Background: NEMO is a known ubiquitin-binding protein that functions as a key regulator of NF-κB activation.
Results: WA is able to covalently modify NEMO to induce a gain-of-function binding to long Lys-48-linked polyubiquitin chains.
Conclusion: WA can change the NEMO specificity for polyubiquitin chain interaction.
Significance: This study reveals a novel drug class to target the UBD:ubiquitin interaction.
Keywords: NF-kappa B (NF-κB), Polyubiquitin Chain, Recombinant Protein Expression, Small Molecule, Zinc Finger, NF-κB Essential Modulator, Withaferin A
Abstract
Post-translational modification by ubiquitin plays important roles in multiple physiological and pathological processes. Ubiquitin-binding proteins play a critical role in recognizing and relaying polyubiquitin-based signaling. NEMO (NF-κB Essential Modulator) is a central player in canonical NF-κB signaling whose major function is to bind to Lys-63- and/or M1- (or linear) linked polyubiquitin chains generated in response to cell stimulation. Here we show that Withaferin A (WA), a steroidal lactone, causes a change in NEMO's interaction with specific types of polyubiquitin chains in vitro. WA induces full-length recombinant NEMO to bind to long Lys-48-linked polyubiquitin chains but not tetra-ubiquitin species. Significantly, the UBAN (ubiquitin binding in ABIN and NEMO) domain, essential for the ability of NEMO to bind M1/Lys-63-linked polyubiquitin, is dispensable for the WA-induced gain-of-function activity. Mass spectrometric analysis demonstrated that WA covalently modifies NEMO on a cysteine residue within the C-terminal zinc finger (ZF) domain. Point mutations to the ZF can reverse the WA-induced Lys-48-polyubiquitin binding phenotype. Our study demonstrates the feasibility of directly altering the ubiquitin interaction properties of an ubiquitin-binding protein by a chemical compound, thereby shedding light on a novel drug class to potentially alter polyubiquitin-based cellular processes.
Introduction
Ubiquitin plays a crucial role in the regulation of multiple biological processes, including DNA repair, cell cycle regulation, and intracellular signaling (1–3). While its initial discovery characterized ubiquitin as a degradative signal, recent studies have shown a plethora of non-degradative signaling functions within the cell (4). Ubiquitin is a 76 amino acid protein whose signaling is carried out through its covalent attachment (ubiquitylation) to lysine residues on target substrates, either as a single molecule (monoubiquitylation) or as a polyubiquitin chain (polyubiquitylation). Polyubiquitin chains are formed through any of the 7 lysine residues on ubiquitin (Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, or Lys-63), as well as more recently recognized chains via the N-terminal methionine (M1/linear), to generate a “ubiquitin code” within the cell (5). Different ubiquitin arrangements are then recognized by ubiquitin binding domains (UBDs)2 residing in ubiquitin-binding proteins, which confer their specificity to result in distinctive signaling outcomes (6).
The main function of UBD-containing proteins is to read and decode the “ubiquitin code” and thus, there is a considerable interest in understanding the underlying mechanisms that regulate the specificity of ubiquitin-binding proteins for different species of ubiquitin. There are currently over 20 families of UBDs; however, based on their structures, UBDs can be categorized into two main types containing either α-helices or zinc fingers (ZF) (7). UBDs primarily recognize hydrophobic patches presented on the surface of ubiquitin. In general, the formation of different polyubiquitin chain linkages alters the availability of the hydrophobic patches that are presented to interact with UBDs. As the interpretation of in vivo ubiquitin binding studies can be complex, in vitro ubiquitin binding studies have been used to improve our understanding of the specificity of ubiquitin recognition by different UBDs.
NEMO (NF-κB essential modulator) is one of the most well characterized ubiquitin binding proteins due to its significance in canonical NF-κB signaling (8). In response to certain canonical NF-κB inducers, such as tumor necrosis factor (TNF) α or interleukin-1 (IL-1) β, the cell organizes a vast array of different polyubiquitin chains near the cell surface receptor (9–13). The ubiquitin signals are then transmitted through the recruitment of the IKK (inhibitor of κB kinase) complex through NEMO interaction with these polyubiquitin chains to allow for subsequent NF-κB activation. There are two known UBDs in NEMO, which consist of both structural classes of UBDs. The first is the α-helical UBAN (ubiquitin binding in ABIN and NEMO) domain, which has two known co-crystal structures with M1 and Lys-63-di-ubiquitin (14, 15). Abolishing the ubiquitin binding function of the UBAN domain via point mutations has been shown to severely attenuate NF-κB activation (14, 16–18). The other UBD in NEMO is the C-terminal ZF domain, which has M1- and Lys-63-linked polyubiquitin chain binding capabilities as well as a proposed model of this interaction (19). While the ZF domain is not generally necessary for NF-κB activation by canonical inducers (20), it does appear to be required for a full signaling response to TNFα, IL-1β, and bacterial lipopolysaccharide (LPS) (21).
Because of the diverse role that ubiquitin plays and its particularly important role in mediating NF-κB signaling, there has been an increasing interest in developing strategies to disrupt ubiquitin-UBD interactions (8, 22). While such outcomes can be experimentally achieved by mutagenesis of UBDs, small molecule inhibitors of ubiquitin-UBD interaction have not been widely developed. An example of these types of inhibitors was reported by Verma et al. in which ubistatins bound to Lys-48-linked polyubiquitin chains thereby disrupting degradation of substrates via the ubiquitin-dependent 26 S proteasome pathway (23). Similarly, Chiaravalli et al. showed that a peptide termed UBI (ubiquitin binding inhibitor), which spans the UBAN region of NEMO, was able to disrupt binding to Lys-63-linked but not M1-linked tetra-ubiquitin chains in vitro (24). Both of these strategies focused on abolishing ubiquitin binding; however, currently, there are no known natural or synthetic compounds that can change the ubiquitin binding specificity of a ubiquitin-binding protein. In this study we show that a chemical compound termed Withaferin A (WA), a steroidal lactone, can covalently modify NEMO to induce a gain-of-function phenotype to bind Lys-48-linked polyubiquitin chains in vitro and in vivo. Furthermore, we identified the critical domains of NEMO that participate in this gain-of-function phenotype as well as mapped the residue which is covalently modified by WA in vitro. Lastly, we generated several NEMO deletion and point mutants that both abolish and enhance this WA-induced Lys-48-linked polyubiquitin binding phenotype. Our findings identify WA as the first chemical compound of a potentially novel drug class of ubiquitin binding modulators.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Monoclonal α-Ub (PD41) and α-NEMO (FL419) antibodies purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Lys-63- (D7A11) and Lys-48- (D9D5) linkage-specific rabbit monoclonal antibodies purchased from Cell Signaling Technology (Danvers, MA) and α-NEMO purchased from BD Biosciences (San Jose, CA) were used in immunoprecipitation and immunoblotting experiments. Recombinant Lys-63-Ub3–7, Lys-48-Ub3–7, M1-Ub4, Lys-63-Ub4, and Lys-48-Ub4 polyubiquitin chains were purchased from Boston Biochem (Cambridge, MA). M1-Ub8 and Ub10 were purchased from ENZO Life Sciences (Farmingdale, NY). Gel Code® Blue Stain was purchased from Thermo Scientific (Waltham, MA). Withaferin A was purchased from EMD Millipore (Billerica, MA). Withanolide A, (4bS,5aS)-4a,6a-dimethyl-6,6a,8,9,9a,9b,10,11octahydrocyclopenta[7,8] phenanthro [4b,5-b] oxirene-2,7(4aH,5aH)-dione Cat#PH006594, 1,4,4a,5,8,8a-hexahydro-5-hydroxy-8-oxo-1 naphthalenecarboxylic acid Cat no. S956066, 2,3-diisopropoxyspiro[4.5]deca-2,7-diene-1,4,6,9-tetrone Cat#T272299, (4bS,5aS,7R,8R)-7-glycoloyl-7-hydroxy-4a,6a,8-trimethyl-5a,6,6a,7,8,9,9a,9b,10,11-decahydrocyclopenta[7,8]phenanthro[4b,5-b]oxiren-2(4aH)-one Cat. No. PH004604, and Bis(4-tert-butylphenyl) iodonium perfluoro-1-butanesulfonate Cat#T194999 were purchased from Sigma. DMSO (Sigma) was used in all reactions as a control solution. Catalogue number will be referred to for all results and figures.
