Background: SLC25A33 and SLC25A36 are two human uncharacterized proteins encoded by the mitochondrial carrier SLC25 genes.
Results: Recombinant SLC25A33 and SLC25A36 transport cytosine, uracil, and thymine (deoxy)nucleotides with different efficiency.
Conclusion: SLC25A33 and SLC25A36 are mitochondrial transporters for pyrimidine (deoxy)nucleotides.
Significance: SLC25A33 and SLC25A36 are essential for mitochondrial DNA and RNA metabolism; other two members of the SLC25 superfamily responsible for 12 monogenic diseases were thoroughly characterized.
Keywords: Liposome, Membrane Transporter Reconstitution, Mitochondria, Mitochondrial Transport, Nucleotide, SLC25 Family, SLC25A33, SLC25A36, Mitochondrial Carrier
Abstract
The human genome encodes 53 members of the solute carrier family 25 (SLC25), also called the mitochondrial carrier family, many of which have been shown to transport inorganic anions, amino acids, carboxylates, nucleotides, and coenzymes across the inner mitochondrial membrane, thereby connecting cytosolic and matrix functions. Here two members of this family, SLC25A33 and SLC25A36, have been thoroughly characterized biochemically. These proteins were overexpressed in bacteria and reconstituted in phospholipid vesicles. Their transport properties and kinetic parameters demonstrate that SLC25A33 transports uracil, thymine, and cytosine (deoxy)nucleoside di- and triphosphates by an antiport mechanism and SLC25A36 cytosine and uracil (deoxy)nucleoside mono-, di-, and triphosphates by uniport and antiport. Both carriers also transported guanine but not adenine (deoxy)nucleotides. Transport catalyzed by both carriers was saturable and inhibited by mercurial compounds and other inhibitors of mitochondrial carriers to various degrees. In confirmation of their identity (i) SLC25A33 and SLC25A36 were found to be targeted to mitochondria and (ii) the phenotypes of Saccharomyces cerevisiae cells lacking RIM2, the gene encoding the well characterized yeast mitochondrial pyrimidine nucleotide carrier, were overcome by expressing SLC25A33 or SLC25A36 in these cells. The main physiological role of SLC25A33 and SLC25A36 is to import/export pyrimidine nucleotides into and from mitochondria, i.e. to accomplish transport steps essential for mitochondrial DNA and RNA synthesis and breakdown.
Introduction
The human genome harbors as many as 53 SLC25 genes encoding a superfamily of transport proteins called solute carrier 25 (SLC25)3 family or mitochondrial carrier family (MCF) (1, 2). The members of this family are characterized by three tandem repeated sequences of ∼100 amino acids, each folded into two transmembrane α-helices and containing a distinct signature motif (PROSITE PS50920, PFAM PF00153, and IPR00193). In recent years many of these transporters have been investigated at the molecular and biochemical levels. They transport inorganic anions, amino acids, carboxylic acids, nucleotides, and coenzymes across the inner mitochondrial membrane and in a few cases across other membranes (1, 3–5). The importance of mitochondrial carriers is demonstrated by their wide distribution in all eukaryotes, their role in numerous metabolic pathways and cell functions, and the identification of several diseases caused by alterations of their genes (1, 2, 6–8). However, despite the research efforts of many laboratories, the transport properties and the substrate(s) transported by ∼20 human mitochondrial carriers remain as yet unknown.
The two human mitochondrial carriers SLC25A33 and SLC25A36, encoded by the SLC25A33 and SLC25A36 genes, respectively, are the close relatives of the Saccharomyces cerevisiae Rim2p. Deletion of RIM2 causes total loss of mtDNA and lack of growth on non-fermentative carbon sources (9). Furthermore, Rim2p has been demonstrated by direct assays in reconstituted liposomes to transport all pyrimidine ribo- and deoxyribonucleotides with similar efficiency as well as guanine ribo- and deoxyribonucleotides but not adenine nucleotides (10). Most information concerning SLC25A33, which was previously named PNC1, derives from studies in human cells in which PNC1 was silenced or overexpressed. PNC1 overexpression enhanced cell size, mitochondrial TTP levels, and diminished reactive oxygen species, whereas its knockdown caused depletion of mtDNA, reduced oxidative phosphorylation, cell size, and mitochondrial UTP levels, and increased reactive oxygen species levels (11–13). Furthermore, analysis of nucleotide flow in cells with down-regulation of PNC1 and treated with labeled uridine or thymidine revealed a slower mitochondrial uptake of uridine triphosphate and a slower release of thymine nucleotides to the cytoplasm (13). Finally, it has recently been found that Drosophila S2R+ cells, silenced for drim2 (the Drosophila melanogaster homolog of the S. cerevisiae Rim2p), contained markedly reduced pools of both purine and pyrimidine dNTPs in mitochondria, whereas cytosolic pools were unaffected (14). Until now the biochemical characterization of purified PNC1 has been addressed in a single publication in which the recombinant SLC25A33, reconstituted into liposomes, was shown to transport pyrimidine nucleoside triphosphates with preference for UTP (11). However, in this study only a limited number of potential substrates have been tested, and, for example, it is unknown whether nucleoside mono- and di-phosphates are transported by SLC25A33.
The high homology existing between SLC25A33 and SLC25A36 has suggested that these two members of the MCF might have similar functions (1, 2). However, the substrate(s) transported by SLC25A36 has not yet been discovered, and its function is still completely elusive. This study reports the identification and functional characterization of SLC25A36 as well as a much more in-depth characterization of SLC25A33 transport activity as compared with that reported by Floyd et al. (11). The SLC25A33 and SLC25A36 genes were overexpressed in Escherichia coli, and the gene products were purified and reconstituted into liposomes. Recombinant SLC25A33 was shown to transport uracil, thymine, and cytosine (deoxy)nucleoside di- and tri-phosphates and SLC25A36 cytosine and uracil (deoxy)nucleoside mono-, di-, and triphosphates. Both SLC25A33- and SLC25A36-catalyzed transports were saturable and inhibited by mercurial compounds. The green fluorescent protein (GFP) fused to SLC25A36 was found to be targeted to mitochondria as previously shown for SLC25A33 (11). Furthermore, expression of SLC25A33 or SLC25A36 proved to restore the phenotypes of S. cerevisiae cells lacking RIM2, encoding the yeast well characterized mitochondrial pyrimidine nucleotide carrier (10). As pyrimidine (deoxy)nucleotide transporters, SLC25A33 and SLC25A36 are essential for the synthesis and breakdown of mitochondrial DNA and RNA by providing the precursors and removing the products of these processes.
EXPERIMENTAL PROCEDURES
Materials
Radioactive compounds were supplied from PerkinElmer Life Sciences. All (deoxy)nucleotides were obtained from Sigma.
Sequence Search and Analysis
Protein and genomic data bases (www.ncbi.nlm.nih.gov) were screened with the protein sequence of SLC25A33 (accession number NP_115691.1) and SLC25A36 (accession number NP_001098117.1) using BLASTP and TBLASTN. The amino acid sequences were aligned with ClustalW (Version 1.8).
Construction of Expression Plasmids
The coding sequences of SLC25A33 and SLC25A36 (accession numbers NM_032315.2 and NM_001104647.1, respectively) were amplified by PCR from human testis and brain cDNA, respectively. The oligonucleotide primers were synthesized corresponding to the extremities of the coding sequences, with additional NdeI and HindIII (for SLC25A33) or BamHI and EcoRI (for SLC25A36) restriction sites as linkers. The amplified products were cloned into the pRUN (SLC25A33) and pMW (SLC25A36) vectors for expression in E. coli.
The RIM2-pRS42H plasmid was constructed by cloning a DNA fragment consisting of the 381 bp upstream of the RIM2 open reading frame (ORF), the RIM2 ORF, and the 352 bp downstream of the RIM2 ORF (amplified from S. cerevisiae genomic DNA by PCR using primers with additional BamHI and SacI sites) into the episomal vector pRS42H. For the preparation of the SLC25A33-pRS42H and SLC25A36-pRS42H plasmids, a chimera consisting of the 381 bp upstream of the RIM2 ORF, the coding sequence of SLC25A33 or SLC25A36 with a His6 tag before the termination codon and the 352 bp downstream of the RIM2 ORF was constructed. Each of the three fragments was amplified using long primers consisting of the sequences corresponding to the extremities of the template to be amplified and an additional sequence corresponding to the extremity of the fragment to be linked. After amplification and purification, the three fragments were mixed together and used in an overlapping PCR. The first 5 cycles of PCR were run without primers to generate the complete overlapping template, then forward primers (first 25 nucleotides of the chimera with HindIII site) and reverse primers (last 25 nucleotides of the chimera with SacI site) were added, and further 25 cycles of PCR were run. The two chimeras containing either the coding sequence of SLC25A33 or SLC25A36 were checked for correct overlapping and sequence errors, then they were digested with HindIII and SacI and cloned into the pRS42H plasmid. All the plasmids, prepared as above, were transformed into E. coli DH5α cells. Transformants were selected on 2X YT plates containing ampicillin (100 μg/ml) and screened by direct colony PCR and by restriction digestion of purified plasmids. The sequences of the inserts were verified.
Bacterial Expression and Purification of Recombinant SLC25A33 and SLC25A36
The expression of recombinant proteins was carried out at 37 °C in E. coli strain BL21(DE3) (15, 16). Control cultures with the empty vector were processed in parallel. Inclusion bodies were purified on a sucrose density gradient (17) and washed at 4 °C first with TE buffer (10 mm Tris/HCl, 1 mm EDTA, pH 7.0) then twice with a buffer containing 3% (w/v) Triton X-114, 1 mm EDTA, and 10 mm PIPES-NaOH, pH 7.0, and last with TE buffer, pH 7.0. Finally, the recombinant SLC25A33 and SLC25A36 were solubilized in 1.2% (w/v) lauric acid and diluted 1:3 with 3% (w/v) Triton X-114, 10 mm PIPES-NaOH, pH 7.0, and 1 mm EDTA in the presence (SLC25A36) or absence (SLC25A33) of 20 mm Na2SO4. Eventual small residues were removed by centrifugation (20,800 × g for 20 min at 4 °C).
