Abstract
Effects of the supercharging reagents m-NBA and sulfolane on sodium ion adduction to protein ions formed using native mass spectrometry were investigated. There is extensive sodium adduction on protein ions formed by electrospray ionization from aqueous solutions containing millimolar concentrations of NaCl, which can lower sensitivity by distributing the signal of a given charge state over multiple adducted ions and can reduce mass measuring accuracy for large proteins and non-covalent complexes for which individual adducts cannot be resolved. The average number of sodium ions adducted to the most abundant ion formed from ten small (8.6–29 kDa) proteins for which adducts can be resolved is reduced by 58% or 80% on average, respectively, when 1.5% m-NBA or 2.5% sulfolane are added to aqueous solutions containing sodium compared to without the supercharging reagent. Sulfolane is more effective than m-NBA at reducing sodium ion adduction and at preserving non-covalent protein-ligand and protein-protein interactions. Desalting with 2.5% sulfolane enables detection of several glycosylated forms of 79.7 kDa holo-transferrin and NADH bound to the 146 kDa homotetramer LDH, which are otherwise unresolved due to peak broadening from extensive sodium adduction. Although sulfolane is more effective than m-NBA at protein ion desalting, m-NBA reduces salt clusters at high m/z and can increase the signal-to-noise ratios of protein ions by reducing chemical noise. Desalting is likely a result of these supercharging reagents binding sodium ions in solution, thereby reducing the sodium available to adduct to protein ions.
Introduction
The effects that different salts can have on protein stability have been known since the late 1800’s, when Franz Hofmeister reported an ordering of cations and anions based on their ability to salt in or salt out proteins,1 and both non-specific and specific salt-protein interactions can strongly influence protein structure and function.1–8 Phosphate and Tris buffers as well as sodium chloride are often added to aqueous protein solutions in order to stabilize native or native-like protein structure by mimicking the environment inside the cell, which has an ionic strength of ~150 mM.9 Specific salts and other small molecules are essential in the enzyme-cofactor or protein-ligand interactions in such varied and vital processes as electron transport,10, 11 ion pumping across cell membranes,12, 13 and drug interactions.14, 15
Some protein structural methods, such as X-ray crystallography and NMR, are not adversely affected by high salt concentrations, whereas high salt concentrations can be detrimental to the performance of electrospray ionization (ESI) and to a lesser extent matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS). Salts can increase baseline noise due to the formation of ionic clusters and can cause ion suppression.16–21 For example, alcohol dehydrogenase tetramer ions formed by nanoelectrospray ionization (nESI) from 50 mM ammonium acetate can be measured with excellent signal-to-noise ratios (S/N), whereas the tetramer is undetectable from the same solution with 10 mM Tris or HEPES buffers.16 Similarly, addition of 10 mM CsCl to solutions of lysozyme in 1:1 methanol:water decreases the total ion abundance for the protein by 130-fold.17 Salt adduction to protein ions distributes the protein signal over multiple adducted ions, reducing the S/N of each protein ion, and broadens mass spectral peaks of large proteins and protein complexes for which individual adducts cannot be resolved.19, 21–25 Peak broadening decreases mass measurement accuracy for these large proteins and can also inhibit the identification of covalent (glycosylations, phosphorylation, or other post-translational modifications) or non-covalent (specific ion or ligand binding) protein modifications.23 McLuckey and coworkers found that the extent of sodium ion adduction on a protein ion formed by ESI is related to the protein pI, solution pH, and charge state.18, 26 There is more sodium ion adduction to low charge states,18, 21, 25–27 but there is more adduction of trivalent metal ions to high charge states.28
To reduce the adverse effects of many salts on MS performance, salts are often removed by dialysis,29, 30 ion exchange chromatography,16, 31 or diafiltration32–34 prior to analysis by MS, and a myriad of products for fast filtration and online chromatographic desalting of protein solutions have been developed. However, removing even low concentrations of salts can significantly change the structure of some proteins and protein complexes. For example, NtrC4 (a σ activator protein from Aquifex aeolicus) requires millimolar concentrations of certain salts, e.g., Mg2+, BeF3−, and ADP, in order to assemble into an active hexamer.35, 36 Several techniques for reducing sodium ion adduction to proteins in ESI have been developed that do not require removal of the salts from solution prior to ion formation. Buffer loading,21, 22 in which high concentrations of a volatile buffer, typically ammonium acetate, is added to solution, reduces sodium ion adduction to proteins and reduces the number of salt ion clusters formed. The addition of 7 M ammonium acetate to aqueous solutions containing 20 mM NaCl results in an increase in the S/N of the most abundant ion of cytochrome c and ubiquitin by more than 6-fold and 11-fold, respectively, compared to solutions without the buffer added.21 Buffer loading also works well for proteins that require high concentrations of salt to function or assemble, like concanavalin A.22 Other ammonium buffer salts can also effectively reduce sodium ion adduction to proteins, and can do so at much lower concentrations. For example, 25 mM ammonium bromide added to an aqueous 1 mM NaCl solution containing ubiquitin decreases the average number of sodium ions adducted to the most abundant charge state of the protein from 6.0 to 0.4 and increases the S/N of this ion by a factor of 66.25, 27 The ability of different salts to desalt proteins in the ESI droplet is related to the proton affinities of the anions, where anions with low proton affinities lead to less sodium adduction.27 However, anions with low proton affinities also tend to adduct to the protein as an acid molecule and form ion clusters,27 which can decrease the protein ion signal. Konermann and coworkers suggested that some salts, such as citrate and tartrate, may chelate ions in solution, such as calcium, that can non-specifically adduct to proteins.37
Supercharging reagents can be used to produce high charge-state ions by ESI from both denaturing38–45 and native solutions.46–55 Supercharging reagents are high-boiling point compounds that are typically added in small amounts (1–2%) to sample solutions prior to ESI.38–40, 45–55 For aqueous solutions, the low concentration of the supercharging reagents does not significantly affect the protein structure prior to ESI.50–52 However, the concentration of supercharging reagent increases in the droplet as solvent evaporation occurs, and the native structure of the protein can be chemically/thermally destabilized in the electrospray droplet, resulting in partial or extensive loss of folded structure with a concomitant increase in the number of charges on the gaseous protein ions that are formed.48–53 Protein-protein or protein-ligand complex dissociation may also occur as a result of this protein destabilization.46, 48, 50, 51, 53 Supercharging reagents can also increase protein charging from denaturing solutions in which the protein is initially unfolded,38–45 a result attributable to many factors, including the high surface tension of supercharging reagents compared to water-methanol-acid solutions.40 Droplets with higher surface tension can support more charge, and this high charge density can result in the formation of high charge-state ions. Loo and coworkers showed that the presence of m-NBA or sulfolane in the spray solvent in desorption electrospray ionization (DESI) experiments performed on HPLC column effluents containing trifluoroacetic acid (TFA) decreases TFA cluster ion intensity and increases protein S/N compared to when no supercharging reagent is in the DESI solvent.20 The authors suggested that this result is likely due to the supercharging reagent binding to TFA anions, thereby inhibiting TFA cluster formation, or preventing TFA dissociation into TFA anion and a free proton in solution, reducing the amount of free TFA anion in solution.20
Here, we show that m-NBA and sulfolane, two of the most common supercharging reagents, reduce sodium adduction to protein ions formed by nESI from native solutions. Sulfolane is more effective than m-NBA at reducing sodium ion adduction while still preserving non-covalent protein-protein and protein-ligand interactions. However, the use of m-NBA to desalt protein ions can lead to up to a 7-fold increase in protein ion S/N as a result of fewer clusters and lower chemical noise. This new method for desalting protein ions in nESI can improve the mass measuring accuracy for large proteins and protein complexes and can be used to resolve different glycoforms or ligand-adducted forms of proteins that are otherwise obscured by peak broadening resulting from extensive sodium adduction. m-NBA and sulfolane bind to sodium ions, suggesting that sodium sequestration by supercharging reagents may be the origin of the desalting effect of these supercharging reagents.
