Abstract
We introduce a new method to detect ancient selective sweeps centered on a candidate site. We explored different patterns produced by sweeps around a fixed beneficial mutation, and found that a particularly informative statistic measures the consistency between majority haplotypes near the mutation and genotypic data from a closely related population. We incorporated this statistic into an approximate Bayesian computation (ABC) method that tests for sweeps at a candidate site. We applied this method to simulated data and show that it has some power to detect sweeps that occurred more than 10,000 generations in the past. We also applied it to 1,000 Genomes and Complete Genomics data combined with high-coverage Denisovan and Neanderthal genomes to test for sweeps in modern humans since the separation from the Neanderthal–Denisovan ancestor. We tested sites at which humans are fixed for the derived (i.e., nonchimpanzee allele) whereas the Neanderthal and Denisovan genomes are homozygous for the ancestral allele. We observe only weak differences in statistics indicative of selection between functional categories. When we compare patterns of scaled diversity or use our ABC approach, we fail to find a significant difference in signals of classic selective sweeps between regions surrounding nonsynonymous and synonymous changes, but we detect a slight enrichment for reduced scaled diversity around splice site changes. We also present a list of candidate sites that show high probability of having undergone a classic sweep in the modern human lineage since the split from Neanderthals and Denisovans.
Keywords: selective sweeps, modern humans, Neanderthal, Denisova, approximate Bayesian computation
Introduction
The sequencing of high-coverage archaic human genomes (Meyer et al. 2012; Prüfer et al. 2014) has permitted the identification of nearly all single-nucleotide changes (SNCs) that are fixed derived in present-day humans but ancestral in Denisovans and Neanderthals. However, the question of which of these changes have been driven to fixation by natural selection remains unresolved. In total, 109 of them were identified as leading to amino acid changes in Ensembl genes. However, a change need not have fixed due to selection, and could have instead risen in frequency due to genetic drift or draft (Gillespie 2000). Here, we investigate whether any of the genic or regulatory motif changes that are fixed derived in present-day humans shows population genetic signatures consistent with selection.
Signatures of ongoing selective sweep events include patterns of extended homozygosity (Sabeti et al. 2002; Voight et al. 2006) and reduced linkage disequilibrium (LD) (McVean 2007). However, statistics reliant on a reduction in haplotype homozygosity lose power as the selected allele reaches fixation (Sabeti et al. 2007) and statistics based on the increase in LD around the beneficial mutation (Kim and Nielsen 2004) or on patterns of single-nucleotide variation (Tajima 1989; Fay and Wu 2000) do not persist for long after the sweep ends (Przeworski 2002). This makes it difficult to detect patterns created by ancient selection in modern humans, meaning selection that occurred soon after the separation of modern humans from Neanderthals.
Prüfer et al. (2014) used a hidden Markov model (HMM) to find long tracks of the genome where Neanderthals fall outside of present-day human variation. These regions are likely to have undergone ancient selective sweeps. However, this method does not provide information about which sites were selected. Additionally, the regions inferred to have been selected are not enriched for changes predicted to be highly disruptive based on their biochemical properties (Prüfer et al. 2014).
Przeworski (2003) developed a Bayesian approach to estimate the posterior support for a selective sweep at a fixed candidate site and to estimate the time since fixation. This method uses the number of segregating sites, the number of distinct haplotypes, and Tajima’s D measured on a nearby 104-bp region. Simulations showed that this method was able to detect selective sweeps that occurred within the past 10,000 generations in humans. Hernandez et al. (2011) used a different approach to testing for ancient sweeps. They compared human diversity scaled by human–macaque divergence to look for signatures of selection around fixed human–chimpanzee differences. They found that classic selective sweeps were not abundant during human evolution (but see Enard et al. [2014]). Here, we will exploit similar patterns of homozygosity and haplotype diversity in the linked neutral region surrounding a favored allele that fixed soon after the separation of Neanderthals and modern humans, roughly 12,000–20,000 generations ago (Prüfer et al. 2014).
First, we apply the method used in Hernandez et al. (2011) to different categories of fixed modern-human-specific derived mutations. Then, we explore the performance of several statistics in detecting ancient selective sweeps around a candidate site (see Materials and Methods). We use these statistics to attempt to detect sweeps in different categories of modern-human-specific SNCs. We apply an approximate Bayesian computation (ABC) method to candidate sites listed in Prüfer et al. (2014). To account for local differences in levels of background selection and mutation rates across the genome, we not only scale all our statistics by the ratio of divergences between regions near and far from the site but also compare results obtained in our test regions with results from regions that have similar genomic characteristics, including functional density, recombination rate, and average human–chimpanzee divergence, but that are far from the candidate regions, following Enard et al. (2014).
Results
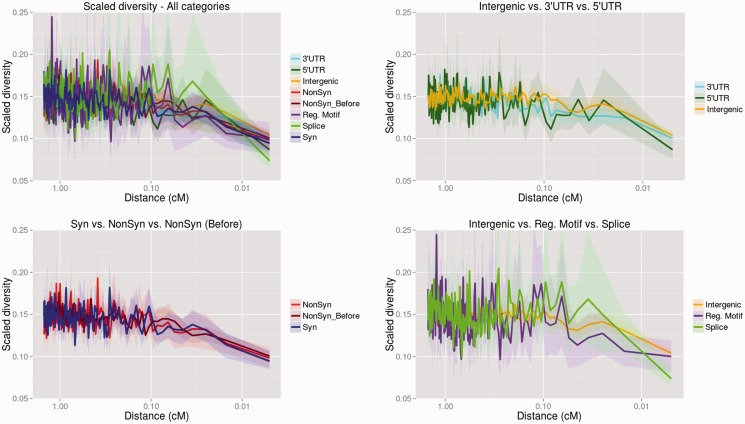
We first looked at human diversity per site scaled by divergence to the human–chimpanzee ancestor in nonoverlapping windows of size 0.01 cM around different types of modern-human-specific SNCs. To represent present-day humans, we used a panel of 200 Yoruba and Luhya phased haploid genomes from the 1000 Genomes Project (1000G; 1000 Genomes Project Consortium et al. 2010; Abecasis et al. 2012), as well as a panel of 13 Yoruba and Luhya high-coverage diploid genomes produced by Complete Genomics (CG; Drmanac et al. 2010) that were phased using Beagle (Browning BL and Browning SR 2013), to obtain 26 phased haploid genomes. The choice of data here seems to make little difference: We observe similar patterns in scaled diversity between functional categories using the 1000G data (fig. 1) and the CG data (supplementary fig. S1, Supplementary Material online). We also observe similar patterns when using smaller windows of size 0.005 cM (supplementary fig. S2 for 1000G data and supplementary fig. S3 for CG data, Supplementary Material online)
Fig. 1.
Human diversity per site (calculated in the 1000G panel) scaled by divergence of the human reference to the human–chimpanzee ancestor around different classes of fixed modern-human-specific single-nucleotide changes where Altai Neanderthal and Denisova are homozygous ancestral. The statistic was calculated in windows of 0.01 cM and the x axis shows distance of the window midpoint to the fixed change on a log-scale. The upper left panel shows all functional categories tested, whereas the other panels show different subsets of these for ease of comparison.
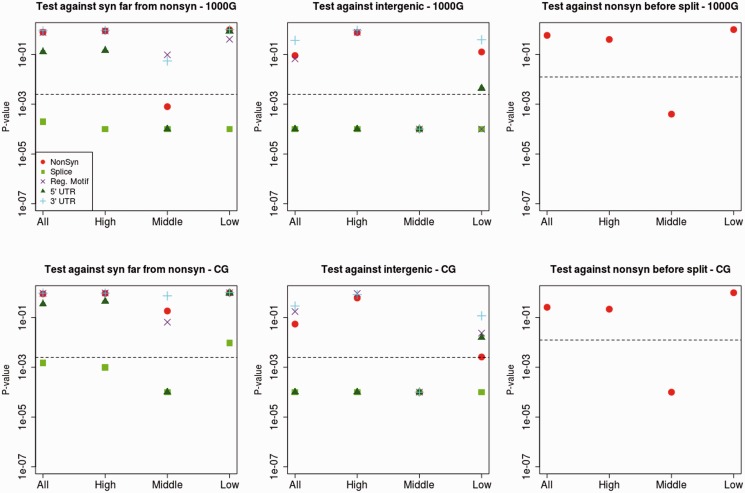
We used a bootstrap-based test (Hernandez et al. 2011) to test for significant troughs (after accounting for multiple testing) in scaled diversity in a 0.02-cM region around nonsynonymous, splice site, untranslated region (UTR), or regulatory motif changes. We compared each category against two categories which are presumably neutral: 1) Synonymous changes located far (>1 Mb) from any nonsynonymous change and 2) intergenic changes. Because we do not expect to see large differences in the entire distribution of changes, we divided the data within each category by different quantiles: All changes, sites in the lowest third quantile of scaled diversity, changes in the middle third quantile of scaled diversity, and changes in the highest third quantile of scaled diversity. We then tested for differences between the same quantiles of each of the two categories under comparison. We were concerned that clustering between sites would somehow bias our results. To address this, we subsampled the changes within each functional category, so that each SNC was more than 100 kb from any other SNC in the same category.
