Abstract
It has been suggested that deficient protein trafficking to the cell membrane is the dominant mechanism associated with type 2 Long QT syndrome (LQT2) caused by Kv11.1 potassium channel missense mutations, and that for many mutations the trafficking defect can be corrected pharmacologically. However, this inference was based on expression of a small number of Kv11.1 mutations. We performed a comprehensive analysis of 167 LQT2-linked missense mutations in four Kv11.1 structural domains and found that deficient protein trafficking is the dominant mechanism for all domains except for the distal C-terminus. Also, most pore mutations—in contrast to intracellular domain mutations —were found to have severe dominant-negative effects when co-expressed with wild type subunits. Finally, pharmacological correction of the trafficking defect in homomeric mutant channels was possible for mutations within all structural domains. However, pharmacological correction is dramatically improved for pore mutants when co-expressed with wild type subunits to form heteromeric channels.
Introduction
Kv11.1 is a voltage-gated K+ channel encoded by the human ether-a-go-go-related gene (hERG or KCNH2) and is associated with the pathophysiology of inherited cardiac arrhythmia diseases including type 2 long QT syndrome (LQT2)1, sudden infant death syndrome (SIDS)2, and short QT syndrome (SQTS)3, as well as various forms of cancer4, epilepsy5 and schizophrenia6. Nearly 500 KCNH2 mutations have been linked to LQT2 - which is characterized by a prolonged time duration from ventricular depolarization to repolarization (QT interval on an ECG) - and increased risk for sudden cardiac death. These loss-of-function mutations have been classified into four molecular mechanisms; class 1: abnormal transcription/translation, class 2: deficient protein trafficking, class 3: abnormal channel gating/kinetics, and class 4: altered channel permeability, in all of which the repolarizing outward K+ current, IKv11.1 or IKr, is reduced7. While a fraction of KCNH2 mutations are nonsense and postulated to invoke a class 1 mechanism due to nonsense-mediated mRNA decay (NMD)8, the majority are missense mutations with most postulated to invoke a class 2 mechanism due to protein misfolding and endoplasmic reticulum-associated degradation (ERAD)9–12.
Interestingly, for some trafficking deficient mutations, the defect can be corrected pharmacologically (usually with high affinity Kv11.1 channel blockers), with reduced culture temperature, or by RNAi10,13–16, suggesting therapeutic potential for LQT2 carriers, although application of these findings to LQT2 patients remains a major challenge. In addition, of the >300 missense mutations most remain functionally uncharacterized and are spread throughout the Kv11.1 multidomain protein, which contains voltage sensor (VSD, S1–4) and pore (S5–6) domains comprising the transmembrane domain (TMD)17, a N-terminus containing the PerArntSim domain (PASD) with PAS-cap collectively making up a conserved ‘EAG’ domain present in the EAG family of Kv channels18, a C-terminus containing the cyclic-nucleotide-binding homology domain (CNBHD)19 along with a distal C-terminal ER retention signal (RXR)20 and coiled-coil domain (CCD)21. Furthermore, carriers of LQT2 mutations are heterozygous making co-assembly dynamics (dominant negative, haploinsufficiency) of the tetrameric channel another factor contributing to disease complexity22. While much has been learned about the molecular basis underlying LQT223, many important gaps remain, three of which we address in this paper.
Not all mutations characterized in heterologous expression systems show a loss-of-function phenotype suggesting that some reported mutations may be benign sequence variants or single nucleotide polymorphisms (SNPs)10,24. This emphasizes the need to functionally express and analyze individual mutations.
The location of a mutation within the Kv11.1 protein may be important but the molecular basis for this is unknown. Mutations in the pore clinically have a more severe phenotype25,26. Several lines of evidence suggest that TMD (including pore) and CNBHD mutations invoke a class 2 (trafficking deficient) mechanism10,27 versus intracellular (‘core’) mutations that may traffic normally and exert a class 3 (abnormal gating) mechanism28. ‘Core’ interactions between the PASD, S4–S5-linker, and CNBHD regulate Kv11.1 gating29–35 and many engineered and LQT2-linked ‘core’ mutations exhibit more rapid deactivation18,32,36, and a recent study reported that some PASD mutations traffic normally28. Alternatively, differences may be attributed to differences in wild type (WT)-mutant subunit interactions. Several intracellular mutations reduce IKv11.1 in a partially dominant-negative manner or through haploinsufficiency10,37–39, while most pore mutations have a strong dominant-negative interaction with WT subunits producing little to no current10,14,40,41. Unfortunately, most LQT2 mutations remain functionally uncharacterized and the disease mechanisms are unknown.
Studies using mostly homomeric channels show that culture in the presence of pore blocking drugs like E4031, which involve π-cation, π - π stacking and hydrophobic interactions with aromatic residues in TMD S642,43 can correct defective protein trafficking of some mutations in the TMD (pore and VSD) and PASD10,38,44. Second-site S6 mutations can also correct some pore mutations45. These findings and the observation that E4031 corrected channels are more resistant to proteases suggest that E4031 correction works by stabilizing the pore, which can also facilitate cooperative folding between domains/subunits resulting in more tightly packed channels that evade ERAD46. However, E4031 correction seems limited primarily to pore domain mutations along with a few in the PASD and none in the CNBHD10,28. E4031-dependent pharmacological correction has been studied for relatively few homomeric and even fewer heteromerically (with WT) expressed mutations, thus a comprehensive analysis of mutations co-expressed with WT subunits across multiple structural domains is needed to better understand pharmacological correction as a potential therapy.
In the present work, we undertook a large-scale analysis of LQT2 missense mutations in different structural domains of the Kv11.1 protein to generate new insights about LQT2 disease mechanisms. We studied missense mutations because they are the most commonly identified genetic abnormality in patients with LQT2. We took a data-driven approach employing heterologous mammalian expression to generate immunoblot and electrophysiology data, combined with computational structural models, to characterize both homotetrameric (mutant or WT) and heterotetrameric (mutant + WT) channels. We generated and studied 167 LQT2 mutations as well as three KCNH2 SIDS mutations and two SNPs. We show that 88% of LQT2 missense mutations in the PASD, C-linker/CNBHD and pore domains invoke a class 2 (trafficking deficient) mechanism, but with some major differences. Over 70% of LQT2 pore mutations were strictly dominant negative whereas PASD or C-linker/CNBHD mutations were not. In addition, LQT2 mutations in all domains were pharmacologically (E4031) correctable (PASD>pore>C-linker/CNBHD), and this improved dramatically when mutations where co-expressed with WT subunits (pore>PASD>C-linker/CNBHD) making pharmacological correction much more common than previously thought. We also mapped each mutation to structural models to analyze structure/(dys)function relationships. We then combined these results with previously published studies (see supplementary tables) and discuss a simplified model for the different Kv11.1 trafficking phenotypes. To our knowledge, this is the largest mutational study for any congenital disease, which should serve as a valuable resource and help spark further inquiry into this ion channelopathy.
Results
Trafficking phenotype of homomeric EAGD mutations
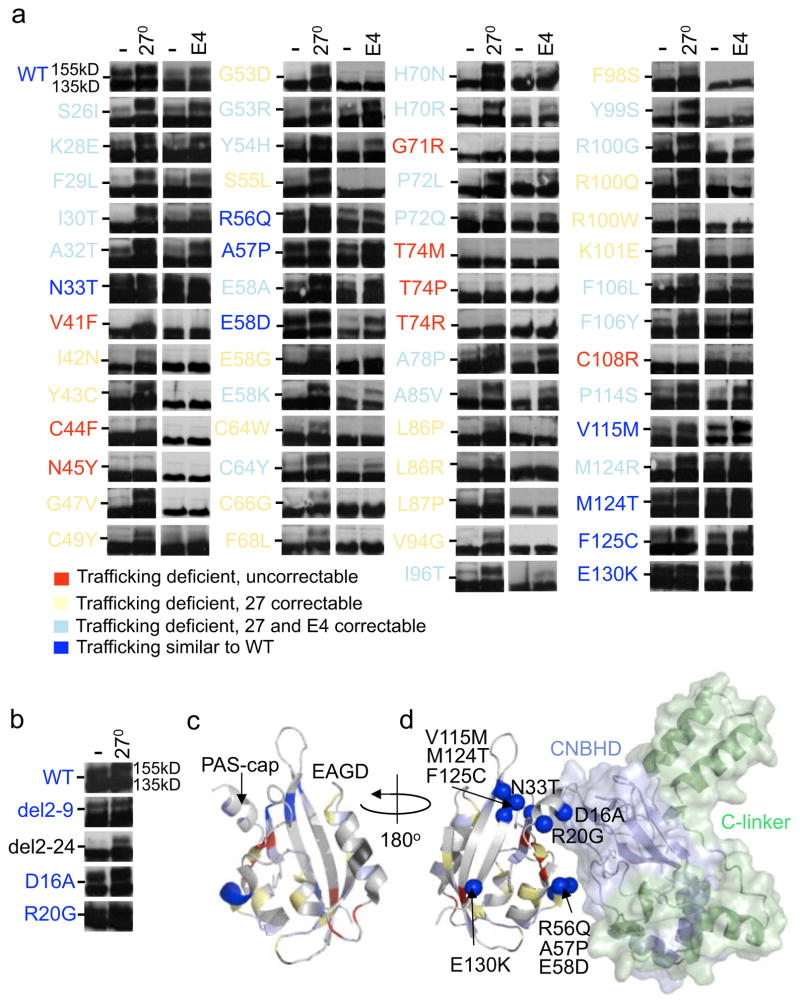
We compiled a list of 291 LQT2 and 10 SIDS-linked KCNH2 mutations as well as 28 SNPs (see methods, Fig. 1 and Supplementary Table 1). While the TMD contains 1/2 of the LQT2 missense mutations, the PASD is another “hotspot” with at least 62 (61 LQT2, 1 SIDS) mostly uncharacterized mutations at 47 sites. Two more are located in the N-terminal proximal to the PASD in the PAS-cap amphipathic helix (amino acids 13–23) (Fig 2.c) 47. To determine their trafficking phenotype, immunoblot was performed on transiently transfected HEK293 cells expressing homomeric Kv11.1 channels cultured at physiological temperature (37°C) as well as reduced culture temperature (27°C for 24hrs) or with 10μM E4031 in the culture medium (24hrs) to test for correction. Kv11.1 protein (132kD) is synthesized in the ER where it undergoes N-linked core-glycosylation (135kD) and then traffics to the golgi for N-linked complex glycosylation (155kD) before reaching the plasma membrane. A doublet on immunoblot (135kD and 155kD bands) indicates cell surface trafficking whereas a lack or decrease of the 155kD band indicates deficient trafficking9,10. Figure 2a shows representative immunoblots (n≥2) for 57 PASD mutations, which exhibited four different trafficking phenotypes: trafficking deficient and uncorrectable (red), trafficking deficient but correctable with culture at 27°C (yellow), trafficking deficient but correctable with culture at 27°C or with E4031 (light blue), or normal trafficking (blue). 49/57 (86%) lacked or had a diminished 155kD band on immunoblot compared to WT under control conditions. 40 (82%) of those were temperature correctable and 23 (47%) were E4031 correctable. Interestingly, 17 were only temperature correctable but none were only E4031 correctable. So, in contrast to a previous study28, most PASD mutations are trafficking deficient and correctable. These data combined with several mutations already reported in the literature is summarized in Supplementary Table 2.
