Abstract
Many bacteria assemble extracellular amyloid fibers on their cell surface. Secretion of proteins across membranes and the assembly of complex macromolecular structures must be highly coordinated to avoid the accumulation of potentially toxic intracellular protein aggregates. Extracellular amyloid fiber assembly poses an even greater threat to cellular health due to the highly aggregative nature of amyloids and the inherent toxicity of amyloid assembly intermediates. Therefore, temporal and spatial control of amyloid protein secretion is paramount. The biogenesis and assembly of the extracellular bacterial amyloid curli is an ideal system for studying how bacteria cope with the many challenges of controlled and ordered amyloid assembly. Here, we review the recent progress in the curli field that has made curli biogenesis one of the best-understood functional amyloid assembly pathways.
Keywords: Curli, functional amyloid, aggregate, biofilm, nucleation-precipitation, Type VIII secretion
1. Curli are functional amyloids
Amyloids have been intensely studied for decades and are hallmarks of many human illnesses including Alzheimer’s, Parkinson’s and Huntington’s diseases [1–3]. More recently, a new class of amyloids—the so-called ‘functional’ amyloids—has been described, and new members of this class are rapidly being discovered. Functional amyloids are now believed to be ubiquitous and they perform an enormous diversity of biological functions [4–8]. Numerous functional amyloids are produced by a variety of microbes with the majority of them playing roles in adhesion to surfaces and structural integrity during the establishment and persistence of biofilm communities [9].
Curli, the first identified functional amyloid, are extracellular protein fibers produced by many enteric bacteria including Escherichia coli and Salmonella species (Fig. 1A) [10–13]. Curli are the major proteinaceous component of E. coli biofilms and are important for surface colonization and interacting with host factors and the host immune system [10, 14–21]. Curli production is easily scored in the lab by plating cells on agar plates containing the amyloid binding dye Congo red [17, 22]. Like all amyloids, the major curli subunit CsgA is capable of self-polymerizing in vitro into β-sheet-rich amyloid fibers that bind to the amyloid specific dye Thioflavin T (ThT) and can be visualized using transmission electron microscopy (TEM) [4, 23, 24]. In vitro assembled CsgA amyloid fibers are indistinguishable from fibers formed in vivo [23].
Figure 1.
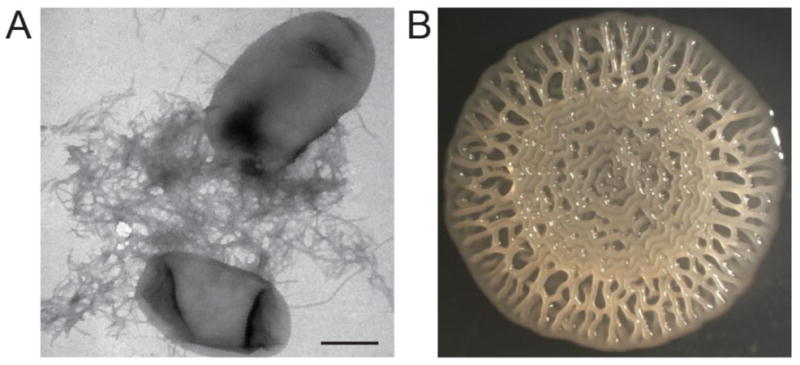
Curli production contributes to E. coli biofilms. A. E. coli k-12 strain BW25113 grown on a low salt agar plate at 26°C produce cell surface associated curli fibers that are visible by transmission electron microscopy. Scale bar is 500nm. B. The uropathogenic E. coli strain UTI89 develops a complex rugose colony morphology that is dependent on curli.
The ability of curli to act as a strong scaffolding agent in biofilm formation stems from properties inherent to all amyloids. Curli are highly stable 6–12 nm wide non-branching fibers (Fig. 1A) that are resistant to degradation by proteases and denaturation by detergents [4].
Pretreatment with a strong denaturant such as formic acid or hexafluoroisopropanol is required to depolymerize curli fibers so that monomers of the major subunit protein, CsgA, can be resolved on an SDS-PAGE gel [4]. CsgA amyloids, like other amyloids, are β-sheet rich and assemble into a highly stable cross-β structure stabilized in large part by tight “steric zipper” interactions between side-chains on adjacent β-sheets [4, 25–27]. The amyloid characteristics of curli fibers are clearly important for their biological function and afford many useful techniques for rapid in vivo and in vitro study of curli biogenesis, biofilm formation and integrity, and amyloid assembly [28, 29].
2. Curli expression is highly coordinated
Curli gene expression is a highly regulated and coordinated event both on the cellular level and within a biofilm community. Curli are the major protein component of E. coli biofilms yet the production of curli within a biofilms is restricted to a distinct subpopulation [14, 30–33]. Expression is controlled by several environmental signals and chemical gradients including temperature, osmolarity and oxygen [34–36]. Within a rugose, or rough, colony biofilm (Fig. 1B) curliated cells localize to the air-colony interface [30, 33]. This bimodal population development can be triggered by oxidative stress, and in turn, rugose biofilms are protected from oxidative damage resulting from exposure to hydrogen peroxide [30]. Bacteria within Salmonella enterica curli- and cellulose-dependent rugose colonies are also protected from desiccation and are more resistant to sodium hypochlorite treatment (or bleach) when compared to non-curliated colonies or planktonic cells, respectively [37]. Successful biofilm formation requires spatial and temporal regulation of curli assembly, which relies on elaborate coordination of gene expression and protein production (Table 1).
Table 1.