Reconstitution of SLC25A33 and SLC25A36 into Liposomes
The recombinant proteins in lauric acid were reconstituted into liposomes by cyclic removal of the detergent with a hydrophobic column of Amberlite beads (Fluka), as previously described (18, 19) with some modifications. The composition of the initial mixture used for reconstitution was 50 μl of purified protein (∼20 μg of SLC25A33 or 30 μg of SLC25A36), 60 μl of 10% Triton X-114, 100 μl of 10% phospholipids (l-α-phosphatidylcholine from egg yolk; Sigma) in the form of sonicated liposomes, 10 mm substrate except where otherwise indicated, 0.6 mg (SLC25A33) or 1 mg (SLC25A36) of cardiolipin, 10 mm PIPES-NaOH, pH 7.0 (SLC25A33) or 5 mm MES-NaOH, 5 mm PIPES-NaOH, pH 6.25 (SLC25A36), and water to a final volume of 700 μl. After vortexing, this mixture was recycled 13-fold through an Amberlite column (3.2 × 0.5 cm) pre-equilibrated with a buffer containing 10 mm PIPES-NaOH at pH 7.0 (SLC25A33) or 5 mm MES-NaOH, 5 mm PIPES-NaOH at pH 6.25 (SLC25A36) and the substrate at the same concentration as in the starting mixture.
Transport Measurements
External substrate was removed from proteoliposomes on Sephadex G-75 columns pre-equilibrated with 50 mm NaCl and 10 mm PIPES-NaOH at pH 7.0 for SLC25A33 (buffer A) or 50 mm NaCl and 5 mm MES-NaOH, 5 mm PIPES-NaOH at pH 6.25 for SLC25A36 (buffer B). Transport at 25 °C was started by adding the indicated labeled substrates to substrate-loaded proteoliposomes (exchange) or to empty proteoliposomes (uniport). In both cases transport was terminated by the addition of 30 mm pyridoxal 5′-phosphate and 20 mm bathophenanthroline, which in combination and at high concentrations inhibit the activity of several mitochondrial carriers rapidly and completely (see, for example, Refs. 20–23). In controls, the inhibitors were added at the beginning together with the radioactive substrate. Finally, the external radioactivity was removed from each sample of proteoliposomes by a Sephadex G-75 column pre-equilibrated with buffer A or buffer B for SLC25A33 and SLC25A36, respectively, and the entrapped radioactivity was measured. The experimental values were corrected by subtracting control values. The initial transport rate was calculated from the radioactivity taken up by proteoliposomes after 1.5 min (SLC25A33) or 3 min (SLC25A36) (in the initial linear range of substrate uptake). For efflux measurements, proteoliposomes containing 2 mm substrate were labeled with 5 μm radioactive substrate by carrier-mediated exchange equilibration (18, 24). After 40 min, the external radioactivity was removed by passing the proteoliposomes through Sephadex G-75 columns pre-equilibrated with buffer A or buffer B for SLC25A33 or SLC25A36, respectively. Efflux was started by adding unlabeled external substrate or buffer alone (buffer A for SLC25A33 and buffer B for SLC25A36) to aliquots of proteoliposomes and terminated by adding the inhibitors indicated above.
Subcellular Localization
For the subcellular localization of SLC25A36 in Chinese hamster ovary (CHO) cells, these cells were co-transfected with 4 μg of mtEBFP/pcDNAI (where EBFP stands for enhanced blue fluorescent protein) and 4 μg of a modified pcDNA3 plasmid containing the coding sequence of SLC25A36 fused with the enhanced green fluorescent protein (EGFP) sequence at the C terminus (25). EGFP and EBFP fluorescence were detected as described (25).
Yeast Strains, Media, and Growth Conditions
The strains used in this study are all in W303 genetic context (his3-11,15; ade2-1; leu2-3,112; ura3-1; trp1-1; can1-100). The wild-type W303 rho° haploid strain was produced as follows. Cells were grown at a density of 1 × 106 cells/ml on YPD medium for 24 h, then 0.05 m phosphate buffer, pH 6.5, and 50 μg/ml ethidium bromide were added to 1 ml of culture. This culture was incubated at 28 °C for 24 h, then the cells were washed twice with H2O and plated on YPD medium. The lack of rho° cell growth was assessed on glycerol as the carbon source and the absence of mtDNA by 4′,6-diamidino-2-phenylindole (DAPI; Sigma) staining. RIM2Δ diploid strain (RIM2/RIM2::kanMX) was generated using the PCR-mediated gene disruption technique (26) by replacing one of the two wild-type RIM2 copies with the kanMX cassette in wild-type W303 diploid strain (EUROSCARF). This strain was sporulated to obtain the wild-type and the RIM2Δ haploid strains. To obtain RIM2Δ strain-containing mtDNA, the RIM2Δ haploid strain was transformed with the RIM2-pRS42H, SLC25A33-pRS42H, or SLC25A36-pRS42H plasmid and crossed with the wild-type haploid strain of the opposite mating type to obtain the diploid strains. Cell crossing was generated by bringing the cells near on plate with the needle of the Singer micromanipulator. The generation of zygotes was usually produced in 2 h. All the transformed RIM2Δ diploid strains were used for tetrad dissection to obtain transformed RIM2Δ haploid strains containing mtDNA (named RIM2Δ + RIM2, RIM2Δ + A33, RIM2Δ + A36).
Wild-type, wild-type rho°, and deletion strains were grown at 28 °C in rich medium containing 1% Bacto-peptone and 1% yeast extract supplemented with 2% glucose (YPD) or 3% glycerol (YPG). 2.2% Bacto agar (Difco) was also present in solid media. In all cases the final pH was adjusted to 4.5. The pre-sporulation medium contained 1% yeast extract, 1% Bacto-peptone, 0.17% yeast nitrogen base, 1% potassium acetate, 0.5% ammonium sulfate, and 0.05 m potassium phthalate, pH 5, and the sporulation medium contained 1% potassium acetate, pH 7.6.
Procedures for Sporulation and Dissection
After 48 h of growth on YPD plate at 28 °C, diploid strains were inoculated in pre-sporulation medium; after 24 h, cells were collected, washed 2 times in H2O, and inoculated in the sporulation medium. To avoid plasmid loss, hygromycin (100 μg/ml) was added to the pre-sporulation medium of the diploid strains containing the plasmid RIM2-pRS42H, SLC25A33-pRS42H, or SLC25A36-pRS42H. The formation of tetrads was followed by microscopic observation, and diploid strains were treated 18 min with cytohelicase (Sigma); the spores of asci were separated with the needle of the Singer micromanipulator. After 3 days, tetrad analysis was performed using marker selection; RIM2Δ spores were Geneticin (G418)-resistant; transformed RIM2Δ spores were hygromycin-resistant.
Microscopy
The microscope used was a Zeiss Axio Imager Z1 Fluorescence Microscope with AxioVision 4.8 Digital Image Processing System, and the objective lens used was 63× oil. The fluorescence was observed using filter sets for DAPI (365-nm excitation and 445/450-nm emission), DASPMI (2-(4-(dimethylamino)styryl)-1-methylpyridinium iodide; Sigma; 550/25-nm excitation and 605/670-nm emission), and GFP (470/40-nm excitation and 525/50-nm emission).
Measurements of Oxygen Consumption
Intact cell respiration was determined at 30 °C using an Oxygraph-2 k system (Oroboros, Innsbruck, Austria) equipped with two chambers, and the data were analyzed using DatLab software (27). Exponentially growing cultures in YPD or YPG were harvested at an A600 of 0.7–1, centrifuged at 3000 × g for 5 min at 4 °C, and resuspended in YP at a density of ∼5 A600 units. 50 μl of this suspension corresponding to ∼5 × 106 cells/ml were added to each chamber containing 2 ml of YP. The chambers were closed, and respiration was recorded. The highest rate of respiration was determined by adding 0.5% ethanol and then 2.5–10 μl of 2 mm carbonyl cyanide m-chlorophenylhydrazone (Sigma). The addition of 2 μm antimycin A (Sigma) accounted for non-mitochondrial oxygen consumption.
Other Methods
Proteins were analyzed by SDS-PAGE and stained with Coomassie Blue dye. The identity of purified SLC25A33 and SLC25A36 was assessed by matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry of trypsin digests of the corresponding band excised from a Coomassie-stained gel (28, 29). The amount of pure SLC25A33 and SLC25A36 was estimated by laser densitometry of stained samples using carbonic anhydrase as the protein standard. To assay the protein incorporated into liposomes, the vesicles were passed through a Sephadex G-75 column, centrifuged at 300,000 × g for 30 min, and delipidated with organic solvents as described in Capobianco et al. (30). Then the SDS-solubilized protein was determined by comparison with carbonic anhydrase in SDS gels. The share of incorporated SLC25A33 or SLC25A36 was ∼18% of the protein added to the reconstitution mixture. For fluorescence microscopy analyses cells were grown in YPD liquid medium at 28 °C, then exponential phase cells were fixed with 1% formaldehyde and treated with 1 μg/ml for DAPI staining or 1 mm for DASPMI staining.
RESULTS
Subcellular Localization of SLC25A36
Because the presence of MCF proteins is by no means restricted to mitochondrial membranes (see Refs. 7 and 31 and references therein) and the subcellular localization of SLC25A36 has not yet been determined, the intracellular localization of SLC25A36 was investigated. CHO cells were transfected with the pcDNA3-SLC25A36-EGFP plasmid, and images were acquired 40–45 h after transfection. ∼35% of cells were transfected, and green fluorescence revealed typical mitochondrial localization of SLC25A36 (Fig. 1). Furthermore, the green fluorescence of the GFP-tagged protein completely overlapped with the blue fluorescence of a mitochondria-targeted BFP co-expressed in the same cells. Thus SLC25A36, which lacks a canonical mitochondrial targeting N-terminal extension, contains in its amino acid sequence the structural information for import into mitochondria in accordance with data available for other mitochondrial carriers (32, 33).
FIGURE 1.
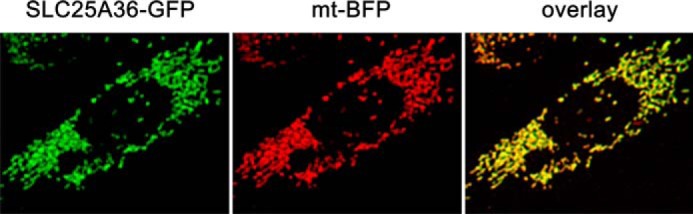
Subcellular localization of SLC25A36. CHO cells were transiently co-transfected with pcDNA3 vector carrying the DNA sequence coding SLC25A36 in-frame with the GFP DNA sequence and pcDNAI plasmid carrying the DNA sequence of a mitochondrial-targeted BFP (see “Experimental Procedures”). Images were acquired by a fluorescence microscope equipped with appropriate filters. SLC25A36-GFP, fluorescence of GFP fused to SLC25A36; mt-BFP, fluorescence of the mtBFP (here shown in red); overlay, merged image of mtBFP fluorescence with SLC25A36-GFP fluorescence.
