Abstract
Interventions that slow aging and prevent chronic disease may come from an understanding of how dietary restriction (DR) increases lifespan. Mechanisms proposed to mediate DR longevity include reduced mTOR signaling, activation of the NAD+-dependent deacylases known as sirtuins, and increases in NAD+ that derive from higher levels of respiration. Here, we explored these hypotheses in Caenorhabditis elegans using a new liquid feeding protocol. DR lifespan extension depended upon a group of regulators that are involved in stress responses and mTOR signaling, and have been implicated in DR by some other regimens [DAF-16 (FOXO), SKN-1 (Nrf1/2/3), PHA-4 (FOXA), AAK-2 (AMPK)]. Complete DR lifespan extension required the sirtuin SIR-2.1 (SIRT1), the involvement of which in DR has been debated. The nicotinamidase PNC-1, a key NAD+ salvage pathway component, was largely required for DR to increase lifespan but not two healthspan indicators: movement and stress resistance. Independently of pnc-1, DR increased the proportion of respiration that is coupled to ATP production but, surprisingly, reduced overall oxygen consumption. We conclude that stress response and NAD+-dependent mechanisms are each critical for DR lifespan extension, although some healthspan benefits do not require NAD+ salvage. Under DR conditions, NAD+-dependent processes may be supported by a DR-induced shift toward oxidative metabolism rather than an increase in total respiration.
Keywords: aging, dietary restriction, C. elegans, stress response, sirtuins, NAD+
Introduction
The reduction of food consumption without malnutrition, termed dietary restriction (DR), is the most conserved intervention known to increase lifespan. DR promotes longevity in essentially all eukaryotes, including yeast, rotifers, spiders, Drosophila,C. elegans, many strains of mice, and possibly non-human primates (Fontana et al., 2010; Haigis & Sinclair, 2010; Guarente, 2013; Colman et al., 2014). In mammals, DR improves many physiological parameters that are associated with aging, and delays aging-associated disorders ranging from neurodegenerative disease to cancer. An understanding of how DR confers these benefits may allow development of interventions that mimic the effect of DR, but circumvent the difficulties of severely reducing nutrient intake.
The response to DR is thought to be an evolutionary adaptation to survive periods of low food availability, predicting that mechanisms through which DR extends lifespan are likely to be conserved. Indeed, genetic analyses of the model organisms S. cerevisiae,D. melanogaster, and C. elegans have implicated essential nutrient-sensing mechanisms in the benefits of DR. Considerable evidence indicates that DR acts on the nutrient-sensing mechanistic target of rapamycin complex 1 (mTORC1) kinase, which is activated by amino acid availability, oxygen, and growth signaling (Fontana et al., 2010; Johnson et al., 2013). Lower levels of mTORC1 activity, as would be expected to be encountered in DR, result in reduced protein and lipid synthesis, increased autophagy and stress-defense activity, enhanced regenerative capacity, and in longer life in various organisms.
Sirtuins, which modulate transcription and numerous metabolic processes, comprise another set of energy-sensing mechanisms involved in DR (Haigis & Sinclair, 2010; Guarente, 2013). Sirtuins are protein deacylases or ADP-ribosyltransferases that convert nicotinamide adenine dinucleotide (NAD+) into nicotinamide (Nam), which in turn inhibits sirtuins. It has been proposed that DR promotes health and longevity by activating the nuclear sirtuin Sir2 (SIRT1 in mammals) (Haigis & Sinclair, 2010; Guarente, 2013). In mammals, SIRT1 and other sirtuins mediate DR effects on parameters such as mitochondrial biogenesis, metabolism, body weight, and glucose tolerance, and in mice overexpression of SIRT1 in the brain increases lifespan (Haigis & Sinclair, 2010; Guarente, 2013; Satoh et al., 2013). While these data suggest a major role for sirtuins in DR, the involvement of sirtuins in DR lifespan extension has remained a subject of debate. In lower organisms, the requirement for SIRT1 for DR longevity depends upon the experimental conditions, and in C. elegans, the SIRT1 ortholog sir-2.1 was required for lifespan extension by a weak mutation in eat-2, a genetic DR model, but not in other eat-2 experiments or DR methods that utilize bacterial food dilution (Mair et al., 2009; Kenyon, 2010; Burnett et al., 2011; Guarente, 2013).
From yeast to mammals, sirtuin activity is increased by conditions that increase NAD+ availability or reduce Nam levels (Haigis & Sinclair, 2010; Guarente, 2013). Interventions that boost NAD+ levels increase C. elegans lifespan and restore mitochondrial function in aging mice, in each case dependent upon sir-2.1/SIRT1 (Gomes et al., 2013; Mouchiroud et al., 2013b). C. elegans lifespan was also increased in a sir-2.1-dependent manner by reduced activity of PARPs (poly (ADP-ribose) polymerases, PARPs), which consume NAD+ (Mouchiroud et al., 2013a). Together, these findings support the idea that higher NAD+ levels promote longevity by activating SIR-2.1/SIRT1. In yeast, DR longevity requires the nicotinamidase Pnc1, which acts in a salvage pathway that allows Nam generated by sirtuins and other NAD+ consumers to be recycled to NAD+ (Fig. S1, Supporting Information) (Anderson et al., 2003). The importance of NAD+ salvage in DR has not been assessed in a metazoan, but overexpression of the PNC-1 ortholog D-NAAM increases Drosophila lifespan in a Sir2-dependent manner (Balan et al., 2008). In yeast, DR shifts metabolism away from glycolysis and toward respiration, which generates NAD+ by consuming NADH (Guarente, 2013; Schleit et al., 2013). While DR reduces caloric availability, it has been proposed that DR paradoxically increases respiration by inducing this metabolic shift, leading to higher NAD+ levels that drive lifespan extension (Bishop & Guarente, 2007; Guarente, 2013).
Caenorhabditis elegans provides a powerful metazoan model for studying DR because it has a short lifespan and is amenable to genetic disruption of processes that might be essential in mammals. The genetic requirements for lifespan extension vary among C. elegans DR regimens (Greer & Brunet, 2009; Mair et al., 2009). This variation may arise because of differences in culture conditions, including differences between liquid and solid culture protocols with respect to oxygen exposure and the extent to which the animals move. This diversity has allowed identification of a wider range of DR-associated mechanisms than would be possible with one ‘standard’ protocol (Greer & Brunet, 2009; Mair et al., 2009). Here, we investigated requirements for stress defense and NAD+-associated mechanisms, using a new liquid feeding DR method that robustly extends lifespan and minimizes maintenance. DR lifespan extension depended upon stress-defense regulators that are regulated by mTOR and growth pathways, and also upon SIR-2.1. The NAD+ salvage pathway enzyme PNC-1 was required for DR lifespan extension but not some healthspan benefits, providing the first evidence in a metazoan that implicates NAD+ salvage in DR. Independently of pnc-1, DR reduced total oxygen consumption but increased the proportion of respiration devoted to ATP production. Apparently, DR drives the activity of key NAD+-associated mechanisms through a shift toward oxidative metabolism, but does not necessarily increase overall respiration rates.