Direct Regulators of the Curli Operons.
| Regulator | PcsgBAC | PcsgDEFG | Evidence | Reference(s) | |
|---|---|---|---|---|---|
| Transcriptional Regulators | RpoS | + | + | Expression analysis, consensus binding site | [42, 60, 102, 103] |
| Crl | + | + | Interacts with RpoS and stabilizes binding to csg promoters | [41, 42] | |
| CsgD | + | + | Expression analysis, consensus binding site | [17, 47] | |
| MlrA | + | Expression analysis, consensus binding site | [43, 44] | ||
| Cra (FruR) | + | Expression analysis, consensus binding site | [49] | ||
| Crp | + | Expression analysis | [51] | ||
| RcdA | + | Expression analysis, consensus binding site | [50] | ||
| DNA Modifying Enzymes | IHF | + | Expression analysis, consensus binding site | [58, 59] | |
| H-NS | −/+ (E. coli/S. typhimurium) | Expression analysis, consensus binding site | [59, 60, 102]/[58] | ||
| Two-Component Systems | CpxA/R | − | − | Expression analysis, consensus binding site | [52, 53, 59] |
| EnvZ/OmpR | + | Expression analysis, consensus binding site | [53, 55, 57–59, 104] | ||
| RcsA/B | − | − | Expression analysis | [53, 54, 105] | |
| RstB/A | −/+ (basic/acidic conditions) | Expression analysis, consensus binding site | [59, 106] | ||
| ArcA/B | + | Expression analysis, consensus binding site | [107] | ||
| BasS/R | + | Expression analysis, consensus binding site | [108] | ||
| Small Regulatory RNAs | OmrA/B | − | Antisense binding to 5′UTR | [61] | |
| McaS | − | Antisense binding to 5′UTR | [62, 64] | ||
| GcvB | − | Antisense binding to 5′UTR | [62] | ||
| RprA | − | Antisense binding to 5′UTR | [62, 63] | ||
| ArcZ | + | Expression analysis post-transcriptional regulation | [65] | ||
| SdsR | + | Expression analysis, transcriptional regulation | [65] |
Regulators of the csgBAC and csgDEFG promoters. Positive and negative regulation of each promoter is indicated by “+” or “−”, repectively.
The curli specific genes (csg) are found in two divergently transcribed operons, the intergenic region of which is the 7th largest in E. coli and subject to extensive regulation [17, 38, 39]. The csgDEFG promoter is recognized as one of the most complexly regulated promoters in E. coli [40]. Curli are primarily expressed during stationary phase and at low temperature (below 30°C), although some clinical isolates can express curli at 37°C [10, 11, 35, 41]. Both curli promoters are regulated by the stationary phase alternative sigma factor σS, which is assisted by the thermo-sensitive protein Crl (Table 1) [10, 41, 42]. Stationary phase expression of the csgDEFG promoter is likewise positively regulated by the stationary phase transcription factor MlrA (Table 1) [43, 44]. Curli expression is also internally regulated by the first gene product of the csgDEFG operon: CsgD (Table 1). CsgD is a member of the FixJ/LuxR family of transcriptional regulators that coordinates the expression of multiple biofilm components including curli and cellulose, while repressing expression of flagellar genes [17, 45–47]. CsgD activity and stability is modulated by phosphorylation of an N-terminal aspartic acid residue (DNA binding is decreased by in vitro phosphorylation) [48]. Several additional transcriptional regulators modulate expression of the csgDEFG promoter: the catabolite repressor/activator protein Cra, the cAMP receptor protein CRP, and the recently identified protein RcdA (Table 1) [43, 44, 49–51]. At least two known two-component systems negatively regulate the csgDEFG operon (CpxA/R and RcsA/B) and one positively (OmpR/EnvZ) (Table 1) [52–57]. The two global DNA organization protein complexes IHF and H-NS impact curli gene expression inversely: IHF contributes to promoter curli gene expression while H-NS represses curli expression in E. coli (Table 1) [58–60]. Both positive and negative regulation involves simultaneous binding of regulatory proteins as well as competitive binding between positive and negative regulators. For instance, H-NS and IHF act in competition for binding upstream of the csgDEFG promoter while CpxR and H-NS can bind simultaneously to negatively regulate the csgDEFG promoter and OmpR and RstA can bind simultaneously with IHF to positively regulate the csgD promoter [59]. Finally, the csgDEFG transcript is also subject to negative post-transcriptional regulation by five small regulatory RNAs: OmrA, OmrB, McaS, GcvB and RprA (Table 1) [61–64]. The small RNAs ArcZ and SdsR also positively regulate csgD in S. enterica, but whether this is a direct effect has yet to be determined (Table 1) [65]. For a more comprehensive review of the role of small RNAs in regulating the transition between biofilm and sessile growth see Mika and Hengge, 2013 [66].
3. Building a curli fiber on the cell surface
3.1. CsgA amyloid assembly by design
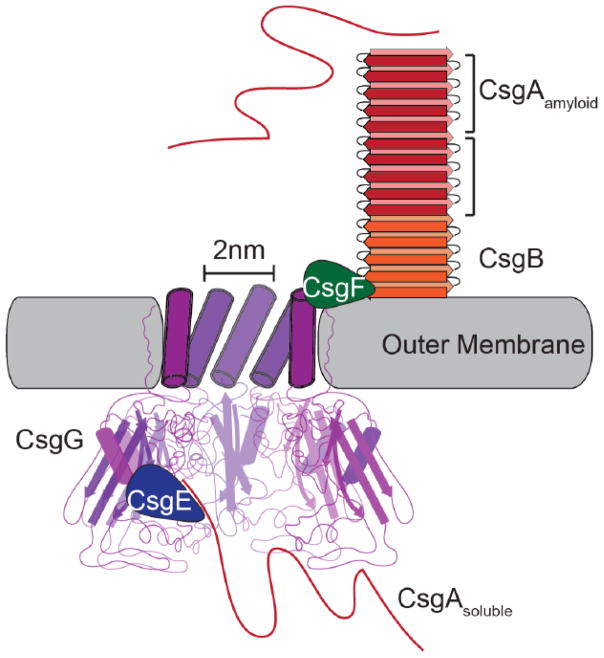
A striking aspect of curli biogenesis is that, despite CsgA’s propensity to aggregate, amyloid formation is so faithfully restricted to the cell surface. The major and minor curli subunits, CsgA and CsgB, respectively, are encoded by the csgBAC operon [17, 38, 67]. CsgA is secreted across the outer membrane as an unstructured soluble peptide (Fig. 2) [68]. Soluble CsgA is then templated into an amyloid on the cell surface by the nucleator and minor curli subunit CsgB (Fig. 2) [69, 70]. This mechanism of secretion is termed nucleation-precipitation or Type VIII secretion [71]. A third, and less well understood, gene csgC is located downstream of csgA. CsgC is a small periplasmic protein that is proposed to play a role in subunit secretion [68, 72]. CsgC is a β-sheet rich protein with an immunoglobulin-like fold and a conserved CxC motif that shares a high degree of structural similarity with the N-terminus of the redox-active DsbD [72]. Whether the other curli proteins are redox substrates of CsgC still unclear; the curli pore protein CsgG, however, is an attractive substrate candidate as it is the only other curli protein that contains a solvent exposed cysteine residue [72]. Additionally, the presence of CsgC renders the csgBA mutant more sensitive to bile salts compared to the csgBAC mutant suggesting that CsgC may keep the pore open or ungated [72]. Furthermore, an S. Enteritidis csgC mutant produces wider curli fibers (~20nm) suggesting that CsgC may also function in CsgA folding and curli fiber structure [68]. The csgE, csgF and csgG genes encode the proteins that comprise the unique outermembrane curli secretion apparatus (Fig. 2). CsgG assembles into a pore-like structure in the outermembrane while CsgE and CsgF play chaperone-like functions assisting in secretion and cell surface attachment of the curli fibers (Fig. 2) [73–75]. CsgD is a positive regulator that promotes biofilm formation by positively regulating curli and cellulose production (Table 1) [17, 45–47].
