SUMMARY
Depending on endoplasmic reticulum (ER) stress levels, the ER transmembrane multi-domain protein IRE1α promotes either adaptation or apoptosis. Unfolded ER proteins cause IRE1α lumenal domain homo-oligomerization, inducing trans auto-phosphorylation that further drives homo-oligomerization of its cytosolic kinase/ endoribonuclease (RNase) domains to activate mRNA splicing of adaptive XBP1 transcription factor. However, under high/chronic ER stress, IRE1α surpasses an oligomerization threshold that expands RNase substrate repertoire to many ER-localized mRNAs, leading to apoptosis. To modulate these effects, we developed ATP-competitive IRE1α Kinase Inhibiting RNase Attenuators—KIRAs—that allosterically inhibit IRE1α’s RNase by breaking oligomers. One optimized KIRA, KIRA6, inhibits IRE1α in vivo and promotes cell survival under ER stress. Intravitreally, KIRA6 preserves photoreceptor functional viability in rat models of ER stress-induced retinal degeneration. Systemically, KIRA6 preserves pancreatic β-cells, increases insulin, and reduces hyperglycemia in Akita diabetic mice. Thus, IRE1α powerfully controls cell fate, but can itself be controlled with small molecules to reduce cell degeneration.
INTRODUCTION
Secreted and transmembrane proteins fold and assemble in the endoplasmic reticulum (ER) through reactions catalyzed by ER-resident activities. When these reactions are saturated or corrupted, cells experience “ER stress,” and unfolded protein accumulation in the ER triggers intracellular signaling pathways termed the unfolded protein response (UPR). The UPR induces transcription of genes encoding ER chaperones, oxidoreductases, and ER-associated degradation (ERAD) components (Travers et al., 2000), while inhibiting translation (Harding et al., 2000). These outputs are adaptive because they enhance ER protein-folding capacity, reduce secretory protein load, and promote degradation of ER unfolded proteins.
However, if ER stress remains irremediably high and adaptive outputs are overwhelmed, alternative “Terminal UPR” signals trigger apoptosis. While cell death under high ER stress may protect organisms from exposure to improperly folded secretory proteins, many human degenerative diseases, such as diabetes mellitus and retinopathies, may be caused by excessive ER stress-induced cell death (Shore et al., 2011). Mechanistic understanding of critical Terminal UPR signaling events may lead to effective therapies for such conditions.
Unfolded ER proteins activate three ER transmembrane sensors, PERK, ATF6, and IRE1α, by changing their oligomerization state in the ER membrane (Kohno, 2007). IRE1α, the most ancient of these components, senses unfolded proteins either directly or indirectly through an ER lumenal domain that becomes oligomerized during stress (Credle et al., 2005; Zhou et al., 2006). Subsequently, IRE1α’s bifunctional kinase/endoribonuclease (RNase) activities become juxtaposed on its cytosolic face, allowing monomers to trans-autophosphorylate. Kinase autophosphorylation conformationally activates IRE1α’s RNase to site-specifically cleave the XBP1 mRNA. Religation and translation of XBP1 mRNA in an alternate open reading frame produces the XBP1s transcription factor whose targets encode proteins that enhance ER protein folding and quality control (Calfon et al., 2002; Lee et al., 2003; Yoshida et al., 2001). Thus, IRE1α promotes adaptation via XBP1s.
However, under high ER stress, IRE1α’s RNase relaxes its substrate specificity to endonucleolytically cleave many other mRNAs that localize to the ER membrane as their encoded proteins undergo co-translational translocation (Han et al., 2009; Hollien et al., 2009). IRE1α’s RNase also cleaves precursors of apoptosis-inhibitory microRNAs (Lerner et al., 2012; Upton et al., 2012).
Here we show that multiple Terminal UPR outputs, including cell proliferation blocks, sterile inflammation, and apoptosis result from kinase-driven increases in the oligomerization state of IRE1α’s cytosolic domains that hyperactivate its RNase. These destructive events are prevented by breaking IRE1α oligomerization through rational mutations or somatic mutations found in the Ire1α gene of various human cancers. To test physiological effects of pharmacologically inhibiting IRE1α, we developed small molecule kinase inhibitors that prevent oligomerization and allosterically inhibit its RNase. One such IRE1α kinase inhibitor preserves viability and function in ER-stressed cells, pancreatic islet explants, and rodent models of ER stress-induced retinitis pigmentosa and diabetes.
RESULTS
IRE1α’s kinase is a rheostat that employs self-association to control RNase activity
Previously, we proposed that IRE1α’s kinase regulates catalytic activity of its adjoining RNase in a graduated manner to impact cell fate in mammals, yet the mechanistic basis for the rheostatic control remained unclear (Han et al., 2009). Here, we hypothesized that the degree of self-association of kinase/RNase subunits on the cytosolic face connects IRE1α phosphorylation status to RNase activation levels. Increasing phosphorylation of the IRE1α kinase may proportionally increase the oligomeric state of kinase/RNase subunits past a critical threshold, thereby driving RNase activity into a hyperactive state. Consequently, IRE1α RNase expands its specificity past its canonical XBP1 mRNA substrate, endonucleolytically cleaving many ER-localized mRNAs and pushing cells into apoptosis. In this view, IRE1α’s lumenal domains are responsive to protein-folding conditions in the ER, but rheostatic control by the kinase over the RNase ultimately determines cell fate. If these predictions are correct, genetic and small molecule control over IRE1α kinase oligomerization should enable cell fate control, irrespective of upstream ER stress.
To begin testing this hypothesis, we employed IRE1α recombinant proteins and cell lines. Artificial ER stress agents are widely used to acutely activate the UPR, but saturating doses that have no natural pathophysiological correlate are often employed. To test cytoprotection later in the work, we established dose-response regimes using three ER stress agents that dose-dependently push rat insulinoma (INS-1) cells, which have a well-developed ER and secrete insulin, past a stress threshold, and in switch-like manner, into apoptosis. The percentage of INS-1 cells entering apoptosis due to the ER Ca2+ pump inhibitor thapsigargin (Tg) depends aggregately on two variables: concentration and duration of exposure (Figure S1A,B). Similar results hold for the glycosylation inhibitor tunicamycin (Tm) and the anterograde trafficking inhibitor brefeldin A (BFA) (Figure S1C,D). Preceding apoptosis, increasing ER stress agent levels progressively increases IRE1α phosphorylation, XBP1 mRNA splicing, endonucleolytic decay of the ER-localized mRNA, Ins1 mRNA (which encodes proinsulin), induction of thioredoxin-interacting protein (TXNIP) mRNA (whose product activates the NLRP3 inflammasome), and downstream c-Jun terminal kinase phosphorylation (JNKs) (Figure S1E,F).
All these Terminal UPR signature events, culminating in apoptosis, can be simulated, without imposing ER stress, by conditionally over-expressing wild-type (WT) IRE1α in INS-1 stable lines (Han et al., 2009). Induced with doxycycline (Dox), the transgenic IRE1α (WT) proteins spontaneously self-associate, trans-autophosphorylate, and trigger XBP1 mRNA splicing (Figure 1A,B) (Han et al., 2009). Increasing [Dox] causes progressive decay of Ins1 mRNA, elevation of TXNIP mRNA, and apoptosis (Figure 1C,D). Thus, as with ER stress agents, dose escalation of transgenic IRE1α (WT) allows graduated control over the Terminal UPR and is sufficient to push cells, in switch-like manner, into apoptosis (Figure 1E).
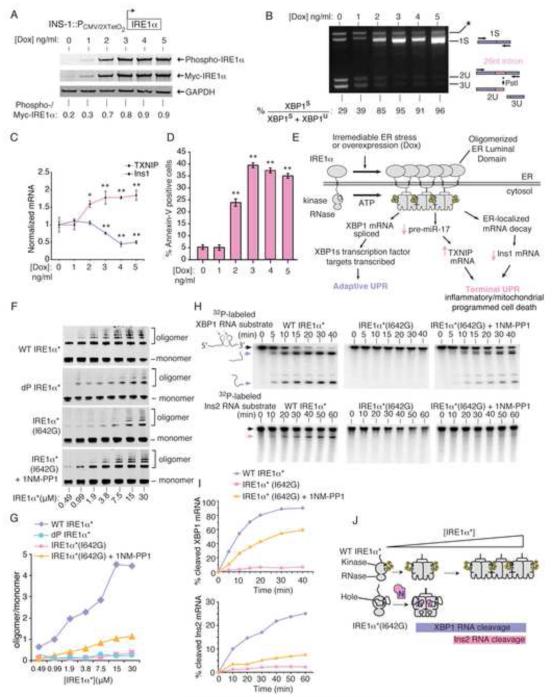
Figure 1. IRE1α’s kinase uses homo-oligomerization as a rheostat to control RNase activity and apoptosis.