Results
A simple liquid protocol for C. elegans dietary restriction
Among the approaches used to restrict C. elegans dietary intake, liquid DR protocols (Table1, Table S1, Supporting Information) provide the advantage of allowing for: (i) variation of bacterial food availability, (ii) monitoring of food concentration over time, (iii) utilization of standard C. elegans strains, and (iv) scaling up for biochemical analyses. As C. elegans, liquid DR methods are generally labor intensive, we developed a protocol that requires relatively low maintenance (Fig.1A). On day one of adulthood, worms are transferred to NGM plates seeded with bacteria that have been maintained in antibiotics for 1 week at 4 °C (treated OP50). On day three of adulthood, these worms are transferred to liquid cultures that contain treated OP50 at different concentrations (Fig.1A). The treatment step prevents bacteria from re-entering growth phase (Fig. S2), eliminating the need for frequent food replacement.
Table 1.
Comparison of C. elegans liquid DR methods
| Liquid DR method | Age at DR onset | Food source | Antibiotics present | Average lifespan of DR animals (days) | Average DR lifespan extension for WT at 20 °C (%) | Epistasis analysis |
|---|---|---|---|---|---|---|
| Klassa | 48 hr. post-hatching | OP50 | None | 26 | 73 | NA |
| Vanfleterenb | L4 | E. coli 9001 | None, 50uM FUdR | 12* | 140τ* | daf-2 independent, daf-16 partially required |
| Dillincd | Day-2 adult | OP50 | 50 µg mL−1 Carb, 1 µg mL−1 Tet, 10 µg mL−1 Kan, 100 µg mL−1 FUdR | 42 | 60 (26 in smg–1 ts)γ | pha-4 required in smg-1 ts, daf-16 partially required, aak-2 not required, sir-2.1;sir-2.3 not required |
| Brunete | Day-2 adult | OP50-1 | 50 µg mL−1 Amp, 1 µg mL Tet, 10 µg mL−1 Kan, 100 mg L−1 FUdR | 42† | 51† | Requirement for aak-2 and daf-16 |
| Guarentef | L4/young adult | HT115 | 1 mg mL−1 Erythro, 50 µg mL−1 Amp, 12.5 µg mL−1 FUdR, 1mM IPTG | 33 | 22 | skn-1 required |
| Sinclair, Hart, Blackwell | Day-3 adult | OP50 | 50 µg mL−1 Amp, 10 µg mL−1 Kan, 1 µg mL−1 Tet, NYS, 100 µg mL−1 FUdR | 39 (42 DD in WT) (39 DD in smg–1 ts) | 60 (76 DD in WT) (45 DD in smg-1 ts) | See figures |
This table compares our DR method with other C. elegans liquid DR methods.
Klass (1977)).
Houthoofd et al. (2007).
Panowski et al. (2007).
Mair et al. (2009).
Greer & Brunet (2009).
Bishop & Guarente (2007).
Values approximated from graph.
Average calculated from all presented data. τ Animals were grown at 17 °C from hatching, at L4 were switched to 24 °C for remainder of lifespan. γ animals were grown at 25 °C from hatching, at first day of adulthood switched to 20 °C for 1 day, then 15 °C for remained of lifespan.
Figure 1.
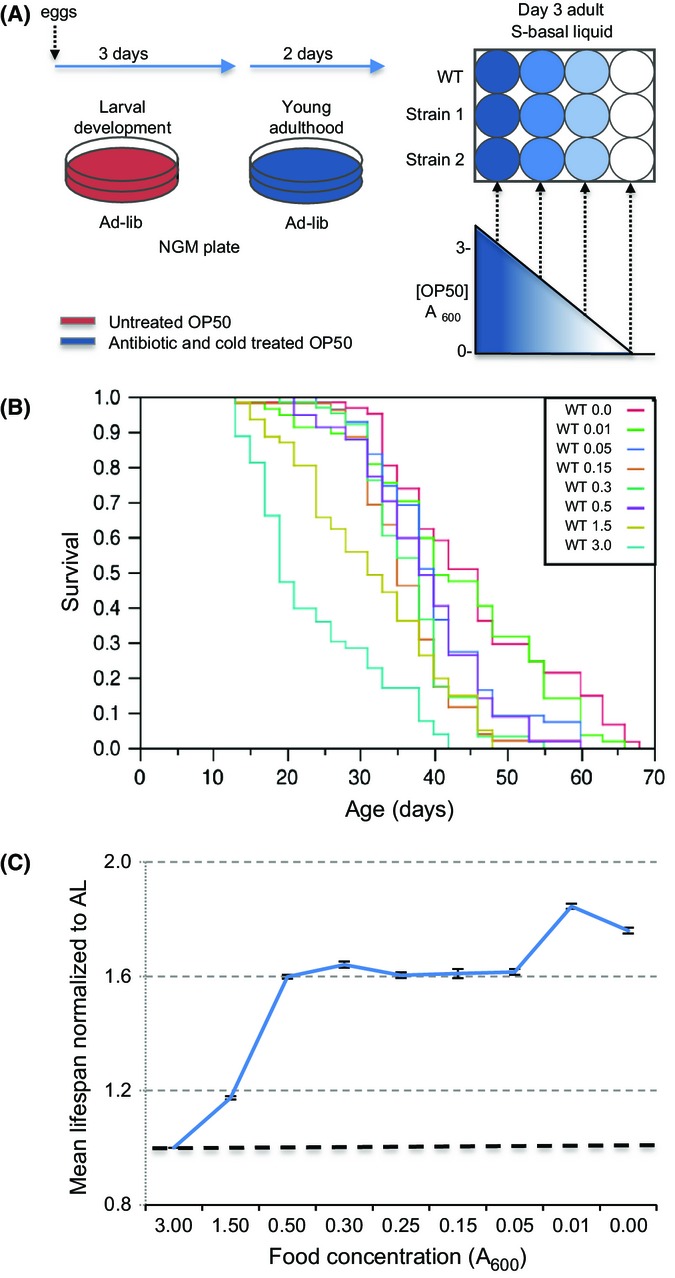
Lifespan extension from liquid-based DR and DD. (A) Description of the liquid DR method. C. elegans were allowed to develop under standard C. elegans conditions, then moved to NGM plates seeded with treated bacterial food (strain OP50) as day-one adults. These animals were moved to 12-well plates containing 2.5 mL of different concentrations of treated OP50 after 2 days of AL feeding, then moved to fresh cultures every 2–3 weeks. (B) Representative lifespans of wild-type (N2) worms that were fed different concentrations of treated bacteria (indicated as bacterial OD) and examined in parallel. Mean values and analysis of this experiment are presented in Table S12 (experiment 1C). (C) Effect of food concentration on mean lifespan. This graph describes a composite of 21 independent experiments, each of which examined a range of serially diluted treated bacteria in parallel. Lifespans were normalized to AL by dividing the mean lifespan at each food concentration by the AL value determined in parallel. DR and DD increased lifespan by 59.4% and 76%, respectively. SEM is shown. Compared to AL, a two-tailed t-test for all A600 values, P < 0.001. Individual experiments are presented in Table S12.