Nonsynonymous changes do not have significantly lower scaled diversity than synonymous changes (P = 0.78 for 1000G data, P = 0.89 for CG data), echoing observations in Hernandez et al. (2011) for human–chimpanzee fixed differences. We observe no significant differences in any quantile comparison, with the exception of the middle third quantile of the 1000G data (fig. 2). Intriguingly, splice site changes have significantly lower scaled diversity than synonymous changes when using the 1000G data (P < 0.0025) for all quantile tests, even after accounting for multiple testing. When using the CG data, splice sites remain significant in three of the four quantiles that are significant when using the 1000G data (fig. 2). We find no reduction in scaled diversity around regulatory motif positions or UTR changes relative to synonymous changes, except for 5′-UTR changes in the middle third quantile. However, the number of regulatory motif changes available for testing is small (n = 21), which may reduce power when testing that particular category.
Fig. 2.
P values from bootstrap-based test (Hernandez et al. 2011) comparing various genomic classes to look for significant differences in modern human diversity per site scaled by divergence to the human–chimpanzee ancestor in a 0.02-cM region around modern-human-specific changes. We tested putatively functional categories (nonsynonymous, splice site and UTR changes) against putatively neutral categories: 1) Synonymous changes far from any nonsynonymous change (left panels) and 2) intergenic changes (middle panels). We also compared nonsynonymous modern-specific changes against nonsynonymous changes that fixed before the modern-Neandertal human population split (right panels). The top panels were produced using the 1000 Genomes Project (1000G) data and the bottom panels were produced using the CG data. The x axis denotes the partitioning of scaled diversity values into quantiles (all sites, highest third, middle third, and lowest third) in each of the two categories under comparison. Black dashed lines denote the Bonferroni-corrected P values (0.05/20 = 0.0025 for left and middle panels; 0.02/4 = 0.0125 for right panels).
When comparing different categories against intergenic changes, we find similar patterns to the test against synonymous changes, with splice site changes and 5′-UTR changes having significantly reduced scaled diversity at most quantiles (fig. 2). However, unlike the test against synonymous changes, the test against intergenic changes need not necessarily reflect patterns of positive selection, as scaled diversity has been found to be reduced around functional regions in humans due to background selection (Hernandez et al. 2011).
When comparing nonsynonymous changes that occurred after and before the modern human–Neanderthal split, we observe a slight reduction in scaled diversity in the “after” category, but this difference is only significant in one quantile (fig. 2).
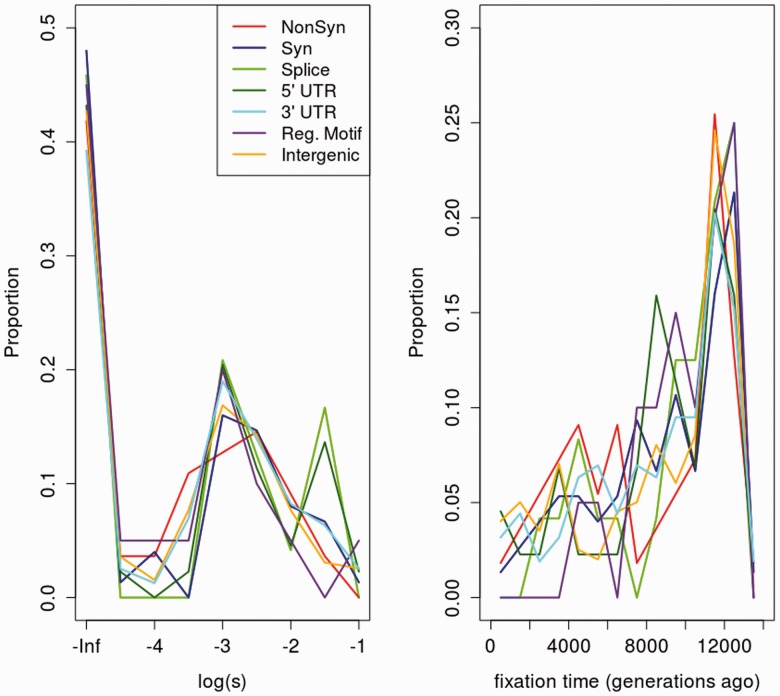
To explore whether we could obtain more information using other signals that are produced by selection, we developed an ABC approach that uses msms (Ewing and Hermisson 2010) to sample from various selective sweep and neutral models. We explored a variety of statistics that were found to be indicative of a selective sweep around a candidate site. Some of these were particularly useful for testing for ancient selection using simulations, especially those relying on the consistency between haplotypes and genotypes in two different populations (see Materials and Methods). We plot the density of estimated posterior modes of the log of the selection coefficient () and the time of fixation of the derived allele (tS) for different classes of fixed modern-human-specific derived SNCs in figure 3. Here, we assume a constant effective population size Ne of 10,000. We observe a slight relative abundance of SNCs with strong estimated selection strength (large s) in splice site changes and, to a lesser extent, 5′-UTR changes. Figure 3 also suggests the majority of fixed changes appear to be neutral or only weakly advantageous () and ancient (large fixation time), regardless of their genomic category.
Fig. 3.
Overlapped histograms of ABC-estimated posterior modes for (left) and the fixation time (right) across different genomic classes, using the 1000G data (PLS = 10). Sites with BF <1 in favor of selection were assigned s = 0, whereas those with BF >1 were assigned the posterior mode of the distribution of s. For the time of fixation, we show the posterior mode inferred from the best-supported model (neutral or selection), based on the same BF cutoff.
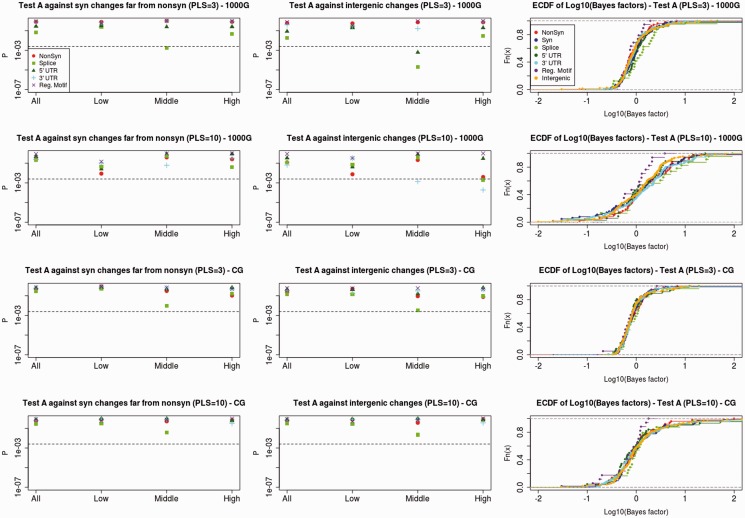
We tested for significantly higher Bayes factors (BF) in favor of a selective sweep model relative to a neutral model at particular genomic categories. We compared the BF distribution of putatively functional SNCs (nonsynonymous, splice site, UTR, and regulatory motif changes) with the BF distribution of putatively neutral SNCs (synonymous and intergenic changes), using a one-tailed Wilcoxon rank-sum test (WRT). As before, because we do not necessarily expect to see differences in the entire distribution of changes, we also partitioned the data within each category by different quantiles and tested for differences in the same quantiles for each of the two categories under comparison. We distinguish two tests: Test A, in which we compare only sites that are good model fits (P > 0.05 for the neutral or the selection model, fig. 4), and Test B, in which we also include sites that are poor model fits (P < 0.05 for both models, supplementary fig. S4, Supplementary Material online). We also used both a small number (3) of Partial Least Squares Discriminant Analysis (PLSDA) components (see Materials and Methods) and a large number (10) of components to check the robustness of our results to the number of components used (see Materials and Methods). We subsampled the SNCs within each category as described above to prevent any effects that could be produced by clustering.