Experimental
Experiments were performed using a Waters Quadrupole-Time-of-Flight (Q-TOF) Premier (Waters, Milford, MA) mass spectrometer. Protein ions were formed by nanoelectrospray from borosilicate capillaries (1.0 mm o.d./0.78 mm i.d., Sutter Instruments, Novato, CA, USA) that were pulled to a tip i.d. of ~1 μm with a Flaming/Brown micropipette puller (Model P-87, Sutter Instruments, Novato, CA, USA) and positioned ~2 mm from the capillary inlet to the Q-TOF instrument. Nanoelectrospray was initiated by applying a potential of about +1.0 kV to a 0.127 mm diameter platinum wire inserted into the capillary and in contact with the sample solution. The nanoelectrospray potential was adjusted to optimize protein ion S/N for each tip. Values of the average number of sodium ions adducted are from four replicate measurements using four different nanospray emitters to account for tip-to-tip variation in adduction levels. All comparative S/N measurements were made using the same nanoelectrospray capillary to eliminate tip-to-tip variability, and the capillary was washed with methanol and water between each solution. All proteins were purchased as lyophilized powders from Sigma except for barnase and barstar, which were expressed in E. coli and purified as described previously.56 Ammonium bicarbonate, sodium chloride, platinum wire, and supercharging reagents were obtained from Sigma (St. Louis, MO, USA).
Results and Discussion
Effects of supercharging reagents on protein charge and sodium ion adduction
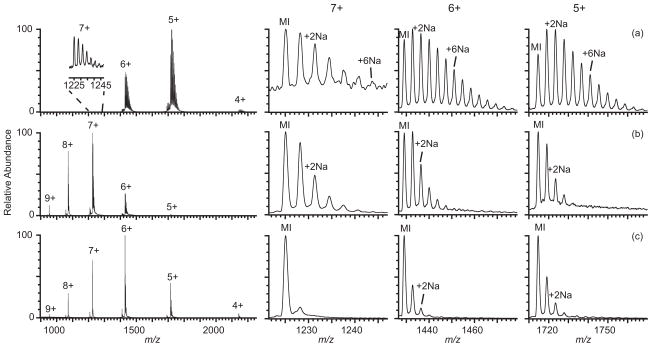
The average charge of ubiquitin ions formed by nESI from 5 μM ubiquitin in 10 mM ammonium bicarbonate and 1 mM sodium chloride (pH 7.8) is 5.33 ± 0.01+ (Figure 1a). There is extensive sodium ion adduction to the 5+ – 7+ charge states, with more adduction on lower charge states. The average number of sodium ions adducted to these charge states are 3.5 ± 0.1, 3.5 ± 0.1, and 2.0 ± 0.4, respectively (Table 1 and Table S-1). Mass spectra obtained from the same solution with either 1.5% m-NBA or 2.5% sulfolane are shown in Figures 1b and 1c, respectively. The average charge of ubiquitin formed from these respective solutions is 7.1 ± 0.1+ and 6.3 ± 0.1+. The average number of sodium ions adducted to the 5+ – 7+ charge states is significantly lower when either supercharging reagent is present. The average number of sodium ions adducted to the most abundant charge state decreases from 3.5 ± 0.1 in the spectrum obtained without a reagent to 1.2 ± 0.2 with m-NBA and 0.44 ± 0.03 with sulfolane. m-NBA is not soluble in aqueous solution above ~1.5%, and at the concentration used for both supercharging reagents, there should be little effect on the protein structure in solution prior to droplet formation by nESI.46, 51, 52 The presence of the 1 mM sodium chloride in these protein solutions has only a small effect on the charge of the protein ions formed with supercharging reagents. For example, the average charge of ubiquitin ions produced by nESI from solutions containing both added sodium chloride and m-NBA is only about one charge lower compared to that with the same amount of m-NBA but without sodium chloride added. The slightly lower charging with sodium chloride is likely due to stabilization of folded protein structure in the ESI droplet as a result of the higher ionic strength.
Figure 1.
Mass spectra of ubiquitin (5 μM) formed by nESI from aqueous solutions containing 10 mM ammonium bicarbonate and 1 mM NaCl (a) with no supercharging reagent, (b) with 1.5% m-NBA, and (c) with 2.5% sulfolane. Expansions showing adduction of sodium ions to the 5–7+ molecular ions are shown on the right; MI indicates the protonated molecular ion, (M+nH)n+.
Table 1.