To determine whether a gene is required for DR and does not simply alter the optimal response to DR, lifespan must be examined over a range of bacterial food concentrations (Mair et al., 2009). We designated 3 × 109 C.F.U. mL−1 (3.0 A600) as ad libitum (AL) feeding, because at this concentration wild-type (WT) lifespan most closely resembled published observations on plates and in liquid (Fig.1B,C, Table S3). A plot of mean lifespan vs. food would be parabolic if concentrations below that optimal for DR resulted in starvation-like effects (Mair et al., 2009). However, combined analyses of wild-type (WT) worms resulted in a plateau-shaped plot that exhibited bimodal lifespan maxima (Fig.1B,C; Table S3). Even complete removal from food increased lifespan robustly, as was seen in dietary deprivation (DD) protocols in which C. elegans adults were maintained on plates without food beginning at day 2 (Kaeberlein et al., 2006; Lee et al., 2006). By initially keeping adults on plates with food for 3 days, longer than other liquid protocols (Table1), we apparently made it possible for the animals to live longer even if they subsequently relied only on stored nutrients. We refer to the first local maximum on the lifespan extension curve as DR. This occurred at 5 × 108 C.F.U. mL−1 (0.5 A600), a six-fold reduction in food compared to AL (Fig.1B,C, Table S3). The second local maximum occurred at 1 × 107 C.F.U. mL−1 (0.01 A600) (Fig.1B,C, Table S3), which is close to complete DD. Over the range of bacteria concentrations, our regimen increased mean lifespan by 61–84.5%, (Fig.1C, Table S3), one of the largest extensions seen in C. elegans DR (Table1).
Genetic analysis of DR-associated mechanisms
We compared our protocol to other methods by investigating whether lifespan extension required a set of regulators that were needed in some but not necessarily all C. elegans DR protocols in which they were examined. We first looked at the transcription factors DAF-16/FOXO, SKN-1/Nrf 1,2,3, and PHA-4/FOXA, each of which is regulated by mTORC1 signaling (Fig. S4) (Johnson et al., 2013). DAF-16/FOXO is required for longevity from reduced activity of insulin/IGF-1 signaling (IIS) or mTORC1 (Fig. S4) (Kenyon, 2010; Robida-Stubbs et al., 2012). DAF-16 is completely or partially required for lifespan extension by some DR protocols (Greer et al., 2007; Greer & Brunet, 2009; Honjoh et al., 2009), but appears to be dispensable in other DR regimens (summarized in Greer & Brunet, 2009). Our data are consistent with the former view: a lack of DAF-16 substantially reduced lifespan extension from either DR (from 58.2% to 26.3%) or DD (from 93.4% to 39%) (Fig.2A,B, Table S4). SKN-1/Nrf has important functions in stress and starvation responses, proteostasis, and metabolism, and contributes to lifespan extension from downregulation of either IIS or mTORC1 (Fig. S4) (Tullet et al., 2008; Robida-Stubbs et al., 2012; Glover-Cutter et al., 2013). skn-1 was required for longevity in a liquid DR protocol that was less robust than this one, but not in a plate DR regimen (Table1) (Bishop & Guarente, 2007; Greer & Brunet, 2009). Here, lack of skn-1 greatly impaired the lifespan increase from DR (from 48.3% to 20.9%) or DD (from 91% to 13.3%) (Fig.2C,D, Table S5). PHA-4/FOXA is involved in autophagy and is required for lifespan extension from reduced TOR activity, the genetic DR model eat-2, and a liquid DR method (Panowski et al., 2007; Sheaffer et al., 2008), although it was not required in a DR protocol that involved solid media (Greer & Brunet, 2009). pha-4 was needed for DR lifespan extension in our protocol, although DR was less effective in the smg-1(ts) background in which the pha-4 mutation is maintained (Panowski et al., 2007; Sheaffer et al., 2008) (23.2% lifespan increase in smg-1(ts) vs. 77.3% in WT from DR, and 44.6% vs. 77.3% from DD) (Fig.2E,F, Table S6).
Figure 2.
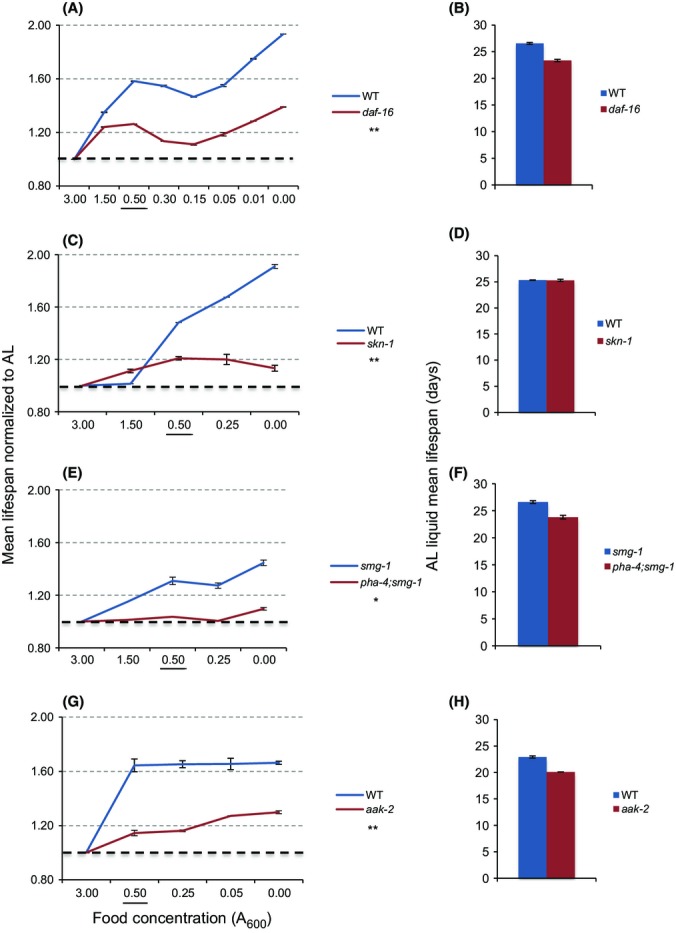
Importance of stress response mechanisms for DR lifespan extension. (A) Lack of DAF-16 impaired DR lifespan extension. AL lifespans obtained in parallel are shown in (B). (C,D) skn-1 was required for DR to extend lifespan. Whereas WT worms experienced an average of 48.3% and 91.0% increase in lifespan upon DR and DD, respectively, these increases were only 20.9% and 13.3% in predicted null skn-1 mutants. (D) Under AL liquid conditions, skn-1 mutants’ mean lifespan was equal to that of WT, in contrast to results obtained on plates (Tullet et al., 2008). (E, F) pha-4(zu225) mutation eliminated DR longevity in the control smg-1(cc546ts) background. smg-1 lifespan was increased 33.2% by DR and 38.8% by DD, respectively. (G,H) The AMPK subunit AAK-2 is required for full DR and DD lifespan extension. Composites of all analyses are shown, with individual experiments presented in Table S13 (Fig.2A,B), Table S14 (Fig.2C,D), Table S15 (Fig.2E,F), and Table S16 (Fig.2G,H). Two-way ANOVA analysis across the bacterial gradient (compared to control): *P = 0.0004, **P = 0.0001. The food concentration 0.50 A600 is underlined to facilitate comparison of results.
We also looked at the 5′ AMP-activated kinase (AMPK), which coordinates an adaptive response to low-energy availability that enhances oxidative metabolism, mitochondrial biogenesis, and autophagy, and is inhibited by mTORC1 in mammals (Mair et al., 2011). Increased activity of the AMPK catalytic subunit AAK-2 increases C. elegans lifespan (Apfeld et al., 2004; Mair et al., 2011) by inhibiting the transcriptional co-activator CRTC (Mair et al., 2011) and activating DAF-16/FOXO (Greer et al., 2007; Greer & Brunet, 2009). AMPK was important for DR lifespan extension in Drosophila (Stenesen et al., 2013), and in C. elegans was required in some DR methods but not others (Greer et al., 2007; Greer & Brunet, 2009; Mair et al., 2009). In our system, aak-2 loss reduced lifespan extension from DR (from 64.4% to 14.5%) or DD (From 66.4% to 30%) (Fig.2G,H, Table S7).
