Abstract
Background
Linezolid is one of the most effective treatments against Gram-positive pathogens. However, linezolid-resistant/intermediate strains have recently emerged in worldwide. The purpose of this study was to analyse the prevalence and resistance mechanisms of linezolid-resistant/intermediate staphylococci and enterococci in Shanghai, China.
Results
Thirty-two linezolid-resistant/intermediate strains, including 14 Staphylococcus capitis, three Staphylococcus aureus, 14 Enterococcus faecalis and one Enterococcus faecium clinical isolates, were collected in this study which displayed linezolid MICs of 8 to 512 μg/ml, 8–32 μg/ml, 4–8 μg/ml and 4 μg/ml, respectively. All linezolid-resistant S. capitis isolates had a novel C2131T mutation and a G2603T mutation in the 23S rRNA region, and some had a C316T (Arg106Cys) substitution in protein L4 and/or harboured cfr. Linezolid-resistant S. aureus isolates carried a C389G (Ala130Gly) substitution in protein L3, and/or harboured cfr. The cfr gene was flanked by two copies of the IS256-like element, with a downstream orf1 gene. Linezolid-resistant/intermediate enterococci lacked major resistance mechanisms. The semi-quantitative biofilm assay showed that 14 linezolid-resistant E. faecalis isolates produced a larger biofilm than linezolid-susceptible E. faecalis strains. Transmission electron microscopy showed the cell walls of linezolid-resistant/intermediate strains were thicker than linezolid-susceptible strains.
Conclusion
Our data indicated that major resistance mechanisms, such as mutations in 23S rRNA and ribosomal proteins L3 and L4, along with cfr acquisition, played an important role in linezolid resistance. Secondary resistance mechanisms, such as biofilm formation and cell wall thickness, should also be taken into account.
Keywords: Linezolid resistance, Mutations of 23S rRNA and ribosomal proteins, cfr, Biofilm production, Cell wall thickness
Background
Gram-positive cocci pose a worldwide threat to human health. The emergence of antibiotic resistance in Gram-positive cocci, including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant staphylococci (VRS) and vancomycin-resistant enterococci (VRE), has created a clinical demand for effective novel therapeutic agents. Linezolid (LZD), the first member of the oxazolidinone class of antibiotics, was approved for clinical use in 2000 and has a broad spectrum of activity against a variety of Gram-positive pathogens. It acts by inhibiting protein synthesis via binding to the peptidyl transferase centre of the 50S ribosomal subunit, and preventing formation of the fMet-tRNA-30S ribosome-mRNA initiation complex [1]. Because of its unique antimicrobial mechanism, linezolid has been widely applied in the treatment of clinically-important Gram-positive bacteria, including aerobic and anaerobic Gram-positive cocci, aerobic and anaerobic Gram-positive bacilli, and nocardia and mycobacteria species. However, linezolid-resistant (LR) staphylococcus was first reported in peritonitis patients undergoing oral linezolid treatment during peritoneal dialysis in 2001 [2]. Since then, the occurrence of LR strains has been reported worldwide [3-5].
The major mechanism of resistance to linezolid is caused by mutations in the V domain of the 23S rRNA gene, with a G2576T substitution (Escherichia coli numbering) occurring most frequently. C2104T, G2447T, T2500A, A2503G, T2504A, G2603T and G2631T substitutions have also been found in LR strains [6-9]. Another resistance mechanism is horizontal acquisition of cfr, which encodes a methyltransferase and modifies adenosine at A2503 in the 23S rRNA. cfr is usually plasmid-located and confers cross-resistance to phenicol, lincosamide, oxazolidinone, pleuromutilin and streptogramin A (known as the PhLOPSA phenotype) [10,11]. Alterations in the ribosomal proteins L3, L4 and L22, encoded by rplC, rplD and rplV, respectively, have also been associated with increased resistance to linezolid [12-14]. In addition, secondary resistance mechanisms, such as biofilm formation and cell wall thickening, can enhance resistance to antibiotics as well [15,16].
As the global emergence of LR isolates has increased [2-7], LR Gram-positive cocci have also become a problem in China [8,17-19]. To comprehensively understand the current prevalence and resistance mechanisms among LR clinical isolates in China, we analysed 32 linezolid-resistant/intermediate strains, including 14 Staphylococcus capitis, three S. aureus, 14 Enterococcus faecalis and one Enterococcus faecium isolates.
Methods
Ethics statement
The collection of the linezolid-resistant/intermediate bacterial isolates from patients and the related information of patients were approved by the ethics committee of Huashan Hospital, Shanghai Medical College, Fudan University and the ethics committee of Renji Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China. All subjects provided written informed consent before their inclusion in the study.
Bacterial isolates
Thirty-two non-duplicated linezolid-resistant/intermediate isolates were collected from patients of two Shanghai comprehensive teaching hospitals in China from 2009–2013. One of the hospitals was Huashan Hospital, which is a tertiary care hospital affiliated with Fudan University, located in the centre of Shanghai. It is one of the largest (1300 beds) teaching hospitals in china, handling approximately 8000 admissions per day. The other was Renji Hospital, a tertiary care hospital affiliated with Shanghai Jiaotong University, which is located in the east of Shanghai, and is also one of the largest (1800 beds) teaching hospitals in china, handling about 9000 admissions per day. The studied isolates comprised 14 S. capitis (from Huashan Hospital), three S. aureus (from Huashan hospital), 14 E. faecalis (12 isolates from Huashan Hospital and two from Renji Hospital) and one E. faecium (from Huashan Hospital). Among these 32 isolates, 15, 10, 3, 2, 1 and 1 were recovered from patients with bacteraemia, urinary tract infection, pneumonia, wound infection, biliary tract infection and prostate infection, respectively (Table 1). Isolates were identified using a VITEK 2 compact system (bioMérieux, Marcy l’Etoile, France) and a molecular method based on analysis of the 16S rRNA gene sequence. S. aureus RN4220 was used as the recipient strain for transformation experiments. S. aureus ATCC 29213, E. faecalis ATCC 29212 (both with linezolid MIC of 2 μg/ml, American type culture collection, USA), S. capitis HS12-102 and E. faecium HS13-194 (both with linezolid MIC of <2 μg/ml, from Huashan Hospital) were included as linezolid-susceptible (LS) strains. All strains were stored at −70°C until use and were incubated overnight on blood agar at 37°C.
Table 1.