Average charge and average number of sodium ions adducted to the most abundant protein charge state formed by nESI from aqueous solutions containing 5 μM protein, 10 mM ammonium bicarbonate, and 1 mM NaCl with no supercharging reagent, with 1.5% m-NBA, or with 2.5% sulfolane.
| Protein | Average charge | Average # Na adducts to (most abundant charge state) | Protein size | Protein pI | # Disulfide bonds |
|---|---|---|---|---|---|
| Ubiquitin | 5.33 ± 0.01 | 3.5 ± 0.1 (5+) | 8.6 kDa | 6.6 | 0 |
| m-NBA | 7.1 ± 0.1 | 1.2 ± 0.2 (7+) | |||
| sulfolane | 6.3 ± 0.1 | 0.44 ± 0.03 (6+) | |||
|
| |||||
| Barstar | 5.5 ± 0.1 | 3.5 ± 0.4 (5+) | 10.2 kDa | 4.7 | 0 |
| m-NBA | 7.1 ± 0.1 | 1.4 ± 0.2 (7+) | |||
| sulfolane | 5.7 ± 0.1 | 1.0 ± 0.4 (6+) | |||
|
| |||||
| Cytochrome c | 6.5 ± 0.1 | 2.5 ± 0.2 (7+) | 12.3 kDa | 9.6 | 0 |
| m-NBA | 9.42 ± 0.04 | 0.93 ± 0.01 (9+) | |||
| sulfolane | 8.6 ± 0.2 | 0.17 ± 0.01 (9+) | |||
|
| |||||
| Ribonuclease A | 6.9 ± 0.1 | 4.5 ± 0.2 (7+) | 13.7 kDa | 9.6 | 4 |
| m-NBA | 9.16 ± 0.03 | 1.2 ± 0.2 (9+) | |||
| sulfolane | 7.7 ± 0.1 | 0.5 ± 0.1 (8) | |||
|
| |||||
| α-Lactalbumin | 6.8 ± 0.1 | 3.3 ± 0.5 (7+) | 14.2 kDa | 4.8 | 4 |
| m-NBA | 8.9 ± 0.1 | 1.7 ± 0.1 (9+) | |||
| sulfolane | 7.2 ± 0.2 | 1.0 ± 0.3 (8+) | |||
|
| |||||
| Lysozyme | 7.5 ± 0.1 | 1.7 ± 0.1 (8+) | 14.3 kDa | 11.4 | 4 |
| m-NBA | 9.9 ± 0.1 | 0.60 ± 0.03 (10+) | |||
| sulfolane | 8.8 ± 0.3 | 0.4 ± 0.1 (9+) | |||
|
| |||||
| holo-Myoglobin | 8.2 ± 0.1 | 3.9 ± 0.4 (8+) | 17.6 kDa | 7.4 | 0 |
| m-NBA | 11.2 ± 0.2 | 0.79 ± 0.02 (11+) | |||
| sulfolane | 9.8 ± 0.4 | 0.3 ± 0.1 (10+) | |||
|
| |||||
| apo-Myoglobin | -- | -- | 17.0 kDa | 7.4 | 0 |
| m-NBA | 12.4 ± 0.6 | 0.71 ± 0.04 (11+) | |||
| sulfolane | 9.2 ± 0.3 | 0.28 ± 0.01 (9+) | |||
|
| |||||
| β-lactoglobulin | 8.13 ± 0.04 | 4.0 ± 0.2 (8+) | 18.3 kDa | 4.8 | 3 |
| m-NBA | 10.41 ± 0.03 | 2.88 ± 0.03 (10+) | |||
| sulfolane | 8.5 ± 0.2 | 1.9 ± 0.2 (9+) | |||
|
| |||||
| Carbonic | 9.8 ± 0.1 | 4.5 ± 0.6 (10+) | 29.0 kDa | 6.4 | 0 |
| Anhydrase | 13.5 ± 0.3 | 3.0 ± 0.5 (13+) | |||
| m-NBA | 11.7 ± 0.1 | 0.7 ± 0.2 (12+) | |||
| sulfolane | |||||
The same experiments were performed with nine other proteins for which sodium adducts could be resolved. The results are given in Table 1 for the most abundant charge state in each spectrum and in Table S-1 for every charge state. The presence of m-NBA or sulfolane in aqueous solutions containing sodium decreases the average number of sodium ions adducted to both the most abundant charge state and any given charge state in the mass spectra for all proteins. For these ten proteins, the number of sodium ions adducted to the most abundant charge state decreases by an average of 58% with m-NBA, and by 80% with sulfolane compared to solutions without supercharging reagents. Thus, adding small quantities of supercharging reagents is an effective way to desalt protein ions in aqueous solutions containing sodium and also increases the average charge state of protein ions formed from these native solutions. Sulfolane at 2.5% is more effective than 1.5% m-NBA at reducing sodium adduction to protein ions. The effectiveness of m-NBA and sulfolane at reducing sodium adduction to protein ions in nESI is protein-dependent. However, the extent of adduct reduction with supercharging reagents does not correlate well with any of several protein characteristics, such as protein size, pI, or number of disulfide bonds (Table 1).