Importance of SIR-2.1 and NAD+ salvage in DR
SIRT1 has been implicated in DR in various organisms, but was required for DR lifespan extension in only one of several previous C. elegans studies (Introduction; Table1). SIRT1/SIR-2.1 functions both upstream and downstream of AMPK and activate DAF-16/FOXO (Canto et al., 2009; Rizki et al., 2011; Price et al., 2012; Guarente, 2013; Mouchiroud et al., 2013a), suggesting that it might be important in our DR regimen. Accordingly, a null sir-2.1 mutation significantly reduced lifespan extension from either DR (from 69.5% to 40.5%) or DD (From 89.4% to 47.4%) (Fig.3A,B, Table S8). The importance of sir-2.1 for DR-associated longevity was particularly striking because the sir-2.1 mutant lived slightly longer than WT under AL conditions (Fig.3B, Table S8).
Figure 3.
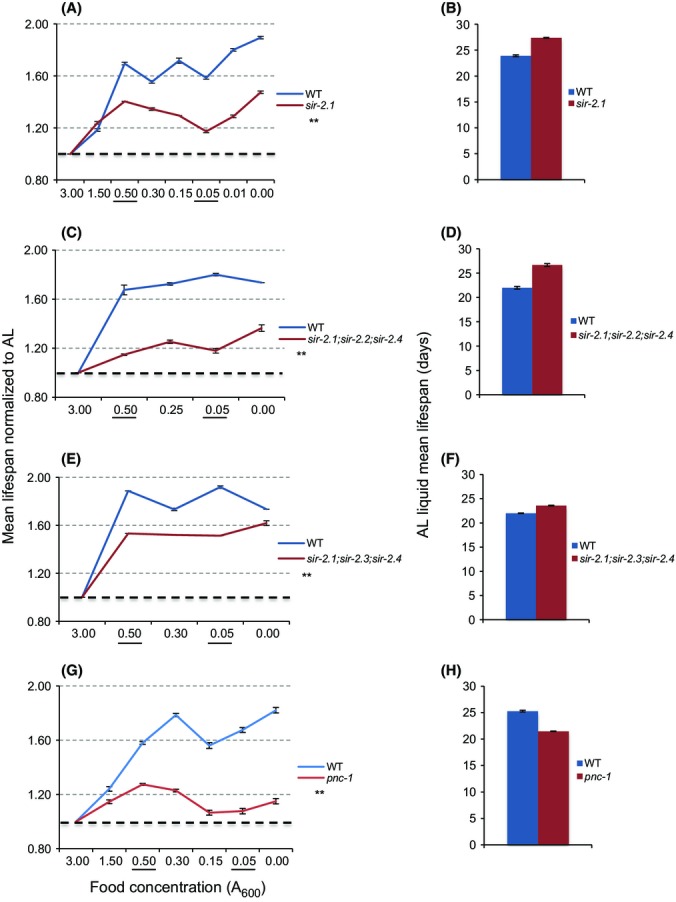
The NAD+ salvage pathway and NAD+-dependent SIRT1/sir-2.1 regulate DR lifespan. (A, B) Reduced DR response in sir-2.1(ok434) mutants. Note that the sir-2.1 mutants live longer than WT worms under AL feeding. (C-F) Impairment of DR lifespan extension in sir-2.1;sir-2.2;sir-2.4 (C, D) and sir-2.1;sir-2.3;sir-2.4 (E, F) triple mutants. (G, H) pnc-1 is required for DR lifespan extension. Composites are shown, with data from individual experiments presented in Table S17 (Fig.3A,B), Table S18 (Fig.3C,D), Table S19 (Fig.3E,F), and Table S20 (Fig.3G,H). Two-way ANOVA analysis: **P = 0.0001.
We considered the possibility that the three additional C. elegans sirtuin genes might have DR-related functions that overlap with those of sir-2.1. sir-2.2 and sir-2.3 are nearly identical orthologs of the mitochondrial sirtuin SIRT4, and sir-2.4 is orthologous to the nuclear sirtuins SIRT6 and SIRT7. Each of these sirtuins modulates critical metabolic processes (Haigis & Sinclair, 2010), and in C. elegans, SIR-2.4 acts non-catalytically to promote DAF-16 activity under stress conditions (Chiang et al., 2012). As sir-2.2 and sir-2.3 are located less than one kilobase apart (wormbase.org), making it difficult to disrupt them simultaneously, we analyzed the triple-null mutants sir-2.1; sir-2.2; sir-2.4 and sir-2.1; sir-2.3; sir-2.4. Each of these mutants responded to DR and DD comparably to sir-2.1 (Fig.3C–F, Tables S9 and S10). The data do not reveal any additive requirement for these sirtuins and sir-2.1, but do not exclude the possibility that redundancy might mask a role for sir-2.2 and sir-2.3.
The importance of sir-2.1 for DR lifespan extension predicts that NAD+ availability would be a critical factor. Moreover, NAD+ has critical sirtuin-independent functions that could be important in DR (Pollak et al., 2007). NAD+ serves as a co-enzyme in numerous metabolic electron transfer reactions, and its reduced form NADH plays a central role in mitochondrial electron transport. NAD+ is also the precursor to NADP+ (NAD phosphate), the reduced form of which (NADPH) is essential for cellular oxidative defense and other reductive detoxification reactions (Pollak et al., 2007). We investigated the role of NAD+ in DR by examining requirements for PNC-1, the C. elegans ortholog of the NAD+ salvage nicotinamidase Pnc1 (Fig. S1) (Vrablik et al., 2009). A predicted null pnc-1 mutation dramatically reduced the lifespan increase associated with either DR (from 57.2% to 32.1% at OD 0.5 and 77.2 to 27 at OD 0.3) or DD (from 82% to 15%) (Fig.3G,H, Table S11), suggesting that the NAD+ salvage pathway plays a major role in C. elegans DR.
In metazoa, DR not only extends lifespan but improves many parameters associated with health and resistance to chronic disease and slows their decline during aging (Fontana et al., 2010; Haigis & Sinclair, 2010; Guarente, 2013). This beneficial effect on ‘healthspan’ has been described not only in mammals but also in C. elegans, in which DR increases muscle activity (Greer et al., 2007). We investigated the importance of the NAD+ salvage pathway for muscle activity by comparing how DR affects movement in WT and pnc-1 animals. In pnc-1 mutants, body-wall muscle function is impaired (Vrablik et al., 2011), and under AL conditions their rate of spontaneous body bending was reduced compared to WT (Fig.4A, Table S21). Surprisingly, however, DR comparably increased the frequency of bending in older WT and pnc-1 animals (Fig.4A,B, Table S21). As a second healthspan measure, we examined thermotolerance, which is characteristically increased by DR in C. elegans (Kenyon, 2010). In WT animals, heat resistance declined with age, but this decline was reversed by day 15 under DR (Fig.4C,D, Table S22). In pnc-1 mutants, DR increased thermotolerance similarly (Fig.4C,D, Table S22). These improvements in healthspan parameters suggest that pnc-1 mutants are not refractory to DR lifespan extension because they are simply sick. Apparently, in C. elegans, the NAD+ salvage pathway is required for DR to increase lifespan, but not to confer these healthspan benefits.
Figure 4.