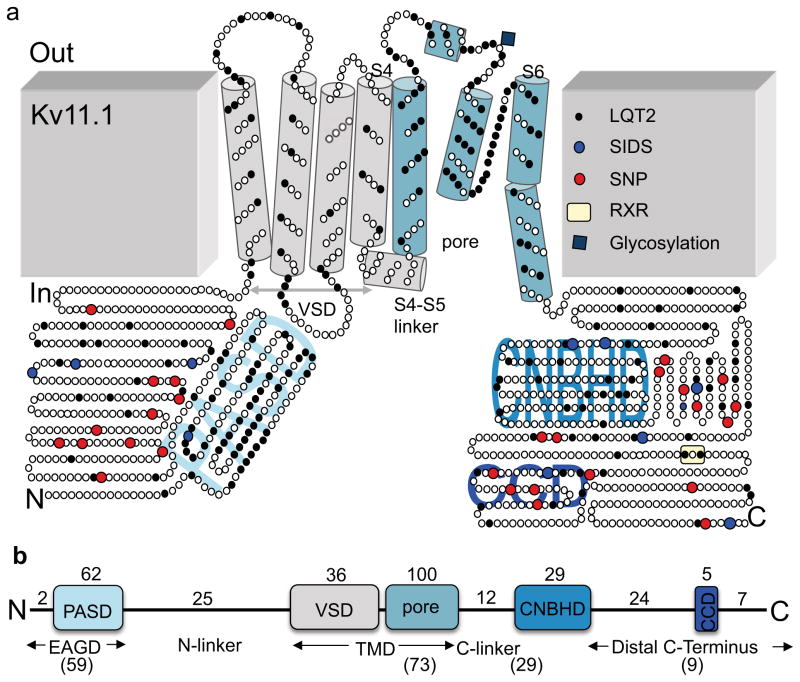
Figure 1. Topology of Kv11.1.
(a) Cartoon of Kv11.1 with circles representing amino acids and cylinders representing TMD segments 1–6 and the S4–S5 linker. S1–S4 make up the VSD and the pore is located between S5 and S6. The intracellular PASD, CNBHD, and CCD are labeled in light blue. Black and blue circles show the location of all reported LQT2 and SIDS mutations, respectively. Red circles show the location of all reported SNPs. The ER retention signal RXR is highlighted in yellow and the glycosylation site at N598 is shown with a blue diamond. (b) Linear representation of the different Kv11.1 domains (to scale) showing the number and domain location of all LQT2-linked and SIDS mutations identified to date. The number characterized in this study are in parentheses (59 of 64 mutations for the EAGD, 73 of 100 mutations for the pore, 29 of 41 mutations for the C-linker and CNBHD, and 9 of 36 mutations for the Distal C-terminus).
Figure 2. Trafficking phenotype and structural context of LQT2-linked EAGD missense mutations.
(a, b) Representative immunoblots of transiently transfected HEK293 cells comparing trafficking under control culture conditions at 37°C, at reduced temperature (27°C, 24hrs) or in E4031 (E4, 24hrs). Dashes (-) indicate the 140kD molecular weight marker. Mutations are color-coded as follows: trafficking deficient and uncorrectable in red, trafficking deficient but correctable at 27°C in yellow, trafficking deficient but correctable at 27°C or with E4031 in light blue, and those that traffic similar to WT in blue. (c) Crystal structure of the EAGD (PDB: 4HP9). Mapped LQT2 residues correspond to panel A and uncharacterized mutations (I96V and D102A) are black. (d) Representation of the EAGD (PDB: 4HP9) complexed with the C-linker/CNBHD model from alignments to the structure of the EAGD-CNBHD complex from mouse (PDB: 4LLO). C-linker region is shown in green, CNBHD in blue, and intrinsic ligand in magenta. Mutations that are trafficking competent (blue balls) are labeled. An example of a full length immunoblot can be found in Supplementary figure 2.
Quickened deactivation from N-terminus truncated channels and LQT2-mutations can be restored by adding back the EAG domain or just the first 16 residues of the PAS-cap18,31,48. Binding of the PASD to the CNBHD is thought to position the amphipathic helix and in turn the N-terminal residues near the bottom of S6 to modulate Kv11.1 gating30,34,35. To see if the PAS-cap is also important for trafficking, we found that deletion of residues 2–9 had no effect on trafficking but deletion of residues 2–24 containing the amphipathic helix did impair trafficking. The LQT2 mutations D16A and R20G were both trafficking-competent (Fig. 2b).
Structural and functional properties of EAGD mutations
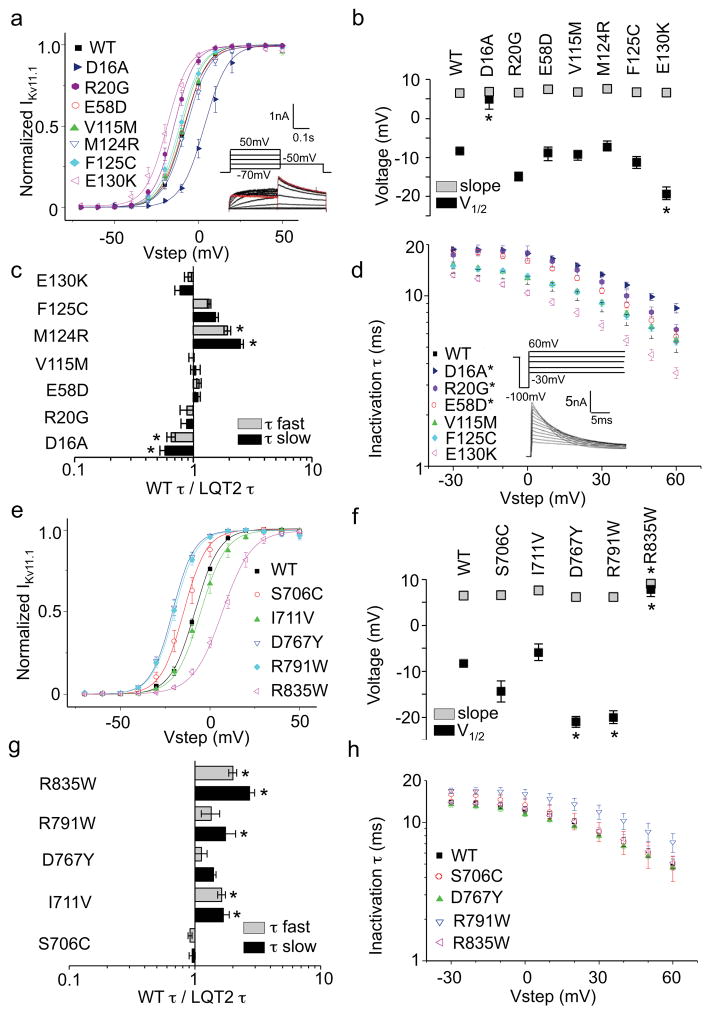
EAGD mutations were mapped onto its structure (PDB: 4HP9)49, which has a fold consisting of a central antiparallel β-sheet flanked by several α-helices (Fig. 2c). While the different trafficking deficient phenotypes are scattered showing no correlation with location, most trafficking competent mutations lie near its hydrophobic surface which interfaces with the CNBHD (PDB 4LLO)35; a region important for regulating Kv11.1 deactivation18,50,51 (Fig. 2d; Supplementary Fig. 1a, b, g, h). To determine their electrophysiological properties, stably transfected cells were generated for the trafficking competent D16A, R20G, E58D, V115M, F125C, E130K as well the trafficking deficient M124R located near the hydrophobic surface. Representative western blots and peak tail IKv11.1 densities are shown in Supplementary Fig. 2a. The greatest change in V1/2 was for D16A and E130K, shifted +13mV and −10mV, respectively (Fig. 3a, b). Deactivation measured at −50mV was slower for D16A and faster for M124R compared to WT consistent with previous reports (Fig. 3c)47,52, and D16A, R20G, and E58D all exhibited slightly slower inactivation measured at 0mV (Fig. 3d). In contrast, the functional properties of E58D, V115M, and F125C were similar to WT suggesting that they may be benign variants (Table 1; Fig. 3a–d). In addition to M124R, stable cell lines of F106L also exhibited the same trafficking phenotype as transient transfections showing a weak 155kD band on immunoblot, which increased with culture at reduced temperature or with E4031 (Supplementary Fig. 2d).
Figure 3. Electrophysiological properties of LQT2-linked EAGD and C-linker/CNBHD missense mutations.