Figure 2.
The curli biogenesis system possesses a unique outer membrane secretion apparatus. The CsgG pore is composed of eight subunits (the front three have been removed to improve clarity) with a 2nm wide central pore. Both CsgE and CsgF interact directly with the CsgG pore. CsgE is periplasmic and required for directing soluble CsgA to the CsgG pore for secretion while CsgF is surface exposed and contributes to the assembly of CsgB into a surface associated, amyloid-templating conformation. Once outside the cell, CsgA interacts with CsgB and assembles into an amyloid fiber.
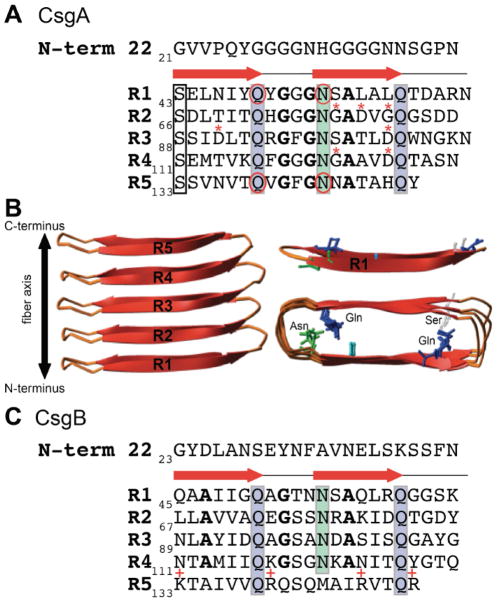
CsgA is the major subunit of the curli fiber and is secreted across the outermembrane as an unstructured and soluble monomer (Fig. 2) [4, 17, 68, 76, 77]. The mature CsgA protein is comprised of an N-terminal 22 amino acids (N-term22) required for outermembrane secretion, and a C-terminal amyloid core domain (Fig. 3AB) [25, 26, 75]. The amyloid core of CsgA is composed of five imperfect repeating units: R1 through R5 (Fig. 3AB) [23, 77]. Each repeating unit contains a Ser-X5-Gln-X-Gly-X-Gly-Asn-X-Ala-X3-Gln motif and forms a β-sheet-turn-β-sheet secondary structure with an overall β-helical, cross-β structure (Fig. 3AB) [25, 26]. The arrangement of the repeating units in the cross-β structure aligns the Gln and Asn residues in each repeating unit in stacks that stabilize the amyloid fold (Fig. 2AB)[78]. It is also worth noting that the placement and stacking of the Gly and Ala residues in the consensus sequence is highly conserved in Gammaproteobacteria and likely play an important role in amyloid assembly [13].
Figure 3.
The molecular details of CsgA and CsgB. A. The mature CsgA protein is composed of an N-terminal 22 amino acids and 5 amyloid repeating units, each with a β-sheet-turn-β-sheet (indicated by the red arrows). Ser, Gln and Asn residues (boxes) in each repeating unit align in stacks that stabilize the amyloid conformation. The Gln residues at positions 49 and 139 and the Asn residues at positions 54 and 144 (circled) are essential for amyloid formation. Repeating unites R2, R3 and R4 contain “gatekeeper” residues (*) that temper amyloid formation. The conserved glycine and alanine residues are indicated in bold font. B. A cartoon representation of the predicted structure of CsgA shows the five repeating units assemble into a β-helical, cross-β structure (side view, left). The Gln, Asn and Ser residues of R1 (upper right) are shown as sticks and the overhead view of CsgA (lower right) shows the alignment of these residues within the predicted structure. C. The mature CsgB protein is composed of an N-terminal 22 amino acids and 5 amyloid repeating units with similarly conserved Gln and Asn stacks (boxes). The conserved glycine and alanine residues in R1-R4 are indicated in bold font. Repeating unit 5 lacks one of the Asn repeats, but instead contains four charged residues (indicated by +) that may be important for tethering CsgB to the cell surface via R5.
Purified CsgA self-assembles into amyloid fibers that bind ThT and can be visualized by transmission electron microscopy (TEM) in vitro. CsgA polymerization in vitro is very robust: CsgA polymerizes into indistinguishable amyloid fibers over large pH (3.0–9.0) and ionic (0–500mM NaCl) ranges [24]. CsgA polymerization is concentration dependent and can be divided into three phases: a lag phase, an elongation phase and a stationary phase. At CsgA concentrations between 5 and 30 μM, the lag phase is approximately 2 hours [23, 24]. During the lag and elongation phases, a CsgA folding intermediate that is conserved in most amyloid-forming proteins can be detected with a conformation-specific antibody [23, 79]. Although purified CsgA readily forms amyloid in vitro, CsgA amyloid formation in vivo requires the nucleator protein CsgB [69, 70]. CsgB nucleated polymerization of CsgA can be recapitulated in vitro in a process called “seeding”. When added to soluble CsgA, preformed CsgB or CsgA fibers (also referred to as seeds) accelerate the polymerization of CsgA by eliminating the lag phase [78]. Interestingly, curli subunits from closely related species exhibit a cross-seeding promiscuity. Curli subunits from E. coli and the closely related bacteria Citrobacter koseri and Salmonella enterica are capable of cross-seeding both in vivo and in vitro [80].