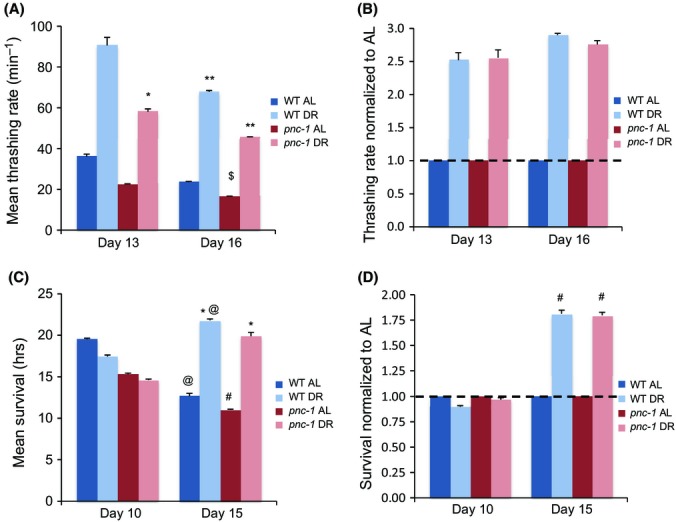
DR increases movement and stress resistance independently of NAD+ salvage. (A,B) DR increased the rate of spontaneous movement comparably in aging WT and pnc-1(pk9605) animals. Body bends per minute were scored. Note that pnc-1 mutation did not affect the percentage increase associated with DR. (C,D) DR comparably increased thermotolerance (survival at 38 °C) in WT and pnc-1 animals. The time points indicated refer to days after hatching. Composites of all analyses are shown, with individual experimental data, mean, standard error, percent change, and statistical analysis presented in Table S21 (Fig.4A,B), and Table S22 (Fig.4C,D). *t-test vs. AL,P < 0.05; **t-test vs. AL,P < 0.001; $t-test vs. age 13, P < 0.01; @t-test vs. age 10, P < 0.065; #t-test vs. age 10, P < 0.025.
DR reduces total oxygen consumption but proportionally increases productive respiration
Another source of NAD+ is conversion from NADH during mitochondrial respiration. Based upon data from yeast, it has been proposed that DR elevates NAD+ levels by increasing respiration (Guarente, 2013). However, two C. elegans studies reached opposite conclusions with respect to DR effects on respiration, and an analysis of Drosophila did not see an increase in respiration with DR (Hulbert et al., 2004; Bishop & Guarente, 2007; Houthoofd et al., 2007). Previous C. elegans studies of DR measured oxygen consumption in liquid culture using the Clark electrode, which analyzes large samples that are difficult to generate in an age-matched fashion. We circumvented this issue by measuring oxygen consumption rate (OCR) using the Seahorse XF24 Analyzer, which can precisely examine tens of animals per replicate.
We investigated how aging, DR, and the NAD+ salvage pathway influence respiration by examining WT and pnc-1 animals under AL and DR conditions at 9,12, and 16 days after hatching (3, 6, and 9 days of DR). Under AL conditions, the OCR per worm decreased with age in WT animals (Fig.5A, Tables S23 and S24), in close agreement with a recent study that used the XF24 (Mouchiroud et al., 2013b). C. elegans shrink in size post-reproductively, however, and with age exhibit profound sarcopenia in pharyngeal and body-wall muscles (Herndon et al., 2002). Respiration was therefore modestly elevated in older animals on a per-protein mass basis (Day 16; Fig.5B, Tables S25 and S26). Surprisingly, DR markedly reduced the OCR per worm at each day examined on either a per-worm or per-mass basis (Fig.5C,D, Tables S27 and S28). Similar trends were apparent in pnc-1 mutants. Under our conditions, therefore, DR lowered overall oxygen consumption, and the NAD+ salvage pathway was not required to maintain respiration rates during DR.
Figure 5.
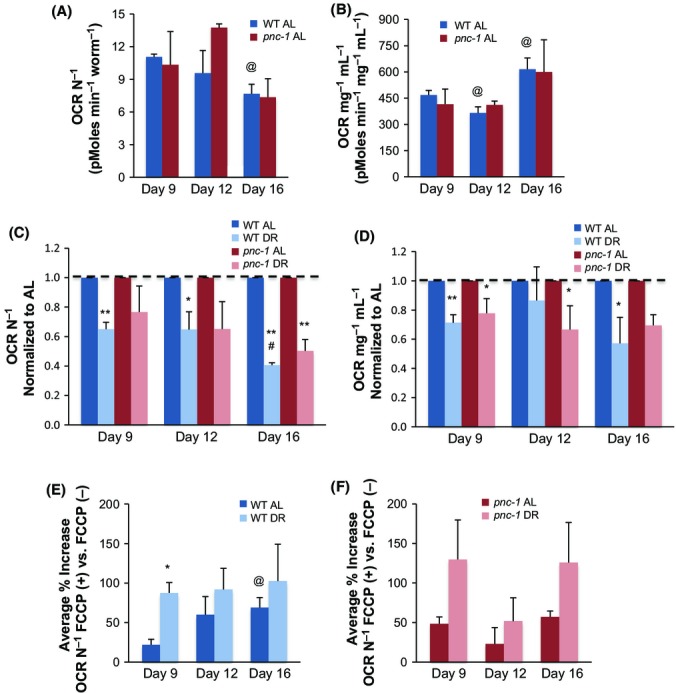
Differential effects of DR on overall and productive respiration. (A) Oxygen consumption per animal decreases with age. One-way ANOVA WT day 9 vs. day 12 vs. day 16, P < 0.0001, one-way ANOVA pnc-1 day 9 vs. day 12 vs. day 16, P < 0.0001. (B) Oxygen consumption per protein mass is elevated in older animals. One-way ANOVA WT day 9 vs. day 12 vs. day 16, P < 0.001, one-way ANOVA pnc-1 day 9 vs. day 12 vs. day 16, P < 0.0001. (C,D) DR reduces the overall respiration rate. One-way ANOVA WT AL vs. DR,P < 0.0001, two-way ANOVA WT AL/DR and age, P < 0.0003, one-way ANOVA pnc-1AL vs. DR,P < 0.032, two-way ANOVA pnc-1AL/DR and age, P < 0.0025. OCR is shown as normalized to worm number (C) and protein (D). Note that OCR values and trends are comparable in WT and pnc-1(pk9605) animals. AL data for WT and pnc-1(pk9605) that were used for normalization in (C,D) are shown in (A,B) and were obtained and analyzed in parallel to DR data. (E,F) DR increases the productive fraction of respiration, as detected by the increase in OCR seen upon administration of the mitochondrial uncoupler FCCP. Note that this trend was similar in WT and pnc-1(pk9605) animals. One-way ANOVA WT AL vs. DR,P < 0.023, two-way ANOVA WT AL vs. DR and age, P < 0.0635, one-way ANOVA pnc-1AL vs. DR,P < 0.011, two-way ANOVA pnc-1AL vs. DR and age, P < 0.0208, two-way ANOVA WT/pnc-1 and age, P < 0.5548, two-way ANOVA WT/pnc-1 and AL vs. DR,P < 0.0021. OCR is shown as normalized to worm number. Similar trends are seen in data normalized to protein content, presented in Fig. S3. In each experiment, samples were assayed in 3–5 replicates per experiment. Wells that did not respond were censored. Individual experimental data, mean, standard error, percent change, and statistical analysis are presented in (A) Table S23, S24, (B) Table S25, S26, (C) Table S27, (D) Table S28, (E) Table S23, S24, (F) Table S23, S24. *t-test vs. AL,P < 0.06; **t-test vs. AL,P < 0.01; @t-test vs. age 9, P < 0.055; #t-test vs. age 9, P < 0.01.