Bacterial Expression of SLC25A33 and SLC25A36
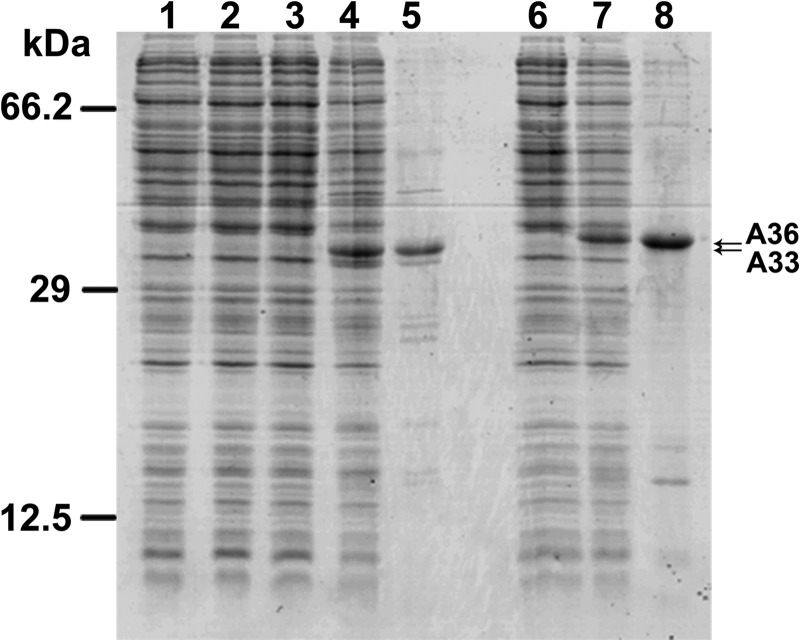
Reconstitution of recombinant proteins in liposomes is a method frequently used to identify transport properties of carrier proteins. Therefore, the open reading frames of SLC25A33 and SLC25A36 were expressed in E. coli BL21(DE3) cells (Fig. 2, lanes 4 and 7, respectively). The gene products accumulated as inclusion bodies and were purified by centrifugation and washing (Fig. 2, lanes 5 and 8, respectively). The apparent molecular masses of the purified proteins were ∼36.3 and 36.8 kDa for SLC25A33 and SLC25A36, respectively, in good agreement with their respective molecular masses (calculated values with initiator methionine, 35.374 and 34.281 kDa, respectively). The identities of both recombinant proteins were confirmed by MALDI-TOF mass spectrometry, and the yield of the purified proteins was ∼40 mg/liter of culture for both SLC25A33 and SLC25A36. The recombinant proteins were not detected in bacteria harvested immediately before induction of expression (Fig. 2, lane 2, for SLC25A33) nor in cells harvested after induction but lacking the coding sequence in the expression vector (Fig. 2, lane 3 for SLC25A33 and lane 6 for SLC25A36).
FIGURE 2.
Expression in E. coli and purification of SLC25A33 and SLC25A36. Proteins were separated by SDS-PAGE and stained with Coomassie Blue dye. Lanes 1–5, SLC25A33; lanes 6–8, SLC25A36; markers (bovine serum albumin, bovine carbonic anhydrase, and cytochrome c) in the left column. Lanes 1–4, 6, and 7, E. coli BL21(DE3) containing the expression vector, without (lanes 1, 3, and 6) and with the coding sequence of SLC25A33 (lanes 2 and 4), and with the coding sequence of SLC25A36 (lane 7). Samples were taken immediately before induction (lanes 1 and 2) and 5 h later (lanes 3, 4, 6, and 7). The same number of bacteria was analyzed in each sample. Lanes 5 and 8, purified SLC25A33 (2 μg) and purified SLC25A36 (4 μg) originated from bacteria shown in lanes 4 and 7, respectively.
Functional Characterization of SLC25A33 and SLC25A36
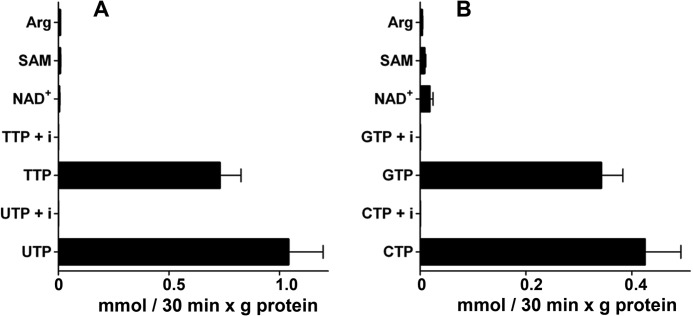
In the search for potential substrates of SLC25A33 and SLC25A36, we based our choice on the fact that SLC25A33 was previously identified as a pyrimidine nucleotide carrier transporter of primarily UTP, and both SLC25A33 and SLC25A36 are related to Rim2p, which has been thoroughly characterized and shown to be the pyrimidine nucleotide transporter of mitochondria from S. cerevisiae (10). In homo-exchange experiments (i.e. with the same substrate inside and outside), liposomes reconstituted with recombinant and purified SLC25A33 catalyzed active [3H]UTP/UTP and [3H]TTP/TTP exchanges and liposomes reconstituted with recombinant and purified SLC25A36 catalyzed active [3H]CTP/CTP and [3H]GTP/GTP exchanges (Fig. 3). These homo-exchanges were completely inhibited by a mixture of pyridoxal 5′-phosphate and bathophenanthroline. In contrast, despite the long incubation period (i.e. 30 min), neither SLC25A33- nor SLC25A36-reconstituted liposomes catalyzed homo-exchanges of NAD+, S-adenosylmethionine, and arginine (Fig. 3). Importantly, no [3H]UTP/UTP and [3H]TTP/TTP exchange activities in liposomes reconstituted with SLC25A33 and no [3H]CTP/CTP and [3H]GTP/GTP exchange activities in liposomes reconstituted with SLC25A36 were detected when SLC25A33 or SLC25A36 was inactivated by boiling before incorporation into liposomes or when liposomes were reconstituted with lauric acid-solubilized protein from bacterial cells either lacking the expression vector or harvested immediately before induction of expression (results not shown). Furthermore, the above-mentioned homo-exchanges were nil using pure liposomes, i.e. without incorporated protein (results not shown).
FIGURE 3.
Homo-exchange activities in liposomes reconstituted with SLC25A33 (A) or SLC25A36 (B). Transport was initiated by adding the indicated radioactive substrate (final concentration, 0.2 mm) to proteoliposomes preloaded internally with the same substrate (concentration, 10 mm). Where indicated (UTP + i, TTP + i, CTP + i, and GTP + i), the radioactive substrate was added together with 30 mm pyridoxal 5′-phosphate and 20 mm bathophenanthroline. The reaction was terminated after 30 min. The values are the means ± S.D. of at least three independent experiments. Differences between the UTP/UTP and TTP/TTP (A) and the CTP/CTP and GTP/GTP (B) homo-exchanges and the NAD+/NAD+, S-adenosylmethionine (SAM)/S-adenosylmethionine and arginine/arginine homo-exchanges were significant (p < 0.01, one way ANOVA and the Bonferroni t test). The difference between the UTP/UTP and the TTP/TTP homo-exchanges in A and the difference between the CTP/CTP and the GTP/GTP homo-exchanges in B were significant (p < 0.05, one way ANOVA and the Bonferroni t test).
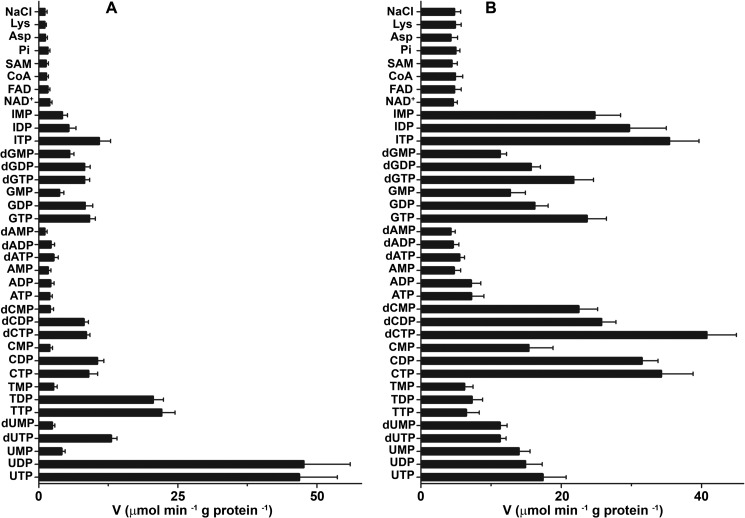
The substrate specificities of reconstituted SLC25A33 and SLC25A36 were examined in-depth by measuring the rates of [3H]UTP and [3H]CTP into SLC25A33- and SLC25A36-reconstituted liposomes, respectively, that had been preloaded with various potential substrates (Fig. 4). The highest activities of SLC25A33-mediated [3H]UTP uptake into proteoliposomes were found with internal UTP, UDP, TTP, and TDP (Fig. 4A). To a lesser extent [3H]UTP also exchanged with internal dUTP, CTP, CDP, dCTP, dCDP, GTP, GDP, dGTP, dGDP, and ITP. In contrast, the uptake of [3H]UTP was negligible or very low in the absence of internal substrate (NaCl present, uniport) or in the presence of the internal substrates UMP, dUMP, TMP, CMP, dCMP, ATP, ADP, AMP, dATP, dADP, dAMP, GMP, dGMP, NAD+, FAD, coenzyme A, S-adenosylmethionine, phosphate, aspartate, lysine (Fig. 4A) and adenosine 5′-phosphosulfate, cAMP, FMN, thiamine monophosphate, thiamine diphosphate, sulfate, succinate, malate, citrate, carnitine, glutamate, glutamine, arginine, cysteine, and glutathione (not shown). The (deoxy)nucleoside monophosphates of the U, T, and C bases were either not exchanged with external [3H]UTP or exchanged at a much lower rate than the corresponding (deoxy)nucleoside di- and triphosphates. The latter were transported at approximately the same rate by reconstituted SLC25A33.
FIGURE 4.