The molecular features of both CsgA and CsgB that contribute to amyloid formation and nucleation have been thoroughly dissected. The repeating units of CsgA have distinct characteristics that either promote or temper amyloid formation (Fig. 3AB) [78, 81]. Repeating units R1, R3, and R5 are amyloidogenic, while R2 and R4 are non-amyloidogenic: synthetic peptides of R1, R3 and R5 rapidly assemble into amyloid fibers in vitro while R2 and R4 do not [23, 82]. Furthermore, CsgA ΔR1 and ΔR5 do not assemble into curli fibers in vivo and cannot be seeded by either CsgA or CsgB in vitro [82]. R1 and R5 are therefore not only important for amyloid assembly, but are also the domains that mediate CsgA-CsgA and CsgA-CsgB interactions. The Gln and Asn residues in R1 and R5 (at positions 49, 54, 139 and 144) are critical for amyloid formation (Fig. 3A) [78]. Mutation of the Gln and Asn in R1 and R5 results in a CsgA variant, aptly named CsgAslowgo, that polymerizes with a significantly extended (~100x longer) lag phase compare to WT CsgA [23, 78]. While R1, R3 and R5 synthetic peptides all polymerize into amyloids in vitro, R3 polymerizes more slowly [82]. R1 and R5 are interchangeable with one another (R12341 and R52345 CsgA chimeras produce WT curli in vivo), while replacement of either R1 or R5 with R3 (R32345 or R12343) results in little to no curli [82].
Certain residues in the CsgA repeating units appear to temper or slow CsgA aggregation into amyloid [81]. Asp residues at positions 91 and 104 within R3 have been identified as “gatekeeper” residues that reduce the amyloidogenicity of R3 (Fig. 3A) [81]. Mutations to either of these Asp residues in such a way that the peptide more closely resembles R1 or R5 restore curli assembly of the R12343 chimera in vivo and polymerization in vitro [81]. Conversely, substitution of Asn136 and His149 within R5 for Asp, thus rendering R5 more similar to R3, abolishes curli assembly in vivo [81]. The non-amyloidogenic repeating units R2 and R4 also contain “gatekeeper” residues (Fig. 3A). Mutating Gly78/Asp80/Gly82 in R2 and Gly123/Asp127 in R4 to the corresponding amino acids in R1 and R5, respectively, restored curli assembly to the CsgA ΔR1 and ΔR5 truncations in vivo [81]. Mutation of all of the “gatekeeper” residues in R2, R3 and R4 to the corresponding residues in R1 or R5 in full-length CsgA results in a variant (CsgA*) that polymerizes without a lag phase in vitro [81].
Furthermore, CsgA* is capable of forming curli in vivo in a CsgB/CsgF-independent manner and exhibits a toxic effect when overexpressed [81]. These “gatekeeper” residues enable a level of intramolecular control over CsgA amyloid assembly that is important in avoiding amyloid-associated cytotoxicity.
3.2. Amyloid assembly is nucleation-dependent in vivo
In addition to keeping CsgA polymerization in check with intramolecular “gatekeeping” residues, curli assembly is also controlled by dividing the tasks of nucleation and polymerization between two proteins, CsgB and CsgA, respectively. CsgB is dispensable for polymerization of CsgA in vitro but is required for curli assembly on the cell surface in vivo [4, 23, 69, 70, 82]. In the absence of the surface-associated CsgB, CsgA is secreted away from the cell and remains SDS-soluble [69, 83, 84]. However, soluble CsgA either supplemented exogenously or from a nearby csgB mutant donor cell can be engaged on the surface of cells presenting CsgB in a process referred to as interbacterial complementation (Fig. 4) [4, 69, 82]. Donor and recipient strains streaked on an agar plate as far apart as three millimeters can still participate in interbacterial complementation, owing to the incredible efficiency of the nucleation-precipitation mechanism of curli (Fig. 4) [69].
Figure 4.
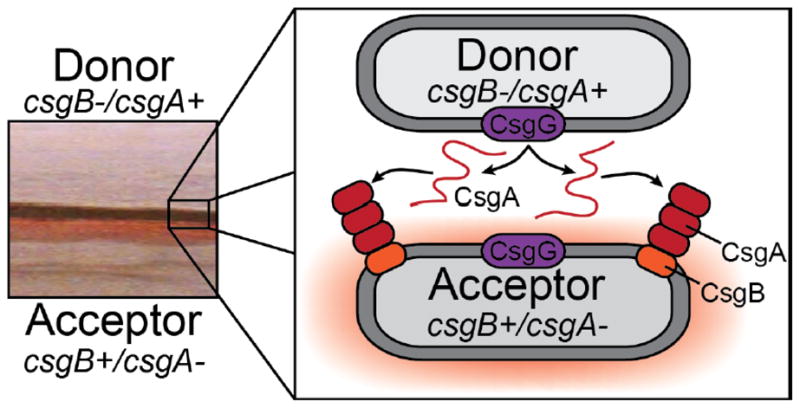
Curli subunit sharing between adjacent cells, or interbacerial complementation, is made possible by the nucleation-precipitation mechanism of curli fiber assembly. A csgB mutant (or csgA+ donor strain) secretes soluble CsgA into the extracellular milieu. CsgA from a csgB mutant can polymerize on a csgA mutant (or csgB+ acceptor) that is presenting CsgB on the cell surface when grown in close proximity to one another on a plate. The left panel shows a csgB mutant streaked near a csgA mutant, and on the edge of the CsgB-expressing streak there is a Congo red staining region.
Like CsgA, CsgB is a glutamine and asparagine-rich protein comprised of a SEC secretion signal sequence, an N-terminal domain and a core domain (Fig. 3C). CsgB shares 30% sequence identity with CsgA and has an amyloid-like core similar to CsgA [70, 85]. Each of the first four repeating units of CsgB contains an Ala-X3-Gln-X-Gly-X2-Asn-X-Ala-X3-Gln motif similar to the repeat motif of CsgA. Repeating unit 5, however, lacks several of these residues and instead contains four positively charged amino acids that are absent in the other CsgB repeating units: Lys133, Arg140, Arg147 and Arg151 (Fig. 3C). Deletion of repeating unit 5 results in a truncated version of CsgB (CsgBtrunc) that is no longer cell surface associated [86]. Moreover, CsgBtrunc self-assembles into amyloid fibers in vitro and can seed the polymerization of CsgA consistent with the hypothesis that CsgB also assembles into an amyloid core [70]. Repeating units 1–3 appear to be largely dispensable for in vitro polymerization and localization, leaving R4 and R5 of primary importance for localization and nucleation of CsgA [86]. However, it is also possible that some of the CsgB repeating units function redundantly. These observations suggest a model where CsgB associates with the cell membrane via the fifth repeating unit while R1-R4 assume an amyloid fold that can template soluble CsgA into amyloid fibers. How CsgB is associated with the outermembrane is still largely unknown; however, CsgB localization requires at least one component of the curli secretion apparatus, CsgF [74].