The evidence that DR promotes oxidative metabolism in yeast (Guarente, 2013; Schleit et al., 2013) predicts that this might also be true in C. elegans, even if DR reduces total oxygen consumption. This is an attractive model, because it seems logical that the efficiency of ATP production might need to be increased when food availability is reduced. To test this idea, we used the mitochondrial respiration uncoupler FCCP (Brand & Nicholls, 2011) to assess the extent to which oxygen consumption increases when it is uncoupled from ATP production. This increase reflects the proportion of respiratory capacity that was unused prior to uncoupling (Brand & Nicholls, 2011). The greater this increase, the more efficient the mitochondria were in generating ATP under basal conditions. At each day of life examined, the response to FCCP was dramatically higher in the DR group in either WT or pnc-1 animals (Fig.5E,F, Tables S23 and S24). This suggests that under DR conditions, a greater proportion of oxygen consumption was devoted to ATP generation. Thus, although the overall respiration rate was not increased, DR induced a shift toward oxidative metabolism that might underlie the functions of NAD+-dependent mechanisms.
Discussion
It has been proposed that DR lifespan extension is mediated through reduction of mTORC1 signaling, and activation of sirtuins and other NAD+-dependent mechanisms through an increase in respiration (Guarente, 2013; Johnson et al., 2013). Here, we used a new C. elegans liquid DR protocol to examine the importance of mechanisms that are associated with these two models. Our findings are consistent with some key predictions of each hypothesis, but suggest an important revision to the second model.
Importance of stress defense and nutrient-sensing pathways in DR
We found that DR lifespan extension involved stress-defense regulators that are required in other longevity interventions (Kenyon, 2010), but were not necessarily essential in other C. elegans DR methods. One of these was DAF-16/FOXO (Figs2A and 6), which some but not all other studies implicated in DR (Greer et al., 2007; Greer & Brunet, 2009; Honjoh et al., 2009). Interestingly, the response to DR is improved by impairment of the insulin/IGF-1 receptor DAF-2, which inhibits DAF-16 (Bishop & Guarente, 2007). Together, these findings suggest that IIS, which senses nutrients, might be important in DR (Fig.6). Our DR lifespan extension also depended upon SKN-1/Nrf (Figs2C and 6), which is inhibited by IIS in parallel to DAF-16 (Tullet et al., 2008). Another study indicated that DR induces SKN-1 to increase respiration (Bishop & Guarente, 2007) but we observed that DR reduces oxygen consumption (Fig.5C,D), suggesting that SKN-1 has additional functions in DR. PHA-4/FOXA was also required for DR longevity in our protocol, although the background smg-1(ts) mutation substantially impaired the response to DR (Fig.2E). The SMG-1 kinase functions in nonsense-mediated decay, in which translationally stalled mRNAs are degraded (Sheaffer et al., 2008). Perhaps, clearing of stalled mRNAs by nonsense-mediated decay is important in DR.
Figure 6.
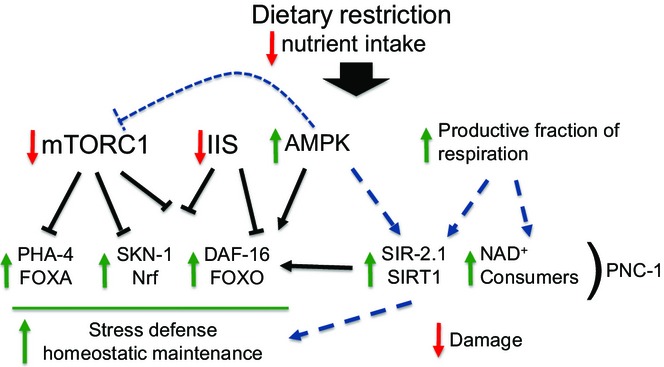
Importance of stress defense and metabolic mechanisms in DR. As described in the text, genetic experiments implicate the indicated growth-regulated stress-defense pathways and NAD+-associated mechanisms in DR. Other mechanisms that have been implicated in DR in C. elegans are consistent with this overall model. For example, both AMPK and PHA-4 promote autophagy (Hansen et al., 2008; Egan et al., 2011).
Each of these transcription factors is functionally inhibited by mTORC1 signaling (Fig.6) and required for longevity that results from inhibiting this pathway (Sheaffer et al., 2008; Robida-Stubbs et al., 2012). The proposed importance of mTORC1 in DR (Johnson et al., 2013) also fits with the importance of the low-energy sensor AMPK (Greer et al., 2007; Greer & Brunet, 2009) (Figs2G and 6), which inhibits mTORC1 in many species (Johnson et al., 2013). Our findings support the view that DR extends lifespan in part by strengthening stress defenses through a reduction in mTORC1 signaling, and possibly IIS (Fig.6). The importance of daf-16,skn-1, and aak-2 that we observed differed from results of some C. elegans studies that examined these genes individually (see above). Perhaps, these mechanisms may control overlapping and possibly compensatory processes. It is also possible that our DR method has more stringent requirements for lifespan extension than some other protocols because the animals spend more time on AL feeding prior to DR (Table1).
NAD+-dependent mechanisms, respiration, and DR
Our data also indicate the importance of NAD+-dependent mechanisms in DR and represent the first time that SIR-2.1/SIRT1 was required for DR lifespan extension in a C. elegans feeding protocol (Fig.3A,B). DR increased lifespan in sir-2.1 mutants, which were slightly long-lived, but this lifespan extension was consistently reduced compared to WT across four experiments that were performed in our three laboratories (Table S8). The importance of SIR-2.1 in our DR regimen is consistent with the evidence that SIRT1 mediates many metabolic effects of DR in mammals (Haigis & Sinclair, 2010; Guarente, 2013), and strongly supports the idea that SIR-2.1/SIRT1 plays a major role in DR.
DR lifespan extension appeared to be less effective in pnc-1 than sir-2.1 mutants, revealing for the first time in a metazoan that the NAD+ salvage pathway is critical in DR, and suggesting that its importance might reflect the activity of NAD+ consumers besides SIR-2.1/SIRT1 (Fig.3A,G; Table2). However, DR improved two healthspan parameters independently of pnc-1 (Fig.4; Tables S20 and S21), indicating that pnc-1 mutants were not simply sick or completely refractory to DR. This result also indicates that some DR benefits may not require NAD+-mediated signals; therefore, that some effects of DR on healthspan can be uncoupled mechanistically from its longevity effects. Using C. elegans genetics, it may be possible to unravel how DR influences different parameters associated with health, and to elucidate how they contribute to long life.
Table 2.
