Walter et al., 2012; Wilmott et al., 2012).
These results also have important clinical implications for MM clinical trials. For example, BRAF inhibitors are being explored in MM harboring BRAF mutation, and the first patient with BRAF V600E–positive MM who experienced a durable response to BRAF inhibition has just been reported (Andrulis et al., 2013). However, if a BRAF mutation is not clonal, suboptimal clinical benefit would be expected. In principle, treating patients harboring subclonal BRAF mutations with BRAF inhibitors may stimulate the growth of BRAF-wildtype tumor cells. Combined BRAF and MEK inhibition might mitigate this effect, but this remains to be demonstrated in vivo.
The clonal heterogeneity observed in this study offers a generally sobering view of prospects for predicting the effects of targeted therapy for cancer in general. Therapy targeting a mutation present in only a fraction of tumor cells would be expected to affect only that subclone, leading to limited clinical benefit. At worst, targeted therapy might have a paradoxically stimulatory effect on the subclones lacking the relevant mutation. At a minimum, we suggest that it will not be sufficient to simply document the presence or absence of mutations in the diagnostic setting. Rather, it will be important to enumerate the extent of clonal heterogeneity in patients being evaluated for targeted therapy, and to interpret the results of subsequent therapy in light of such genetic heterogeneity. Effective targeted therapy will require either drug combinations targeting distinct subclones, or more likely, deploying targeted therapies only in patients for whom the drug target is entirely clonal.
EXPERIMENTAL PROCEDURES
Sample selection and quality assessment of DNA
Bone marrow aspirates and peripheral blood samples were collected at Multiple Myeloma Research Consortium (MMRC) institutions from patients diagnosed with MM and then shipped to the MMRC Tissue Bank for processing as previously described (Ahmann et al., 2008). The studies were approved by the Committee on the Use of Humans as Experimental Subjects of MIT, protocol #0803002647. All patients provided written informed consent under institutional review board approval. Sample processing was slightly modified from previous reports (Salhia et al., 2010) as described in Supplemental Experimental Procedures.
Whole exome, whole genome sequencing, and detection of copy number variations
Whole-exome capture libraries were constructed from 100ng of tumor and normal DNA following shearing, end repair, phosphorylation and ligation to barcoded sequencing adapters (Fisher et al., 2011; Gnirke et al., 2009). Ligated DNA was size-selected for lengths between 200–350bp and subjected to exonic hybrid capture using SureSelect v2 Exome bait (Agilent). Samples were multiplexed and sequenced on multiple Illumina HiSeq flowcells (paired end 76bp reads) to average depth of coverage of 89x and 88x for tumor and normals, respectively. For whole-genome sequencing library construction was done with 1–3 micrograms of native DNA from primary tumor and germline samples for each patient. The DNA was sheared to a range of 101–700 bp using the Covaris E210 Instrument, and then phosphorylated and adenylated according to the Illumina protocol. Adapter ligated purification was done by preparatory gel electrophoresis, and size was selected by excision of two bands (500–520 bp and 520–540 bp respectively) yielding two libraries per sample with average of 380 bp and 400 bp respectively. The libraries were then sequenced with the Illumina GA-II or Illumina HiSeq sequencer with 76 or 101 bp reads, achieving an average of ~30X coverage depth. The resulting data was analyzed with the current Illumina pipeline, which generates data files (BAM files), which contain the reads and quality parameters. Copy number variations (CNV) of 153 patients of the sequencing cohort were determined by Affymetrix SNP 6.0 array. Sequencing data are available in the dbGaP database (www.ncbi.nlm.nih.gov/gap) under accession number phs000348.
Analysis of whole genome and whole exome sequencing data
Described in detail in Supplemental Experimental Procedures.
Myeloma cell lines, MM cell proliferation and BRAF inhibition
A human derived BRAF-mutant cell line (U266) and three BRAF-wildtype cell lines (OPM2, MM1S, and SKMM1) were plated at 1.0 × 10(6) cells/mL (total 2.0 × 10(6) in 2 mL) in 6-well plates. For western blot, cells were then treated with concentrations of 0 µM, 2.5 µM, or 10 µM of the BRAF-inhibitor, PLX4720 for one hour or 24 hours. Following treatment, cells were harvested and lysed on ice for 5 minutes with 300 uL of a modified NP40 lysis buffer (1% NP-40, 50 mM Tris-ph-7.5, 150 mM NaCl, 2mM EDTA-ph-8, 1 mg/mL NaF and deuterium depleted water) for Western blot analysis. To determine the proliferation of MM cell lines in the presence of the BRAF inhibitor dabrafenib, and the MEK-inhibitor trametinib, 3000 cells per well were plated in quadruplicate in a 384 well plate, in the presence of the indicated drug concentrations.
