Cytoplasmic dynein light intermediate chains are required for the maintenance of centriole cohesion and the formation of a bipolar spindle in both human cells and Xenopus embryos.
Abstract
Cytoplasmic dynein 1 (dynein) is a minus end–directed microtubule motor protein with many cellular functions, including during cell division. The role of the light intermediate chains (LICs; DYNC1LI1 and 2) within the complex is poorly understood. In this paper, we have used small interfering RNAs or morpholino oligonucleotides to deplete the LICs in human cell lines and Xenopus laevis early embryos to dissect the LICs’ role in cell division. We show that although dynein lacking LICs drives microtubule gliding at normal rates, the LICs are required for the formation and maintenance of a bipolar spindle. Multipolar spindles with poles that contain single centrioles were formed in cells lacking LICs, indicating that they are needed for maintaining centrosome integrity. The formation of multipolar spindles via centrosome splitting after LIC depletion could be rescued by inhibiting Eg5. This suggests a novel role for the dynein complex, counteracted by Eg5, in the maintenance of centriole cohesion during mitosis.
Introduction
Cytoplasmic dynein 1 (dynein) is a motor protein that carries many cargoes toward microtubule minus ends (Allan, 2011). Dynein is essential for cell division in metazoans, where it plays multiple roles. Bipolar spindle assembly requires the separation of the duplicated centrosomes in late G2/prophase, and this is driven by dynein on the nuclear envelope (NE) and at the cell cortex and by the plus end–directed motor Eg5 (a kinesin-5), which generates antiparallel sliding of overlapping microtubules (Robinson et al., 1999; Tanenbaum et al., 2008; Tanenbaum and Medema, 2010; Raaijmakers et al., 2012, 2013). At the end of prophase, dynein assists NE breakdown by pulling on the nuclear membrane (Beaudouin et al., 2002; Salina et al., 2002). In the spindle, Eg5 pushes the centrosomes apart, aided by kinesin-12, chromosome-associated kinesins, and forces provided by the polymerization of kinetochore fibers (K fibers; Tanenbaum and Medema, 2010; Maiato and Logarinho, 2014). Dynein counteracts these outward forces (Gaglio et al., 1996; Mitchison et al., 2005; Tanenbaum et al., 2008; Ferenz et al., 2009; Florian and Mayer, 2012), perhaps by driving antiparallel microtubule sliding (Tanenbaum et al., 2013).
Dynein keeps microtubules tightly focused at the spindle poles by working together with nuclear mitotic apparatus (NuMA; Gaglio et al., 1996; Merdes et al., 2000; Silk et al., 2009; Raaijmakers and Medema, 2014). Dynein at the kinetochore generates the initial lateral attachment to and translocation of chromosomes along microtubules (Yang et al., 2007), working in opposition to the kinetochore-associated kinesin CENP-E (Kapoor et al., 2006). After end-on interactions form between the kinetochore and the K-fiber microtubules, dynein contributes to the polewards movement of sister chromatids by causing sliding of K fibers toward the poles (Elting et al., 2014; Sikirzhytski et al., 2014). Kinetochore dynein may also participate in cell cycle control by removing spindle assembly checkpoint (SAC) components from correctly aligned kinetochores (Hoffman et al., 2001; Howell et al., 2001; Wojcik et al., 2001; Mische et al., 2008; Sivaram et al., 2009), although this is controversial (Raaijmakers et al., 2013). Finally, cortical dynein controls the orientation of the spindle (Raaijmakers and Medema, 2014).
When dynein activity is compromised, spindles become multipolar and disorganized, chromosomes fail to attach properly to the spindle, and cell cycle progression is slowed (Robinson et al., 1999; Wojcik et al., 2001; Maiato et al., 2004; Mische et al., 2008; Tanenbaum et al., 2008; Firestone et al., 2012; Iwakiri et al., 2013; Raaijmakers et al., 2013). Impaired pole focusing and a lack of opposition to Eg5-driven forces undoubtedly contribute to aberrant spindle assembly. However, centrosomes themselves may not function normally because dynein is implicated in the accumulation of several pericentriolar material (PCM) components (Doxsey et al., 2005), including PCM-1 (Kubo et al., 1999; Dammermann and Merdes, 2002) and pericentrin (PC; Purohit et al., 1999; Tynan et al., 2000b). Dynein may also contribute directly to microtubule attachment to the centrosome (Heald et al., 1997; Burakov et al., 2008).
Each dynein complex contains two motor subunits, dynein heavy chain (DHC; DYNC1H1), along with two intermediate chains (ICs; DYNC1I1 and 2), two light ICs (LICs; DYNC1LI1 and 2), and several light chains (Allan, 2011). Although the LICs are essential for proper dynein function (Yoder and Han, 2001; Lee et al., 2005; Mische et al., 2008; Palmer et al., 2009; Sivaram et al., 2009; Horgan et al., 2010a,b; Tan et al., 2011; Raaijmakers et al., 2013), their role within the complex is not well understood. Only vertebrates have two isoforms, and, unlike other dynein subunits, LIC is not well conserved outside metazoans (Lee et al., 2005; Pfister et al., 2006; Zhang et al., 2009). Vertebrate LIC1 is phosphorylated by Cdk1 in mitosis, which leads to dynein’s release from membranes (Niclas et al., 1996; Dell et al., 2000; Addinall et al., 2001) and promotes its association with the SAC components Mad1/2 and ZW10 (Sivaram et al., 2009).
Because vertebrate dynein complexes contain either LIC1 or LIC2, but not both (Tynan et al., 2000a), an attractive idea is that they recruit specific cargoes. Indeed, LIC1, but not LIC2, binds to PC (Tynan et al., 2000b), whereas only LIC2 interacts with Par3 (Schmoranzer et al., 2009). However, both LICs can bind to the recycling endosome (RE) component FIP3 (Horgan et al., 2010a,b) and Rab-interacting lysosomal protein (Scherer et al., 2014), and the data for specific roles for LIC1 or 2 in membrane traffic are contradictory (Palmer et al., 2009; Tan et al., 2011; Hunt et al., 2013; Scherer et al., 2014). Furthermore, LICs act redundantly in centrosome anchoring to the NE in late G2 and in mitotic chromosome alignment, mitotic progression, and bipolar spindle maintenance (Raaijmakers et al., 2013).
Here, we investigate the role of LICs in cell division in cultured cells and in an organismal context using early embryos of Xenopus laevis. We show that although LICs are not needed for dynein’s motor activity in vitro, they are important for mitotic progression and the formation and maintenance of bipolar spindles. Upon depletion of LICs, spindle poles split apart in a process requiring the activity of Eg5, giving poles that often contain a single centriole. Because mother and daughter centrioles normally remain closely associated throughout mitosis, and only disengage during G1 (Mardin and Schiebel, 2012), this work reveals a novel role for dynein in maintaining centrosome integrity during mitosis.
Results
LICs are dispensable for motor activity in vitro
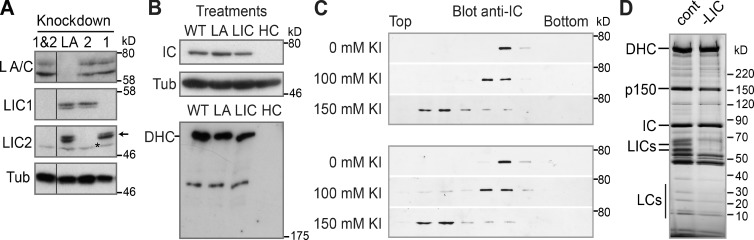
We depleted LIC1, LIC2, or both LICs from HeLaM cells (Fig. 1 A) using LIC1- or LIC2-specific siRNAs (Palmer et al., 2009). Comparable results were obtained using SMARTpools for LICs 1 and 2 (unpublished data). As previously reported (Tan et al., 2011), we found that depletion of both LICs did not affect human dynein complex integrity, because IC was stable (Fig. 1 B) and dynein migration on sucrose density gradients was unaffected (Fig. 1 C). In addition, dynein was similarly sensitive to the chaotropic agent KI, which disrupts interactions between dynein subunits (Fig. 1 C; King et al., 2002; Ori-McKenney et al., 2010).
Figure 1.
Loss of LICs does not affect dynein complex stability. (A) LIC KnD in HeLaM cells was assessed by immunoblotting with isoform-specific LIC antibodies (asterisk, a cross-reacting band; arrow, LIC2) with lamin A/C depletion (LA) as a control. Black line indicates that intervening lanes have been spliced out. Tub, tubulin. (B) Extracts made from untreated (wild type [WT]) or HeLaM cells depleted of lamin A/C, both LICs (LIC), or DHC (HC), were immunoblotted for IC and DHC, with tubulin as a loading control. (C) KI-treated extracts from control (cont.; top) or LIC1- and 2-depleted (bottom) HeLaM cells were run on sucrose gradients containing 0, 100, or 150 mM KI and immunoblotted for IC. (D) Silver-stained SDS-PAGE of dynein isolated from control and LIC1- and 2-depleted cells. Molecular mass markers (in kilodaltons) are shown.
We tested whether lack of LICs led to a loss of motor function in vitro. Dynein was purified from cells depleted of both LICs using microtubule binding and release followed by sucrose gradient sedimentation to separate dynein from kinesins (Fig. S1, A and B). Dynein purified from LIC-depleted or control cells appeared identical, save for the loss of the LICs (Fig. 1 D and Fig. S1 A). Dynein motor activity was assessed using in vitro microtubule gliding assays, in which multiple dyneins work together in a system with minimal drag. Control and LIC-free dynein both supported microtubule gliding (Video 1), and the frequency of movements was equally sensitive to dilution of the motor preparations (not depicted). Hence, their concentrations of active motor were approximately equivalent. Moreover, the mean speed of microtubule gliding was 0.57 µm/s ± 0.012 (SEM) for control and 0.56 µm/s ± 0.014 for LIC-free dynein (P = 0.632; Fig. S1 C). Microtubule gliding was completely inhibited by 50 µM sodium orthovanadate, which preferentially inhibits dynein (Cohn et al., 1989; Shimizu et al., 1995), whereas kinesin fractions were unaffected (unpublished data). Hence, LICs do not obviously influence dynein motility in vitro when multiple motors work together in the absence of cargo.
LIC depletion causes multipolar spindle formation and slows cell cycle progression in HeLaM cells
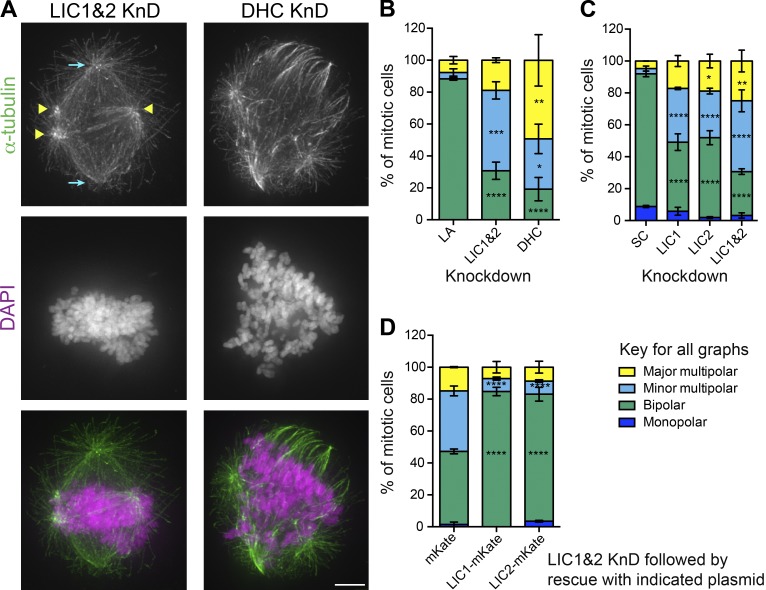
Although dynein lacking LICs drives microtubule gliding, previous studies have shown that LIC depletion inhibits many dynein functions in vivo, as described in the Introduction. We investigated the effect of LIC knockdown (KnD) on mitotic spindle assembly and stability in detail. Loss of both LICs led to ∼70% of mitotic cells having multipolar spindles (Fig. 2, A and B). These fell into two categories: cells with three or more poles with fairly similar densities of microtubules (major multipolar; Fig. S2 A), and those with a primarily bipolar spindle that had auxiliary poles (minor multipolar; Figs. 2 A and S2 A). Similar defects were seen in human embryonic kidney (HEK; Fig. S2 B) and U2OS cells (Fig. S4 A). Depletion of DHC in HeLaMs had more severe effects (P < 0.05, compared with LICs), generating multipolar spindles with often unfocused poles and highly disorganized chromosomes (Fig. 2, A and B).
Figure 2.