(A) Anti-phospho-IRE1α and anti-Myc immunoblots (ratiometric quantitation, normalized to GAPDH). (B) Agarose gel of PstI-digested XBP1 cDNA amplicons (ratiometric quantitation of spliced to total XBP1 cDNAs). (C) Q-PCR for Insulin1 (Ins1) and TXNIP mRNAs. (D) %Annexin-V positive staining. (A-C) utilized INS-1::IRE1α (WT) cells under increasing [Dox] at 24hr, whereas (D) is at 72 hr. (E) Model of how IRE1α promotes both adaptive and apoptotic outputs. (F) Immunoblots of increasing concentrations of IRE1α*(WT), dP-IRE1α*(WT), and IRE1α*(I642G) −/+ 1NM-PP1 (10 μM) followed by disuccinimidyl suberate (DSS) (250 μM) crosslinking, with oligomer/monomer quantification (G). (H) Time course urea PAGE of cleavage of α32P-labeled XBP1 RNA and Insulin2 (Ins2) RNA by IRE1α*(WT) and IRE1α*(I642G) −/+ 1NM-PP1 (10 μM), with quantification (I). (J) Model of oligomerization-dependence of RNase activity against XBP1 and Ins2 RNAs by IRE1α*(WT) and IRE1α*(I642G). Three independent biological samples were used for XBP1 splicing, Q-PCR and Annexin V experiments. Data plotted as mean value ± SD. P-values: *<0.05 and ** <0.01, ns=not significant. Also see Figure S1.
To study how IRE1α autophosphorylation impacts oligomeric state and RNase substrate selectivity, we expressed and purified a recombinant soluble mini-protein containing the kinase/RNase domains—called IRE1α*. Salt bridges formed through phospho-amino groups in neighboring IRE1α kinases contribute to kinase/RNase homo-oligomerization (Ali et al., 2011; Korennykh et al., 2009). IRE1α* is basally autophosphorylated and spontaneously homo-oligomerizes as its concentration is raised (Figure 1F,G). Dephosphorylation of IRE1α* with λ-phosphatase—(dP-IRE1α*)— reduces the oligomer/monomer ratio, confirming that phosphorylation drives oligomerization.
We next tested the impact of IRE1α*oligomerization on RNase activity against in vitro-transcribed XBP1 RNA and a less efficient substrate, Ins2 RNA, derived from one of the two mRNA isoforms encoding rodent proinsulin (Figure 1H,I). We also utilized an IRE1α* variant whose oligomeric state can be controlled with a small molecule. Mutation of IRE1α* at the isoleucine (I) gatekeeper residue to glycine (G) in its kinase ATP-binding pocket creates a “hole”—IRE1α* (I642G); in the full-length protein the I642G mutation cripples autophosphorylation (Han et al., 2009). As with dP-IRE1α*, IRE1α* (I642G) has reduced oligomerization compared to IRE1α* (Figure 1F,G). 1NM-PP1 is a “bumped” kinase inhibitor that selectively binds mutant kinases that contain glycine gatekeeper residues. Working as a ligand, 1NM-PP1 increases IRE1α* (I642G) oligomerization, but to levels well below those of equimolar IRE1α*.
Consistent with partial increases in oligomeric state, RNase activity is revived in 1NM-PP1-bound IRE1α* (I642G), but with activity largely confined to XBP1 RNA (Figure 1H,I). Therefore, both IRE1α* and 1NM-PP1-bound IRE1α* (I642G) efficiently cleave XBP1 RNA, but only IRE1α* surpasses the oligomerization threshold needed to catalyze the more sluggish Ins2 RNA cleavage reaction (Figure 1J). Thus, oligomerization state directly impacts IRE1α’s RNA substrate specificity.
We next tested effects in vivo. Upon self-association of its overexpressed lumenal domains, an INS-1 line expressing IRE1α (I642G) fully splices XBP1 mRNA under 1NM-PP1, without ER stress, and without causing Ins1 mRNA decay or apoptosis (Figure S1G). In fact, without 1NM-PP1, IRE1α (I642G) even reduces apoptosis under ER stress, acting as a strong dominant-negative (Figure S1H). Another kinase-dead mutant, IRE1α (K599A) (which is also RNase-dead), and an RNase-dead mutant, IRE1α (N906A) are also strongly dominant-negative for apoptosis (Figure S1H).
We previously showed that by pre-emptively producing XBP1s, 1NM-PP1–activated IRE1α (I642G) provides a metastable degree of cytoprotection against subsequent ER stress (Han et al., 2009; Han et al., 2008), as does forced expression of XBP1s, shown here (Figure S1I). However, without a window of sufficient time to permit adaptive pre-conditioning, simultaneous provision of 1NM-PP1 and ER stress agents rescues Ins1 mRNA decay and apoptosis, in 1NM-PP1 dose-dependent manner (Figure S1J,K).
Further supporting the notion that IRE1α triggers apoptosis using its RNase, a “holed”-RNase-dead double mutant—IRE1α (I642G/N906A)—remains dominant-negative under 1NM-PP1 (Figure S1L). In aggregate, the chemical-genetic studies show that the oligomeric state of IRE1α kinase/RNase subunits impacts both RNA substrate selection and cell fate, and that discrete, intermediate activation states are available to the effector catalytic domains (Figure S1M).
Divergent allosteric modulation of IRE1α oligomeric state and RNase activity with distinct kinase inhibitors
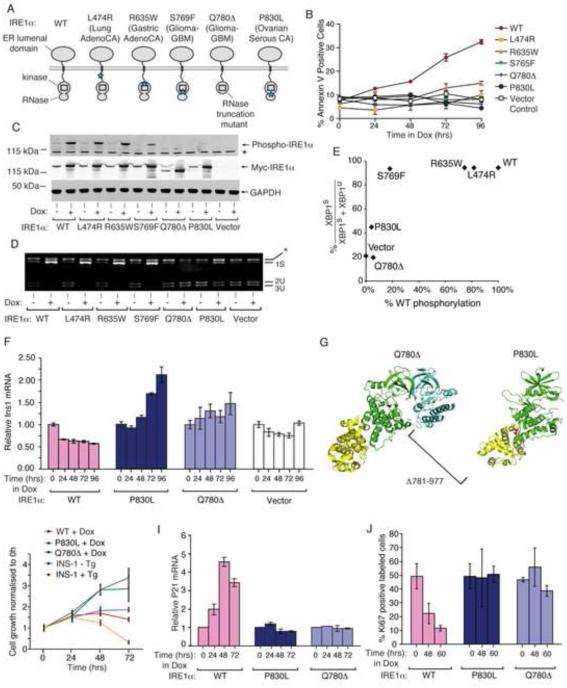
As with the rationally-engineered mutants, we find that intermediate activation states in IRE1α occur naturally through rare somatic Ire1α gene mutations found in human cancers, including glioblastoma, lung adenocarcinoma and serous ovarian cancer (Greenman et al., 2007). We predicted that five mutations spanning the kinase and RNase should affect function: four are missense, and one, Q780Δ, that is nonsense, amputates the entire RNase (Figure 2A). Expressed conditionally in isogenic INS-1 lines, the human IRE1α cancer mutants are all compromised for apoptosis (Figure 2B). Normalized to wild-type, the mutations significantly abrogate auto-phosphorylation and XBP1 splicing (Figure 2C-E). Expression of severely crippled IRE1α (Q780Δ) or IRE1α (P830L) actually increases Ins1 mRNA levels (Figure 2F), suggesting that some basal decay may even be blocked. Cells expressing IRE1α (Q780Δ) or IRE1α (P830L) proliferate well, in contrast to those expressing IRE1α (WT) or parental lines under ER stress (Figure 2H) (Movies S2A-F). The mRNA encoding cyclin-dependent kinase inhibitor, p21, is strongly induced in cells expressing IRE1α (WT), but not IRE1α (Q780Δ) or IRE1α (P830L) (Figure 2I). Marking cycling cells, Ki67 sharply declines upon expression of IRE1α (WT), but not IRE1α (Q780Δ) or IRE1α (P830L) (Figure 2J).
