Abstract
• Aims Plants and animals represent the first two kingdoms recognized, and remain the two best-studied groups in terms of nuclear DNA content variation. Unfortunately, the traditional chasm between botanists and zoologists has done much to prevent an integrated approach to resolving the C-value enigma, the long-standing puzzle surrounding the evolution of genome size. This grand division is both unnecessary and counterproductive, and the present review aims to illustrate the numerous links between the patterns and processes found in plants and animals so that a stronger unity can be developed in the future.
• Scope This review discusses the numerous parallels that exist in genome size evolution between plants and animals, including (i) the construction of large databases, (ii) the patterns of DNA content variation among taxa, (iii) the cytological, morphological, physiological and evolutionary impacts of genome size, (iv) the mechanisms by which genomes change in size, and (v) the development of new methodologies for estimating DNA contents.
• Conclusions The fundamental questions of the C-value enigma clearly transcend taxonomic boundaries, and increased communication is therefore urged among those who study genome size evolution, whether in plants, animals or other organisms.
Keywords: Body size, cell size, complexity, C-value enigma, C-value paradox, database, development, DNA content, genome size, introns, physiology, selfish DNA, sequencing, transposable elements
INTRODUCTION
In the beginning (of taxonomy), there were two kingdoms of life: green things were plants and moving things were animals. Today, thanks in no small part to comparative genomic analyses, four kingdoms of eukaryotes are commonly recognized (Animalia, Plantae, Fungi, Protozoa), all of them contained in the Eukarya, which many authors argue is just one of three ‘empires’ (along with Bacteria and Archaea). The process of splitting is still not complete, given that ‘protists’ probably comprise two kingdoms of their own (Protozoa and Chromista) while also having representatives in the others (Cavalier-Smith, 1998). To be sure, an acknowledgement of the deeply branching diversity of living things has been a great achievement in evolutionary biology, but somehow the equally important underlying unity—most notably, the simple fact that everything alive has a genome which can be compared—has been forgotten.
It is a mere truism that for every genome there is a size, defined by either mass (in picograms, pg) or number of base pairs (bp). It is far from a given, however, that these sizes should be (mostly) constant species-specific characters, that they should vary over several orders of magnitude among eukaryotes, or that they should bear no connection to organismal complexity (Gregory, 2001a). In fact, each of these features came as a major surprise to early researchers. Thus, Vendrely and Vendrely (1948) were struck by ‘a remarkable constancy in the nuclear DNA content of all the cells in all the individuals within a given animal species’ (my translation), and Comings (1972) lamented that ‘the lowly liverwort has 18 times as much DNA as we, and the slimy, dull salamander known as Amphiuma has 26 times our complement of DNA’.
The C-value enigma: a cross-kingdom puzzle
As part of a defence of the Vendrelys' ‘DNA constancy hypothesis’, Hewson Swift (1950a, b) studied DNA contents in different tissues of both animals (frog, mouse and grasshopper) and plants (Tradescantia and Zea) and developed the concept of the ‘C-value’ in reference to the haploid, or 1C, DNA amount. (In diploid organisms, including the vast majority of animals but probably a minority of plants, ‘genome size’ and ‘C-value’ will be identical; in recent polyploids, the situation is more complex because the C-value will comprise more than one genome.) DNA constancy, upon which the C-value concept was based, was in turn taken as evidence that DNA, and not the highly variable proteins, serves as the hereditary material (e.g. Swift, 1950a).
The expression of the combination of demonstrable DNA constancy (within species) and profound genome size variation (among species) as a ‘C-value paradox’ (Thomas, 1971) is easy to comprehend. However defined—as ‘simple’ organisms having more DNA than ‘complex’ ones, in terms of some closely related species displaying highly divergent DNA contents, or by noting that any given organism contains more DNA than would be expected based on its presumed number of genes—the basic ‘paradox’ was that DNA amount is constant because it is the stuff of genes, and yet is unrelated to expected gene number.
The solution to the paradox is now well known: most DNA is non-coding, so the size of a genome need not imply anything at all about the number of genes it contains. The term ‘C-value paradox’ persists, but only by virtue of historical entrenchment. Certainly, the discovery of non-coding DNA, which ended the paradox, raised a number of questions of its own. Whence this non-coding DNA? Does it have any phenotypic effects (or even functions)? How is it gained and lost? What are the patterns of its distribution among taxa? Why do some species contain so much of it and others so little?
Because a ‘paradox’ begs a one-dimensional solution, there is a tendency to deal with only one of these questions and to present an answer as ‘the’ solution to the problem of genome size evolution. A much more productive approach is to recognize these issues, taken together, as components of a complex puzzle—the ‘C-value enigma’ (Gregory, 2001a, 2005). Importantly, this distinction makes it immediately clear that the enigma applies to all eukaryotes, but that the particulars will vary according to the biology of the groups in question.
Most of the existing genome size information comes from plants and animals, and much progress has been made in exploring the C-value enigma's various questions. Unfortunately, this work has often proceeded in parallel, with very limited interaction, by botanists on one side of the classic divide and zoologists on the other. However, and as Comings's (1972) disdain for liverwort genomes clearly attests, the puzzle of genome size variation (now past its 50th year in existence) clearly transcends these taxonomic boundaries. The purpose of this paper is to further the construction of bridges across the original taxonomic chasm which still divides departments, literature and, too often, communication.
THE STATE OF KNOWLEDGE REGARDING PLANT AND ANIMAL GENOME SIZES
Plant DNA C-values Database
One of the first broad comparisons of available genome size data was made three decades ago by Sparrow et al. (1972), and included values from plants, animals, fungi and prokaryotes. Around this time, a large number of additional C-values began to be measured and compiled in order to compare DNA content with features of practical interest, such as cell and life cycle duration and geographic distribution. As data began to accumulate, it was recognized that additional published lists could be of great value, and a few years later Bennett and Smith (1976) presented the first in a series of plant DNA C-value compilations that extends to the present day (Bennett et al., 1982; Bennett and Smith, 1991; Bennett and Leitch, 1995, 1997, 2001; Murray, 1998; Bennett et al., 2000a; Voglmayr, 2000).
The first electronic version of the Angiosperm DNA C-values Database was made available in April 1997, in anticipation of the plant genome size meetings at Kew in September of that year. Since 2001, it has been presented as an expanded Plant DNA C-values Database that includes all the major groups of land plants. Release 2.0 of the Plant DNA C-values Database, launched in January of 2003, currently includes data from nearly 4000 species consisting of 3493 angiosperms, 181 gymnosperms, 63 monilophytes (members of the horsetail-fern clade; see Pryer et al., 2001), and 171 byrophytes. This covers about 1·5 % of known land plants, and in gymnosperms in particular the coverage is nearly 25 % (Bennett and Leitch, 2003). Plans are also under way to add more than 200 values for algae (see Kapraun, 2005) within the next year.
The first of ten key recommendations made during the 1997 Kew meeting was to increase the coverage of angiosperms by a further 1 % and to obtain estimates from at least one representative of each family. Following the second Kew meeting in September 2003, revised targets of 75 % familial, 10 % generic and an additional 1 % species coverage for angiosperms, a level of 2 % species representation for pteridophytes, and improved geographic sampling for bryophytes were proposed. This will involve a considerable number of new estimates, but is expected to be completed within the next 5 years. In addition, groups of specific taxonomic and/or biological interest may be targeted to allow detailed comparative study.
Animal Genome Size Database
Unlike the situation with plants, the initial effort of Sparrow et al. (1972) to compile animal genome sizes had not been followed up in any comprehensive way until quite recently. As such, much of the work on animal genome size evolution has necessarily followed in the footsteps of botanists. Like its botanical predecessor, the Animal Genome Size Database began as part of an investigation of the patterns and phenotypic implications of genome size variation. In particular, the initial animal compilation was made for a study of the relationship between genome size and red blood cell size in mammals (Gregory, 2000). This was subsequently expanded to cover birds (Gregory, 2002a), and eventually grew to include all animals. The database was launched in January of 2001, and as of October 2004 contains data for more than 3700 animals, including roughly 2470 vertebrates and 1260 invertebrates (Gregory, 2001b). Obviously, this coverage is highly biased towards the 50 000 or so species of vertebrates, given that it includes 20 % of jawless fishes, about 12 % of cartilaginous fishes, 7 % each of amphibians and mammals, roughly 4 % of ray-finned fishes and reptiles, and nearly 2 % of birds, but an abysmally tiny percentage of invertebrates (which undoubtedly total in the millions). First-time assays of major invertebrate groups over the past few years have begun to lessen this discrepancy, but in truth have only scratched the surface (Gregory, 2005).