With 2.5% sulfolane, the average charge of the proteins increases by an average of 1.0+ from solutions containing no supercharging reagent, whereas with 1.5% m-NBA, the average charge increases by 2.5+. This result is consistent with previous results that showed that sulfolane, on a per volume basis, is a less effective supercharging reagent than m-NBA, and therefore likely disrupts native protein structure to a lesser extent than m-NBA.51
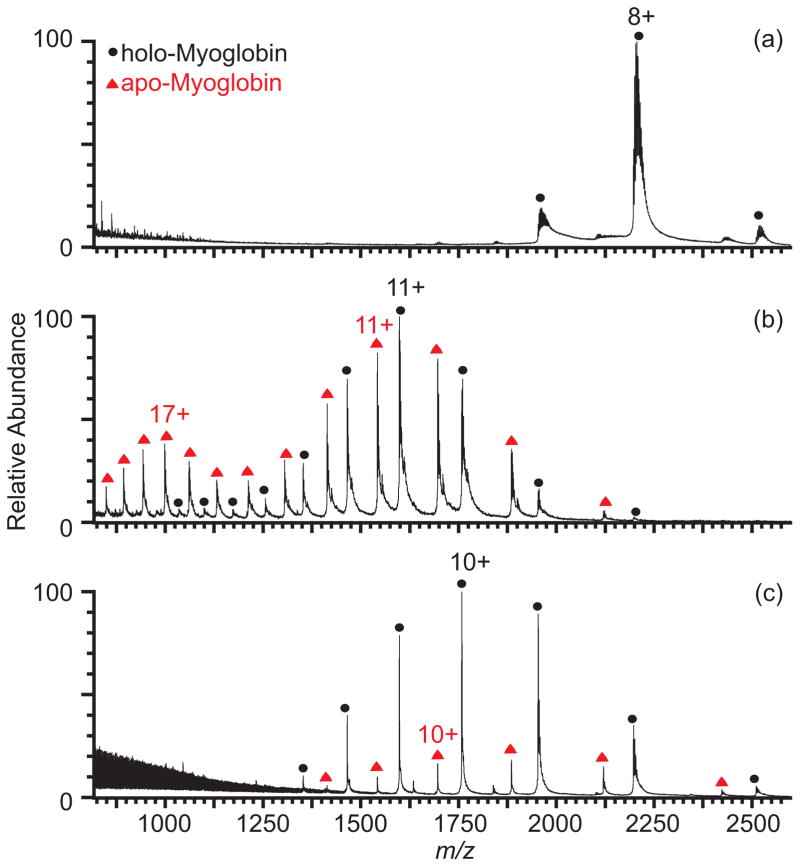
The effectiveness of sulfolane at protein ion desalting without extensively perturbing protein structure is important for desalting non-covalent protein-protein or protein-ligand complexes. Nanoelectrospray of myoglobin (Figure 2, Table 1), which contains a non-covalently bound heme group, from an aqueous solution with 10 mM ammonium bicarbonate and 1 mM NaCl results in 100% holo-myoglobin ions. Only 48% of the ion population is holo-myoglobin with 1.5% m-NBA, and the charge-state distribution is bimodal, indicating that some of the myoglobin is unfolded in the nESI droplet. In contrast, holo-myoglobin is 83% of the ion population observed with 2.5% sulfolane, and the monomodal charge-state distribution indicates that there is less perturbation of the folded form of the protein in the nESI droplet.
Figure 2.
Mass spectra of myoglobin (5 μM) formed by nESI from aqueous solutions containing 10 mM aqueous ammonium bicarbonate and 1 mM NaCl (a) with no supercharging reagent, (b) with 1.5% m-NBA, and (c) with 2.5% sulfolane.
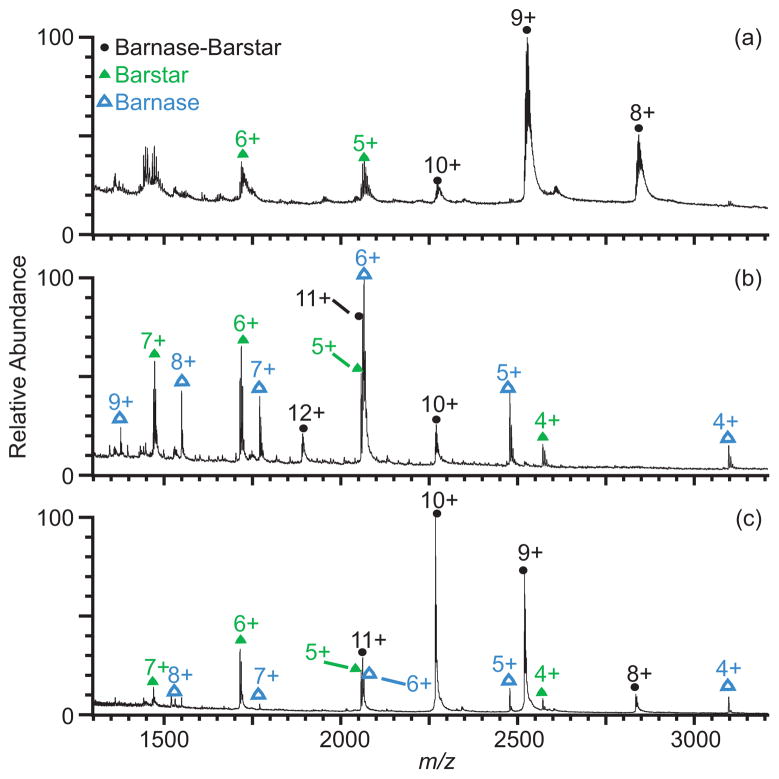
Protein-protein complexes, such as the 22 kDa heterodimer barnase-barstar (Figure 3, Table 2), are also preserved better with 2.5% sulfolane than with 1.5% m-NBA. A nESI mass spectrum of a 6:5 molar ratio mixture of barstar and barnase in sodiated ammonium bicarbonate solution shows the 8+ through 10+ charge-state ions of the barnase-barstar complex and ions of barstar, which is the excess reagent (Figure 3a). Because the barnase-barstar complex has a very low dissociation constant (25 ± 5 pM in aqueous ammonium bicarbonate solution, pH 7.2, 1% glycerol57), nearly all of the barnase, the limiting reagent, is present in the complex, and ions of free barnase are not observed in the mass spectrum. The most abundant charge state of the complex is 9+, and there is an average of 4.0 sodium ions adducted. In a mass spectrum of the same solution containing 1.5% m-NBA (Figure 3b), the charge-state distribution is shifted to higher charge, and the appearance of barnase ions indicates that some solution-phase unfolding and partial dissociation of the complex occur in the nESI droplet. The most abundant charge state of the complex shifts to 11+ (an average of 0.4 sodium ions adducted), and only 38% of the complex remains intact. In comparison, the charge of the most abundant ion of the complex increases to only 10+, and 84% of the barnase-barstar complex remains intact in the mass spectrum of the same solution with sodium and 2.5% sulfolane (Figure 3c). The average number of sodium ions adducted to the most abundant charge state with sulfolane is 0.6, similar to that obtained from the m-NBA solution. The examples of myoglobin and barnase-barstar illustrate that using sulfolane as a desalting agent can be particularly useful in native nESI experiments where measurement of ions from the intact complex is desired, such as when complex stoichiometry or the presence of ligand binding is being determined, although caution in interpreting these data is necessary owing to destabilization and possible dissociation of the complexes in the nESI droplet.
Figure 3.
Mass spectra of barnase-barstar (5 μM) with excess barstar formed by nESI from aqueous solutions containing 10 mM ammonium bicarbonate and 1 mM NaCl (a) with no supercharging reagent, (b) with 1.5% m-NBA, and (c) with 2.5% sulfolane.
Table 2.