3.3. Curli are secreted by a dedicated outermembrane secretion apparatus
The curli system possesses a unique outermembrane secretion apparatus comprised of at least three proteins: CsgG, CsgE and CsgF (Fig. 2) [4, 17, 84]. CsgG is a lipoprotein that spans the outermembrane and assembles into an oligomeric annular-shaped structure with an inner diameter of approximately 2 nm [72, 75, 84]. High-resolution electron microscopy and modeling studies predict that the transmembrane domain of CsgG is alpha-helical while the periplasmic domain is predominantly random coil and β-sheet rich with a single α-helix (Fig. 2) [72]. Overexpression of CsgG results in increased sensitivity to the small antibiotic erythromycin (740Da) but not the larger vancomycin (1440Da) suggesting that CsgG has a pore-like activity [75]. Overexpression of CsgE complements the erythromycin sensitivity phenotype of a CsgG overexpressing strains [31, 73]. CsgE may therefore act, in part, by gating CsgG to prevent the influx of environmental chemicals and molecules through the pore.
The CsgG oligomers are not uniformly distributed around the cell but are instead spatially restricted to discrete regions of the cell that co-localize with what appears to be the primary curli fiber attachment site [31]. The mechanism of CsgG localization, however, and how the curli secretion apparatus is spatially localized to discrete spots around the cell remains unknown. Because subcellular localization is important for many biological functions, including macromolecular organelle assembly in bacteria, it is not unreasonable to hypothesize that spatial restriction of the curli secretion apparatus is also important for curli assembly [87, 88]. Diffusion of protein subunits into the environment is a significant challenge of the nucleation-precipitation/Type VIII secretion system [71, 89]. Localized secretion of CsgA may lead to high local concentrations that promote efficient curli formation on the cell surface. Interestingly, CsgA and CsgB are both required for stable oligomerization of CsgG suggesting that either subunit secretion or active curli polymerization on the cell surface somehow facilitates stable pore assembly [31].
Stable oligomerization of CsgG is dependent on both of the major and minor curli subunits, CsgA and CsgB, as well as the accessory proteins CsgE and CsgF [31]. Conversely, CsgG is required for the stability and secretion of CsgA, CsgB and CsgF across the outermembrane [74, 84]. CsgG interacts with CsgE and CsgF as well as fusion proteins in which the N-term22 of CsgA has been fused to the N-terminus of a substrate protein [75]. CsgF is extracellular and interacts with CsgG on the cell surface where it stabilizes and localizes CsgB to cell-surface [74].
In the absence of CsgF, both CsgB and CsgA remain predominately SDS-soluble and are secreted away from the cell [74]. Overexpression of CsgB in a csgF mutant restores the cell-surface localization of CsgB but it does not restore WT cell-surface associated curli fibers [74]. CsgF therefore may play a more important role in remodeling CsgB into an amyloid-templating conformation rather than actually anchoring CsgA fibers.
Ensuring that curli amyloid assembly occurs at the correct time and place is a feat that is at least in part achieved by molecular chaperones [73, 90]. Nenninger et al. described CsgF as having chaperone-like activity: CsgF appears to assist in directing the assembly of CsgA into an amyloid on the cell surface [74]. CsgE, on the other hand, functions earlier in the curli biogenesis pathway by inhibiting premature amyloid formation within the periplasm, but also by directing CsgA subunits to CsgG for secretion [73]. CsgE inhibits CsgA amyloid formation and interacts with CsgA amyloid fibers in vitro [73, 91]. Two lines of evidence using fusion protein constructs with the N-terminal domain of CsgA implicate the N-terminal 22 amino acids of CsgA as the interacting domain with both CsgE and CsgG [73, 75]. First, the large periplasmic protein PhoA coimmunoprecipitated with CsgG when PhoA was fused to the N-terminal 42 amino acids of CsgA (including the SEC secretion signal sequence) [75]. Second, expression of CsgE specifically permits the secretion of the second domain of PapD and CpxP only when C-terminally fused to the N-terminal 22 amino acids of CsgA [73]. However, overexpression of CsgG results in unchecked secretion indicating that secretion through CsgG doesn’t necessarily require CsgE [73, 75]. Together, these results suggest a coordinated chaperone/delivery and secretion mechanism. CsgE putatively has three functions: 1) bind to CsgG and gate the outer membrane secretion pore, 2) bind to and inhibit amyloid formation of curli subunits within the periplasm, and 3) deliver these subunits to CsgG for secretion.
4. The curli system can be exploited for studying other amyloids
Other bacteria may have also adapted the CsgG-like amyloid secretion pore. The Caulobacter crescentus holdfast protein HfaA shares the amyloid-like characteristics of being resistant to heat and SDS denaturation [92]. Like CsgA, the secretion and stability of HfaA depends on an outermembrane lipoprotein, HfaB, which shares 31% identity and 42% similarity with CsgG [93]. It is attractive to hypothesize that the curli secretion mechanism is representative of a distinct class of apparatuses for secreting amyloid subunits across membranes.
The unique curli secretion apparatus may be ideally suited for transporting generic unfolded amyloid proteins across the outer membrane. CsgA is unstructured upon secretion and the CsgG pore, only about 2 nm (or 20 Å) wide, is not large enough to secrete CsgA in its folded conformation, suggesting that CsgA is maintained in an unstructured and soluble conformation prior to secretion [68, 75, 77]. The N-terminal 22 amino acids of the mature CsgA not only direct its secretion across the outermembrane, but when fused to unrelated amyloid proteins, can also direct their secretion [29, 73, 75, 94]. E. coli expressing fusions of unrelated amyloids with the N-terminal domain of CsgA produce Congo red binding colonies on agar plates and SDS-insoluble fibers [29, 94]. Harnessing the curli secretion system to assemble heterologous amyloids on the bacterial cell surface may pave the way for new high throughput screening methods for modulators of amyloid assembly.