Clinical characteristics of patients with linezolid-resistant Staphylococcus or Enterococcus infection
| Strains | Organism | Underlying diseases | Infection type | Ward | Prior antibiotic use | Outcome |
|---|---|---|---|---|---|---|
| HS09-206 | S. capitis | Haematencephalon,Hypertension, Renal failure | Bacteraemia | Intensive care unit (ICU) | SCF.AMK.MTZ | Discharge |
| HS09-17 | S. capitis | Severe craniocerebral injury, Infective shock | Bacteraemia, Pneumonia, | Department of Cerebral Surgery | MEN.LZD.SCF. DOX | Discharge |
| HS10-204 | S. capitis | Craniocerebral injury, | Bacteraemia, Pneumonia | ICU | SXT.VAN | Discharge |
| HS10-24 | S. capitis | Hip joint surgery, | Bacteraemia, Diffuse peritonitis | ICU | TZP.MEN.LZD. LVX.SCF | Discharge |
| HS11-203 | S. capitis | Bladder surgery, Infective shock | Bacteraemia | ICU | MEN.VAN | Discharge |
| HS11-44 | S. capitis | Subarachnoid haemorrhage | Bacteraemia | ICU | LZD | Discharge |
| HS12-49 | S. capitis | Craniocerebral injury, | Bacteraemia, Pneumonia | ICU | VAN.SCF | Discharge |
| HS12-201 | S. capitis | Ventricular fibrillation, Epilepsy | Bacteraemia | ICU | MEN.SCF.AMK. TZP | Discharge |
| HS12-51 | S. capitis | Circulatory failure, | Bacteraemia, Pneumonia | ICU | IPM.LZD.VAN. FEP.TEC.CAZ. LVX.AMK | Discharge |
| HS12-53 | S. capitis | Craniocerebral injury | Bacteraemia | Department of Cerebral Surgery | MEN.AMK.SAM.VAN | Discharge |
| HS12-55 | S. capitis | Acute respiratory failure, Nephrosis syndrome | Bacteraemia, Pneumonia, | ICU | IPM.FOF.MEN. SCF.DOX | Discharge |
| HS12-58 | S. capitis | Subarachnoid haemorrhage | Bacteraemia | ICU | CXM.SCF | Discharge |
| HS13-60 | S. capitis | Brain tumour operation, Respiratory failure | Bacteraemia | ICU | MEN.LZD.SCF. DOX | Discharge |
| HS13-207 | S. capitis | Subarachnoid haemorrhage | Bacteraemia | ICU | IPM.LZD.VAN. FEP.TEC.CAZ. LVX.AMK | Discharge |
| HS09-205 | S. aureus | COPD, Coronary disease | Pneumonia | General ward | SCF | Discharge |
| HS11-202 | S. aureus | Hematencephalon | Pneumonia | ICU | SCF | Discharge |
| HS12-56 | S. aureus | Craniocerebral injury | Pneumonia | ICU | VAN.SCF | Discharge |
| HS10-311 | E. faecalis | Urinary tract infection | Urinary infection | Department of Urology (outpatient) | N/day | Infection cleared |
| HS11-304 | E. faecalis | Hepatocellular carcinoma after operation | Bacteraemia, Infectious endocarditis | Department of Infectious Disease | VAN.FOF.SAM. AMK.MTZ | Discharge |
| HS11-305 | E. faecalis | Urinary tract infection | Urinary infection | Department of Urology (outpatient) | N/D | Infection cleared |
| HS11-307 | E. faecalis | Chronic subdural hematoma | Urinary infection, Pneumonia | Department of Cerebral Surgery | MEN.LZD.SCF. DOX | Discharge |
| HS12-309 | E. faecalis | Urinary tract infection | Urinary infection | Department of Urology (outpatient) | N/D | Infection cleared |
| HS12-308 | E. faecalis | Urinary tract infection | Urinary infection | Department of Urology (outpatient) | N/D | Infection cleared |
| HS12-310 | E. faecalis | Femoral fracture | Wound infection | Department of Orthopaedics | MEN | Discharge |
| HS13-301 | E. faecalis | Urinary tract infection | Urinary infection | Department of Urology (outpatient) | N/D | Infection cleared |
| HS13-302 | E. faecalis | Scrofula | Biliary tract infection | Department of Infectious Disease | MXF.SCF.IHY.PYR. | Discharge |
| HS13-303 | E. faecalis | Drug fever | Urinary infection | Department of Infectious Disease | AZM | Discharge |
| RJ13-314 | E. faecalis | Juvenile arthritis | Urinary infection | Department of Urology (outpatient) | N/D | Infection cleared |
| RJ13-315 | E. faecalis | Chronic prostatitis | Prostate infection | Department of Urology (outpatient) | N/D | Infection cleared |
| HS13-312 | E. faecalis | Psoriasis | Wound infection | Department of Dermatology | Not used | Discharge |
| HS13-313 | E. faecalis | Urinary tract infection | Urinary infection | Department of Urology (outpatient) | N/D | Infection cleared |
| HS11-306 | E. faecium | Lithangiuria | Urinary infection | Department of Urology | TZP.SAM.VAN.LVX.CXM | Discharge |
HS, isolate from Huashan Hospital; RJ, isolate from Renji Hospital; COPD, chronic obstructive pulmonary disease; SCF, cefoperazone/sulbactam; AMK, amikacin; MTZ, metronidazole; MEM, meropenem; LZD, linezolid; DOX, doxycycline; SXT, sulfamethoxazole/trimethoprim; VAN, vancomycin; TZP, piperacillin/tazobactam; LVX, levofloxacin; IPM, imipenem; FEP, cefepime; TEC, teicoplanin; CAZ, ceftazidime; SAM, ampicillin/sulbactam; FOF, fosfomycin; CXM, cefuroxime; MXF, moxifloxacin; IHY, isonicotinyl hydrazide; PYR, pyrazinamide; AZM, Azithromycin. N/D, not determined.
Antibiotic susceptibility testing
The antimicrobial agents tested were linezolid, vancomycin, teicoplanin, oxacillin, cefoxitin, trimethoprim-sulfamethoxazole, erythromycin, clindamycin, chloramphenicol, tetracycline, ciprofloxacin, penicillin, ampicillin and high-level gentamicin. The MIC of each antimicrobial agent was determined by the broth microdilution MIC method and interpretation of MIC results was based on 2013 Clinical and Laboratory Standards Institute guidelines [20]. S. aureus ATCC 29213 was tested concurrently for quality control.
Multilocus sequence typing (MLST) analysis
S. aureus, E. faecalis and E. faecium were screened using a previously described method [21-23] to detect the seven housekeeping genes. For S. aureus, there are carbamate kinase (arcC), shikimate dehydrogenase (aroE), glycerol kinase (glp), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL). For E. faecalis, there are glucose-6-phosphate dehydrogenase (gdh), glyceraldehydes-3-phosphate dehydrogenase (gyd), phosphate ATP binding cassette transporter (pstS), glucokinase (gki), shikimate-5-dehydrogenase (aroE), xanthine phosphoribosyltransferase (xpt), and acetyl-CoA acetyltransferase (yiqL). For E. faecium, there are adenylate kinase (adk), ATP synthase, alpha subunit (atpA), d-alanine:d-alanine ligase (ddl), glyceraldehyde-3-phosphate dehydrogenase (gyd), glucose-6-phosphate dehydrogenase (gdh), phosphoribosylaminoimidazol carboxylase ATPase subunit (purK), and phosphate ATP-binding cassette transporter (pstS). Each sequence was submitted to the MLST website (http://www.mlst.net) to determine the allelic profile and sequence type (ST) of each isolate [24].
PFGE
PFGE was performed according to a previously described method [25] with some modifications. The staphylococci were treated with lysostaphin (Sigma-Aldrich, Saint Louis, MO, USA) and genomic DNA was prepared in agarose blocks and then digested with the restriction enzyme SmaI (NEB, Hitchin, UK). The DNA fragments were separated using a CHEF-DR II PFGE system (Bio-Rad, Hercules, CA, USA) for 20 h at 6 V/cm and 14°C, with a pulse angle of 120° and pulse times from 5–25 s. PFGE banding patterns were analysed visually.
Molecular detection of resistance genes and mutations
Isolates were screened for the presence of cfr and mutations in the 23S rRNA and the L3, L4 and L22 ribosomal proteins by PCR and DNA sequencing, as previously described [26,27]. Amplicons were sequenced on both strands and were compared with those from S. aureus ATCC 29213, E. faecalis ATCC 29212, LS S. capitis and LS E. faecium, obtained from Shanghai Huashan Hospital during the study period, using the Lasergene software package (DNAStar; Madison, WI, USA).
Gene dosage
Gene dosage was determined according to a previously described method [28,29] with some modifications. Isolates of LR S. capitis containing mutations in the central loop of the 23S rRNA gene were amplified using primers based on the S. capitis 23S rRNA gene (primer F, 5′-AAGGCGTAACGATTTGGG-3′; primer R, 5′-CAGCACTTATCCCGTCCA-3′; expected PCR product size: 720 bp). Thermal cycler conditions were 94°C for 5 min, followed by 35 cycles of 94°C (30 s), 55°C (30 s) and 72°C (30 s), with a final extension at 72°C (7 min). The DNA concentration of PCR amplicons was normalized to 0.3 pmol prior to cloning. Then, the amplification products were ligated to plasmid pMD-18 T (Takara Biotechnology, Dalian, China) and transformed into E. coli DH5α cells (Sangon Biotech, Shanghai, China). Each LR S. capitis was cloned and 30 clones from each strain were sequenced.