Average charge state, average number of sodium adducts to the most abundant charge state, and fractional population of barnase-barstar and free barnase formed by nESI from aqueous solutions containing 6 μM barstar, 5 μM barnase, 10 mM ammonium bicarbonate, and 1 mM NaCl with no supercharging reagent, with 1.5% m-NBA, or with 2.5% sulfolane.
| Protein | Average charge | Average # Na adducts to most abundant charge state | % Total protein ion population |
|---|---|---|---|
| Barnase-Barstar | 8.8+ | 4.0 | 100% |
| + m-NBA | 11.0+ | 0.4 | 38% |
| + sulfolane | 9.6+ | 0.6 | 84% |
|
| |||
| Free Barnase | -- | -- | 0% |
| + m-NBA | 6.4+ | 0.7 | 62% |
| + sulfolane | 5.7+ | 0.3 | 16% |
Improving mass accuracy for large proteins and protein complexes
The charge-state distributions of large proteins and protein complexes in native ESI shift to higher m/z with increasing molecular weight.58 The inability to resolve individual sodium ion adducts at high m/z can reduce mass measuring accuracy and obscure the presence of different populations of covalently and non-covalently modified forms of the protein. Robinson and coworkers showed that retention of solvent and buffer molecules by large proteins and protein complexes in ESI results in measured masses much higher than calculated masses. For example, the measured mass of the 685 kDa 20S proteasome lacking an α-subunit is ~7 kDa higher than the calculated mass.23
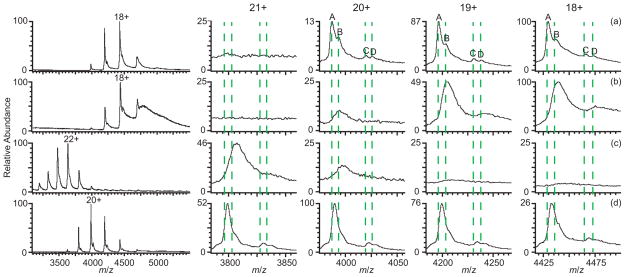
A native nESI mass spectrum of human holo-transferrin (79.7 kDa) in 10 mM ammonium bicarbonate with no sodium salt added is shown in Figure 4a. Four peaks corresponding to four different glycoforms of the protein are evident in the 18–20+ charge states. These glycoforms were determined from the masses measured from a denaturing solution of the protein (Figure S-1) to correspond well with the two oligosaccharide (one diantennary and one triantennary) glycosylations to this protein that were identified by van Halbeek and coworkers59 both with and without a fucosylation site on the oligosaccharide. The glycoforms are labelled as A) two diantennary oligosaccharides; B) A with one fucosylation; C) one di- with one triantennary oligosaccharide; and D) C with one fucosylation. The average masses measured for each of these four glycoforms formed by nESI from native solution (Figure 4a, Table 3) are on average ~6 Da higher than the measured masses of these ions formed from denaturing solution (the calculated m/z is indicated by the dashed lines in Figure 4), indicating that on average, less than one sodium ion is adducted to the protein. With 1 mM NaCl in the same solution (Figure 4b), the centroids of each peak shift to higher m/z so that the masses of ions A and C increase by ~168 Da and ~189 Da, respectively. This additional mass corresponds to an average of 8.3 and 9.3 sodium ions adducted to these two glycoforms, respectively, and the peak widths in the spectrum are broad due to sodium adduction such that the fucosylated ions B and D are no longer evident in the mass spectrum. Sodium adducts also lead to a significant increase in the baseline at higher m/z. With 1.5% m-NBA (Figure 4c), the charge-state distribution is shifted to higher charge, the most abundant charge state being 22+, and all of the peaks in the ion distribution are still broad as a result of sodium adduction. The fucosylated ions B and D are not resolved, and even ion C is evident only as a shoulder on the main A peak. Ions A and C have masses that are ~230 Da and ~300 Da higher than those formed from nESI from aqueous solutions without sodium, and ~62 Da and ~111 Da higher than those from aqueous solutions with sodium. The higher mass of these ions compared to those formed from solutions containing sodium without supercharging reagents may be due to m-NBA adduction to the protein, which has been observed before by Loo and coworkers for smaller proteins.46 With 2.5% sulfolane (Figure 4d), the most abundant charge state is 20+, and all glycosylated ions are evident in the mass spectrum, with ions A, C, and D resolved, and ion B distinguishable as a shoulder on the peak of ion A. The increase in mass for each glycosylated form of the protein is ≤~69 Da, or ~3 sodium ions. Thus, addition of sulfolane not only enables a more accurate mass measurement from a solution containing sodium, but it also reveals two additional fucosylated species present in solution that were unresolvable in the mass spectrum from this same solution without sulfolane present (Figure 4b).
Figure 4.
Mass spectra of human holo-transferrin (79.7 kDa) formed by nESI from aqueous solutions containing 10 mM ammonium bicarbonate with (a) no additional salt added, (b) 1 mM NaCl, (c) 1 mM NaCl with 1.5% m-NBA, and (d) 1 mM NaCl with 2.5% sulfolane. Expansions of the 18–21+ charge states are shown to the right. Dashed lines indicate the calculated average m/z for each glycoform without adducts.
Table 3.
Measured masses and average sodium ion adduction for four different glycoforms of holo-transferrin formed by nESI from aqueous solutions containing 10 mM ammonium bicarbonate (ABC) and 1 mM NaCl with no supercharging reagent, with 1.5% m-NBA, or with 2.5% sulfolane.