In absolute terms, the plant and animal databases are fairly similar in size, but clearly the relative coverage is far superior in plants (in part simply because there are already five times as many described animal species as plants). There are currently no plans to gather data from 1 % of animals, which would require an enormous effort—another 2500 estimates will be required just to get 1 % of beetles, counting only described species. For the foreseeable future, the primary goal with the animal dataset will be simply to fill in some of the more glaring gaps, including several classes (or even phyla!) that remain largely or entirely unknown, as well as numerous orders and/or families of insects, mammals, fishes and birds that have not yet been studied (as a quick reminder, the zoological taxonomic hierarchy is: Kingdom, Phylum, Class, Order, Family, Genus, Species). Of course, the identification of these gaps itself represents a major step forward that was not possible prior to the assembly of an animal database comparable to that for plants.
PATTERNS OF VARIATION IN PLANTS AND ANIMALS
Interspecific variation
The angiosperms encompass the entire range of C-values found among land plants, in total around a 1000-fold variation. Other groups of land plants vary considerably less: the monilophytes about 95-fold, lycophytes around 75-fold, gymnosperms roughly 14-fold and bryophytes only 12-fold (Bennett and Leitch, 2003). Taking the Chlorophyta, Phaeophyta and Rhodophyta together, algae display a level of variation exceeding 1300-fold, although within any of these groups the range is between only 9- and 200-fold (Kapraun, 2005). From the smallest alga to the largest angiosperm, plants as a whole range nearly 8500-fold in their C-values.
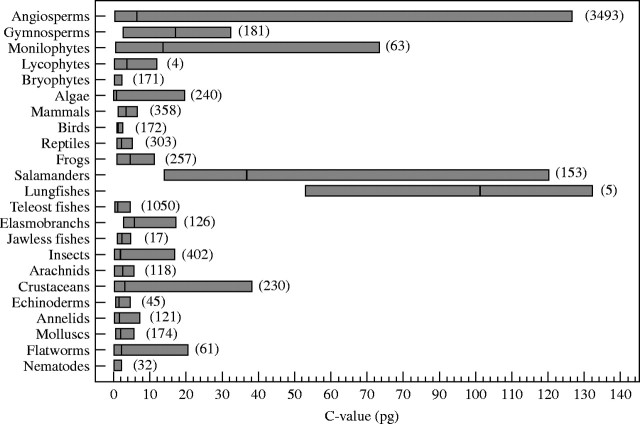
Simple ranges can be somewhat misleading, however, since the majority of plants in all of these groups have relatively small genomes. Thus, the modal genome sizes in all but the gymnosperms (∼10 pg) and monilophytes (∼8 pg) are 0·6 pg or less, including for the hypervariable angiosperms and algae. This tendency for small genome sizes is also apparent in Fig. 1, which shows the C-value ranges and means for the major groups of plants and animals so far studied. With the exception of the gymnosperms, in all cases the mean is near the bottom end of the overall range.
Fig. 1.
Ranges in C-value (haploid nuclear DNA contents, in pg) for the major groups of plants and animals that have been studied to date. The number of species in each group for which data are available (as of November 2003) is given in parentheses. Mean C-value is indicated by the vertical line within the bars, and is obviously close to the low end of the distribution in most of these groups. This indicates that most C-values in both plant and animal taxa are small, with only a few subgroups extending to larger values. The monilophytes refers to the monophyletic clade of horsetails and ferns (see Pryer et al., 2001). Data are from the Plant DNA C-values Database (Bennett and Leitch, 2003), the Animal Genome Size Database (Gregory, 2001b), and Kapraun (2005).
Interspecific variation is higher in animals than in the land plants, with an overall range of about 3300-fold (Gregory, 2001b). The smallest animal genome size (0·04 pg) is found in the placozoan Trichoplax adhaerens, which, being constructed of only four cell types and essentially resembling a giant ciliated amoeba, is also by far the simplest member of the kingdom. The largest animal genome size so far reported (∼132 pg) is that of the marbled lungfish, Protopterus aethiopicus. The tunicate Oikopleura dioica has a genome size of about 0·07 pg, making the range in chordates around 1800-fold, and several pufferfishes of the family Tetraodontidae exhibit C-values around 0·4 pg, for a vertebrate range of roughly 330-fold. Thus, even the vertebrates alone are considerably more variable than any one group of plants besides angiosperms. Ranges among some invertebrate groups may also approach this level, as with flatworms (340-fold), crustaceans (240-fold) and insects (190-fold), but in many cases are considerably smaller, as among annelids (125-fold), arachnids (70-fold), nematodes (40-fold), molluscs (15-fold) and echinoderms (9-fold).
The general pattern among animals, as with plants, is for most members of each major group to be rather constrained in their genome size variation, with only one or a few subset(s) exhibiting large genomes (Fig. 1). In plants, certain ferns, monocots and many gymnosperms tend to fit in this category. Among vertebrates, only the cartilaginous fishes, lungfishes and amphibians (especially salamanders) possess exceptionally large C-values. Mammals, birds, reptiles and teleost fishes, despite much higher species numbers, are all remarkably limited in terms of genome size variation, and even within the Amphibia there is no overlap in genome size between frogs and salamanders (Fig. 1). In insects the Orthoptera (especially grasshoppers), and in crustaceans the Decapoda (especially caridean shrimps), Stomatopoda (mantis shrimps) and calanoid Copepoda, are the only groups to far exceed a typically small range. The most speciose insect orders like the Coleoptera (beetles), Diptera (flies) and Lepidoptera (moths and butterflies) tend to have small genome sizes with very few or no exceptions. Molluscs, the second most diverse invertebrate phylum behind the arthropods, display no C-values larger than 6 pg.
Quantum shifts in genome size
In 1976, Sparrow and Nauman suggested that the minimum genome sizes of groups as wide-ranging as viruses, bacteria, plants, fungi and animals varied discontinuously by following a doubling series within and among taxa. Since this apparent series of multiples did not correspond to differences in chromosome numbers, they considered this to represent a process of ‘cryptopolyploidy’ (as they put it, ‘polyploidy results in more chromosomes; cryptopolyploidy results in larger chromosomes’; Sparrow and Nauman, 1976). Overall, this pattern is very rough, and since it applies only to minimum genome sizes is of limited interest to the C-value enigma.
In a less expansive (and more realistic) context, quantum shifts in genome size have been reported within numerous genera of plants (see Sparrow and Nauman, 1973; Narayan, 1985, 1988, 1998) and also in algae (Maszewski and Kolodziejczyk, 1991). In these cases, it is not minimal genome size across broad groups that varies by a series of doublings, but rather the C-values of congeneric species that differ by multiples of the lowest genome in the group. For example, in a sample of 20 species of the plant genus Tephrosia, Raina et al. (1986) found genome sizes to vary from 1·3 to 7·4 pg by increments of about 0·74 pg (approximately half the value of the smallest genome in the group). In some cases, these discontinuous patterns have been explained as an artifact of incomplete taxonomic sampling (e.g. Ohri et al., 1998; Greilhuber and Obermayer, 1999), but it would require a remarkably fortuitous series of collections to account for all the known examples. Other authors have ascribed genome size discontinuity to the existence of ‘steady states’ in DNA content maintained by stabilizing selection (Narayan, 1998), but the question remains as to why the steady states should be found at exact multiples.