Plasmid analysis and cfr location
Plasmid DNA was extracted from all cfr-carrying isolates using a Plasmid DNA Midi Kit (QIAGEN, Hilden, Germany) and separated on 1% (w/v) agarose gel. The migration distances of DNA bands were measured directly from gel photographs in relation to the reference plasmids in E. coli V517 (sizes, 54, 5.6, 5.1, 3.9, 3.0, 2.7, and 2.1 kb) marker. Plasmid DNA bands were transferred onto a nylon membrane by Southern blotting. A cfr-specific labelled probe (primer F, 5′-GAAGCTCTAGCCAACCGTCA-3′; primer R, 5′-TCTACCTGCCCTTCGTTTGC3′; expected PCR product size: ~600 bp) was generated using a non-radioactive DIG High Prime DNA Labelling and Detection Kit (Roche Applied Sciences, Mannheim, Germany) according to the manufacturer’s instructions, and was hybridised to the membrane. To determine the genetic environment of cfr, the partial plasmid DNA sequence of all cfr-carrying isolates was determined using the following primers, designed based on the sequence of pSS-01 (GenBank accession no. JQ041372): 1 F 5′- CACTTCCTTTATTATTTTTC-3′; 1R 5′- CGACTGAATCAAGAAGTACG-3′; 2 F 5′-TGAACCATAACCTTTGTC-3′, 2R 5′-TGTTTCTAGCCTGACTGA-3′; 3 F 5′-ACTCGATGGTCCTCACGG-3′, 3R 5′-TCAGGCTCATTATTACTTC-3′; 4 F 5′-TTACCACTAGAGCAAATT′, 4R 5′-GACCACAAGCAGCGYCAA-3′; 5 F 5′-AATGGACTACTGGGTGGA-3′, 5R 5′-ATGATTCAGAAAGCAGAT-3′; 6 F 5′-CTTTCCGAATGGACTAAT-3′, 6R 5′-TGAGCGCAGCTTTACGAC-3′ (PCR product sizes: 1000 ~ 2500 bp). Amplification products from the ten cfr-carrying plasmids were sequenced on both strands and were compared with plasmid pSS-01 using the Lasergene software package (DNAStar; Madison, WI, USA). In addition, the purified plasmids extracted from each of the ten original strains were electrotransformed (2kv, 25 μF, 1000Ω) into the S. aureus recipient strain RN4220. Transformants were incubated in fresh tryptic soy broth (TSB; Oxoid, Basingstoke, UK) at 37°C with shaking at 200 rpm for 1 h and then were selected by incubation for 24 h at 37°C on TSB agar supplemented with 6 μg/ml linezolid. The linezolid MICs of transformants were measured by the broth microdilution method. Plasmid DNA was extracted from all the transformants and separated on 1% agarose gel. Southern hybridisation was then used to confirm whether the plasmids were cfr-positive as described previously [17].
Detection of biofilm production
All LR strains were divided into four groups by species (S. capitis, S. aureus, E. faecalis and E. faecium). S. capitis group contained 14 LR isolates and 14 LS isolates. S. aureus group contained 3 LR isolates and 15 LS isolates. E. faecalis group contained 14 LR isolates and 14 LS isolates. E. faecium group contained 1 LR isolate and 15 LS isolates. A semi-quantitative biofilm assay was conducted using flat-bottomed, tissue culture-treated, 96-well assay plates (Costar 3599; California, USA). Four wells in each assay plate contained untreated cultures, which served as negative controls. Bacteria were prepared by making a 1:100 dilution of an overnight culture grown in TSB containing 5 g/L glucose. Assay plates were inoculated with 200 μL of culture per well (OD600 = 0.01) and were incubated at 37°C for 24 h. The medium were removed from assay plates. Then assay plates were washed four times with PBS. Biofilms were fixed by Bouin’s fixation for at least 1 h then assay plates were washed four times with PBS. Following crystal violet staining, the biofilms were measured using a Synergy H1 Hybrid Multi-Mode Microplate Reader (Biotek, Vermont, USA) at OD570.
Transmission electron microscopy (TEM)
Isolates for TEM were grown from a single colony in 10 mL of liquid brain-heart infusion (BHI) medium with 0.5% (w/v) beef extract. Four groups were studied. The S. capitis group contained strains HS12-102 (LS S. capitis strain HS12-55), HS09-17 (MIC 16 μg/ml), HS09-17 + LZD (strain HS09-17 grown in BHI broth containing 4 μg/ml linezolid), HS12-55 (MIC >256 μg/ml) and HS12-55 + LZD (strain HS12-55 grown in BHI broth containing 32 μg/ml linezolid). The S. aureus group contained S. aureus ATCC29213 (LS S. aureus), HS11-202 (MIC 16 μg/ml) and HS11-202 + LZD (strain HS11-202 grown in BHI broth containing 2 μg/ml linezolid). The E. faecalis group contained E. faecalis ATCC29212 (LS E. faecalis), HS11-304 (MIC 4 μg/ml), HS12-309 (MIC 8 μg/ml) and HS12-309 + LZD (strain HS12-309 grown in BHI broth containing 2 μg/ml linezolid). Finally, the E. faecium group contained strains HS13-194 (LS E. faecium) and HS11-306 (MIC 4 μg/ml). The cells were grown at 37°C with shaking at 200 rpm for approximately 4–5 h to mid-exponential phase (OD600 ~ 0.7–0.8). An 8-mL volume of each culture was pelleted by centrifugation at 4000 g for 15 min and suspended in 1 mL of 2.5% (v/v) glutaraldehyde in 0.1 M phosphate buffer, and stored at 4°C for at least 2 h. The cells were then post-fixed with 1% (w/v) osmium tetroxide in the same buffer. Following dehydration, cells were infiltrated with and embedded in Eponate 12 resin for 24 h. Ultrathin sections were cut at 70 nm, stained with uranyl acetate and lead citrate, and then examined on a JEM-1230 80 kV TEM (JEOL, Tokyo, JAPAN) equipped with a Gatan Orius SC200W camera. Images were captured at 30,000× magnification and measurements were made using ImageJ software [30]. Thirty cell walls were measured per sample, and means and standard deviations were calculated in Excel (Microsoft, San Francisco, USA).
Results
Incidence of linezolid resistance
From 2009–2013, 3446 Staphylococcus and 1713 Enterococcus non-duplicated isolates were tested for susceptibility to linezolid. The incidence of linezolid resistance in our study was 1.18% of coagulase-negative staphylococci (CONS) (14/1188 isolates), 0.13% of S. aureus (3/2258 isolates), 1.72% of E. faecalis (14/812 isolates) and 0.11% of E. faecium (1/877 isolates) isolates.
Antimicrobial susceptibility testing
The susceptibility profiles of 14 S. capitis isolates, three S. aureus isolates and 14 E. faecalis isolates were similar (Table 2). The S. capitis isolates showed various levels of linezolid resistance, with MIC values of 8–512 μg/ml. The S. aureus isolates were resistant to linezolid, with MIC values of 8–32 μg/ml. Other than S. aureus HS09-205, all staphylococcal strains were methicillin-resistant and exhibited high-level resistance to most of the antibiotics tested, except for vancomycin, teicoplanin and tetracycline. Moreover, 17 staphylococcal isolates showed different levels of trimethoprim-sulfamethoxazole susceptibility data, with MIC values of 0.12/2.37–8/152 μg/ml. The fifteen enterococcal isolates demonstrated either linezolid-intermediate (LI) or low-level linezolid resistance (MICs 4–8 μg/ml). These strains were resistant to erythromycin, chloramphenicol, ciprofloxacin and tetracycline (isolate HS11-307 was susceptible to tetracycline, with a MIC of 1 μg/ml), but susceptible to vancomycin (MICs, 0.5–2 μg/ml), teicoplanin (MICs, 0.12–0.25 μg/ml), penicillin (MICs, 0.5–4 μg/ml) and ampicillin (MICs, <0.5–4 μg/ml). Eight enterococcal isolates (HS11-304, HS12-309, HS12-308, HS12-310, HS13-301, HS13-303, RJ13-314 and HS11-306) were not high-level resistant to gentamicin, whereas the remaining strains were resistant.
Table 2.