| Protein mass (Da) | |||||
|---|---|---|---|---|---|
| Protein glycoforms | 10 mM ABC | 10 mM ABC 1 mM NaCl |
+1.5% m-NBA | +2.5% sulfolane | Calculated mass |
| A | 79,714 ± 4 | 79,882 ± 21 | 79,944 ± 12 | 79,764 ± 5 | 79,700 Da |
| B | 79,848 ± 10 | 79,910 ± 15 | 79,846 Da | ||
| C | 80,372 ± 7 | 80,561 | 80,675 ± 21 | 80,425 ± 10 | 80,356 Da |
| D | 80,505 ± 12 | 80,567 ± 20 | 80,502 Da | ||
| Average # Na adducts | |||||
| 10 mM ABC |
10 mM ABC 1 mM NaCl |
+1.5% m-NBA | +2.5% sulfolane | ||
| A | 0.6 ± 0.2 | 8.3 ± 1.0 | 11.1 ± 0.6 | 2.9 ± 0.2 | |
| B | 0.1 ± 0.5 | 2.9 ± 0.7 | |||
| C | 0.7 ± 0.3 | 9.3 | 14.5 ± 1.0 | 3.1 ± 0.5 | |
| D | 0.1 ± 0.6 | 3.0 ± 0.9 | |||
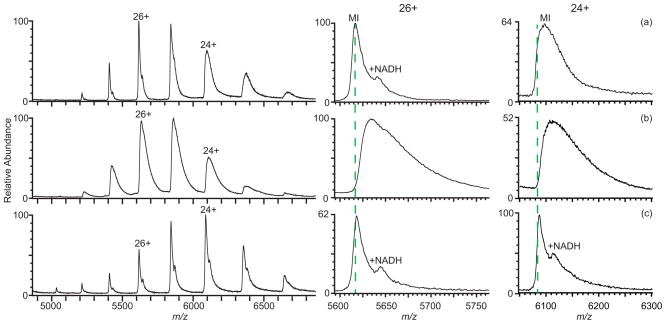
A mass spectrum of the 146 kDa homotetramer of rabbit lactate dehydrogenase (LDH) formed by nESI from 10 mM ammonium bicarbonate is shown in Figure 5a (dashed lines are the calculated m/z values). The measured mass of the tetramer is 146,004 ± 18 Da, which is close to the calculated mass of 145,996 Da based on the monomer mass determined from a spectrum of LDH obtained under denaturing conditions (Figure S-2). The 8 Da mass difference corresponds to an average of ~0.4 sodium ions adducted. There is also a distribution of LDH tetramer with a mass of 146,619 ± 14 Da corresponding to bound NADH, a cofactor of LDH that has a mass of 664 Da.60 With NaCl added at 5 mM concentration to this solution (Figure 5b), the LDH tetramer peaks are broader due to sodium ion adduction, and the NADH adduct is unresolved. The measured mass of the tetramer is 146,521 ± 23 Da, and corresponds to a mass increase of ~525 Da, or an average of ~23.9 sodium ions adducted. The peaks in a mass spectrum of rabbit LDH in the same solution containing NaCl but with 2.5% sulfolane (Figure 5c) are significantly narrower compared to spectra without the supercharging reagent (Figure 5b). The non-covalently bound NADH is clearly resolved on all of the tetramer peaks, except for the lowest charge state, and no dissociation of the NADH from the complex or increase in the average charge of the protein complex is evident, indicating that sulfolane does not significantly disrupt the structure of this protein complex. The addition of sulfolane also reduces the error in mass measurement associated with sodium adduction. The measured masses for the tetramer with and without NADH bound are 146,025 ± 26 Da and 146,763 ± 13 Da, respectively, with sulfolane, ~29 Da and ~103 Da higher than the calculated mass, corresponding to an average of only ~1.3 and ~4.7 sodium ions adducted, respectively. There is no data shown for LDH with 1.5% m-NBA because the protein precipitates out of solution upon addition of the supercharging reagent as evidenced by solution clouding, suggesting that the stabilities of some proteins can be affected by even low concentrations of supercharging reagents.
Figure 5.
Mass spectra of rabbit LDH tetramer (146 kDa) formed by nESI from aqueous solutions containing 10 mM ammonium bicarbonate with (a) no additional salt added, (b) 5 mM NaCl, and (c) 5 mM NaCl with 2.5% sulfolane. Expansions of the 26+ and 24+ charge states are shown to the right. Dashed lines indicate the calculated average m/z of the tetramer without adducts.
Effects on S/N and detection limits
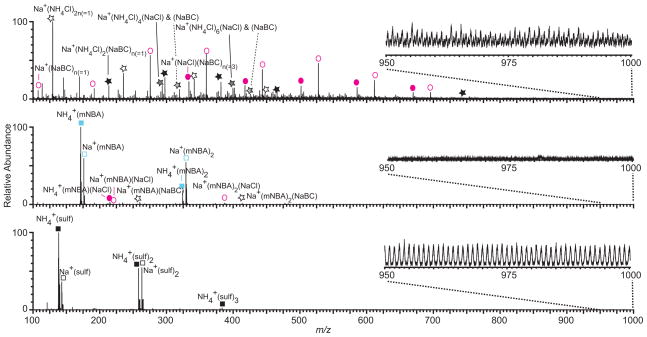
Loo and coworkers reported binding of m-NBA to ammonium and sodium cations at low m/z, but no protonated m-NBA was observed.46 It is possible that supercharging reagents reduce sodium ion adduction to proteins by binding to sodium ions in solution. Mass spectra of 5 μM barstar formed by nESI from aqueous solutions containing 10 mM ammonium bicarbonate and 1 mM NaCl with and without supercharging reagents are shown in Figure 6. In the expanded region below m/z = 700, many clusters containing sodium, chloride, bicarbonate, and ammonium are observed without supercharging reagents (Figure 6a), resulting in significant chemical noise at high m/z where protein ion signal is often observed (Table S-3). This can lower the protein ion S/N (Table S-3). With m-NBA (Figure 6b), the dominant low m/z ions are ammonium and sodium ions complexed with one or two m-NBA molecules, but there are also some low abundance ions containing sodium, chloride, bicarbonate, and ammonium, as well as sodium chloride and sodium bicarbonate cluster ions bound to m-NBA. Thus, m-NBA appears to inhibit the formation of salt cluster ions. There are no clusters above m/z ~700 (Figure 6b, inset), resulting in very low chemical noise in the higher m/z region of the spectrum where barstar ions are observed (Table S-3). With sulfolane, sodium and ammonium complexed to one, two, or three sulfolane molecules are most abundant (Figure 6c). Clusters extend to m/z ~1600, resulting in high chemical noise in the region where protein ions are often observed that is similar to the chemical noise without supercharging reagents (Figure 6a and c insets; Table S-3).
Figure 6.
Mass spectra of barstar (5 μM) formed by nESI from aqueous solutions containing 10 mM ammonium bicarbonate and 1 mM NaCl (a) with no supercharging reagent, (b) with 1.5% m-NBA, and (c) with 2.5% sulfolane. The low m/z region of the spectra are expanded. Spectra are normalized to the most abundant ion, but insets are on the same absolute scale for comparison.