Plasmids and Construction of NEMO Mutants
All genes are from corresponding human clones unless otherwise noted. GST-NEMO, GST-TAB2 525–693, GST-TAB2 663–693, and GST-TAB3 682–712 constructs were generated in the pGEX6p-1 vector system (GE Life Sciences) using BamHI/EcoRI restriction sites. Site-directed mutagenesis was conducting using QuikChangeII (Stratagene) using pGEX6p-1 NEMO WT as a template. GST-NEMO ΔZF (1–394aa) was generated via PCR as described previously (20) and subcloned into pGEX6p-1 using the following primers (Forward: 5′-CACACAGGATCCATGAATAGGCACCTCTGGAAGAGCCAACTG-3′ and Reverse: 5′-CACAGAATTCCTAGTCAGGTGGCTCCTC-3′). GST-TAB2 and -TAB3 constructs were generated using the following primers (TAB2 525aa Forward: 5′-GAGAGGATCCATGGGATCTGATGATGCTGCCTACAC-3′,TAB2–663aa Forward: 5′-GAGAGGATCCGATGAGGGAGCTCAGTGGAATTGTAC-3′ TAB2 693aa Reverse: 5′-GAGAGAATTCTCAGAAATGCCTTGGCATCTCACACTG-3′, TAB3 Forward: 5′-GAGAGGATCCATGGCGCAAAGCAGCCCACAGC-3′, TAB3 Reverse: 5′-GAGAGAATTCTCAGGTGTACCGTGGCATCTCG-3′) using pGEX6p-1 TAB2 and pGEX6p-1 TAB3 as templates, respectively. NEMO WT and ΔZF were generated in the pBABE-puro vector system using EcoRI and SalI sites. The integrity of the sequences was confirmed by direct sequencing. PreScission protease (GE Healthcare; Pittsburg, PA) was verified in collaboration with Dr. Anjon Audhya.
Cell Culture and Generation of Stable Cell Lines
HEK293 and NEMO−/− MEF cells were incubated in a 5% CO2 humidified incubator (Forma) and maintained in DMEM (Corning) medium supplemented with 10% fetal bovine serum (Seradigm) 100 IU of penicillin G (Sigma), and 0.1 mg of streptomycin sulfate (Sigma) per ml. U20S and RPE cells were grown in DMEM/F12 (Hyclone) supplemented with 10% FBS and penicillin/streptomycin. NEMO−/− MEF cells were retrovirally infected with NEMO WT or ΔZF constructs using the pBABE-puro retroviral system. Virus was produced by transiently transfecting NEMO WT, CC/SS, ΔZF, or vector alone and p-VSV-G into Phoenix HEK293 cells via calcium phosphate. Virus was harvested 48 h after transfection and placed through a 0.4-μm filter. MEF cells were incubated with virus for 12 h in cultured medium containing 6 μg/ml polybrene (Thermo Fisher Scientific). Cells were selected using 1.5 μg/ml of puromycin (Sigma).
Purification and Expression of Full-length GST-NEMO
Detailed protocols for purification of GST-NEMO and GST-PreScission protease will be described elsewhere. Briefly, pGEX6p-1 NEMO was transformed into BL21 Rosetta 2 Escherichia coli strain and purified via IPTG induction followed by lysis in GST-Lysis buffer (1× PBS, 250 mm NaCl, 0.5 mm EDTA, 0.5 mm EGTA, 10% glycerol, 0.1% Tween, pH 7.4) containing 1 μg/ml aprotinin, 1 μm leupeptin, and 1 mm PMSF protease inhibitors, and loaded onto GSH-agarose beads (Pierce). The column was then extensively rinsed with GST-Wash buffer (1xPBS, 250 mm NaCl, 10% glycerol, 0.1% Tween, 1 mm DTT) containing 1 mm PMSF and GST-NEMO fusion protein was eluted from the column with GST-Elution buffer (50 mm Tris-Cl, 75 mm NaCl, 10 mm reduced glutathione, pH 8.5). The fractions containing GST-NEMO, as detected by an SDS-PAGE gel and visualized by Gel Code (Pierce), were pooled and extensively dialyzed against GST-Dialysis buffer (20 mm Tris-Cl, 75 mm NaCl, 10% glycerol). For cleaved recombinant NEMO, 1 mg of GST-NEMO on GSH-agarose beads was incubated with 20 μg of GST-Prescission protease overnight at 4 °C while tumbling end-over-end. PreScission was purified similarly to GST-NEMO with the following changes. Following IPTG induction, bacterial cell pellets were lysed by sonication in PreScission Resuspension buffer (50 mm Tris, 150 mm NaCl, 10 mm EDTA, 20% glycerol). Protein was bound and eluted from GSH-agarose as above and extensively dialyzed against PreScission-Dialysis buffer (50 mm Tris-Cl, 150 mm NaCl, 10 mm EDTA, 20% glycerol, 1 mm DTT).
In Vitro Ubiquitin Binding Assay
5 μg of GST-NEMO WT and mutants and 1 μg of Lys-63-Ub3–7 or Lys-48-Ub3–7 polyubiquitin chains were rocked in the presence of indicated amounts of WA or DMSO control in a 200-μl total volume of GST-Lysis buffer at room temperature for 20 min. Subsequently, 10 μl of GSH-agarose beads (pre-washed in GST-Lysis buffer) were added to each reaction to capture GST-NEMO and rocked for 20 min at room temperature. The beads were washed 1× in 500 μl of GST-Wash buffer and transferred to a new 1.5-ml microcentrifuge tube to reduce background nonspecific signals. The GSH beads were then washed an additional three times and then heated at 70 °C for 5 min in SDS Laemmli sample buffer and run on an SDS-PAGE gel. For tetra-ubiquitin species 210 ng of GST-NEMO and 100 ng of each tetra-ubiquitin species were used to avoid binding saturation of the assay, which was separately determined by titration studies.
Covalent Modification of NEMO by WA
2 μg of recombinant NEMO cleaved of its GST tag by Prescission protease was incubated with increasing doses of WA in a 40 μl reaction at room temperature for 40 min. Samples were subsequently heated at 100 °C for 5 min in SDS Laemmli sample buffer and run on an SDS-PAGE gradient gel (Invitrogen) and visualized by Gel Code Coomassie stain.