LICs 1 and 2 act redundantly in bipolar spindle assembly in HeLaM cells. (A) HeLaM cells depleted of both LICs (LIC1&2) or DHC were labeled with anti–α-tubulin and DAPI. Maximum projections of deconvolved z series are shown. Cyan arrows show major poles; yellow arrowheads show minor poles. Bar, 5 µm. (B–D) Quantitation of HeLaM spindle morphology (see Results for details). (B) Both LICs, DHC, or Lamin A/C (LA) were depleted. (C) KnDs with scrambled (SC), LIC1, LIC2, or both LIC1 and 2 siRNAs. For B and C, ≥100 cells were scored per condition; the mean of three independent experiments ± SEM is shown. (D) HeLaM cells were depleted of both LICs for 48 h and then transfected RNAi-resistant LIC1-mKate or LIC2-mKate, with mKate as a control (115, 112, and 235 cells were scored, respectively, from three independent experiments; means ± SEM). Two-way ANOVA results versus controls: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Because each dynein complex contains either LIC1 or LIC2 (Tynan et al., 2000b), we tested the effect of removing LICs individually. Loss of either LIC greatly increased the incidence of multipolar spindles in HeLaM cells but to a lesser extent than for the double depletion (Fig. 2 C, P ≤ 0.05 for LIC1 vs. LIC1&2 KnD and P ≤ 0.001 for LIC2 vs. LIC1&2 KnD). Importantly, the spindle defects in cells depleted of both LICs were rescued equally well by expression of RNAi-resistant LIC1-mKate or LIC2-mKate (Fig. 2 D), revealing that the two LICs act redundantly in spindle assembly in HeLaM cells.
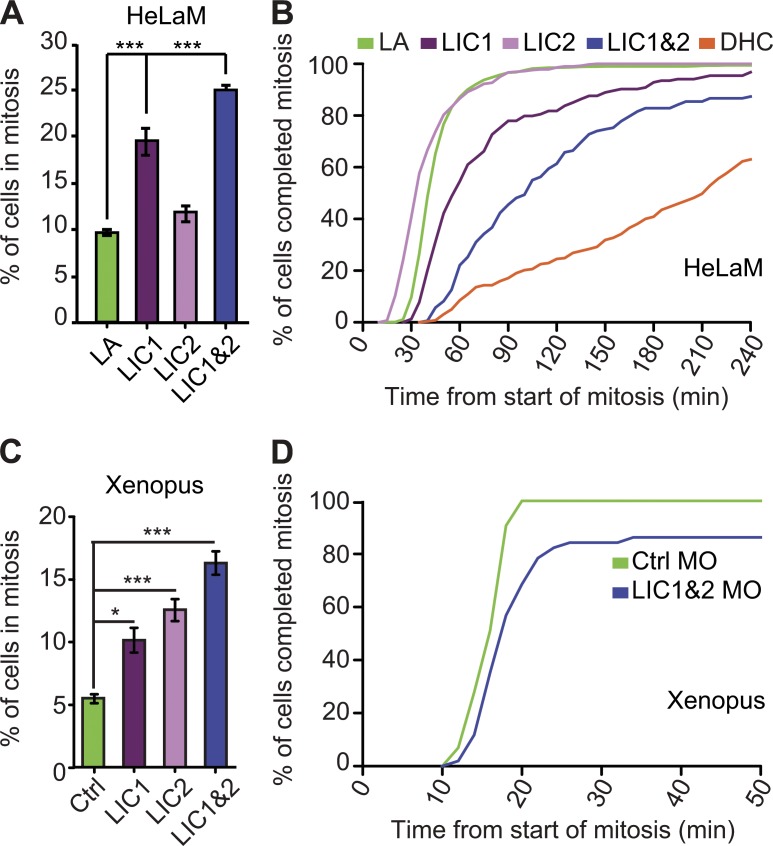
To test the effects of depleting LICs on mitotic progression in HeLaM cells, we determined the mitotic index and duration of mitosis. The mitotic index increased 2–2.5-fold after depletion of both LICs, or LIC1 alone, whereas removal of LIC2 had a small but statistically insignificant effect (Fig. 3 A), as recently reported (Raaijmakers et al., 2013). Moreover, depletion of LIC1 prolonged the duration of mitosis (P = 0.048), as previously seen (Sivaram et al., 2009), whereas loss of LIC2 had no effect (P = 0.815; Fig. 3 B), even though it caused spindle multipolarity (Fig. 2 C). Strikingly, depletion of both LICs slowed mitosis far more (P < 0.001) than LIC1 KnD, and 13% of cells failed to complete mitosis within 4 h (Fig. 3 B). However, DHC-depleted cells were more profoundly retarded (P < 0.001). In keeping with delayed mitotic progression, unaligned chromosomes were common after depletion of both LICs (Figs. S2 A and S3), and they displayed high levels of the SAC proteins Bub1 and BubR1 (Fig. S2 C). The chromosome misalignment was not caused by a loss of dynein from kinetochores (Fig. S2 D). Furthermore, the kinetochores of chromosomes at the metaphase plate were under tension, as judged by the extended centromeres and reduced Bub1 and BubR1 labeling (Fig. S2 C). There was also no evidence of premature loss of chromosome cohesion (cohesion fatigue), which can occur after prolonged metaphase arrest (27 cells from three experiments; Daum et al., 2011; Stevens et al., 2011). These results suggest that depletion of both LICs affects initial chromosome alignment but not the end-on K fiber–kinetochore attachments and associated SAC silencing, as previously reported (Raaijmakers et al., 2013).
Figure 3.
Mitosis is prolonged when both LICs are depleted. (A) Mitotic index of HeLaMs after KnDs (>731 cells in total per condition, three independent experiments; means ± SEM. ***, P < 0.001, Student’s t test). (B) The duration of mitosis after depleting LICs or DHC or lamin A/C (LA) was determined from phase contrast time-lapse videos. The plot shows the percentage of cells completing mitosis in a given time (158–1,133 cells per treatment combined from three independent experiments; see Results for survival analysis p-values). (C) Mitotic index of epithelial cells in Xenopus embryos injected with a control MO (Ctrl) or MOs targeting LIC 1, 2, or both LICs (*, P < 0.05; ***, P < 0.001; one-way ANOVA, n = 3 independent experiments; means ± SEM). (D) Mitotic progression in Xenopus embryos injected with control or LIC1 and LIC2 MOs was assessed by time-lapse confocal imaging and plotted as a cumulative frequency (n = 43 and 51 spindles analyzed in four and five independent experiments for control MO and LIC1 plus LIC2 MOs, respectively).
Multipolar spindles form in Xenopus embryos upon depletion of LICs
To test the role of LICs in an organismal context, we used morpholino (MO) oligonucleotides to knock down the LICs in Xenopus early embryos. Depletion of each LIC individually led to an increased mitotic index in gastrula stage embryos, but loss of both LICs had a greater effect (Fig. 3 C). Time-lapse confocal imaging of embryos expressing GFP–α-tubulin and mCherry-H2B (mCherry-histone2B) revealed a slight increase in mitosis duration after depletion of both LICs (Fig. 3 D). The mean time taken to complete mitosis was 26 min in LIC morphants compared with 15 min in controls (48 and 43 cells, respectively, from five independent experiments). However, some LIC morphant cells were particularly strongly affected, with 14% of cells failing to complete mitosis within 50 min (Fig. 3 D).
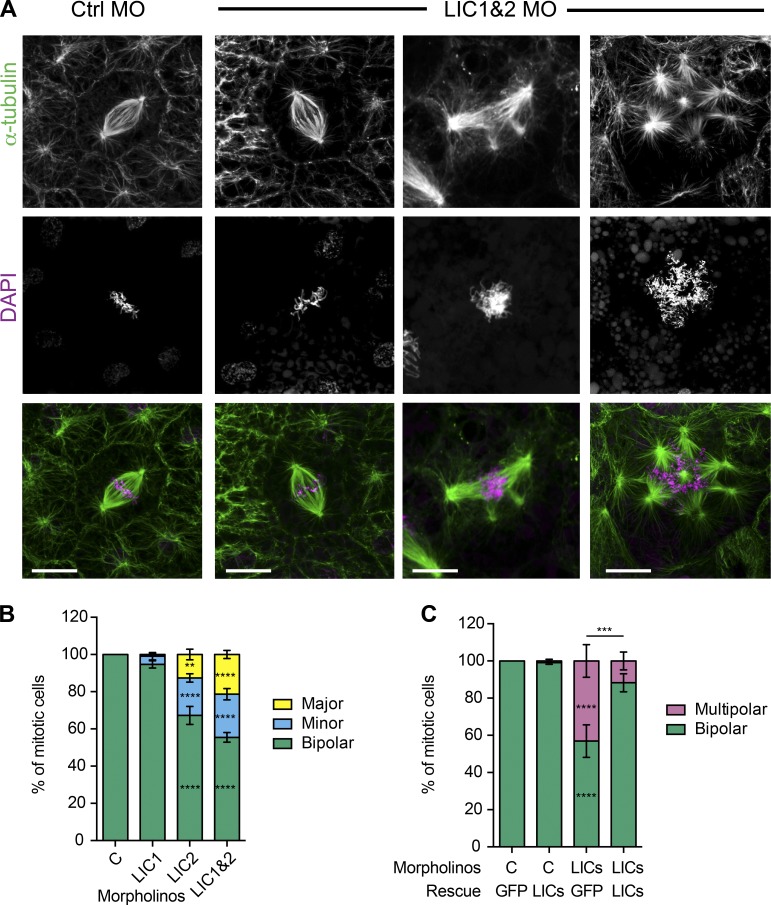
Depletion of LICs in Xenopus embryos disrupted spindle morphology, with multipolar spindles found in 44% of mitotic cells (Fig. 4, A and B) by developmental stage 10.5 (gastrulation), whereas no multipolar spindles were seen in controls. Unlike the multipolar spindles seen in the HeLaM cells, where the minor poles often had far fewer microtubules than the main poles and frequently overlapped the bipolar spindle, all auxiliary poles in embryo spindles were more easily distinguished (Fig. 4 A). We therefore categorized the multipolar phenotype in embryos by counting the number of auxiliary poles. Around half of the aberrant spindles had one or two extra poles (minor multipolar), whereas the rest had three or more additional poles (major multipolar; Fig. 4 B). Although depletion of LIC1 alone had only a slight effect on spindle morphology compared with LIC2 loss, depletion of both LICs gave a stronger phenotype than depletion of either LIC alone (Fig. 4 B), as seen in HeLaM cells. Importantly, the multipolar spindle phenotype was rescued by expression of GFP-tagged LICs (Fig. 4 C). Collectively, these data show that the LICs have overlapping activities during mitosis in two very different vertebrate cell systems but that the relative importance of LIC1 versus LIC2 differs.
Figure 4.
Depletion of LICs leads to multipolar spindles in Xenopus embryos. (A) Confocal images of spindles in embryos injected with control (Ctrl MO) or LIC 1 and 2 MOs (LIC1&2 MO) labeled with anti–α-tubulin and DAPI. Bars, 10 µm. (B) Quantification of spindle morphology in Xenopus embryos after depletion of LIC1, LIC2, or both LICs. Spindles categories: two poles = bipolar; three to four poles = minor multipolar; more than four poles = major multipolar. (n = 4 independent experiments, ≥27 embryos scored in total, 240–400 spindles categorized for each condition, means ± SEM). (C) Embryos injected with control or LIC1 and 2 MOs were rescued with a GFP plasmid or a mixture of plasmids encoding LICs 1 and 2 and scored as in B (a total of 98–159 mitotic cells from 13–18 embryos scored per condition, means ± SEM). Two-way ANOVA analysis versus controls: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. For other comparisons, see Results. C, control.
Two routes for forming multipolar spindles in Xenopus cells lacking LICs
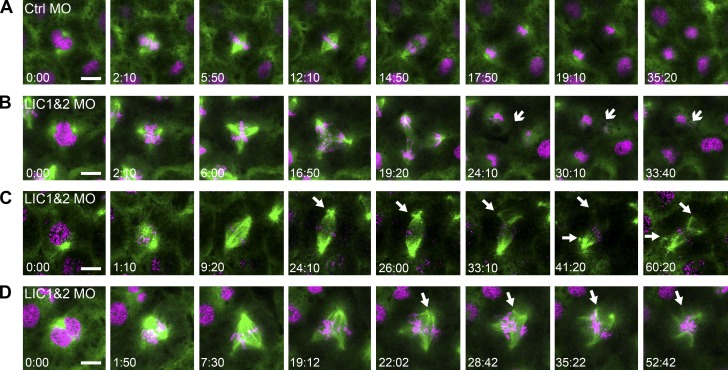
To investigate how the multipolar spindles arise, we imaged the mitotic spindle in living Xenopus embryos. Embryos were injected with mRNA encoding GFP–α-tubulin and mCherry-H2B along with standard control (Fig. 5 A and Video 2) or LIC1- and 2-targeted MO oligos (Fig. 5, B–D; and Videos 3–5). Live imaging of LIC morphants revealed that multipolar spindles arose by two different routes. We found that 40% of multipolar spindles in LIC morphants (8/20 from five independent experiments) had additional poles from the very start of mitosis, forming as the spindle assembled (Fig. 5, B and D; and Videos 3 and 5). However, the other 60% of multipolar spindles assembled first as bipolar spindles and then underwent a pole fragmentation event, in which an additional pole formed by breaking away from the bona fide spindle pole (Fig. 5 C and Video 4). We also saw instances where spindles assembled as multipolar spindles but then underwent pole fragmentation to form spindles with five or more poles (Fig. 5 D and Video 5). The presence of multiple poles affected the ability of cells to complete cytokinesis, which failed entirely or showed regression of one or more cytokinetic furrows in 10 out of 20 multipolar cells, leading to multinucleate cells (e.g., Fig. 5 B, arrows).
Figure 5.