Dearden and coworkers61 showed that alkali cations can be removed from peptides and small protein ions in the gas phase by crown ether molecules, leading to a concomitant decrease in the peptide or protein charge as the cations are removed. The average charge of protein ions formed from solutions containing m-NBA and sulfolane with sodium chloride is typically the same or higher than that obtained without these reagents. This indicates that protein desalting with m-NBA and sulfolane is not solely due to these supercharging reagents stripping sodium ions from proteins in the gas phase. Supercharging reagents must be influencing the amount of sodium ion adduction to proteins in solution, most likely in the nESI droplet, where enrichment of both sodium ions and supercharging reagent occurs. It is likely that sequestration of sodium ions by supercharging reagents is the cause of the reduced sodium ion adduction to protein ions observed with m-NBA and sulfolane. Other groups have reported adduction of supercharging reagents to protein ions,46, 54, 62 although no obvious adduction was observed in our study. It is plausible that the supercharging reagents may also block sites on the protein where sodium ions bind.
Addition of m-NBA or sulfolane to aqueous solutions containing sodium substantially increases the S/N of some protein ions owing to reduced chemical noise and fewer sodiated species over which protein signal is spread (Table S-3, Figure S-3). For example, the S/N of the most abundant ion of barstar formed from a 5 μM solution is 7-fold higher with m-NBA than without this supercharging reagent, and there is a 2-fold enhancement with sulfolane (Table S-3). Supercharging reagents can also decrease the limit of detection (LOD) for these protein ions formed from solutions containing sodium. The LOD for barstar from sodiated solutions, determined from protein concentration dependent measurements, is almost an order of magnitude lower with m-NBA than without (0.02 μM versus 0.12 μM) (Table S-3). However, the effect of supercharging reagents on the S/N and detection limit is protein dependent, and an improvement was not observed for all proteins (Table S-3). This may be due to differences in the chemical and salt impurities in the protein samples.
Conclusions
Supercharging reagents can increase the charge of protein ions formed by nESI from both native and denaturing solutions, but they also have the added benefit of effectively reducing sodium ion adduction to protein and protein complex ions formed from aqueous solutions. Addition of <3% of m-NBA or sulfolane to aqueous solutions containing millimolar concentrations of sodium can reduce sodium adduction to protein ions by an average of ~58% and ~80%, respectively. The presence of m-NBA in protein solutions containing sodium reduces chemical noise caused by clusters, resulting in up to a factor of 7 enhancement in S/N for some protein ions, whereas the presence of sulfolane has little effect on protein ion S/N due to salt cluster formation that extends to high m/z. Sodium adduction to both m-NBA and sulfolane occurs, and this may sequester a sufficient number of sodium ions such that sodium adduction to proteins is significantly reduced. In the future, it would be interesting to investigate whether the use of supercharging reagents for desalting protein ions formed by nESI is effective for other ions that commonly bind nonspecifically to proteins, such as potassium and calcium, or for anions, such as sulfate and phosphate, which are often added as buffers to native protein solutions.
Supplementary Material
Acknowledgments
The authors thank the National Institutes of Health (Grant No. R01GM097357) and the National Science Foundation (Graduate Research Fellowship for CAC; Grant No. DGE1106400) for financial support.
References
- 1.Hofmeister F. Archiv f experiment Pathol u Pharmakol. 1888;24:247–260. [Google Scholar]
- 2.Lo Nostro P, Ninham BW. Chem Rev. 2012;112:2286–2322. doi: 10.1021/cr200271j. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin RL. Biophys J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Bremer ETJ, Jiskoot W, James R, Moore GR, Kleanthous C, Heck AJR, Maier CS. Protein Sci. 2002;11:1738–1752. doi: 10.1110/ps.0200502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu MM, Rempel DL, Zhao J, Giblin DE, Gross ML. Biochemistry. 2003;42:15388–15397. doi: 10.1021/bi035188o. [DOI] [PubMed] [Google Scholar]
- 6.Maret W. J Inorg Biochem. 2012;111:110–116. doi: 10.1016/j.jinorgbio.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 7.Fritz G. In: Calcium-Binding Proteins and RAGE. Heizmann CW, editor. Vol. 963. Humana Press; 2013. pp. 87–97. ch 6. [Google Scholar]
- 8.Zhang Y, Cremer PS. Annu Rev Phys Chem. 2010;61:63–83. doi: 10.1146/annurev.physchem.59.032607.093635. [DOI] [PubMed] [Google Scholar]
- 9.Creighton TE. Proteins: Structures and Molecular Properties. 2. W. H. Freeman and Co; New York: 1993. [Google Scholar]
- 10.Weber KA, Achenback LA, Coates JD. Nat Rev Micro. 2006;4:752–764. doi: 10.1038/nrmicro1490. [DOI] [PubMed] [Google Scholar]
- 11.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Biochim Biophys Acta. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Toyoshima C, Nomura H. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 13.Gamba G. Physiol Rev. 2005;85:423–493. doi: 10.1152/physrev.00011.2004. [DOI] [PubMed] [Google Scholar]
- 14.Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, Curry S. J Mol Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 15.McCammon MG, Scott DJ, Keetch CA, Greene LH, Purkey HE, Petrassi HM, Kelly JW, Robinson CV. Structure. 2002;10:851–863. doi: 10.1016/s0969-2126(02)00771-2. [DOI] [PubMed] [Google Scholar]
- 16.Hernández H, Robinson CV. Nat Protocols. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Cole RB. Anal Chem. 1994;66:3702–3708. [Google Scholar]
- 18.Pan P, McLuckey SA. Anal Chem. 2003;75:5468–5474. doi: 10.1021/ac034344u. [DOI] [PubMed] [Google Scholar]