Antimicrobial susceptibility profiles and genetic resistance markers of linezolid-resistant Staphylococcus and Enterococcus isolates
| Strain ID | Source | ST | MIC ( μg/ml) | Genetic resistance markers | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LZD | VAN | TEC | OXA | FOX | ERY | CLI | CHL | TET | CIP | PEN | AMP | GNH | SXT | 23S rRNA mutation (percentage of clones with mutation) | cfr | L3 | L4 | L22 | |||
| S. capitis | |||||||||||||||||||||
| HS09-206 | blood | N/D | 8 | 1 | 0.25 | >256 | R | >256 | >256 | 32 | 4 | 64 | 64 | N/D | N/D | 2/38 | C2131T (43.3%) | - | - | - | - |
| G2603T (70.0%) | |||||||||||||||||||||
| HS09-17 | blood | N/D | 16 | 1 | 0.25 | 256 | R | >256 | 256 | 32 | 4 | 64 | 32 | N/D | N/D | 2/38 | C2131T (46.7%) | - | - | - | - |
| G2603T (83.3%) | |||||||||||||||||||||
| HS10-204 | blood | N/D | 32 | 1 | 0.5 | >256 | R | >256 | >256 | 128 | 4 | 64 | 64 | N/D | N/D | 4/76 | C2131T (36.7%) | - | - | - | - |
| G2603T (66.7%) | |||||||||||||||||||||
| HS10-24 | blood | N/D | 64 | 1 | 0.25 | 256 | R | >256 | >256 | 32 | 4 | 64 | 32 | N/D | N/D | 8/152 | C2131T (53.3%) | - | - | - | - |
| G2603T (80.0%) | |||||||||||||||||||||
| HS11-203 | blood | N/D | 256 | 1 | 0.25 | >256 | R | >256 | >256 | 128 | 4 | 64 | 64 | N/D | N/D | 2/38 | C2131T (60.0%) | + | - | - | - |
| G2603T (80.0%) | |||||||||||||||||||||
| HS11-44 | blood | N/D | 256 | 1 | 0.25 | >256 | R | >256 | 256 | 64 | 4 | 64 | 32 | N/D | N/D | 2/38 | C2131T (53.3%) | + | - | - | - |
| G2603T (80.0%) | |||||||||||||||||||||
| HS12-49 | blood | N/D | 256 | 1 | 0.5 | >256 | R | >256 | >256 | 128 | 4 | 64 | 32 | N/D | N/D | 4/76 | C2131T (70.0%) | + | - | - | - |
| G2603T (70.0%) | |||||||||||||||||||||
| HS12-201 | blood | N/D | 512 | 1 | 0.25 | >256 | R | >256 | >256 | 128 | 4 | 64 | 64 | N/D | N/D | 4/76 | C2131T (60.0%) | + | - | C316T(Arg106 Cys) | - |
| G2603T (100%) | |||||||||||||||||||||
| HS12-51 | blood | N/D | 128 | 1 | 0.5 | 256 | R | >256 | 256 | 32 | 4 | 64 | 64 | N/D | N/D | 2/38 | C2131T (36.7%) | + | - | - | - |
| G2603T (63.3%) | |||||||||||||||||||||
| HS12-53 | blood | N/D | 512 | 1 | 0.25 | >256 | R | >256 | 256 | 128 | 4 | 64 | 32 | N/D | N/D | 8/152 | C2131T (60.0%) | + | - | - | - |
| G2603T (80.0%) | |||||||||||||||||||||
| HS12-55 | blood | N/D | 512 | 1 | 0.25 | >256 | R | >256 | >256 | 128 | 4 | 64 | 32 | N/D | N/D | 4/76 | C2131T (50.0%) | + | - | C316T(Arg106 Cys) | - |
| G2603T (86.7%) | |||||||||||||||||||||
| HS12-58 | blood | N/D | 512 | 1 | 0.25 | >256 | R | >256 | >256 | 64 | 2 | 32 | 8 | N/D | N/D | 4/76 | C2131T (60.0%) G2603T (100%) |
+ | - | - | - |
| HS13-60 | blood | N/D | 256 | 1 | 0.5 | >256 | R | >256 | >256 | 64 | 4 | 64 | 64 | N/D | N/D | 4/76 | C2131T (60.0%) | + | - | - | - |
| G2603T (76.7%) | |||||||||||||||||||||
| HS13-207 | blood | N/D | 32 | 1 | 0.25 | >256 | R | >256 | >256 | 32 | 4 | 64 | 32 | N/D | N/D | 8/152 | C2131T (83.3%) | - | - | - | - |
| G2603T (100%) | |||||||||||||||||||||
| S. aureus | |||||||||||||||||||||
| HS09-205 | sputum | 88 | 8 | 1 | 0.5 | <0.25 | S | >256 | >256 | 32 | 0.5 | 64 | 32 | N/D | N/D | 0.12/2.37 | - | - | - | - | - |
| HS11-202 | sputum | 239 | 16 | 1 | 1 | >256 | R | >256 | >256 | 32 | 4 | 128 | 128 | N/D | N/D | 0.25/4.75 | - | - | C389G(Ala130Gly) | - | - |
| HS12-56 | sputum | 5 | 32 | 1 | 1 | >256 | R | >256 | >256 | 32 | 8 | 128 | 8 | N/D | N/D | 0.12/2.37 | - | + | - | - | - |
| E. faecalis | |||||||||||||||||||||
| HS10-311 | urine | 16 | 4 | 1 | 0.12 | N/D | N/D | >256 | N/D | 32 | 128 | 64 | 2 | <0.5 | R | N/D | - | - | - | - | - |
| HS11-304 | blood | 16 | 4 | 1 | 0.5 | N/D | N/D | >256 | N/D | 32 | 64 | 16 | 2 | <0.5 | S | N/D | - | - | - | - | - |
| HS11-305 | urine | 476 | 4 | 1 | 0.25 | N/D | N/D | >256 | N/D | 32 | 128 | 16 | 4 | <0.5 | R | N/D | - | - | - | - | - |
| HS11-307 | urine | 4 | 4 | 2 | 0.12 | N/D | N/D | >256 | N/D | 32 | 1 | 64 | 0.5 | <0.5 | R | N/D | - | - | - | - | - |
| HS12-309 | urine | 69 | 8 | 2 | 0.12 | N/D | N/D | >256 | N/D | 32 | 64 | 64 | 0.5 | <0.5 | S | N/D | - | - | - | - | - |
| HS12-308 | urine | 480 | 8 | 1 | 0.25 | N/D | N/D | >256 | N/D | 32 | 32 | 64 | 4 | <0.5 | S | N/D | - | - | - | - | - |
| HS12-310 | wound | 300 | 8 | 1 | 0.25 | N/D | N/D | >256 | N/D | 32 | 64 | 64 | 2 | 4 | S | N/D | - | - | - | - | - |
| HS13-301 | urine | 476 | 8 | 1 | 0.25 | N/D | N/D | >256 | N/D | 64 | 128 | 64 | 2 | <0.5 | S | N/D | - | - | - | - | - |
| HS13-302 | bile | 27 | 8 | 0.5 | 0.125 | N/D | N/D | >256 | N/D | 32 | 128 | 64 | 2 | <0.5 | R | N/D | - | - | - | - | - |
| HS13-303 | urine | 480 | 8 | 1 | 0.125 | N/D | N/D | >256 | N/D | 32 | 64 | 16 | 0.5 | <0.5 | S | N/D | - | - | - | - | - |
| RJ13 -314 | urine | 69 | 8 | 1 | 0.12 | N/D | N/D | >256 | N/D | 32 | 64 | 16 | 1 | <0.5 | S | N/D | - | - | - | - | - |
| RJ13 -315 | urine | 4 | 8 | 1 | 0.25 | N/D | N/D | >256 | N/D | 32 | 128 | 64 | 2 | <0.5 | R | N/D | - | - | - | - | - |
| HS13-312 | wound | 37 | 4 | 1 | 0.25 | N/D | N/D | >256 | N/D | 32 | 64 | 16 | 4 | 4 | R | N/D | - | - | - | - | - |
| HS13-313 | urine | 480 | 4 | 1 | 0.25 | N/D | N/D | >256 | N/D | 64 | 128 | 64 | 2 | <0.5 | R | N/D | - | - | - | - | - |
| E. faecium | |||||||||||||||||||||
| HS11-306 | urine | 650 | 4 | 2 | 0.25 | N/D | N/D | >256 | N/D | 32 | 128 | 16 | 4 | 2 | S | N/D | - | - | - | - | - |
ST, sequence typing; R, resistant; S, susceptible; LZD, linezolid; VAN, vancomycin; OXA, oxacillin; FOX, cefoxitin; ERY, erythromycin; CLI, clindamycin; CHL, chloramphenicol; TET, tetracycline; CIP, ciprofloxacin; PEN, penicillin; AMP, ampicillin; GNH, high-level gentamicin; SXT, trimethoprim-sulfamethoxazole; N/D, not determined.