Figure 2. IRE1α cancer mutants are disabled for apoptosis.
(A) Cancer-associated mutations in human IRE1α. (B) Time course Annexin-V staining of INS-1 cells stably expressing human IRE1α (WT), (L474R), (R635W), (S765F), (Q780*), and (P830L) under saturating Dox (1μg/ml). (C) Anti-phospho-IRE1α and anti-Myc immunoblots, and (D) agarose gel of PstI-digested XBP1 cDNA amplicons from INS-1 cells expressing human IRE1α (WT) and mutants with Dox (1μg/ml) for 24hr. (E) XBP1 splicing from (D) as a function of IRE1α phosphorylation from (C). (F) Time course Q-PCR of Ins1 mRNA from INS-1 cells expressing IRE1α (WT) and mutants under Dox (1μg/ml). (G) Cartoon of monomeric human IRE1α (P830L) (right panel) and IRE1α (Q780*) dimerized with a IRE1α (WT) subunit (left panel) based on PDB: 3P23. (H) Time course MTT staining of INS-1 cells expressing IRE1α (WT), IRE1α (P830L) or IRE1α (Q780*) −/+ Dox (1μg/ml), or parental INS-1 cells −/+ 100 nM Tg. (I,J) Time-course Q-PCR for p21 mRNA, and Ki67 staining, from INS-1 IRE1α (WT), IRE1α (P830L) or IRE1α (Q780*) cells under Dox (1μg/ml). Three independent biological samples were used for Q-PCR, Ki67, and Annexin V experiments. Data plotted as mean +/− SD. Also see Figure S2.
Lack of the RNase in IRE1α (Q780Δ) converts it into a dominant-negative (Figure S2A-D). The P830L mutation, which occurs at the kinase/RNase junction (Figure 2G), may destabilize a dimerization interface (Xue et al., 2011). We predicted and tested that RNase activity in IRE1α (P830L) can be rescued with a kinase inhibitor, as IRE1α (I642G) can with 1NM-PP1.
We previously employed two distinct classes of kinase inhibitors—types I and II—to stabilize alternate kinase active site conformations in IRE1α (Wang et al., 2012). APY29 is a type I kinase inhibitor of IRE1α that stabilizes an active kinase domain conformation, which is typically adopted by ATP-bound kinases. By stabilizing the active kinase conformation, type I inhibitors act as ligands that allosterically activate IRE1α’s RNase; e.g., 1NM-PP1 is a type I inhibitor of IRE1α (I642G).
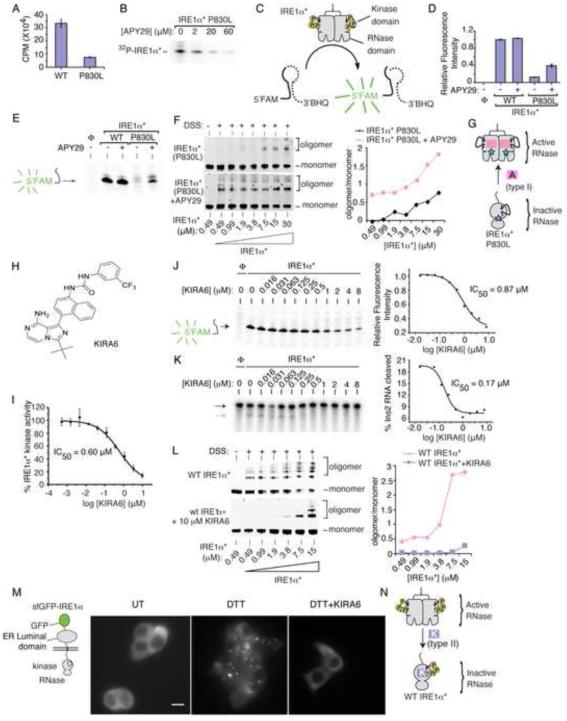
Compared to IRE1α* (WT), IRE1α* (P830L) has reduced kinase activity (Figure 3A), as the full-length protein does in vivo (Figure 2C). APY29 dose-dependently suppresses residual autophosphorylation of IRE1α* (P830L) (Figure 3B). IRE1α* (P830L) cannot cleave a FRET-quenched XBP1 RNA mini-substrate (Han et al., 2009) (Figure 3C-E), consistent with reduced RNase activity in vivo (Figure 2D). But opposite to effects on kinase activity, APY29 increases IRE1α* (P830L)’s oligomeric state to rescue RNase activity (Figure 3D-G).
Figure 3. Divergent modulation of IRE1α RNase activity using distinct classes of kinase inhibitors.
(A) Phosphorimager analysis of human IRE1α* (25 nM) and IRE1α* (P830L) (25 nM) kinase activity against peptide substrate (PAKtide, 2 μM) in the presence of 32Pγ-ATP. (B) Autoradiogram of IRE1α* (P830L) autophosphorylation under increasing [APY29]. (C) 5’FAM-3’BHQ XBP1 minisubstrate to measure RNase activity. (D) RNase activities of IRE1α* and IRE1α* (P830L) −/+ APY29 (20 μM) per (C). (E) Urea PAGE of XBP1 cleavage products from (D). (F) Immunoblots of increasing IRE1α* (P830L) after incubation with DMSO or APY29 (200 μM) and DSS, with oligomer/monomer quantification. (G) Model of APY29 rescue of oligomerization and RNase activity in IRE1α* (P830L). (H) Structure of KIRA6. (I) KIRA6 inhibition of IRE1α* kinase activity. IC50 values by fitting percent kinase activity per assay in (A) (n = 3). (K) Urea PAGE of competition cleavage by IRE1α* of XBP1 RNA mini-substrate (J) and α32P-labeled Ins2 RNA (K), under indicated [KIRA6]; IC50s by fitting in-gel fluorescence intensities (XBP1) and phosphorimager (Ins2). (L) Immunoblots of increasing [IRE1α*] incubated with DMSO or KIRA6 (10 μM) and DSS, with oligomer/monomer quantification. (M) Left: cartoon of sfGFP-IRE1α reporter. Right: Images of sfGFP-IRE1α induced with (sub-apoptotic) 1ng/ml Dox for 24hr in INS-1 cells −/+ DTT (5 mM) for 1hr −/+ KIRA6 (1 μM). Scale bar is 5 μm. (N) Model for how KIRA6 lowers oligomeric status and RNase activity of IRE1α*. Data plotted as mean +/− SD. Also see Figure S3.
If, as all preceding results suggest, kinase-driven oligomerization of IRE1α hyperactivates its RNase to trigger apoptosis, then kinase inhibitors that block oligomerization should prevent apoptosis under ER stress. To this end, we employed type II kinase inhibitors that stabilize an inactive ATP-binding site conformation in IRE1α. We previously developed a subset of type II kinase inhibitors designated KIRAs, for Kinase-Inhibiting RNase-Attenuators, that inhibit IRE1α’s RNase activity by breaking oligomers (Wang et al., 2012). Since our original report, we have identified KIRA6 as a more potent version (Figure 3H). KIRA6 dose-dependently inhibits IRE1α* (WT) kinase activity, XBP1 RNA cleavage, Ins2 RNA cleavage (with lower IC50 than for XBP1 RNA in a competition assay), and oligomerization (Figure 3I-L).