Mass Spectrometry Analysis
Bands were excised from the gel, rinsed with 100 mm ammonium bicarbonate, and dried. Cysteines were reduced using 10 mm dithiothreitol in 100 mm ammonium bicarbonate at 56 ºC and alkylated with 55 mm iodoacetamide. Proteins were digested overnight at room temperature with 12.5 ng/μl trypsin. The proteins were eluted from the gel by alternating four times between 50% acetonitrile, 5% formic acid, and 50 mm sodium bicarbonate and an additional three times alternating between 100% acetonitrile and 50 mm NH4CO3. Eluted peptides were combined and dried. Peptides were separated using a NanoAcquity ultra high-pressure liquid chromatography system (Waters) coupled to LTQ-Velos Pro mass spectrometer (Thermo Fisher Scientific). Peptides were loaded onto a precolumn (75 μm inner diameter, packed with Magic C18 100Å 5 μm particles, Bruker) at a flow rate of 1 μl min−1. Peptides were eluted through an analytical column (75 μm inner diameter, packed with Magic C18 100 Å 5 μm particles, Bruker) using a 90-min linear gradient from 7.5 to 28% acetonitrile with 0.2% formic acid and a flow rate of 300 nl min−1 with monitoring of effluent by MS/MS. For the LTQ Velos Pro, a survey scan performed in the ion trap (MS1) was used to select ten precursors for tandem mass spectrometry (MS2) analysis. Selected peptides were fragmented by CID (NCE = 35) and analyzed in the ion trap. A small window (±1.5 m/z) was used to exclude a maximum of 500 selected precursors for 45 s with a repeat count of 1. Automatic gain control was used at target values of 40,000 for MS1 analysis and 10,000 for ion trap MS2 analysis. To analyze the data, the mass of Withaferin A was added to the precursor mass of tryptic peptides containing cysteine. If a peptide contained more than one cysteine, carbamidomethylation modifications were also looked for. Spectra containing the targeted masses were manually annotated to verify the Withaferin A modification.
Immunoprecipitation and Immunoblotting Analysis
Cells were grown to confluence on a 10-cm culture dish and treated with TNFα (10 ng/ml for 7 min.) or WA (10 μm for 30 min.) and harvested in 1× PBS then pelleted by centrifugation at 1000 rpm at 4 °C for 5 min. Cells were lysed in IP lysis buffer (20 mm Tris, pH 7.5, 250 mm NaCl, 3 mm EDTA, 3 mm EGTA, 0.5% Nonidet P-40, and 5% glycerol plus protease inhibitors, 10 mm NaF, 100 mm n-ethylmaleimide, and 20 mm iodoacetamide) for 30 min at 4 °C. Samples were spun down at 13,000 × g for 45 min at 4 °C. 500 μg of total soluble protein was used for immunoprecipitation with 5 μg of α-NEMO (FL419) in a total volume of 5 ml. Samples were tumbled at 4 °C for 16 h and then washed 4× in IP buffer containing 1 mm DTT and boiled in 2× SDS sample buffer for 5 min. Samples were run on a denaturing SDS-PAGE gel and transferred to a nitrocellulose membrane. Membranes were boiled in H2O for 10 min before incubating with primary antibodies. To visualize immunoprecipitated NEMO by Western blot we used α-NEMO (BD Bioscience) raised in mouse to prevent visualization of the antibody heavy chain. We also noticed that WA induced a shift and loss of signal of NEMO by FL419 antibody both in vitro and in vivo, which we attributed to potential masking of the epitope by WA (data not shown). Therefore, we used the α-NEMO (BD Bioscience) antibody for NEMO recognition of our inputs as well.
RESULTS
Full-length Recombinant NEMO Is Able to Selectively Bind M1- and Lys-63-linked Polyubiquitin Chains in Vitro
Using GST-tagged recombinant NEMO we set up and conducted an in vitro ubiquitin binding assay to assess the function and specificity of the purified recombinant protein (Fig. 1, A and C). It has previously been reported that NEMO has a binding preference for Lys-63- but not Lys-48-linked polyubiquitin chains (17). Therefore, we incubated recombinant Lys-63- or Lys-48-linked polyubiquitin chains with GST-NEMO to determine the specificity of ubiquitin binding. We found that GST-NEMO strongly prefers Lys-63-linked polyubiquitin chains to Lys-48-linked chains with further preference for longer polyubiquitin chains (Fig. 1B). NEMO has also been reported to bind to M1-linked (linear) polyubiquitin chains (14). Therefore, we also tested GST-NEMO ability to bind M1-linked tetra-ubiquitin using varying ratios of ubiquitin chain to GST-NEMO and found that our recombinant NEMO was efficiently able to bind M1-linked ubiquitin chains (Fig. 1D).
FIGURE 1.
GST-NEMO pulldown of Lys-63- and Lys-48-linked polyubiquitin chains. A, schematic representation of the in vitro ubiquitin binding assay. B, indicated amounts of Lys-63- or Lys-48-linked polyubiquitin chains (Boston Biochem) were incubated with 5 μg of GST-NEMO or GST. C, Coomassie stain of GST-NEMO purified protein run on SDS-PAGE gel. D, 100 ng of M1-linked tetra-ubiquitin was added to increasing amounts of GST-NEMO or GST. Samples were run on through SDS-PAGE and transferred to a nitrocellulose membrane. Membranes were stained with Ponceau S stain to verify loading of GST-fusion proteins and then were subsequently probed with a pan α-Ub antibody.
Withaferin A Selectively Induces the Gain-of-Function Ability for GST-NEMO to Bind Lys-48-linked Polyubiquitin Chains
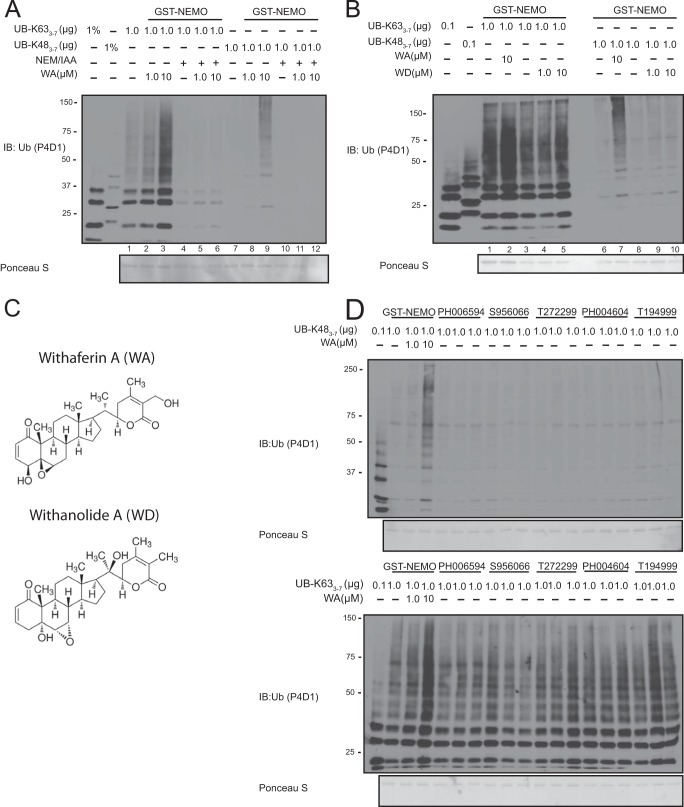
Previous reports have implicated WA in the disruption of NF-κB signaling through various mechanisms including attenuation of IKK activation (25). Recently, it was hypothesized that WA could disrupt the formation of the IKK complex in silico; however, this appears to not be the case based on previous literature (26, 27). Instead, we decided to test the hypothesis that WA could disrupt the NEMO-ubiquitin interaction. Therefore, we incubated recombinant GST-NEMO with WA in the presence of recombinant polyubiquitin chains to assess a change in ubiquitin binding. Contrary to our hypothesis, WA induced increased binding to longer Lys-63-linked ubiquitin chains; however, WA failed to increase binding to longer M1 chains of 8 and 10 molecule lengths (Fig. 2, A lanes 1–4, C). As a negative control we also tested Lys-48-linked chains, which have not been reported to interact with full-length NEMO. Surprisingly, WA also induced NEMO binding to longer chain Lys-48-linked ubiquitin chains (Fig. 2A, lanes 5–8). The presence of Lys-48 and Lys-63 linkages in these longer polyubiquitin chains was verified using Lys-48- and Lys-63-linkage specific antibodies, respectively (Fig. 2B). Using these antibodies we observed that indeed, WA induced an increase to long Lys-48- and Lys-63-linked polyubiquitin chains (Fig. 2D). These results revealed that WA has an unusual ability to confer NEMO the gain-of-function binding to Lys-48-linked polyubiquitin chains in vitro.