Similar patterns of discontinuous variation have been reported for groups of animals as diverse as aphids (Finston et al., 1995), polychaete annelids (Sella et al., 1993; Gambi et al., 1997) and turbellarian flatworms (Gregory et al., 2000). In one of the most striking animal examples, genome sizes in copepod crustaceans of the genera Calanus and Pseudocalanus vary by intervals of about 2 pg, from 2·25 to 12·5 pg (McLaren et al., 1988, 1989), and similar patterns may hold in other copepods as well (Gregory et al., 2000). Certainly, quantum shifts are not the dominant mode of change in either plants or animals, but are nevertheless sufficiently common in both groups to be of special interest.
Intraspecific variation: real and artifactual
Even if it proceeds by less than quantum jumps, genome size change may still be restricted in its occurrence to speciation events (i.e. punctuational). However, it is also conceivable that much change occurs in a truly gradualistic (anagenetic) mode, beginning with differences among conspecifics and ending with differences across species. Pronounced intraspecific variation would, of course, pose a major challenge to the DNA constancy hypothesis upon which the C-value concept and all indirect DNA quantification methods (see below) are based, and is therefore of both theoretical and pragmatic importance in genome size study.
The genomes of plants have frequently been labelled as ‘fluid’, ‘dynamic’, and ‘in constant flux’, due in large part to the seemingly common observation of pronounced intraspecific variation in their nuclear DNA contents. In some cases, real variation within species can be explained by the differential presence of supernumerary B chromosomes. Strictly speaking, this does not refute the notion of DNA constancy because the A chromosome complement remains unchanged. In other examples, however, intraspecific variation in DNA content can be attributed to recognizable polymorphisms in the A chromosomes themselves, as with heterochromatic knobs in maize (e.g. Poggio et al., 1998) or differentially deleted transposable element remnants in barley (Kalendar et al., 2000).
Over the past few years, it has become necessary to abandon many of the most celebrated examples of intraspecific variation in plants as they have been attributed to experimental error (e.g. Greilhuber, 1988, 1997, 1998, 2005; Greilhuber and Obermayer, 1998; Bennett and Leitch, 2005). Even the most careful study can be subject to unanticipated sources of error, as illustrated by the case of apparent environmentally induced variation in DNA content in the sunflower, Helianthus annuus. Although it initially appeared that differences in light exposure could alter DNA content (Price and Johnston, 1996), it was later realized that sunflowers generate compounds in the presence of light which interfere with propidium iodide staining and therefore give a false impression of DNA content variation (Price et al., 2000). Similar effects have since been observed in coffee (Noirot et al., 2002), and the presence of stain inhibitors has become a major concern for DNA estimation in plants (see below). In other cases, extensive sampling has revealed striking stability in plant genome sizes. Most notably, it has recently been demonstrated that populations of the onion (Allium cepa), a species often used as a standard in plant studies, maintain a constant genome size across four continents (Bennett et al., 2000b).
Fluidity has also been attributed to the genomes of salamanders, although more because of their large size than any detailed evidence of dynamic behaviour (Vignali and Nardi, 1996). Examples of intraspecific variation in genome size have been reported for many animal groups, including insects (e.g. Kumar and Rai, 1990), mammals (e.g. Garagna et al., 1999), copepod crustaceans (Escribano et al., 1992), fishes (e.g. Johnson et al., 1987; Lockwood and Bickham, 1991, 1992; Lockwood and Derr, 1992; Collares-Pereira and Moreira da Costa, 1999), molluscs (Rodriguez-Juiz et al., 1996) and reptiles and amphibians (Lockwood et al., 1991; MacCulloch et al., 1996). Specimens of fish and amphibians taken from radioactively contaminated or otherwise polluted areas also show some apparent fluctuations in DNA content (Lingenfelser et al., 1997; Dallas et al., 1998; Vinogradov and Chubinishvili, 1999).
These examples, although interesting, do not provide conclusive evidence of serious violations of DNA constancy. Some of these cases could be based on the presence of cryptic subspecies (Lockwood and Bickham, 1992; MacCulloch et al., 1996); in most instances, the variation in genome size is associated with differences in geography, either at the inter-populational level, or at least along some geographic cline. This issue of inadequate species delineation has also been pointed out for some apparent cases of intraspecific variation in plants (e.g. Ebert et al., 1996). The possibility of experimental error is also ever present in animals, as with plants. For example, Thindwa et al. (1994) reported that specimens of the aphid Schizaphis graminum raised on sorghum had lower DNA contents than individuals from the same biotypes reared on wheat or johnsongrass, but the fact that the animals were simply homogenized prior to flow cytometric analysis raises the possibility that some botanical compound in their food confused staining with propidium iodide.
It must also be noted that most of the examples of intraspecific variation in fishes involve either salmonids or cyprinids, which are well known to exhibit more dynamic chromosome-level changes than most teleost groups. For example, differences in chromosome numbers can be observed among populations of rainbow trout (Thorgaard, 1983). Thus, these families may be atypical in this regard (Johnson et al., 1987; Lockwood and Bickham, 1991), making them of special interest in their own right, but not generally indicative of the situation in fishes (let alone animals or all eukaryotes, as some authors suggest). Perhaps tellingly, a detailed sampling from various wild and domesticated stocks of the channel catfish, Ictalurus punctatus (family Ictaluridae), revealed a high level of genome size stability (Tiersch et al., 1990).
In some groups of animals, it is entirely unclear whether genomes are remarkably flexible or highly stable. For example, Escribano et al. (1992) found significant variation within the copepod crustaceans Calanus glacialis and Pseudocalanus acuspes based on geographical differences, between P. elongatus reared for 96 generations in the laboratory versus wild-caught specimens (and also according to season in one year, but not in others), and within these various species according to differences in rearing conditions including food availability and temperature. On the other hand, no disparity in C-values was found among populations of the cyclopoid copepod Mesocyclops edax collected along the east coast of North America from Nova Scotia to Florida (Wyngaard and Rasch, 2000). In this instance it is worthy of note that the cyclopoid copepods, which apparently have very stable C-values intraspecifically, do not vary greatly among species, whereas the calanoids, which may be more flexible within species, also exhibit marked interspecific variability (Wyngaard and Rasch, 2000).
In any event, B chromosomes are known from many animals (Camacho et al., 2000), and groups with chromosomal sex determination systems are expected to evince differences among males and females in absolute DNA content, although this variation is usually (but not always) minor. Heterochromatic polymorphisms in sex chromosomes may also provide some real intraspecific variation in certain cases (e.g. Garagna et al., 1999). It therefore seems prudent to maintain a certain level of agnosticism regarding the extent of intraspecifc variation in genome size for both plants and animals. Obviously, DNA contents must change by some mechanism(s), whether strictly gradualistic or more punctuated. As with most important mechanistic questions in evolutionary biology, this is probably an issue of relative frequencies, not exclusive absolutes.
MECHANISMS OF GENOME SIZE CHANGE IN PLANTS AND ANIMALS
There are many ways in which genome sizes can change over time. In general, each of the most commonly recognized mechanisms applies to both plants and animals, although their specific nature and importance may vary considerably both within and between the two groups. The more notable mechanisms common to plants and animals are discussed in the following sections, with special emphasis on the points of divergence between the kingdoms.
Transposable elements
Under the classic ‘selfish DNA’ theory, sequences such as transposable elements (TEs), which are capable of their own propagation, spread within the genome until their activity is halted by selection against replicational costs (Doolittle and Sapienza, 1980; Orgel and Crick, 1980). Although the modern view of transposable elements is much more complex, with TE–host interactions recognized as ranging from parasitic to mutualistic (Kidwell and Lisch, 2001), the basic notion that TEs contribute substantially to eukaryotic genomes has been borne out. For example, roughly 45 % of the human genome (International Human Genome Sequencing Consortium, 2001), and more than 60 % of some plant genomes (Bennetzen, 2002), is comprised of TEs and (mostly) their defunct remnants. In a most striking example, it appears that the genome of maize has doubled in size over only a few million years by a surge in TE activity, indicating that this can be a mechanism of rapid change nearly on par with whole-scale duplication (SanMiguel and Bennetzen, 1998).