To follow IRE1α oligomerization in vivo, we first tested a reporter called IRE1-3F6HGFP that contains an EGFP domain positioned near the kinase (Li et al., 2010), but found that it has attenuated XBP1 splicing and fails to induce apoptosis (Figure S3A,B). To avoid potential steric effects on the kinase, we constructed a superfolder green fluorescent protein (sfGFP) N-terminally fused to the ER lumenal domain. Expressed isogenically in INS-1 cells, sfGFP-IRE1α retains apoptotic activity and gathers into discrete fluorescent foci in the ER membrane under the ER stress agent dithiothreitol (DTT) (Figure 3M, S3A,B). An (I642G) version of sfGFP-IRE1α fully splices XBP1 mRNA under 1NM-PP1 without forming foci (Figure S3C,D). In fact, without 1NM-PP1, sfGFP-IRE1α (I642G) resists forming foci under DTT, suggesting that without its ligand it adopts an inactive kinase conformation and explaining dominant-negative effects of IRE1α (I642G)(Figure S1H). Similar to apoptosis, foci formation by sfGFP-IRE1α (I642G) requires both ER stress and 1NM-PP1, further supporting the tight link between IRE1α oligomerization—shown in vivo through foci—and apoptosis. Thus, using sfGFP-IRE1α, which faithfully recapitulates cytosolic events, we tested and found that KIRA6 prevents foci formation by DTT (Figure 3M). Hence, KIRAs fulfill their design principle of breaking kinase/RNase oligomers to inhibit the RNase (Figure 3N).
KIRA6 inhibits IRE1α in vivo to preserve cell viability and function in diverse cells and rodent tissues experiencing ER stress
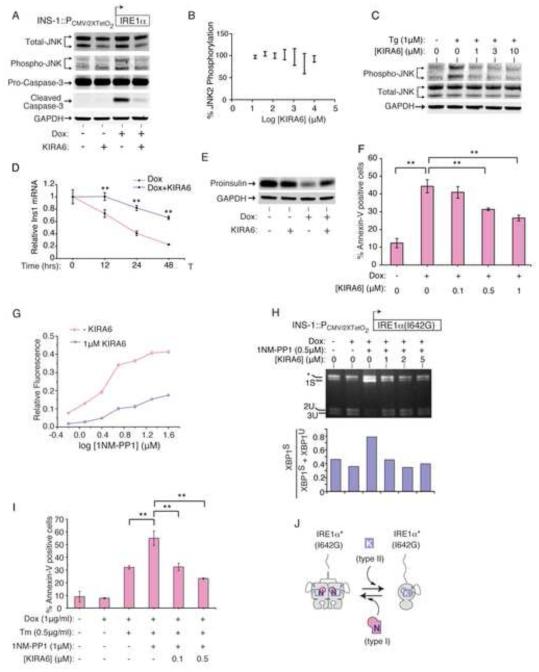
The remainder of our work focused on testing physiological effects of IRE1α kinase inhibition. APY29 showed pleiotropic toxicity, including proliferative blocks at low micromolar concentrations, precluding further in vivo testing of ON-target effects (Figure S4A). In contrast, KIRA6 had negligible toxicity up to 10 μM (Figure S4A), providing a favorable therapeutic index to test cytoprotection. INS-1 lines confirmed ON-target effects: Pro-Caspase-3 cleavage upon IRE1α (WT) expression is prevented by KIRA6 (Figure 4A). Moreover, despite its inability to directly inhibit JNK activity in vitro, KIRA6 strongly inhibits JNK phosphorylation from IRE1α hyperactivation or ER stress (Figure 4A-C). Also, KIRA6 dose-dependently inhibits Ins1 mRNA decay, proinsulin depletion, and apoptosis from IRE1α hyperactivation (Figure 4D-F).
Figure 4. KIRA6 inhibits IRE1α Terminal UPR outputs and apoptosis.
(A) Anti-total JNK, anti-phospho-JNK, and anti-Pro- and Cleaved Caspase-3 immunoblots of INS-1 IRE1α (WT) cells treated with Dox (5 ng/ml) −/+ 1μM KIRA6 for 72hr. (B) JNK2α1 phosphorylation under indicated [KIRA6] by in vitro ELISA-based anti-phospho-JNK assay. (C) Anti-total and phospho-JNK immunoblots of INS-1 cells pretreated for 1hr with indicated [KIRA6], then 1μM Tg for 2hr. (D) Q-PCR for Ins1 mRNA in INS-1 IRE1α (WT) cells treated with Dox (5 ng/ml) −/+ KIRA6 (1 μM). (E) Anti-Proinsulin immunoblot of samples in (A). (F) % Annexin V staining in INS-1 IRE1α (WT) cells after 72hr in Dox (5 ng/ml) and indicated [KIRA6]. (G) Competition between indicated [1NM-PP1] and KIRA6 (1 μM) for IRE1α* (I642G) RNase. (H) Agarose gel of PstI-digested XBP1 cDNA amplicons from INS-1 cells IRE1α (I642G) cells induced by Dox (1 μg/ml) for 24hr, then 1NM-PP1 (0.5 μM) −/+ indicated [KIRA6] for 3hr, with quantitation. (I) Annexin V staining of INS-1 IRE1α (I642G) cells after 72hr with Dox (1 μg/ml), Tm (0.5 μg/ml), 1NM-PP1 (1 μM) and indicated [KIRA6]. (J) Model of 1NM-PP1 and KIRA6 competition of oligomerization and RNase activity in IRE1α* (I642G). Data plotted as mean +/− SD. P-values: *<0.05, ** <0.01. Also see Figure S4.
Chemical-genetic tools enabled ON-target competition tests. KIRA6: [1] reduces 1NM-PP1-induced XBP1 RNA cleavage by IRE1α* (I642G) in vitro (Figure 4G); [2] antagonizes 1NM-PP1-induced XBP1 splicing by IRE1α (I642G) in vivo (Figure 4H); and [3] reduces 1NM-PP1 potentiation of Ins1 mRNA decay and apoptosis during ER stress, in dose-dependent manner (Figure 4I, S4B,C). KIRA6 does not inhibit the activity of a panel of Ser/Thr kinases (including JNK2 and 3) in vitro (Figure S4D). Moreover, KIRA6 does not inhibit nor secondarily promote eIF2α phosphorylation by PERK, the other UPR kinase (Figure S4E).
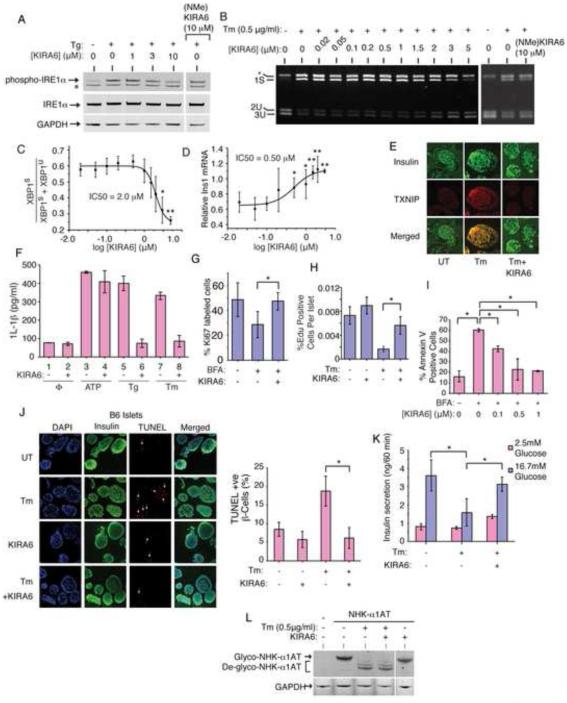
Having confirmed that KIRA6 has ON-target effects, we next tested efficacy against endogenous IRE1α using the established ER stress regimes in their linear ranges straddling the apoptotic trigger point (Figure S1A). In INS-1 cells, KIRA6 inhibits IRE1α auto-phosphorylation by Tg and XBP1 mRNA splicing by Tm in a dose-dependent manner (Figure 5A-C); whereas, a control analog, (NMe)KIRA6, incapable of binding to the kinase hinge region, inhibits neither output at 10 μM (Figure 5A,B, S5A,B).
Figure 5. KIRA6 reduces ER stress-induced death of cultured cells and in pancreatic islet explants.