FIGURE 2.
WA can induce a change in the ubiquitin binding properties of NEMO. A, GST-NEMO and Lys-63- or Lys-48-linked polyubiquitin chains were incubated in the presence of the indicated amounts of WA and run through the in vitro ubiquitin binding assay. Samples were run through SDS-PAGE and probed with an α-Ub antibody. B, recombinant Lys-63- or Lys-48-linked polyubiquitin chains were run in duplicate and run through SDS-PAGE. The samples were probed with Lys-63- or Lys-48-polyubiquitin link specific antibodies. C, Ub8 and Ub10 M1-linked polyubiquitin chains were incubated with GST-NEMO as in A. The samples were probed with an α-Ub antibody to detect ubiquitin chains. D, same reaction as in A was run with GST-NEMO and GST and probed with chain specific antibodies as indicated.
Witherfin A Fails to Induce Increased Ubiquitin Binding of NEMO on Short Polyubiquitin Chains
The NEMO and ubiquitin interaction has been extensively studied in vitro as well as via structural analyses using short ubiquitin chains, such as di-ubiquitin (22). WA appeared to induce a stronger impact on NEMO's ability to bind longer polyubiquitin chains over shorter chains (Fig. 2A). To directly test whether WA could also change NEMO specificity for short ubiquitin chains, we tested the ability of WA to induce increased NEMO binding to tetra-ubiquitin chains. Interestingly, WA failed to induce an interaction between NEMO and Lys-48-linked tetra-ubiquitin chains (Fig. 3, lanes 7–9). WA also failed to induce increased binding for M1- and Lys-63-linked tetra-ubiquitin species (Fig. 3, lanes 1–6). These results further suggested that WA induces NEMO binding only to longer polyubiquitin chains and Lys-48-linked tetra-ubiquitin is insufficient to show the gain-of-function binding to NEMO following WA treatment.
FIGURE 3.
WA fails to induce Lys-48-linked tetra-ubiquitin binding of GST-NEMO. Top: GST-NEMO or GST was incubated with tetra-ubiquitin containing Lys-63-, Lys-48-, or M1-linkages in the presence of increasing amounts of WA. Bottom: GST was incubated with Lys-63-, Lys-48-, and M1-linked tetra-ubiquitin chains in the presence and absence of WA. Samples were run on through SDS-PAGE and probed with a pan α-Ub antibody.
The Zinc Finger (ZF) but Not the UBAN Domain of NEMO Is Necessary for Its WA-induced Lys-48-linked Ubiquitin Chain Interaction
NEMO has two known ubiquitin binding domains (UBD) termed the UBAN (ubiquitin binding in ABIN and NEMO, also known as NOA or NUB) and the ZF (zinc finger) domains (17, 28, 29). The current literature suggests that the UBAN of NEMO is necessary for canonical NF-κB activation; however, the ZF domain is not generally required (20). To test which of these domains plays a role in WA-induced Lys-48-linked ubiquitin chain binding, we generated a Y308S point mutation in the UBAN, previously shown to inhibit Lys-63-linked ubiquitin binding (17), and a truncation mutant that is missing the C-terminal ZF domain (ΔZF). As expected the Y308S point mutation in NEMO abolished nearly all Lys-63-linked ubiquitin chain interaction in the absence of WA (Fig. 4A, bottom panel: comparing lanes 1 and 4). Interestingly, WA still increased Lys-63-ubiquitin binding (Fig. 4A, bottom panel: lanes 4–6). As expected, the Y308S mutation had no impact on basal Lys-48-linked polyubiquitin binding; however, significantly, this mutation failed to abolish the WA-induced Lys-48-linked ubiquitin binding. Furthermore, a deletion of the NEMO ZF abolished the WA-induced Lys-48-linked ubiquitin chain interaction with GST-NEMO (Fig. 4A, top panel: lanes 7–9). These results identify the C-terminal ZF domain, but not the UBAN domain, as the required NEMO domain for WA-induced gain-of-function phenotype.
FIGURE 4.

Determining the Ubiquitin Binding Domain (UBD) required for WA induced Lys-48-linked polyubiquitin chain binding. A, top: GST and GST-NEMO WT, Y308S and ΔZF and Lys-48-linked polyubiquitin chains were incubated with the indicated concentration of WA in vitro. Bottom: same as above except with Lys-63-linked polyubiquitin chains. Samples were run on through SDS-PAGE and probed with a pan α-Ub antibody. B, Coomassie stain of GST fusion proteins.
WA Likely Modifies Cysteine Residue(s) of NEMO and Shows Specificity Over Other Similar Compounds
Previous reports have shown that WA is able to covalently modify cysteine resides of various proteins (30, 31). To test whether WA could also covalently modify NEMO on cysteine residue(s), we incubated GST-NEMO with two cysteine reactive compounds, NEM (N-ethylmaleimide) and IAA (iodoacetamide) in the presence and absence of WA. We observed that pre-treatment of NEM/IAA prevented the WA-induced Lys-48-linked ubiquitin chain interaction of GST-NEMO (Fig. 5A, lanes 10–12). Interestingly, this pre-treatment also showed a decrease in Lys-63-linked ubiquitin binding of GST-NEMO (Fig. 5A, lanes 4–6). Next, we tested a WA-like compound, Withanolide A (WD), for its ability to induce a change in NEMO's ubiquitin binding properties. While the structures of these compounds are extremely similar (Fig. 5C), WD failed to induce NEMO interaction with Lys-48-linked ubiquitin chains (Fig. 5B, lanes 8–10). We also tested 5 other chemical compounds that appeared to have similar reactive groups as WA; however, all of these compounds failed to induce an interaction between NEMO and Lys-48-linked polyubiquitin chains (Fig. 5D).
FIGURE 5.
Lys-48-linked polyubiquitin chain binding is specific to WA and cysteine modifiers can prevent WA effects. A, 5 μg of GST-NEMO was pre-incubated with 1 mm NEM and 1 mm IAA in GST-Lysis buffer for 20 min at room temperature. Recombinant Lys-48- or Lys-63-linked polyubiquitin chains were then added to the reaction in the presence of the indicated amounts of WA and run through the in vitro ubiquitin binding assay. B, 5 μg GST-NEMO was incubated with Lys-63- or Lys-48-linked polyubiquitin chains in the presence of the indicated amounts of WA or WD. Samples were run on through SDS-PAGE and probed with a pan α-Ub antibody. C, chemical structures of WA and WD from Sigma. D, top: GST-NEMO and Lys-48-linked polyubiquitin chains were incubated with the indicated chemical compound (see “Experimental Procedures” for full chemical name) in an in vitro ubiquitin binding assay. Samples were run through SDS-PAGE and probed with α-Ub antibody. Bottom: same as top except using Lys-63-linked polyubiquitin.