5. Folding intermediates can be probed using antibodies and small molecules
The discovery of conformation specific antibodies has contributed immensely to our understanding of amyloid assembly [79, 95, 96]. We now have the tools to detect intermediate oligomeric and fibrillar species during the amyloid assembly process. The conformation-specific antibody A11 was raised against an oligomeric intermediate of Aβ, the amyloid protein associated with Alzheimer’s disease, and reacts with the oligomeric intermediates of many other disease-associated amyloids [79]. CsgA is also transiently recognized by A11 in vitro indicating that CsgA functional amyloid formation proceeds by the same mechanism of assembly as disease-associated amyloids in vitro [23]. Antibodies have also been raised that specifically recognize the amyloid fiber conformation and also broadly react with unrelated amyloid fibers [95, 96]. Conformation specific antibodies have motivated the development of a new class of antibodies, gammabodies (for grafted amyloid motif antibodies), that not only react specifically with the fiber conformation, but also have a high degree of specificity for protein sequence [97, 98]. Gammabodies have been demonstrated to act in distinct mechanisms, depending on the grafted sequence, to inhibit amyloid formation of several disease-associated amyloids [97]. Gammabodies have been engineered against each repeating unit of CsgA and have been show to exhibit the same sequence and conformation specificity as gammabodies designed to interact with disease-associated amyloids [91, 98]. It has yet to be determined if similar inhibitory interactions are possible between gammabodies and functional amyloids.
Peptidomimetic compounds are also attractive candidates for modulating protein-protein interactions and amyloid formation. A screen of compounds originally designed to inhibit the elaboration of E. coli P pili yielded a set of small molecules capable of inhibiting amyloid formation of Aβ [99, 100]. These compounds have since been subject to extensive chemical modification and assayed for biological activity against several amyloid substrates including curli [28, 91, 101]. This extensive library of 2-pyridone compounds, or curlicides, exhibit varying degrees of CsgA amyloid inhibition and curli-dependent biofilm inhibition [28, 91, 101]. Interestingly, the compound FN075 inhibits both Aβ and CsgA amyloid formation but accelerates amyloid formation of the Parkinson’s Disease-associated protein α-synuclein [101]. FN075 promotes the oligomerization of both CsgA and α-synuclein, however, the α-synuclein oligomers are competent for amyloid formation (on-pathway) while the CsgA oligomers are not (off-pathway) [101]. These provocative results suggest that the 2-pyridone compounds may be used as tools for probing and distinguishing between on- and off-pathway protein folding intermediates. Andersson et al. have now identified other 2-pyridone compounds that are capable of accelerating CsgA polymerization [91]. Further analysis of curli assembly in the presence of these curlicide and accelerator compounds may be instrumental in dissecting early stages of function amyloid assembly.
6. Conclusions
The amyloid field has undergone a sea change since curli were characterized as an amyloid and the functional amyloid field was born. Curli biology is ideal for studying protein secretion and folding in the context of bacterial community behavior and, remarkably, can be amended for studying of protein folding and misfolding in the context of many human diseases. Elaborate mechanisms are in place to ensure that curli are assembled only at the right time, at the right place and with remarkable efficiency to overcome large diffusion rates inherent in a nucleation-precipitation model. It seems only natural that the unique properties of curli fibers are accompanied by an equally unique biogenesis system.
Highlights.
Curli are a bacterial functional amyloid.
Regulation of curli gene expression is extraordinarily complex.
Curli amyloid assembly is controlled by intra- and intermolecular interactions.
Curli have a unique secretion apparatus that can be adapted to unrelated amyloids.
Curli folding intermediates can be probed using antibodies and small molecules.
Acknowledgments
We would like to thank W. H. DePas and D. A. Hufnagel for critical reading of this manuscript. This work was supported by the National Institutes of Health (AI073847 to M. R. C.), the University of Michigan Cellular Biotechnology Training Program (NIH NRSA T32 GM008353) and the University of Michigan 2013–2014 Rackham Graduate School Predoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Breydo L, Wu JW, Uversky VN. Alpha-synuclein misfolding and Parkinson’s disease. Biochim Biophys Acta. 2012;1822:261–285. doi: 10.1016/j.bbadis.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 4.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheibel T, Kowal AS, Bloom JD, Lindquist SL. Bidirectional amyloid fiber growth for a yeast prion determinant. Curr Biol. 2001;11:366–369. doi: 10.1016/s0960-9822(01)00099-9. [DOI] [PubMed] [Google Scholar]
- 7.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 9.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 11.Zogaj X, Bokranz W, Nimtz M, Romling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 13.Dueholm MS, Albertsen M, Otzen D, Nielsen PH. Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One. 2012;7:e51274. doi: 10.1371/journal.pone.0051274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCrate OA, Zhou X, Reichhardt C, Cegelski L. Sum of the Parts: Composition and Architecture of the Bacterial Extracellular Matrix. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pawar DM, Rossman ML, Chen J. Role of curli fimbriae in mediating the cells of enterohaemorrhagic Escherichia coli to attach to abiotic surfaces. J Appl Microbiol. 2005;99:418–425. doi: 10.1111/j.1365-2672.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 16.Boyer RR, Sumner SS, Williams RC, Pierson MD, Popham DL, Kniel KE. Influence of curli expression by Escherichia coli 0157:H7 on the cell’s overall hydrophobicity, charge, and ability to attach to lettuce. J Food Prot. 2007;70:1339–1345. doi: 10.4315/0362-028x-70.6.1339. [DOI] [PubMed] [Google Scholar]