To be sure, the exact types of TE sequences present may vary considerably among genomes, even within kingdoms. For example, while LTR retrotransposons have been suggested to predominate in plant genomes (Kumar and Bennetzen, 1999), this may in fact be restricted to grasses (Wendel and Wessler, 2000). Likewise, LTR retrotransposons may be among the most common sequences in fruit fly and mosquito genomes (Adams et al., 2000; Holt et al., 2002), but LINEs and SINEs are much more common in mammals and pufferfishes (International Human Genome Sequencing Consortium, 2001; Aparicio et al., 2002; Mouse Genome Sequencing Consortium, 2002), while DNA transposons predominate in nematodes (C. elegans Sequencing Consortium, 1998). Nevertheless, it is evident that TEs in general play a very important role in shaping genome size diversity among eukaryotes. In broad terms, among those eukaryotes whose genomes have been sequenced in detail (mostly animals, but also some plants) there is an approximately linear relationship between genome size and the total amount of transposable element DNA present, although TEs generally contribute a higher percentage of DNA in larger genomes (Kidwell, 2002).
Introns and other sequences
The abundances of many other sequence types also correlate positively with genome size in eukaryotes, which is in keeping with the notion of global genomic forces acting to shape DNA content (Gregory and Hebert, 1999; Petrov, 2001). Simple sequence repeat content (Hancock, 2002), rDNA gene copy number (Prokopowich et al., 2003), and intron size (Vinogradov, 1999) are all positively associated with genome size across taxonomic samples. However, again these overall similarities persist despite important differences. For example, the relationship with rDNA multiplicity is slightly weaker in plants than in animals, and plants clearly maintain more rDNA gene copies per unit genome size than animals (Prokopowich et al., 2003).
The differences between plants and animals may be especially pronounced with regard to introns. Whereas correlations between intron size and genome size are apparent among related animals like Drosophila species (Moriyama et al., 1998), such relationships appear to be absent in plants such as Gossypium (Wendel et al., 2002a). Perhaps most significantly, it has been claimed that the majority of non-coding DNA is intronic in animals, but that this is not true of plants, in which most non-coding DNA is apparently intergenic (Wong et al., 2000).
Polyploidy
Technically, polyploidy itself does not represent a change in genome size per se because it actually involves the addition of a second genome. That is to say, so long as the two concellular genomes remain distinct (i.e. until rediploidization occurs), C-value will change but genome size will not. As it turns out, the new polyploid C-value may be less than the expected sum of the parental genomes (e.g. Ozkan et al., 2003; Bennett and Leitch, 2005), but obviously this would nevertheless be a means of adding significant amounts of DNA to the nucleus. In any case, variation in C-value (and probably genome size) has been much more greatly affected by polyploidy in plants than in animals (Gregory and Mable, 2005; Tate et al., 2005). It is often stated that polyploidy is common in plants, but the frequency with which it occurs actually varies considerably among taxa. For example, most (perhaps all) angiosperms and ferns and about 60 % of mosses have polyploidy in their ancestry, whereas it is rather uncommon in liverworts and gymnosperms (e.g. Averett, 1980; Delevoryas, 1980; Masterson, 1994; Otto and Whitton, 2000; Wendel, 2000). And although its occurrence is usually under-appreciated in animals (given that examples can be found in every major phylum), it is true that cases are comparatively quite scarce among the metazoa. Nevertheless, an ancient polyploidization event appears to have played an important role in early vertebrate genome evolution (e.g. McLysaght et al., 2002). In teleost fishes, there may have been a second round of genome duplication (van de Peer et al., 2003), and it is notable that in general the only extant teleosts that exceed a fairly small genome size range are those such as the salmonids, cyprinids and catostomids whose more recent ancestry includes polyploidy. On the other hand, polyploidy is much more common in frogs than in salamanders, despite the invariably larger genomes of the latter.
DNA loss
Until recently, all of the known mechanisms of genome size change had involved DNA gain. This fact provided cause to wonder whether plants have ‘a one-way ticket to genomic obesity’ (Bennetzen and Kellogg, 1997a). While it is likely that the ancestral genome size was small in plants (Leitch et al., 1998, 2005), phylogenetic reconstructions of specific taxa have shown both increases and decreases to have occured (e.g. Watanabe et al., 1999; Wendel et al., 2002b). The same can be said of animals, so there is a clear need for a mechanism of DNA loss in both groups. Fortunately, mutational mechanisms of DNA loss have been increasingly emphasized of late (Hartl, 2000; Petrov, 2001; Bennetzen, 2002). Unfortunately, in some cases these have been taken to an undue extreme by the development of purely neutralist models in which differences in DNA loss rate are considered the prime determinants of variation in genome size (Petrov, 2002).
The mutational mechanisms in question operate on (at least) three very different scales. The first, which forms the basis of the ‘mutational equilibrium model’, is based on a predominance of deletions over insertions on scales less than 400 bp. All of the relatively limited data presented in support of this mechanism are derived from animals, but it has been suggested that this should also apply to plants (Petrov, 1997). However, at this scale the deletional mechanism is extremely weak, and is unlikely to play a major role in the large-scale genome size evolution in either plants or animals (Bennetzen and Kellogg, 1997b; Gregory, 2003a, 2004). As a prime example, by this mechanism it would take more than 600 million years to delete half of the newly acquired TEs in the maize genome, provided that no other insertions or duplications took place in the meantime (Gregory, 2003a, 2004). Since the entire family Poaceae is only about 75 million years old and exhibits particularly dynamic genomes (Gaut, 2002), maize's recent genomic expansion would hardly seem to be ‘noise around the long-term equilibrium value’ as Petrov (2002) suggests.
The second scale of deletional mechanisms is considerably more powerful, and involves recombination between homologous copies of the long terminal repeats characteristic of LTR retrotransposons. In this case, all of the available evidence comes from plants, most notably Hordeum spp. (Kalendar et al., 2000). When this recombination occurs, most of the element is lost, leaving behind only a ‘solo LTR’. Of course, this mechanism can only slow genomic growth by TE insertions, because at the very least a solo LTR is retained each time (Devos et al., 2002).
The third scale, which also involves LTR retrotransposons and is also so far only studied in plants, is the only one of the three capable of producing extensive genomic shrinkage over reasonable timescales. Here, it is not recombination between homologous LTRs, but rather ‘illegitimate recombination’ between LTRs either on the same or different chromosomes that leads to DNA loss. And since these are non-homologous elements, the DNA lost will be all that located in between the two elements and may therefore involve much larger amounts than are added by each TE insertion (Bennetzen, 2002, Bennetzen et al., 2005). In theory, this mechanism should apply to both plants and animals, although a great deal more data are needed before any general conclusions can be drawn regarding its actual role in genome size evolution. Moreover, when such large deletions are involved, the role of selection cannot be so easily dismissed as in the mutational equilibrium model.
PARALLEL IMPACTS ON PLANT AND ANIMAL PHENOTYPES
Cell size
A general association between nucleus size and cell size in vertebrate red blood cells has long been recognized (e.g. Gulliver, 1875), and even in one of the very first comparative surveys of animal genome size, Mirsky and Ris (1951) noted that ‘in the nucleated red cells of vertebrates…there is an approximately direct relationship between cell mass and DNA content’. This general relationship applies to plants and unicellular eukaryotes as well, and, as noted by Cavalier-Smith (1982) more than 20 years ago, is perhaps ‘the most reliably established fact about genome evolution’.
In plants, seed size and genome size are linked by something of a triangular relationship, meaning that small genomes can be associated with small or large seeds, but that large genomes are not found in small seeds (Bennett, 1987; Thompson, 1990; Knight and Ackerly, 2002). Egg size, the zoological counterpart of seed size, has not been nearly as well studied, but is known to correlate positively with genome size in cladoceran crustaceans (which all have very small genomes; Beaton, 1995) and plethodontid salamanders (which have large genomes; data from Jockusch, 1997). Given that egg size, like seed size, often has important fitness consequences, this is a parameter worthy of much more study.