Fig. 4.
We subsampled SNCs within each genomic category so that each SNC was more than 100 kb away from any other. We then tested whether changes in different presumably functional sites have higher BF in favor of selection relative to synonymous changes that are far (>1 Mb) from any nonsynonymous change (left panels) or relative to intergenic changes (middle panels), using a one-tailed WRT. The x axis denotes the partitioning of BF into quantiles (all sites, lowest third, middle third, and highest third) in each of the two categories under comparison. The dashed lines denote the P values cutoff after correcting for multiple testing (P = 0.05/20 = 0.0025). We also show empirical cumulative distribution functions of BF for each category tested (right panels). First row from top: Test A (excluding poor model fits) using 1000G data and first three PLSDA components. Second row: Test A using 1000G data and first ten PLSDA components. Third row: Test A using CG data and first three PLSDA components. Bottom row: Test A using CG data and first ten PLSDA components.
We find no significant increase in BF in favor of a selective sweep model for nonsynonymous changes relative to synonymous changes that are far from any nonsynonymous change (P > 0.05 for all quantile partitions), regardless of which data set or test we use. Splice sites show somewhat elevated signatures of selection, but this is only significant at specific quantiles after accounting for multiple testing. UTR and splice site changes show significantly elevated signatures of selection when tested against intergenic changes at medium and high quantiles (P < 0.0025) when using the 1000G data. However, we cannot exclude weaker background selection in intergenic regions as a possible cause for this.
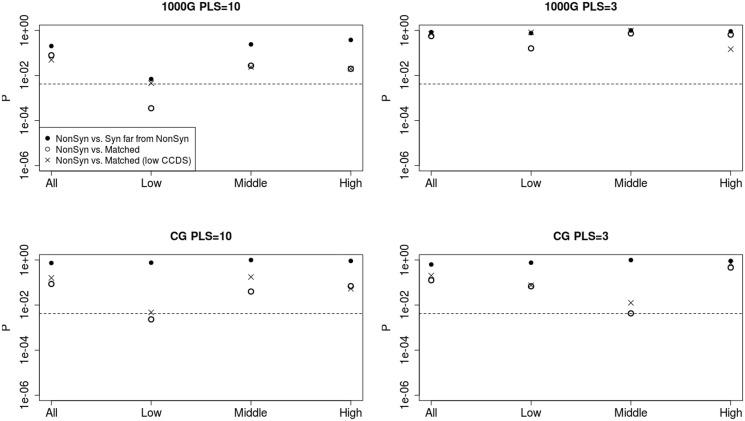
We explored whether we could see significantly larger BF for nonsynonymous changes when comparing their surrounding regions with regions sampled to resemble them in a variety of genomic properties (see Materials and Methods), following Enard et al. (2014) (fig. 5). We compared regions with nonsynonymous changes with their corresponding matched regions. We also filtered for low functional density (i.e., density of conserved coding DNA sequence [CDS] smaller than median of all regions with nonynonymous SNCs) and compared only these regions with their corresponding matched regions. We see that, in general, P values are smaller than in the synonymous versus nonsynonymous test, but only a few quantiles are significant after multiple-test correction (fig. 5). We note, however, that the number of sites used for testing is considerably smaller than the number of sites used in Enard et al. (2014), when looking at selection along the entire human lineage since the split from chimpanzees, and so our power to distinguish positive from background selection is much lower.
Fig. 5.
P values from one-tailed WRTs to look for significantly higher BF at nonsynonymous changes relative to regions matched to the regions containing the nonsynonymous changes in a variety of genomic properties (open circles) and matched regions filtered to have low CCDS density (crosses), using either the first 10 (left) or 3 (right) PLS-DA components of the data. The x axis denotes the partitioning of BF into quantiles (all sites, lowest third, middle third, and highest third). Upper row: Test using 1000 Genomes data. Lower row: Test using CG data. In all panels, we also show P values corresponding to nonsynonymous changes tested against synonymous changes far from any nonsynonymous change (filled circles), for comparison. The dashed black lines correspond to the Bonferroni-corrected P value for each test (P = 0.05/12 = 0.0042). PLS, number of PLS-DA components used for BF estimation.
In supplementary table S1, Supplementary Material online, we list all putatively functional (nonsynonymous, splice site, regulatory motif, and UTR) SNCs that have BF >10 in favor of selection using either the 1000G or the CG data sets. We also require that P > 0.05 for the selection model for both data sets. No regulatory motif SNC passes these cutoffs. Many of the changes in this list are close to each other in the genome and so share signatures of population variation, which may be due to only one causative change in a given region (they were pruned when subsampling the data to test for differences between functional categories). In table 1, we present a reduced version of supplementary table S1, Supplementary Material online, showing only the change with the highest BF for each gene in supplementary table S1, Supplementary Material online. We also reiterate that BF for these changes are only based on comparing a model of a classic selective sweep against a model of neutrality for the candidate site, without explicitly modeling background selection, soft sweeps, or other forms of selection that may be operating in the region.
Table 1.
Modern-Human-Specific Changes That Lead to an Amino Acid Replacement, Affect a Splice Site or Are Located in an UTR, and That: 1) Have BF > 10 in Favor of Selection Using Either the 1000G and CG Data Sets and 2) Are a Good Fit (P > 0.05) to the Selection Model Using Both the 1000G and CG Data Sets.
| Position | Log(BF) | Log(s) (1K) | Log(s) (CG) | tS (1K) | tS (CG) | Class | Gene |
|---|---|---|---|---|---|---|---|
| chr1:38423232 | 1.05 | −1.95 | −2.6 | 9,476 | 10,322 | 3′-UTR | SF3A3 |
| chr1:78183739 | 1.96 | −1.03 | −1.99 | 11,596 | 7,777 | Splice | USP33 |
| chr1:114516356 | 4.76 | −1.47 | −0.62 | 5,094 | 11,878 | 3′-UTR | HIPK1 |
| chr1:162750208 | 1.21 | −1.95 | −3.85 | 11,737 | 9,333 | 3′-UTR | DDR2 |
| chr3:9428211 | 1.44 | −1.59 | −3.77 | 6,648 | 8,767 | 3′-UTR | THUMPD3 |
| chr3:28503157 | 1.55 | −1.35 | −2.04 | 11,596 | 4,384 | 3′-UTR | ZCWPW2 |
| chr3:47316797 | 1.16 | −1.99 | −0.58 | 11,313 | 12,302 | 3′-UTR | KIF9 |
| chr3:47386060 | 1.05 | −2.08 | −0.66 | 12,303 | 11,029 | 3′-UTR | KLHL18 |
| chr3:52009091 | 1.41 | −1.59 | −2.48 | 11,737 | 3,535 | 5′-UTR | ABHD14B |
| chr3:52109349 | 1.21 | −1.87 | −2.36 | 11,879 | 11,171 | 3′-UTR | POC1A |
| chr4:103936040 | 1.17 | −1.39 | −3.37 | 8,486 | 2,828 | 5′-UTR | SLC9B1 |
| chr4:139983298 | 2.52 | −2.28 | −0.66 | 10,182 | 11,736 | 5′-UTR | ELF2 |
| chr4:73930626 | 1.06 | −1.23 | −3.45 | 10,041 | 7,212 | Splice | COX18 |
| chr5:86564477 | 1.14 | −1.27 | −1.99 | 10,748 | 11,171 | NonSyn | RASA1 |
| chr7:73113999 | 2.18 | −1.47 | −1.19 | 7,638 | 9,474 | 3′-UTR | STX1A |
| chr9:127282609 | 1.23 | −1.71 | −1.91 | 10,324 | 10,888 | 3′-UTR | NR6A1 |
| chr10:102724515 | 1.17 | −2.4 | −3.81 | 11,879 | 12,160 | 3′-UTR | FAM178A |
| chr10:15254162 | 1.01 | −2.16 | −3.77 | 9,900 | 12,302 | 3′-UTR | FAM171A1 |
| chr11:64900743 | 1.17 | −1.47 | −2.32 | 11,455 | 9,333 | 5′-UTR | SYVN1 |
| chr11:66406503 | 1.17 | −1.39 | −1.91 | 8,345 | 4,667 | 5′-UTR | RBM4 |
| chr11:66453702 | 1.3 | −1.27 | −1.95 | 7,073 | 8,060 | 3′-UTR | SPTBN2 |
| chr11:129769974 | 1.64 | −1.19 | −1.47 | 12,161 | 10,322 | 3′-UTR | PRDM10 |
| chr13:41132149 | 1.06 | −2.36 | −3.13 | 12,161 | 9,191 | 3′-UTR | FOXO1 |
| chr13:52301811 | 1.48 | −1.19 | −1.63 | 6,083 | 9,757 | Splice | WDFY2 |
| chr16:66947064 | 1.14 | −3.09 | −1.55 | 10,465 | 7,212 | NonSyn | CDH16 |
| chr16:66968760 | 1.3 | −2.48 | −1.75 | 8,062 | 7,212 | 5′-UTR | CES2 |
| chr17:27959258 | 2 | −4.01 | −2.04 | 11,879 | 8,060 | NonSyn | SSH2 |
| chr20:33337529 | 2.24 | −3.69 | −2.76 | 6,648 | 11,029 | NonSyn | NCOA6 |
| chr20:35412323 | 1.43 | −1.11 | −1.39 | 8,203 | 6,505 | 3′-UTR | SOGA1 |
| chr22:40724058 | 2.34 | −1.83 | −1.99 | 9,052 | 6,646 | 3′-UTR | TNRC6B |
| chr22:40760978 | 1.08 | −1.95 | −0.94 | 6,790 | 6,080 | NonSyn | ADSL |
Note.—Parameters listed are the posterior modes inferred using ABC. The BF shown for each site is the maximum across the two data sets. When two or more SNCs pass our cutoffs and are located in the same gene, we only show the SNC with the highest BF here, but show all SNCs in supplementary table S1, Supplementary Material online. tS is in generations. All logs are base 10. 1K, 1000 Genomes.