MLST and molecular typing
All of the isolates, except for those from S. capitis, were typed by MLST. The STs of the three S. aureus isolates were ST5, ST88 and ST239. The 14 E. faecalis isolates were classified into eight STs: two isolates each belonged to ST4, ST16, ST69, and ST476, while three were ST480 and three others belonged to different STs. The E. faecium isolate belonged to ST650. PFGE analysis showed that nine S. capitis isolates (HS11-203, HS11-44, HS12-49, HS12-201, HS12-51, HS12-53, HS2-55, HS12-58, HS13-60) exhibited identical band pattern, and four other isolates also shared the same band pattern (HS09-206, HS09-17, HS10-204, HS10-24). HS13-207 showed a unique band pattern (Figure 1). In addition, E. faecalis isolates from the same ST also exhibited different band patterns.
Figure 1.
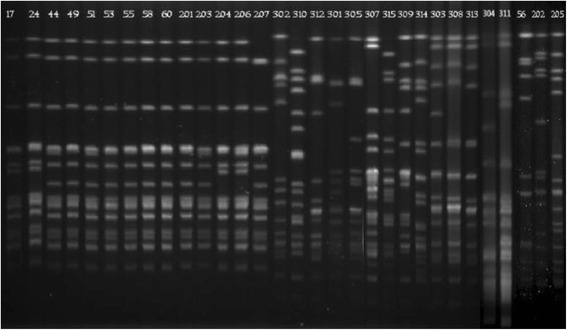
PFGE of Sma I-digested chromosomal DNA of linezolid-resistant isolates. Lane 17–207, fourteen isolates of LR S. capitis. 17, HS09-17; 24, HS10-24; 44, HS11-44; 49, HS12-49; 51, HS12-51; 53, HS12-53; 55, HS12-55; 58, HS12-58; 60, HS13-60; 201, HS12-201; 203, HS11-203; 204, HS10-204; 206, HS09-206; 207, HS13-207. Lane 302–311, fourteen isolates of LR E. faecalis. 302, HS13-302; 310, HS12-310; 312, HS13-312; 301, HS13-301; 305, HS11-305; 311, HS10-311; 307, HS11-307; 315, RJ13-315; 309, HS12-309; 314, RJ13-314; 303, HS13-303; 308, HS12-308; 313, HS13-313; 304, HS11-304; 311, HS10-311. Lane 56–205, three isolates of LR S. aureus. 56, HS12-56; 202, HS11-202; 205, HS09-205.
Resistance genes and mutations
A novel C2131T mutation and a previously reported G2603T mutation, both in domain V of the 23S rRNA gene, were identified in all S. capitis isolates (Table 2). All S. capitis isolates, except HS09-206, HS09-17, HS10-204, HS10-24 and HS13-207, were positive for cfr. Furthermore, S. capitis HS12-201 and HS12-55 had C316T (Arg106Cys) mutations in ribosomal protein L4, but no mutations were detected in ribosomal proteins L3 or L22 in any isolates. Among S. aureus isolates, only HS12-56 was positive for cfr. S. aureus HS11-202 had a C389G (Ala130Gly) mutation in ribosomal protein L3, but proteins L4 and L22 were wild-type. No mutation was identified in any of the studied genes in the E. faecalis and E. faecium isolates, nor was cfr detected.
Gene dosage
Gene dosage tests were performed on fourteen LR S. capitis isolates that contained mutations in domain V of the 23S rRNA gene. Isolates with different linezolid resistance levels displayed diverse C2131T and G2603T mutation rates (Table 2). The relationship between linezolid resistance levels and the percentage of mutated 23S rRNA genes in S. capitis is shown in Figure 2. Clones derived from S. capitis (linezolid MIC 8–64 μg/ml) had mutation rates of 36.7–83.3% (C2131T) and 63.3–100% (G2603T), and those derived from S. capitis (linezolid MIC 256–512 μg/ml) had mutation rates of 50.0–70.0% (C2131T) and 70.0–100% (G2603T).
Figure 2.
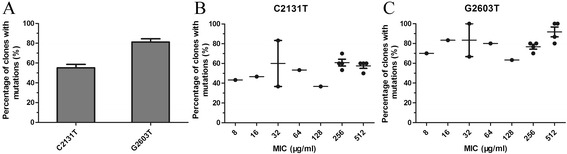
Mutations in the 23S rRNA gene and linezolid resistance levels in LR S. capitis isolates. A. The percentages of clones with C2131T and G2603T mutations. B. The percentage of clones with C2131T mutations in isolates with various linezolid resistance levels. C. The percentage of clones with G2603T mutations in isolates with various linezolid resistance levels. Error bars represent the mean ± SEM.
Genetic environment of cfr in plasmids
Southern hybridisation indicated that cfr resided on plasmids of similar sizes (ca. 54 kb) in all ten cfr-positive isolates (Figure 3). The cfr-carrying plasmids were extracted from the isolates and transformed into S. aureus RN4220. Plasmids were then extracted from all the transformants and were confirmed by Southern hybridisation (data not shown). The linezolid MICs of ten transformants were 32 μg/ml. The genetic environment surrounding cfr in pHS (plasmid from Huashan Hospital) was shown to be identical in all ten cfr-carrying plasmids and showed 99% identity to the corresponding region of plasmid pSS-01 (GenBank accession no. JQ041372, Figure 4). The cfr gene was located downstream of aminoglycoside resistance gene aacA-aphD and was flanked by two copies of the IS256-like element, with a downstream orf1 gene.
Figure 3.
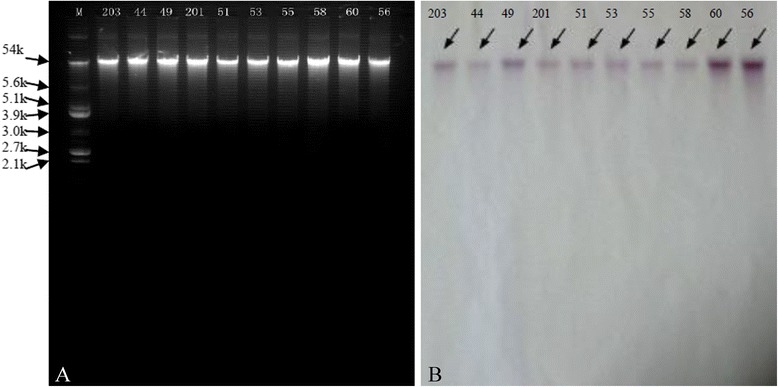
Analysis of plasmids in cfr -positive S. capitis isolates. Plasmids of ca.54 kb were detected in all ten cfr -positive isolates. A. Southern hybridisation of S. capitis isolates with a cfr probe. B. M, E. coli V517 marker; 203, HS11-203; 44, HS11-44; 49, HS12-49; 201, HS12-201; 51, HS12-51; 53, HS12-53; 55, HS12-55; 58, HS12-58; 60, HS13-60; 56, HS12-56.
Figure 4.
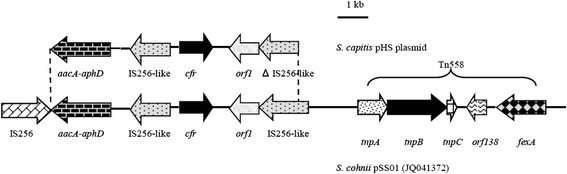
Schematic representation of the genetic environment of cfr in pHS and pSS-01. The arrows indicate the positions and directions of the transcription of the genes. The regions of homology between pHS and pSS-01 (GenBank accession no. JQ041372) are indicated by dashed lines and grey shading. Δ indicates a truncated gene.
Biofilm production
The semi-quantitative biofilm assay showed that the mean biofilm production in LR E. faecalis strains was significantly higher than that in LS E. faecalis strains (P < 0.0001, one-way ANOVA), whereas there were no significant differences between LR and LS isolates from the other three species (Figure 5).
Figure 5.
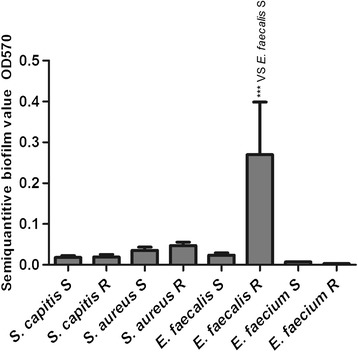
Semi-quantitative biofilm analysis of S. capitis, S. aureus, E. faecalis, E. faecium groups. LS strains were used as controls. Comparisons are vs. LS E. faecalis strains for LR E. faecalis strains. ***P < 0.0001 (one-way ANOVA).