Western blot analysis of MAPK pathway following PLX4720 treatment
Lysate protein concentrations were obtained using the BIO-RAD DC Protein Assay kit, and concentrations were subsequently adjusted to 1 ug/uL final concentrations. Twelve micrograms of protein from each cell lysate was run per well on NuPAGE 4–12% Bis-Tris Midi Gels (Life Technologies WG1403BX10). The gel was blotted onto nitrocellulose membrane paper (Invitrogen LC2001), using the iBlot gel transfer device (Life Technologies IB1001). The membrane was subsequently blocked (LiCor Blocking Buffer 927–40000) for one hour, and stained as described in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Deep sequencing reveals significant genetic events in multiple myeloma
Intra-tumor genetic heterogeneity is common in multiple myeloma
Recurrent mutations can occur either early or late in the evolution of a tumor
Genetic heterogeneity in cancer may limit effectiveness of targeted therapy
SIGNIFICANCE.
A vision for precision cancer medicine calls for the deployment of molecularly-targeted therapeutics in genetically-defined patient populations. A first step in that process involves a description of the genetic landscape of cancer. We describe here a more comprehensive characterization of the MM genome, identifying recurrently mutated genes, copy number alterations and signaling pathways. We find evidence for extensive clonal heterogeneity in the disease, a finding that may complicate the interpretation of genome-inspired clinical trials for MM. More generally, our findings indicate a need for the delineation of clonal heterogeneity in genome-based diagnostic approaches to cancer.
ACKNOWLEDGEMENTS
This project was funded by a grant from the Multiple Myeloma Research Foundation. This work was also supported by NIH grant 5P50CA100707-09 (DF/HCC SPORE in Multiple Myeloma) and a Conquer Cancer Foundation Young Investigator Award (J.G.L.). We are grateful to members of the Broad Institute’s Genomics Platform, without whom this work would not have been possible, and to Jadwiga Grabarek and the members of the Golub lab for helpful discussions and critical review of the manuscript. The investigators of the Multiple Myeloma Research Consortium are: Kenneth C. Anderson, Paul Richardson, Amrita Krishnan, Sagar Lonial, Jonathan Kaufman, David S. Siegel, David H. Vesole, Vivek Roy, Candido E. Rivera, S. Vincent Rajkumar, Shaji Kumar, Rafael Fonseca, Greg J. Ahmann, P. Leif Bergsagel, A. Keith Stewart, Craig C. Hofmeister, Yvonne A. Efebera, Sundar Jagannath, Ajai Chari, Suzanne Trudel, Donna Reece, Jeffrey Wolf, Thomas Martin, Todd Zimmerman, Cara Rosenbaum, Andrzej J. Jakubowiak, Daniel Lebovic, Ravi Vij, Keith Stockerl-Goldstein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: J.G.L. designed and performed experiments, and analyzed data.
P.S., S.L.C. conceived and implemented computational methods for data analysis.
J.G., G.S., K.C., A.M., S.E.S., M.R., and M.S.L. provided analytical support.
J.G.L., S.L.C., P.S., G.G., and T.R.G. wrote the manuscript.
P.C.G. performed experiments and analyzed data.
D.A., C.S. performed sample and data management and gave conceptual advice. B.K. designed experiments and analyzed data.
M.A.C., R.S., designed experiments. J.L., L.M.P performed analysis of clinical data. J.J.K.
performed sequencing analysis.
The Multiple Myeloma Research Consortium contributed patient samples.
G.G., T.R.G designed the experimental strategy, supervised the analysis. All authors discussed the results and implications and reviewed the manuscript.
REFERENCES
- Ahmann GJ, Chng WJ, Henderson KJ, Price-Troska TL, DeGoey RW, Timm MM, Dispenzieri A, Greipp PR, Sable-Hunt A, Bergsagel L, Fonseca R. Effect of tissue shipping on plasma cell isolation, viability, and RNA integrity in the context of a centralized good laboratory practice-certified tissue banking facility. Cancer Epidemiol Biomarkers Prev. 2008;17:666–673. doi: 10.1158/1055-9965.EPI-07-2649. [DOI] [PubMed] [Google Scholar]
- Andrulis M, Lehners N, Capper D, Penzel R, Heining C, Huellein J, Zenz T, von Deimling A, Schirmacher P, Ho AD, et al. Targeting the BRAF V600E Mutation in Multiple Myeloma. Cancer Discov. 2013;3:862–869. doi: 10.1158/2159-8290.CD-13-0014. [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata CM, Hernandez L, Davis RE, Zingone A, Lamy L, Lam LT, Hurt EM, Shaffer AL, Kuehl WM, Staudt LM. A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood. 2011;117:2396–2404. doi: 10.1182/blood-2010-04-278788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang S, Zhou Y, Wu X, Entin I, Epstein J, Yaccoby S, Xiong W, Barlogie B, Shaughnessy JD, Jr., Zhan F. Identification of early growth response protein 1 (EGR-1) as a novel target for JUN-induced apoptosis in multiple myeloma. Blood. 2010;115:61–70. doi: 10.1182/blood-2009-03-210526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirstea D, Hideshima T, Santo L, Eda H, Mishima Y, Nemani N, Hu Y, Mimura N, Cottini F, Gorgun G, et al. Small-molecule multi-targeted kinase inhibitor RGB-286638 triggers P53-dependent and -independent anti-multiple myeloma activity through inhibition of transcriptional CDKs. Leukemia. 2013 doi: 10.1038/leu.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comino-Mendez I, Gracia-Aznarez FJ, Schiavi F, Landa I, Leandro-Garcia LJ, Leton R, Honrado E, Ramos-Medina R, Caronia D, Pita G, et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat Genet. 2011;43 doi: 10.1038/ng.861. 663-U189. [DOI] [PubMed] [Google Scholar]