(a, e) Activation current-voltage (I-V) relationships. From a holding potential of −80mV, cells were depolarized to voltages between −70mV and 50mV in 10mV increments for 3s to quantify tail current. I-V relations were determined by normalizing peak tail currents (Itail) from each step to the maximal peak Itail. (b, f) V1/2 and slope factors. The voltage at which peak IKv11.1 was half-maximal (V1/2) and the slope factor (k) were determined by fitting the normalized I-V relationship with the Boltzmann function. (c, g) WT-to-mutant time constant ratios (speeding factor) for deactivation at −50mV. The fast (taufast) and slow (tauslow) time constants of channel deactivation were determined with a double exponential fit of the Itail decay from 50mV to −50mV (red trace).. (d, h) Inactivation time constants determined at 0mV. From a holding potential of −80mV, cells were depolarized to 50mV for 1.5s to open and inactivate channels followed by a short 10ms step to −100mV to remove inactivation without allowing enough time for the channels to deactivate. This was followed by test pulses from −30mV to 60mV in 10mv increments. Inactivation time constants for each step were fit with a single exponential. Error bars are SEM. Asterisks indicate statistical significance (p<0.05). n = 3 to 9 HEK293 cells for each experiment.
Table 1.
Electrophysiological properties of Kv11.1 missense mutations.
| LQT2 | Current Density | Activation | Deactivation @ −50mV | Inactivation @ 0mV | ||
|---|---|---|---|---|---|---|
|
| ||||||
| pA/pF | V1/2 (mv) | slope (mv/e-fold) | tau slow (s) | tau fast (ms) | tau (ms) | |
| WT | 97.7±6.2 (9) | −8.3±0.4 (5) | 6.5 ±0.4 (5) | 5.3±0.3 (5) | 835±32 (5) | 12.4±0.7 (3) |
| D16A | 57.3±12.4 (4) | 4.9±2.5* (4) | 6.9±0.4 (4) | 9.3±0.8* (4) | 1281±105* (4) | 17.7± 0.1* (4) |
| R20G | 48.5±7.7 (4) | −15.0±1.2 (4) | 6.6±0.2 (4) | 6.1±0.1 (4) | 943±119 (4) | 17.8±1.8* (4) |
| E58D | 62.2±9.7 (8) | −8.8±1.5 (7) | 7.5±0.3 (7) | 4.9±0.3 (8) | 746±37 (8) | 16.1±0.6* (4) |
| V115M | 73.1±15.3 (9) | −9.2±0.7 (8) | 6.8±0.2 (8) | 5.1±0.5 (4) | 863±48 (4) | 12.7±0.1 (3) |
| M124R | 21.6±5.3 (7) | −7.3±1.6 (4) | 7.6±0.3 (4) | 2.2±0.1* (5) | 430±28* (5) | n/a |
| F125C | 54.1±13.7 (9) | −11.2±1.4 (9) | 6.7±0.1 (9) | 3.4±0.2 (9) | 615±33 (9) | 13.0±0.5 (4) |
| E130K | 43.2±6.1 (7) | −19.4±1.9* (9) | 6.6±0.3 (9) | 6.9±0.7 (7) | 922±65 (7) | 10.4±0.3 (6) |
| S706C | 97.7±6.2 (6) | −14.4±2.3 (5) | 6.6±0.4 (5) | 5.6±0.7 (5) | 914±65 (5) | 13.4±1.6 (3) |
| I711V | 45.5±4.0 (4) | −5.9±1.8 (4) | 7.6±0.6 (4) | 3.2±0.2* (4) | 512±16* (4) | n/a |
| D767Y | 73.1±9.7 (5) | −21.0±1.2* (5) | 6.1±0.2 (5) | 3.8±0.3 (4) | 741±82 (4) | 11.4±0.4 (4) |
| R791W | 105±11 (4) | −20.5±1.4* (4) | 6.1±0.2 (4) | 3.1±0.6* (4) | 620±104 (4) | 16.0±2.6 (4) |
| R835W | 17.2±1.8 (5) | 7.8±1.6* (5) | 9.0±0.6* (5) | 1.9±0.2* (4) | 416±31* (4) | 12.6±0.2 (3) |
| R1005Q | 97.7±6.2 (7) | −14.6±0.5* (7) | 6.4±0.3 (7) | 4.7±0.3 (6) | 823±43 (6) | 15.1±0.6 (5) |
| L1049P | 34.6±4.5 (7) | −17.0±2.5* (5) | 7.7±0.6 (5) | 11.3±1.0* (5) | 1440±24* (5) | 10.6+0.5 (5) |
| L1066V | 74.3±8.7 (6) | −0.5±0.8* (5) | 7.1±0.4 (5) | 4.0±0.2 (4) | 719±29 (4) | 18.9±1.1* (4) |
| V644L | 62.0±6.4 (12) | −10.6±1.7 (7) | 6.6±0.4 (7) | 4.8±0.3 (4) | 790±87 (4) | 8.9±0.3* (5) |
| I662T | 73.1±10.0 (8) | −8.4±1.0 (6) | 7.3±0.1 (6) | 7.5±0.6 (5) | 822±57 (5) | 10.6±0.6 (4) |
Values with an asterisk indicate statistical significance (P<0.05) compared with WT assessed using one-way analysis of variance followed by the Tukey post hoc test.
Trafficking of homomeric CNBHD mutations
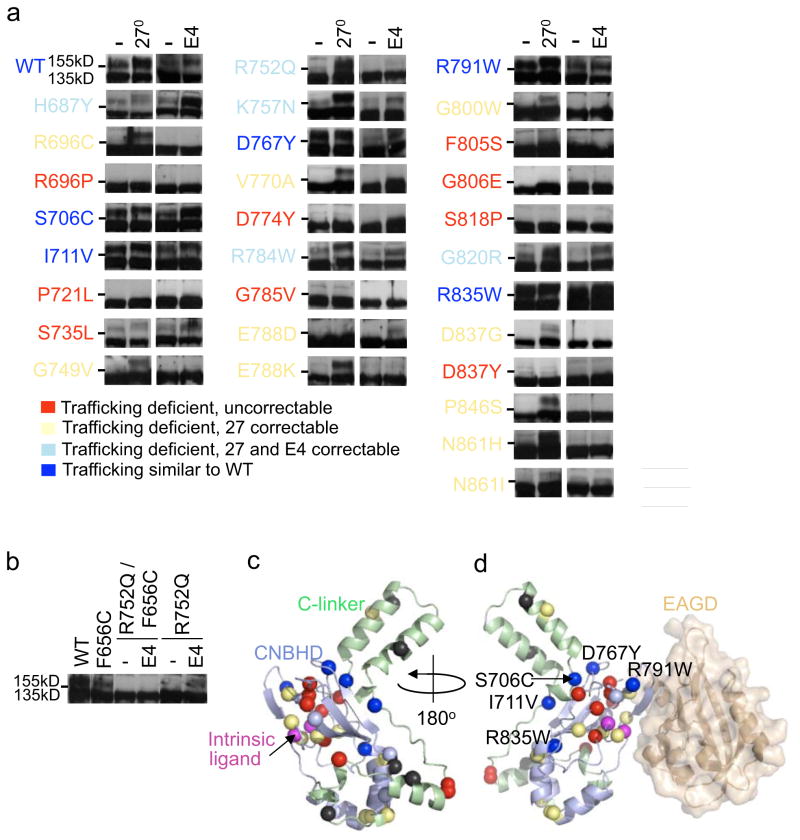
The C-terminus of Kv11.1 contains a CNBHD coupled to the pore through a Clinker region (C-linker/CNBHD, PDB: 3UKN)19. This domain contains at least 41 (39 LQT2, 2 SIDS) mostly uncharacterized mutations at 33 sites. To determine their trafficking phenotype we used the same immunoblot assay and color-code presentation described for the EAGD. Figure 4a shows representative immunoblots (n≥2) for 29 mutations. 24(82%) lacked or had a diminished 155kD band on immunoblot compared to WT for control (37°C) culture conditions. 15 of the 24 (62%) were correctable with culture at 27°C and 5 (17%) were correctable with culture in E4031. Similar to the EAGD, 10 mutations were only temperature correctable but no mutations were only E4031 correctable. These data combined with several mutations already reported in the literature is summarized in Supplementary Table 3. Since this is the first report of an E4031 correctable C-terminal mutation, we tested to see if it acts through the same pore drug binding-site42,43. Indeed, F656C, which traffics normally but attenuates E4031 drug block of IKv11.1, abolished E4031 correction of R752Q (Fig. 4b).
Figure 4. Trafficking phenotype and structural context of LQT2-linked Clinker/CNBHD LQT2 missense mutations.
(a, b) Representative immunoblots of transiently transfected HEK293 cells comparing trafficking under control conditions at 37°C, at reduced temperature (27°C, 24hrs) or in E4031 (E4, 24 hrs). Dashes (-) indicate the 140kD molecular weight marker. Mutations are color-coded as follows: trafficking deficient and uncorrectable in red, trafficking deficient but correctable at 27°C in yellow, trafficking deficient but correctable at 27°C or with E4031 in light blue and those that traffic similar to WT in blue. (c) Model of the C-linker/CNBHD with C-linker region in green, CNBHD in blue, and intrinsic ligand in magenta. Mapped LQT2 residues correspond to panel A and uncharacterized mutations (L678P, L693P, I728F) are black. (d) Representation of the EAGD (colored wheat) (PDB: 4HP9) complexed with the C-linker/CNBHD model from alignments to the structure of the EAGD-CNBHD complex from mouse (PDB: 4LLO).