Electron microscopy
TEM showed differences in cell wall thickness between LS, LI and LR strains grown with and without linezolid (Figure 6, Table 3). The overall cell diameter had not significant difference between any of the strains of each species (data not shown), but the cell walls of LI and LR strains were thicker than those of LS strains (P < 0.0001, two-tailed t-test),except for LI E. faecium HS11-306. Furthermore, the cell wall of LR strains grown with linezolid were thicker than that without linezolid (P < 0.0001, two-tailed t-test), except for LR E. faecalis HS12-309 (P ≤ 0.001, two-tailed t-test) (Figure 7).
Figure 6.
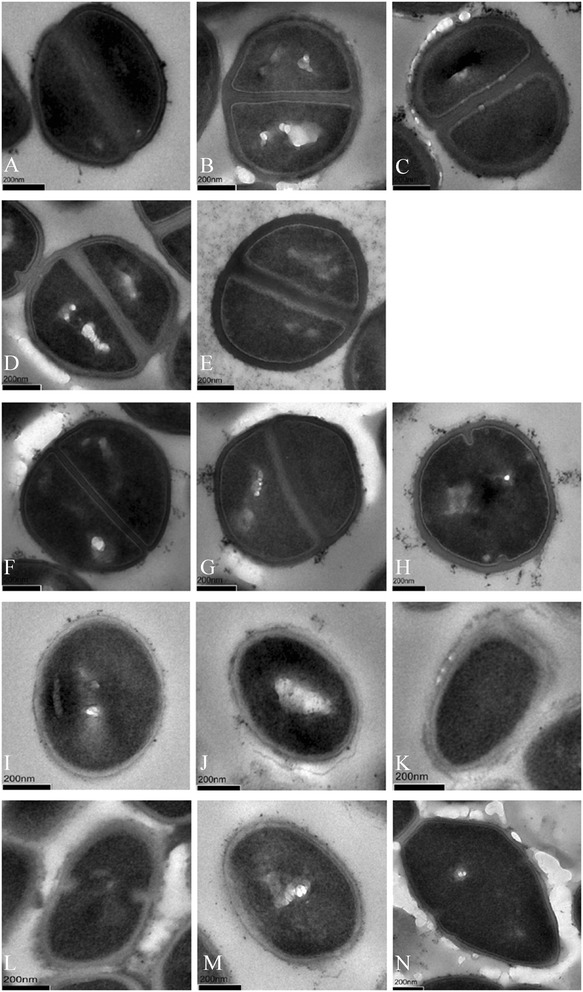
Transmission electron micrographs (TEM) showing cell wall thickness in bacterial isolates. TEM images show LS S. capitis HS12-102 (A), LR S. capitis HS09-17 (MIC 16 μg/ml) grown without (B) and with (C) 4 μg/ml linezolid, LR S. capitis HS12-55 (MIC >256 μg/ml) grown without (D) and with (E) 32 μg/ml linezolid, wild-type ATCC29213 S. aureus (F), LR S. aureus HS11-202 (MIC 16 μg/ml) grown without (G) and with (H) 2 μg/ml linezolid, wild-type ATCC29212 E. faecalis (I), LI E. faecalis HS11-304 (MIC 4 μg/ml) grown without linezolid (J), LR E. faecalis HS12-309 (MIC 8 μg/ml) grown without (K) and with (L) 2 μg/ml linezolid, LS E. faecium HS13-194 (M), LI E. faecium HS11-306 (MIC 4 μg/ml) grown without linezolid (N). Cell wall thicknesses are given in Table 3. Scale bars indicate 200 nm.
Table 3.
Cell wall thickness measured by TEM
| Species | Strains | Cell wall thickness a |
|---|---|---|
| S. capitis | HS12-102 | 30.76 ± 4.10 |
| S. capitis | HS09-17 | 40.12 ± 6.04 |
| S. capitis | HS09-17 + 4 μg/ml linezolid | 57.54 ± 7.75 |
| S. capitis | HS12-55 | 41.13 ± 6.22 |
| S. capitis | HS12-55 + 32 μg/ml linezolid | 50.52 ± 7.78 |
| S. aureus | ATCC 29213 | 25.69 ± 2.17 |
| S. aureus | HS11-202 | 31.74 ± 5.73 |
| S. aureus | HS11-202 + 2 μg/ml linezolid | 43.67 ± 6.30 |
| E. faecalis | ATCC 29212 | 31.01 ± 5.57 |
| E. faecalis | HS11-304 | 40.96 ± 7.41 |
| E. faecalis | HS12-309 | 40.44 ± 5.38 |
| E. faecalis | HS12-309 + 2 μg/ml linezolid | 46.14 ± 7.55 |
| E. faecium | HS13-194 | 32.42 ± 4.60 |
| E. faecium | HS11-306 | 34.79 ± 5.38 |
ameasurements (in nm) are means ± standard deviations from 30 cells per sample.
Figure 7.
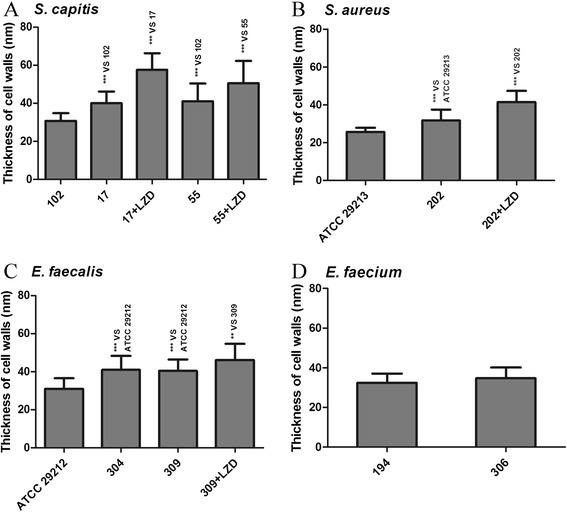
Comparison between bacterial cell wall thickness in LS, LR and LI isolates. There are S. capitis (A), S. aureus (B), E. faecalis (C) and E. faecium (D) isolates. 102, HS12-102; 17, HS09-17; 17 + LZD, HS09-17 grown with 4 μg/ml linezolid; 55, HS12-55; 55 + LZD, HS12-55 grown with 32 μg/ml linezolid; ATCC29213, ATCC29213 LS S. aureus; 202, HS11-202; 202 + LZD, HS11-202 grown with 2 μg/ml linezolid; ATCC29212, ATCC29212 LS E. faecalis; 304, HS11-304; 309, HS12-309; 309 + LZD, HS12-309 grown with 2 μg/ml linezolid; 194, HS13-194; 306, HS11-306 (MIC 4 μg/ml). LS reference strains HS12-102 (S. capitis), ATCC29213 (S. aureus), ATCC29212 (E. faecalis) and HS13-194 (E. faecium) were used as controls for the respective groups. 17 versus 102, 55 versus 102, 17 + LZD versus 17, 55 + LZD versus 55, 202 versus ATCC29213, 202 + LZD versus 202, 304 versus ATCC29212, 309 versus ATCC29212, 309 + LZD versus HS12-309 showed significantly differences (**P ≤ 0.001; ***P < 0.0001, two-tailed t-test).
Discussion
The prevalence of MRSA, VRS and VRE strains has presented a new challenge in antimicrobial medication. Linezolid is one of the few treatment options that is highly active against Gram-positive pathogens. However, LR strains have been increasingly reported in worldwide [2-7,9-14] and now have also emerged in China [8,17-19]. Although the incidence of linezolid resistance among Gram-positive organisms remains low, the emergence of LR strains is still of great concern.
Surveillance data in our study suggested that all the LR S. capitis were recovered from bacteraemia patients, whereas LR S. aureus (LRSA) came from respiratory tract specimens. Linezolid-resistant/intermediate enterococci were collected from blood, urine, wound, bile and prostatic fluid. All of the patients infected with LR staphylococci were critically ill, which is consistent with findings from previous studies [8,17-19]. A study in Spain [31] described that LR strains emerged after patients received three courses of linezolid for 600 mg every 12 h for 14 days. Interestingly, as shown in Table 1, only seven hospitalised patients (HS09-17, HS10-24, HS11-44, HS12-51, HS13-60, HS13-207 and HS11-307) in our study had received linezolid therapy (range 13–40 days) prior to the appearance of LR strains. The remaining patients, other than outpatients whose antibiotic drug histories were not determined, had not been exposed to linezolid. So, except for linezolid exposure, if there were other factors involved worth considering.