Multipolar spindles form by two mechanisms in LIC1 and 2 morphant embryos. Mitotic spindles were imaged live in Xenopus embryos using GFP–α-tubulin (green) and mCherry-H2B (magenta). (A) Bipolar spindles assembled and proceeded normally through mitosis and cytokinesis in controls (Ctrl). (B) An LIC morphant that formed a multipolar spindle at the onset of mitosis and subsequently failed cytokinesis (arrows). (C) An LIC morphant that assembled a bipolar spindle, which became multipolar when spindle poles fragmented (arrows). (D) Pole fragmentation was also seen in multipolar LIC MO spindles (arrows). Bars, 10 µm; time stamps display minutes and seconds.
The pole fragmentation events were particularly intriguing, as they suggested a role for the LICs in the maintenance of centrosome integrity. Importantly, the fragmentation was not simply caused by a prolonged prometaphase arrest (e.g., Sluder and Rieder, 1985; Hinchcliffe et al., 1998; Hut et al., 2003) because it occurred on average only 17 min (±2 min, SEM; n = 12 spindles, in five independent experiments) after the start of mitosis.
Depletion of LICs results in premature centriole disengagement
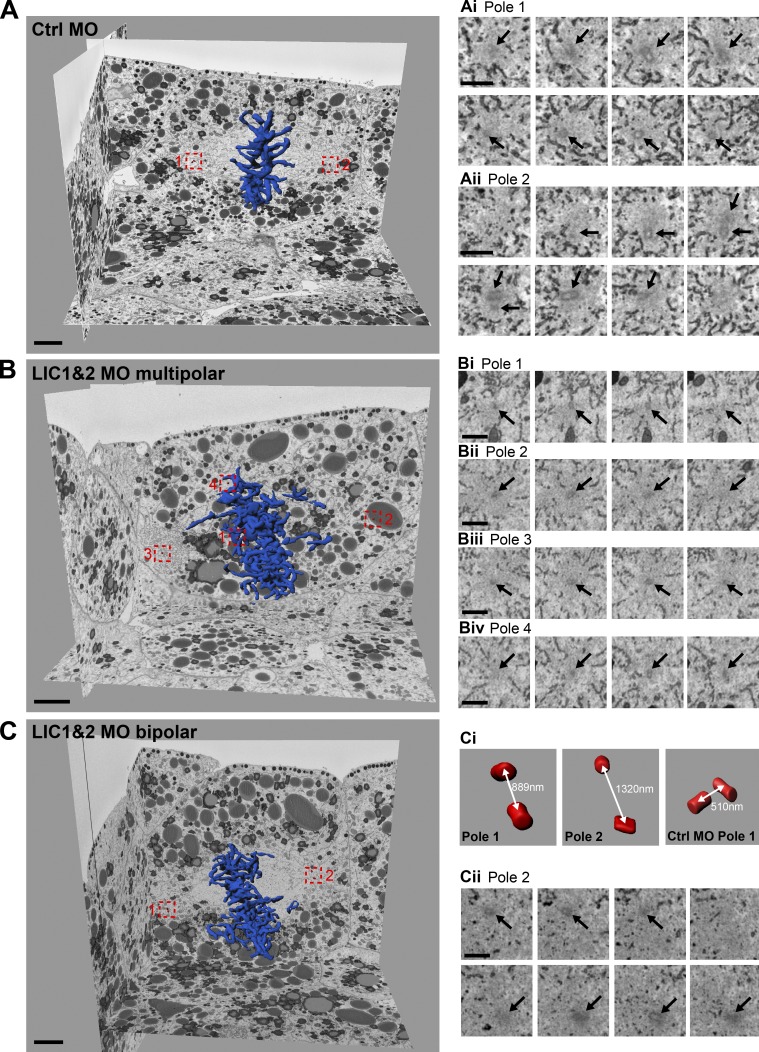
Spindle pole fragmentation can result from PCM fragmentation or premature disengagement of mother and daughter centrioles (Maiato and Logarinho, 2014). We therefore investigated whether the poles of multipolar spindles contained centrioles and, if so, whether these were pairs or single centrioles. We used 3View EM to assess cellular ultrastructure over a large z volume with sufficient resolution to distinguish paired centrioles in the Xenopus embryo. Although single microtubules could not be resolved, spindle poles were easily identified by following the trajectory of spindle-associated membranes (Tsai et al., 2006; Ma et al., 2009), which were unaffected by LIC depletion. In control cells, centriole pairs could be clearly seen at both poles of the bipolar spindle (Fig. 6, Ai and Aii; and Video 6). However, in LIC morphants the poles of multipolar spindles contained single centrioles (22 single centrioles and 12 pairs in 34 poles counted; Fig. 6, Bi–Biv; and Video 7). The presence of single centrioles suggested that premature centriole disengagement was occurring. In keeping with this, we found that in LIC morphant cells with bipolar spindles (Fig. 6, C–Cii), the distance between mother and daughter centrioles was significantly increased from 440 ± 35 nm (SEM; two cells and three centriole pairs) in controls to 696 ± 100 nm (SEM; 7 cells and 11 centriole pairs: P < 0.05) in LIC morphants.
Figure 6.
Multipolar LIC1 and 2 morphant spindle poles contain single centrioles. (A–C) Spindle pole composition was analyzed in control (Ctrl; A) and LIC1 and 2 morphant (B and C) embryos using 3View EM. Chromosomes (blue) and centrioles (red) were reconstructed using Imaris software. Sequential z slices of spindle poles showing a pair of centrioles in control (Ai and Aii) and single centrioles in a multipolar LIC morphant spindle (Bi–Bv). (Ci and Cii) An LIC morphant cell with a bipolar spindle has an increased distance (white double-headed arrows) between centrioles compared with control. Bars: (A, B, and C) 5 µm; (Ai, Aii, Bi–Biv, and Cii) 1 µm. Black arrows mark centrioles.
To compare the fate of centrioles after LIC depletion in human cells, we used a HeLa cell line stably expressing GFP-tagged centrin-1 as a marker for mother and daughter centrioles (Piel et al., 2000). Analysis of mitotic cells with minor multipolar spindles revealed premature centriole disengagement (Fig. 7 A), with 58% of poles having a single centriole (304 poles scored in 69 cells from two independent experiments). We could also rule out that centrioles were being overduplicated in interphase after LIC depletion (Fig. 7 B). The small increase in cells with more than four centrioles likely reflects cells that had previously failed cytokinesis. Indeed, LIC2 depletion leads to failed abscission (Palmer et al., 2009), and LIC1 is found at the midbody (Horgan et al., 2011). Labeling of HeLaM cells with antibodies to the centriole component CETN3 revealed that premature centriole disengagement occurred in cells depleted of LIC1 or 2 individually, as well as in combination (Fig. 7 C). The LIC2 KnDs confirm that cohesion fatigue plays no part in the pole fragmentation, as this treatment does not affect cell cycle progression (Fig. 3 B). We conclude that the LICs play a key role in vertebrate centriole cohesion because knockdown of the LICs leads to premature centriole disengagement and the formation of multipolar spindles.
Figure 7.
Premature centriole disengagement and PCM fragmentation occurs after LIC depletion in HeLa cells. (A, B, and D) Centrin-1–GFP HeLa cells treated with scrambled (control) or LIC 1 and 2 siRNAs were labeled with anti–α-tubulin, anti-PC, and DAPI. (A) Minor multipolar spindles in centrin-1–GFP HeLa cells. White arrowheads, mother centrioles; red arrowheads, daughter centrioles. PC spots without associated centrioles are common at minor poles (cyan arrows) and rare at major poles (yellow arrow). Bars, 5 µm. (B) Centriole number in interphase centrin-1–GFP HeLa cells was scored (>200 cells scored per condition in each of three to four independent experiments, means ± SEM). (C) HeLaM KnD cells were stained with anti-CETN3 to detect centrioles. Centriole number was counted per pole in 100 mitotic cells, in three independent experiments (means ± SEM). (D) Centriole and PC distribution at each major and minor spindle pole in LIC1- and 2-depleted centrin-1–GFP HeLa cells was scored (69 cells, from two independent experiments). ***, P < 0.001, Student’s t test.
Fragmentation of PCM contributes to multipolar spindle formation
One possible explanation for the effect of LIC KnD on spindle morphology is that dynein without LICs fails to recruit cargoes needed for spindle pole organization. Because PC is required for centriole cohesion (Matsuo et al., 2012) and interacts with LIC1 (Tynan et al., 2000b), we tested whether depletion of LICs led to loss of PC from the centrosome, so making the centriole–centriole linkage fragile. Surprisingly, we found that depletion of both LICs had little effect on PC localization in multipolar spindles in HeLaM (Fig. S3) or centrin-GFP HeLa cells (Fig. 7 A). Quantitation of PC levels showed no change in the total amount of PC at spindle poles versus controls (P = 0.15, n ≥ 41 HeLaM cells from four independent experiments).
We analyzed the distribution of PC in LIC-depleted centrin-GFP HeLa cells to determine whether each PC spot was associated with a centriole because the fragmentation of PCM can contribute to multipolar spindle formation (Maiato et al., 2004). Cells with the minor multipolar phenotype have a bipolar spindle with two major poles and a variable number of minor extra poles. PC was found at all major poles, regardless of whether they contained a single centriole or centriole pair, but only rarely did a major pole possess PC but no centriole (Fig. 7, A [yellow arrows] and D). Minor poles with single or paired centrioles always possessed PC. However, it was also common for minor poles to be PC positive but lack centrioles (Fig. 7, A [blue arrows] and D): PCM fragmentation was therefore taking place in addition to premature centriole disengagement. Notably, PC spots were almost always a focus for microtubules (Figs. 7 A and S3), suggesting that they act as nucleation/anchoring sites. Similar centriole-free PC spots with associated microtubules have been observed after depletion of a kinesin-3 (STARD9) and protein phosphase 2 (Torres et al., 2010, 2011). Both major poles were positive for PC in 98% of HEK cells and 93% of HeLaM cells (Fig. S4 B). In contrast, this was true in only 20% of U2OS cells (Fig. S4, A and B), suggesting a third route for multipolar spindle formation in this cell line, which generates acentrosomal poles. The centrosomes in LIC-depleted U2OS cells were often mislocalized to the very cell periphery in late G2/prophase (Fig. S4, C and D), as previously reported (Raaijmakers et al., 2013), but this was much less common in HeLaM cells (Fig. S4 E). Such peripheral centrosomes are likely not to contribute to bipolar spindle formation (Ferenz et al., 2009).
Localization of spindle assembly factors in LIC-depleted spindles
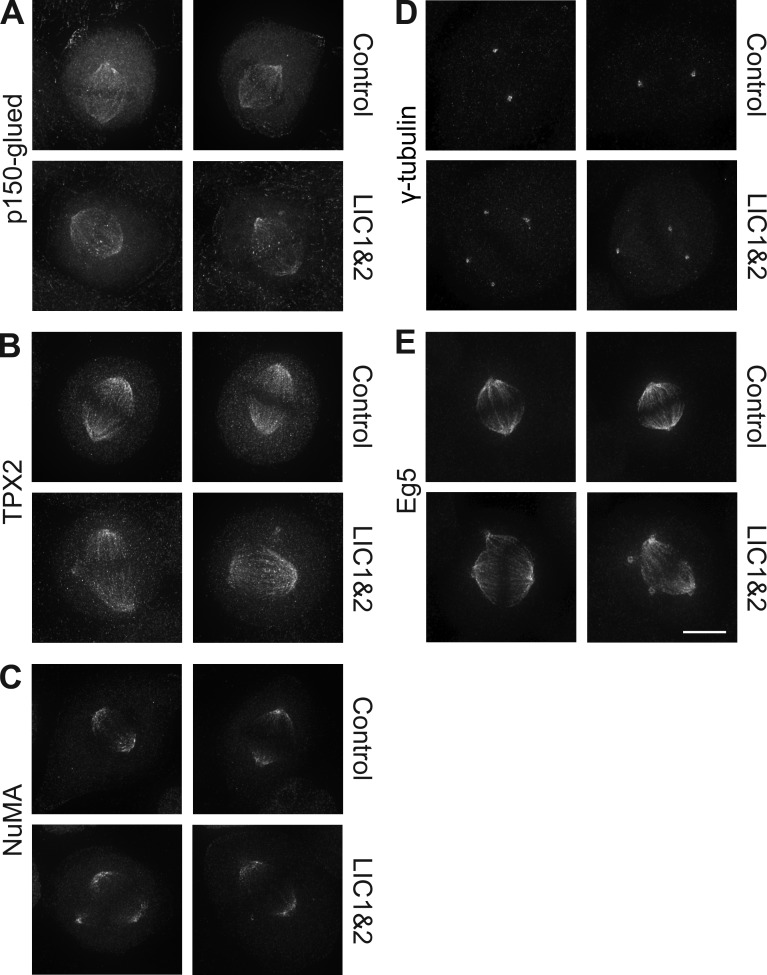
We next investigated whether dynein and dynactin were normally localized in spindles after LIC depletion. We could not visualize dynein in HeLaM spindles with available antibodies, but dynein localization in Xenopus embryo cells was unchanged in LIC morphants (Fig. S5 A). Dynactin (p150-glued) distribution was also unaffected in most LIC-depleted Xenopus (Fig. S5 B) and HeLaM cells (Fig. 8 A, P = 0.51 for individual pole intensity in control vs. LIC KnD, n ≥ 28 cells and three independent experiments). Dynactin was prominent at kinetochores in some LIC-depleted cells (Fig. 8 A), in keeping with defective chromosome alignment.
Figure 8.