We verified that sites with high BF in favor of selection did not lie in regions with high probability of having been introgressed from Neandertals into modern humans, as one would not expect this for modern-human-specific selective events. To do so, we retrieved the inferred probability of Neandertal ancestry (PNA) in a panel of European and Asian present-day humans from Sankararaman et al. (2014) at the nearest informative SNP of each tested fixed SNCs. We plotted PNA as a function of each SNC’s inferred selective coefficient or its BF (supplementary fig. S5, Supplementary Material online). Though we did not use Eurasians in our calculation of summary statistics, sites with large BF have low probability of Neandertal ancestry in Eurasians: PNA < 0.047 for all SNCs with BF > 10, and the mean PNA at SNCs with BF > 10 is equal to 28–88% of the mean PNA at all tested fixed SNCs and 23–39% of the mean PNA at all informative SNPs (depending on the data set and number of PLS-DA components used).
The largest BF in favor of the selection model is found in a 3′-UTR SNC in the HIPK1 gene, coding for a kinase that is involved in antioxidative stress response (Ecsedy et al. 2003; Sekito et al. 2006) and the regulation of eyeball size and retinal formation during embryonic development. Another 3′-UTR with a large BF in favor of selection is located in STX1A, a gene encoding a syntaxin involved in ion channel regulation and synaptic exocytosis (Hu et al. 2002; Stein et al. 2009). We also find the 5′-UTR SNCs with large BF in RBM4, coding for a protein involved in the response to hypoxia (Uniacke et al. 2012).
Among the nonsynonymous changes, we find an SNC with large BF that leads to an amino acid change (Ala-to-Val) in the C-terminal domain of adenylosuccinate lyase (ADSL), coding for an enzyme involved in purine metabolism (Šebesta et al. 1997; Gitiaux et al. 2009). This gene has been previously identified as belonging to the Human Phenotype Ontology (Robinson and Mundlos 2010) categories “aggressive behavior” and “hyperactivity,” which are particularly enriched for amino acid replacements in the modern human lineage (including the one in ADSL) (Castellano et al. 2014). Additionally, we observe a nonsynonymous SNC in RASA1, which has been involved in vascular malformations (Hershkovitz et al. 2008) and a splice site SNC in WDFY2, which has an important role in endocytosis (Hayakawa et al. 2006). We also observe a change with high BF in a splice site found in USP33, coding for a deubiquinating enzyme that may play a role in centrosome duplication (Li et al. 2013).
Discussion
We tried to find differences in signatures of positive selection around different categories of modern-human-specific SNCs, where Neanderthals and Denisovans carry the ancestral allele. We evaluated the sensitivity and specificity of a variety of different statistics and implemented them in an ABC method. We attempted to correct for differences in mutation rates and background selection by scaling our statistics by the divergence to an outgroup species (Hernandez et al. 2011; Sattath et al. 2011) and using carefully matched regions (Enard et al. 2014), but did not explicitly model differences in background selection across the genome. A future avenue of research could be to include these differences into our modeling approach. We also only focused on signatures of selection predicted to be left by hard sweeps and so did not consider cases of soft sweeps or polygenic adaptation. Finally, we have not explored more complex demographic scenarios in the modern human population, due to the impossibility of generating selective allele trajectories in msms that allow for population size changes and migration, while conditioning on the time of fixation.
We do not detect a significant difference in patterns of positive selection between nonsynonymous and synonymous changes, regardless of whether we merely look at differences in scaled diversity or whether we use the more sophisticated ABC method. There are three possible reasons for this: a) Hard selective sweeps at nonsynonymous sites were not a predominant adaptive process in the modern human lineage, as has been argued with respect to the entire human lineage since the human–chimpanzee ancestor (Hernandez et al. 2011); b) hard sweeps were common but selection was too weak to be detectable with our method; or c) strong variation in the intensity of background selection along the genome is occluding the signal. Enard et al. (2014) argue a comparison between regions centered on nonsynonymous and synonymous changes will be biased against finding evidence for positive selection, because regions with synonymous changes will be enriched for genes under strong constraint and therefore under strong background selection. Given that fixations that are exclusive to the modern human lineage had a small period of time to rise in frequency, it is likely that a large proportion of nonsynonymous changes arose in regions of low constraint. Taking background selection into account may thus be especially important in this case.
We found that when controlling for patterns of background selection, a slight enrichment for positive selection at nonsynonymous sites becomes more apparent, though only marginally significant at specific quantiles, after accounting for multiple testing. This lends some support to hypothesis (c), but we do not think we have enough data to reject the null hypothesis of rarity of classic sweeps in the lineage that is specific to modern humans.
Splice site SNCs show significantly reduced scaled diversity relative to both intergenic and synonymous changes, suggesting a possibly important role for alternative splicing in recent human evolution. Our ABC approach echoes this pattern, but yields significant results only in a few of the quantiles tests. Additionally, regulatory motif positions appear not to show reduced scaled diversity, suggesting either that our sample size for these regions is too low or that other types of regulatory changes may need to be tested to look for selection at nongenic sequences.
Among the changes with highest BF in favor of the selection model, we find sites in genes involved in various biological processes including metabolism, heart development, and ion channel regulation. These changes are promising candidates for selection in the modern human lineage. However, further computational and experimental analyses will be needed to verify whether any of them was important in recent human evolution.
Materials and Methods
Data
We sought to look for signatures of positive selection around autosomal candidate SNCs that were fixed derived in 1000G present-day humans (1000 Genomes Project Consortium et al. 2010) and homozygous ancestral in Denisova and Altai Neanderthal (Meyer et al. 2012; Prüfer et al. 2014) and that passed quality filters detailed in Prüfer et al. (2014). We filtered for sites that were 5 cM away from any centromeric or telomeric boundary. We classified these sites by different types of genomic consequences using the Ensembl Variant Effect Predictor v2.5 (McLaren et al. 2010), yielding 83 nonsynonymous SNCs, 103 synonymous SNCs, 35 SNCs in splice sites, 295 SNCs in 3′-UTR, 73 SNCs in 5′-UTR, and 21 SNCs in regulatory motif positions. As a negative control, we also tested 300 randomly sampled modern-human-specific SNCs in intergenic regions, where we expect selection to be less prominent than in genic or regulatory regions. We also tested 300 randomly sampled nonsynonymous changes that are fixed derived in present-day humans, Denisovans, and Neanderthals and that are far from any modern-human-specific nonsynonymous SNC to determine whether signatures of selection after the split are significantly stronger than before the split, due to the recency of postsplit sweeps.