- 19.Mirza UA, Chait BT. Anal Chem. 1994;66:2898–2904. doi: 10.1021/ac00090a017. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Miao Z, Lakshmanan R, Loo RRO, Loo JA, Chen H. Int J Mass Spectrom. 2012;325–327:161–166. doi: 10.1016/j.ijms.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iavarone AT, Udekwu OA, Williams ER. Anal Chem. 2004;76:3944–3950. doi: 10.1021/ac049724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sterling HJ, Batchelor JD, Wemmer DE, Williams ER. J Am Soc Mass Spectrom. 2010;21:1045–1049. doi: 10.1016/j.jasms.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. J Am Chem Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 24.Freeke J, Robinson CV, Ruotolo BT. Int J Mass Spectrom. 2010;298:91–98. [Google Scholar]
- 25.Flick TG, Cassou CA, Chang TM, Williams ER. Anal Chem. 2012;84:7511–7517. doi: 10.1021/ac301629s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan P, Gunawardena HP, Xia Y, McLuckey SA. Anal Chem. 2004;76:1165–1174. doi: 10.1021/ac035209k. [DOI] [PubMed] [Google Scholar]
- 27.Flick T, Merenbloom S, Williams E. J Am Soc Mass Spectrom. 2011;22:1968–1977. doi: 10.1007/s13361-011-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flick T, Williams ER. J Am Soc Mass Spectrom. 2012;23:1885–1895. doi: 10.1007/s13361-012-0463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benkestock K, Edlund PO, Roeraade J. Rapid Commun Mass Spectrom. 2002;16:2054–2059. doi: 10.1002/rcm.832. [DOI] [PubMed] [Google Scholar]
- 30.Xiang F, Lin Y, Wen J, Matson DW, Smith RD. Anal Chem. 1999;71:1485–1490. doi: 10.1021/ac981400w. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y, Hofstadler SA. Anal Biochem. 2003;316:50–57. doi: 10.1016/s0003-2697(03)00024-1. [DOI] [PubMed] [Google Scholar]
- 32.Borges-Alvarez M, Benavente F, Barbosa J, Sanz-Nebot V. Rapid Commun Mass Spectrom. 2010;24:1411–1418. doi: 10.1002/rcm.4530. [DOI] [PubMed] [Google Scholar]
- 33.DeMarco ML, Burnham CAD. Am J Clin Path. 2014;141:204–212. doi: 10.1309/AJCPQYW3B6JLKILC. [DOI] [PubMed] [Google Scholar]
- 34.Prodanov M, Garrido I, Vacas V, Lebrón-Aguilar R, Dueñas M, Gómez-Cordovés C, Bartolomé B. Anal Chim Acta. 2008;609:241–251. doi: 10.1016/j.aca.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 35.Batchelor JD, Sterling HJ, Hong E, Williams ER, Wemmer DE. J Mol Biol. 2009;393:634–643. doi: 10.1016/j.jmb.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batchelor JD, Doucleff M, Lee CJ, Matsubara K, De Carlo S, Heideker J, Lamers MH, Pelton JG, Wemmer DE. J Mol Biol. 2008;384:1058–1075. doi: 10.1016/j.jmb.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 37.Pan J, Xu K, Yang X, Choy WY, Konermann L. Anal Chem. 2009;81:5008–5015. doi: 10.1021/ac900423x. [DOI] [PubMed] [Google Scholar]
- 38.Iavarone AT, Jurchen JC, Williams ER. Anal Chem. 2001;73:1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iavarone AT, Williams ER. Int J Mass Spectrom. 2002;219:63–72. [Google Scholar]
- 40.Iavarone AT, Williams ER. J Am Chem Soc. 2003;125:2319–2327. doi: 10.1021/ja021202t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iavarone AT, Williams ER. Anal Chem. 2003;75:4525–4533. doi: 10.1021/ac034144i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sze SK, Ge Y, Oh H, McLafferty FW. Proc Natl Acad Sci U S A. 2002;99:1774–1779. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madsen JA, Brodbelt JS. J Am Soc Mass Spectrom. 2009;20:349–358. doi: 10.1016/j.jasms.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 44.Kjeldsen F, Giessing AMB, Ingrell CR, Jensen ON. Anal Chem. 2007;79:9243–9252. doi: 10.1021/ac701700g. [DOI] [PubMed] [Google Scholar]
- 45.Miladinovi SM, Fornelli L, Lu Y, Piech KM, Girault HH, Tsybin YO. Anal Chem. 2012;84:4647–4651. doi: 10.1021/ac300845n. [DOI] [PubMed] [Google Scholar]
- 46.Lomeli SH, Peng IX, Yin S, Loo RRO, Loo JA. J Am Soc Mass Spectrom. 2010;21:127–131. doi: 10.1016/j.jasms.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lomeli SH, Yin S, Loo RRO, Loo JA. J Am Soc Mass Spectrom. 2009;20:593–596. doi: 10.1016/j.jasms.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sterling HJ, Kintzer AF, Feld GK, Cassou CA, Krantz BA, Williams ER. J Am Soc Mass Spectrom. 2012;23:191–200. doi: 10.1007/s13361-011-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sterling HJ, Cassou CA, Trnka MJ, Burlingame AL, Krantz BA, Williams ER. Phys Chem Chem Phys. 2011;13:18288–18296. doi: 10.1039/c1cp20277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sterling HJ, Prell JS, Cassou CA, Williams ER. J Am Soc Mass Spectrom. 2011;22:1178–1186. doi: 10.1007/s13361-011-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sterling HJ, Daly MP, Feld GK, Thoren KL, Kintzer AF, Krantz BA, Williams ER. J Am Soc Mass Spectrom. 2010;21:1762–1774. doi: 10.1016/j.jasms.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sterling HJ, Williams ER. Anal Chem. 2010;82:9050–9057. doi: 10.1021/ac101957x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sterling HJ, Williams ER. J Am Soc Mass Spectrom. 2009;20:1933–1943. doi: 10.1016/j.jasms.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hogan CJ, Loo RRO, Loo JA, de la Moraa JF. Phys Chem Chem Phys. 2010;12:13476–13483. doi: 10.1039/c0cp01208d. [DOI] [PubMed] [Google Scholar]
- 55.Erba EB, Ruotolo BT, Barsky D, Robinson CV. Anal Chem. 2010;82:9702–9710. doi: 10.1021/ac101778e. [DOI] [PubMed] [Google Scholar]
- 56.Krishnaswamy SR, Williams ER, Kirsch JF. Protein Sci. 2006;15:1465–1475. doi: 10.1110/ps.062083406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartley RW. Methods Enzymol. 2001;341:599–611. doi: 10.1016/s0076-6879(01)41179-7. [DOI] [PubMed] [Google Scholar]
- 58.de la Mora JF. Anal Chim Acta. 2000;406:93–104. [Google Scholar]
- 59.Fu D, van Halbeek H. Anal Biochem. 1992;206:53–63. doi: 10.1016/s0003-2697(05)80010-7. [DOI] [PubMed] [Google Scholar]
- 60.Burgner JW, Ray WJ. Biochemistry. 1984;23:3636–3648. doi: 10.1021/bi00311a010. [DOI] [PubMed] [Google Scholar]
- 61.Pope RM, Shen N, Hofstadler SA, Dearden DV. Int J Mass Spectrom. 1998;175:179–186. [Google Scholar]
- 62.Douglass K, Venter A. J Am Soc Mass Spectrom. 2012;23:489–497. doi: 10.1007/s13361-011-0319-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.