- D'Costa K, Emslie D, Metcalf D, Smyth GK, Karnowski A, Kallies A, Nutt SL, Corcoran LM. Blimp1 is limiting for transformation in a mouse plasmacytoma model. Blood. 2009;113:5911–5919. doi: 10.1182/blood-2008-08-172866. [DOI] [PubMed] [Google Scholar]
- Dahlman KB, Xia J, Hutchinson K, Ng C, Hucks D, Jia P, Atefi M, Su Z, Branch S, Lyle PL, et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov. 2012;2:791–797. doi: 10.1158/2159-8290.CD-12-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood. 2010;115:3541–3552. doi: 10.1182/blood-2009-09-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, Sullivan K, Vijayakrishnan J, Wang Y, Pittman AM, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S, Middha S, Asmann Y, Schmidt J, Braggio E, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120:1060–1066. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, Morgan G, Van Ness B, Chesi M, Minvielle S, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holien T, Vatsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood. 2012;120:2450–2453. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- Jovanovic B, Egyhazi S, Eskandarpour M, Ghiorzo P, Palmer JM, Bianchi Scarra G, Hayward NK, Hansson J. Coexisting NRAS and BRAF mutations in primary familial melanomas with specific CDKN2A germline alterations. J Invest Dermatol. 2010;130:618–620. doi: 10.1038/jid.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Sakata-Yanagimoto M, Sanada M, Sato-Otsubo A, Enami T, Suzukawa K, Kurita N, Nishikii H, Yokoyama Y, Okoshi Y, et al. Identification of unbalanced genome copy number abnormalities in patients with multiple myeloma by single-nucleotide polymorphism genotyping microarray analysis. Int J Hematol. 2012;96:492–500. doi: 10.1007/s12185-012-1171-1. [DOI] [PubMed] [Google Scholar]
- Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, Van Wier S, Blackburn PR, Baker AS, Dispenzieri A, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120:1067–1076. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Getz G, Wu CJ. Clonal evolution in hematological malignancies and therapeutic implications. Leukemia. 2013a doi: 10.1038/leu.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013b;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FR, Kuo HK, Ying HY, Yang FH, Lin KI. Induction of apoptosis in plasma cells by B lymphocyte-induced maturation protein-1 knockdown. Cancer Res. 2007;67:11914–11923. doi: 10.1158/0008-5472.CAN-07-1868. [DOI] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahindra A, Laubach J, Raje N, Munshi N, Richardson PG, Anderson K. Latest advances and current challenges in the treatment of multiple myeloma. Nat Rev Clin Oncol. 2012;9:135–143. doi: 10.1038/nrclinonc.2012.15. [DOI] [PubMed] [Google Scholar]
- Mandelbaum J, Bhagat G, Tang H, Mo T, Brahmachary M, Shen Q, Chadburn A, Rajewsky K, Tarakhovsky A, Pasqualucci L, Dalla-Favera R. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhia B, Baker A, Ahmann G, Auclair D, Fonseca R, Carpten J. DNA methylation analysis determines the high frequency of genic hypomethylation and low frequency of hypermethylation events in plasma cell tumors. Cancer Res. 2010;70:6934–6944. doi: 10.1158/0008-5472.CAN-10-0282. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, Dewald G, Kirsch IR, Bergsagel PL, Kuehl WM. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci U S A. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai YT, Fulciniti M, Hideshima T, Song W, Leiba M, Li XF, Rumizen M, Burger P, Morrison A, Podar K, et al. Targeting MEK induces myeloma-cell cytotoxicity and inhibits osteoclastogenesis. Blood. 2007;110:1656–1663. doi: 10.1182/blood-2007-03-081240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC, Fenwick K, Kozarewa I, Gonzalez D, Lord CJ, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120:1077–1086. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, Larson DE, McLellan MD, Dooling D, Abbott R, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmott JS, Tembe V, Howle JR, Sharma R, Thompson JF, Rizos H, Lo RS, Kefford RF, Scolyer RA, Long GV. Intratumoral Molecular Heterogeneity in a BRAF-Mutant, BRAF Inhibitor-Resistant Melanoma: A Case Illustrating the Challenges for Personalized Medicine. Mol Cancer Ther. 2012;11:2704–2708. doi: 10.1158/1535-7163.MCT-12-0530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.