(A) Immunoblots for total and phospho-IRE1α in INS-1 cells pretreated for 1hr with indicated [KIRA6], or 10 μM (NMe)KIRA6, then Tg (1 μM) for 2hr. (B) Agarose gel of XBP1 cDNA amplicons from INS-1 cells pre-treated with indicated [KIRA6] for 1hr, or 10 μM (NMe)KIRA6, followed by 0.5 μg/ml Tm for 8hr. (C) Ratios of XBP1S over (XBP1S + XBP1U) from (B). (D) Q-PCR for Ins1 mRNA (normalized to no Tm) in INS-1 cells pretreated for 1hr with indicated [KIRA6], then 12hr in Tm (0.5 μg/ml). (E) Immunofluorescence: Insulin (green) and TXNIP (red) in islets of C57BL/6 mice under 0.5μg/ml Tm −/+ 0.5 μM KIRA6 for 16hr. (F) IL-1β secretion from THP-1 cells after 4hr −/+ 0.5 μM KIRA6, 5 μg/ml Tm, 1 μM Tg, or 5 mM ATP. (G) Ki67+ INS1 cells under 0.25 μg/ml BFA −/+ 0.5 μM KIRA6 for 48hr. (H) Proliferating mouse islet β-cells under 0.5 μg/mL Tm −/+ 0.5 μM KIRA6 for 48hr (nuclei double-positive for EdU and β-cell nuclear marker, Nkx6.1, over total Nkx6.1 positive nuclei). (I) Annexin-V staining of INS-1 cells treated with 0.25 μg/ml BFA and indicated [KIRA6] for 72hr. (J) Immunofluorescence images of C57BL/6 islets treated with 0.5 μg/mL Tm −/+ 0.5 μM KIRA6 for 16 hr. Co-stained for DAPI (blue), insulin (green), and TUNEL (red). Quantification of TUNEL+ β-cells (white arrows) normalized to DAPI+ cells. (K) Glucose-stimulated insulin secretion (GSIS) by C57BL/6 islets after 0.5μg/mL Tm −/+ 0.5 μM KIRA6 for 16hr; [Glucose] was 2.5 mM or 16.7 mM for 60 min. (L) Immunoblots for alpha-1 anti-trypsin in HEK293 cells transfected with pCDNA3.1-α1hAT-NHK, then treated with KIRA6 (1 μM) −/+ Tm (0.5 μg/ml) for 20hr. Three independent biological samples were used for XBP1 splicing, Q-PCR, Annexin V and immunofluorescence experiments. Data plotted as mean +/− SD. P-values: *<0.05, ** <0.01. Also see Figure S5.
We next tested multiple Terminal UPR endpoints and found that KIRA6: [1] Inhibits Ins1 and Ins2 mRNA decay by Tm in INS-1 cells in dose-dependent manner (Figure 5D, S5C). We noted that the in vivo IC50 of KIRA6 for Ins1 mRNA rescue is lower than that for inhibiting XBP1 splicing, and Ins2 mRNA levels recover even at 20 nM KIRA6 and exceed basal, untreated levels in dose-dependent manner. Furthermore, KIRA6: [2] Inhibits TXNIP induction by Tm in murine C57BL/6 pancreatic islets (Figure 5E); [3] Inhibits IRE1α-dependent activation of a TXNIP 3’UTR luciferase reporter containing its two miR-17 binding sites (Figure S5D); [4] Prevents 1L-1β secretion by Tm and Tg (but not ATP) in THP1 macrophage lines (Figure 5F); [5] Prevents loss of INS-1 Ki67-positive cells and C57BL/6 pancreatic islet double-positive Nkx6.1/EdU β–cells under ER stress (Figure 5G,H, S5E)(Movies S5A-C); [6] Dose-dependently inhibits apoptosis of INS-1 cells under BFA (Figure 5I); [7] Reduces TUNEL staining of β–cells in C57BL/6 and human islets under Tm (Figure 5J, S5F); and [8] preserves glucose-stimulated insulin secretion (GSIS) in C57BL/6 islets under Tm (Figure 5K).
We also tested effects of STF-083010, a small molecule tool compound that reactively modifies Lysine 907 in the RNase active site (Papandreou et al., 2011)(Figure S5G). As with KIRA6, STF-083010 (at 50 μM) also decreases Ins1 mRNA decay under IRE1α hyperactivation and apoptosis by Tm (Figure S5H,I). Moreover, when used in combination at doses that are sub-therapeutic individually, STF-083010 (1μM) and KIRA6 (50 nM) afford significant cytoprotection under Tm (Figure S5J). Together, these data further implicate IRE1α’s RNase in promoting apoptosis, in this case by showing that the RNase can even be inhibited combinatorially through two distinct sites in IRE1α for cytoprotection.
To rule out the possibility that KIRA6 defeats ER stress agents upstream of IRE1α, we tested whether blocks to ER post-translational modification still persist under KIRA6. A test substrate, the null Hong Kong variant of alpha-1 anti-trypsin (NHK-A1AT), normally glycosylated and ER-retained, is deglycoslyated under Tm. NHK-A1AT clearly remains deglycoslyated under both Tm and KIRA6 (Figure 5L).
Encouraged by clear and convincing evidence that KIRA6 preserves cell viability and function in multiple cell and explant systems under diverse ER stress regimes, we next applied a higher evidentiary standard by testing disease-relevant animal models. Given compelling evidence that ER stress contributes to photoreceptor loss in many retinal diseases, including retinitis pigmentosa (RP) (Zhang et al., 2014), we tested KIRA6 in two rodent models. Transgenic rats expressing a misfolded Rhodopsin mutant (P23H) exhibit spontaneous photoreceptor degeneration and are a model of autosomal dominant RP (Gorbatyuk et al., 2010). Retinas of hemizygous P23H rats develop normally but lose photoreceptors beginning on postnatal day (P) 10; by P40, the outer nuclear layer (ONL), representing photoreceptor nuclei, is reduced to ~50% of the thickness of wild-type rats (Pennesi et al., 2008). We intravitreally injected KIRA6 or carrier into either eye of individual P23H rats at P9 and P15. ONL thickness at P40 revealed partial, yet statistically significant, protection from photoreceptor loss in KIRA6-treated eyes (Figure S6A,B).
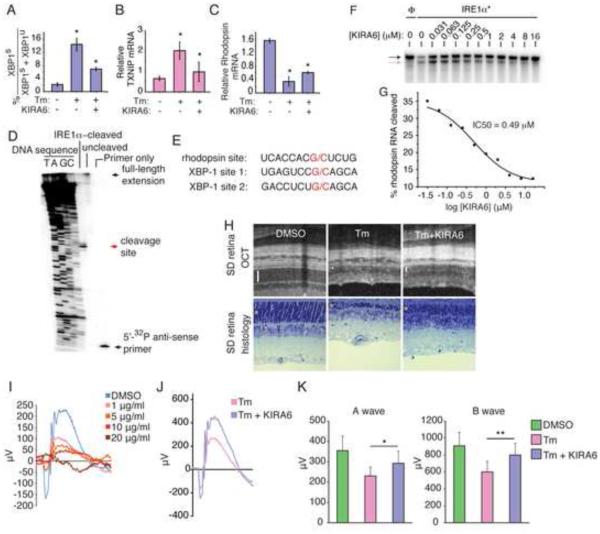
Given rapid clearance of intravitreally injected small molecules (half-life <60h), we were unable to maintain sufficient KIRA6 in the vitreous over the ~30d progression of retinal degeneration in P23H rats to test for functional protection. Therefore, we used a model of acute photoreceptor loss occurring over 7 days from single intravitreal injection of Tm into adult rats (Shimazawa et al., 2007). Intravitreal co-injection of KIRA6 with Tm significantly reduces XBP1 splicing, TXNIP induction, and decay of the ER-localized photoreceptor-specific Rhodopsin mRNA (Figure 6A-C). Rhodopsin mRNA may be an IRE1α RNase substrate since Rhodopsin RNA is cleaved by IRE1α*, but not RNase-dead IRE1α* (N906A), at a G/C site with flanking similarity to scission sites in XBP1 (Figure S6C; 6D,E). KIRA6 dose-dependently inhibits Rhodopsin RNA cleavage by IRE1α* (Figure 6F,G). Concomitant with blockage of Terminal UPR outputs, co-injection of KIRA6 in the Tm model reduces photoreceptor loss by Optical Coherence Tomography (OCT) and histology (Figure 6H).