It can be difficult to find correlations between C-value and the sizes of gametes besides eggs in both kingdoms because both pollen and sperm can be greatly modified according to specific reproductive requirements. In plants, a correlation is observed so long as similar types (e.g. wind-dispersed) of pollen are compared across related species (e.g. Bennett, 1972). In animals, the situation may be even more complex, with a correlation appearing when comparing very closely related samples, but not across larger groups. For example, the red viscacha rat (Tympanoctomys barrerae), the only polyploid mammal known, has considerably larger sperm than its diploid relatives (Gallardo et al., 1999), and even the sizes of X- vs Y-chromosome carrying sperm differ in direct proportion to their varying DNA contents (about 3–4 %) in both bulls and humans (Cui, 1997; van Munster et al., 1999). However, across mammals at large there is no relationship between genome size and sperm size (Gage, 1998), presumably because sperm morphology is adaptively modified to a considerable degree in mammals while genome size remains constrained. Other groups with larger genome size ranges may reveal a correlation with sperm size, but this has yet to be investigated.
Relationships with somatic cells are well established in both plants and animals, although obviously the pertinent cell types differ greatly between the two groups. In plants, the best studied cell type from this perspective is meristems (Price et al., 1973), although leaf guard cell size is also associated with DNA content (Masterson, 1994). Cell size is also correlated with genome size in algae (Holm-Hansen, 1969; Kapraun and Dunwoody, 2002). The sizes of various cell types, including neurons, liver cells (hepatocytes) and epithelial cells, all appear to correlate positively with genome size in vertebrates (Gregory, 2001b). However, by far the best known relationship in animals involves red blood cells (erythrocytes), which differ fundamentally from plant meristems in that they are highly compact and non-dividing (Gregory, 2001b, c). Yet, in all of these cell types from both kingdoms, genome size and cell size are probably linked causally by the influence of DNA content on the cell cycle, such that larger genomes delay division and result in the production of larger daughter cells (Gregory, 2001b, c). The fact that erythrocyte size correlates positively with genome size in mammals, even though their mature red blood cells are enucleated (i.e. genome-free), strongly supports this hypothesis (Gregory, 2000, 2001b, c, 2005).
Morphology
Because bodies are composed of cells, the obvious possibility exists that a change in genome size will result in a change in body size. However, a general correlation between genome size and overall body size has not been reported to occur in plants. Instead, the sizes of various structures, such as leaves, tend to correlate with genome size. Even here the situation is somewhat complex, given that leaf size may actually correlate positively or negatively with genome size, depending on the taxonomic sample (Knight et al., 2005). A similar situation exists in animals, with positive correspondences found between genome size and body size in numerous groups of invertebrates, such as aphids (Finston et al., 1995), flies (Ferrari and Rai, 1989), copepod crustaceans (McLaren et al., 1988; Gregory et al., 2000), polychaete annelids (Soldi et al., 1994) and turbellarian flatworms (Gregory et al., 2000). In nematodes, body size is influenced not by genome size per se, but by the level of somatic endopolyploidy (Flemming et al., 2000). On the other hand, such relationships are not observed in groups like oligochaete annelids (Gregory and Hebert, 2002), moths (Gregory and Hebert, 2003), spiders (Gregory and Shorthouse, 2003) or within the few families of beetles that have been studied to date (Juan and Petitpierre, 1991; Petitpierre and Juan, 1994; Gregory et al., 2003). In most vertebrates, cell (and therefore genome) size does not correlate in any way with body size; notably, the difference in mass between the smallest shrew and the blue whale, which covers more than seven orders of magnitude, is due almost entirely to variation in cell number. However, in birds and rodents, where cell number variation is much more limited, cell and genome size do correlate positively with body size (Gregory, 2002a, b). Some authors have suggested a negative correlation with body size within the beetle genus Pimelia (Palmer et al., 2003), but this is only a phylogenetic correlation and may not have much real biological significance (Gregory et al., 2003).
Physiology
The primary role of red blood cells in vertebrates is in gas exchange, a process strongly dependent on cellular surface area to volume ratios. As such, it has long been suggested that genome sizes are assorted according to metabolic parameters, with ‘wasteful’ groups like mammals and birds having small cells and genomes and ‘frugal’ ones like amphibians having large cells and genomes (Szarski, 1983). Indeed, there is a significant negative correlation between mass-corrected oxygen consumption rate and genome size in both mammals (Vinogradov, 1995) and birds (Gregory, 2002a). However, and contrary to many previous assumptions, this has not been found to extend to amphibians except insofar as frogs as a group are more active and have smaller genomes than salamanders (Gregory, 2003b).
In the simplest terms, the respiratory physiology of plants is the opposite of that in animals. However, this may nonetheless relate to genome size, given that stomatal size and specific leaf area correlate positively with DNA content (Masterson, 1994; Chung et al., 1998; Knight et al., 2005), just as do erythrocyte sizes in vertebrates. In fact, it has recently been shown that photosynthetic rate in plants, like metabolic rate in homeothermic vertebrates, correlates negatively with C-value (Knight et al., 2005).
With regard to physiological stress, it has been noted that increased tolerance to both droughts (e.g. Castro-Jimenez et al., 1989; Wakamiya et al., 1993, 1996) and frost (e.g. MacGillivray and Grime, 1995) are associated with larger genome size in plants. As an interesting parallel, Shahbasov and Ganchenko (1990) reported a positive association between DNA content and non-specific thermal and hypoxic stress tolerance in frogs and salamanders. On the other hand, it is becoming apparent that large-genomed plants are generally excluded from the extremes of climatic ranges, whether cold or hot (Knight and Ackerly, 2002; Knight et al., 2005). At least one example of this is found in animals as well: In polychaete annelids, macrobenthic species inhabiting stable environments have larger genome sizes, while those found in harsh interstitial environments invariably have small C-values (Sella et al., 1993; Soldi et al., 1994; Gambi et al., 1997).
Development
It is not only cell size, but also both mitotic and meiotic division rates that correlate with genome size. Most of the demonstrations of this inverse correlation have come from plants, but this has also been documented experimentally in some animals (reviewed in Gregory, 2001a). At the organismal level, this often translates into a negative correlation between genome size and developmental rate. Indeed, such correlations have been found within and among numerous groups of plants (e.g Bennett, 1972; Grime et al., 1985; Mowforth and Grime, 1989). There is even evidence that experimental selection for earlier flowering time may result in a reduction in genome size (Rayburn et al., 1994). More generally, there are the well-known patterns whereby plants with large genomes cannot adopt an annual or ephemeral lifestyle and in which weeds tend to have small genomes (Bennett, 1987; Bennett et al., 1998). However, this developmental correlation is not universal in plants, and in some cases a positive relationship can be observed, again depending on the taxa being compared (Knight et al., 2005).
As with plants, genome size and developmental rate are inversely correlated in many but not all animal taxa. Thus, negative correlations are found in amphibians, insects and crustaceans (see Gregory, 2002c), but not in mammals and birds (Gregory, 2002b). In amphibians and insects, there is a direct parallel between the annual versus perennial lifestyle threshold seen in plants, in this case involving constraints related to metamorphosis. To appreciate this, it is necessary to draw a distinction between developmental rate (the time taken to develop) and developmental complexity (the amount of developing to be done in a limited amount of time), which are in fact two sides of the same coin (Gregory, 2002c, 2005). Since metamorphosis involves a strongly time-limited period of intense tissue differentiation, it requires rapid cell divisions and therefore small genomes. Thus, frogs inhabiting ephemeral pools have the smallest amphibian genomes (∼1 pg) whereas obligately non-metamorphosing (neotenic) salamanders have the largest (up to 120 pg). In like fashion, in insects with complete metamorphosis (holometabolous development), genome sizes almost never exceed 2 pg, whereas those with no or only incomplete metamorphosis (ametabolous or hemimetabolous development) may have genomes up to 17 pg (Gregory, 2001b, 2002a).
There are clearly numerous parallels between animals and plants in terms of the correlations between genome size and cell size and division rate, and in the influence of these on the organismal phenotype. The particular biology of the organisms in question is important, of course, and there are major differences in the expression of these relationships between plants and animals. In fact, this is also the case within kingdoms, as for example with mammals and birds, in which metabolism is important but development is not, versus amphibians, in which the opposite is true. The key point to recognize is that while the specific expression of the phenomena differ among groups, the underlying mechanisms are the same whether in plants or animals.