Dependence on internal substrate of the transport properties of liposomes reconstituted with recombinant SLC25A33 (A) and SLC25A36 (B). Proteoliposomes were preloaded internally with various substrates (each concentration 10 mm). Transport was started by adding 0.16 mm [3H]UTP or 0.2 mm [3H]CTP to liposomes reconstituted with SLC25A33 or SLC25A36, respectively. The reaction time was 1.5 min (SLC25A33) and 3 min (SLC25A36). The data are the means ± S.D. of at least three independent experiments. In A differences between the activities of UTP uptake with internal UTP, UDP, dUTP, TTP, TDP, CTP, CDP, dCTP, dCDP, GTP, GDP, dGTP, dGDP, and ITP and the activity with internal NaCl and no substrate were significant (p < 0.05, one way ANOVA and the Bonferroni t test). Differences between the activities of UTP uptake with internal substrate other than those mentioned above and the activity with internal NaCl were not significant (p > 0.05, one way ANOVA and the Bonferroni t test). In B differences between the activities of CTP uptake with internal UTP, UDP, UMP, dUTP, dUMP, CTP, CDP, CMP, dCTP, dCDP, dCMP, GTP, GDP, GMP, dGTP, dGDP, dGMP, ITP, IDP and IMP and the activity with internal NaCl were significant (p < 0.05, one way ANOVA and the Bonferroni t test). Differences between the activities of CTP uptake with internal substrate other than those mentioned above and the activity with internal NaCl were not significant (p > 0.05, one way ANOVA and the Bonferroni t test). SAM, S-adenosylmethionine; Pi, phosphate.
In liposomes reconstituted with SLC25A36 (Fig. 4B) [3H]CTP exchanged efficiently with the (deoxy)nucleotides of the bases C, U, I, and G. In contrast, the SLC25A36-mediated [3H]CTP uptake in the presence of internal thymine nucleotides and adenine (deoxy)nucleotides was not significantly higher than that found in the presence of NaCl. Similarly, the amount of radioactivity taken up in the presence of internal NAD+, FAD, coenzyme A, S-adenosylmethionine, phosphate, aspartate, lysine (Fig 4B) and adenosine 5′-phosphosulfate, cAMP, FMN, thiamine monophosphate, thiamine diphosphate, sulfate, succinate, malate, citrate, carnitine, glutamate, glutamine, arginine, cysteine, and glutathione (not shown) was virtually the same as that taken up by uniport. At variance with the SLC25A33-mediated [3H]UTP transport (Fig. 4A), the nucleoside monophosphates of the C, U, I, and G bases were clearly transported by reconstituted SLC25A36 although at a lower rate than the corresponding nucleoside tri- and diphosphates. The deoxynucleotides of C, U, and G were transported nearly at the same rate than the corresponding nucleotides.
Effects of Inhibitors on SLC25A33 and SLC25A36 Activities
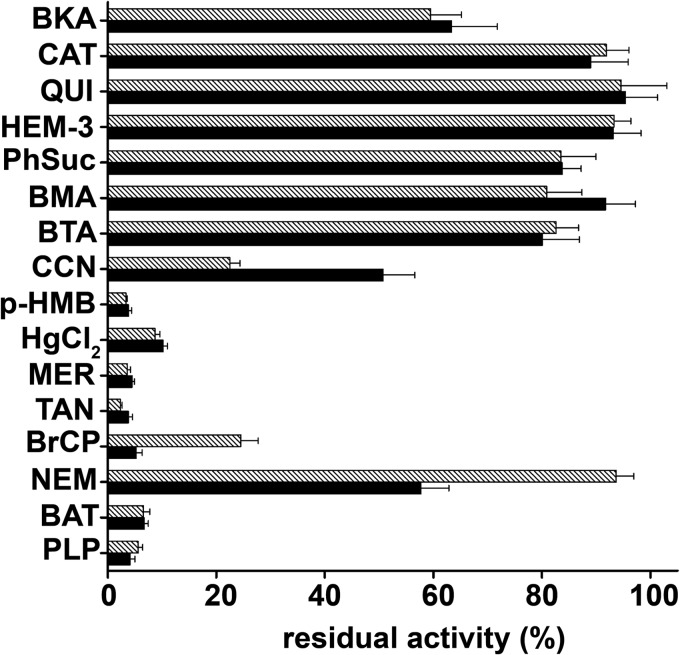
The effects of other mitochondrial carrier inhibitors on the activities of SLC25A33 and SLC25A36 were examined using [3H]UTP and [3H]CTP, respectively. Both the [3H]UTP/UTP and [3H]CTP/CTP exchange reactions catalyzed by reconstituted SLC25A33 and SLC25A36, respectively, were inhibited strongly by pyridoxal 5′-phosphate, bathophenanthroline, bromcresol purple, tannic acid, and mercurials (HgCl2, mersalyl, p-hydroxymercuribenzoate) (Fig. 5). The SLC25A33-mediated [3H]UTP exchange was inhibited partially by α-cyano-4-hydroxycinnamate and N-ethylmaleimide. In contrast, the SLC25A36-mediated [3H]CTP exchange was inhibited markedly by α-cyano-4-hydroxycinnamate and very poorly by N-ethylmaleimide. Furthermore, little inhibition was observed with 1,2,3-benzenetricarboxylate, butyl malonate, phenyl succinate, hemicholinium-3, and quinine. Notably, bongkrekic acid inhibited the activities of SLC25A33 and SLC25A36 partially (36.7 and 40.5%, respectively), whereas carboxyatractyloside had a much lower effect at a concentration (10 μm) that completely inhibits the ADP/ATP carrier (34). The inhibitor sensitivity of SLC25A33 and SLC25A36, therefore, resembles that of yeast Rim2p but is not identical.
FIGURE 5.
Effect of inhibitors on the [3H]UTP/UTP and [3H]CTP/CTP exchanges by reconstituted SLC25A33 and SLC25A36, respectively. Transport was initiated by adding 0.16 mm [3H]UTP to SLC25A33-reconstituted liposomes preloaded internally with 10 mm UTP (black bars) or 0.2 mm [3H]CTP to SLC25A36-reconstituted liposomes preloaded with 10 mm CTP (hatched bars). The reaction time was 1.5 min (SLC25A33) and 3 min (SLC25A36). Thiol reagents were added 2 min before the labeled substrate; the other inhibitors were added together with the labeled substrate. The final concentrations of the inhibitors were 20 mm (PLP, pyridoxal 5′-phosphate; BAT, bathophenanthroline), 0.2 mm (MER, mersalyl; p-HMB, p-hydroxymercuribenzoate; HgCl2), 2 mm (BMA, butyl malonate; BTA, 1,2,3-benzenetricarboxylate; PhSuc, phenyl succinate), 1 mm (NEM, N-ethylmaleimide; CCN, α-cyano-4-hydroxycinnamate), 0.4 mm (HEM-3, hemicholinium-3; QUI, quinine), 0.3 mm (BrCP, bromcresol purple), 0.1% (TAN, tannic acid), and 10 μm (BKA, bongkrekic acid; CAT, carboxyatractyloside). The data expressed as the percentage of residual activity of SLC25A33-mediated [3H]UTP/UTP or SLC25A36-mediated [3H]CTP/CTP exchange are the means ± S.E. of at least three independent experiments for each inhibitor and each carrier. The control values for uninhibited [3H]UTP/UTP and [3H]CTP/CTP exchanges were 45.9 ± 2.6 and 31.2 ± 1.5 μmol/min × g of protein, respectively.
Kinetic Characteristics of Recombinant SLC25A33 and SLC25A36
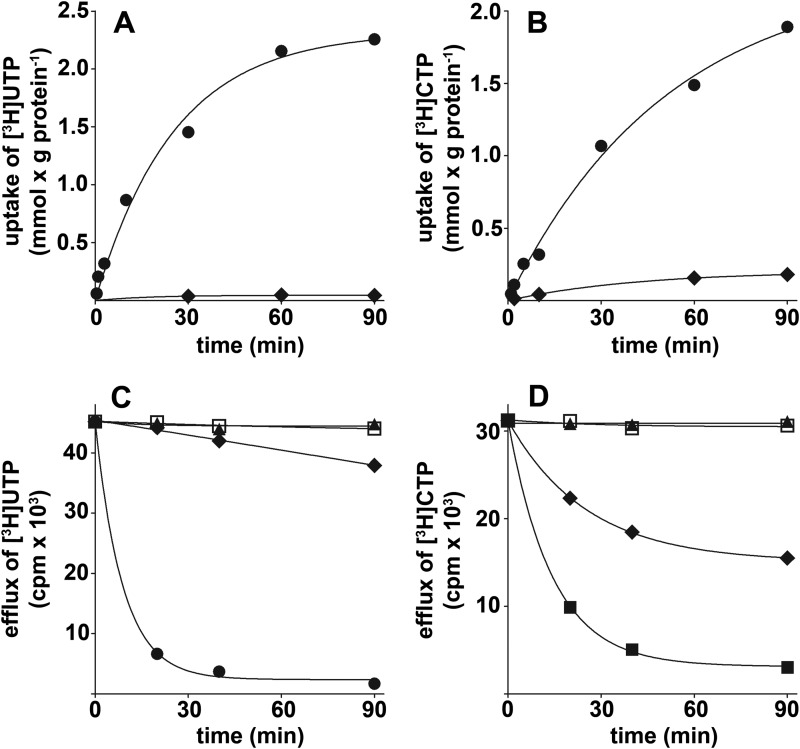
In Fig. 6, A and B, the time-courses of 1 mm [3H]UTP or [3H]CTP uptake into liposomes reconstituted with SLC25A33 and SLC25A36, respectively, were compared either as uniport (in the absence of internal substrate) or as exchange (in the presence of 10 mm UTP or CTP, respectively). The exchange reactions catalyzed by both SLC25A33 and SLC25A36 followed first-order kinetics, with isotopic equilibrium being approached exponentially. The rate constants and the initial rates of the exchanges deduced from the time-courses (18) were 0.04 and 0.02 min−1 and 91 and 44 μmol/min × g protein for SLC25A33 and SLC25A36, respectively. In contrast, the uniport uptake of [3H]UTP by SLC25A33 was negligible and that of [3H]CTP by SLC25A36 was very low. The uniport mode of transport was further investigated by measuring the efflux of [3H]UTP or [3H]CTP from proteoliposomes preloaded with these compounds because this experimental approach provides a more sensitive assay for unidirectional transport (18). As shown in Fig. 6, C and D, in the absence of external substrate both the rate and extent of radioactive substrate efflux was very low from SLC25A33-reconstituted liposomes and substantial from SLC25A36-reconstituted liposomes. Of note, with both reconstituted carriers a rapid and extensive efflux of [3H]UTP or [3H]CTP occurred upon the addition of external UTP or CTP, respectively. Furthermore, both effluxes, i.e. with and without external substrate, were completely prevented if the inhibitors pyridoxal 5′-phosphate and bathophenanthroline were present from the beginning of the proteoliposome incubation (at time 0). The kinetic constants of recombinant SLC25A33 and SLC25A36 were determined from the initial transport rate of homo-exchanges at various external labeled substrate concentrations in the presence of a fixed saturating internal substrate concentration of 10 mm (Table 1). The specific activity (Vmax) of SLC25A33 for the UTP/UTP exchange at 25 °C was 2-fold greater than that of SLC25A36 for the CTP/CTP exchange, whereas the half-saturation constants (Km) of SLC25A33 for external UTP and of SLC25A36 for external CTP were nearly the same and similar to the physiological concentrations of UTP and CTP in human cells (35). Several external substrates were competitive inhibitors of SLC25A33 and SLC25A36, as they increased the apparent Km without changing the Vmax of the UTP/UTP and CTP/CTP exchange, respectively (not shown). The inhibitory constants (Ki) of these compounds for SLC25A33 were 174 ± 20 μm (UTP), 195 ± 23 μm (CTP), 318 ± 28 μm (GTP), and 333 ± 35 μm (ITP), and those for SLC25A36 were 1.3 ± 0.2 mm (UTP), 224 ± 19 μm (CTP), 276 ± 26 μm (GTP), and 181 ± 25 μm (ITP). These results show that UTP and CTP are the best substrates of SLC25A33 and SLC25A36, respectively.