- 17.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 18.Collinson SK, Doig PC, Doran JL, Clouthier S, Trust TJ, Kay WW. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen A, Wick MJ, Morgelin M, Bjorck L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect Immun. 1998;66:944–949. doi: 10.1128/iai.66.3.944-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kai-Larsen Y, Luthje P, Chromek M, Peters V, Wang X, Holm A, Kadas L, Hedlund KO, Johansson J, Chapman MR, Jacobson SH, Romling U, Agerberth B, Brauner A. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010;6:e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dueholm MS, Nielsen SB, Hein KL, Nissen P, Chapman M, Christiansen G, Nielsen PH, Otzen DE. Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry. 2011;50:8281–8290. doi: 10.1021/bi200967c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collinson SK, Parker JM, Hodges RS, Kay WW. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J Mol Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 26.Shewmaker F, McGlinchey RP, Thurber KR, McPhie P, Dyda F, Tycko R, Wickner RB. The functional curli amyloid is not based on in-register parallel beta-sheet structure. J Biol Chem. 2009;284:25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivanathan V, Hochschild A. Generating extracellular amyloid aggregates using E. coli cells. Genes Dev. 2012;26:2659–2667. doi: 10.1101/gad.205310.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DePas WH, Hufnagel DA, Lee JS, Blanco LP, Bernstein HC, Fisher ST, James GA, Stewart PS, Chapman MR. Iron induces bimodal population development by Escherichia coli. Proc Natl Acad Sci U S A. 2013;110:2629–2634. doi: 10.1073/pnas.1218703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epstein EA, Reizian MA, Chapman MR. Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J Bacteriol. 2009;191:608–615. doi: 10.1128/JB.01244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grantcharova N, Peters V, Monteiro C, Zakikhany K, Romling U. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar typhimurium. J Bacteriol. 2010;192:456–466. doi: 10.1128/JB.01826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio. 2013;4:e00103–13. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerstel U, Romling U. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ Microbiol. 2001;3:638–648. doi: 10.1046/j.1462-2920.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- 35.Olsen A, Arnqvist A, Hammar M, Normark S. Environmental regulation of curli production in Escherichia coli. Infect Agents Dis. 1993;2:272–274. [PubMed] [Google Scholar]
- 36.Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White AP, Gibson DL, Kim W, Kay WW, Surette MG. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J Bacteriol. 2006;188:3219–3227. doi: 10.1128/JB.188.9.3219-3227.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. Characterization of the agfBA fimbrial operon encoding thin aggregative fimbriae of Salmonella enteritidis. Adv Exp Med Biol. 1997;412:247–248. doi: 10.1007/978-1-4899-1828-4_37. [DOI] [PubMed] [Google Scholar]
- 39.Rudd KE. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28:60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishihama A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol Rev. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- 41.Arnqvist A, Olsen A, Pfeifer J, Russell DG, Normark S. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol Microbiol. 1992;6:2443–2452. doi: 10.1111/j.1365-2958.1992.tb01420.x. [DOI] [PubMed] [Google Scholar]
- 42.Bougdour A, Lelong C, Geiselmann J. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J Biol Chem. 2004;279:19540–19550. doi: 10.1074/jbc.M314145200. [DOI] [PubMed] [Google Scholar]
- 43.Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss Rr. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol. 2001;41:349–363. doi: 10.1046/j.1365-2958.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- 44.Ogasawara H, Yamamoto K, Ishihama A. Regulatory role of MlrA in transcription activation of csgD, the master regulator of biofilm formation in Escherichia coli. FEMS Microbiol Lett. 2010;312:160–168. doi: 10.1111/j.1574-6968.2010.02112.x. [DOI] [PubMed] [Google Scholar]
- 45.Romling U, Rohde M, Olsen A, Normark S, Reinkoster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 46.Brombacher E, Dorel C, Zehnder AJ, Landini P. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology. 2003;149:2847–2857. doi: 10.1099/mic.0.26306-0. [DOI] [PubMed] [Google Scholar]
- 47.Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol. 2011;193:2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zakikhany K, Harrington CR, Nimtz M, Hinton JC, Romling U. Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2010;77:771–786. doi: 10.1111/j.1365-2958.2010.07247.x. [DOI] [PubMed] [Google Scholar]
- 49.Reshamwala SM, Noronha SB. Biofilm formation in Escherichia coli cra mutants is impaired due to down-regulation of curli biosynthesis. Arch Microbiol. 2011;193:711–722. doi: 10.1007/s00203-011-0708-7. [DOI] [PubMed] [Google Scholar]
- 50.Shimada T, Katayama Y, Kawakita S, Ogasawara H, Nakano M, Yamamoto K, Ishihama A. A novel regulator RcdA of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli. Microbiologyopen. 2012;1:381–394. doi: 10.1002/mbo3.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dorel C, Vidal O, Prigent-Combaret C, Vallet I, Lejeune P. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol Lett. 1999;178:169–175. doi: 10.1111/j.1574-6968.1999.tb13774.x. [DOI] [PubMed] [Google Scholar]
- 53.Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, Dorel C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol. 2005;187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrieres L, Clarke DJ. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol. 2003;50:1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x. [DOI] [PubMed] [Google Scholar]
- 55.Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, Landini P, Dorel C. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol. 2001;183:7213–7223. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerstel U, Kolb A, Romling U. Regulatory components at the csgD promoter--additional roles for OmpR and integration host factor and role of the 5′ untranslated region. FEMS Microbiol Lett. 2006;261:109–117. doi: 10.1111/j.1574-6968.2006.00332.x. [DOI] [PubMed] [Google Scholar]
- 58.Gerstel U, Park C, Romling U. Complex regulation of csgD promoter activity by global regulatory proteins. Mol Microbiol. 2003;49:639–654. doi: 10.1046/j.1365-2958.2003.03594.x. [DOI] [PubMed] [Google Scholar]
- 59.Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology. 2010;156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- 60.Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 61.Holmqvist E, Reimegard J, Sterk M, Grantcharova N, Romling U, Wagner EG. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jorgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev. 2012;27:1132–1145. doi: 10.1101/gad.214734.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, Pruteanu M, Hengge R. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol Microbiol. 2012;84:51–65. doi: 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomason MK, Fontaine F, De Lay N, Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monteiro C, Papenfort K, Hentrich K, Ahmad I, Le Guyon S, Reimann R, Grantcharova N, Romling U. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 2012;9:489–502. doi: 10.4161/rna.19682. [DOI] [PubMed] [Google Scholar]
- 66.Mika F, Hengge R. Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella. Int J Mol Sci. 2013;14:4560–4579. doi: 10.3390/ijms14034560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibson DL, White AP, Rajotte CM, Kay WW. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella enteritidis. Microbiology. 2007;153:1131–1140. doi: 10.1099/mic.0.2006/000935-0. [DOI] [PubMed] [Google Scholar]