WA Covalently Modifies NEMO on Cysteine Residue 397
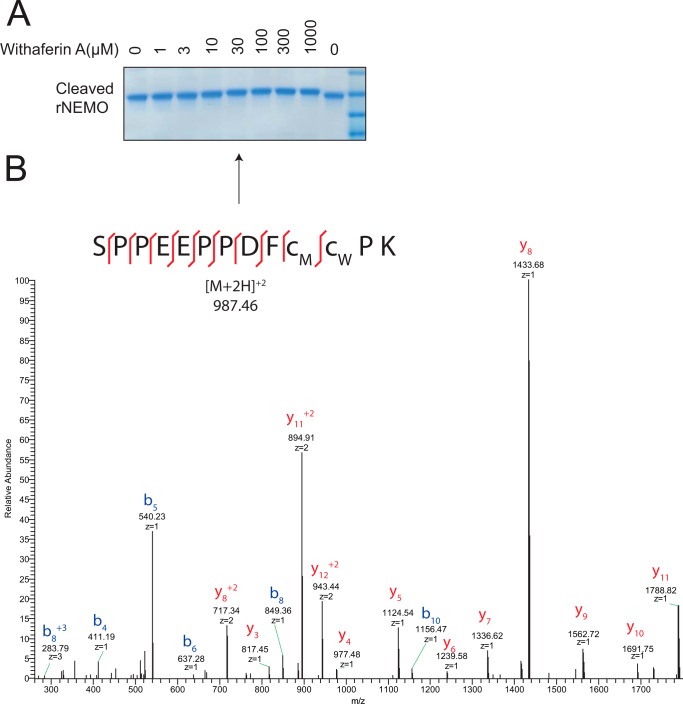
To confirm that WA was able to covalently modify a cysteine residue of NEMO, we incubated recombinant NEMO cleaved of its GST tag with increasing doses of WA. We observed that WA was able to induce a molecular weight shift in NEMO as measured by Coomassie stain (Fig. 6A). Recombinant NEMO treated with 30 μm WA was then excised from the gel and enzymatically digested with trypsin. Eluted peptides were separated via NanoAcquity HPLC and monitored via MS/MS. The mass of WA was added to cysteine containing peptides and analyzed for modification. Spectra were manually annotated to verify WA covalent modification of NEMO and found that cysteine residue 397 was the site of WA modification (Fig. 6B).
FIGURE 6.
WA covalently modifies NEMO on cysteine residue 397. A, 2 μg of recombinant NEMO (rNEMO) was incubated with the indicated amounts of WA for 40 min at room temperature with gentle agitation of the reaction mixture every 5 min. Samples were run on a gradient SDS-PAGE gel (Invitrogen) and Coomassie stained with Gel Code. Arrow indicates the band that was excised for mass spectrometry analysis. B, mass spectra for the identification of WA modified rNEMO on cysteine reside 397.
Point Mutations to the NEMO ZF Can Both Enhance and Abolish the WA-induced Gain-of-Function Phenotype of NEMO
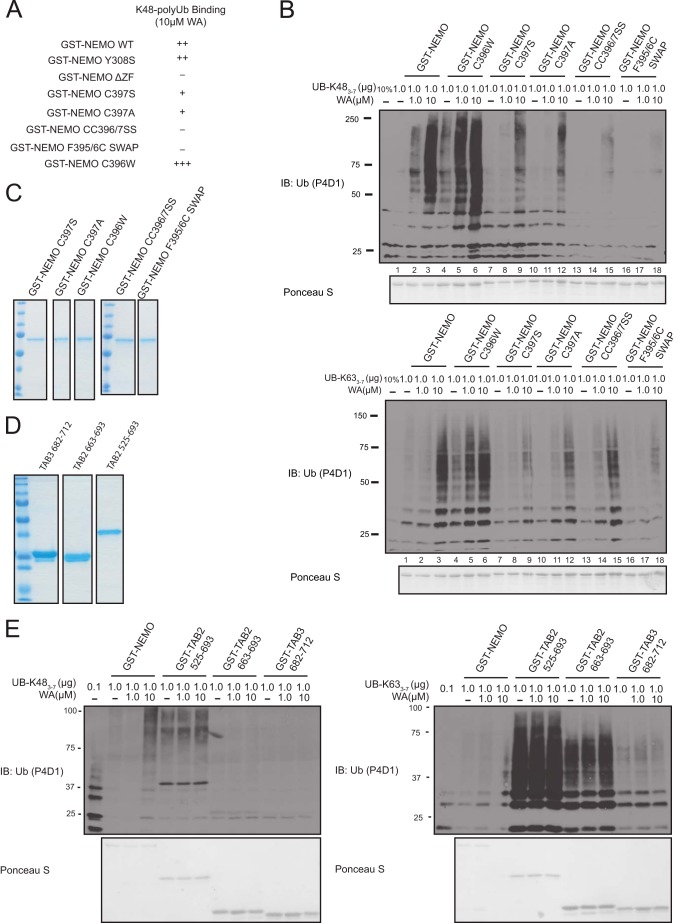
To test whether the cysteine residue 397 was critical for WA ability to increase GST-NEMO interaction with Lys-48-linked polyubiquitin chains, we mutated this residue to both serine (C397S) and alanine (C397A) and performed the ubiquitin binding assay as above with corresponding GST-NEMO versions. We observed that both C397S and C397A mutants showed a marked decrease in WA-induced Lys-48-linked ubiquitin chain binding compared with WT (Fig. 7B, top panel: compare lanes 3–9 and 12). Unique to NEMO ZF is an extra cysteine on residue 396 next to the presumed zinc-chelating cysteine 397 (32). We hypothesized that Cys-396 may have some compensatory effects for interacting with WA when Cys-397 is mutated. Therefore, we generated a double cysteine to serine mutant (C396S/C397S) and assessed its WA-induced Lys-48-linked polyubiquitin binding. The C396S/C397S NEMO mutant showed a further decrease in Lys-48-linked ubiquitin chain binding compared with the single C397S and C397A point mutants in the presence of WA (Fig. 7B, top panel: compare lanes 3 to 9, 11, and 15).
FIGURE 7.
Critical point mutations in the ZF of GST-NEMO disrupt the WA-induced Lys-48-linked polyubiquitin interaction. A, summary of NEMO constructs and their effect on WA-induced Lys-48-linked polyubiquitin chain interaction. B, top: GST-NEMO WT, C396W, C397S, C397A, C396S/C397S (CC396/7SS), or F395C/F396C (F395/396C) SWAP and Lys-48-linked polyubiquitin chains were incubated with the indicated concentration of WA and run through the in vitro ubiquitin binding assay. Bottom: same as above except with Lys-63-linked polyubiquitin chains. Samples were run on through SDS-PAGE and probed with a pan α-Ub antibody. C, Coomassie stain of GST fusion proteins. D, Coomassie stain of GST fusion proteins. E, left: GST-NEMO, GST-TAB2 (525–693aa), GST-TAB2 (663–693aa), or GST-TAB3 (682–712aa) and Lys-48-linked polyubiquitin chains were incubated with the indicated concentrations of WA in vitro. Right: same as above except with Lys-63-linked polyubiquitin chains. Samples were run through SDS-PAGE and probed with α-Ub antibody.
To reduce any potential compensatory effects of Cys-396 for Cys-397 we also swapped Cys-396 with phenylalanine 395 (Phe-F395) to generate a Cys-395/Phe-396 NEMO mutant. Surprisingly, this swap mutant had a dramatic reduction in WA-induced Lys-48-linked ubiquitin binding (Fig. 7B, top panel: lanes 16–18). Conversely, the addition of a tryptophan residue to replace Cys-396 (C396W) showed an increase in both basal and WA-induce Lys-48-linked ubiquitin binding (Fig. 7B, top panel: compare lanes 1–3 and 4–6). We also observed that all NEMO mutants, except the Cys-395/Phe-396 swap, had very little effect on Lys-63-linked ubiquitin binding suggesting that these residues are specifically involved in increasing Lys-48-linked polyubiquitin chain interaction. A summary of all NEMO mutants Lys-48-linked ubiquitin binding in the presence of WA can be found in Fig. 7A. We also tested the effects of WA on the UBDs of TAB2 and TAB3, which are components of TAK1 kinase complex that is thought to recruit TAK1 to the IKKβ to mediate NF-κB signaling (33). These UBDs termed Npl4 zinc fingers (NZFs) have a known specificity for Lys-63-linked polyubiquitin chain binding (34). We observed no change in either TAB2 or TAB3 NZF preference for Lys-48-linked polyubiquitin chains (Fig. 7, D and E). Collectively, our results demonstrate that WA acts directly on Cys-397 of NEMO to alter the specificity of its ubiquitin binding to long-chain Lys-48-linked polyubiquitin chains in vitro. Mutations of Cys-397 or residues near this residue can alter the responsiveness of NEMO to WA.