FIGURE 6.
Kinetics of [3H]UTP or [3H]CTP transport in liposomes reconstituted with SLC25A33 or SLC25A36, respectively. Liposomes were reconstituted with recombinant SLC25A33 (A and C) or SLC25A36 (B and D). A and B, uptake of [3H]UTP and [3H]CTP. 1 mm [3H]UTP in A or 1 mm [3H]CTP in B was added to proteoliposomes containing 10 mm UTP in A or CTP in B (●, exchange) or 10 mm NaCl and no substrate (♦, uniport). C and D, efflux of [3H]UTP and [3H]CTP. The internal substrate of proteoliposomes (2 mm UTP in C and 2 mm CTP in D) was labeled with [3H]UTP in C and [3H]CTP in D by carrier-mediated exchange equilibration. After removal of the external substrate by Sephadex G-75, the efflux of [3H]UTP (C) or [3H]CTP (D) was started by adding buffer alone (buffer A for SLC25A33 or buffer B for SLC25A36) (♦) or in buffer A 5 mm UTP (●), 30 mm pyridoxal 5′-phosphate, and 20 mm bathophenanthroline (□) and 5 mm UTP, 30 mm pyridoxal 5′-phosphate, and 20 mm bathophenanthroline (▴) or in buffer B 5 mm CTP (■), 30 mm pyridoxal 5′-phosphate and 20 mm bathophenanthroline (□) or 5 mm CTP, 30 mm pyridoxal 5′-phosphate and 20 mm bathophenanthroline (▴). The values are the means of at least three independent experiments.
TABLE 1.
Kinetic parameters of recombinant SLC25A33 and SLC25A36
Km and Vmax were calculated from double-reciprocal plots of the rates of [3H]UTP, [3H]CTP, [3H]TTP, or [3H]GTP uptake at various concentrations into proteoliposomes containing the same substrates (concentration, 10 mm). The data represent the means ± S.D. of at least four independent experiments.
| Homo-exchanges | SLC25A33 |
SLC25A36 |
||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| mm | μmol/min × g protein | mm | μmol/min × g protein | |
| [3H]UTP/UTP | 0.16 ± 0.01 | 98.5 ± 10.2 | 1.19 ± 0.15 | 30.7 ± 4.4 |
| [3H]CTP/CTP | 0.18 ± 0.02 | 22.5 ± 3.8 | 0.21 ± 0.02 | 51.2 ± 5.7 |
| [3H]TTP/TTP | 0.08 ± 0.01 | 50.8 ± 6.1 | >2.5 | <8 |
| [3H]GTP/GTP | 0.30 ± 0.02 | 15.7 ± 2.4 | 0.23 ± 0.04 | 23.6 ± 2.9 |
SLC25A33 and SLC25A36 Genes Are Able to Complement the Absence of RIM2 in the S. cerevisiae RIM2 Null Mutant
To investigate whether the two human genes can replace the Rim2p function in vivo, the RIM2Δ haploid strain was transformed with the yeast plasmid pRS42H containing the SLC25A33 or SLC25A36 gene under the yeast RIM2 promoter (see “Experimental Procedures”). The RIM2Δ haploid strain was unable to grow on non-fermentable carbon sources like glycerol (see the next section), because this strain looses mtDNA at very high frequency, as visualized with DAPI staining (Fig. 7A and Ref. 9). As a consequence, to perform a gene complementation assay, it was necessary to reintroduce the mtDNA in the RIM2Δ strain transformed with SLC25A33 or SLC25A36. First we verified whether the RIM2Δ haploid strain transformed with the S. cerevisiae RIM2 gene could reintroduce the mtDNA. To this end this transformed strain was crossed with the wild-type W303 haploid strain, so that during meiosis the mtDNA segregated in all four spores, including the two RIM2Δ haploid strains. After sporulation and tetrad analysis, the transformed RIM2Δ haploid strains (named RIM2Δ + RIM2) were selected; these strains were able to grow on glycerol (not shown). Furthermore, when stained with DAPI, the RIM2Δ + RIM2 strain was found to contain mtDNA (Fig. 7A), showing that RIM2 is essential to maintain mtDNA. To check whether stabilization of mtDNA in the RIM2Δ + RIM2 strain was due to the presence of RIM2 gene, a plasmid loss assay was performed, and the resulting cells without plasmid were devoid of mtDNA (not shown).
FIGURE 7.
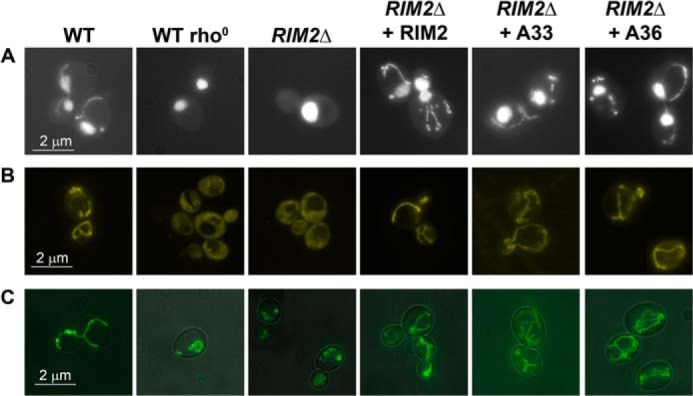
The SLC25A33 and SLC25A36 human genes are able to complement the absence of RIM2 in S. cerevisiae cells rescuing the mtDNA, the mitochondrial membrane potential, and the tubular mitochondrial morphology. Wild-type W303, wild-type W303 rho°, RIM2Δ and RIM2Δ transformed with the RIM2-pRS42H, SLC25A33-pRS42H, or SLC25A36-pRS42H plasmid (shown as WT, WT rho°, RIM2Δ, RIM2Δ + RIM2, RIM2Δ + A33 and RIM2Δ + A36, respectively) haploid strains were grown until exponential phase in liquid YPD medium at 28 °C and stained with DAPI (A), DASPMI (B), mito-gfp (C).
Having verified that the presence of the RIM2 gene in RIM2Δ cells reintroduced the mtDNA, we applied the same procedure to investigate the effect of complementing RIM2Δ cells with SLC25A33 or SLC25A36. Fig. 7A shows that the S. cerevisiae RIM2Δ strains upon transformation with the SLC25A33 or SLC25A36 gene, crossing with the wild-type strain, sporulation, and tetrad dissection (named RIM2Δ + A33, RIM2Δ + A36 strains, respectively) contained mtDNA, proving that both the two human genes are able to maintain mtDNA as does the S. cerevisiae RIM2 gene. The stabilization of mtDNA in RIM2Δ + A33 and RIM2Δ + A36 was due to the presence of the human genes because (i) after the loss of plasmid containing SLC25A33 or SLC25A36, these strains were unable to grow on glycerol and (ii) no mtDNA was observed when the RIM2Δ strain was transformed with the empty plasmid (data not shown). To verify whether the mitochondria were functional, all the strains under investigation were incubated with DASPMI, a vital dye that stains only the membranes of energized mitochondria with a membrane potential positive outside (36). As shown in Fig. 7B, the mitochondrial membranes of the RIM2Δ strain as well as that of the wild-type rho° strain were not stained by DASPMI; by contrast, the mitochondrial membranes of the RIM2Δ + RIM2, RIM2Δ + A33, and RIM2Δ + A36 strains were stained as the wild-type strain, indicating that the mitochondria of the latter strains are functional. Furthermore, the mitochondrial morphology of the various S. cerevisiae strains was investigated by using mito-gfp, a mitochondria-targeted green fluorescent protein (37). The RIM2Δ strain transformed with mito-gfp exhibited a fragmented mitochondrial morphology similar to that of wild-type rho° cells (Fig. 7C). In contrast, the RIM2Δ + RIM2, RIM2Δ + A33, and RIM2Δ + A36 strains transformed with mito-gfp displayed a typical tubular mitochondrial morphology as the wild-type strain. These results demonstrate that the human genes SLC25A33 and SLC25A36 are able to complement the absence of RIM2 in S. cerevisiae cells restoring mtDNA, the mitochondrial membrane potential, and the tubular mitochondrial morphology.