Figure 6. Intravitreal KIRA6 preserves photoreceptor cell numbers and function under ER stress.
(A) % XBP1 splicing in SD rat retinas 72hr post-intravitreal—and Q-PCR for TXNIP mRNA (B) and Rhodopsin mRNA (C) 96hr post-intravitreal—injection of 20 μg/ml Tm −/+ 10 μM KIRA6. (D) Primer extension mapping of IRE1α cleavage site in Rhodopsin RNA with alignment of Rhodopsin and XBP1 mRNA (E). Urea PAGE of cleavage of 32P-labeled Rhodopsin mRNA by IRE1α* with indicated [KIRA6], with IC50 (F); black arrow: intact RNA; red arrow: cleaved RNA. (H) OCT images and histological sections of SD rats 7d post-intravitreal injection of 20 μg/ml Tm −/+ 10 μM KIRA6; bars and asterisks denote ONLs. (I) SD rats intravitreally injected at P21with 2 μl Tm or DMSO to achieve indicated [Tm]; ERG measurements at a light intensity of 0 dB recorded at P28. (J) Representative scotopic ERG at a light intensity of 0 dB from a SD rat treated with Tm (3μg/ml) +/− KIRA6 (10 μM) at P21 and analyzed at P28. (K) Quantified a- and b-wave amplitudes of 0 dB scotopic ERGs from SD rats treated with DMSO or Tm (3μg/ml) +/− KIRA6 (10 μM) at P21 and analyzed at P28. Also see Figure S6.
Next, to test whether KIRA6 also provides functional protection, we established a dose-response curve to determine threshold [Tm] that cause functional retinal damage as measured by scotopic electroretinograms (ERG) (Figure 6I). Based on the results, we injected Tm at 3 μg/ml. In this regime, co-injection with KIRA6 substantially protects against loss of ERG responsiveness, significantly preserving both a- and b-wave amplitudes (Figure 6J,K, S6D).
Finally, to test in vivo efficacy of systemic KIRA6, we chose the Ins2+/Akita (Akita) mouse, which expresses a mutant (C96Y) proinsulin unable to complete oxidative folding, thus causing chronic ER stress, β-cell apoptosis, and diabetes in infancy (Lerner et al., 2012). The pharmacokinetic profile of KIRA6 in BALB/c mice intraperitoneally (i.p.) dosed at 10 mg/kg showed good drug plasma AUC levels (AUC 0-24h = 14.3 μM*h) with moderate clearance (22.4 mL/min/kg). Drug half-life was 3.90 hours, Cmax was 3.3 μM, and plasma levels at 4 and 8hr were 1.2 μM and 0.33 μM, respectively. Initial systemic studies utilized a Tm i.p. challenge in C57BL/6 mice, with and without KIRA6 co-injection, and UPR markers measured in liver. Low dose Tm (2 μg/kg) elevates liver XBP1 splicing without decay of ER-localized Blos1 mRNA (Hollien et al., 2009), while KIRA6 co-provision reduces XBP1 splicing (Figure S7A,B). Escalation of Tm to 100 μg/kg further increases XBP1 splicing and triggers Blos1 mRNA decay, with both markers attenuated by KIRA6 (Figure S7C,D).
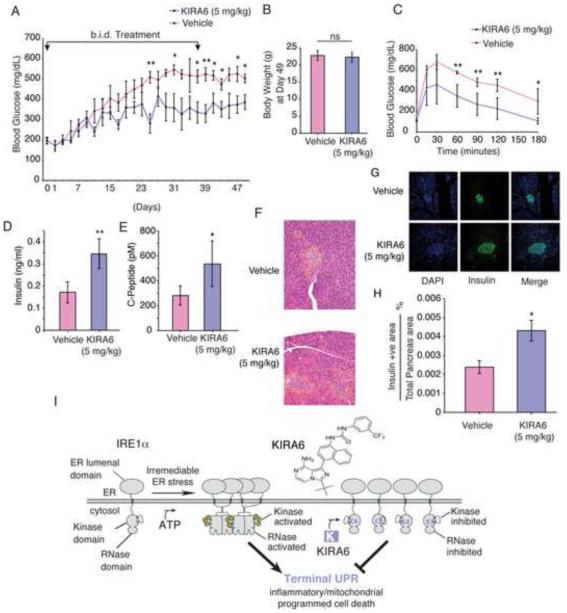
Based on low micromolar KIRA6 needed for protection in cell culture, we chose i.p. dosing regimens of 5 or 10 mg/kg b.i.d for Akita chronic efficacy studies to provide similar exposure. We injected KIRA6 into randomized 3-week old male Akita mice when their random blood glucose levels were at prediabetic range (~200 mg/dl). In both dosing regimes, we observed significant amelioration of random glucose levels over several weeks in KIRA6-treated mice compared to vehicle, both fed ad lib (Figure 7A, S7E). TXNIP mRNA levels decline in islets of KIRA6-treated mice within one week, without compensatory increase of CHOP mRNA (downstream of PERK) (Figure S7G,H). KIRA6-treated mice appeared healthy even after 49 days from initial injection and displayed no significant differences in weight from vehicles (Figure 7B, S7F). Even 12 days after stopping injections, the 5mg/kg KIRA6-treated mice show significantly improved random blood glucose levels and glucose tolerance tests (GTT) (Figure 7C). Even 21 days after stopping injections, KIRA6-treated mice display statistically significant doubling in both plasma insulin and C-peptide levels (Figure 7D,E). H&E and insulin staining of whole pancreas sections reveals increased islet size in KIRA6 treated-animals (Figure 7F,G). Insulin-positive islet areas remained significantly higher in the KIRA6-treated group 18 days after stopping injections (Figure 7H).
Figure 7. Systemic KIRA6 attenuates β-cell functional loss, increases insulin levels, and ameliorates hyperglycemia in the Akita mouse.
(A) Random AM blood glucose (BG) levels in male Ins2+/Akita mice intraperitoneally (i.p) injected for 37 days b.i.d. with KIRA6 (5 mg/kg)(n=6) or vehicle (n=6) starting at P21 (i.e., Day 1). BGs (mean +/− SEM), also analyzed by Two-way RM ANOVA; p-value = 0.0122. (B) Cohort body weights at Day 49. (C) Glucose tolerance tests on Day 49 (12d post injections) of O/N fasted Ins2+/Akita mice (P53) after i.p. (2 g/kg) glucose (KIRA6 n=6, Vehicle n=3). (D-E) Random insulin and C-peptide levels in Ins2+/Akita mice on Day 58 (21d post injections). KIRA6 (5 mg/kg)(n=5) and vehicle (n=4). (F) Whole pancreatic histological sections from Ins2+/Akita mice on Day 53 (15d post injections). Islets delineated by dashed outline. Immunofluorescence micrographs of samples in (F): co-stained for DAPI (blue), insulin (green), with merge. (H) Total β-cell area as a percentage of total pancreas area on Day 55 (18d post injections). KIRA6 (5 mg/kg)(n=6) and vehicle (n=3). (I) Model of how KIRA6 prevents the terminal UPR by inhibiting IRE1α oligomers. Data plotted as mean +/− SD. P-values: *<0.05, ** <0.01. Also see Figure S7.
DISCUSSION
In the baker’s yeast S. cerevisiae, the UPR is a homeostatic signaling pathway controlled by IRE1-mediated splicing of an mRNA encoding an adaptive transcription factor called Hac1 (Cox and Walter, 1996). Following this paradigm from this unicellular eukaryote, reports have suggested that the signaling outputs of mammalian IRE1α are likewise solely restricted to restoring homeostasis and promoting survival under ER stress (Lin et al., 2007). Furthermore, these models posit that when ER stress becomes irremediable, IRE1α’s pro-survival signaling through XBP1 splicing circumstantially wanes (through an unknown mechanism), even as apoptotic outputs from PERK rise to promote cell death, without further opposition by IRE1α (Lin et al., 2009). These arguments therefore predict that sustained IRE1α activation (even if artificially imposed) should universally promote cell survival under ER stress; whereas genetic or pharmacological inhibition of IRE1α should hasten cell death. Through forcibly activating and inhibiting IRE1α in a variety of cell systems and animal models, here we generated extensive data that refute these aforementioned predictions to instead support opposite conclusions.