WA Can Induce Increased Association between NEMO and Ubiquitin in Cells
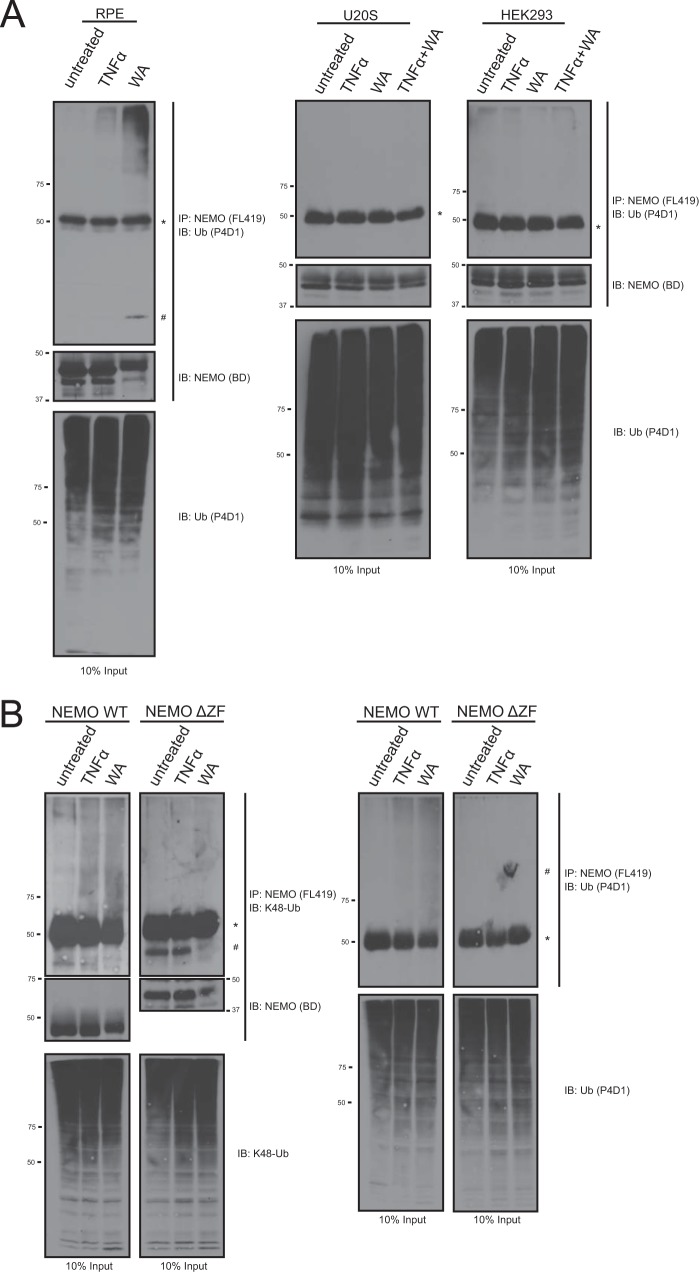
To determine if this phenotype could affect endogenous NEMO we next treated multiple cell lines with WA to determine if it could induce NEMO gain-of-function phenotype to interact with polyubiquitin chains. HEK293, U2OS, and RPE cells were treated with the indicated amount of TNFα or WA and then harvested and lysed. Endogenous NEMO was immunoprecipitated with α-NEMO antibody to evaluate the presence of co-immunoprecipitated ubiquitin from the cell lysate detected by Western blotting. In RPE cells we observed an increase in polyubiquitin chain interaction in cells treated with WA (Fig. 8A). As a positive control we treated cells with TNFα, which has previously been shown to induce the formation of multiple polyubiquitin chains, including Lys-48-linked ubiquitin, at the TNFR (12), and this also showed an increase association with polyubiquitin chains (Fig. 8A). Surprisingly, we also noticed that in WA-treated lanes NEMO ran at a higher molecular weight. This suggests that NEMO is modified in the cell; however, it remains to be tested if this NEMO band is modified by WA in vivo (Fig. 8A, RPE, middle panel). Interestingly, we failed to observe any shift in NEMO band and association between endogenous NEMO and ubiquitin chains in either HEK293 or U2OS cells treated with TNFα or WA (Fig. 8A, right panels).
FIGURE 8.
WA is able to induce an association between both endogenous and exogenous NEMO and polyubiquitin chains in a ZF-dependent manor. A, RPE, U20S, and HEK293 cells were treated with TNFα (10 ng/ml) or WA (10 μm) and subsequently harvested and lysed. Endogenous NEMO was immunoprecipitated with FL419 NEMO antibody. Co-immunoprecipitated polyubiquitin chains were identified by running co-IP samples on an SDS-PAGE gel and probing with an α-Ub antibody. Additionally, NEMO was also probed for using an α-NEMO antibody raised in mouse. B, NEMO−/− MEF cell lines infected with either NEMO WT or NEMO ΔZF were generated and assayed as in A. Samples were probed with a Lys-48-chain specific ubiquitin antibody (left panel) or α-Ub (P4D1) antibody (right panel). Additionally, NEMO was also visualized by probing with an α-NEMO antibody raised in mouse. * denotes heavy chain that was identified by the α-Ub (P4D1) antibody. # denotes a nonspecific band.
To assess if the cysteine point mutations in NEMO could attenuate the WA induced association between NEMO and ubiquitin in vivo we first generated NEMO−/− MEF stable cell lines reconstituted with NEMO WT or C396S/C397S mutant. Treatment of both TNFα and WA were able to induce an increase in association between NEMO WT and Lys-48-linked polyubiquitin chains (Fig. 8B). Interestingly, mutagenesis of C396S/C397S into NEMO failed to attenuate the WA-induced interactions with polyubiquitin chains (data not shown). This result is not entirely inconsistent with our in vitro data which showed that the CC/SS mutant attenuated but did not completely abolish the effects of WA (Fig. 7B compare lanes 3 and 15). Therefore, we next reconstituted the NEMO−/− MEF cells with NEMO ΔZF which was able to completely abolish the WA-induced association with polyubiqution chains in vitro (Fig. 4A). The deletion of the NEMO ZF was able to completely abolish its WA gain-of-function interaction with Lys-48-polyubiquitin chains (Fig. 8B). Probing with a pan-ubiquitin antibody verified this result for all types of ubiquitin chains present in the IP. It should be noted that probing for NEMO we can see a molecular weight shift in cells treated with WA for NEMO WT and a reduced shift for NEMOΔZF. However, we have not verified if this shift is due to a covalent modification by WA.
DISCUSSION
The specific interaction between ubiquitin moieties and UBDs is thought to transmit intracellular ubiquitin signals, which control many different biological outcomes that are often dysregulated in human diseases (6). Modulating this interaction could therefore have biological and therapeutic implications. About a decade ago Verma et al. discovered a novel set of chemicals, called Ubistatins, which are able to interfere with the proteasome's recognition of Lys-48-linked polyubiquitylated substrates. There are several UBD containing proteins that selectively recognize Lys-48-polyubiquitylated substrates on the 19 S cap of the 26 S proteasome that help to facilitate protein degradation. Ubistatins were shown to directly interact with ubiquitin at the Lys-48-linkage between two ubiquitin molecules to disrupt the Ile-44 hydrophobic patch known to interact with UBDs. The binding of Ubistatins to Lys-48-linked ubiquitin molecules ultimately disrupted the Lys-48-linked ubiquitin:UBD interaction on the proteasome to prevent protein degradation (23). To date, this is one of the only publications that has observed a way to chemically modulate the ubiquitin:UBD interaction. Here we observed that a chemical compound, WA, is able to covalently modify NEMO to alter its ubiquitin binding properties. This represents a potentially novel strategy to regulate the ubiquitin:UBD interaction by changing the way a UBD-containing protein is able to recognize ubiquitin, and thus potentially alter the biological outcome of the ubiquitin signal.