Structural and functional properties of CNBHD mutations
Mutations were mapped onto a homology model of the C-linker/CNBHD (residues 666–749) described in the methods (Fig. 4c; Supplementary Fig. 1c, d). Each monomer consists of a β-roll motif containing a short β-strand with F860 and L862 forming an ‘intrinsic ligand’ rather than a modulatory ligand (PDB: 3UKN)19,35,53. While the different trafficking deficient phenotypes are scattered showing no correlation with location, most of the uncorrectable mutations (red) lie inside the β-roll and near the intrinsic ligand suggesting that disruption of this subdomain is highly destabilizing. (Fig. 4c, d). Previous studies have shown that a hydrophobic patch that interfaces with the EAGD as well as an acidic patch are important for regulating Kv11.1 deactivation30,36 (Fig. 4d; Supplementary Fig. 1e–h). Surprisingly, of the trafficking competent mutations, only R791W lies close to a gating microdomain at the interface with the EAGD. S706C, I711V and R835W are largely buried but near the C-linker/CNBHD dimer interface while D767Y is not located near any known gating microdomain (Supplementary Fig. 1c, g, h). To determine their electrophysiological properties, stable cell lines were generated for each of these with representative immunoblots and IKv11.1 densities shown in Supplementary Figure 2A. The functional properties of S706C were similar to WT in contrast to a previous report showing small changes in gating (Table 1; Fig. 3e–h) 36. The V1/2 of D767Y and R791W were both shifted by −12mV, while R835W was shifted by +16mV (Fig. 3e, f) and deactivation measured at −50mV for I711V, R791W, and R835W was faster compared to WT (Fig. 3g). To our knowledge, these are the first reports of class 3 (altered gating) LQT2-linked CNBHD mutations studied in a mammalian expression system.
Trafficking of homomeric pore mutations
At least 100 pore missense mutations have been linked to LQT2, yet the majority remain uncharacterized. To determine their trafficking phenotype, we used the same immunoblot assay and color-code presentation described for the EAGD with the exception of orange indicating E4031 correctable only. Figure 5a shows representative immunoblots (n≥2) for 71 mutations. 60 (85%) lacked a 155kD band on immunoblot compared to WT under control culture conditions. 12 of those 60 (20%) were correctable with culture at 27°C and 20 (33%) were correctable with culture in E4031. Interestingly, 8 were only E4031 correctable and none were only temperature correctable, in contrast to the PASD and CNBHD. These data, combined with several mutations already reported in the literature, are summarized in Supplementary Table 4.
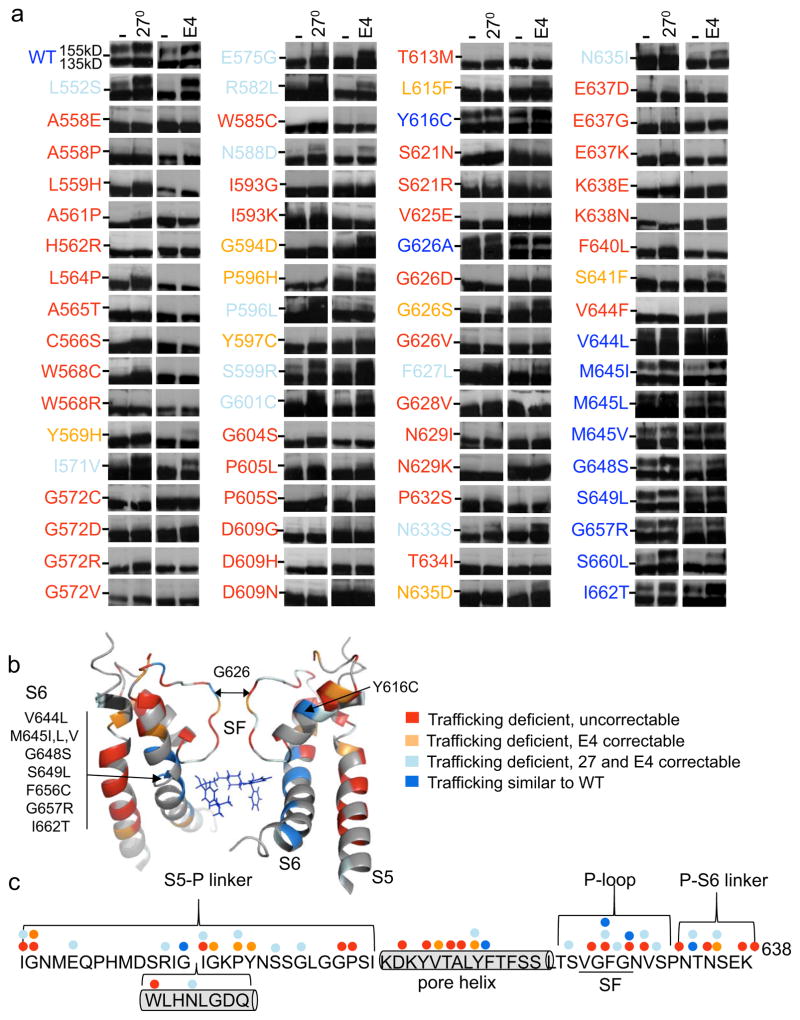
Figure 5. Trafficking phenotype of LQT2-linked pore missense mutations.
(a) Representative immunoblots of transiently transfected HEK293 cells comparing trafficking under control conditions at 37°C, at reduced temperature (27°C) or in E4031 (E4). Dashes (-) indicate the 140kD molecular weight marker. Mutations are color-coded as follows: trafficking deficient and uncorrectable in red, trafficking deficient but correctable at 27°C or with E4031 in light blue, trafficking deficient but only correctable with E4031 in orange and those that traffic similar to WT in blue. (b) Structural model of the pore with Terfenadine (blue molecule) modeled into the putative drug-binding domain54. Mapped LQT2 residues correspond to panel A and uncharacterized mutations (M574R and L622F) are black. (c) Linear representation of the S5–S6 pore linker with helices represented as cylinders and colored dots representing the different trafficking phenotypes associated with each residue (e.g. G626).
Structural and functional properties of pore mutations
Mutations were mapped onto a previously published model of the open state (Fig. 5b)54. A few patterns emerge. First, most of the trafficking competent mutations are located in S6. Second, most S5 mutations have a severe trafficking phenotype with all 14 trafficking deficient and only two correctable. Finally, most trafficking deficient but correctable mutations lie in the pore linker between S5 and S6 (Fig. 5b, c). Similar to the PASD and C-linker/CNBHD, some residues contain more than one LQT2-linked mutation with different trafficking phenotypes (Fig. 5c). For example, G626A in the selectivity filter traffics normally whereas G626D and G626V are trafficking deficient and uncorrectable and G626S is E4031 correctable only (Fig. 5a, b).
To determine if the trafficking competent pore mutations express currents, stably transfected cell lines were generated for V644L, S649L, G657R (a conserved glycine) and I662T, all located in S6 as well as Y616C located in the pore helix. All show a 155kD band but only V644L, G657R and I662T express large amplitude IKv11.1 (Supplementary Fig. 2a). By contrast, stable cell lines for Y616C (6.0±1.0 pA/pF, n=8) and S649L (2.5±0.6 pA/pF, n=8) showed minimal IKv11,1 suggesting a class 4 (altered permeation) mechanism. For the G657R mutation, depolarizing steps to −70 to 50mV also resulted in activation of no outward IKv11,1, whereas a small amplitude inward tail current was elicited by a voltage step to −120mV suggesting a class 3 mechanism (Supplementary Fig. 2b, c). The functional properties of I662T behaved similar to WT but V644L exhibited faster inactivation measured at 0mV consistent with a class 3 (abnormal gating) mechanism (Table 1; Fig. 6c–f).
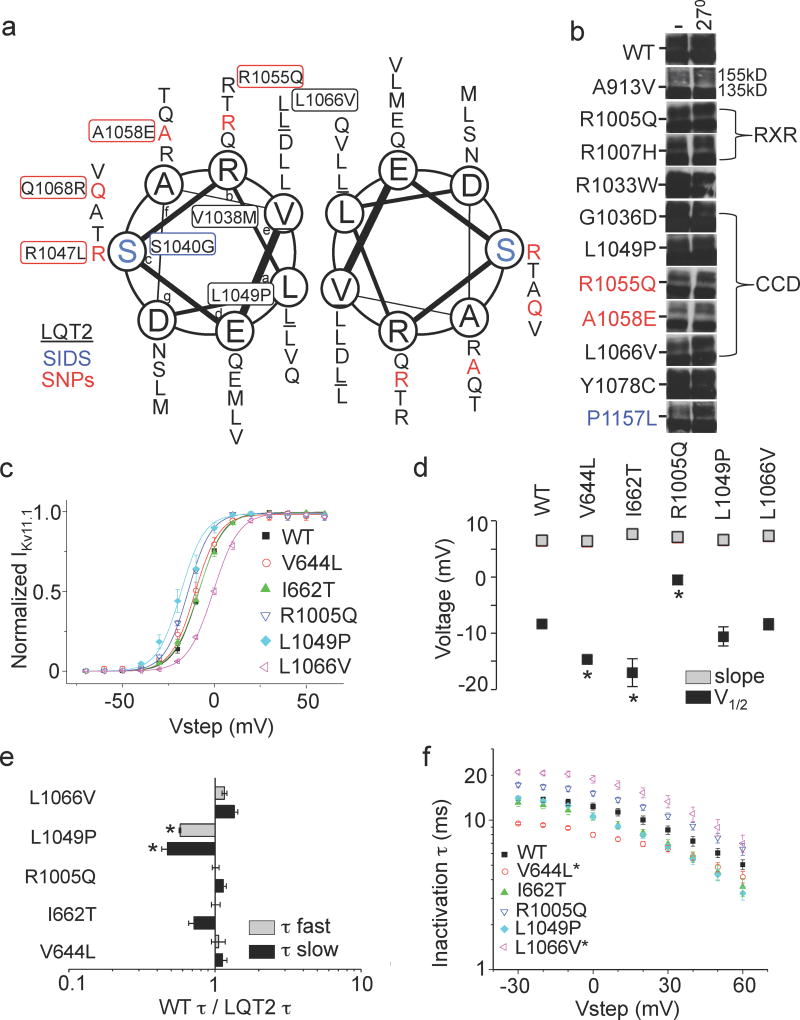
Figure 6. Trafficking phenotype of LQT2-linked distal C-terminus missense mutations.