To represent present-day humans in the calculation of summary statistics, we used the genomes of human individuals who belong to populations that show little to no evidence of Neanderthal or Denisovan introgression, unlike Eurasians (Green et al. 2010) and Melanesians (Reich et al. 2010). We obtained phased genotypes from two different data sets. First, we used a panel of 100 phased Yoruba sequences and 100 phased Luhya sequences from Phase 1 of the 1000G (1000 Genomes Project Consortium et al. 2010; Abecasis et al. 2012). These sequences were obtained by combining low-coverage whole-genome shotgun sequencing and high-coverage exome capture sequencing.
Second, we used a panel of nine Yoruba diploid genomes and eight Luhya diploid genomes produced by whole-genome sequencing (Drmanac et al. 2010), and made available by CG (http://www.completegenomics.com/public-data/, last accessed February 25, 2014). We computationally phased these data using Beagle 4 (Browning BL and Browning SR, 2013) to obtain a total of 26 phased haploid genomes. These sequences have high coverage (51–89×) and low error rates (one miscalled variant per 100 kb) (Drmanac et al. 2010). To improve accuracy in phasing, we used all 54 diploid genomes from across the globe that belong to the published CG panel, but restricted to the phased Yoruba and Luhya genomes for subsequent analyses. Although the 1000G data set contains a larger number of individuals than the CG data set, the 1000G data set may cause biases in the calculation of summary statistics due to increased coverage at exonic regions, whereas the latter data should not produce such biases.
To account for variability in recombination rates, we transformed distances in base pairs to distances in centimorgans using the HapMap II recombination map (Myers et al. 2005).
Diversity Scaled by Divergence
We first applied the method developed in Sattath et al. (2011) and Hernandez et al. (2011) to look at signatures of selection in different classes of modern-human-specific SNCs. Briefly, for a sample of size n sequences, with major allele frequency p, we calculated diversity per site () scaled by the divergence per site from the human reference to the human–chimpanzee ancestor and analyzed in nonoverlapping windows of size 0.01 or 0.005 cM, throughout a 3-cM region centered around the candidate changes. Divergence was calculated using Ensembl EPO primate alignments (Paten, Herrero, Beal, et al. 2008; Paten, Herrero, Fitzgerald, et al. 2008). To increase power, we produced a folded version of the plots from Sattath et al. (2011), combining windows that were equidistant from the candidate site but on opposite sides of it (fig. 1 and supplementary fig. S2 for 1000G data, figs. S1 and S3 for CG data, Supplementary Material online). We performed 100 bootstraps in each genomic category to obtain 95% confidence intervals.
To test for significant differences in scaled diversity in the immediate neighborhood of the candidate sites among different genomic categories, we computed pairwise one-tailed P values based on 10,000 bootstraps of presumably neutral (e.g., synonymous) changes tested against putatively functional classes of changes, as described in Hernandez et al. (2011), in a 0.02-cM-wide region centered on the candidate site. To increase power, we divided each region at the position of the fixed SNC, treating the two 0.01-cM-wide regions on opposite sides of a fixed SNC as distinct observations (effectively folding the signal as above). To prevent biases caused by clustering of SNCs of the same type, we subsampled the changes within each functional category, so that each SNC was more than 100 kb from any other SNC in the same category.
P values were estimated as , where N is the total number of bootstraps and i is the number of bootstraps in which the scaled diversity around neutral (e.g., synonymous) SNCs was lower than the scaled diversity around presumably functional (e.g., nonsynonymous) SNCs. Because we used 10,000 bootstraps, the minimum possible P value is therefore 0.00009998. As we expect only a small proportion of sites within each category to be positively selected, if at all, we also repeated these tests after filtering for different quantiles of scaled diversity in each of the two categories under comparison (fig. 2).
Simulations
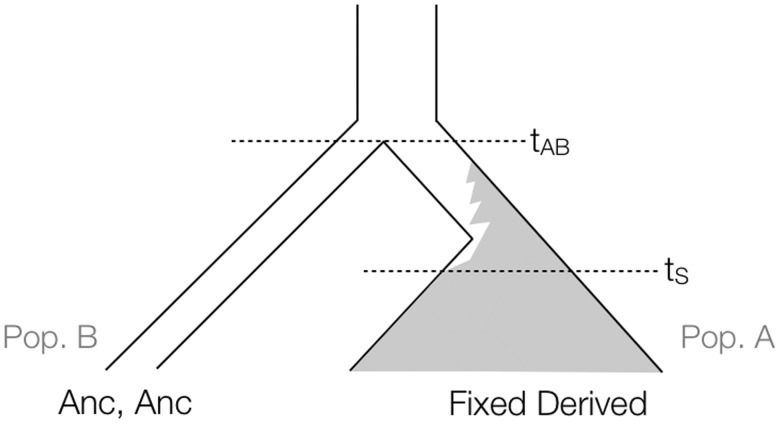
We explored how well different statistics perform in detecting signatures of ancient hard selective sweeps. We used msms (Ewing and Hermisson 2010) to simulate a history of two populations (A and B) with a selective sweep event exclusive to population A, conditioned on the time of completion of the sweep (fig. 6). The mutation rate was set to μ at per base-pair per generation and the recombination rate to ρ at per base-pair per generation. We also assumed that
Fig. 6.
Tree representing msms runs to simulate a change in a site that is homozygous ancestral in an archaic human (Pop. B) and rises to fixation in modern humans (Pop. A). tAB, modern-archaic split time; tS, derived allele fixation time.
1) the split time between the two populations is known;
2) the selected site is fixed derived in population A; and
3) two copies of the candidate site have been sampled from population B and they are both ancestral.
These conditions are meant to reflect a situation in which a candidate site of interest is fixed derived in a population with a large number of sequenced individuals—for example present-day humans—but also is homozygous ancestral in a closely related population from which only one high-quality (unphased) genome is available—for example Neanderthals. Both populations are of constant size, , and the number of sampled haploid individuals from population A is equal to 200 (1000G-like simulation) or 26 (CG-like simulation).
Because msms does not allow for backward simulations containing both a population split and a selective sweep conditioned on the time the sweep ends, we used a combination of simulations to generate the desired gene genealogies. First, we produced a trajectory under selection in population A, specifying the magnitude of the selection coefficient (s) and the time the selected allele reached fixation (tS) in units of generations. Then, we simulated another trajectory for the allele in population B without selection, starting from the time the two populations split and setting the initial frequency of the derived allele equal to the frequency of the derived allele in population A at the time the two populations split. Finally, we simulated a two-population history forward-in-time using the two trajectories generated before-hand, under constant population sizes. For a given set of parameters, we used rejection sampling to condition on having observed two copies of the ancestral allele at the candidate site in population B.
We note that this method allows for cases in which the selected allele arises before the split time, if the fixation time is set sufficiently far in the past. In such a case, selection would have operated both in population A and in the ancestral population, but not in population B after the separation from A. Thus, the derived allele would have been either lost during B’s history or segregating in B but not sampled in the present.
Statistics
We simulated a 5-cM region around a candidate site and observed the behavior of different statistics in a smaller core region surrounding the site. We define four summary statistics calculated on blocks of a particular number X of SNPs, which we use to detect footprints of ancient selection for a favored allele that is fixed in population A.
HE: Population diversity () per SNP in population A, averaged over a block of X adjacent SNPs.
HM: Haplotype majority frequency in a block of X adjacent SNPs in population A
HS: Haplotype frequency sample skewness in a block of X adjacent SNPs in population A. Sample skewness was calculated as where m3 is the sample third central moment of haplotype counts and m2 is the sample variance.
HI: Inconsistency of the majority haplotype in a block of X adjacent SNPs in population A with the diploid genotype corresponding to the same set of SNPs observed in the two (unphased) sequences from population B (equal to 0 if the majority haplotype in A can be obtained from the diploid genotype in B and equal to 1 otherwise).
HE, HM, HS, and HI were calculated on blocks of X SNPs with a (X − 1) SNP overlap with the immediately adjacent blocks on either side. We tested a range of numbers for the size X of the block: 1, 2, 4, or 8 SNPs. We averaged the values of each statistic for all blocks within nonoverlapping 0.1-cM windows in the neighborhood of the selected site (2.5 cM downstream and 2.5 cM upstream). We explored a range of selection regimes (), times since fixation (), and number of present-day human sequences sampled (200 to mimic the 1000G data and 26 to mimic the CG data). We then observed the behavior of the average per-window value of the statistics in 200 simulations all run under the same selection coefficient and time since fixation (supplementary fig. S6, Supplementary Material online). t is measured in units of generations, so with Ne = 10,000, t = 0.325 corresponds to 13,000 generations. We assumed populations A and B split 16,000 generations ago.