MEASURING GENOME SIZES IN PLANTS AND ANIMALS
Methodology: similarities and differences
As with all of the issues discussed in this review, the methods employed in genome size measurement are far more similar between plants and animals than they are different. In both groups, genome sizes have traditionally been assessed either by Feulgen microdensitometry or flow cytometry (usually using propidium iodide or DAPI). More recently, and essentially simultaneously, the technique of Feulgen image analysis densitometry has been developed for use with both plant (Vilhar et al., 2001; Vilhar and Dermastia, 2002) and animal (Hardie et al., 2002) specimens.
All of the same basic issues arise when using these techniques regardless of whether the specimen is a plant or an animal. For Feulgen-based methods, the primary issues in both groups are the preparation of a suitable monolayer of cells, the temperature and duration of acid hydrolysis, and the quality of the Schiff reagent used (Greilhuber and Temsch, 2001; Hardie et al., 2002; Bennett and Leitch, 2005). In flow cytometry, the preparation of individual nuclei, the choice of dye and the staining protocols are equally important in both groups.
One of the most notable differences between plant and animal studies is that the former typically involve dividing cells like root or shoot tips or leaves, while the latter almost always make use of non-dividing cells such as blood cells or sperm. In practical terms, this means that plant studies involve a challenge in finding 2C or 4C nuclei among the entire range of partially replicated genomes, whereas in animal studies the cells are all either 2C (blood) or 1C (sperm). The use of blood or sperm also alleviates the problem of endopolyploidy, which is common to both animals and plants. However, with many small invertebrates whole bodies or large segments thereof must be used in flow cytometry, and in this case the problem of locating cells of known ploidy is just as prominent as it is in plants.
Plant and animal studies also tend to differ in the methods of tissue preparation as a result of the very different cell types used. In Feulgen-based methods, plant tissues are often fixed in the field and later prepared as squashes. In animals, blood smears can be made in the field and simply allowed to air-dry, or else dissections must be performed on fresh material in the laboratory (e.g. to get insect sperm); the dehydration caused by fixation makes it impossible to acquire blood cells from either vertebrates or invertebrates and greatly complicates dissection. By contrast, flow cytometry often involves the preparation of fresh tissues in plants, but almost invariably uses fixed or frozen tissues (including blood) for animals. The fixatives used may also differ among kingdoms, since 3 methanol : 1 acetic acid is commonly used in plants, whereas 85 methanol : 10 formalin : 5 acetic acid is recommended for most animal preparations (Hardie et al., 2002). However, with some invertebrates (e.g. copepod crustaceans), the fixation and squash protocol employed may be essentially the same as that used for plants (e.g. Wyngaard and Rasch, 2000).
As noted above, many plants produce phytochemicals that may interfere with either Feulgen or fluorescent staining and produce artifactual examples of intraspecific variation (Greilhuber, 1986; Price et al., 2000; Noirot et al., 2002). In animals, no such problematic chemicals have been identified to date, and are not likely to be found when tissues such as blood are used. However, the possibility does exist that stain inhibitors may be present when using whole-body specimens of invertebrates, and once again zoologists would be well advised to take a lesson from their botanical colleagues.
Choice of standards
One of the largest sources of error in genome size measurements, in both plants and animals and whether using either Feulgen methods or flow cytometry, involves differences in DNA compaction levels, which directly affect the level of stain uptake. This can be dealt with in several ways, but the best by far is to simply choose a standard of the same cell type as the unknown being measured. Even within animals, this is very important (Hardie et al., 2002), and this becomes even more critical when comparing across kingdoms. Animal standards (especially chicken or trout blood) have been used many times for plant studies (Bennett and Leitch, 2003), and very occasionally plant standards have been employed in animal measurements (Greilhuber et al., 1983).
Chicken erythrocytes are not considered a suitable standard for comparison with epithelial cells, leukocytes or sperm from fellow vertebrates, and cannot even be compared accurately to liver cells from the same animal (Hardie et al., 2002). Comparisons with plant cells would obviously be much more problematic than this, and it is therefore not surprising that the use of animal standards in plant studies was strongly discouraged at both the 1997 and 2003 Kew meetings. There is one exception to this that was noted at the most recent meeting, which is that it would be advisible to use a completely sequenced genome whose size is known with absolute certainty to calibrate a series of ‘gold standards’ for use in future plant studies. Since the only genome that has truly been sequenced in its entirety is that of the nematode Caenorhabditis elegans, this remains the only choice currently available. Fortunately, it appears that C. elegans can be used in flow cytometric comparisons to plants with small genomes, such as Arabidopsis (Bennett et al., 2003).
THE FUTURE OF GENOME SIZE
Links with sequencing projects
While the measurement and compilation of plant genome sizes have consistently exceeded the efforts made for animals, the opposite is true in terms of genome sequencing. To date, roughly five times as many animal genomes have been sequenced as compared with plants. This discrepancy is only likely to increase in the future, given that numerous additional animal projects are under way, whereas only very few such programmes are under development for plants. Moreover, while there is much current talk of attempting to acquire genome sequences for at least one representative of every major animal phylum in addition to all the major experimental models and species of economic and medical importance, the upcoming sequencing (or only mapping) efforts for plants will be mainly restricted to species of agricultural interest (e.g. wheat, barley, maize, soybean, oat, banana and tomato).
In any case, it is well appreciated that one of the first steps in deciding on a subject and strategy for sequencing is to determine genome size. For an unknown reason, there has been a large disconnect between the sequencing and genome size communities for both plant and animal projects. As a result, an incorrect genome size was assumed by the Arabidopsis Genome Initiative (2000), even though the C-value of this species had been estimated in ten different publications before the sequencing results were published (see Bennett et al., 2003). These estimates would have been easily available from the Plant DNA C-values Database. Similarly, the Drosophila genome sequence paper of 2000 made no reference to the five different genome size estimates that had already been published (Adams et al., 2000). Granted, the Animal Genome Size Database did not exist in 2000, but it still should have been possible to locate well-known papers like that by Rasch et al. (1971), which provided a very careful and accurate estimate using Feulgen densitometry. Obviously, a stronger link between sizers and sequencers should help to avoid these problems in the future. More importantly, an improved awareness of the Plant DNA C-values Database will greatly facilitate the selection of the next series of plant sequencing subjects once the obvious choice of crop species is exhausted.
Biologists interested in the evolution of genome size would also benefit from such an integration with sequencing projects. Most obviously, whole-scale genome sequencing information can be used to assess the relative contribution of different sequence types to variation in genome size. This has already been done in a very preliminary way with regard to transposable elements, in this review and elsewhere (Kidwell, 2002). As more sequences become available, it will finally become possible to address the crucial component of the C-value enigma dealing with mechanisms of genome size change. Conversely, an integration with existing information on patterns and implications of genome size variation will allow the data of sequencing projects to be placed in a real biological context.
Talk across the trenches
It is unlikely that genome sequencing programmes will pay due attention to existing studies of genome size so long as there is no unified front presented by botanists and zoologists. On the other hand, once it is recognized that plant and animal genome size are components of a single overarching puzzle, the general importance of the issue will probably become much clearer to those in related fields.
Increased talk across the taxonomic trenches would benefit both sides in other ways as well. Since most (if not all) of the patterns and consequences of genome size variation transcend taxonomic boundaries, the work being carried out by one side cannot help but illuminate that of the other. Correlations with cell size, as one clear example, can be best studied comparatively and mechanistically using both plants and animals (Gregory, 2001a). Based on the survey presented above, it would seem that morphology, physiology, development, methodology and mechanisms of change also fall into this category of mutual overlap. On practical grounds, zoologists would benefit from increased interaction simply because botanists have led the way on many of these issues, making it unnecessary to re-invent the conceptual wheel. Botanists, for their part, should recognize the basic fact that zoologists are often asked to review plant genome size papers (while the reverse rarely occurs), suggesting an obvious pragmatic benefit of increased understanding between the two groups.