LIC depletion does not affect the distribution of motor-associated components or γ-tubulin. HeLaM cells treated with scrambled or LIC1 and 2 siRNAs were fixed and labeled with the indicated antibodies, and cells with minor multipolar spindles were imaged using the same exposure for control and LIC-depleted cells. Maximum projections of deconvolved z series are shown. Bar, 5 µm.
Dynein and dynactin are involved in recruiting both NuMA and TPX2 (targeting protein for Xklp2) to spindles, which in turn contribute to correct spindle assembly (Merdes et al., 2000; Radulescu and Cleveland, 2010; Neumayer et al., 2014). However, loss of LICs in HeLaM cells or Xenopus embryos did not alter the localization of either TPX2 or NuMA to spindle poles and microtubules (Fig. 8, B and C; and Fig. S5, C and D). In addition, γ-tubulin, which is part of the microtubule nucleating γ-TuRC complex (Mardin and Schiebel, 2012), was tightly focused at all poles (Fig. 8 D and Fig. S5 E), in keeping with the ability of the additional poles to nucleate and anchor microtubules. Finally, Eg5 accumulated at normal levels at all spindle poles after LIC depletion (Fig. 8 E). Importantly, these observations were reflected in quantitation of HeLaM staining: for NuMA, γ-tubulin, and Eg5, intensity at individual poles was not significantly different between control and LIC KnD (P = 0.86, 0.31, and 0.89, respectively, n ≥ 25 cells, three independent experiments). TPX2 was slightly different, in that intensity was reduced when measured at individual poles (P < 0.0001, n ≥ 27 cells, three independent experiments) but not if the sum intensity of all poles in each cell was assessed, indicating that intensity is only reduced as a result of pole fragmentation.
The clustering of Rab11-positive REs has been reported to contribute to spindle and pole integrity (Hehnly and Doxsey, 2014), and because REs in interphase cells redistribute after LIC depletion (Palmer et al., 2009), alterations in RE positioning might contribute to the spindle phenotype we see. However, we found that only 6/236 control mitotic HeLaM cells had RE clustered at spindle poles, and furthermore, the mitotic phenotypes seen after loss of REs from spindle poles (Hehnly and Doxsey, 2014) are clearly distinct from those we observe for LIC depletion. The premature centriole splitting seen upon LIC KnD in HeLaM cells and Xenopus embryos is therefore unlikely to be caused by the loss of PC, TPX2, NuMA, γ-tubulin, Eg5, dynein, or dynactin from the spindle pole or to redistribution of REs.
Source of the forces driving centriole separation after LIC depletion
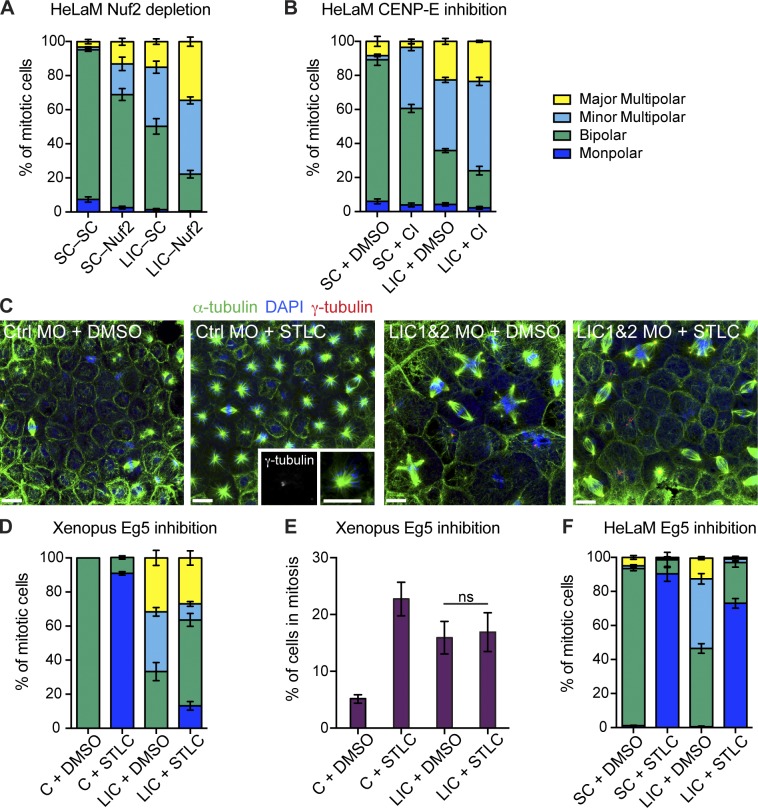
Because key spindle pole proteins were localized normally, we next examined whether the LIC KnD phenotype was caused by an alteration of forces within the spindle. For example, the forces exerted on spindle poles via K fibers play a part in spindle organization and function. Dynein pulls K fibers toward the poles (Goshima et al., 2005; Elting et al., 2014; Sikirzhytski et al., 2014), so generating inward tension, while kinesin-12, CENP-E, and chromokinesins push poles apart (Tanenbaum and Medema, 2010; Maiato and Logarinho, 2014). To test the role of K fiber–mediated forces in spindle assembly after LIC depletion, we codepleted the kinetochore component Nuf2 to prevent K-fiber formation (Manning and Compton, 2007; O’Connell et al., 2009; Toso et al., 2009; Logarinho et al., 2012). Nuf2 KnD alone generated multipolar spindles (Fig. 9 A and Tables S1 and S2 for statistical analysis; Logarinho et al., 2012) and in combination with LIC depletion led to >80% of cells having multipolar spindles. Because CENP-E inactivation also suppresses premature centriole splitting in some cases (Mattiuzzo et al., 2011; Logarinho et al., 2012), we inhibited CENP-E using GSK-923295 (Wood et al., 2010). This generated minor multipolar spindles in scrambled-treated cells, as previously seen (Thein et al., 2007), and also worsened the spindle defect in LIC-depleted cells (Fig. 9 B). Altogether, these results suggest that forces acting on poles via kinetochores and K fibers do not contribute to pole fragmentation after LIC depletion, but instead, their loss exacerbates the spindle defects.
Figure 9.
Inhibition of Eg5, but not loss of K fibers, prevents the formation of minor multipolar spindles. (A, B, and F) HeLaM cells were treated as indicated, labeled, and scored for mitotic phenotype (>100 cells in each of four independent experiments, means ± SEM). (A) Scrambled (SC) or LIC siRNA-treated HeLaM cells were incubated for 48 h and then transfected with scrambled or Nuf2 siRNAs and fixed after a further 24-h incubation. (B) Depleted cells were treated with DMSO or 100 nM CENP-E inhibitor (GSK-923295; CI) for 90 min before fixation. (C) Confocal images of mitotic spindles in control (Ctrl) or LIC1 and 2 Xenopus morphants treated with DMSO or STLC to inhibit Eg5. Spindles were stained with anti–α-tubulin, DAPI, and anti–γ-tubulin. Insets show zoom in of a monopolar spindle: two adjacent γ-tubulin foci indicate a single pole. Bar, 20 µm. (D) Quantitation of spindle morphology in control and LIC1 and 2 morphants treated with DMSO or STLC (means ± SEM, six independent experiments). (E) The mitotic index of control (C) and LIC morphants was determined ± STLC treatment (ns = not significant, Student’s t test, means ± SEM, n = 6 independent experiments). (F) Scrambled or LIC siRNA-treated HeLaM cells were treated with DMSO or 2 µM STLC for 2 h. See Tables S1 and S2 for two-way ANOVA results for A, B, D, and E.
We next tested the role of Eg5, which drives centrosome separation during bipolar spindle assembly. Inhibiting Eg5 with monastrol or S-trityl-l-cysteine (STLC) generates monopolar spindles (Mayer et al., 1999; Skoufias et al., 2006). Depletion or inhibition of dynein prevents the formation of monopolar spindles after Eg5 inhibition, indicating that Eg5 and dynein-driven forces normally function antagonistically to separate the centrosomes (Tanenbaum et al., 2008; Ferenz et al., 2009; Florian and Mayer, 2012; Raaijmakers et al., 2013). We examined spindle morphology in embryos treated with STLC or DMSO as a control. Embryos injected with control MO and treated with STLC developed monopolar spindles in >90% of mitotic cells (Fig. 9, C and D; and Tables S1 and S2). In contrast, LIC morphants formed few monopolar spindles after STLC treatment. Monopolar spindle formation upon Eg5 inhibition was also reduced in LIC-depleted HeLaM cells compared with controls (Fig. 9 F). Together, these results indicate that dynein without LICs is less able to generate the force necessary to collapse the spindle to a monopolar structure when Eg5 is inhibited.
Strikingly, treatment with STLC also significantly reduced the formation of spindles with one or two additional poles in LIC morphants and concomitantly increased the proportion of bipolar spindles (Fig. 9, C and D; and Tables S1 and S2). This was not caused by a change in the number of mitotic cells because STLC treatment did not significantly alter the mitotic index in LIC morphants (Fig. 9 E). Inhibition of Eg5 in HeLaM cells also rescued both the minor and major multipolar phenotypes in LIC-depleted cells (Fig. 9 F), although the rescued cells were more likely to have monopolar than bipolar spindles. Eg5-driven forces are therefore required to split the centrioles prematurely when LICs are depleted.
Discussion
Dynein plays crucial roles in the spindle in higher eukaryotes, and we find that many of these are affected by LIC depletion. A clear consequence of LIC loss is the formation of multipolar spindles, which we show occurs via two distinct major routes. One of these—the assembly of multipolar spindles in cells containing extra centrosomes—is commonly seen after reducing dynein activity using a range of approaches (Robinson et al., 1999; Wojcik et al., 2001; Maiato et al., 2004; Goshima et al., 2005; Morales-Mulia and Scholey, 2005; Quintyne et al., 2005; Tanenbaum et al., 2008; Firestone et al., 2012; Raaijmakers et al., 2013). The second main route involves the active fragmentation of poles after spindles have assembled, by a combination of premature centriole disengagement and PCM fragmentation. This phenotype, not previously described for dynein disruption, identifies a novel role for dynein in maintaining centrosomal structure during mitosis.
We find that Eg5 is the major driving force behind this pole splitting, because inhibiting Eg5 profoundly reduced the formation of minor additional poles. Although forces exerted via K fibers contribute to pole splitting in some situations (Maiato and Logarinho, 2014), this is not the case for LIC-depleted cells, because inhibition of CENP-E or the removal of K fibers by Nuf2 depletion both worsened spindle pole fragmentation, rather than rescuing the defect. Indeed, it is likely that K fibers normally contribute to spindle stability because dynein can mediate K-fiber sliding toward the poles (Goshima et al., 2005; Elting et al., 2014; Sikirzhytski et al., 2014), which would generate inwards pulling forces (Tanenbaum and Medema, 2010). Our data also suggest that the Eg5 forces that lead to fragmentation are exerted via the interpolar microtubules rather than the K fibers. Moreover, this excludes a role for kinesin-12 (and its interactor TPX2) in pole fragmentation because it is lost from the spindle after Nuf2 depletion, whereas Eg5 is not (Sturgill and Ohi, 2013). Importantly, both Eg5 and TPX2 are normally localized after LIC depletion, and the kinetochores of aligned chromosomes are under tension.
Exertion of a force sufficient to cause premature centriole disengagement requires that both centrioles have firmly attached microtubules. Although the mother centriole is the primary site of microtubule nucleation during interphase and mitosis (Wang et al., 2011), single daughter centrioles can clearly recruit PCM components and nucleate microtubules during late G2/prophase (Sluder and Rieder, 1985; Hut et al., 2003; Thein et al., 2007; Stevens et al., 2011; Kleylein-Sohn et al., 2012; Logarinho et al., 2012). Because dynein activity is needed for microtubule anchoring at the centrosome (Heald et al., 1997; Burakov et al., 2008), this might explain why DHC depletion does not lead to premature centriole separation but rather gives multipolar spindles with broad, disorganized poles that often consist of small microtubule bundles that lack associated γ-tubulin (Robinson et al., 1999; Wojcik et al., 2001; Maiato et al., 2004; Morales-Mulia and Scholey, 2005; Tanenbaum et al., 2008; Raaijmakers et al., 2013). Such spindles typically possess only two centrosomes, which are often dissociated from the spindle itself. In contrast, microtubules are clearly anchored to both mother and daughter centrioles in LIC-depleted cells and to the PC spots that are not associated with centrioles, which would allow Eg5 to drive pole fragmentation. The LICs are therefore not required for dynein’s microtubule anchoring activity. The microtubules are also well focused, suggesting that components such as NuMA and TPX2 are not only localized normally but are fully functional.
A key question is why LIC loss should allow Eg5 to drive apart centrioles and fragment the PCM. Centrosomes normally resist the forces exerted on them during spindle assembly, and the centrioles only come apart in late mitosis or early G1. In vertebrates, this involves separase-mediated cleavage of centrosomal cohesin (Tsou and Stearns, 2006; Ban et al., 2007; Wang et al., 2008; Nakamura et al., 2009; Tsou et al., 2009). One possibility is that centriole disengagement is triggered too early after LIC depletion, perhaps as a result of reduced recruitment of cohesin or proteins, such as Emi, that block cohesin cleavage (Ban et al., 2007; Wang et al., 2008; Nakamura et al., 2009). Indeed, cohesin and Emi interact with NuMA (Ban et al., 2007; Kong et al., 2009), whereas Emi, NuMA, and dynactin rely on each other for centrosomal localization (Ban et al., 2007). However, because LIC depletion does not affect NuMA, dynactin, or dynein localization, the centriole disengagement we see is unlikely to be caused by loss of Emi. Astrin and kinastrin also protect cohesin from premature separase cleavage, and their depletion leads to multipolar spindles with single centrioles at poles (Thein et al., 2007; Dunsch et al., 2011). Although both proteins are part of a complex that includes the dynein light chain DYNLL1 (Schmidt et al., 2010; Dunsch et al., 2011), this complex is needed for spindle positioning rather than centriole cohesion (Dunsch et al., 2012). Importantly, the centriole disengagement seen after manipulation of cohesin cleavage usually occurs after a long prometaphase arrest and premature sister chromatid separation (Maiato and Logarinho, 2014), which we do not see.