HE and HM are meant to measure the reduction in SNP and haplotype diversity, as a consequence of a completed selective sweep. HS is meant to account for the fact that mutations occurring some time after the sweep may decrease the frequency of the majority haplotype (lowering HM), but will increase the skewness in the haplotype frequency distribution, due to an abundance of singleton and low-frequency haplotypes. As predicted by deterministic and coalescent theory (Maynard-Smith and Haigh 1974; Kaplan et al. 1989), the observed signature of reduced genomic variation extends for a region of approximately bp in size, so, for example, in the case of s = 0.1, the reduction in HE can be seen in a region approximately bp long soon after the sweep completes (supplementary fig. S6, Supplementary Material online).
The statistic HI is particularly interesting because it uses information from the recently diverged population in which the sweep did not occur (population B). In the case of selection exclusive to modern humans, population B corresponds to archaic humans, for example, Neanderthals. This statistic has most power at intermediate values of tS. We hypothesize the reason is that an ancient selective sweep creates a star-like genealogy early in the history of population A. Consequently, the majority haplotype will resemble the ancestral haplotype, because most mutations occurring after the sweep will be private to distinct lineages within population A and thereby not contribute much to the majority haplotype. In contrast, a recent sweep will drive a single haplotype that may have already accumulated some mutations specific to population A to high-frequency and this haplotype will therefore not resemble the ancestral haplotype. Thus, this statistic allows us to gain information otherwise not available about the time since completion of the sweep. When calculating HI on real data, we used the Altai Neanderthal genome to obtain the archaic genotype.
For parameter inference methods described below, we standardized the values of the statistics in the core sweep region relative to local patterns of variation. We calculated the difference between the average value of each statistic X in an internal region (Int[X]) that extends 0.02 cM to either side of the candidate site and the average value in an external region (Ext[X]) that extends from 0.6 cM up to 2.5 cM on either side of the candidate site. We then divided this difference by the standard deviation (SD) of the statistic in the external region. In addition, we multiplied this ratio by the ratio of the divergence of the human reference to the human–chimpanzee ancestor in the internal region (Int[DNC]) over the same divergence in the external region (Ext[DNC]). In this way, we aim to control for differences in mutation rates between the external and internal regions. As an example, the standardized version of the HE statistic, , is obtained as follows:
| (1) |
Equivalent transformations were made to HM, HS and HI to obtain , and .
We also took simple ratios of over for each statistic, controlling for divergence to the human–chimpanzee ancestor in the internal region (by either multiplying or dividing by the divergence ratio, depending on the statistic), but without accounting for the SD of these values in the external region. We labeled this simple ratio as , for a given statistic X. For example:
| (2) |
All and statistics and their expected behavior under positive selection are listed in table 2.
Table 2.
Summary Statistics Mentioned in Main Text.
| Name | Formula | Behavior Near Positively Selected Allele (relative to neutrality) |
|---|---|---|
| Positive | ||
| Positive | ||
| Negative | ||
| Positive or negative depending on s, tS and distance from selected site | ||
| Larger than 1 | ||
| Larger than 1 | ||
| Smaller than 1 | ||
| Larger or smaller than 1 depending on s, ts and distance from selected site |
Note.—Only the top four were used in the ABC analysis. See main text for explanation of abbreviations.
Performance in Rejecting Neutrality
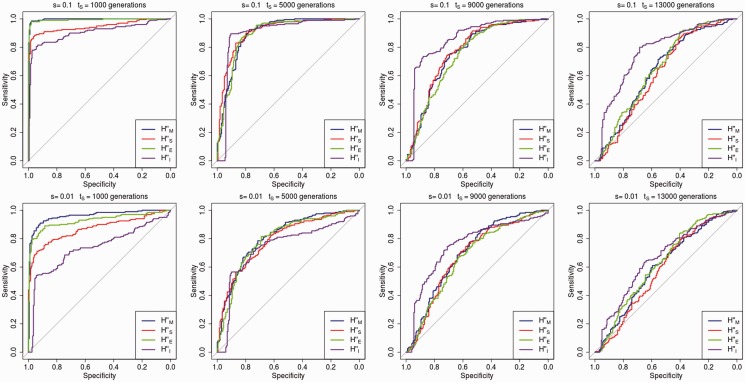
We tested the power of each of the statistics to reject neutrality using simulations. We calculated the fraction of selective sweep simulations (out of 200) where the statistic of interest reaches more extreme values than 90% of the values reached by the same statistic in 200 simulations under neutrality. For the case when 200 sequences are available (like in the 1000G panel), supplementary figure S7, Supplementary Material online, shows power curves comparing simulations under selection with particular fixation times (x axis) against simulations under neutrality in which the neutral allele fixed at the same time. Supplementary figure S8, Supplementary Material online, shows a slightly different way to compute power, where instead of comparing selective and neutral simulations with the same fixation time, we compared selective simulations with particular fixation times against a combination of neutral simulations where the allele may have fixed recently or anciently. Supplementary figures S9 and S10, Supplementary Material online, show the corresponding power curves for the case when 26 sequences are available (like in the CG panel). Though the diversity, skewness and haplotype majority statistics perform well for recent sweeps, the statistic appears to be the best performing statistic when the sweep is ancient (especially for blocks of size 4 and 8 SNPs). This suggests might be useful in distinguishing ancient from recent sweeps, as it reaches its maximum value at an intermediate value of tS.
In all analyses below, we chose to use the normalized statistics () rather than the ratio statistics () because accounting for the SD of the statistics over a neutral region serves to control for regional differences in mutation rates which we did not model in our simulations. We calculated receiving operator characteristic (ROC) curves to compare the specificity and sensitivity of these statistics under different parameters, comparing selective and neutral simulations with the same fixation times. Figure 7 (1000G-like data) and supplementary figure S11 (CG-like data), Supplementary Material online, show that, for recent sweeps, and perform best, but their performance is worse than that of when the sweep is ancient (approximately generations).
Fig. 7.
ROC curves showing performance in rejecting neutrality for different statistics (with SNP blocks of size 4) under different selection coefficients and times since fixation, when 200 modern human sequences are available (like in the 1000G data). Note that the specificity and sensitivity of H″I (relative to the other statistics) are higher than the specificity and sensitivity of other statistics when the sweep is ancient.
Parameter Estimation Using ABC
We wanted to estimate two parameters of interest: The time since fixation in population A in coalescent units (tS) and the logarithm base 10 of the selection coefficient of the favored allele (). We implemented an ABC method of parameter estimation and model testing, similar to Peter et al. (2012) and Garud et al. (2013), using msms and the package ABCtoolbox (Wegmann et al. 2010). We assumed a human–chimpanzee population split time tHC = 5 coalescent units and a modern–archaic human population split time tHN = 0.5 coalescent units.
We used uniform prior distributions to sample parameters of interest:
Here, θ equals , where Ne is the effective population size and μ is the mutation rate per generation in a bp region around the selected site. The statistics we use are, however, largely insensitive to the overall mutation rate, because we only look at relative differences in variation between two regions, controlling for the SD in variation for a given θ. We fixed the recombination rate at per base pair per generation, so that the total simulated region is equivalent to 5 cM.
For each set of sampled parameters, we simulated using rejection sampling until we observed two copies of the ancestral allele at the candidate site in population B. The upper bound on the prior for tS is a heuristic limit meant to keep the sampling step from becoming inconveniently long. As tS increases and approaches the split time of populations A and B, it becomes very hard to sample neutral or weakly selected allele trajectories conditional on them being ancestral in population B. In other words, neutral or weakly selected alleles that are ancestral in at least two members of population B (and are therefore either segregating or fixed ancestral in the ancestral presplit population) are very unlikely to go to fixation fast enough in population A. Consequently, it takes a very long time to obtain trajectories where the sweep finishes shortly after the population split time. Furthermore, figure 7 shows that the upper bound we use for the time since fixation tS (14,000 generations) coincides with the time at which the sensitivity to distinguish selection from neutrality becomes small, for any of the statistics we consider.