One surprising feature of WA modulation of the NEMO:Ubiquitin interaction is that it does not require a functional UBAN domain (Fig. 4). Previous studies have shown that disruption of the NEMO ubiquitin binding properties via point mutations in the UBAN domain severely attenuate NF-κB activation and are associated with anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) (35). Therefore, we anticipated the requirement of the UBAN domain for NEMO's WA-induced interaction with Lys-48-linked polyubiquitin chains. Instead we found that the WA-induced Lys-48-binding phenotype required the ZF domain of NEMO for covalent modification. Agou et al. have published a series of studies that implicated the ZF of NEMO as a bona fide UBD (29, 36). Most recently, Ngadjeua et al. have modeled the NEMO ZF with Lys-63-linked di-ubiquitin, through extensive mutational and genetic complementation studies. Interestingly, in their analysis they implicated Phe-395 as a critical reside for interacting with the Ile-44 patch of the proximal Lys-63-linked di-ubiquitin (19). We observed that swapping Phe-395 and Cys-396 to generate Cys-395/Phe-396 nearly abolished all WA-induced increased polyubiquitin chain binding (Fig. 7). The major way NEMO is able to interact with M1- and Lys-63-linked ubiquitin chains is through linkage context. Different ubiquitin linkages present specific combinations of hydrophobic patches that can be recognized by UBDs. In the case of NEMO, while it cannot detect the direct M1- or Lys-63-linked peptide bond, its UBAN is able to “read” the Ile-44 patch on one ubiquitin molecule and either the phenylalanine 4 or Ile-44 patch of the adjacent ubiquitin molecule, respectively (14, 15). The conformation and structure of free Lys-48-linked ubiquitin chains is more compact than either M1- or Lys-63-linked ubiquitin and therefore will likely have different interaction surfaces. However, there are currently no co-crystal structures of UBDs with Lys-48-linked ubiquitin chains to validate this hypothesis. Moreover, the structure of full-length NEMO is currently also unknown, which poses a second technical hurdle in modeling binding of NEMO with Lys-48-linked ubiquitin chains. Thus, further investigation is needed to elucidate how WA-modified NEMO selectively binds to long Lys-48-linked polyubiquitin chains.
We also found that WA induced a Lys-48-linked ubiquitin binding phenotype onto NEMO but not TAB2 or TAB3 UBDs, suggesting that WA has specificity of reactivity toward the NEMO ZF domain. Interestingly, NEMO ZF contains two consecutive cysteine residues (Cys-396 and Cys-397) that are missing in all other known UBD ZF domains, including those of TAB2/3. Several chemical studies have implicated the 2(3)-en-1-one moiety in the unsaturated A-ring at the 3rd carbon position as the major reactive group on WA (37). Moreover, WA has been shown to covalently modify cysteine residues of other molecules (30, 31) most likely through this chemical group. We have shown that a closely related family member, Withanolide A (WD), which lacks this chemically reactive moiety failed to induce a change in the NEMO ubiquitin binding properties (Fig. 5). Mere modification by any cysteine reactive compounds, such as NEM and IAA, is also insufficient to induce Lys-48-linked polyubiquitin binding. This suggests that the chemical structure of WA is critical for its specificity and reactivity to induce NEMO ZF cysteine modification and subsequent long Lys-48-linked ubiquitin chain binding. Additional investigation into WA-related chemicals that retain the cysteine-reactive moiety present in other family members/analogues of Withania somnifera (e.g. derivatives shown in Ref. 37) will help elucidate the chemical mechanism of how WA is able to modify NEMO and alter its ubiquitin binding properties. These studies along with further ZF domain analysis could reveal a way to target only NEMO in a complex mixture of cellular environment and allow for improved drug development of this potentially novel class of ubiquitin binding modifiers.
In conclusion, we have shown that WA is able to directly modify Cys-397 on the NEMO ZF and alter its ability to interact with polyubiquitin chains. While the precise mechanism of how WA-modified NEMO is able to recognize long Lys-48-linked polyubiquitin chains remains elusive, we have identified the first chemical compound that can alter the ubiquitin binding properties of a UBD containing protein in vitro. Furthermore, we were able to show that WA can be treated onto cells to induce an increase in the NEMO affinity for Lys-48-linked polyubiquitin chains in vivo as measured by co-immunoprecipitiation analysis. Surprisingly, this phenotype was not completely attenuated via mutagenesis of critical cysteine sites suggesting a potential alternative mechanism in cells. However, it is equally possible that the co-IP assay presents a completely saturated condition where the concentration of ubiquitin is significantly greater than in the in vitro pulldown assay. Regardless, we were able to show that the ZF of NEMO is a critical domain for this phenotype as deletion of this domain completely abolished WA effect on the NEMO and Lys-48-linked ubiquitin chain interaction. Taken together this study presents a potentially novel strategy for regulating the “ubiquitin code” by directly targeting the ubiquitin binding protein of interest, which may subsequently permit a modulation of the biological outcome of the ubiquitin signal.
Acknowledgments
We thank Dr. Anjon Audhya for help in the purification of his PreScission protease. We would like to thank Drs. Richard Burgess, Anjon Audhya, Byounghoon Hwang, Kevin McCool, and graduate student Adam Johnson for help and expertise in protein purification. We would also like to thank Drs. Christopher C. Oberley and Eric Strieter for their thoughtful discussions on experiments and interpretations.
This work was supported by National Institutes of Health Grants R01 GM083681 and R01 CA077474.
- UBD
- ubiquitin binding domain
- NF-κB
- nuclear factor-κB
- NEMO
- NF-κB essential modulator
- M1
- linear ubiquitin
- WA
- Withaferin A
- UBAN
- ubiquitin binding in ABIN and NEMO
- ZF
- zinc finger
- TNF α
- tumor necrosis factor α
- IL-1β
- interleukin-1 β
- IKK
- IκB kinase
- LPS
- lipopolysaccharide
- UBI
- ubiquitin binding inhibitor
- GSH
- glutathione
- NOA
- NEMO-OPTINEURON-ABIN
- NUB
- NEMO ubiquitin binding
- NEM
- N-ethylmaleimide
- IAA
- iodoacetamide
- WD
- Withanolide A
- NZF
- npl4 zinc finger
- TAB
- TGF-β-activated kinase 1-binding protein
- EDA-ID
- ectodermal dysplasia with immunodeficiency.