Nowadays, linezolid susceptibility of Gram-positive clinical isolates is mainly monitored through two surveillance programmes, the Zyvox Annual Appraisal of Potency and Spectrum (ZAAPS) Program in Europe and the Linezolid Experience and Accurate Determination of Resistance (LEADER) Program in USA. In 2014, the ZAAPS Program analysed the linezolid activity of 7972 g-positive clinical isolates collected over 9 years (2004–2012) from 73 medical centres in 33 countries on five continents [32]. Data showed that 0.11% of CoNS (8/6909 isolates), 0.02% of S. aureus (4/25148 isolates), 0.92% of E. faecalis (4/434 isolates) and 0.03% of E. faecium (1/333 isolates) isolates were resistant to linezolid. Gu et al. [33] summarized the linezolid susceptibility profile of staphylococci from the LEADER Program over 7 years (2004–2010). Data showed that 1.4% of CoNS (73/5202 isolates) and 0.05% of S. aureus (13/23077 isolates) isolates were resistant to linezolid. Our data showed that the incidence of LR strains in China was higher than that of the two surveillance programs, except for LRCoNS which was similar to that of the LEADER program. In particular, the incidence of LRCoNS in China was nine times higher than that of LRSA, and the incidence of LR E. faecalis was 15 times higher than that of LR E. faecium, suggesting that ongoing surveillance is necessary.
Recently, multifocal outbreaks of LR staphylococci have been reported, including S. epidermidis (France, Spain, Mexico, Italy, Brazil, China), S. cohnii (Mexico, China), S. haemolyticus (Mexico, Brazil, China), S. sciuri (China), S. capitis (Greece, China), S. hominis (Brazil), S. lugdenesis (Brazil) and S. aureus (Italy, Hong Kong, Brazil), and linezolid-resistant/intermediate enterococci, including E. faecalis (Poland, Ireland, China, Taiwan), E. faecium (Germany, Ireland, China) and E. avium (Poland) [8,18,19,32,34]. In our study, we detected multiple LR S. capitis, S. aureus, E. faecalis and E. faecium isolates. Notably, the clinically-derived LRSA isolates characterised in our investigation are the first to be reported in China. Previous studies [8,18,19] described that most of LRCoNS in China exhibited high-level resistance (MIC 256 μg/ml), with very few low- and medium-level LR isolates, whereas in other countries [32,34], low to medium-level LRCoNS (8–32 μg/ml) were more common, with a lack of high-level LRCoNS strains. In contrast, the 14 LRCoNS isolates in our study exhibited a wide range of linezolid resistance levels, with MICs from 8 to 512 μg/ml. Three LRSA isolates exhibited linezolid resistance (MIC 8–32 μg/ml), which were higher than LRSA isolates from Italy, Hong Kong and Brazil (MIC 4–8 μg/ml). Thereinto, two of the LRSA isolates were methicillin-resistant and multidrug-resistant, whereas HS09-205 LRSA was methicillin-susceptible. Moreover, low-level resistance in linezolid-resistant/intermediate enterococci (MIC 4–8 μg/ml) was similar to previous studies [8,18,19,32,34]. MLST and PFGE results showed that LR S. capitis, LR S. aureus, E. faecalis and E. faecium were polyclonal, suggesting diverse STs were all vulnerable to linezolid exposure.
Known mechanisms of linezolid resistance in Gram-positive cocci include mutations in the 23S rRNA gene, acquisition of cfr and mutations of ribosomal proteins L3, L4 and L22. The novel mutation C2131T and the previously-described mutation G2603T in the V domain of the 23S rRNA were identified in all 14 LR S. capitis clones. Although G2603T was first reported in 2009 [9], this is the first report of this mutation in China. The novel mutation C2131T, whether it plays a role in linezolid resistance is uncertain and needs a further confirmation. But this revelation may provide new information for investigating the mechanism of linezolid resistance caused by mutations in the V domain of 23S rRNA gene. Because staphylococci possess five or six copies of the 23S rRNA gene, linezolid resistance caused by 23S rRNA mutations develop slowly. Besier et al. indicated that accumulation of single point mutations might be associated with gradually-increasing levels of resistance, in a mechanism known as “gene dosage” [28]. To gain insight into the relationship between gene dosage and 23S rRNA mutations, we examined the proportion of cloned PCR products with mutations. Our data showed that the percentage of clones with G2603T mutation was significantly higher than that with the C2131T mutation. Independent of other resistance mechanisms, various levels of the 23S rRNA mutations mediated low- to medium-level linezolid resistance (HS09-206, HS09-17, HS10-204, HS10-24, HS13-207; MICs: 8–64 μg/ml). Moreover, the synergistic action of mutations in the 23S rRNA gene and ribosomal proteins L3 and L4, as well as acquisition of cfr gene mediated high-level linezolid resistance (MICs: 128–512 μg/ml). However, the complicated background of resistance mechanisms involved and the biological characteristics of the clinical bacteria overlaid the relationship between gene dosage and linezolid resistance levels.
The cfr gene is often located on transferable plasmid and is thus considered to promote horizontal spread of linezolid resistance. In our study, 10 LR staphylococcal isolates (nine S. capitis and one S. aureus) harboured cfr gene and all the S. aureus RN4220 transformants showed that cfr-carrying pHS mediate medium-level linezolid resistance (MIC 32 μg/ml). Wang et al. had indicated the two copies of IS256-like elements played an important role in the dissemination of cfr in animal isolates [17]. Subsequently, similar structures were found in LR S. capitis from Zhejiang [18], LR S. cohnii from Beijing [19], and now LR S. capitis and LR S. aureus from Shanghai in the current study. Notably, the pHS with cfr flanked by IS256-like elements is the first to be reported in a LRSA strain (HS12-56) in this study. Moreover, compared with plasmid S. capitis MHZ (GenBank accession no. JX232067) from Zhejiang [18], S. capitis pHS possessed additional upstream aminoglycoside resistance gene aacA-aphD. Considering the first cfr-positive LR S. capitis strain was detected in 2011 and HS12-56 LRSA which carried the same plasmid emerged in 2012, it suggested that the pHS was most probably transferred from LRCoNS to LRSA. This finding presented significant concerns about the possibility of cfr-positive LRCoNS acting as reservoirs for linezolid resistance. Based on the information above, cfr flanked by two IS256-like elements may be transmitted from animal-derived isolates to human-derived isolates and spread horizontally within, or even between, bacterial species. Therefore, ongoing surveillance is essential to avoid the dissemination of linezolid resistance.
Mutations in ribosomal proteins L3, L4 and L22 of the peptidyltransferase centre also contribute to decreased susceptibility to linezolid [7,12-14]. In our study, mutation C316T (Arg106Cys) in L4 was first detected in two S. capitis clones (HS12-201, HS12-55) and mutation C389G (Ala130Gly) in L3 was first detected in S. aureus (HS11-202). None of the LR isolates contained mutations in L22. Because no other mutation was detected in S. aureus HS11-202, which has a linezolid MIC of 16 μg/ml, suggesting that the Ala130Gly mutation in L3 might be associated with low-level linezolid resistance.
Biofilm could provide a microenvironment for bacteria to survive. Bacteria in a biofilm aggregated into micelles which could withstand attacks by innate host defence mechanisms and evaded the threats of antibiotics [15]. We performed a semi-quantitative biofilm assay, which only showed that biofilm formation was significantly higher in LR E. faecalis isolates than in LS E. faecalis isolates. This suggested that formation of biofilm might increase low-level resistance to linezolid in LR E. faecalis (MICs 4–8 μg/ml).