Cohesin and separase are not the only components that maintain centriole cohesion (Daum et al., 2011; Stevens et al., 2011; Oliveira and Nasmyth, 2013): structural PCM components may also play an important role (Cabral et al., 2013), some of which are transported by dynein (Doxsey et al., 2005). Of these, PC is particularly interesting, as it interacts with LIC1 but not LIC2 (Tynan et al., 2000b) and is cleaved by separase (Matsuo et al., 2012). However, although depleting LICs led to PC being redistributed to extra poles, the total amount of PC at poles was not affected. Furthermore, depletion of LIC2 alone led to spindle pole fragmentation.
Alternatively, pole fragmentation may result from an imbalance of forces within the spindle poles, primarily between dynein and Eg5. This would be analogous to the well-known opposing roles these motors play in setting up the bipolar spindle, where both motors exert force on antiparallel microtubules nucleated by the two centrosomes. This would occur primarily on interpolar microtubules, as discussed earlier. We propose that a similar situation exists between microtubules nucleated by mother and daughter centrioles during mitosis and within the PCM surrounding each centrosome, with dynein pulling the structures closer together and Eg5 pushing/pulling them apart. After LIC depletion, Eg5 forces likely outweigh those generated by dynein. This is not because of dynein or dynactin mislocalization, however. Instead, we suggest that the loss of LICs reduces dynein activity in a crucial way in a cellular context. Perhaps it generates less force, is less processive, or is less able to interact with regulatory molecules. In all these scenarios, the ability of dynein to drive cargoes under high loads, as in the spindle, will be reduced.
An observation that supports our proposal that LIC depletion reduces dynein activity in vivo comes from cells that enter mitosis with additional centrosomes. Many cancer cell lines with such extra centrosomes can nevertheless form bipolar spindles as a result of dynein and KIFC1 cooperating to cluster the centrosomes into two spindle poles, counteracting the Eg5 and K fiber–dependent forces that push centrosomes apart (Quintyne et al., 2005; Kwon et al., 2008; Leber et al., 2010; Drosopoulos et al., 2014). However, when dynein or KIFC1 is inactivated, such cells form multipolar spindles. We see the same phenotype in LIC-depleted cells with extra centrosomes, demonstrating that dynein’s ability to counter Eg5-generated forces is compromised.
Reduced force generation in cells with normal centrosome number is also revealed when we inhibit Eg5. This normally leads to the formation of monopolar spindles, caused in part by dynein’s ability to cause antiparallel microtubule sliding (Ferenz et al., 2009; Tanenbaum et al., 2013), but when dynein is also inactivated, bipolar spindles can form (Tanenbaum et al., 2008; Ferenz et al., 2009; Florian and Mayer, 2012; Raaijmakers et al., 2013). The fact that LIC depletion rescues bipolarity in the presence of STLC (Fig. 9; Raaijmakers et al., 2013) strongly suggests that loss of LICs compromises dynein function. Furthermore, bipolar chromosome alignment on the spindle is delayed, despite dynein and dynactin being present at kinetochores. In addition, the detachment of centrosomes from the nucleus in prophase (this work; Raaijmakers et al., 2013) suggests that the dynein at the NE has reduced activity.
How does this proposal fit with our finding that purified dynein can drive microtubule gliding in vitro normally? Recent work has shown that although purified dynein is active in gliding assays in which many motors act together, single dynein molecules are not able to move along microtubules unless they are in a complex with dynactin and one of several adaptor molecules (McKenney et al., 2014; Schlager et al., 2014). The LIC1 and 2 interactor FIP3 is one such adaptor molecule. Perhaps the lack of LICs hinders the formation of a subset of these complexes, impairing certain dynein functions but leaving others intact, such as microtubule anchoring and focusing of spindle poles. Interestingly, although LIC1 and 2 have some distinct interactors and functions, we find that the LICs act redundantly in spindle formation because depletion of either LIC causes premature centriole disengagement, and overexpression of either LIC will rescue spindle bipolarity. Redundant roles for LICs in cell cycle progression, chromosome alignment, and centrosome–NE anchoring have also been reported (Raaijmakers et al., 2013). Given that invertebrates have only a single LIC gene, the redundancy in mitotic LIC functions in vertebrates suggests that these are key ancestral roles for metazoan dynein.
Altogether, our results indicate that, as well as counteracting each other in the spindle to maintain centrosome separation, Eg5 and dynein also function antagonistically within the centrosome to maintain centriole cohesion and that the LICs are key for dynein’s role in this process. This novel role has become apparent because LIC depletion does not disrupt the attachment of microtubules to the centrioles, unlike DHC depletion. Our findings are consistent with a model whereby Eg5 produces an outward force that drives the centrioles apart, which is counteracted by an inward LIC-dependent dynein-driven force. This balance of forces could help hold mother and daughter centrioles together during mitosis: a slight change in the balance could then assist centriole disengagement once mitosis is complete.
Materials and methods
Reagents
This project used mouse antibodies to dynactin p150 (BD), dynein IC (IC74; EMD Millipore), GM130 mouse (BD), NuMA (Oncogene), phospho–histone H3 (EMD Millipore), TAT1 (K. Gull, University of Oxford, Oxford, England, UK), or DM1A (Sigma-Aldrich). The following rabbit antibodies were used: DHC (Sigma-Aldrich), KIF5B (R. Vale, University of California, San Francisco, San Francisco, CA), PC (Abcam), KIF1B-β (Bethyl Laboratories, Inc.), KIF1C (Abcam), KIF11 (Eg5; Sigma-Aldrich), LIC2 (R. Vallee, Columbia University Medical Center, New York, NY; Tan et al., 2011); NuMA (A. Merdes, Paul Sabatier University, Toulouse, France), TPX2 (C. Wiese, University of Wisconsin-Madison, Madison, WI; O’Brien and Wiese, 2006), and γ-tubulin (Keating and Borisy, 2000). The following sheep antibodies were used: Bub1, BubR1, and CENP-F (S. Taylor, University of Manchester, Manchester, England, UK; Taylor et al., 2001; Hussein and Taylor, 2002), dynein N-96 anti–Xenopus IC (Lane and Allan, 1999), and lamin A/C (Santa Cruz Biotechnology, Inc.). In addition, rat anti–α-tubulin, YOL-1 (J. Murray, University of Pennsylvania, Philadelphia, PA), human anticentromere antibodies (S. Taylor), and chicken LIC1 (R. Vallee; Tan et al., 2011) were used. Fluorescently labeled secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. or Invitrogen. IRDye 700CW– and 800C-labeled secondary antibodies were obtained from LI-COR Biosciences. HRP-conjugated secondary antibodies were purchased from Dako.
Cell culture and transfection
HeLaM and HEK cells were grown in DMEM and 10% FCS at 37°C and 8% CO2. HeLa cells stably expressing centrin-1–GFP (Piel et al., 2000) were grown in the same medium but with the addition of G418. U2OS (human osteosarcoma cells) cells were grown in McCoy’s 5A and 10% FCS at 37°C with 5% CO2. siRNA transfections used INTERFERin (MP Biomedicals). siRNAs were obtained from Eurofins MWG Operon, except siGENOME lamin A/C, scrambled control, and Nuf2 (Thermo Fisher Scientific). For LICs, cells were analyzed 48 h after transfection with 20 nM (mitosis experiments) or 72 h after transfection with 5–20 nM oligonucleotide (other experiments). For DHC, three siRNAs (6.67 nM each) were used together, and cells were analyzed after 72 h. The following sequences were used: LIC1, 5′-AGAUGACAGUGUAGUUGUA-3′ (Palmer et al., 2009); LIC2, 5′-ACCUCGACUUGUUGUAUAA-3′ (Palmer et al., 2009); DHC 1a, 5′-ACAUCAACAUAGACAUUCA-3′; DHC 1b, 5′-GAGAGGAGGUUAUGUUUAA-3′; and DHC 1c, 5′-GCAAGAAUGUCGCUAAAUU-3′. For Nuf2 codepletion, cells were treated with scrambled or LIC1 and 2 siRNAs and then 48 h later were treated with either scrambled or a Nuf2 SMARTpool and fixed after a further 24 h.
Human LIC1 and LIC2 (GenBank accession nos. AF078849 and AF035812) were extracted from a HeLa cDNA library and cloned into pTurboFP635-C (mKate; Evrogen). Noncoding mutations resulting in siRNA-resistant LIC1 and LIC2 were generated by PCR-based mutagenesis using the following primers: LIC1, 5′-CAAAGATGACAGTGTCGTCGTACCTCTGGGTGCG-3′; and LIC2, 5′-GAGAAAAACCTCGACTTGCTATACAAGTATATTGTTC-3′. For rescues, cells were transfected with scrambled or LIC1 and 2 siRNAs for 48 h and then transfected in fresh media with siRNA-resistant LIC1-mKate or LIC2-mKate using FuGENE 6 (Promega) and fixed 24 h later.
Xenopus microinjection and MOs
Female Xenopus frogs were preprimed 4–7 d in advance with 50 U of pregnant mare serum gonadotropin (Intervet UK) and then primed with 500 U of human chorionic gonadotropin (Intervet UK) 18 h before use. Frogs were transferred to Marc’s modified Ringer’s (MMR; 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM Hepes, pH 7.4) for egg collection. In vitro fertilization was performed by adding macerated testis to the eggs. After 30 min, embryos were dejellied in 2% cysteine (in 0.1× MMR) and rinsed five times in 0.1× MMR.
Embryos were microinjected into each cell at the two- or four-cell stage in 0.1× MMR plus 5% Ficoll using a Picospritzer III (Parker Instrumentation). The same conditions were used throughout this work. The needle volume was set to 5 or 2.5 nl, respectively. MOs to LIC1 and LIC2, stored as a stock solution of 1 mM diluted in water, were heated for 5 min at 65°C before being further diluted in standard control (MO sequence, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; Gene Tools, LLC) to the desired concentration. Translation-blocking MOs were designed against Xenopus LICs as follows: LIC1 (MO sequence, 5′-CATCCTCTACCACCCGTGCTCTT-3′; Gene Tools, LLC) and LIC2 (MO sequence, 5′-GCAGCTTCCTCTCCCCCGCCATCTT-3′; Gene Tools, LLC) and were injected at a needle concentration of 0.25 mM. After microinjection, embryos were incubated at 16°C.
For rescue experiments, Xenopus LIC1 and LIC2 were cloned from IMAGE (Integrated Molecular Analysis of Genomes and their Expression) cDNA clones (7011679 and 8074560, respectively) and N-terminally tagged with EGFP by insertion into a custom pCS2 ± N-EGFP vector. Full-length LIC1 was used, but LIC2 was slightly truncated at the N terminus to remove the first five amino acids, which corresponded to the LIC2 MO sequence, and was also truncated by 109 amino acids at the C terminus (LIC2aa6–378). Rescue experiments were performed by microinjecting both LIC1 and LIC2 translation-blocking MOs into each cell at the two-cell stage and then injecting capped mRNA encoding LIC1 and LIC2aa6–378 into all cells of the four-cell stage embryo.
Drug treatment
For Xenopus experiments, STLC (Sigma-Aldrich; 50 mM stock in DMSO) was diluted to 1 mM in PBS and injected into the blastocoel of MO-treated embryos 21.5 h after fertilization (16°C). The needle volume was set to 18 nl, and each embryo was injected twice, into each side of the blastocoel. Control embryos were injected with the same volumes of DMSO diluted 1:50 in PBS. Embryos were incubated at RT for 2 h before fixation.
For HeLaM drug treatments, cells were transfected with scrambled or LIC1 and 2 siRNAs and incubated for 70–72 h. Drug treatments were performed in DMEM + 10% FCS at 37°C followed by fixation and immunofluorescence analysis. Eg5 was inhibited with 2 µM STLC for 2 h. CENP-E was inhibited with 100 nM GSK-923295 (10 mM stock in DMSO; Medchem Express; Wood et al., 2010) for 90 min.