We used , , , and (calculated over 4 and 8-SNP blocks) as summary statistics around the candidate site in two regions of different length (0–0.02 cM on either side and 0–0.2 cM on either side), in addition to two other versions of these statistics calculated by defining interior regions located away from the site: 0.02–0.04 cM on either side, 0.04–0.06 cM on either side. As before, the external regions are defined to extend 0.6–2.5 cM away from the candidate site, on either side. SDs for all statistics were calculated over all 4-SNP blocks in the external regions. The values of , , , and throughout the entire parameter space explored are shown in supplementary figure S12 as a function of tS and in supplementary figure S13 as a function of , Supplementary Material online. One can clearly observe that does not decrease monotonically in absolute value as tS increases but tends to be negative for recent sweeps and positive for ancient sweeps. also shows a small increase at slightly older sweeps relative to very recent sweeps, presumably due to the increase in haplotype skewness as a consequence of singleton haplotypes occurring some time after the sweep.
We linearized all statistics using Box–Cox transformation (Box and Cox 1964). We extracted the first ten orthogonal components that best explained the variance in parameter space using Partial Least-Squares (PLS) regression (Tenenhaus 1998) trained on 1,000 simulations (Boulesteix and Strimmer 2007; Wegmann et al. 2009). Supplementary figure S14, Supplementary Material online, shows that only an extremely small decrease in root mean squared error can be gained by using more components. This figure also shows that our statistics are sensitive to the parameters of interest, but insensitive to θ (as expected), so we chose not to try to estimate the latter. For model choice, we used the first ten PLSDA components instead (Tenenhaus 1998; Lê Cao et al. 2009; Peter et al. 2012). We also reran all our tests but using a smaller number (3) of PLSDA and PLS components to test the robustness of our results to the number of components used.
We produced 10,000 simulations under the specified priors and, for each site we considered, kept the best 100 simulations with the smallest Euclidean distance to the observed PLS components. To estimate parameters, we used the “standard” estimation method implemented in ABCtoolbox, with a postsampling regression adjustment (Leuenberger and Wegmann 2009; Wegmann et al. 2010). In order to reject neutrality, we also ran 10,000 simulations under the same priors except for s, which was set to 0. For each site tested, we calculated a BF, defined as the ratio of the marginal probability of the observed data under selection over the marginal probability of the observed data under neutrality, assuming a prior hypothesis of equal probability for the two models. We kept population sizes constant across all populations because of the impossibility of generating variable population size trajectories for population A conditioned on the time since fixation in msms.
We also repeated inferences but assuming a smaller (5× reduced) size for population B, relative to population A, starting immediately after the population split, which is roughly consistent with heterozygosity patterns and pairwise sequentially Markovian coalescent demographic inferences obtained using the Neanderthal and Denisovan genomes in Prüfer et al. (2014). Under this model, we observe qualitatively similar trends to the constant-size model, but focus on results from the latter in the Results and Discussion.
We applied the ABC method developed above to the modern-human-specific SNCs in each category. We excluded from our analysis any changes:
a) That were located within centromeres or telomeres or within less than 5 cM from their boundaries.
b) Whose corresponding central or nearby interior regions lacked information about the chimpanzee-ancestor allele state or had low local constraint or high local mutation rate (), as they artificially inflate the magnitude of our statistics beyond the values simulated in our ABC method.
In one version of our testing procedure (Test A), we also excluded sites that were bad fits to both the selection and the neutral models (i.e., changes with P < 0.05 for both models). This amounted to the exclusion of between 4% and 22% of the sites that passed filter b), depending on the functional category considered. In a different version (Test B), we also include these sites.
Evaluation of ABC Performance
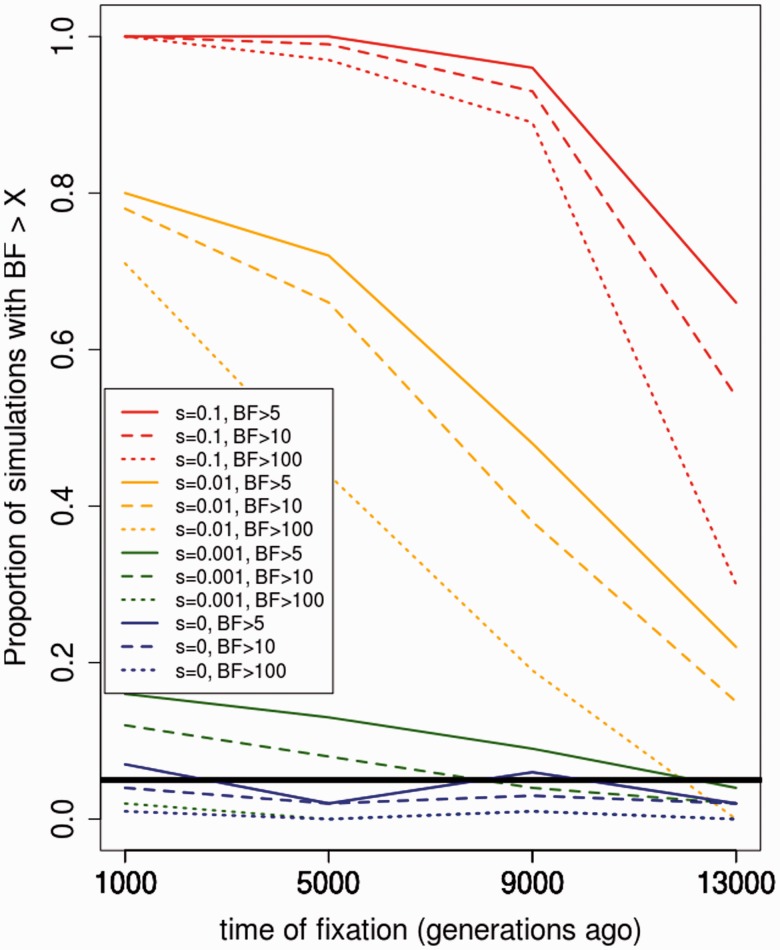
We evaluated the performance of the ABC method by generating sets of 100 simulations under known parameters, in all cases with θ fixed at 3,700 for the entire 5-Mb region, and then running the ABC pipeline to both obtain BF in favor of selection and infer parameters of interest: s and tS. Predictably, BF are generally positive when s is large and tS is small and then decrease for weaker selection and older sweeps (fig. 8 for the case when 200 sequences are available, supplementary figure S15 for the case when 26 sequences are available, supplementary figure S16 for the case when two data sets are available, Supplementary Material online—one with 200 sequences and one with 26 sequences, as in table 1). Importantly, the proportion of simulations with large BF is very small in the case of neutrality (<0.05 for a BF cutoff of >10 or >100), meaning that the proportion of false positives under neutrality should also be small. The accuracy of inferred parameters is similarly dependent on the strength and recency of selection, as can be seen in supplementary figure S17 for and in supplementary figure S18 for tS, Supplementary Material online, assuming 200 sequences are available. We note that the distribution of estimated values of selection when s = 0.001 looks very similar to the neutral distribution, suggesting that we cannot distinguish weak selection from neutrality. Supplementary figures S19 and S20, Supplementary Material online, show equivalent plots for the case when 26 sequences are available.
Fig. 8.
Sets of 100 simulations were run through the ABC pipeline to obtain BF in favor of selection (vs. neutrality) under different known parameters (PLSDA = 10). The colored lines show the proportion of the simulations that have a BF larger than the specified cutoffs, when 200 present-day human sequences are available. The thick black line denotes the 0.05 significance cutoff. s, selection coefficient; t, time since derived allele fixation, in generations.
We also wished to verify whether we were picking up similar signatures of selection as in the HMM selective sweep screen of Prüfer et al. (2014). To do so, we obtained the 100 most disruptive modern-human-specific SNCs in the HMM regions and the 100 most disruptive modern-human-specific SNCs genome–wide. Diruptiveness was determined using a combined annotation score developed in Kircher et al. (2014) and used in Prüfer et al. (2014). As expected, when comparing the two lists, our ABC method infers significantly larger BF in favor of positive selection in the HMM SNCs, relative to the genome–wide SNCs (supplementary fig. S21 when using the first three PLS/PLSDA components, supplementary fig. S22 when using the first ten components, Supplementary Material online).