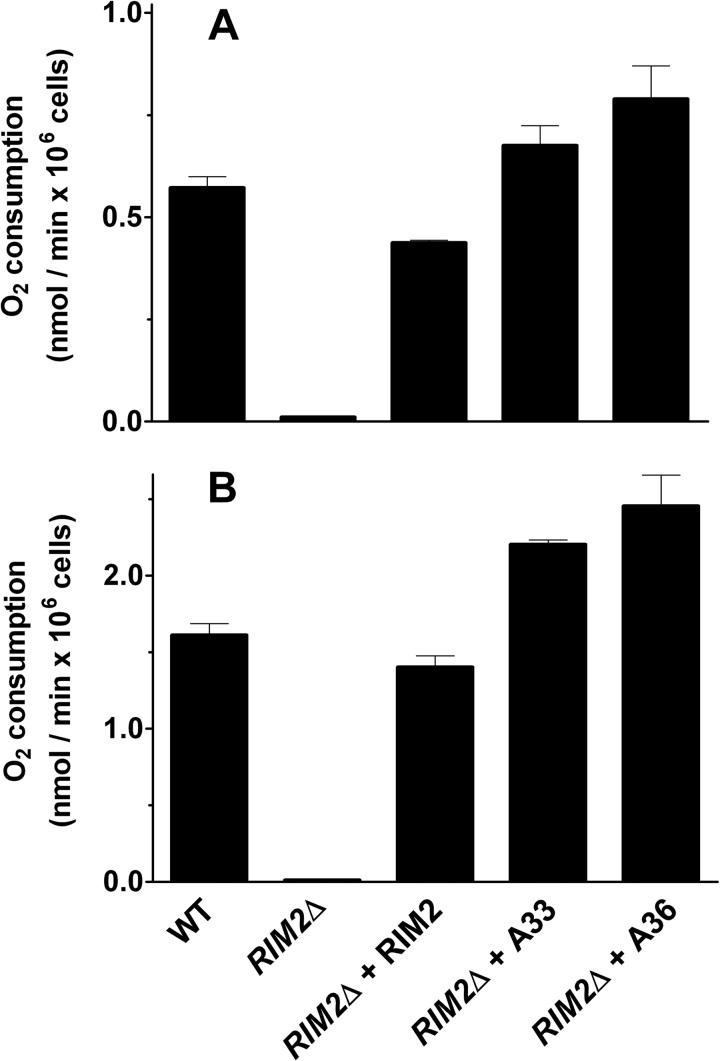
In another set of experiments the mitochondrial-dependent oxygen consumption of the wild type, RIM2Δ, RIM2Δ + RIM2, RIM2Δ + A33, and RIM2Δ + A36 strains was measured in a high resolution oxygraph. With the exception of the RIM2Δ strain grown in YPD, all the other strains were grown in YPD (Fig. 8A) or YPG (Fig. 8B), and respiration was determined in YP medium. The oxygen consumption rate of the wild-type strain was 0.57 nmol of O2 × min−1 × 106 cells (Fig. 8A) and 1.61 nmol O2 × min−1 × 106 cells (Fig. 8B), values that are close to those measured by other investigators under similar conditions (38, 39). As shown in Fig. 8, A and B, independently from the fermentative or non-fermentative carbon source used for growth, the expression of RIM2 or each of the two human genes in RIM2Δ cells restored respiration to wild-type or higher levels. Virtually the same large differences between the wild-type, RIM2Δ, RIM2Δ + RIM2, RIM2Δ + A33, and RIM2Δ + A36 were found when measuring the highest rate of respiration in the presence of the uncoupler m-chlorophenylhydrazone; indeed, the m-chlorophenylhydrazone-stimulated respiration/basal respiration ratio was ∼2.1 for all the strains grown both in YPD and YPG.
FIGURE 8.
Mitochondrial respiration of wild-type W303, RIM2Δ, RIM2Δ + RIM2, RIM2Δ + A33, RIM2Δ + A36 strains. With the exception of the RIM2Δ cells, which were grown in YPD, the other cells were grown in YPD (A) or YPG (B), washed, resuspended in YP, and tested for oxygen consumption. Respiration rates by the indicated cells were corrected for the oxygen consumption measured in the presence of antimycin A. Data represent the means ± S.D. of three independent experiments. In A and B, differences between the respiratory rates of wild-type, RIM2Δ + RIM2, RIM2Δ + A33, and RIM2Δ + A36 cells and the respiratory rate of RIM2Δ cells were significant (p < 0.01, one-way ANOVA, and the Bonferroni t test). In A the differences between the respiratory rates of RIM2Δ + A33 and RIM2Δ + A36 cells and the respiratory rate of wild-type cells were not significant (p > 0.05, one-way ANOVA and the Bonferroni t test). In B the differences between the respiratory rates of RIM2Δ + A33 and RIM2Δ + A36 cells and the respiratory rate of wild-type cells were significant (p < 0.05, one-way ANOVA and the Bonferroni t test), whereas the difference between the respiratory rate of RIM2Δ + RIM2 cells and that of wild-type cells was not significant (p > 0.05, one-way ANOVA, and the Bonferroni t test).
SLC25A33 and SLC25A36 Are Able to Rescue the Lack of Growth of the S. cerevisiae RIM2 Null Mutant
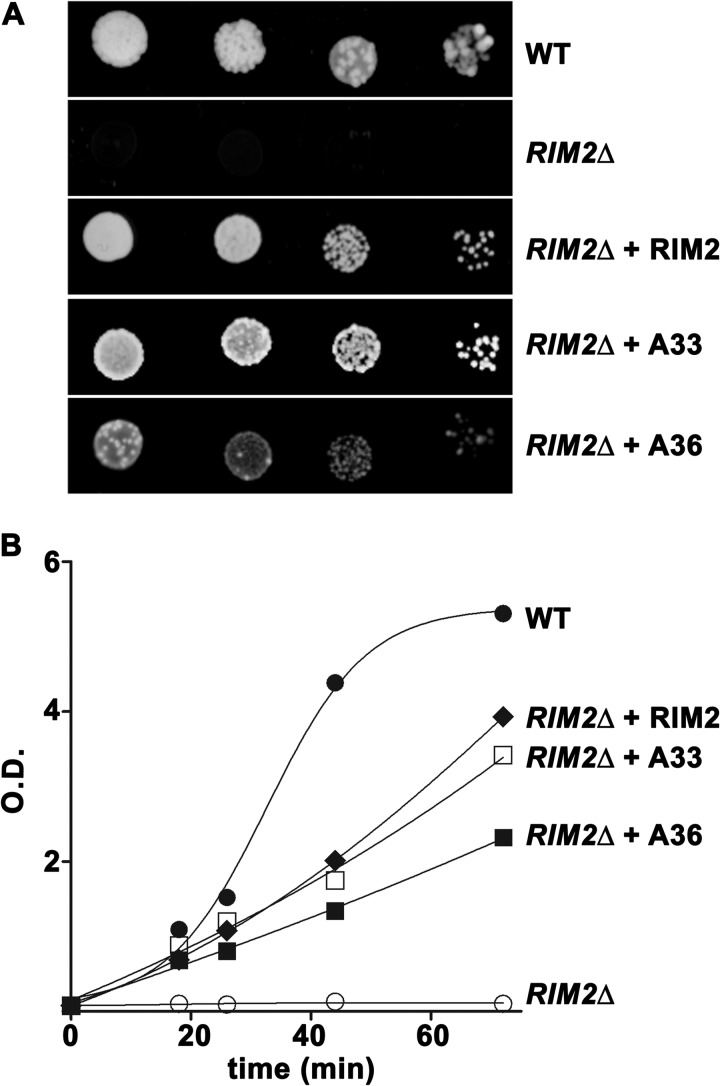
The lack of growth of RIM2Δ cells on glycerol was largely restored by expressing SLC25A33, SLC25A36, or as a control, RIM2 in mtDNA-containing RIM2Δ cells both in solid (Fig. 9A) and liquid (Fig. 9B) YPG medium. In contrast, when the RIM2Δ cells were transformed with the empty vector, no growth restoration was observed (not shown). Similar results were obtained using ethanol and pyruvate as non-fermentative carbon sources instead of glycerol in solid and liquid media (data not shown).
FIGURE 9.
Effect of complementing S. cerevisiae RIM2Δ cells with human SLC25A33 or SLC25A36 on growth. Wild-type W303, RIM2Δ, and RIM2Δ transformed with the RIM2-pRS42H, SLC25A33-pRS42H, or SLC25A36-pRS42H plasmid (shown as WT, RIM2Δ, RIM2Δ + RIM2, RIM2Δ + A33, RIM2Δ + A36, respectively) haploid strains were spotted on solid YPG medium for 48 h at 30 °C as 4-fold serial dilution (A) or were inoculated in liquid YPG medium at 30 °C (B). The values of absorbance (O.D.) at 600 nm refer to cell cultures after the indicated times of growth. The values are means of three independent experiments. (●), WT; (○), RIM2Δ; (♦), RIM2Δ + RIM2; (□), RIM2Δ + A33; and (■), RIM2Δ + A36 strains.
DISCUSSION
In humans several proteins have been identified at the molecular and biochemical level as mitochondrial carriers for adenine nucleotides (see Refs. 1 and 2 and references therein), but none transported pyrimidine ribo- or deoxynucleotides at any appreciable rate. These nucleotides are synthesized in the cytosol by the de novo pathway (absent in the mitochondria) and are required inside the mitochondrial matrix for DNA and RNA synthesis. Mitochondria contain the nucleotide salvage pathway that synthesizes nucleotides from the corresponding nucleosides that are transported into the organelles by the equilibrative nucleoside transporters hENT1 and hENT3 (40, 41). As the contribution of the salvage pathway to mtDNA synthesis is insufficient in quiescent cells and even more in cycling cells (42–44), pyrimidine nucleotides must be transported from the cytosol to the mitochondrial matrix. In the past a partial purification from human mitochondria of a carrier that displayed an efficient transport activity for dCTP has been achieved (45). Furthermore, in studies with isolated mitochondria evidence for a specific and saturable transporter for thymidine monophosphate has been provided (46). However, the molecular nature of these carriers has not been defined.
The results of this study including the transport properties and kinetic characteristics of recombinant SLC25A33 and SLC25A36 together with the localization to mitochondria of SLC25A36, presented here, and SLC25A33, reported previously (11), demonstrate that these proteins are mitochondrial transporters for pyrimidine ribo- and deoxynucleotides. These carriers share 59.8% identical amino acids. However, it is not possible to make reliable assumptions on the substrate specificity or on the transport modes on basis of the amino acid similarity. Therefore, we decided to analyze the biochemical properties of both proteins in a reconstituted system. Similar to other members of the MCF, SLC25A33 and SLC25A36 appear as inclusion bodies after recombinant synthesis in E. coli. Such inclusions, however, are advantageous as they allow the purification of the heterologously expressed carriers by centrifugation and washing.