Immunofluorescence
Typically, Xenopus embryos were fixed for immunofluorescence 21.5 h after fertilization (16°C; stage 10.5) and processed as described previously (Danilchik et al., 1998), omitting the methanol postfix step: embryos were fixed by overnight incubation at RT in microtubule fix (3.7% paraformaldehyde, 0.25% glutaraldehyde, and 0.2% Triton X-100 in microtubule assembly buffer [80 mM Pipes, 5 mM EGTA, and 1 mM MgCl2, buffered to pH 6.8 using KOH]). Fixed embryos were then bisected along the sagittal axis, quenched for ≥1 h in 100 mM sodium borohydride and bleached for 90 min in 10% hydrogen peroxide (both made up in PBS). After bleaching, samples were blocked overnight at 4°C in TBSN/BSA (TBS [155 mM NaCl and 10 mM Tris-Cl, pH 7.4], 0.1% Nonidet P-40, and 10 mg/ml BSA). For dynein and dynactin localization, using the N-96 and p150 antibodies, respectively, and for NuMA staining, embryos were fixed in Dent’s fixative (80% vol/vol MeOH and 20% vol/vol DMSO) for 2 h at RT or overnight at −20°C. All fixed embryos were incubated in primary and secondary antibodies in TBSN/BSA overnight at 4°C, with five 1-h washes with TBSN/BSA after each incubation. To stain DNA, DAPI (Invitrogen), at a final concentration of 10 µg/ml, was added to one of the final TBSN washes and incubated for 30 min at RT. After staining, embryos were dehydrated in increasingly more concentrated methanol/TBSN washes and cleared and mounted in Murray’s clear (2:1 benzyl benzoate/benzyl alcohol; Klymkowsky and Hanken, 1991). Bisected embryos were mounted with the cut edge adjacent to the microscope slide.
For human cell lines, cells were grown on coverslips. For labeling with antibodies to tubulin and PC, cells were fixed for 2–10 min at 37°C in freshly diluted 3% formaldehyde/0.075% glutaraldehyde with 1% Triton X-100 in buffer A (80 mM K-Pipes, 1 mM MgCl2, 5 mM EGTA, 0.05 mM EDTA, and 4% [wt/vol] polyethylene glycol 8000, pH 6.8) followed by 15 min in 3% formaldehyde/0.075% glutaraldehyde in buffer A at RT. They were permeabilized for 30 min at RT in 1% Triton X-100 in buffer A, quenched in 1 mg/ml sodium borohydride, and then labeled. Alternatively, cells were fixed in −20°C methanol and labeled with anticentromere antibodies or antibodies to BubR1, CENP-F, dynactin p150Glued, dynein IC, Eg5, γ-tubulin, NuMA, phospho–histone H3, or TPX2. For kinetochore labeling, cells were transferred to cold DMEM + 10% FCS containing 10 µM nocodazole, returned to the 37°C incubator for 60 min, and then fixed in −20°C methanol. Secondary antibodies labeled with Alexa Fluor 488, Alexa Fluor 594, Cy3, or Cy5 were used along with 1 µg/ml DAPI, and samples were mounted in ProLong Gold (Invitrogen).
Microscopy and image analysis
Fixed, labeled HeLaM cells were imaged using 60×, 1.4 NA Plan Apochromat or 100×, 1.35 NA U Plan Fluor objectives on a microscope (IX70; Olympus) equipped for optical sectioning microscopy (DeltaVision; Applied Precision) using the Sedat filter set (89000; Chroma Technology Corp.) and a camera (CoolSNAP HQ; Photometrics). Each z series (0.2-µm intervals) was deconvolved and projected using SoftWoRx (Applied Precision) with the exception of Fig. S5 C, in which the z series was reconstructed using the ImageJ (National Institutes of Health) extended depth of field plugin. In Figs. 7 A and S2 C, a single deconvolved DAPI image from the center of the z stack is shown. For quantification of the localization of spindle pole proteins in control and LIC KD cells (Fig. 8), z projections were analyzed in ImageJ by drawing around spindle poles by hand and measuring total intensity (IntDen) using the Measure function. The IntDen values for each pole in a spindle were then summed to give a spindle total intensity value. To allow multiple experiments to be compared, each spindle total intensity was normalized by dividing it by the mean spindle total intensity of control spindles from the same experiment. A microscope (BX60; Olympus), with 60×, 1.4 NA Plan Apochromat or 100×, 1.35 NA U Plan Fluor objectives and a camera (CoolSNAP ES), using MetaVue (Molecular Devices), was used for manual scoring of cell phenotypes. For scoring the localization of centrioles and PC in centrin-1–GFP HeLa cells, 21 DeltaVision images were used along with visual scoring of 48 cells from two independent experiments.
Fixed Xenopus embryos were mounted in Murray’s Clear (2:1 benzyl benzoate/benzyl alcohol) and imaged at RT using an inverted confocal (TCS SP5 AOBS; Leica) with 40×, 1.25 NA HCX Plan Apochromat or 63×, 1.4 NA HCX Plan Apochromat oil objectives and LAS AF acquisition software (Leica). The confocal settings were as follows: pinhole of 1 airy unit, sequential scans, and 512 × 512 format. When acquiring 3D optical stacks, z sections were collected at 0.5- or 1-µm intervals. Only the maximum intensity projections of these 3D stacks are shown in the results. All images were scaled using linear transformations in Photoshop CS (Adobe), MetaVue, or ImageJ; Photoshop CS and Illustrator CS were used to construct the final figures.
Live-cell imaging
To score mitotic index, HeLaM cells grown on coverslips were imaged on a simple phase-contrast microscope without fixation, and cells in mitosis and interphase were counted in several fields. To image the duration of mitosis in HeLaMs, cells were grown in glass-bottomed 24-well plates (VWR International), transfected with siRNA duplexes, and imaged 42 h later at 37°C in Ham’s F12 media with 10% FCS on a microscope (IX81; Olympus) fitted with a H117 stage (Proscan; Prior Scientific). Phase-contrast images were acquired, using point visiting, every 5 min for 17 h, using a 20×, 0.50 NA Plan Fluotar phase objective, a light-emitting diode (λ = 530 nm; Cairn Research), and an electron-multiplying charge-coupled device camera (Cascade; Photometrics). Mitosis duration was defined as the interval between the first sign of cell rounding to the first sign of cytokinesis.
For live imaging of mitotic spindles in Xenopus embryos, both cells of two-cell embryos were injected with MO before being microinjected with 2.5 nl of mRNA for EGFP–α-tubulin (needle concentration of 0.5 mg/ml) or mCherry–H2B (0.1 mg/ml) into each cell of the four-cell embryo. Embryos were incubated for 20 h (postfertilization) at 16°C and then mounted for live imaging in 0.1× MMR, using a ring of vacuum grease to contain the embryos and support a glass coverslip (Woolner et al., 2009). Imaging took place at developmental stages 10–11, so covering the 10.5 gastrula stage used in our fixed analysis. Single focal plane live-cell images of spindles were collected at RT (∼21°C) using a confocal microscope (FluoView FV1000; Olympus) with FluoView acquisition software (Olympus) and a 60×, 1.35 NA U Plan S Apochromat objective. Time-lapse videos were constructed from the single focal plane images using ImageJ.
Scanning EM
Embryos were injected with standard control or LIC1 and LIC2 MOs and fixed in 3.7% paraformaldehyde and 0.25% glutaraldehyde in BRB80 (80 mM Pipes, 1 mM MgCl2, and 1 mM EGTA, pH 6.8) 21.5 h after fertilization (incubated at 16°C). Samples were then processed using a high density staining method suitable for block face imaging (see supplementary protocol in Williams et al., 2011). In brief, the samples were fixed for 1 h in 2% (wt/vol) osmium tetroxide and 1.5% (wt/vol) potassium ferrocyanide in cacodylate buffer followed by 20 min in freshly prepared 1% (wt/vol) thiocarbohydrazide and then 30 min in 2% (wt/vol) osmium tetroxide followed by 1% (wt/vol) uranyl acetate overnight at 4°C. The samples were then stained with freshly prepared Walton’s lead aspartate (0.02 M in lead nitrate and 0.03 M in aspartic acid, adjusted to pH 5.5) for 30 min followed by dehydration, embedding in Epon 812 (hard formulation), and trimming on a standard microtome. The samples were examined using a microtome (3View; Gatan) within a scanning electron microscope (Quanta 250 FEG; FEI). The imaging conditions were as follows: indicated quadrant magnification of 1,600×, accelerating voltage of 3.8 kV, pressure at 0.33 Torr, image at 4,000 × 5,000 pixels, and dwell time of 10 µs. Raw data were converted to an MRC file stack using IMOD (Kremer et al., 1996; procedures discussed in detail in Starborg et al., 2013). Imaging noise was removed using 2D Gaussian smoothing with a 3 × 3 kernel to aid manual segmentation. Regions of interest were cropped out using IMOD for examination and segmentation with Imaris (Bitplane). Chromosomes and centrioles were reconstructed with Imaris using the Surfaces tool. To measure the distance between centriole pairs, the Measurement points tool in Imaris was used to select the center of the reconstructed surface for each centriole, and the distance between pairs was measured.
Biochemical analysis
Total HeLaM cell extracts for immunoblotting analysis were prepared by lysis in hot SDS sample buffer. For sucrose gradient analysis after KI treatment, a 10-cm dish of HeLaM cells was used per condition. Cells were trypsinized, washed in complete medium, and then washed twice in BRB80. Cells were lysed for 15 min on ice in 0.5 ml BRB80 plus 0.5% Triton X-100 containing Protease Inhibitor Cocktail III (Sigma-Aldrich). Lysates were centrifuged at 14,000 rpm for 10 min at 4°C and then incubated on ice for 1 h after the addition of 0, 100, or 150 mM KI (Ori-McKenney et al., 2010). Supernatants (45 µl) were applied to 5–20% (wt/vol) continuous sucrose gradients in BRB80 or BRB80 plus 100 or 150 mM KI, as appropriate, prepared in 650-µl centrifuge tubes (Beckman Coulter). Samples were centrifuged in a rotor (SW 55; Beckman Coulter) with adaptors, for 5 h at 38,000 rpm, 4°C, and 75-µl fractions were collected.
For dynein purification, four 15-cm dishes of HeLaM cells were transfected with LIC1/2 siRNAs, cultured overnight, and then split onto 12 dishes. Control dynein was prepared from eight 15-cm dishes of untransfected cells. Cells were trypsinized, washed in complete medium, and then washed twice in BRB80. Cells were lysed for 15 min on ice in 2 vol BRB80 with 1% Triton X-100, 1 mM DTT, 0.2 mM PMSF, 10 µg/ml protease inhibitors (leupeptin, aprotinin, pepstatin, and chymostatin), and 10 µg/ml cytochalasin D. Lysates were centrifuged at 2,500 rpm for 15 min at 4°C, and then, the supernatants were spun at 110,000 gav for 30 min at 4°C. The resulting supernatants were supplemented with 10 U/ml hexokinase, 20 mM glucose, 400 µM AMP-PNP (adenosine 5′-[β,γ-imido]triphosphate), 20 µM taxol, 1 mM DTT, 0.5 µg/ml cytochalasin D, and 100 µg/ml taxol-stabilized microtubules prepared from bovine brain tubulin (Cytoskeleton, Inc.). After 20 min at 30°C, microtubules were pelleted at 70,000 gav in a rotor (TLS-55; Beckman Coulter) at 25°C for 10 min through a 20% (wt/vol) sucrose cushion in BRB80, 1 mM DTT, 4 µM taxol, 1 µg/ml cytochalasin D, and 2.5 µg/ml protease inhibitors. Pellets were washed in BRB80, 1 mM DTT, 20 µM taxol, 10 µg/ml cytochalasin D, and 10 µg/ml protease inhibitors and spun as in the previous step. Pellets were resuspended in 50 µl of the same buffer containing 5 mM MgATP and incubated for 25 min at 30°C to release motors. Samples were then spun for 10 min at 70,000 gav at 22°C (TLS55). Supernatants were collected and loaded onto 600 µl 8–20% (wt/vol) sucrose continuous density gradients in BRB80 containing 2.5 µg/ml protease inhibitors and 1 mM DTT. Gradients were centrifuged at 38,000 rpm for 6 h at 4°C in a rotor (MLS-50; Beckman Coulter) with adaptors, and then, 50-µl fractions were collected and analyzed by SDS-PAGE on 4–15% or 5–15% gels followed by silver staining or immunoblotting using antibodies against kinesins and used for motility assays.
In vitro motility assays
Microscope flow chambers were made (Allan, 1993) using coverslips cleaned by sonication for 10 min in 5% Lipsol detergent at 70°C followed by at least four washes in deionized water at 70°C with sonication. After air drying, coverslips were plasma cleaned for 30 s at 50 mA in a glow discharger (Emitech K100X; Quorum Technologies). Flow cells were coated for 5 min with dynein motility buffer (DMB; 10 mM K-Pipes, 50 mM K-Acetate, 2 mM MgSO4, 1 mM EGTA, and 10% glycerol, pH 6.9, plus 1 mM DTT and 0.2% casein) and then incubated for 5 min with 4–6 µl of motor fraction diluted to 12 µl with DMB. After flowing through 40 µl DMB containing 1 mM ATP, 10 µM taxol, and 8–10 µg/ml taxol-stabilized microtubules, microtubule gliding was visualized by video-enhanced differential interference contrast microscopy. This was performed on a microscope (BX60) equipped with a 60×, 1.40 NA Plan Apochromat objective and a 1.40 NA aplanatic oil immersion universal condenser, with low shear Wollaston prisms (U-DICV). Illumination was provided by a 100-W Hg lamp coupled to the microscope by a 1-mm fiber optic scrambler (Technical Video), passed through 546-nm (20-nm bandwidth), heat, and UV filters. Images were passed through a 5× photo projection lens and captured by a video camera (Newvicon; Hamamatsu Photonics), and background subtraction and two-frame rolling averaging were performed by an image processor (Argus-10; Hamamatsu Photonics). Sequences were recorded onto DVD and then digitized using a FlashBus Spectrim-Pro card (Aegis Electronic Group) using StreamPix4 software (Norpix), and rates of gliding were obtained using ImageJ. Videos were made from cropped sequences by taking every fourth image and then performing a two-frame rolling averaging using the ImageJ RunningZprojector plugin. QuickTime videos were made using QuickTime 7 Pro with H.264 compression (Apple).