We performed TEM to determine whether the decreased susceptibility of LI and/or LR strains was related to the thickness of the cell wall. We found that the cell walls of LI and LR strains were significantly thicker than LS strains. Also, the cell walls of LR strains grown with linezolid were thicker than those of LR strains grown without linezolid. These thicked cell wall bacteria were probably selected by the linezolid pressure and might be able to avoid clearance in patients undergoing antibiotic therapy. There was no significant differences in cell wall thickness between LI and LR strains. Data suggested that the thickness of the cell wall might decrease susceptibility to linezolid to a certain degree, but not play a major role in linezolid resistance. Furthermore, no significant differences in cell wall thickness were observed between LS HS13-194 and LI HS11-306 E. faecium isolates, however, we observed a thick transparent substance around the bacteria. Whether this substance might hinder linezolid absorption and result in resistance is unknown.
Conclusion
In summary, various levels of LR strains have emerged in Shanghai, China in our study. Mutiple resistance mechanism were involved in these LR strains. Although the prevalence of resistance to linezolid remains low, the emergence of LR staphylococcal and enterococcal clinical isolates should prompt increased attention, especially for the horizontal dissemination of cfr, and surveillance of linezolid resistance in Gram-positive bacteria is of increasing importance.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grants 81322025, 81171623 and 81371875), Outstanding Young Talent Plan of Shanghai (XYQ2011039), and Shanghai Shuguang Talent Project (12SG03).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YT performed all experiments. TL assisted in MLST and Gene dosage experiments. YZ assisted in PFGE experiment. BW and XZ assisted in antimicrobial susceptibility testing. ML and YT conceived the study and analyzed the results. ML supervised the study and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yueru Tian, Email: vicky8025@126.com.
Tianming Li, Email: litianming1989@yahoo.cn.
Yuanjun Zhu, Email: zhuyuanj@126.com.
Bei Wang, Email: wa0261008@hotmail.com.
Xue Zou, Email: 250030015@qq.com.
Min Li, Email: ruth_limin@126.com.
References
- 1.Swaney SM, Aoki H, Ganoza MC, Shinabarger DL. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob Agents Chemother. 1998;42:3251–3255. doi: 10.1128/aac.42.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsiodras S, Gold HS, Sakoulas G, Eliopoulos GM, Wennersten C. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet. 2001;358:207–208. doi: 10.1016/S0140-6736(01)05410-1. [DOI] [PubMed] [Google Scholar]
- 3.Wilson P, Andrews JA, Charlesworth R, Walesby R, Singer M. Linezolid resistance in clinical isolates of Staphylococcus aureus. J Antimicrobial Chemotherapy. 2003;51:186–188. doi: 10.1093/jac/dkg104. [DOI] [PubMed] [Google Scholar]
- 4.Meka VG, Pillai SK, Sakoulas G, Wennersten C, Venkataraman L. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23SrRNA gene and loss of a single copy of rRNA. J Infect Dis. 2004;190:311–317. doi: 10.1086/421471. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz S, Werckenthin C, Kehrenberg C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother. 2000;44:2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livermore DM, Warner M, Mushtaq S, North S, Woodford N. In vitro activity of the oxazolidinone RWJ-416457 against linezolid -resistant and -susceptible staphylococci and enterococci. Antimicrob Agents Chemother. 2007;51:1112–1114. doi: 10.1128/AAC.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livermore DM, Mushtaq S, Warner M, Woodford N. Activity of oxazolidinone TR-700 against linezolid -susceptible and -resistant staphylococci and enterococci. J Antimicrobial Chemotherapy. 2009;63:713–715. doi: 10.1093/jac/dkp002. [DOI] [PubMed] [Google Scholar]
- 8.Cai JC, Hu YY, Zhang R, Zhou HW, Chen GX. Linezolid-resistant clinical isolates of meticillin-resistant coagulase-negative staphylococci and Enterococcus faecium from China. J Med Microbiol. 2012;61:1568–1573. doi: 10.1099/jmm.0.043729-0. [DOI] [PubMed] [Google Scholar]
- 9.Lincopan N, de Almeida LM, de Araújo MR E, Mamizuka EM. Linezolid resistance in Staphylococcus epidermidis associated with a G2603T mutation in the 23S rRNA gene. Int J Antimicrob Agents. 2009;34:281–282. doi: 10.1016/j.ijantimicag.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 10.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. A new mechanism for chloramphenicol, forfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 11.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locke JB, Hilgers M, Shaw KJ. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob Agents Chemother. 2009;53:5275–5278. doi: 10.1128/AAC.01032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locke JB, Hilgers M, Shaw KJ. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones Linezolid and Torezolid(TR-700) Antimicrob Agents Chemother. 2009;53:5265–5274. doi: 10.1128/AAC.00871-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almeida LM, Araújo MR, Sacramento AG, Pavez M, Souza AG. Linezolid resistance in Brazilian Staphylococcus hominis strains is associated with L3 and 23S rRNA ribosomal mutations. Antimicrob Agents Chemother. 2013;57:4082–4083. doi: 10.1128/AAC.00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 16.Louis BR, x MD. Antimicrobial resistance in Gram-positive bacteria. Am J Med. 2006;119:11–19. doi: 10.1016/j.amjmed.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Zhang W, Wang J, Wu C, Shen Z. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother. 2012;56:1485–1490. doi: 10.1128/AAC.05827-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XJ, Chen Y, Yang Q, Qu TT, Liu LL. Emergence of cfr-harboring coagulase-negative Staphylococci among patients received linezolid therapy in two hospitals of China. J Med Microbiol. 2013;62:845–850. doi: 10.1099/jmm.0.051003-0. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Wu W, Ni M, Liu Y, Zhang J. Linezolid-resistant clinical isolates of enterococci and Staphylococcus cohnii from a multicentre study in China: molecular epidemiology and resistance mechanisms. Int J Antimicrob Agents. 2013;42:317–321. doi: 10.1016/j.ijantimicag.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing; 23rd Informational Supplement M100-S23. Wayne, PA, USA: CLSI; 2013. [Google Scholar]
- 21.Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/S0966-842X(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 22.Nallapareddy SR, Duh RW, Singh KV, Murray BE. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. J Clin Microbiol. 2002;40:868–876. doi: 10.1128/JCM.40.3.868-876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homan WL, Tribe D, Poznanski S, Li M, Hogg G. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol. 2002;40:1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aanensen DM, Spratt BG. The multilocus sequence typing network: mlst.net. Nucleic Acids Res. 2005;33:728–733. doi: 10.1093/nar/gki415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arias CA, Panesso D, Singh KV, Rice LB, Murray BE. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob Agents Chemother. 2009;10:4240–4246. doi: 10.1128/AAC.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendes RE, Deshpande LM, Farrell DJ, Spanu T, Fadda G. Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome, Italy. J Antimicrobial Chemotherapy. 2010;65:2329–2335. doi: 10.1093/jac/dkq331. [DOI] [PubMed] [Google Scholar]
- 27.Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol. 2007;64:1506–1514. doi: 10.1111/j.1365-2958.2007.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Besier S, Ludwig A, Zander J, Brade V, Wichelhaus TA. Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob Agents Chemother. 2008;52:1570–1572. doi: 10.1128/AAC.01098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall SH, Donskey CJ, Hutton-Thomas R, Salata RA, Rice LB. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother. 2002;10:3334–3336. doi: 10.1128/AAC.46.10.3334-3336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Román F, Roldán C, Trincado P, Ballesteros C, Carazo C. Detection of linezolid-resistant Staphylococcus aureus with 23S rRNA and novel L4 riboprotein mutations in a cystic fibrosis patient in Spain. Antimicrob Agents Chemother. 2013;57:2428–2429. doi: 10.1128/AAC.00208-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendes RE, Hogan PA, Streit JM, Jones RN, Flamm RK. Zyvox(R) Annual Appraisal of Potency and Spectrum (ZAAPS) Program: report of linezolid activity over 9 years (2004–12) J Antimicrobial Chemotherapy. 2014;69:1582–1588. doi: 10.1093/jac/dkt541. [DOI] [PubMed] [Google Scholar]
- 33.Gu B, Kelesidis T, Tsiodras S, Hindler J, Humphries RM. The emerging problem of linezolid-resistant Staphylococcus. J Antimicrobial Chemotherapy. 2013;68:4–11. doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flamm RK, Mendes RE, Ross JE, Sader HS, Jones RN. An international activity and spectrum analysis of linezolid: ZAAPS Program results for 2011. Diagn Microbiol Infect Dis. 2013;76:206–213. doi: 10.1016/j.diagmicrobio.2013.01.025. [DOI] [PubMed] [Google Scholar]