Controlling for Fine-Scale Differences in Background Selection
We ran our ABC method on carefully sampled regions that matched the internal regions corresponding to nonsynonymous SNCs in a variety of genomic properties, using a method similar to the one developed in Enard et al. (2014). This way, we aimed to mimic the patterns of background selection found around the nonsynonymous changes. For each region corresponding to a nonsynonymous change, we first sampled 2,000 regions of the genome that did not overlap with the 0.04 cM internal region corresponding to that change but that had the same physical length. We also required that we had human–chimpanzee ancestor information (Ensembl EPO) (Paten, Herrero, Beal, et al. 2008; Paten, Herrero, Fitzgerald, et al. 2008) for more than two-thirds of the bases in each sample region and that the average human–chimpanzee divergence in each sample region be within 75% and 125% of the divergence in the corresponding test region. We then sequentially applied the following filters, removing regions that did not pass them: No overlap with any of the test regions, similar B score (McVicker et al. 2009) (top 10% best-matching), similar GC content (top 25% best-matching), similar recombination rate (top 25% best-matching), similar genomic content (40–400% of the “conserved CDS” [CCDS] density inside the test region [Enard et al. 2014], 33–500% of the UTR density inside the test region, >33% of the CCDS density surrounding the test region). For each of the test regions, we randomly selected three sample regions that passed all filters.
We tested the distributions of the sampled regions against the test regions for significant differences, using a WRT. The distributions for divergence (P = 0.69), B scores (P = 0.65), GC content (P = 0.3), recombination rate (P = 0.85), and genomic content (P = 0.52) are not significantly different. The P value for GC content is somewhat low because of an excess of high-GC regions which is difficult to match. For those criteria that did not involve fixed percent ranges, but that instead consisted in top best-matching criteria, we show the distribution of the genomic property in the sampled and in the test regions, after applying the filter (supplementary fig. S23, Supplementary Material online). We were not able to sample regions that matched all criteria for six regions with nonsynonymous changes, so we excluded these regions from subsequent analyses. We also subsampled both the real and the matching regions before testing, to avoid confounding effects due to clustering.
Supplementary Material
Supplementary figures S1–S23 and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Joshua Schraiber, Benjamin Peter, Melinda Yang, Rasmus Nielsen, Michael Lachmann, Svante Pääbo, Janet Kelso, Aida Andrés, Flora Jay, Cesare de Filippo, Kelley Harris, and two anonymous reviewers for helpful advice and discussions. This work was supported by the National Institutes of Health (R01-GM40282 to M.S.). The authors used the Extreme Science and Engineering Discovery Environment (XSEDE), which was supported by National Science Foundation grant number ACI-1053575.
References
- Abecasis G, Auton A, Brooks L, DePristo M, Durbin R, Handsaker R, Kang H, Marth G, McVean G. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulesteix A-L, Strimmer K. Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Brief Bioinform. 2007;8(1):32–44. doi: 10.1093/bib/bbl016. [DOI] [PubMed] [Google Scholar]
- Box GEP, Cox DR. An analysis of transformations. J R Stat Soc Series B Stat Methodol. 1964;26(2):211–252. [Google Scholar]
- Browning BL, Browning SR. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194(2):459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano S, Parra G, Sánchez-Quinto FA, Racimo F, Kuhlwilm M, Kircher M, Sawyer S, Fu Q, Heinze A, Nickel B, et al. Patterns of coding variation in the complete exomes of three Neandertals. Proc Natl Acad Sci U S A. 2014;111(18):6666–6671. doi: 10.1073/pnas.1405138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327(5961):78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Ecsedy JA, Michaelson JS, Leder P. Homeodomain-interacting protein kinase 1 modulates Daxx localization, phosphorylation, and transcriptional activity. Mol Cell Biol. 2003;23(3):950–960. doi: 10.1128/MCB.23.3.950-960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard D, Messer PW, Petrov DA. Genome-wide signals of positive selection in human evolution. Genome Res. 2014;24:885–895. doi: 10.1101/gr.164822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing G, Hermisson J. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics. 2010;26(16):2064–2065. doi: 10.1093/bioinformatics/btq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu C-I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155(3):1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud NR, Messer PW, Buzbas EO, Petrov DA. Drosophila melanogaster. arXiv preprint arXiv:1303.0906. 2013. Soft selective sweeps were the primary mode of recent adaptation. [Google Scholar]
- doi: 10.1038/nature09534. 1000 Genomes Project Consortium et al. 2010. A map of human genome variation from population-scale sequencing. Nature 467(7319):1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics. 2000;155(2):909–919. doi: 10.1093/genetics/155.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitiaux C, Ceballos-Picot I, Marie S, Valayannopoulos V, Rio M, Verrieres S, Benoist JF, Vincent MF, Desguerre I, Bahi-Buisson N. Misleading behavioural phenotype with adenylosuccinate lyase deficiency. Eur J Hum Genet. 2009;17(1):133–136. doi: 10.1038/ejhg.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. A draft sequence of the Neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa A, Leonard D, Murphy S, Hayes S, Soto M, Fogarty K, Standley C, Bellve K, Lambright D, Mello C, et al. The WD40 and FYVE domain containing protein 2 defines a class of early endosomes necessary for endocytosis. Proc Natl Acad Sci U S A. 2006;103(32):11928–11933. doi: 10.1073/pnas.0508832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Kelley JL, Elyashiv E, Melton SC, Auton A, McVean G, 1000 Genomes Project. Sella G, Przeworski M. Classic selective sweeps were rare in recent human evolution. Science. 2011;331(6019):920–924. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershkovitz D, Bercovich D, Sprecher E, Lapidot M. Rasa1 mutations may cause hereditary capillary malformations without arteriovenous malformations. Br J Dermatol. 2008;158(5):1035–1040. doi: 10.1111/j.1365-2133.2008.08493.x. [DOI] [PubMed] [Google Scholar]
- Hu K, Carroll J, Fedorovich S, Rickman C, Sukhodub A, Davletov B. Vesicular restriction of synaptobrevin suggests a role for calcium in membrane fusion. Nature. 2002;415(6872):646–650. doi: 10.1038/415646a. [DOI] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. The “hitchhiking effect” revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Nielsen R. Linkage disequilibrium as a signature of a selective sweep. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Cao K-A, González I, Dejean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25:2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger C, Wegmann D. Bayesian computation and model selection without likelihoods. Genetics. 2009;184(1):243–252. doi: 10.1534/genetics.109.109058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, D’Angiolella V, Seeley ES, Kim S, Kobayashi T, Fu W, Campos EI, Pagano M, Dynlacht BD. Usp33 regulates centrosome biogenesis via deubiquitination of the centriolar protein cp110. Nature. 2013;495(7440):255–259. doi: 10.1038/nature11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith J, Haigh J. The hitch-hinking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl Api and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. The structure of linkage disequilibrium around a selective sweep. Genetics. 2007;175(3):1395–1406. doi: 10.1534/genetics.106.062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5(5):e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338(6104):222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18(11):1814–1828. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B, Herrero J, Fitzgerald S, Beal K, Flicek P, Holmes I, Birney E. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008;18(11):1829–1843. doi: 10.1101/gr.076521.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BM, Huerta-Sanchez E, Nielsen R. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 2012;8(10):e1003011. doi: 10.1371/journal.pgen.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505(7481):43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M. Estimating the time since the fixation of a beneficial allele. Genetics. 2003;164:1667–1676. doi: 10.1093/genetics/164.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468(7327):1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Mundlos S. The human phenotype ontology. Clin Genet. 2010;77(6):525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507(7492):354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattath S, Elyashiv E, Kolodny O, Rinott Y, Sella G. Pervasive adaptive protein evolution apparent in diversity patterns around amino acid substitutions in Drosophila simulans. PLoS Genet. 2011;7(2):e1001302. doi: 10.1371/journal.pgen.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šebesta I, Krijt J, Kmoch S, Hartmannova H, Wojda M, Zeman J. Adenylosuccinase deficiency: clinical and biochemical findings in 5 Czech patients. J Inherit Metab Dis. 1997;20(3):343–344. doi: 10.1023/a:1005361408031. [DOI] [PubMed] [Google Scholar]
- Sekito A, Koide-Yoshida S, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Dj-1 interacts with hipk1 and affects h2o2-induced cell death. Free Radic Res. 2006;40(2):155–165. doi: 10.1080/10715760500456847. [DOI] [PubMed] [Google Scholar]
- Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal snare complex into the membrane. Nature. 2009;460(7254):525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhaus M. 1998. La Régression PLS: Théorie et Pratique. Paris (France): Technip. [Google Scholar]
- Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, Lee S. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486(7401):126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recentpositive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D, Leuenberger C, Excoffier L. Efficient approximate Bayesian computation coupled with Markov chain Monte Carlo without likelihood. Genetics. 2009;182(4):1207–1218. doi: 10.1534/genetics.109.102509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L. Abctoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics. 2010;11(1):116. doi: 10.1186/1471-2105-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.