Statistical analysis
Statistical tests were performed using SPSS software (SPSS, Inc.), Excel (Microsoft), and Prism (GraphPad Software). For experiments assessing the duration of mitosis, the percentage of cells completing mitosis in a given time was plotted as a cumulative frequency plot and analyzed using the Kaplan–Meier survival analysis test. For microtubule gliding, the mean speed per assay (12–24 microtubules, three independent experiments, 58–67 microtubules per condition in total) was determined from digitized video sequences using ImageJ. The means of these averages were calculated with SEM, and one-way analysis of variance (ANOVA) was performed. For analysis of fixed cultured cells, ≥100 cells per condition were scored per experiment, and the means of three independent experiments are shown, unless otherwise stated. Analysis was performed using two-way ANOVA with Tukey’s test.
To compare the mitotic index in control MO and LIC MO embryos, a one-way ANOVA with Dunnett’s multiple comparison test was performed; n = 3 independent experiments, and 14–16 embryos were analyzed per condition, with total cell counts between 1,097 and 1,992 cells. To test for significance after STLC treatment, Student’s t tests (unpaired, two-tailed, samples of unequal variance) or two-way ANOVA with Tukey’s test were performed; n = 6 independent experiments, and 39–41 embryos were analyzed for each condition, assessing a total of between 375 and 1,175 spindles for spindle morphology and between 3,409 and 7,338 cells for mitotic index.
Online supplemental material
Fig. S1 shows biochemical analysis of microtubule motors prepared from HeLaM cells. Fig. S2 shows the effect of LIC loss on spindle morphology and kinetochores in human cells. Fig. S3 shows PC localization and spindle morphology in single and double LIC-depleted HeLaM cells. Fig. S4 shows the effect of LIC depletion on spindle assembly, pole composition, and prophase centrosome position in U2OS and HEK cells. Fig. S5 shows dynein, dynactin, and spindle assembly factors localize normally to LIC1- and 2-depleted spindles in Xenopus cells. Tables S1 and S2 provide the statistical analysis of the data in Fig. 9. Video 1 shows that loss of LIC does not affect dynein motor function in vitro. Videos 2–5 show mitotic spindle assembly in Xenopus embryos injected with control (Video 2) or LIC1 and 2 (Videos 3–5) MOs. Multipolar spindle assembly is shown in Video 3, with pole fragmentation shown in a bipolar (Video 4) or multipolar spindle (Video 5). Videos 6–8 show 3View EM of a control spindle (Video 6), an LIC KD multipolar spindle (Video 7), and an LIC KD bipolar spindle (Video 8). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408025/DC1.
Supplementary Material
Acknowledgments
We thank colleagues for generously providing reagents and the Faculty of Life Sciences (FLS) EM and Bioimaging facilities staff for their help. The EM facility is supported by equipment grants from The Wellcome Trust. The Bioimaging Facility microscopes were bought with grants from Biotechnology and Biological Sciences Research Council, The Wellcome Trust, and the University of Manchester Strategic Fund.
This work was funded by The Wellcome Trust (WT086077MA to V.J. Allan and N. Papalopulu; the Institutional Strategic Support Fund [ISSF] 097820/Z/11/Z award to V.J. Allan; ISSF 097820/Z/11/Z Stepping Stone Award and Sir Henry Dale Fellowship to S. Woolner), The Royal Society (Sir Henry Dale Fellowship to S. Woolner), The Medical Research Council (Ph.D. studentship to C. Villemant), and FLS, University of Manchester.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- DHC
- dynein heavy chain
- DMB
- dynein motility buffer
- HEK
- human embryonic kidney
- IC
- intermediate chain
- K fiber
- kinetochore fiber
- KnD
- knockdown
- LIC
- light IC
- MMR
- Marc’s modified Ringer’s
- MO
- morpholino
- NE
- nuclear envelope
- NuMA
- nuclear mitotic apparatus
- PC
- pericentrin
- PCM
- pericentriolar material
- RE
- recycling endosome
- SAC
- spindle assembly checkpoint
- STLC
- S-trityl-l-cysteine
References
- Addinall S.G., Mayr P.S., Doyle S., Sheehan J.K., Woodman P.G., and Allan V.J.. 2001. Phosphorylation by cdc2-CyclinB1 kinase releases cytoplasmic dynein from membranes. J. Biol. Chem. 276:15939–15944 10.1074/jbc.M011628200 [DOI] [PubMed] [Google Scholar]
- Allan V.J.1993. Assay of membrane motility in interphase and metaphase Xenopus extracts. Methods Cell Biol. 39:203–226 10.1016/S0091-679X(08)60172-7 [DOI] [PubMed] [Google Scholar]
- Allan V.J.2011. Cytoplasmic dynein. Biochem. Soc. Trans. 39:1169–1178 10.1042/BST0391169 [DOI] [PubMed] [Google Scholar]
- Ban K.H., Torres J.Z., Miller J.J., Mikhailov A., Nachury M.V., Tung J.J., Rieder C.L., and Jackson P.K.. 2007. The END network couples spindle pole assembly to inhibition of the anaphase-promoting complex/cyclosome in early mitosis. Dev. Cell. 13:29–42 10.1016/j.devcel.2007.04.017 [DOI] [PubMed] [Google Scholar]
- Beaudouin J., Gerlich D., Daigle N., Eils R., and Ellenberg J.. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 108:83–96 10.1016/S0092-8674(01)00627-4 [DOI] [PubMed] [Google Scholar]
- Burakov A., Kovalenko O., Semenova I., Zhapparova O., Nadezhdina E., and Rodionov V.. 2008. Cytoplasmic dynein is involved in the retention of microtubules at the centrosome in interphase cells. Traffic. 9:472–480 10.1111/j.1600-0854.2007.00698.x [DOI] [PubMed] [Google Scholar]
- Cabral G., Sans S.S., Cowan C.R., and Dammermann A.. 2013. Multiple mechanisms contribute to centriole separation in C. elegans. Curr. Biol. 23:1380–1387 10.1016/j.cub.2013.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn S.A., Ingold A.L., and Scholey J.M.. 1989. Quantitative analysis of sea urchin egg kinesin-driven microtubule motility. J. Biol. Chem. 264:4290–4297. [PubMed] [Google Scholar]
- Dammermann A., and Merdes A.. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchik M.V., Funk W.C., Brown E.E., and Larkin K.. 1998. Requirement for microtubules in new membrane formation during cytokinesis of Xenopus embryos. Dev. Biol. 194:47–60 10.1006/dbio.1997.8815 [DOI] [PubMed] [Google Scholar]
- Daum J.R., Potapova T.A., Sivakumar S., Daniel J.J., Flynn J.N., Rankin S., and Gorbsky G.J.. 2011. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr. Biol. 21:1018–1024 10.1016/j.cub.2011.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell K.R., Turck C.W., and Vale R.D.. 2000. Mitotic phosphorylation of the dynein light intermediate chain is mediated by cdc2 kinase. Traffic. 1:38–44 10.1034/j.1600-0854.2000.010107.x [DOI] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., and Mikule K.. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15:303–311 10.1016/j.tcb.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Drosopoulos K., Tang C., Chao W.C., and Linardopoulos S.. 2014. APC/C is an essential regulator of centrosome clustering. Nat. Commun. 5:3686 10.1038/ncomms4686 [DOI] [PubMed] [Google Scholar]
- Dunsch A.K., Linnane E., Barr F.A., and Gruneberg U.. 2011. The astrin–kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J. Cell Biol. 192:959–968 10.1083/jcb.201008023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsch A.K., Hammond D., Lloyd J., Schermelleh L., Gruneberg U., and Barr F.A.. 2012. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J. Cell Biol. 198:1039–1054 10.1083/jcb.201202112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elting M.W., Hueschen C.L., Udy D.B., and Dumont S.. 2014. Force on spindle microtubule minus ends moves chromosomes. J. Cell Biol. 206:245–256 10.1083/jcb.201401091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz N.P., Paul R., Fagerstrom C., Mogilner A., and Wadsworth P.. 2009. Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr. Biol. 19:1833–1838 10.1016/j.cub.2009.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestone A.J., Weinger J.S., Maldonado M., Barlan K., Langston L.D., O’Donnell M., Gelfand V.I., Kapoor T.M., and Chen J.K.. 2012. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 484:125–129 10.1038/nature10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian S., and Mayer T.U.. 2012. The functional antagonism between Eg5 and dynein in spindle bipolarization is not compatible with a simple push-pull model. Cell Reports. 1:408–416 10.1016/j.celrep.2012.03.006 [DOI] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Bingham J.B., Hasbani M.J., Gill S.R., Schroer T.A., and Compton D.A.. 1996. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135:399–414 10.1083/jcb.135.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Nédélec F., and Vale R.D.. 2005. Mechanisms for focusing mitotic spindle poles by minus end–directed motor proteins. J. Cell Biol. 171:229–240 10.1083/jcb.200505107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Habermann A., Karsenti E., and Hyman A.. 1997. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138:615–628 10.1083/jcb.138.3.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H., and Doxsey S.. 2014. Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev. Cell. 28:497–507 10.1016/j.devcel.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Cassels G.O., Rieder C.L., and Sluder G.. 1998. The coordination of centrosome reproduction with nuclear events of the cell cycle in the sea urchin zygote. J. Cell Biol. 140:1417–1426 10.1083/jcb.140.6.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., and Salmon E.D.. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan C.P., Hanscom S.R., Jolly R.S., Futter C.E., and McCaffrey M.W.. 2010a. Rab11-FIP3 binds dynein light intermediate chain 2 and its overexpression fragments the Golgi complex. Biochem. Biophys. Res. Commun. 394:387–392 10.1016/j.bbrc.2010.03.028 [DOI] [PubMed] [Google Scholar]
- Horgan C.P., Hanscom S.R., Jolly R.S., Futter C.E., and McCaffrey M.W.. 2010b. Rab11-FIP3 links the Rab11 GTPase and cytoplasmic dynein to mediate transport to the endosomal-recycling compartment. J. Cell Sci. 123:181–191 10.1242/jcs.052670 [DOI] [PubMed] [Google Scholar]
- Horgan C.P., Hanscom S.R., and McCaffrey M.W.. 2011. Dynein LIC1 localizes to the mitotic spindle and midbody and LIC2 localizes to spindle poles during cell division. Cell Biol. Int. 35:171–178 10.1042/CBI20100284 [DOI] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., and Salmon E.D.. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S.D., Townley A.K., Danson C.M., Cullen P.J., and Stephens D.J.. 2013. Microtubule motors mediate endosomal sorting by maintaining functional domain organization. J. Cell Sci. 126:2493–2501 10.1242/jcs.122317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein D., and Taylor S.S.. 2002. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J. Cell Sci. 115:3403–3414. [DOI] [PubMed] [Google Scholar]
- Hut H.M., Lemstra W., Blaauw E.H., Van Cappellen G.W., Kampinga H.H., and Sibon O.C.. 2003. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol. Biol. Cell. 14:1993–2004 10.1091/mbc.E02-08-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri Y., Kamakura S., Hayase J., and Sumimoto H.. 2013. Interaction of NuMA protein with the kinesin Eg5: its possible role in bipolar spindle assembly and chromosome alignment. Biochem. J. 451:195–204 10.1042/BJ20121447 [DOI] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., and Khodjakov A.. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating T.J., and Borisy G.G.. 2000. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat. Cell Biol. 2:352–357 10.1038/35014045 [DOI] [PubMed] [Google Scholar]
- King S.J., Bonilla M., Rodgers M.E., and Schroer T.A.. 2002. Subunit organization in cytoplasmic dynein subcomplexes. Protein Sci. 11:1239–1250 10.1110/ps.2520102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Pöllinger B., Ohmer M., Hofmann F., Nigg E.A., Hemmings B.A., and Wartmann M.. 2012. Acentrosomal spindle organization renders cancer cells dependent on the kinesin HSET. J. Cell Sci. 125:5391–5402 10.1242/jcs.107474 [DOI] [PubMed] [Google Scholar]
- Klymkowsky M.W., and Hanken J.. 1991. Whole-mount staining of Xenopus and other vertebrates. Methods Cell Biol. 36:419–441 10.1016/S0091-679X(08)60290-3 [DOI] [PubMed] [Google Scholar]
- Kong X., Ball A.R. Jr, Sonoda E., Feng J., Takeda S., Fukagawa T., Yen T.J., and Yokomori K.. 2009. Cohesin associates with spindle poles in a mitosis-specific manner and functions in spindle assembly in vertebrate cells. Mol. Biol. Cell. 20:1289–1301 10.1091/mbc.E08-04-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., and Shiina N.. 1999. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147:969–980 10.1083/jcb.147.5.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M., Godinho S.A., Chandhok N.S., Ganem N.J., Azioune A., Thery M., and Pellman D.. 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22:2189–2203 10.1101/gad.1700908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J.D., and Allan V.J.. 1999. Microtubule-based endoplasmic reticulum motility in Xenopus laevis: activation of membrane-associated kinesin during development. Mol. Biol. Cell. 10:1909–1922 10.1091/mbc.10.6.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B., Maier B., Fuchs F., Chi J., Riffel P., Anderhub S., Wagner L., Ho A.D., Salisbury J.L., Boutros M., and Krämer A.. 2010. Proteins required for centrosome clustering in cancer cells. Sci. Transl. Med. 2:33ra38 10.1126/scitranslmed.3000915 [DOI] [PubMed] [Google Scholar]
- Lee W.L., Kaiser M.A., and Cooper J.A.. 2005. The offloading model for dynein function: differential function of motor subunits. J. Cell Biol. 168:201–207 10.1083/jcb.200407036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E., Maffini S., Barisic M., Marques A., Toso A., Meraldi P., and Maiato H.. 2012. CLASPs prevent irreversible multipolarity by ensuring spindle-pole resistance to traction forces during chromosome alignment. Nat. Cell Biol. 14:295–303 10.1038/ncb2423 [DOI] [PubMed] [Google Scholar]