Beyond plants and animals
To date, there have been no significant efforts to compile databases of ‘protist’ or fungus genome sizes, and even with prokaryotes most of the readily available information comes from species being used in sequencing projects. Given that the most informative comparisons in plants and animals have only become possible after the assembly of broad datasets, it is obvious that many important insights remain hidden amongst the scattered data from these other kingdoms. More fungi have been sequenced than plants, and the construction of a database for these organisms will also undoubtedly help with the highly desirable link between sequence and size described above.
Much work remains to be done in assessing the patterns of variation within animals and plants, and there is clearly a need to better share the lessons learned from the study of these two original kingdoms. However, green things and moving things hardly encompass the fullness of life's diversity and, as such, expansion, as well as integration, will be necessary if genome size evolution is to be properly understood. Every living thing has a genome, and for every genome there is a size. Within these simple principles lie the future of genome size study and the eventual resolution of the C-value enigma.
Acknowledgments
The author was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) post-doctoral fellowship and the NSERC Howard Alper Post-Doctoral Prize. Sincere thanks are given to the organisers and participants of the 2003 Kew genome size workshop and discussion meeting, and especially to M. D. Bennett, T. Cavalier-Smith, J. S. Johnston, C. A. Knight, I. J. Leitch, H. J. Price and B. Vilhar for stimulating discussions and helpful comments.
LITERATURE CITED
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. 2000. The genome sequence of Drosophila melanogaster Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia J-m, Dehal P, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes Science 297: 1301–1310. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Averett JE. 1980. Polyploidy in plant taxa: summary. In: Lewis WH, ed. Polyploidy: biological relevance. New York: Plenum Press, 269–273. [Google Scholar]
- Beaton MJ. 1995.Patterns of endopolyploidy and genome size variation in Daphnia. Ph.D. thesis, Department of Zoology, University of Guelph, Guelph, Ontario, Canada. [Google Scholar]
- Bennett MD. 1972. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London, Series B 181: 109–135. [DOI] [PubMed] [Google Scholar]
- Bennett MD. 1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106 (Suppl.): 177–200. [Google Scholar]
- Bennett MD, Bhandol P, Leitch IJ. 2000. Nuclear DNA amounts in angiosperms and their modern uses—807 new estimates. Annals of Botany 86: 859–909. [Google Scholar]
- Bennett MD, Johnston S, Hodnett GL, Price HJ. 2000.Allium cepa L. cultivars from four continents compared by flow cytometry show nuclear DNA constancy. Annals of Botany 85: 351–357. [Google Scholar]
- Bennett MD, Leitch IJ. 1995. Nuclear DNA amounts in angiosperms. Annals of Botany 76: 113–176. [Google Scholar]
- Bennett MD, Leitch IJ. 1997. Nuclear DNA amounts in angiosperms—583 new estimates. Annals of Botany 80: 169–196. [Google Scholar]
- Bennett MD, Leitch IJ. 2001. Nuclear DNA amounts in pteridophytes. Annals of Botany 87: 335–345. [Google Scholar]
- Bennett MD, Leitch IJ. 2003.Plant DNA C-values Database Royal Botanic Gardens, Kew, UK. http://www.rbgkew.org.uk/cval/homepage.html. [Google Scholar]
- Bennett MD, Leitch IJ. 2005. Genome size evolution in plants. In: Gregory TR ed. The evolution of the genome. San Diego: Elsevier, 89–162. [Google Scholar]
- Bennett MD, Leitch IJ, Hanson L. 1998. DNA amounts in two samples of angiosperm weeds. Annals of Botany 82 (Suppl. A): 121–134. [Google Scholar]
- Bennett MD, Leitch IJ, Price HJ, Johnston JS. 2003. Comparisons with Caenorhabditis (∼100 Mb) and Drosophila (∼175 Mb) using flow cytometry show genome size in Arabidopsis to be ∼157 Mb and thus ∼25% larger than the Arabidopsis Genome Initiative estimate of ∼125 Mb. Annals of Botany 91: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Smith JB. 1976. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London, Series B 274: 227–274. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Smith JB. 1991. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London, Series B 334: 309–345. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Smith JB, Lewis Smith RI. 1982. DNA amounts of angiosperms from the Antarctic and South Georgia. Environmental and Experimental Botany 22: 307–318. [Google Scholar]
- Bennetzen JL. 2002. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 115: 29–36. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Kellogg EA. 1997. Do plants have a one-way ticket to genomic obesity? Plant Cell 9: 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Kellogg EA. 1997. Reply to Petrov. Plant Cell 9: 1901–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. 2005. Mechanisms of recent genome size variation in flowering plants. Annals of Botany 95: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho JPM, Sharbel TF, Beukeboom LW. 2000. B-chromosome evolution. Philosophical Transactions of the Royal Society of London, Series B 355: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Jimenez Y, Newton RJ, Price HJ, Halliwell RS. 1989. Drought stress responses of Microseris species differing in nuclear DNA content. American Journal of Botany 76: 789–795. [Google Scholar]
- Cavalier-Smith T. 1982. Skeletal DNA and the evolution of genome size. Annual Review of Biophysics and Bioengineering 11: 273–302. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1998. A revised six-kingdom system of life. Biological Reviews 73: 203–266. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Chung J, Lee J-H, Arumuganathan K, Graef GL, Specht JE. 1998. Relationships between nuclear DNA content and seed and leaf size in soybean. Theoretical and Applied Genetics 96: 1064–1068. [Google Scholar]
- Collares-Pereira MJ, Moreira da Costa L. 1999. Intraspecific and interspecific genome size variation in Iberian Cyprinidae and the problem of diploidy and polyploidy, with review of genome sizes within the family. Folia Zoologica 48: 61–76. [Google Scholar]
- Comings DE. 1972. The structure and function of chromatin. Advances in Human Genetics 3: 237–431. [DOI] [PubMed] [Google Scholar]
- Cui K. 1997. Size differences between human X and Y spermatozoa and prefertilization diagnosis. Molecular Human Reproduction 3: 61–67. [DOI] [PubMed] [Google Scholar]
- Dallas CE, Lingenfelser SF, Lingenfelser JT, Holloman K, Jagoe CH, Kind JA, Chesser RK, Smith MH. 1998. Flow cytometric analysis of erythrocyte and leukocyte DNA in fish from Chernobyl-contaminated ponds in the Ukraine. Ecotoxicology 7: 211–219. [Google Scholar]
- Delevoryas T. 1980. Polyploidy in gymnosperms. In: Lewis WH, ed. Polyploidy: biological relevance. New York: Plenum Press, 215–218. [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis Genome Research 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284: 601–603. [DOI] [PubMed] [Google Scholar]
- Ebert I, Greilhuber J, Speta F. 1996. Chromosome banding and genome size differentiation in Prospero (Hyacinthaceae): diploids. Plant Systematics and Evolution 203: 143–177. [Google Scholar]
- Escribano R, McLaren IA, Klein Breteler WCM. 1992. Innate and acquired variation of nuclear DNA contents of marine copepods. Genome 35: 602–610. [Google Scholar]
- Ferrari JA, Rai KS. 1989. Phenotypic correlates of genome size variation in Aedes albopictus Evolution 43: 895–899. [DOI] [PubMed] [Google Scholar]
- Finston TL, Hebert PDN, Foottit RB. 1995. Genome size variation in aphids. Insect Biochemistry and Molecular Biology 25: 189–196. [Google Scholar]
- Flemming AJ, Shen Z-Z, Cunha A, Emmons SW, Leroi AM. 2000. Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proceedings of the National Academy of Sciences of the USA 97: 5285–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage MJG. 1998. Mammalian sperm morphometry. Proceedings of the Royal Society of London, Series B 265: 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo MH, Bickham JW, Honeycutt RL, Ojeda RA, Köhler N. 1999. Discovery of tetraploidy in a mammal. Nature 401: 341. [DOI] [PubMed] [Google Scholar]