Maximum affect of DR on lifespan across mutant strains
| Genotype | Average max % increase lifespan vs. AL | SEM | Food conc. (A600) | # Indep. expr. | P-value 2-way ANOVA vs. Ctrl. |
|---|---|---|---|---|---|
| N2 | 85 | < 0.0 | 0.01 | 21 | NA |
| daf-16 | 39 | 0.2 | 0.00 | 3 | < 0.0001 |
| skn-1 | 21 | 1.3 | 0.50 | 6 | < 0.0001 |
| aak-2 | 30 | 1.1 | 0.00 | 2 | < 0.0001 |
| sir-2.1 | 47 | 1.3 | 0.00 | 4 | < 0.0001 |
| sir-2.1;sir-2.2;sir-2.4 | 36 | 2.7 | 0.00 | 2 | 0.0001 |
| sir-2.1;sir-2.3;sir-2.4 | 62 | 1.6 | 0.00 | 2 | < 0.0001 |
| pnc-1 | 32 | 0.9 | 0.50 | 10 | < 0.0001 |
| smg-1 | 45 | 2.5 | 0.00 | 4 | NA |
| pha-4;smg-1 | 10 | 1.0 | 0.00 | 4 | 0.0004 |
This table presents the DR food concentration at which each strain experiences its maximal lifespan extension, in comparison to its lifespan on AL food, as well as what the percent increase in lifespan is. The wild-type N2 is the control for all strains except for pha-4;smg-1, whose genetic control is smg-1.
The percent change experienced by the control strains is in bold.
It is an important question which NAD+ consumers besides SIR-2.1/SIRT1 might contribute to DR lifespan extension. Poly-ADP ribose polymerase (PARP) proteins consume NAD+ but are unlikely to play a positive role in DR because reducing their activity increases C. elegans lifespan (Mouchiroud et al., 2013a). However, we cannot exclude an important function for the predicted redundant sirtuins SIR-2.2 and SIR-2.3 (SIRT4; Fig.3C,E), and in some tissues, NAD+ generated by PNC-1 might maintain proper levels of NADP (and NADPH) or activity of the many NAD+-dependent metabolic processes (Pollak et al., 2007). It might also be critical for PNC-1 to metabolize NAM, excess levels of which recapitulate some developmental pnc-1 phenotypes (Vrablik et al., 2009). Administration of exogenous NAM extends C. elegans lifespan by means of metabolites that increase ROS formation (Schmeisser et al., 2013), but a positive role for NAM in DR seems unlikely given that DR requires PNC-1, which metabolizes NAM.
Our surprising finding that DR sharply reduced the overall OCR (Fig.5C,D; Tables S23–S28) would seem to be strong evidence against the view that DR exerts its beneficial effects by increasing respiration (Bishop & Guarente, 2007; Guarente, 2013). Importantly, however, DR increased the proportion of oxygen consumption that is devoted to ATP generation (Fig.5E; Tables S23 and S24). This shift toward oxidative metabolism, which we documented for the first time in a metazoan, seems to be a fitting response to lower food availability because respiration is more efficient than glycolysis. It will be of interest to determine whether this shift is also seen in other C. elegans DR protocols, including those involving solid culture media. Our data suggest a revised version of the respiration-based model for DR, whereby the driving force for activity of NAD+-dependent processes that delay aging is this respiratory shift (Fig.6), not increased respiration per se. This respiratory shift might also influence the mitochondrial unfolded response and possibly other signals from mitochondria that could affect lifespan (Houtkooper et al., 2013; Mouchiroud et al., 2013b; Schleit et al., 2013; Schmeisser et al., 2013). The next challenge will be to test these ideas by identifying mechanisms that are modulated by SIR-2.1 and other key NAD+-dependent processes in the setting of DR, and elucidating how they and other mitochondrial-associated processes are affected by the metabolic changes that are driven by DR.
Experimental procedures
Full methods and experimental procedures are available in Data S1 (Supporting Information).
Acknowledgments
We thank Wendy Hanna-Rose, Leonard Guarente, Robert Horvitz, Max Heiman, Brendan Manning, Jay Mitchell, and Blackwell lab members for helpful discussions or providing strains. Some strains were provided by the CGC, which is funded by the NIH (P40 OD010440). NM was supported by the Joslin Diabetes Center T32 (T32DK007260), TKB by the NIH (GM062891), the Ellison Medical Foundation and an NIDDK DRC grant to the Joslin (P30DK036836), ACH by the Ellison Medical Foundation and NIH (GM078171), and DAS by grants from NIH (AG028730), The Paul F. Glenn Foundation for Medical Research, The United Mitochondrial Disease Foundation, The Juvenile Diabetes Foundation and a gift from the Schulak Family. We declare that no competing interests exist.
Funding
No funding information provided.
Conflict of interest
None declared.
Author contributions
N. M., J. J. C., and E. A. performed experiments, all authors designed and interpreted experiments, A. C. H., D. A. S. and T. K. B. directed the project, and N. M., A. C. H., D. A. S. and T. K. B. wrote the manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site.
Fig. S1 The NAD+ salvage pathway.
Fig. S2 Analysis of treated bacteria.
Fig. S3 Effects of the Mitochondrial Uncoupler FCCP on AL and DR animals.
Fig. S4 Regulation of longevity transcription factors by the IIS and TORC1 pathways.
Comparison of published C. elegans liquid DR methods.
Bacterial concentration conversion table.
Table S3 Statistical analysis of compiled data of wild-type animals on DR shown in Fig. 1B,C.
Table S4 Statistical analysis of compiled data of daf-16 mutant animals on DR shown in Fig. 2A,B.
Table S5 Statistical analysis of compiled data of skn-1 mutant animals on DR shown in Fig. 2C,D.
Table S6 Statistical analysis of compiled data of pha-4;smg-1 mutant animals on DR shown in Fig. 2E,F.
Table S7 Statistical analysis of compiled data of aak-2 mutant animals on DR shown in Fig. 2G,H.
Table S8 Statistical analysis of compiled data of sir-2.1 mutant animals on DR shown in in Fig. 3A,B.
Table S9 Statistical analysis of compiled data of sir-2.1;sir-2.2;sir-2.4 mutant animals on DR shown in Fig. 3C,D.
Table S10 Statistical analysis of compiled data of sir-2.1;sir-2.3;sir-2.4 mutant animals on DR shown in Fig. 3E,F.
Table S11 Statistical analysis of compiled data of pnc-1 mutant animals on DR shown in Fig. 3G,H.
Individual DR experiments performed on wild-type animals shown in Fig. 1B,C.
Individual DR experiments performed on daf-16 animals shown in Fig. 2A,B.
Individual DR experiments performed on skn-1 animals shown in Fig. 2C,D.
Individual DR experiments performed on pha-4;smg-1 animals shown in Fig. 2E,F.
Individual DR experiments performed on aak-2 animals shown in Fig. 2G,H.
Individual DR experiments performed on sir-2.1 animals shown in Fig. 3A,B.
Individual DR experiments performed on sir-2.1;sir-2.2;sir-2.4 animals shown in Fig. 3C,D.
Individual DR experiments performed on sir-2.1;sir-2.3;sir-2.4 animals shown in Fig. 3E,F.
Individual DR experiments performed on pnc-1 animals shown in Fig. 3G,H.
Analyses of thrashing assays shown in Fig. 4A,B.
Analyses of thermotolerance assays shown in Fig. 4C,D.
Respiration under basal conditions per animal shown in Fig. 5A,E,F.
Uncoupled respiration data per animal shown in Fig. 5E,F.
Respiration under basal conditions per protein shown in Fig. 5B, Fig. S3A,B.
Uncoupled respiration per protein shown in Fig. S3A,S3B.
Respiration per animal normalized to ad-lib conditions shown in Fig. 5C.
Respiration per protein normalized to ad-lib conditions shown in Fig. 5D.
Experimental procedures.