- 69.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A. 2007;104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desvaux M, Hebraud M, Talon R, Henderson IR. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 2009;17:139–145. doi: 10.1016/j.tim.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 72.Taylor JD, Zhou Y, Salgado PS, Patwardhan A, McGuffie M, Pape T, Grabe G, Ashman E, Constable SC, Simpson PJ, Lee WC, Cota E, Chapman MR, Matthews SJ. Atomic resolution insights into curli fiber biogenesis. Structure. 2011;19:1307–1316. doi: 10.1016/j.str.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol Microbiol. 2011;81:486–499. doi: 10.1111/j.1365-2958.2011.07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nenninger AA, Robinson LS, Hultgren SJ. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci U S A. 2009;106:900–905. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cherny I, Rockah L, Levy-Nissenbaum O, Gophna U, Ron EZ, Gazit E. The formation of Escherichia coli curli amyloid fibrils is mediated by prion-like peptide repeats. J Mol Biol. 2005;352:245–252. doi: 10.1016/j.jmb.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 78.Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. J Mol Biol. 2008;380:570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 80.Zhou Y, Smith D, Leong BJ, Brannstrom K, Almqvist F, Chapman MR. Promiscuous cross-seeding between bacterial amyloids promotes interspecies biofilms. J Biol Chem. 2012;287:35092–35103. doi: 10.1074/jbc.M112.383737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X, Zhou Y, Ren JJ, Hammer ND, Chapman MR. Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc Natl Acad Sci U S A. 2010;107:163–168. doi: 10.1073/pnas.0908714107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem. 2008;283:21530–21539. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 85.White AP, Collinson SK, Banser PA, Gibson DL, Paetzel M, Strynadka NC, Kay WW. Structure and characterization of AgfB from Salmonella enteritidis thin aggregative fimbriae. J Mol Biol. 2001;311:735–749. doi: 10.1006/jmbi.2001.4876. [DOI] [PubMed] [Google Scholar]
- 86.Hammer ND, McGuffie BA, Zhou Y, Badtke MP, Reinke AA, Brannstrom K, Gestwicki JE, Olofsson A, Almqvist F, Chapman MR. The C-terminal repeating units of CsgB direct bacterial functional amyloid nucleation. J Mol Biol. 2012;422:376–389. doi: 10.1016/j.jmb.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bardy SL, Maddock JR. Polar explorations Recent insights into the polarity of bacterial proteins. Curr Opin Microbiol. 2007;10:617–623. doi: 10.1016/j.mib.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 88.Ebersbach G, Jacobs-Wagner C. Exploration into the spatial and temporal mechanisms of bacterial polarity. Trends Microbiol. 2007;15:101–108. doi: 10.1016/j.tim.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 89.Soto GE, Hultgren SJ. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Evans ML, Schmidt JC, Ilbert M, Doyle SM, Quan S, Bardwell JC, Jakob U, Wickner S, Chapman MR. E. coli chaperones DnaK, Hsp33 and Spy inhibit bacterial functional amyloid assembly. Prion. 2011;5:323–334. doi: 10.4161/pri.5.4.18555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andersson EK, Bengtsson C, Evans ML, Chorell E, Sellstedt M, Lindgren AEG, Hufnagel DA, Bhattacharya M, Tessier PM, Wittung-Stafshede P, Almqvist F, Chapman MR. Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones. Chemistry and Biology. 2013 doi: 10.1016/j.chembiol.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hardy GG, Allen RC, Toh E, Long M, Brown PJ, Cole-Tobian JL, Brun YV. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol. 2010;76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cole JL, Hardy GG, Bodenmiller D, Toh E, Hinz A, Brun YV. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol Microbiol. 2003;49:1671–1683. doi: 10.1046/j.1365-2958.2003.03664.x. [DOI] [PubMed] [Google Scholar]
- 94.Sivanathan V, Hochschild A. A bacterial export system for generating extracellular amyloid aggregates. Nat Protoc. 2013;8:1381–1390. doi: 10.1038/nprot.2013.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O’Nuallain B, Wetzel R. Conformational Abs recognizing a generic amyloid fibril epitope. Proc Natl Acad Sci U S A. 2002;99:1485–1490. doi: 10.1073/pnas.022662599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, Margol L, Wu J, Breydo L, Thompson JL, Rasool S, Gurlo T, Butler P, Glabe CG. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ladiwala AR, Bhattacharya M, Perchiacca JM, Cao P, Raleigh DP, Abedini A, Schmidt AM, Varkey J, Langen R, Tessier PM. Rational design of potent domain antibody inhibitors of amyloid fibril assembly. Proc Natl Acad Sci U S A. 2012;109:19965–19970. doi: 10.1073/pnas.1208797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perchiacca JM, Ladiwala AR, Bhattacharya M, Tessier PM. Structure-based design of conformation- and sequence-specific antibodies against amyloid beta. Proc Natl Acad Sci U S A. 2012;109:84–89. doi: 10.1073/pnas.1111232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Svensson A, Larsson A, Emtenas H, Hedenstrom M, Fex T, Hultgren SJ, Pinkner JS, Almqvist F, Kihlberg J. Design and evaluation of pilicides: potential novel antibacterial agents directed against uropathogenic Escherichia coli. Chembiochem. 2001;2:915–918. doi: 10.1002/1439-7633(20011203)2:12<915::AID-CBIC915>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 100.Aberg V, Norman F, Chorell E, Westermark A, Olofsson A, Sauer-Eriksson AE, Almqvist F. Microwave-assisted decarboxylation of bicyclic 2-pyridone scaffolds and identification of Abeta-peptide aggregation inhibitors. Org Biomol Chem. 2005;3:2817–2823. doi: 10.1039/b503294f. [DOI] [PubMed] [Google Scholar]
- 101.Horvath I, Weise CF, Andersson EK, Chorell E, Sellstedt M, Bengtsson C, Olofsson A, Hultgren SJ, Chapman M, Wolf-Watz M, Almqvist F, Wittung-Stafshede P. Mechanisms of protein oligomerization: inhibitor of functional amyloids templates alpha-synuclein fibrillation. J Am Chem Soc. 2012;134:3439–3444. doi: 10.1021/ja209829m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Arnqvist A, Olsen A, Normark S. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 103.Pratt LA, Silhavy TJ. Crl stimulates RpoS activity during stationary phase. Mol Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- 104.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vianney A, Jubelin G, Renault S, Dorel C, Lejeune P, Lazzaroni JC. Escherichia coli tol and rcs genes participate in the complex network affecting curli synthesis. Microbiology. 2005;151:2487–2497. doi: 10.1099/mic.0.27913-0. [DOI] [PubMed] [Google Scholar]
- 106.Ogasawara H, Hasegawa A, Kanda E, Miki T, Yamamoto K, Ishihama A. Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J Bacteriol. 2007;189:4791–4799. doi: 10.1128/JB.00319-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu X, De Wulf P. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004;279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- 108.Ogasawara H, Shinohara S, Yamamoto K, Ishihama A. Novel regulation targets of the metal-response BasS-BasR two-component system of Escherichia coli. Microbiology. 2012;158:1482–1492. doi: 10.1099/mic.0.057745-0. [DOI] [PubMed] [Google Scholar]