(a) Helical wheel diagram showing two of the four helices forming the coiled coil domain with LQT2 mutations underlined in black, SIDS mutations in blue and SNPs red. (b) Representative immunoblots of transiently transfected HEK293 cells comparing trafficking under control conditions at 37°C and at reduced temperature (27°C). Dashes (-) indicate the 140kD molecular weight marker. (c) I-V relationships. (d) V1/2 and slope factors. (e) WT-to-mutant time constant ratios (speeding factor) for deactivation at −50mV. (f) Inactivation time constants determined at 0mV. Protocols are described in Fig. 3. Error bars are SEM. Asterisks indicate statistical significance (p<0.05). n = 4 to 12_HEK293 cells for each experiment.
Several LQT2 linked mutations introduce a polar residue or proline and may decrease membrane insertion efficiency; a mechanism underlying other membrane protein misfolding diseases55–57. Using the ΔG predictor tool58, we found that 10 of 17 S5 mutations but only 1 in S6 are predicted to insert less efficiently (ΔΔG≥0.5), which might partially explain why most S5 mutations have a severe trafficking phenotype that is not correctable and most in S6 mutations traffic normally. Mutations in S1–S4 were also modeled using the ΔG predictor tool and only 2 of 20 mutations (both in S1) had a ΔΔG≥0.5. Interestingly, several mutations in S2 and S3 and all in S4 occur at charged residues important for voltage sensing but these also may be important for membrane insertion (Supplementary Table 5)59.
Structural and functional properties of C-term mutations
The distal C-terminus (residues beyond the CNBHD) contains at least 36 (31 LQT2,5 SIDS) mostly uncharacterized mutations and 16 SNPs, several of which are located in or near the CCD and ER retention signal (RXR) (Fig. 1a and 6a). Immunoblot analysis (n≥2) of transiently transfected HEK293 cells revealed that all 8 LQT2 mutations, 1 SIDS and 2 SNPs tested traffic similar to WT (Fig. 6b).
To determine their electrophysiological properties, stably transfected HEK293 cells were generated for R1005Q in the RXR as well as L1049P and L1066V, which are at the hydrophobic interface of the CCD (Supplementary Fig. 2a). The V1/2 of R1005Q and L1049P shifted −6mV and −9mV, respectively, and L1066V shifted +8mV. L1049P also exhibited slower deactivation measured at −50mV and L1066V exhibited slower inactivation measured at 0mV (Table 1; Fig. 7c–f). Interestingly, Paircoil2 analysis predicted that L1049P, but not L1066V disrupts the coiled-coil domain, which might explain their slightly different functional characteristics even though they are similarly located.
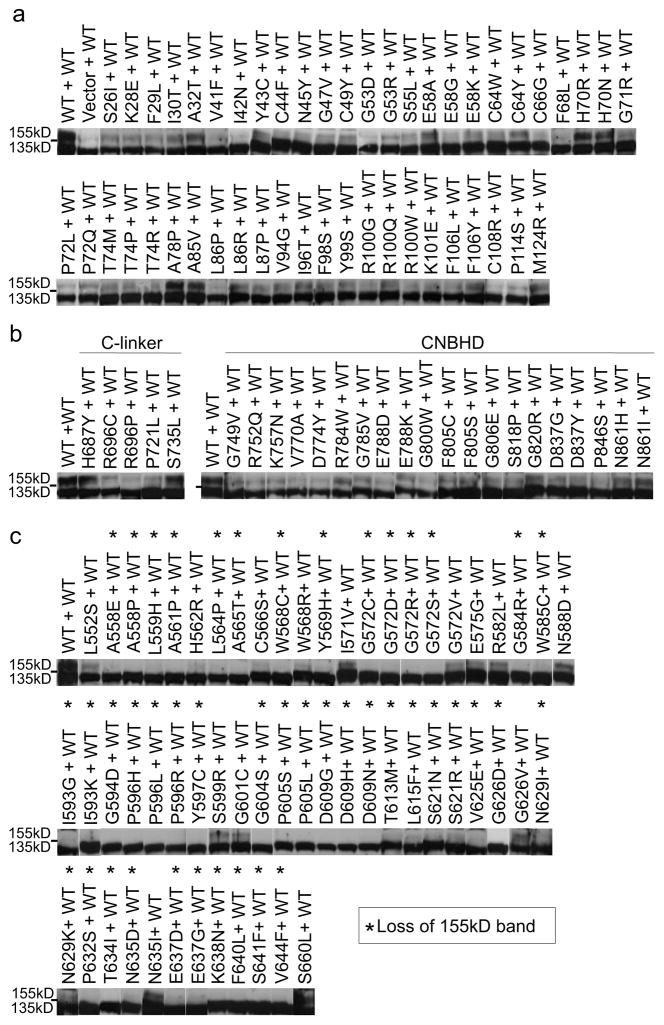
Figure 7. Trafficking phenotype of LQT2 missense mutations co-expressed with WT.
Representative immunoblots of transiently transfected HEK293 cells co-expressing WT and mutant under control conditions. Dashes (-) indicate the 140kD molecular weight marker. Asterisks (*) indicate complete absence of the 155kD band.
Trafficking of LQT2 mutations co-expressed with WT
Since LQT2 is an autosomal dominant disease where only one abnormal allele is present, immunoblots were also performed on HEK293 cells co-transfected with equal amounts of WT and mutant DNA. Immunoblot analysis (n≥2) showed that nearly all missense mutants exhibited a diminished 155kD band compared to WT alone (Fig. 7a–c). A few mutations such as H70R and A78P in the PASD, H687Y and S735L in the CNBD, and S660L in the pore showed a 155kD band similar to WT. However, in contrast to the PASD and CNBHD where nearly every mutation showed at least a weak 155kD band, 44 of 58 (76%) of pore mutations completely lacked a 155kD band (Fig. 8c, indicated as *). Thus, in the pore domain the presence of mutant α-subunit(s) is strictly dominant-negative for the majority of mutations.
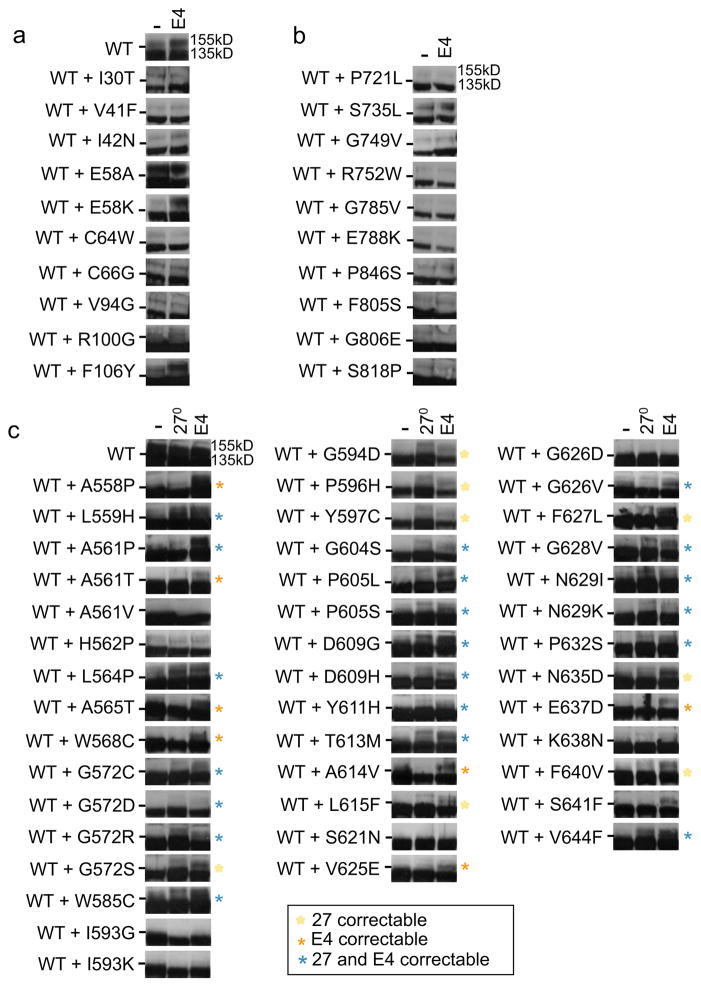
Figure 8. Correction of heteromeric Kv11.1 channels.
Representative immunoblots of transiently transfected HEK293 cells co-expressing WT and mutant alleles as heteromeric channels under control conditions (-), with culture at 27°C (24hrs) or culture in E4031 (E4, 24 hrs). Dashes (-) indicate the 140kD molecular weight marker. Yellow, orange, and blue asterisks indicate correction at 27°C, in E4031, or both, respectively, that were not correctable under those conditions as homomeric channels. H562P is also included, which was not completely dominant negative.
To determine if co-expression of mutant + WT alleles changes the trafficking phenotype of HEK293 cells cultured in E4031, immunoblot analysis (n≥2) was performed on 10 PASD mutations and no differences were found from homomeric channels. Eight mutations remained uncorrectable with culture in E4031 while 2 positive controls (F106Y and E58K) were still correctable (Fig. 8a). Similarly, co-expression of WT + 10 C-linker/CNBHD mutations was tested and no differences in E4031 response was found (Fig. 8b). However, when 42 pore mutations were co-expressed with WT, including four previously reported (A561P, A561T, Y611H, and A614V)10, 35 (83%) showed enhancement of the 155kD band (Fig 8c). Of these 35, 20 demonstrated improvement of the 155kD band with culture at reduced temperature or with E4031 that were not correctable with reduced temperature or E4031 as homomeric channels (blue asterisks). Seven showed improved trafficking with E4031 that were not correctable with E4031 as homomeric channels (orange asterisks) and 8 could be corrected with reduced temperature that were not temperature correctable as homomeric channels (yellow asterisks). Thus, in the pore domain, correction of trafficking deficient mutations depends on α-subunit composition and is enhanced by the addition of WT α-subunits.