Our direct transport measurements show that SLC25A33 and SLC25A36 possess a similar substrate specificity. Indeed, both carriers transport pyrimidine nucleotides, although with different efficiency. They also transport guanine and inosine nucleotides but none of the many other compounds tested including adenine (deoxy)nucleotides and dinucleotides. However, SLC25A33 and SLC25A36 differ in a number of transport properties. Unlike SLC25A36, SLC25A33 does not transport nucleoside monophosphates. This carrier transports (deoxy)nucleoside di- and tri-phosphates (XP) at about the same rate with the following order of efficacy UXP > TXP > CXP. The best substrates of SLC25A36 are the cytosine (deoxy)nucleoside mono-, di-, and tri-phosphates. In comparison with the cytosine (deoxy)nucleotides, the uracil nucleotides are transported by reconstituted SLC25A36 less efficiently and the thymine nucleotides virtually not at all. According to our kinetic data these large differences in the rate of pyrimidine nucleotide transport can be accounted for by the higher Km and lower Vmax values of SLC25A36 for uracil and thymine nucleotides than for cytosine nucleotides. The guanine (deoxy)nucleotides, which are also transported by both SLC25A33 and SLC25A36, have Km similar to those of the respective best substrates but much lower Vmax values. Furthermore, in contrast to SLC25A33 and the great majority of mitochondrial carriers that are obligatory exchangers (1, 47), SLC25A36 catalyzes uniport besides exchange of substrates. In this respect SLC25A36 resembles the carriers for phosphate, glutamate, and carnitine/acylcarnitine, which are also capable of mediating uniport at a much lower rate than exchange (48–50).
The substrate specificity of SLC25A33 and SLC25A36 is distinct from that of the ∼30 members of the MCF characterized until now. Furthermore, SLC25A33 and SLC25A36 do not exhibit significant sequence homology with any other human mitochondrial carrier greater than the homology exiting among the different members of the MCF. It is, therefore, likely that SLC25A33 and SLC25A36 are the only pyrimidine nucleotide carriers of the SLC25 family. As pyrimidine nucleotide carriers, SLC25A33 and SLC25A36 are essential for a number of major processes occurring in the mitochondria, i.e. the synthesis and breakdown of DNA and the various types of RNA including the RNA primers necessary for the initiation of DNA replication and repair. In these processes the pyrimidine (d)NTPs are the precursors of DNA and RNA synthesis, and the pyrimidine (d)NMPs are the products of their breakdown. The biochemical properties of recombinant SLC25A33 (that does not transport (d)NMPs) indicate that the main function of this transporter is to catalyze the exchange of cytosolic pyrimidine (d)NTPs for intramitochondrial pyrimidine (d)NDPs. The latter can be produced inside the mitochondria by the intramitochondrial enzymes that convert pyrimidine (d)NMPs to the corresponding (d)NDPs (51, 52). According to Floyd et al. (11) and Favre et al. (12) SLC25A33 is induced by the insulin-like growth factor signaling pathway to mTor, and its expression is higher in transformed fibroblasts, cancer cell lines, and primary prostate cancer than in normal tissues. It is, therefore, likely that SLC25A33 is operative mainly in cells that grow very rapidly. As regards to SLC25A36, on the basis of our transport measurements the primary physiological function of this carrier (that transports cytosine and uracil (deoxy)nucleoside mono-, di-, and tri-phosphates) is probably to catalyze uptake of pyrimidine (d)NTPs into the mitochondrial matrix in exchange for internal pyrimidine (d)NMPs or, to a lesser extent, (d)NDPs. This exchange may play an important role in mitochondrial nucleic acid metabolism both in quiescent and cycling cells in which the contribution of the salvage pathway is significant and minimal, respectively (42–44). It should be noted that the SLC25A36-mediated import of pyrimidine (d)NDPs in exchange for (d)NMPs should be relatively minor as compared with the uptake of (d)NTPs in exchange for (d)NMPs for thermodynamic reasons. Even if transported into mitochondria in exchange for (d)NMPs, (d)NDPs would recycle across the membrane exchanging with external (d)NTPs, thus accomplishing a net (d)NTPsout/(d)NMPsin exchange. Furthermore, SLC25A36 catalyzes a uniport transport mode besides exchange. The rate of uniport is much lower than that of the exchange. However, the uniport reaction may be necessary under certain conditions, for example during early development when mitochondrial biogenesis and the request for (d)NTPs is particularly high.
Several important findings in vivo, i.e. the rescue in S. cerevisiae RIM2Δ cells of mtDNA, mitochondrial respiration, mitochondrial membrane potential and growth on glycerol and other respiratory substrates, strongly support SLC25A33 and SLC25A36 controlling the uptake of pyrimidine (deoxy)nucleotides into mitochondria. Indeed, these RIM2Δ cell phenotypes can easily be accounted for by insufficient supply of pyrimidine (deoxy)nucleotides to the mitochondrial matrix, where they are indispensable for the synthesis of mtDNA, its transcription, and replication. The altered mitochondrial morphology observed in RIM2Δ cells is similar to that exhibited by the wild-type W303 rho° cells (Fig. 7), indicating that this unusual faulty mitochondrial structure is primarily caused by the loss of mtDNA (53). This phenotype of RIM2Δ cells is also restored by the SLC25A33 and SLC25A36 genes, suggesting a complete suppression of the RIM2Δ cells defects once the mitochondrial DNA is stabilized by the presence of these human proteins. It cannot be excluded, however, that additional changes in RIM2Δ cells may contribute to the observed alteration of the mitochondrial morphology. In this respect it is worth mentioning that Rim2p has been reported to be needed in iron utilization in mitochondria, for example in Fe-S protein maturation and heme synthesis (54), and be capable of transporting iron and other divalent metal ions into the mitochondria in co-transport with pyrimidine nucleotides (55). Future studies are warranted to investigate the effects of SLC25A33 and/or SLC25A36 knock-out in mice to provide further insight on the physiological roles of these mitochondrial transporters. It will also be of great interest to investigate whether antiviral and anticancer nucleoside analogs such as 2′,3′-dideoxycytidine, 2′,3′-dideoxyinosine, and 3′-azido-3′-deoxythymidine, whose toxicity is due to mitochondrial function impairment, are transported by SLC25A33 and/or SLC25A36 from the cytosol, where they are phosphorylated, into the mitochondria where they inhibit the mtDNA polymerase γ.
This work was supported by grants from the Ministero dell'Università e della Ricerca (MIUR), the National Research Council (CNR), Centro di Eccellenza in Genomica Comparata (CEGBA), Apulia Region, and the Italian Human ProteomeNet no. RBRN07BMCT_009 (MIUR).
This work is dedicated to Prof. Ernesto Quagliariello on the occasion of the 10th anniversary of his death.
- SLC25
- solute carrier family 25
- (d)NMP
- (deoxy)nucleoside monophosphate
- NDP
- nucleoside diphosphate
- NTP
- nucleoside triphosphate
- MCF
- mitochondrial carrier family
- BFP
- blue fluorescent protein
- EBFP
- enhanced BFP
- EGFP
- enhanced green fluorescent protein
- YP
- extract/peptone
- YPD
- yeast extract/peptone/dextrose
- YPG
- yeast extract/peptone/glucose
- DASPMI
- 2-(4-(dimethylamino)styryl)-1-methylpyridinium iodide
- ANOVA
- analysis of variance.
REFERENCES
- 1. Palmieri F. (2013) The mitochondrial transporter family SLC25: identification, properties, and physiopathology. Mol. Aspects Med. 34, 465–484 [DOI] [PubMed] [Google Scholar]
- 2. Gutiérrez-Aguilar M., Baines C. P. (2013) Physiological and pathological roles of mitochondrial SLC25 carriers. Biochem. J. 454, 371–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Palmieri F. (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 4. Vozza A., Parisi G., De Leonardis F., Lasorsa F. M., Castegna A., Amorese D., Marmo R., Calcagnile V. M., Palmieri L., Ricquier D., Paradies E., Scarcia P., Palmieri F., Bouillaud F., Fiermonte G. (2014) UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc. Natl. Acad. Sci. U.S.A. 111, 960–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Porcelli V., Fiermonte G., Longo A., Palmieri F. (2014) The human gene SLC25A29, of solute carrier family 25, encodes a mitochondrial transporter of basic amino acids. J. Biol. Chem. 289, 13374–13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palmieri F., Pierri C. L. (2010) Mitochondrial metabolite transport. Essays Biochem. 47, 37–52 [DOI] [PubMed] [Google Scholar]
- 7. Palmieri F., Pierri C. L., De Grassi A., Nunes-Nesi A., Fernie A. R. (2011) Evolution, structure, and function of mitochondrial carriers: a review with new insights. Plant J. 66, 161–181 [DOI] [PubMed] [Google Scholar]
- 8. Palmieri F. (2014) Mitochondrial transporters of the SLC25 family and associated diseases: a review. J. Inherit. Metab. Dis. 37, 565–575 [DOI] [PubMed] [Google Scholar]
- 9. Van Dyck E., Jank B., Ragnini A., Schweyen R. J., Duyckaerts C., Sluse F., Foury F. (1995) Overexpression of a novel member of the mitochondrial carrier family rescues defects in both DNA and RNA metabolism in yeast mitochondria. Mol. Gen. Genet. 246, 426–436 [DOI] [PubMed] [Google Scholar]
- 10. Marobbio C. M., Di Noia M. A., Palmieri F. (2006) Identification of the mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: bacterial expression, reconstitution, and functional characterization. Biochem. J. 393, 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Floyd S., Favre C., Lasorsa F. M., Leahy M., Trigiante G., Stroebel P., Marx A., Loughran G., O'Callaghan K., Marobbio C. M., Slotboom D. J., Kunji E. R., Palmieri F., O'Connor R. (2007) The IGF-I-mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Mol. Biol. Cell 18, 3545–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Favre C., Zhdanov A., Leahy M., Papkovsky D., O'Connor R. (2010) Mitochondrial pyrimidine nucleotide carrier (PNC1) regulates mitochondrial biogenesis and the invasive phenotype of cancer cells. Oncogene 29, 3964–3976 [DOI] [PubMed] [Google Scholar]