References
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, O'Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan V, Miller GS, Kaplun L, Balan K, Chong ZZ, Li F, Kaplun A, VanBerkum MF, Arking R, Freeman DC, Maiese K, Tzivion G. Life span extension and neuronal cell protection by Drosophila nicotinamidase. J. Biol. Chem. 2008;283:27810–27819. doi: 10.1074/jbc.M804681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez-Manrique RP, Orfila AM, Ackerman D, Au C, Vinti G, Riesen M, Howard K, Neri C, Bedalov A, Kaeberlein M, Soti C, Partridge L, Gems D. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Tishkoff DX, Yang B, Wilson-Grady J, Yu X, Mazer T, Eckersdorff M, Gygi SP, Lombard DB, Hsu AL. C. elegans SIRT6/7 homolog SIR-2.4 promotes DAF-16 relocalization and function during stress. PLoS Genet. 2012;8:e1002948. doi: 10.1371/journal.pgen.1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, Anderson RM. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat. Commun. 2014;5:3557. doi: 10.1038/ncomms4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span–from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter KM, Lin S, Blackwell TK. Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. 2013;9:e1003701. doi: 10.1371/journal.pgen.1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Calorie restriction and sirtuins revisited. Genes Dev. 2013;27:2072–2085. doi: 10.1101/gad.227439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Gems D, Johnson TE, Vanfleteren JR. Dietary restriction in the nematode Caenorhabditis elegans. Interdiscip. Top. Gerontol. 2007;35:98–114. doi: 10.1159/000096558. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert AJ, Clancy DJ, Mair W, Braeckman BP, Gems D, Partridge L. Metabolic rate is not reduced by dietary-restriction or by lowered insulin/IGF-1 signalling and is not correlated with individual lifespan in Drosophila melanogaster. Exp. Gerontol. 2004;39:1137–1143. doi: 10.1016/j.exger.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech. Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Panowski SH, Shaw RJ, Dillin A. Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS ONE. 2009;4:e4535. doi: 10.1371/journal.pone.0004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Auwerx J. NAD(+) metabolism: a therapeutic target for age-related metabolic disease. Crit. Rev. Biochem. Mol. Biol. 2013a;48:397–408. doi: 10.3109/10409238.2013.789479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, Guarente L, Auwerx J. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013b;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Pollak N, Dolle C, Ziegler M. The power to reduce: pyridine nucleotides–small molecules with a multitude of functions. Biochem. J. 2007;402:205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, Hubbard BP, Varela AT, Davis JG, Varamini B, Hafner A, Moaddel R, Rolo AP, Coppari R, Palmeira CM, de Cabo R, Baur JA, Sinclair DA. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki G, Iwata TN, Li J, Riedel CG, Picard CL, Jan M, Murphy CT, Lee SS. The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet. 2011;7:e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleit J, Johnson SC, Bennett CF, Simko M, Trongtham N, Castanza A, Hsieh EJ, Moller RM, Wasko BM, Delaney JR, Sutphin GL, Carr D, Murakami CJ, Tocchi A, Xian B, Chen W, Yu T, Goswami S, Higgins S, Jeong KS, Kim JR, Klum S, Liao E, Lin MS, Lo W, Miller H, Olsen B, Peng ZJ, Pollard T, Pradeep P, Pruett D, Rai D, Ros V, Singh M, Spector BL, Wende HV, An EH, Fletcher M, Jelic M, Rabinovitch PS, Maccoss MJ, Han JD, Kennedy BK, Kaeberlein M. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell. 2013;12:1050–1061. doi: 10.1111/acel.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser K, Mansfeld J, Kuhlow D, Weimer S, Priebe S, Heiland I, Birringer M, Groth M, Segref A, Kanfi Y, Price NL, Schmeisser S, Schuster S, Pfeiffer AF, Guthke R, Platzer M, Hoppe T, Cohen HY, Zarse K, Sinclair DA, Ristow M. Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat. Chem. Biol. 2013;9:693–700. doi: 10.1038/nchembio.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Updike DL, Mango SE. The target of rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr. Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenesen D, Suh JM, Seo J, Yu K, Lee KS, Kim JS, Min KJ, Graff JM. Adenosine nucleotide biosynthesis and AMPK regulate adult life span and mediate the longevity benefit of caloric restriction in flies. Cell Metab. 2013;17:101–112. doi: 10.1016/j.cmet.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrablik TL, Huang L, Lange SE, Hanna-Rose W. Nicotinamidase modulation of NAD+ biosynthesis and nicotinamide levels separately affect reproductive development and cell survival in C. elegans. Development. 2009;136:3637–3646. doi: 10.1242/dev.028431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrablik TL, Wang W, Upadhyay A, Hanna-Rose W. Muscle type-specific responses to NAD+ salvage biosynthesis promote muscle function in Caenorhabditis elegans. Dev. Biol. 2011;349:387–394. doi: 10.1016/j.ydbio.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The NAD+ salvage pathway.
Fig. S2 Analysis of treated bacteria.
Fig. S3 Effects of the Mitochondrial Uncoupler FCCP on AL and DR animals.
Fig. S4 Regulation of longevity transcription factors by the IIS and TORC1 pathways.
Comparison of published C. elegans liquid DR methods.
Bacterial concentration conversion table.
Table S3 Statistical analysis of compiled data of wild-type animals on DR shown in Fig. 1B,C.
Table S4 Statistical analysis of compiled data of daf-16 mutant animals on DR shown in Fig. 2A,B.
Table S5 Statistical analysis of compiled data of skn-1 mutant animals on DR shown in Fig. 2C,D.
Table S6 Statistical analysis of compiled data of pha-4;smg-1 mutant animals on DR shown in Fig. 2E,F.
Table S7 Statistical analysis of compiled data of aak-2 mutant animals on DR shown in Fig. 2G,H.
Table S8 Statistical analysis of compiled data of sir-2.1 mutant animals on DR shown in in Fig. 3A,B.
Table S9 Statistical analysis of compiled data of sir-2.1;sir-2.2;sir-2.4 mutant animals on DR shown in Fig. 3C,D.
Table S10 Statistical analysis of compiled data of sir-2.1;sir-2.3;sir-2.4 mutant animals on DR shown in Fig. 3E,F.
Table S11 Statistical analysis of compiled data of pnc-1 mutant animals on DR shown in Fig. 3G,H.
Individual DR experiments performed on wild-type animals shown in Fig. 1B,C.
Individual DR experiments performed on daf-16 animals shown in Fig. 2A,B.
Individual DR experiments performed on skn-1 animals shown in Fig. 2C,D.
Individual DR experiments performed on pha-4;smg-1 animals shown in Fig. 2E,F.
Individual DR experiments performed on aak-2 animals shown in Fig. 2G,H.
Individual DR experiments performed on sir-2.1 animals shown in Fig. 3A,B.
Individual DR experiments performed on sir-2.1;sir-2.2;sir-2.4 animals shown in Fig. 3C,D.
Individual DR experiments performed on sir-2.1;sir-2.3;sir-2.4 animals shown in Fig. 3E,F.
Individual DR experiments performed on pnc-1 animals shown in Fig. 3G,H.
Analyses of thrashing assays shown in Fig. 4A,B.
Analyses of thermotolerance assays shown in Fig. 4C,D.
Respiration under basal conditions per animal shown in Fig. 5A,E,F.
Uncoupled respiration data per animal shown in Fig. 5E,F.
Respiration under basal conditions per protein shown in Fig. 5B, Fig. S3A,B.
Uncoupled respiration per protein shown in Fig. S3A,S3B.
Respiration per animal normalized to ad-lib conditions shown in Fig. 5C.
Respiration per protein normalized to ad-lib conditions shown in Fig. 5D.
Experimental procedures.