Discussion
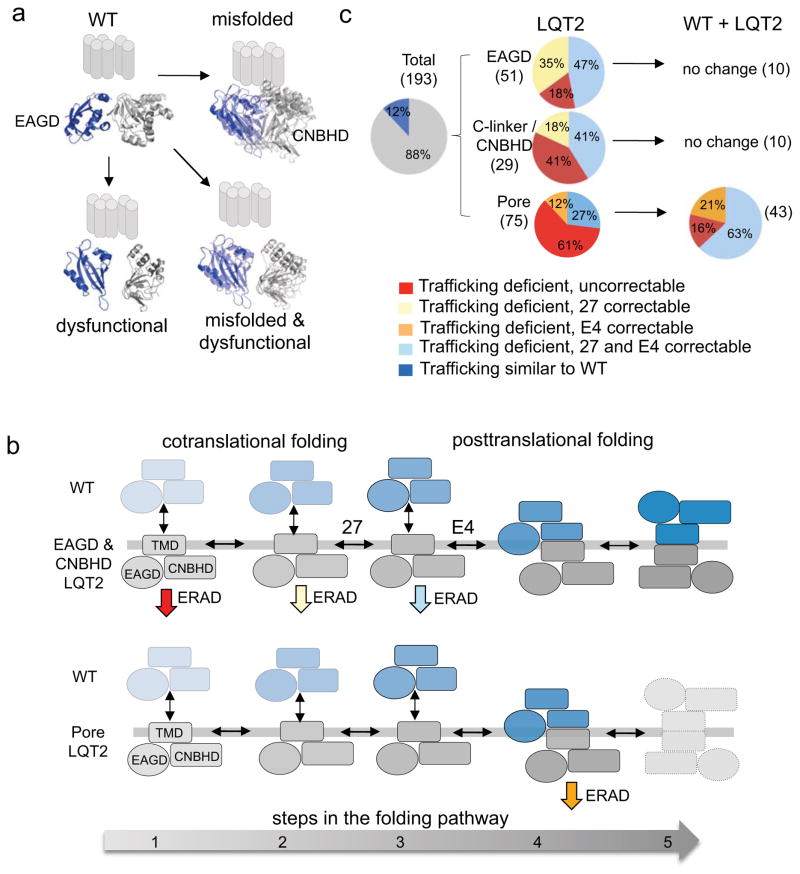
Our data driven approach provides new insights into LQT2 that would not have otherwise been possible. First we identified a loss-of-function mechanism for most of the LQT2-linked missense mutations in three structural domains. Including previously published findings with our present findings (Supplementary Tables 1–4), 193 PASD, Clinker/CNBHD, and pore mutations have been studied and 169 (88%) demonstrate a class 2 (trafficking deficient) mechanism. This is in contrast to a smaller study of 10 PASD mutations that suggested most traffic normally28, underscoring the importance of this comprehensive large-scale analysis. Of mutations studied electrophysiologically, 9 show evidence for being class 3 (abnormal gating) including several shown for the first time in the CNBHD. Four were trafficking competent and generated minimal or no current suggesting either a severe class 3 or a class 4 (altered permeation) mechanism. Several “mutations” behave similar to WT including most in the distal C-terminus and it is unclear whether they are true LQT2 disease-causing mutations or benign variants.
Not surprisingly, we found that C-linker/CNBHD mutations behave similarly to PASD mutations. Since class 3 (abnormal gating) PASD mutations have been shown to disrupt ‘core’ interactions with the CNBHD62, class 3 CNBHD mutations likely act similarly. Indeed, to our knowledge we identified the first class 3 CNBHD mutations as well as the first class 2 (trafficking deficient) CNBHD mutations that can be corrected with culture in E4031. Figure 9A illustrates these similarities where most mutations are destabilizing resulting in misfolded channels with reduced IKv11.1 density but nearly normal gating (e.g. G53R, H70R, and A78P)50. Other mutations traffic normally but are located near gating microdomains resulting in dysfunctional channels but with nearly normal IKv11.1 density (e.g. R56Q, N33T, R835W, R791W)16. Finally, other mutations are both destabilizing (misfolded) and located in gating microdomains (dysfunctional) (e.g. K28E, F29L, M124R, R784W, and E788K)36,51. This model might also help explain why G53R, H70R, A78P all exhibit faster deactivation in Xenopus oocytes but not in HEK293 cells50,51. Misfolding mutations lying outside gating microdomains will still fail to form the ‘core’ interactions necessary for proper function but are not marked for degradation in Xenopus oocytes, which are cultured at reduced temperature. However, the opposite is true for S706C, which was reported to have faster deactivation in Xenopus oocytes but not in this study using HEK293 cells36. Clearly, differences between expression models need to be considered when evaluating LQT2 mutations and perhaps studies in more native model systems would be useful60,61.
Figure 9. Model of Kv11.1 biogenesis and correction.
(a) Model illustrating class 2 and/or class 3 loss-of-function phenotypes for EAGD and C-linker/CNBHD mutations. Mutations can either be destabilizing resulting in misfolding and ERAD (top right, out of focus), disrupt protein-protein interactions resulting in quicker deactivation (bottom left, separated) or both (bottom right). (b) Summary of the trafficking phenotypes by domain for Kv11.1 homomeric channels and channels co-expressed with WT. Number of mutations analyzed are in parentheses (see also Supplementary Tables 2,3,4). (c) Simplified five step folding model illustrating the different correction phenotypes based on data reported here and elsewhere (see discussion). The most destabilizing PASD and C-linker/CNBHD mutations fail to make it to step 2 and undergo ERAD (red). These are uncorrectable at 27° or with E4. Less destabilizing mutations make it further along the folding pathway (step 2) and are amenable to 27°C temperature correction but not E4 pharmacological correction (yellow). Mildly destabilizing mutations make it to step 3 where E4031, which acts postranslationally, can facilitate cooperative interactions between subunits allowing for correction13. (d) Pore mutations (orange) behave differently. Most are stable enough to make it to step 4 but result in severe dominant-negative effects causing both WT and mutant to undergo ERAD. These heteromeric channels fail to make it to step 5 (dashed outlines) required for ER exit but can undergo improved folding and ER exit with pharmacological correction strategies.
Several lines of evidence suggest that correction of trafficking deficient intracellular PASD and CNBHD mutations works at different steps along the folding pathway as illustrated in Fig. 9b. This model is supported by: 1) correction strategies typically work only for missense mutations with a diminished 155kD band but not for mutations completely lacking a 155kD band, 2) the trafficking phenotype for several PASD mutations has been shown to correlate with domain stability28,63, 3) all PASD and C-linker/CNBHD mutations that are E4031 correctable are also temperature correctable suggesting that E4031 works further along the folding pathway where the pore is intact, and 4) E4031 acts posttranslationally13. Since the majority of mutations are correctable and therefore likely only slightly destabilizing, strategies that improve domain stability could help many LQT2- Kv11.1 channels evade ERAD64.
Pore mutations are strikingly different than intracellular domain mutations. 2/3rd act in a completely dominant negative manner when co-expressed with WT in contrast to intracellular mutations (Fig. 9b). Furthermore, heteromeric channels are much more amenable to pharmacological correction than homomeric channels (Fig. 9c) These results provide an explanation for the increased clinical severity of pore mutations but also show more promise for pharmacological chaperones as a possible therapeutic approach65.
While these studies have provided several new molecular insights into LQT2, many important questions remain. Since co-expression of WT can improve mutant Kv11.1 channel dysfunction66, functional studies of co-expressed channels are needed. For example, Kv11.1 G657R appears to behave similar to a Shaker channel mutation that converts it from a K+ channel passing outward current to a K+ channel passing inward current at negative voltages67 What loss-of-function mechanism(s), if any, underlie the other Kv11.1 structural regions like the VSD68 or the linker regions? Interestingly, as shown in Fig 1, all reported SNPs lie in linker regions of the Kv11.1 protein lacking highly ordered structure except for the CCD, thus these areas may be more tolerant of amino acid substitutions. Using the sequence-based prediction tools SIFT, SNP&Go and KvSNP, only 21–73% of mutations and 11–46% of SNPs were predicted to be pathogenic compared to 80–99% in structural domains (see Supplementary Table 1 and 6). What are the structural bases for Kv11.1 misfolding? This is a largely unexplored area but especially important since LQT2 is predominantly a misfolding disease. For example, do LQT2 missense mutations disrupt intra (local) and/or interdomain (global) interactions69? Do TMD mutations insert less efficiently as predicted, which has been demonstrated for other channelopathies55,56? Does L1049P disrupt the CCD as predicted? As a final example, it was surprising that LQT2 mutations in the ER retention signal (R1005Q and R1007H) both traffic normally. This suggests that these mutations do not disrupt binding of distal C-terminus, which is thought to mask the RXR signal20. These questions and many others remain unresolved but this large-scale mutational analysis has shed new light on LQT2 disease mechanisms and may help efforts towards developing rational approaches to predicting the pathogenic mechanism(s) of newly discovered Kv11.1 variants and correction strategies as clinically useful therapeutic options70.
Methods
Mutation database and mutagenesis
LQT2-associated Kv11.1 missense mutations were identified predominantly from the Inherited Arrhythmias Database (www.fsm.it/cardmoc) and published genotyping from several literature sources listed in Supplementary Table 1. All missense mutations were made using the QuikChange II XL kit from Agilent (Santa Clara, CA) using primers designed with their primer design program and obtained from Integrated DNA Technologies (Coralville, IA). A pcDNA3 WT HERG1a expression construct previously published was used as the template10. Restriction digest analysis was used to test the integrity of all constructs and all mutations were verified by sequencing at the UW-Biotechnology Center.