- 13. Franzolin E., Miazzi C., Frangini M., Palumbo E., Rampazzo C., Bianchi V. (2012) The pyrimidine nucleotide carrier PNC1 and mitochondrial trafficking of thymidine phosphates in cultured human cells. Exp. Cell Res. 318, 2226–2236 [DOI] [PubMed] [Google Scholar]
- 14. Da-Rè C., Franzolin E., Biscontin A., Piazzesi A., Pacchioni B., Gagliani M. C., Mazzotta G., Tacchetti C., Zordan M. A., Zeviani M., Bernardi P., Bianchi V., De Pittà C., Costa R. (2014) Functional characterization of drim2, the Drosophila melanogaster homolog of the yeast mitochondrial deoxynucleotide transporter. J. Biol. Chem. 289, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fiermonte G., Walker J. E., Palmieri F. (1993) Abundant bacterial expression and reconstitution of an intrinsic membrane transport protein from bovine mitochondria. Biochem. J. 294, 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agrimi G., Russo A., Pierri C. L., Palmieri F. (2012) The peroxisomal NAD+ carrier of Arabidopsis thaliana transports coenzyme A and its derivatives. J. Bioenerg. Biomembr. 44, 333–340 [DOI] [PubMed] [Google Scholar]
- 17. Palmieri L., Picault N., Arrigoni R., Besin E., Palmieri F., Hodges M. (2008) Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem. J. 410, 621–629 [DOI] [PubMed] [Google Scholar]
- 18. Palmieri F., Indiveri C., Bisaccia F., Iacobazzi V. (1995) Mitochondrial metabolite carrier proteins: purification, reconstitution, and transport studies. Methods Enzymol. 260, 349–369 [DOI] [PubMed] [Google Scholar]
- 19. Palmieri L., Arrigoni R., Blanco E., Carrari F., Zanor M. I., Studart-Guimaraes C., Fernie A. R., Palmieri F. (2006) Molecular identification of an Arabidopsis thaliana S-adenosylmethionine transporter: analysis of organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Plant Physiol. 142, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fiermonte G., Dolce V., Palmieri L., Ventura M., Runswick M. J., Palmieri F., Walker J. E. (2001) Identification of the human mitochondrial oxodicarboxylate carrier: bacterial expression, reconstitution, functional characterization, tissue distribution, and chromosomal location. J. Biol. Chem. 276, 8225–8230 [DOI] [PubMed] [Google Scholar]
- 21. Palmieri L., Lasorsa F. M., Iacobazzi V., Runswick M. J., Palmieri F., Walker J. E. (1999) Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett. 462, 472–476 [DOI] [PubMed] [Google Scholar]
- 22. Fiermonte G., Paradies E., Todisco S., Marobbio C. M., Palmieri F. (2009) A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme A and adenosine 3′,5′-diphosphate in human mitochondria. J. Biol. Chem. 284, 18152–18159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agrimi G., Russo A., Scarcia P., Palmieri F. (2012) The human gene SLC25A17 encodes a peroxisomal transporter of coenzyme A, FAD and NAD+. Biochem. J. 443, 241–247 [DOI] [PubMed] [Google Scholar]
- 24. Marobbio C. M., Agrimi G., Lasorsa F. M., Palmieri F. (2003) Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 22, 5975–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lasorsa F. M., Pinton P., Palmieri L., Fiermonte G., Rizzuto R., Palmieri F. (2003) Recombinant expression of the Ca2+-sensitive aspartate/glutamate carrier increases mitochondrial ATP production in agonist-stimulated Chinese hamster ovary cells. J. Biol. Chem. 278, 38686–38692 [DOI] [PubMed] [Google Scholar]
- 26. Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leadsham J. E., Gourlay C. W. (2010) cAMP/PKA signaling balances respiratory activity with mitochondria-dependent apoptosis via transcriptional regulation. BMC Cell Biol. 11, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Palmieri L., Agrimi G., Runswick M. J., Fearnley I. M., Palmieri F., Walker J. E. (2001) Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J. Biol. Chem. 276, 1916–1922 [DOI] [PubMed] [Google Scholar]
- 29. Hoyos M. E., Palmieri L., Wertin T., Arrigoni R., Polacco J. C., Palmieri F. (2003) Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J. 33, 1027–1035 [DOI] [PubMed] [Google Scholar]
- 30. Capobianco L., Bisaccia F., Mazzeo M., Palmieri F. (1996) The mitochondrial oxoglutarate carrier: sulphydryl reagents bind to cysteine 184 and this interaction is enhanced by substrate binding. Biochemistry 35, 8974–8980 [DOI] [PubMed] [Google Scholar]
- 31. Palmieri F., Rieder B., Ventrella A., Blanco E., Do P. T., Nunes-Nesi A., Trauth A. U., Fiermonte G., Tjaden J., Agrimi G., Kirchberger S., Paradies E., Fernie A. R., Neuhaus H. E. (2009) Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J. Biol. Chem. 284, 31249–31259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zara V., Palmieri F., Mahlke K., Pfanner N. (1992) The cleavable presequence is not essential for import and assembly of the phosphate carrier of mammalian mitochondria, but enhances the specificity and efficiency of import. J. Biol. Chem. 267, 12077–12081 [PubMed] [Google Scholar]
- 33. Palmisano A., Zara V., Hönlinger A., Vozza A., Dekker P. J., Pfanner N., Palmieri F. (1998) Targeting and assembly of the oxoglutarate carrier: general principles for biogenesis of carrier proteins of the mitochondrial inner membrane. Biochem. J. 333, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klingenberg M. (1989) Molecular aspects of the adenine nucleotide carrier from mitochondria. Arch. Biochem. Biophys. 270, 1–14 [DOI] [PubMed] [Google Scholar]
- 35. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 36. Bereiter-Hahn J. (1976) Dimethylaminostyrylmethylpyridiniumiodine (daspmi) as a fluorescent probe for mitochondria in situ. Biochim. Biophys. Acta 423, 1–14 [DOI] [PubMed] [Google Scholar]
- 37. Westermann B., Neupert W. (2000) Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 38. Volejníková A., Hlousková J., Sigler K., Pichová A. (2013) Vital mitochondrial functions show profound changes during yeast culture ageing. FEMS Yeast Res. 13, 7–15 [DOI] [PubMed] [Google Scholar]
- 39. McDonald B. M., Wydro M. M., Lightowlers R. N., Lakey J. H. (2009) Probing the orientation of yeast VDAC1 in vivo. FEBS Lett. 583, 739–742 [DOI] [PubMed] [Google Scholar]
- 40. Lai Y., Tse C.-M., Unadkat J. D. (2004) Mitochondrial Expression of the human equilibrative nucleoside transporter 1 (hENT1) results in enhanced mitochondrial toxicity of antiviral drugs. J. Biol. Chem. 279, 4490–4497 [DOI] [PubMed] [Google Scholar]
- 41. Govindarajan R., Leung G. P., Zhou M., Tse C.-M., Wang J., Unadkat J. D. (2009) Facilitated mitochondrial import of antiviral and anticancer nucleoside drugs by human equilibrative nucleoside transporter-3. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G910–G922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pontarin G., Gallinaro L., Ferraro P., Reichard P., Bianchi V. (2003) Origins of mitochondrial thymidine triphosphate: dynamic relations to cytosolic pools. Proc. Natl. Acad. Sci. U.S.A. 100, 12159–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pontarin G., Ferraro P., Håkansson P., Thelander L., Reichard P., Bianchi V. (2007) p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. J. Biol. Chem. 282, 16820–16828 [DOI] [PubMed] [Google Scholar]
- 44. Gandhi V. V., Samuels D. C. (2011) Enzyme kinetics of the mitochondrial deoxyribonucleoside salvage pathway are not sufficient to support rapid mtDNA replication. PLoS Comput. Biol. 7, e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bridges E. G., Jiang Z., Cheng Y. (1999) Characterization of a dCTP transport activity reconstituted from human mitochondria. J. Biol. Chem. 274, 4620–4625 [DOI] [PubMed] [Google Scholar]
- 46. Ferraro P., Nicolosi L., Bernardi P., Reichard P., Bianchi V. (2006) Mitochondrial deoxynucleotide pool sizes in mouse liver and evidence for a transport mechanism for thymidine monophosphate. Proc. Natl. Acad. Sci. U.S.A. 103, 18586–18591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Monné M., Palmieri F. (2014) Antiporters of the mitochondrial carrier family. Curr. Top. Membr. 73, 289–320 [DOI] [PubMed] [Google Scholar]
- 48. Fiermonte G., Dolce V., Palmieri F. (1998) Expression in Escherichia coli, functional characterization, and tissue distribution of isoforms A and B of the phosphate carrier from bovine mitochondria. J. Biol. Chem. 273, 22782–22787 [DOI] [PubMed] [Google Scholar]
- 49. Fiermonte G., Palmieri L., Todisco S., Agrimi G., Palmieri F., Walker J. E. (2002) Identification of the mitochondrial glutamate transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J. Biol. Chem. 277, 19289–19294 [DOI] [PubMed] [Google Scholar]
- 50. Indiveri C., Iacobazzi V., Tonazzi A., Giangregorio N., Infantino V., Convertini P., Console L., Palmieri F. (2011) The mitochondrial carnitine/acylcarnitine carrier: function, structure and physiopathology. Mol. Aspects Med. 32, 223–233 [DOI] [PubMed] [Google Scholar]
- 51. Xu Y., Johansson M., Karlsson A. (2008) Human UMP-CMP kinase 2, a novel nucleoside monophosphate kinase localized in mitochondria. J. Biol. Chem. 283, 1563–1571 [DOI] [PubMed] [Google Scholar]
- 52. Chen Y. L., Lin D. W., Chang Z. F. (2008) Identification of a putative human mitochondrial thymidine monophosphate kinase associated with monocytic/macrophage terminal differentiation. Genes Cells 13, 679–689 [DOI] [PubMed] [Google Scholar]
- 53. Rak M., Tetaud E., Godard F., Sagot I., Salin B., Duvezin-Caubet S., Slonimski P. P., Rytka J., di Rago J.-P. (2007) Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J. Biol. Chem. 282, 10853–10864 [DOI] [PubMed] [Google Scholar]
- 54. Yoon H., Zhang Y., Pain J., Lyver E. R., Lesuisse E., Pain D., Dancis A. (2011) Rim2, a pyrimidine nucleotide exchanger, is needed for iron utilization in mitochondria. Biochem. J. 440, 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Froschauer E. M., Rietzschel N., Hassler M. R., Binder M., Schweyen R. J., Lill R., Mühlenhoff U., Wiesenberger G. (2013) The mitochondrial carrier Rim2 co-imports pyrimidine nucleotides and iron. Biochem. J. 455, 57–65 [DOI] [PubMed] [Google Scholar]