REFERENCES
- 1. Bergink S., Jentsch S. (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 [DOI] [PubMed] [Google Scholar]
- 2. Teixeira L. K., Reed S. I. (2013) Ubiquitin ligases and cell cycle control. Annu. Rev. Biochem. 82, 387–414 [DOI] [PubMed] [Google Scholar]
- 3. Chen J., Chen Z. J. (2013) Regulation of NF-κB by ubiquitination. Curr. Opin. Immunol. 25, 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kulathu Y., Komander D. (2012) Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13, 508–523 [DOI] [PubMed] [Google Scholar]
- 5. Komander D., Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 6. Husnjak K., Dikic I. (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 [DOI] [PubMed] [Google Scholar]
- 7. Dikic I., Wakatsuki S., Walters K. J. (2009) Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark K., Nanda S., Cohen P. (2013) Molecular control of the NEMO family of ubiquitin-binding proteins. Nat. Rev. Mol. Cell Biol. 14, 673–685 [DOI] [PubMed] [Google Scholar]
- 9. Newton K., Matsumoto M. L., Wertz I. E., Kirkpatrick D. S., Lill J. R., Tan J., Dugger D., Gordon N., Sidhu S. S., Fellouse F. A., Komuves L., French D. M., Ferrando R. E., Lam C., Compaan D., Yu C., Bosanac I., Hymowitz S. G., Kelley R. F., Dixit V. M. (2008) Ubiquitin Chain Editing Revealed by Polyubiquitin Linkage-Specific Antibodies. Cell 134, 668–678 [DOI] [PubMed] [Google Scholar]
- 10. Xu M., Skaug B., Zeng W., Chen Z. J. (2009) A Ubiquitin Replacement Strategy in Human Cells Reveals Distinct Mechanisms of IKK Activation by TNFα and IL-1β. Mol. Cell 36, 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dynek J. N., Goncharov T., Dueber E. C., Fedorova A. V., Izrael-Tomasevic A., Phu L., Helgason E., Fairbrother W. J., Deshayes K., Kirkpatrick D. S., Vucic D. (2010) c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 29, 4198–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerlach B., Cordier S. M., Schmukle A. C., Emmerich C. H., Rieser E., Haas T. L., Webb A. I., Rickard J. A., Anderton H., Wong W. W. L., Nachbur U., Gangoda L., Warnken U., Purcell A. W., Silke J., Walczak H. (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 [DOI] [PubMed] [Google Scholar]
- 13. Ikeda F., Deribe Y. L., Skånland S. S., Stieglitz B., Grabbe C., Franz-Wachtel M., van Wijk S. J. L., Goswami P., Nagy V., Terzic J., Tokunaga F., Androulidaki A., Nakagawa T., Pasparakis M., Iwai K., Sundberg J. P., Schaefer L., Rittinger K., Macek B., Dikic I. (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 15. Yoshikawa A., Sato Y., Yamashita M., Mimura H., Yamagata A., Fukai S. (2009) Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS Lett. 583, 3317–3322 [DOI] [PubMed] [Google Scholar]
- 16. Kensche T., Tokunaga F., Ikeda F., Goto E., Iwai K., Dikic I. (2012) Analysis of nuclear factor-κB (NF-κB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-κB. J. Biol. Chem. 287, 23626–23634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ea C.-K., Deng L., Xia Z.-P., Pineda G., Chen Z. J. (2006) Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22, 245–257 [DOI] [PubMed] [Google Scholar]
- 18. Emmerich C. H., Ordureau A., Strickson S., Arthur J. S. C., Pedrioli P. G. A., Komander D., Cohen P. (2013) Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. U.S.A. 110, 15247–15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ngadjeua F., Chiaravalli J., Traincard F., Raynal B., Fontan E., Agou F. (2013) Two-sided Ubiquitin Binding of NF- B Essential Modulator (NEMO) Zinc Finger Unveiled by a Mutation Associated with Anhidrotic Ectodermal Dysplasia with Immunodeficiency Syndrome. J. Biol. Chem. 288, 33722–33737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang T. T., Feinberg S. L., Suryanarayanan S., Miyamoto S. (2002) The zinc finger domain of NEMO is selectively required for NF-κB activation by UV radiation and topoisomerase inhibitors. Mol. Cell Biol. 22, 5813–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Makris C., Roberts J. L., Karin M. (2002) The Carboxyl-Terminal Region of IκB Kinase γ (IKKγ) Is Required for Full IKK Activation. Mol. Cell Biol. 22, 6573–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lo Y. C., Lin S. C., Rospigliosi C. C., Conze D. B., Wu C.-J., Ashwell J. D., Eliezer D., Wu H. (2009) Structural basis for recognition of diubiquitins by NEMO. Mol. Cell 33, 602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Verma R., Peters N. R., D'Onofrio M., Tochtrop G. P., Sakamoto K. M., Varadan R., Zhang M., Coffino P., Fushman D., Deshaies R. J., King R. W. (2004) Ubistatins Inhibit Proteasome-Dependent Degradation by Binding the Ubiquitin Chain. Science 306, 117–120 [DOI] [PubMed] [Google Scholar]
- 24. Chiaravalli J., Fontan E., Fsihi H., Coic Y.-M., Baleux F., Véron M., Agou F. (2011) Direct inhibition of NF-κB activation by peptide targeting the NOA ubiquitin binding domain of NEMO. Biochem. Pharmacol. 82, 1163–1174 [DOI] [PubMed] [Google Scholar]
- 25. Vanden Berghe W., Sabbe L., Kaileh M., Haegeman G., Heyninck K. (2012) Molecular insight in the multifunctional activities of Withaferin A. Biochem. Pharmacol. 84, 1282–1291 [DOI] [PubMed] [Google Scholar]
- 26. Grover A., Shandilya A., Punetha A., Bisaria V. S., Sundar D. (2010) Inhibition of the NEMO/IKKβ association complex formation, a novel mechanism associated with the NF-κB activation suppression by Withania somnifera's key metabolite withaferin A. BMC genomics 11, S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaileh M., Vanden Berghe W., Heyerick A., Horion J., Piette J., Libert C., De Keukeleire D., Essawi T., Haegeman G. (2007) Withaferin A Strongly Elicits I B Kinase beta Hyperphosphorylation Concomitant with Potent Inhibition of Its Kinase Activity. J. Biol. Chem. 282, 4253–4264 [DOI] [PubMed] [Google Scholar]
- 28. Wu C.-J., Conze D. B., Li T., Srinivasula S. M., Ashwell J. D. (2006) Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected]. Nat. Cell Biol. 8, 398–406 [DOI] [PubMed] [Google Scholar]
- 29. Cordier F., Grubisha O., Traincard F., Véron M., Delepierre M., Agou F. (2009) The Zinc Finger of NEMO Is a Functional Ubiquitin-binding Domain. J. Biol. Chem. 284, 2902–2907 [DOI] [PubMed] [Google Scholar]
- 30. Bargagna-Mohan P., Hamza A., Kim Y.-E., Khuan Abby Ho Y., Mor-Vaknin N., Wendschlag N., Liu J., Evans R. M., Markovitz D. M., Zhan C.-G., Kim K. B., Mohan R. (2007) The tumor inhibitor and antiangiogenic agent withaferin A targets the intermediate filament protein vimentin. Chem. Biol. 14, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ozorowski G., Ryan C. M., Whitelegge J. P., Luecke H. (2012) Withaferin A binds covalently to the N-terminal domain of annexin A2. Biol. Chem. 393, 1151–1163 [DOI] [PubMed] [Google Scholar]
- 32. Cordier F., Vinolo E., Véron M., Delepierre M., Agou F. (2008) Solution structure of NEMO zinc finger and impact of an anhidrotic ectodermal dysplasia with immunodeficiency-related point mutation. J. Mol. Biol. 377, 1419–1432 [DOI] [PubMed] [Google Scholar]
- 33. Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J.-I., Chen Z. J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 34. Kulathu Y., Akutsu M., Bremm A., Hofmann K., Komander D. (2009) Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 16, 1328–1330 [DOI] [PubMed] [Google Scholar]
- 35. Courtois G., Gilmore T. D. (2006) Mutations in the NF-κB signaling pathway: implications for human disease. Oncogene 25, 6831–6843 [DOI] [PubMed] [Google Scholar]
- 36. Laplantine E., Fontan E., Chiaravalli J., Lopez T., Lakisic G., Véron M., Agou F., Israël A. (2009) NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 28, 2885–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wijeratne E. M. K., Xu Y.-M., Scherz-Shouval R., Marron M. T., Rocha D. D., Liu M. X., Costa-Lotufo L. V., Santagata S., Lindquist S., Whitesell L., Gunatilaka A. A. L. (2014) Structure-Activity Relationships for Withanolides as Inducers of the Cellular Heat-Shock Response. J. Med. Chem. 57, 2851–2863 [DOI] [PubMed] [Google Scholar]