Bioinformatics
PASD structural models were created in Pymol using PDB: 1BYW. A Clinker/CNBHD structural model (Kv11.1 amino acids 672–864) was generated by Swiss-Model using the C-terminal structure from the homologous zELK KCNH (PDB: 3UKN) as a template. The quality of the model was evaluated using Molprobity and scored in the 56th percentile (100% being the best). After energy minimization using UCSF Chimera’s minimization function, the model improved to the 91st percentile and was used for all subsequent modeling. Mutation pathogenicity was also evaluated for all mutations using the sequence-based programs SIFT, KvSNP, and SNPs&Go. Membrane insertion efficiency was predicted using the ΔG predictor. This tool predicts the apparent free energy difference for membrane insertion based on an experimentally determined biological hydrophobicity scale. Paircoil 2 was used to predict coiled-coil formation. Web servers and literature sources are listed in Supplementary Table 7.
Kv11.1 trafficking
HEK293 cells of similar confluence were transiently transfected with SuperFect (Qiagen) and grown at 37°C for 24hrs before study. Transfections were done with 3μg of cDNA for homomeric expression or 1.5μg mutant +1.5μg WT cDNA for co-expression. To test for correction of deficient Kv11.1 protein trafficking, cells were grown for an additional 24hrs at 37°C for control, or at 27°C, or in the presence of 10μM E4031 (Alamone). Immature and mature Kv11.1 protein bands were detected by immunoblot analysis of whole-cell lysates. Briefly, lysates were mixed with an equal amount of Laemili sample buffer, separated by 7% SDS-PAGE, and transferred to nitrocellulose paper. Blots were incubated with a rabbit antibody (1:10,000) directed to the distal C-terminus as previously described9,10. Bands were detected with a goat anti-rabbit secondary antibody conjugated to HRP. Uncropped western blots are shown in Supplementary Fig 2e.
Kv11.1 Function
Stable cell lines were generated by transfecting HEK293 cells with mutant HERG pcDNA3 and selecting in G418 as previously described10. Single colonies of G418 resistant cells were then tested for Kv11.1 expression by immunoblot. Cell lines that gave a robust 155kD band on immunoblot were used for electrophysiological analysis. IKv11.1 was measured using the whole-cell patch clamp technique as previously described10. Voltage protocols are described in the Fig. 3 legend and data analysis was done using pCLAMP 8.0 (Axon Instruments) and Origin (6.0 Microcal).
Statistical Analysis
All data are presented as mean ± SE. One-way analysis of variance (ANOVA) was used for statistical analysis followed by the Tukey post hoc test. P<0.05 considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Jennifer Poehls in the Dept. of Medicine and Dr. Krishanu Saha in the Dept. of Biomedical Engineering at UW-Madison for helpful suggestions. We thank Dr. Michael Ackerman (Mayo Clinic) for sharing pre-publication genotype information. This study was supported by the NIH R01 HL060723 (C.T.J), an AHA Midwest Affiliate Predoctoral Fellowship (C.L.A), a grant from the Saving tiny Hearts Society (B.P.D), and a NIH training grant T32 HL07936 (Dr. Jonathan C. Makielski).
Footnotes
Author Contributions. C.L.A., C.E.K., R.R.C., and C.J.H. performed the experiments. C.L.A., B.P.D. and C.T.J. contributed to the study design and manuscript generation.
Competing Financial Interests. Dr. January is a co-founder of Cellular Dynamics International. Dr. Delisle has a research contract with Gilead Scientific. The authors declare no other competing financial interests.
References
- 1.Curran ME, et al. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80(5):795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 2.Tester DJ, Ackerman MJ. Sudden infant death syndrome: How significant are the cardiac channelopathies? Cardiovasc Res. 2005;67:388–396. doi: 10.1016/j.cardiores.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Brugada R, et al. Sudden death associated with short-QT syndrome linked to mutations in HERG. Circulation. 2004;109(1):30–5. doi: 10.1161/01.CIR.0000109482.92774.3A. [DOI] [PubMed] [Google Scholar]
- 4.Crociani O, et al. Cell cyce-dependent expression of HERG1 and HERG1B isoforms in tumor cells. J Biol Chem. 2003;278:2947–2955. doi: 10.1074/jbc.M210789200. [DOI] [PubMed] [Google Scholar]
- 5.Johnson JN, et al. Identification of a possible pathogenic link between long QT syndrome and epilepsy. Neurology. 2009;27:224–231. doi: 10.1212/01.wnl.0000335760.02995.ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huffaker SJ, et al. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med. 2009;15(5):509–18. doi: 10.1038/nm.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delisle BP, Anson BD, Rajamani S, January CT. Biology of cardiac arrhythmias: ion channel protein trafficking. Circ Res. 2004;94(11):1418–28. doi: 10.1161/01.RES.0000128561.28701.ea. [DOI] [PubMed] [Google Scholar]
- 8.Gong Q, Zhang L, Vincent MG, Horne BD, Zhou Z. Nonsense mutations in hERG cause a decrease in mutant mRNA transcripts by nonsense-mediated mRNA decay in human long QT syndrome. Circulation. 2007;116(1):17–24. doi: 10.1161/CIRCULATIONAHA.107.708818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, Gong Q, Epstein ML, January CT. HERG channel dysfunction in human long QT syndrome. Intracelllar transport and functional defects. J Biol Chem. 1998;273(33):21061–21066. doi: 10.1074/jbc.273.33.21061. [DOI] [PubMed] [Google Scholar]
- 10.Anderson CL, et al. Most LQT2 mutations reduce Kv11.1 (hERG) current by a class 2 (trafficking-deficient) mechanism. Circulation. 2006;113:365–373. doi: 10.1161/CIRCULATIONAHA.105.570200. [DOI] [PubMed] [Google Scholar]
- 11.Gong Q, Keeney DR, Molinari M, Zhou Z. Degradation of trafficking-defective long QT syndrome type II mutant channels by the ubiquitin-proteasome pathway. J Biol Chem. 2005;280:19419–19425. doi: 10.1074/jbc.M502327200. [DOI] [PubMed] [Google Scholar]
- 12.Walker VE, et al. Hsp40 chaperones promote degradation of the hERG potassium channel. J Biol Chem. 2010;285:3319–3329. doi: 10.1074/jbc.M109.024000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z, Gong Q, January CT. Correction of defective protein trafficking of a mutant HERG potassium channel in human long QT syndrome. Pharmacological and temperature effects. J Biol Chem. 1999;274(44):31123–31136. doi: 10.1074/jbc.274.44.31123. [DOI] [PubMed] [Google Scholar]
- 14.Lu X, et al. RNA interference targeting E637K mutation rescues the hERG channel currents and restores its kinetic properties. Heart Rhythm. 2013;10(1):128–136. doi: 10.1016/j.hrthm.2012.09.124. [DOI] [PubMed] [Google Scholar]
- 15.Matsa E, et al. Allele-specific RNA interference rescues the long-QT syndrome phenotype in human-induced pluripotency stem cell cardiomyocytes. Eur Heart J. 2013;35(16):1078–1087. doi: 10.1093/eurheartj/eht067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balijepalli SY, Anderson CL, Lin EC, January CT. Rescue of mutated cardiac ion channels in inherited arrhythmia syndromes. J Cardiovasc Pharmacol. 2010;56(2):113–122. doi: 10.1097/FJC.0b013e3181dab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 18.Morais-Cabral JH, Lee A, Cohen SL, Chait BT, IM, Mackinnon R. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 19.Brelidze TI, Carlson AE, Sankaran B, Zagotta WN. Structure of the carboxy- terminal region of a KCNH channel. Nature. 2012;481(7382):530–533. doi: 10.1038/nature10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kupershmidt S, et al. Defective human ether-a-go-go-related gene trafficking linked to an endoplasmic reticulum retention signal in the C-terminus. J Biol Chem. 2002;277:27442–27448. doi: 10.1074/jbc.M112375200. [DOI] [PubMed] [Google Scholar]
- 21.Jenke M, et al. C-terminal domains implicated in the functional expression of potassium channels. EMBO J. 2003;22(3):395–403. doi: 10.1093/emboj/cdg035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong Q, Anderson CL, January CT, Zhou Z. Pharmacological rescue of trafficking defective HERG channels formed by co-assembly of wild-type and long QT mutant N470D subunits. Am J Physiol Heart Circ Physiol. 2004;287(2):H652–658. doi: 10.1152/ajpheart.00052.2004. [DOI] [PubMed] [Google Scholar]
- 23.Vandenberg JI, et al. hERG K+ channels: structure, function, and clinical significance. Physiol Rev. 2012;92:1393–1478. doi: 10.1152/physrev.00036.2011. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita K, et al. A novel missense mutation causing a G487R substitution in the S2–S3 loop of human ether-a-go-go-related gene channel. J Cardiovasc Electrophysiol. 2012;23(11):1246–1253. doi: 10.1111/j.1540-8167.2012.02383.x. [DOI] [PubMed] [Google Scholar]
- 25.Moss AJ, et al. Increased risk of arrhythmic events in long QT syndrome with mutations in the pore region of the human ether-a-go-go-related gene potassium channel. Circulation. 2002;105:794–799. doi: 10.1161/hc0702.105124. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu W, et al. Genotype-phenotype aspects of type 2 long QT syndrome. J Am Coll Cardiol. 2009;54(22):2052–62. doi: 10.1016/j.jacc.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akhavan A, et al. Identifcation of the cyclic-nucleotide-binding domain as a conserved determinant of ion channel cell-surface localization. J Cell Sci. 2005;118:2803–2812. doi: 10.1242/jcs.02423. [DOI] [PubMed] [Google Scholar]
- 28.Harley CA, Jesus CS, Carvalho R, Brito RM, Morais-Cabral JH. Changes in channel trafficking and protein stability caused by LQT2 mutations in the PAS domain of the HERG channel. PLoS One. 2012;7(3):e32654. doi: 10.1371/journal.pone.0032654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, et al. NMR solution structure of the N-terminal domain of hERG and its interaction with the S4-S5 linker. Biochem Biophys Res Comm. 2010;403(1):126–132. doi: 10.1016/j.bbrc.2010.10.132. [DOI] [PubMed] [Google Scholar]