An alternative model that we previously proposed (Han et al., 2009), and mechanistically substantiated here, is that IRE1α switches outputs depending on the level of ER stress. Under low, manageable levels of ER stress, adaptive UPR signaling promotes secretory homeostasis, partly through IRE1α-mediated splicing of XBP1 mRNA and consequent XBP1s outputs. Likewise, pre-emptive PERK activation affords a measure of cytoprotection against subsequent ER stress by attenuating translation (Lu et al., 2004), as does pre-conditioning with 1NM-PP1-bound IRE1α (I642G) to transiently stabilize an intermediate activation mode of the RNase confined to XBP1 splicing (Han et al., 2008).
However, under high ER stress, IRE1α acquires endonucleolytic activity against a large plethora of RNA targets, first identified in D. melanogaster and termed RIDD (Hollien and Weissman, 2006), including ER-localized mRNAs and non-coding RNAs in mammals (Han et al., 2009; Hollien et al., 2009; Lerner et al., 2012; Upton et al., 2012). These extra-XBP1 RNA cleavage events precede and closely track with entry of ER-stressed cells into apoptosis, and we showed here that their amelioration with small molecule inhibitors of IRE1α kinase/RNase inhibits apoptosis. Thus, rather than have the two UPR kinases working in opposition, multiple lines of evidence suggest that a continuum of graded activation states (dependent upon the strength of upstream stress) is available to either IRE1α or PERK, both of which under high activation undergo switch-like conversion from promoting homeostasis to promoting cell death (Han et al., 2009; Lin et al., 2009). Similar switching mechanisms occur in other cell surface death receptors that respond divergently depending on the strength or context of upstream inputs (Festjens et al., 2007; Ofengeim and Yuan, 2013).
Further supporting a model of binary, rather than unitary outputs, three postulates that we posed and tested reasonably establish causality between IRE1α hyperactivation and cell death:
(I) First, forced hyperactivation of IRE1α’s RNase should suffice to lead cells into the Terminal UPR and along a continuum of destructive outcomes, including proliferative blocks, loss of differentiated cell identity, and eventually into apoptosis. In line with this expectation, past a critical oligomerization threshold, IRE1α’s RNase degrades key mediators of specialized cell function, including abundant insulin-encoding mRNAs in pancreatic β-cells and Rhodopsin mRNAs in retinal photoreceptor cells (both ER-localized). Also, as previously shown, mRNAs encoding ER-resident enzymatic activities are also targeted by hyperactive IRE1α RNase, potentially compromising ER function (Han et al., 2009). Thus, at high activation, IRE1α’s adaptive outputs become overshadowed by its destructive outputs and further amplified downstream by TXNIP, causing IL-1β secretion, sterile inflammation/pyroptosis (linked to both types 1 and 2 diabetes)(Lerner et al., 2012; Schroder et al., 2010) and JNK signaling (Urano et al., 2000). Further linking IRE1α to cell death, IRE1α cancer mutants show defective homo-oligomerization and RNase activity, which may allow the Terminal UPR to become disabled or co-opted for survival advantage. Indeed, proliferative blocks normally imposed through IRE1α are defeated in the cancer mutants. Given these results, future studies of mutated IRE1α in cancer are warranted.
(II) Second, a class of IRE1α inhibitors that disrupt oligomerization should reduce RNase activity and Terminal UPR events in tandem. Unique among multi-domain kinases, the mechanistic relationship between IRE1α’s kinase and RNase allows divergent small molecule allosteric control (Wang et al., 2012). Whereas both are ATP-competitive, IRE1α type I kinase inhibitors increase oligomerization to increase RNase activity, while IRE1α type II kinase inhibitors decrease oligomerization to decrease RNase activity. Here we developed and tested the effects of KIRA6, a novel IRE1α type II kinase inhibitor. Given that KIRA6 has a favorable therapeutic index and shows IRE1α ON-target effects, we predicted that it would reduce cell death under ER stress. Remarkably, blocking IRE1α with KIRA6 raises the apoptotic threshold and enhances survival during ongoing upstream ER stress, indicating that destructive signaling rather than a compromised ER micro-environment per se promotes cell death (Figure 7I). While poly-pharmacological toxicity precluded testing ON-target effects of APY29, our results justify development and testing of non-toxic type I kinase inhibitors against IRE1α cancer mutants.
(III) Third, blocking IRE1α with KIRA6 should protect against ER stress-mediated cell degeneration in vivo, leading not only to increased cell survival but also preserved physiological function. Consistent with this, in various cell types and explants, KIRA6 not only reduced cell loss under acute ER stress, but also prevented proliferative blocks and preserved function (e.g., GSIS). Encouraged by these data, we tested KIRA6 in rodent models of chronic ER stress-induced retinal degeneration. P23H rats intravitreally treated with KIRA6 had significantly preserved photoreceptor numbers, and in the Tm co-injection model, functional protection was found. Finally, systemic administration of KIRA6 in Akita diabetic mouse significantly reduced random blood glucose levels, improved glucose tolerance acutely, preserved β-cells, and elevated blood insulin and C-peptide levels. Remarkably, beneficial effects persisted even several weeks after stopping treatment. To our knowledge, this is the first work showing small molecule efficacy in the highly penetrant Akita genetic model.
Thus, we conclude that IRE1α exerts powerful effects on cell fate and function under ER stress, and that its kinase domain presents an attractive target for small molecule modulation. In summary, KIRA6, a novel small molecule kinase inhibitor of IRE1α, reduces cell death in several disease relevant models of ER stress-induced cell degeneration. While homozygous deletion of either Ire1α or Xbp1 impedes embryogenesis and secretory cell development (Reimold et al., 2001; Tirasophon et al., 1998; Urano et al., 2000; Zhang et al., 2005), the ability to titrate down IRE1α’s catalytic activities with a small molecule provides an opportunity to uncouple extra-XBP1 destructive outputs from XBP1-dependent adaptation. From this work, we propose the existence of a natural therapeutic window for IRE1α inhibition owing to the higher oligomeric state needed for extra-XBP1 endonucleolytic activation (RIDD); thus, lower concentrations of IRE1α type II kinase inhibitors block RIDD while maintaining XBP1 splicing (Figure 5D). Subsequent work is necessary to understand the consequences of long-term IRE1α inhibition. While further medicinal chemistry to optimize KIRA6 is beyond the scope of this study, such efforts may lead to first-in-class agents capable of preventing cell loss and affording therapeutic benefit in myriad human degenerative diseases, including retinitis pigmentosa and diabetes.
EXPERIMENTAL PROCEDURES
Tissue Culture, ER stress agents, and small molecules
Generation of isogenic, stable INS-1 lines was described previously (Han et al., 2009); also see Supplemental Experimental Methods. Thapsigargin (Tg), Brefeldin A (BFA), and Dox were purchased from Sigma-Aldrich. Tunicamycin (Tm) was purchased from Millipore. APY29, KIRA6, (NMe)KIRA6 1NM-PP1 and STF-083010 were synthesized in house (see Supplemental Experimental Methods for synthesis, purification and purity determination methods of small molecules).
Western Blots and Antibodies
For protein analysis, cells were lysed in M-PER buffer (Thermo Scientific) plus complete EDTA-free protease inhibitor (Roche) and phosphatase inhibitor cocktail (Sigma). Protein concentration was determined using BCA Protein Assay (Thermo). Western blots were performed using 10% and 12% Bis-Tris precast gels (NuPage) on Invitrogen XCell SureLock® Mini-Cell modules. Gels were run using MES buffer and transferred onto nitrocellulose transfer membrane using an XCell II™ Blot Module. Antibody binding was detected by using near-infrared-dye-conjugated secondary antibodies (Licor) on the LI-COR Odyssey scanner or visualized by capturing on a CL-XPosure film using ECL SuperSignal West Dura Extended Duration Substrate (both from Thermo Scientific). See Supplemental Experimental Methods for details of the antibodies used.