- Ma L., Tsai M.-Y., Wang S., Lu B., Chen R., Iii J.R., Zhu X., and Zheng Y.. 2009. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat. Cell Biol. 11:247–256 10.1038/ncb1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., and Logarinho E.. 2014. Mitotic spindle multipolarity without centrosome amplification. Nat. Cell Biol. 16:386–394 10.1038/ncb2958 [DOI] [PubMed] [Google Scholar]
- Maiato H., Rieder C.L., and Khodjakov A.. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840 10.1083/jcb.200407090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.L., and Compton D.A.. 2007. Mechanisms of spindle-pole organization are influenced by kinetochore activity in mammalian cells. Curr. Biol. 17:260–265 10.1016/j.cub.2006.11.071 [DOI] [PubMed] [Google Scholar]
- Mardin B.R., and Schiebel E.. 2012. Breaking the ties that bind: New advances in centrosome biology. J. Cell Biol. 197:11–18 10.1083/jcb.201108006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K., Ohsumi K., Iwabuchi M., Kawamata T., Ono Y., and Takahashi M.. 2012. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr. Biol. 22:915–921 10.1016/j.cub.2012.03.048 [DOI] [PubMed] [Google Scholar]
- Mattiuzzo M., Vargiu G., Totta P., Fiore M., Ciferri C., Musacchio A., and Degrassi F.. 2011. Abnormal kinetochore-generated pulling forces from expressing a N-terminally modified Hec1. PLoS ONE. 6:e16307 10.1371/journal.pone.0016307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer T.U., Kapoor T.M., Haggarty S.J., King R.W., Schreiber S.L., and Mitchison T.J.. 1999. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 286:971–974 10.1126/science.286.5441.971 [DOI] [PubMed] [Google Scholar]
- McKenney R.J., Huynh W., Tanenbaum M.E., Bhabha G., and Vale R.D.. 2014. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 345:337–341 10.1126/science.1254198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W.C., and Cleveland D.W.. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149:851–862 10.1083/jcb.149.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische S., He Y., Ma L., Li M., Serr M., and Hays T.S.. 2008. Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation. Mol. Biol. Cell. 19:4918–4929 10.1091/mbc.E08-05-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J., Maddox P., Gaetz J., Groen A., Shirasu M., Desai A., Salmon E.D., and Kapoor T.M.. 2005. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell. 16:3064–3076 10.1091/mbc.E05-02-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Mulia S., and Scholey J.M.. 2005. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell. 16:3176–3186 10.1091/mbc.E04-12-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Arai H., and Fujita N.. 2009. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J. Cell Biol. 187:607–614 10.1083/jcb.200906019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumayer G., Belzil C., Gruss O.J., and Nguyen M.D.. 2014. TPX2: of spindle assembly, DNA damage response, and cancer. Cell. Mol. Life Sci. 71:3027–3047 10.1007/s00018-014-1582-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niclas J., Allan V.J., and Vale R.D.. 1996. Cell cycle regulation of dynein association with membranes modulates microtubule-based organelle transport. J. Cell Biol. 133:585–593 10.1083/jcb.133.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien L.L., and Wiese C.. 2006. TPX2 is required for postmitotic nuclear assembly in cell-free Xenopus laevis egg extracts. J. Cell Biol. 173:685–694 10.1083/jcb.200512107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell C.B., Loncarek J., Kaláb P., and Khodjakov A.. 2009. Relative contributions of chromatin and kinetochores to mitotic spindle assembly. J. Cell Biol. 187:43–51 10.1083/jcb.200903076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R.A., and Nasmyth K.. 2013. Cohesin cleavage is insufficient for centriole disengagement in Drosophila. Curr. Biol. 23:R601–R603 10.1016/j.cub.2013.04.003 [DOI] [PubMed] [Google Scholar]
- Ori-McKenney K.M., Xu J., Gross S.P., and Vallee R.B.. 2010. A cytoplasmic dynein tail mutation impairs motor processivity. Nat. Cell Biol. 12:1228–1234 10.1038/ncb2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer K.J., Hughes H., and Stephens D.J.. 2009. Specificity of cytoplasmic dynein subunits in discrete membrane-trafficking steps. Mol. Biol. Cell. 20:2885–2899 10.1091/mbc.E08-12-1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister K.K., Shah P.R., Hummerich H., Russ A., Cotton J., Annuar A.A., King S.M., and Fisher E.M.. 2006. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2:e1 10.1371/journal.pgen.0020001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C.L., and Bornens M.. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit A., Tynan S.H., Vallee R., and Doxsey S.J.. 1999. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 147:481–492 10.1083/jcb.147.3.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne N.J., Reing J.E., Hoffelder D.R., Gollin S.M., and Saunders W.S.. 2005. Spindle multipolarity is prevented by centrosomal clustering. Science. 307:127–129 10.1126/science.1104905 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J.A., and Medema R.H.. 2014. Function and regulation of dynein in mitotic chromosome segregation. Chromosoma. 123:407–422 10.1007/s00412-014-0468-7 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J.A., van Heesbeen R.G., Meaders J.L., Geers E.F., Fernandez-Garcia B., Medema R.H., and Tanenbaum M.E.. 2012. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 31:4179–4190 10.1038/emboj.2012.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J.A., Tanenbaum M.E., and Medema R.H.. 2013. Systematic dissection of dynein regulators in mitosis. J. Cell Biol. 201:201–215 10.1083/jcb.201208098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu A.E., and Cleveland D.W.. 2010. NuMA after 30 years: the matrix revisited. Trends Cell Biol. 20:214–222 10.1016/j.tcb.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Wojcik E.J., Sanders M.A., McGrail M., and Hays T.S.. 1999. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146:597–608 10.1083/jcb.146.3.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D., Bodoor K., Eckley D.M., Schroer T.A., Rattner J.B., and Burke B.. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 108:97–107 10.1016/S0092-8674(01)00628-6 [DOI] [PubMed] [Google Scholar]
- Scherer J., Yi J., and Vallee R.B.. 2014. PKA-dependent dynein switching from lysosomes to adenovirus: a novel form of host–virus competition. J. Cell Biol. 205:163–177 10.1083/jcb.201307116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager M.A., Hoang H.T., Urnavicius L., Bullock S.L., and Carter A.P.. 2014. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33:1855–1868 10.15252/embj.201488792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J.C., Kiyomitsu T., Hori T., Backer C.B., Fukagawa T., and Cheeseman I.M.. 2010. Aurora B kinase controls the targeting of the Astrin–SKAP complex to bioriented kinetochores. J. Cell Biol. 191:269–280 10.1083/jcb.201006129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J., Fawcett J.P., Segura M., Tan S., Vallee R.B., Pawson T., and Gundersen G.G.. 2009. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr. Biol. 19:1065–1074 10.1016/j.cub.2009.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Toyoshima Y.Y., Edamatsu M., and Vale R.D.. 1995. Comparison of the motile and enzymatic properties of two microtubule minus-end-directed motors, ncd and cytoplasmic dynein. Biochemistry. 34:1575–1582 10.1021/bi00005a013 [DOI] [PubMed] [Google Scholar]
- Sikirzhytski V., Magidson V., Steinman J.B., He J., Le Berre M., Tikhonenko I., Ault J.G., McEwen B.F., Chen J.K., Sui H., et al. 2014. Direct kinetochore–spindle pole connections are not required for chromosome segregation. J. Cell Biol. 206:231–243 10.1083/jcb.201401090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk A.D., Holland A.J., and Cleveland D.W.. 2009. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184:677–690 10.1083/jcb.200810091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaram M.V., Wadzinski T.L., Redick S.D., Manna T., and Doxsey S.J.. 2009. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J. 28:902–914 10.1038/emboj.2009.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D.A., DeBonis S., Saoudi Y., Lebeau L., Crevel I., Cross R., Wade R.H., Hackney D., and Kozielski F.. 2006. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J. Biol. Chem. 281:17559–17569 10.1074/jbc.M511735200 [DOI] [PubMed] [Google Scholar]
- Sluder G., and Rieder C.L.. 1985. Centriole number and the reproductive capacity of spindle poles. J. Cell Biol. 100:887–896 10.1083/jcb.100.3.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starborg T., Kalson N.S., Lu Y., Mironov A., Cootes T.F., Holmes D.F., and Kadler K.E.. 2013. Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization. Nat. Protoc. 8:1433–1448 10.1038/nprot.2013.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D., Gassmann R., Oegema K., and Desai A.. 2011. Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS ONE. 6:e22969 10.1371/journal.pone.0022969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill E.G., and Ohi R.. 2013. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr. Biol. 23:1280–1290 10.1016/j.cub.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.C., Scherer J., and Vallee R.B.. 2011. Recruitment of dynein to late endosomes and lysosomes through light intermediate chains. Mol. Biol. Cell. 22:467–477 10.1091/mbc.E10-02-0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., and Medema R.H.. 2010. Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell. 19:797–806 10.1016/j.devcel.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macůrek L., Galjart N., and Medema R.H.. 2008. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 27:3235–3245 10.1038/emboj.2008.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Vale R.D., and McKenney R.J.. 2013. Cytoplasmic dynein crosslinks and slides anti-parallel microtubules using its two motor domains. eLife. 2:e00943 10.7554/eLife.00943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., Hussein D., Wang Y., Elderkin S., and Morrow C.J.. 2001. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114:4385–4395. [DOI] [PubMed] [Google Scholar]
- Thein K.H., Kleylein-Sohn J., Nigg E.A., and Gruneberg U.. 2007. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J. Cell Biol. 178:345–354 10.1083/jcb.200701163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J.Z., Ban K.H., and Jackson P.K.. 2010. A specific form of phospho protein phosphatase 2 regulates anaphase-promoting complex/cyclosome association with spindle poles. Mol. Biol. Cell. 21:897–904 10.1091/mbc.E09-07-0598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J.Z., Summers M.K., Peterson D., Brauer M.J., Lee J., Senese S., Gholkar A.A., Lo Y.-C., Lei X., Jung K., et al. 2011. The STARD9/Kif16a kinesin associates with mitotic microtubules and regulates spindle pole assembly. Cell. 147:1309–1323 10.1016/j.cell.2011.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A., Winter J.R., Garrod A.J., Amaro A.C., Meraldi P., and McAinsh A.D.. 2009. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 184:365–372 10.1083/jcb.200809055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.-Y., Wang S., Heidinger J.M., Shumaker D.K., Adam S.A., Goldman R.D., and Zheng Y.. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 311:1887–1893 10.1126/science.1122771 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., and Stearns T.. 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 442:947–951 10.1038/nature04985 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Wang W.J., George K.A., Uryu K., Stearns T., and Jallepalli P.V.. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell. 17:344–354 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan S.H., Gee M.A., and Vallee R.B.. 2000a. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J. Biol. Chem. 275:32769–32774 10.1074/jbc.M001537200 [DOI] [PubMed] [Google Scholar]
- Tynan S.H., Purohit A., Doxsey S.J., and Vallee R.B.. 2000b. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 275:32763–32768 10.1074/jbc.M001536200 [DOI] [PubMed] [Google Scholar]
- Wang X., Yang Y., Duan Q., Jiang N., Huang Y., Darzynkiewicz Z., and Dai W.. 2008. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev. Cell. 14:331–341 10.1016/j.devcel.2007.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-J., Soni R.K., Uryu K., and Tsou M.F.. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193:727–739 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.E., Wilke S.A., Daggett A., Davis E., Otto S., Ravi D., Ripley B., Bushong E.A., Ellisman M.H., Klein G., and Ghosh A.. 2011. Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron. 71:640–655 10.1016/j.neuron.2011.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaërou F., Karess R., and Hays T.. 2001. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3:1001–1007 10.1038/ncb1101-1001 [DOI] [PubMed] [Google Scholar]
- Wood K.W., Lad L., Luo L., Qian X., Knight S.D., Nevins N., Brejc K., Sutton D., Gilmartin A.G., Chua P.R., et al. 2010. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl. Acad. Sci. USA. 107:5839–5844 10.1073/pnas.0915068107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolner S., Miller A.L., and Bement W.M.. 2009. Imaging the cytoskeleton in live Xenopus laevis embryos. Methods Mol. Biol. 586:23–39 10.1007/978-1-60761-376-3_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tulu U.S., Wadsworth P., and Rieder C.L.. 2007. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 17:973–980 10.1016/j.cub.2007.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J.H., and Han M.. 2001. Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol. Biol. Cell. 12:2921–2933 10.1091/mbc.12.10.2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li S., Musa S., Zhou H., and Xiang X.. 2009. Dynein light intermediate chain in Aspergillus nidulans is essential for the interaction between heavy and intermediate chains. J. Biol. Chem. 284:34760–34768 10.1074/jbc.M109.026872 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.