- 30.Muskett FW, et al. Mechanistic insight into human ether-a-go-go-related gene (hERG) K+ channel deactivation gating from the solution structure of the EAG domain. J Biol Chem. 2011;286(8):6184–6191. doi: 10.1074/jbc.M110.199364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gustina AS, Trudeau MC. A recombinant N-terminal domain fully restores deactivation gating in N-truncated and long QT syndrome mutant hERG potassium channels. Proc Natl Acad Sci. 2009;106(31):13082–13087. doi: 10.1073/pnas.0900180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gustina AS, Trudeau MC. hERG potassium channel gating is mediated by N- and C-terminal region interactions. J Gen Physiol. 2011;137(3):315–325. doi: 10.1085/jgp.201010582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gianulis EC, Liu Q, Trudeau MC. Direct interaction of eag domains and cyclic nucleotide-binding homology domains regulate deactivation gating in hERG channels. J Gen Physiol. 2013;142(4):351–356. doi: 10.1085/jgp.201310995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Peña P, Machín A, Fernández-Trillo J, Domínguez P, Barros F. Mapping of interactions between amino and carboxy termini and the channel core in hERG K+ channels. Biochem J. 2013;451(3):463–474. doi: 10.1042/BJ20121717. [DOI] [PubMed] [Google Scholar]
- 35.Haitin Y, Carlson AE, Zagotta WN. The structural mechanism of KCNH-channel regulation by the eag domain. Nature. 2013;501:444–448. doi: 10.1038/nature12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Al-Owais M, Bracey K, Wray D. Role of intracellular domains in the function of the herg potassium channel. Eur Biophys J. 2009;38:569–576. doi: 10.1007/s00249-009-0408-2. [DOI] [PubMed] [Google Scholar]
- 37.Krishnan Y, Zheng R, Walsh C, Tang Y, McDonald TV. Partially dominant mutant channel defect corresponding with intermediate LQT2 phenotype. Pacing Clin Electrophysiol. 2012;35(1):3–16. doi: 10.1111/j.1540-8159.2011.03222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulussen A, et al. A novel mutation (T65P) in the PAS domain of the human potassium channel HERG results in the long QT syndrome by trafficking deficiency. J Biol Chem. 2002;277(50):48610–48616. doi: 10.1074/jbc.M206569200. [DOI] [PubMed] [Google Scholar]
- 39.Aidery P, et al. Identification and functional characterization of the novel human ether-a-go-go related gene (hERG) R744P mutant associated with hereditary long QT syndrome type 2. Biochem Biophys Res Commun. 2012;418(4):830–835. doi: 10.1016/j.bbrc.2012.01.118. [DOI] [PubMed] [Google Scholar]
- 40.Huo J, et al. The G604S-hERG mutation alters the biophysical properties and exerts a dominant-negative effect on expression of hERG channels in HEK293 cells. Pflugers Arch. 2008;456(5):917–928. doi: 10.1007/s00424-008-0454-0. [DOI] [PubMed] [Google Scholar]
- 41.Zhao JT, et al. Not all hERG pore domain mutations have a severe phenotype: G584S has an inactivation gating defect with mild phenotype compared to G572S, which has a dominant-negative trafficking defect and severe phenotype. Cardiovasc Electrophysiol. 2009;20(8):923–30. doi: 10.1111/j.1540-8167.2009.01468.x. [DOI] [PubMed] [Google Scholar]
- 42.Ficker E, Obejero-Paz CA, Zhao S, Brown AM. The binding site for channel blockers that rescue misprocessed human Long QT Syndrome Type 2 ether-a- go-go-related gene (HERG) mutations. J Biol Chem. 2002;277:4989–4998. doi: 10.1074/jbc.M107345200. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez D, Ghanta A, Kauffman GW, Sanguinetti MC. Physicochemical features of the HERG channel drug-binding site. J Biol Chem. 2004;279(11):10120–10127. doi: 10.1074/jbc.M310683200. [DOI] [PubMed] [Google Scholar]
- 44.Rossenbacker T, et al. Novel mutation in the Per-Arnt-Sim domain of KCNH2 causes a malignant form of long-QT syndrome. Circulation. 2005;111:961–968. doi: 10.1161/01.CIR.0000156327.35255.D8. [DOI] [PubMed] [Google Scholar]
- 45.Delisle BP, et al. Intragenic suppression of trafficking-defective KCNH2 channels associated with long QT syndrome. Mol Pharmacol. 2005;68:233–240. doi: 10.1124/mol.105.012914. [DOI] [PubMed] [Google Scholar]
- 46.Gong Q, Jones MA, Zhou Z. Mechanisms of pharmacological rescue of trafficking-defective hERG mutant channels in human long QT syndrome. J Biol Chem. 2006;281(7):4069–4074. doi: 10.1074/jbc.M511765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng CA, et al. The N-terminal tail of hERG contains an amphipathic a-helix that regulates deactivation. PLoS One. 2011;6(1):e16191. doi: 10.1371/journal.pone.0016191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, Myers CD, Robertson GA. Dynamic control of deactivation gating by a soluble amino-terminal domain in HERG K channels. J Gen Physiol. 2000;115(6):749–758. doi: 10.1085/jgp.115.6.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adaixo R, Harley CA, Castro-Rodrigues AF, Morais-Cabral JH. Structural properties of PAS domains from the KCNH potassium channels. PLoS One. 2013;8(3):e59265. doi: 10.1371/journal.pone.0059265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J, Zhou A, Splawski I, Keating MT, Sanguinetti MC. Long QT syndrome-assoicated mutations in the Per-Arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J Biol Chem. 1999;274(15):10113–10118. doi: 10.1074/jbc.274.15.10113. [DOI] [PubMed] [Google Scholar]
- 51.Gianulis EC, Trudeau MC. Rescue of aberrant gating by a genetically encoded PAS (Per-Arnt-Sim) domain in several long QT syndrome mutant human ether-a-go-go-related gene potassium channels. J Biol Chem. 2011;286(25):22160–22169. doi: 10.1074/jbc.M110.205948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shushi L, et al. Clinical, genetic, and electrophysiologic characteristics of a new PaS-domain HERG mutation (M124R) causing Long QT syndrome. Ann Noninvasive Electrocardiol. 2005;10(3):334–341. doi: 10.1111/j.1542-474X.2005.00643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brelidze TI, Carlson AE, Zagotta WN. Absence of direct cyclic nucleotide modulation of mEAG1 and hERG1 channels revealed with fluorescence and electrophysiological methods. J Biol Chem. 2009;284:27989–27997. doi: 10.1074/jbc.M109.016337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farid R, Day T, Friesner RA, Pearlstein RA. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg Med Chem. 2006;14:3160–3173. doi: 10.1016/j.bmc.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 55.Choi MY, et al. Destabilization of the transmembrane domain induces misfolding in a phenotypic mutant of cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2005;280(6):4968–4974. doi: 10.1074/jbc.M410069200. [DOI] [PubMed] [Google Scholar]
- 56.Gallagher MJ, Ding L, Maheshwari A, MacDonald RL. The GABAA receptor alpha-1 subunit epilepsy mutation A322D inhibits transmembrane helix formation and causes proteasomal degradation. Proc Natl Acad Sci. 2007;104(32):12999–13004. doi: 10.1073/pnas.0700163104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rakoczy EP, Kiel C, McKeone R, Stricher F, Serrano L. Analysis of disease-linked rhodopsin mutations based on structure, function, and protein stability calculations. J Mol Biol. 2010;405(2):584–606. doi: 10.1016/j.jmb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 58.Hessa T, et al. Molecular code for transmembrane-helix recognition by the SEC61 translocon. Nature. 2007;450(7172):1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, et al. Contribution of hydrophobic and electrostatic interactions to the membrane integration of the Shaker K+ channel voltage sensor domain. Proc Natl Acad Sci. 2007;104(20):8263–8268. doi: 10.1073/pnas.0611007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lahti AL, et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5(2):220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itzhaki I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 471:225–230. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 62.Gustina AS, Trudeau MC. HERG potassium channel regulation by the N-terminal eag domain. Cell Signal. 2012;24(8):1592–1598. doi: 10.1016/j.cellsig.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ke Y, et al. Trafficking defects in PAS domain mutant Kv11.1 channels: roles of reduced domain stability and altered domain-domain interactions. Biochem J. 2013;454(1):69–77. doi: 10.1042/BJ20130328. [DOI] [PubMed] [Google Scholar]
- 64.Sampson HM, et al. Identification of a NBD1-binding pharmacological chaperone that corrects the trafficking defect of F508del-CFTR. Chem Biol. 2011;18(2):231–242. doi: 10.1016/j.chembiol.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 65.Peal DS, et al. Novel chemical suppressor of long QT syndrome identified by an in vivo functional screen. Circulation. 2011;123(1):23–30. doi: 10.1161/CIRCULATIONAHA.110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balijepalli SY, et al. Mechanism of loss of Kv11.1 K+ current in mutant T421M-Kv11.1-expressing rat ventricular myocytes: interaction of trafficking and gating. Circulation. 2012;126(24):2809–2818. doi: 10.1161/CIRCULATIONAHA.112.118018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L, Liu K, Hu Y, Li D, Luan S. Single mutations convert an outward K+ channel into an inward K + channel. Proc Natl Acad Sci. 2008;105(8):2871–2876. doi: 10.1073/pnas.0712349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McBride CM, et al. Mechanistic basis for type 2 long QT syndrome caused by KCNH2 mutations that disrupt conserved arginine residues in the voltage sensor. J Membr Biol. 2013;246(5):355–64. doi: 10.1007/s00232-013-9539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhaskara RM, Srinivasan N. Stability of domain structures in multi-domain proteins. Sci Rep. 2011;1:40. doi: 10.1038/srep00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giudicessi JR, et al. Phylogenetic and physiochemical analyses enhance the classification of rare nonsynonymous single nucleotide variants in type 1 and 2 long-QT syndrome. Circ Cardiovasc Genet. 2012;5(5):519–528. doi: 10.1161/CIRCGENETICS.112.963785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.