RNA isolation, Real-Time PCR (Q-PCR), and Primers
RNA was isolated from whole cells using either Qiagen RNeasy kits or Trizol (Invitrogen). TissueLyser II (Qiagen) was used for RNA isolation from liver and retina. For standard mRNA detection, generally 1 μg total RNA was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen). For Q-PCR, we used SYBR green (Qiagen) and StepOnePlus Real-Time PCR System (Applied Biosystems). Gene expression levels were normalized to GAPDH or Actin. See Supplemental Experimental Methods for primer sequences used for Q-PCRs, and for details of XBP1 mRNA splicing.
In vitro IRE1α* crosslinking, RNase and kinase assays
See (Wang et al., 2012) and supplemental Experimental Methods for details of RNAse, kinase and oligomerization assays.
Flow Cytometry
For assaying apoptosis by Annexin V staining, cells were plated in 12-well plates overnight. Cells were then treated with various ER stress agents for indicated times. On the day of analysis, cells were trypsinized and washed in PBS and resuspended in Annexin V binding buffer with Annexin-V FITC (BD Pharmingen™). Flow cytometry was performed on a Becton Dickinson LSRII flow cytometer.
Islet staining
Islets were extracted from C57BL/6 mice using previously reported methods (Szot et al., 2007). Approximately 150 islets were cultured for each condition in triplicate. Non-diabetic human islets were obtained from Prodo Labs (Irvine, CA) and cultured in Prodo Islet Medium (PIM from Prodo Labs). Islets were stained with TUNEL using ApopTag® Red In Situ Apoptosis Detection Kit (Millipore) according to the manufacturer’s instructions. Islets were also co-stained with anti-TXNIP (MBL International), guinea pig anti-insulin (Zymed), DAPI (Sigma), and goat anti-guinea pig secondary (Rockland) before mounting onto slides with VectaShield (Vector Laboratories). At least 10 islets and > 500 β-cell nuclei were counted per group, in triplicate. Also see Supplemental Experimental Methods for details of fixation and staining.
Intravitreal Injections of small molecules
2 μl was injected intravitreally into each eye to achieve indicated final concentrations based on known rat vitreous volumes. Tm (20 μg/μl final) +/− KIRA6 (10 μM final) was injected into SD rats at P21 with equivalent DMSO as vehicle control. Retinas were collected at 72 and 96hr after injections in Trizol (Invitrogen) for Q-PCR. Eyes were examined by optical coherence tomography (OCT) 7 days post injection then collected for morphological analysis. P23H rats were injected with KIRA6 (10 μM final) or DMSO vehicle at P9 and P15, and eyes were examined at P40 by OCT and by morphological analysis. See Supplemental Experimental Methods for details of image guided (OCT), morphological analysis, and electroretinography (ERG).
Mouse systemic injections
Male Ins2+/Akita mice were injected i.p. with KIRA6 in a 2 mg/ml solution made of 3% Ethanol: 7% Tween-80: 90% Saline twice a day (b.i.d). Same solution without KIRA6 is denoted as Vehicle. C567BL/6 mice were also injected with same KIRA6 solution and indicated doses of Tm for liver analysis. See Supplemental Experimental Methods for details of islet explant experiments, GSIS, proliferation studies, blood glucose determination, glucose tolerance tests, insulin and C-peptide measurements, and β-cell area determination.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael Matthes and Doug Yasumura for technical assistance with rat models, Vinh Nguyen for islet isolation, and Oakes and Papa lab members for discussions. The work was supported by NIH: DP2OD001925 (F.R.P), RO1 CA136577 (S.A.O.), RO1 DK080955 (F.R.P), RO1 DK095306 (S.A.O. and F.R.P), RO1 DK100623 (D.J.M. and F.R.P), PO1 HL108794 (F.R.P), P30 DK063720 (F.R.P. and M.S.G.), UO1 DK089541 (F.R.P. and M.S.G.), RO1 DK021344 (M.S.G.), RO1 GM086858 (D.J.M.), R21 CA177402 (D.J.M.), EY019504 (D.B.G), EY01919 (D.B.G), EY06842 (D.B.G), F31-NS083323 (E.S.W.); HHMI Physician-Scientist Early Career Award (S.A.O.); American Cancer Society Research Scholar Award (S.A.O.); Burroughs Wellcome Foundation (F.R.P.); Juvenile Diabetes Research Foundation (F.R.P. and S.A.O.); Harrington Discovery Institute Scholar-Innovator Award (S.A.O. and F.R.P.); Alfred P. Sloan Foundation (D.J.M.); Camille and Henry Dreyfus Foundation (D.J.M.); National Science Foundation (E.S.W.); American Diabetes Association ADA-7-07-MN-22 (H.M.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ali MM, Bagratuni T, Davenport EL, Nowak PR, Silva-Santisteban MC, Hardcastle A, McAndrews C, Rowlands MG, Morgan GJ, Aherne W, et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell's decision to live or die. Cell death and differentiation. 2007;14:400–410. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk MS, Knox T, LaVail MM, Gorbatyuk OS, Noorwez SM, Hauswirth WW, Lin JH, Muzyczka N, Lewin AS. Restoration of visual function in P23H rhodopsin transgenic rats by gene delivery of BiP/Grp78. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5961–5966. doi: 10.1073/pnas.0911991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Upton JP, Hagen A, Callahan J, Oakes SA, Papa FR. A kinase inhibitor activates the IRE1alpha RNase to confer cytoprotection against ER stress. Biochemical and biophysical research communications. 2008;365:777–783. doi: 10.1016/j.bbrc.2007.11.040. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. The Journal of cell biology. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Kohno K. How transmembrane proteins sense endoplasmic reticulum stress. Antioxid Redox Signal. 2007;9:2295–2303. doi: 10.1089/ars.2007.1819. [DOI] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Molecular and cellular biology. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, Igbaria A, Shen S, Nguyen V, Backes BJ, Heiman M, et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012;16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16113–16118. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PloS one. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, Kaufman RJ, Ron D, Harding HP. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. The EMBO journal. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengeim D, Yuan J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nature reviews Molecular cell biology. 2013;14:727–736. doi: 10.1038/nrm3683. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, Solow-Cordero DE, Bouley DM, Offner F, Niwa M, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennesi ME, Nishikawa S, Matthes MT, Yasumura D, LaVail MM. The relationship of photoreceptor degeneration to retinal vascular development and loss in mutant rhodopsin transgenic and RCS rats. Experimental eye research. 2008;87:561–570. doi: 10.1016/j.exer.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Schroder K, Zhou R, Tschopp J. The NLRP3 inflammasome: a sensor for metabolic danger? Science. 2010;327:296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- Shimazawa M, Inokuchi Y, Ito Y, Murata H, Aihara M, Miura M, Araie M, Hara H. Involvement of ER stress in retinal cell death. Molecular vision. 2007;13:578–587. [PMC free article] [PubMed] [Google Scholar]
- Shore GC, Papa FR, Oakes SA. Signaling cell death from the endoplasmic reticulum stress response. Current opinion in cell biology. 2011;23:143–149. doi: 10.1016/j.ceb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szot GL, Koudria P, Bluestone JA. Murine pancreatic islet isolation. J Vis Exp. 2007:255. doi: 10.3791/255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Wang L, Perera BG, Hari SB, Bhhatarai B, Backes BJ, Seeliger MA, Schurer SC, Oakes SA, Papa FR, Maly DJ. Divergent allosteric control of the IRE1alpha endoribonuclease using kinase inhibitors. Nat Chem Biol. 2012;8:982–989. doi: 10.1038/nchembio.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, He Y, Ye K, Gu Z, Mao Y, Qi L. A conserved structural determinant located at the interdomain region of mammalian inositol-requiring enzyme 1alpha. J Biol Chem. 2011;286:30859–30866. doi: 10.1074/jbc.M111.273714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. The Journal of clinical investigation. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SX, Sanders E, Fliesler SJ, Wang JJ. Endoplasmic reticulum stress and the unfolded protein responses in retinal degeneration. Exp Eye Res. 2014;125C:30–40. doi: 10.1016/j.exer.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.