- Gambi MC, Ramella L, Sella G, Protto P, Aldieri E. 1997. Variation in genome size of benthic polychaetes: systematic and ecological relationships. Journal of the Marine Biology Association of the United Kingdom 77: 1045–1057. [Google Scholar]
- Garagna S, Civitelli MV, Marziliano N, Castiglia R, Zuccotti M, Redi CA, Capanna E. 1999. Genome size variations are related to X-chromosome heterochromatin polymorphism in Arvicanthis sp. from Benin (West Africa). Italian Journal of Zoology 66: 27–32. [Google Scholar]
- Gaut BS. 2002. Evolutionary dynamics of grass genomes. New Phytologist 154: 15–28. [Google Scholar]
- Gregory TR. 2000. Nucleotypic effects without nuclei: genome size and erythrocyte size in mammals. Genome 43: 895–901. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Reviews 76: 65–101. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2001.Animal Genome Size Database http://www.genomesize.com. [Google Scholar]
- Gregory TR. 2001. The bigger the C-value, the larger the cell: genome size and red blood cell size in vertebrates. Blood Cells, Molecules, and Diseases 27: 830–843. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2002. A bird's-eye view of the C-value enigma: genome size, cell size, and metabolic rate in the class Aves. Evolution 56: 121–130. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2002. Genome size and developmental parameters in the homeothermic vertebrates. Genome 45: 833–838. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2002. Genome size and developmental complexity. Genetica 115: 131–146. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2003. Is small indel bias a determinant of genome size? Trends in Genetics 19: 485–488. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2003. Variation across amphibian species in the size of the nuclear genome supports a pluralistic, hierarchical approach to the C-value enigma. Biological Journal of the Linnean Society 79: 329–339. [Google Scholar]
- Gregory TR. 2004. Insertion-deletion biases and the evolution of genome size. Gene 324: 15–34. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2005. Genome size evolution in animals. In: Gregory TR ed. The evolution of the genome. San Diego: Elsevier, 3–87. [Google Scholar]
- Gregory TR, Hebert PDN. 1999. The modulation of DNA content: proximate causes and ultimate consequences. Genome Research 9: 317–324. [PubMed] [Google Scholar]
- Gregory TR, Hebert PDN. 2002. Genome size estimates for some oligochaete annelids. Canadian Journal of Zoology 80: 1485–1489. [Google Scholar]
- Gregory TR, Hebert PDN. 2003. Genome size variation in lepidopteran insects. Canadian Journal of Zoology 81: 1399–1405. [Google Scholar]
- Gregory TR, Hebert PDN, Kolasa J. 2000. Evolutionary implications of the relationship between genome size and body size in flatworms and copepods. Heredity 84: 201–208. [DOI] [PubMed] [Google Scholar]
- Gregory TR, Mable BK. 2005. Polyploidy in animals. In: Gregory TR ed. The evolution of the genome. San Diego: Elsevier, 427–517. [Google Scholar]
- Gregory TR, Nedved O, Adamowicz SJ. 2003. C-value estimates for 31 species of ladybird beetles (Coleoptera: Coccinellidae). Hereditas 139: 121–127. [DOI] [PubMed] [Google Scholar]
- Gregory TR, Shorthouse DP. 2003. Genome sizes of spiders. Journal of Heredity 94: 285–290. [DOI] [PubMed] [Google Scholar]
- Greilhuber J. 1986. Severely distorted Feulgen-DNA amounts in Pinus (Coniferophytina) after nonadditive fixations as a result of meristematic self-tanning with vacuole contents. Canadian Journal of Genetics and Cytology 28: 409–415. [Google Scholar]
- Greilhuber J. 1988. Critical reassessment of DNA content variation in plants. In: Brandham PE, ed. Kew chromosome conference III. London: HMSO, 39–50. [Google Scholar]
- Greilhuber J. 1997. The problem of variable genome size in plants (with special reference to woody plants). In: Borzan Z, Schlarbaum SE, eds. Cytogenetic Studies of Forest Trees and Shrub Species. Croatia: Croatian Forests, Inc., Faculty of Forestry, University of Zagreb, 13–34. [Google Scholar]
- Greilhuber J. 1998. Intraspecific variation in genome size: a critical reassessment. Annals of Botany 82 (Suppl. A): 27–35. [Google Scholar]
- Greilhuber J. 2005. Intraspecific variation in genome size in angiosperms—identifying its existence. Annals of Botany 95: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Obermayer R. 1998. Genome size variation in Cajanus cajan (Fabaceae): a reconsideration. Plant Systematics and Evolution 212: 135–141. [Google Scholar]
- Greilhuber J, Obermayer R. 1999. Cryptopolyploidy in Bunias (Brassicaceae) revisited—a flow-cytometric and densitometric study. Plant Systematics and Evolution 218: 1–4. [Google Scholar]
- Greilhuber J, Temsch EM. 2001. Feulgen densitometry: some observations relevant to best practice in quantitative nuclear DNA content determination. Acta Botanica Croatia 60: 285–298. [Google Scholar]
- Greilhuber J, Volleth M, Loidl J. 1983. Genome size of man and animals relative to the plant Allium cepa Canadian Journal of Genetics and Cytology 25: 554–560. [DOI] [PubMed] [Google Scholar]
- Grime JP, Shacklock JML, Band SR. 1985. Nuclear DNA contents, shoot phenology and species co-existence in a limestone grassland community. New Phytologist 100: 435–445. [Google Scholar]
- Gulliver G. 1875. Observations on the sizes and shapes of the red corpuscles of the blood of vertebrates, with drawings of them to a uniform scale, and extended and revised tables of measurements. Proceedings of the Zoological Society of London 1875: 474–495. [Google Scholar]
- Hancock JM. 2002. Genome size and the accumulation of simple sequence repeats: implications of new data from genome sequencing projects. Genetica 115: 93–103. [DOI] [PubMed] [Google Scholar]
- Hardie DC, Gregory TR, Hebert PDN. 2002. From pixels to picograms: a beginners' guide to genome quantification by Feulgen image analysis densitometry. Journal of Histochemistry and Cytochemistry 50: 735–749. [DOI] [PubMed] [Google Scholar]
- Hartl DL. 2000. Molecular melodies in high and low C. Nature Reviews Genetics 1: 145–159. [DOI] [PubMed] [Google Scholar]
- Holm-Hansen O. 1969. Algae: amounts of DNA and organic carbon in single cells. Science 163: 87–88. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Jockusch EL. 1997. An evolutionary correlate of genome size change in plethodontid salamanders. Proceedings of the Royal Society of London, Series B 264: 597–604. [Google Scholar]
- Johnson OW, Utter FM, Rabinovitch PS. 1987. Interspecies differences in salmonid cellular DNA identified by flow cytometry. Copeia 1987: 1001–1009. [Google Scholar]
- Juan C, Petitpierre E. 1991. Evolution of genome size in darkling beetles (Tenebrionidae, Coleoptera). Genome 34: 169–173. [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. 2000. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences of the USA 97: 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapraun DF. 2005. Nuclear DNA content estimates in multicellular eukaryotic green, red and brown algae: phylogenetic considerations. Annals of Botany 95: 7–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapraun DF, Dunwoody JT. 2002. Relationship of nuclear genome size to some reproductive cell parameters in the Florideophycidae (Rhodophyta). Phycologia 41: 507–516. [Google Scholar]
- Kidwell MG. 2002. Tranposable elements and the evolution of genome size in eukaryotes. Genetica 115: 49–63. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. 2001. Transposable elements, parasitic DNA, and genome evolution. Evolution 55: 1–24. [DOI] [PubMed] [Google Scholar]
- Knight CA, Ackerly DD. 2002. Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecology Letters 5: 66–76. [Google Scholar]
- Knight CA, Molinari N, Petrov DA. 2005. The large genome constraint hypothesis: environment, phenotype, and evolution. Annals of Botany 95: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. 1999. Plant retrotransposons. Annual Review of Genetics 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Kumar A, Rai KS. 1990. Intraspecific variation in nuclear DNA content among world populations of a mosquito, Aedes albopictus (Skuse). Theoretical and Applied Genetics 79: 748–752. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Chase MW, Bennett MD. 1998. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Annals of Botany 82 (Suppl. A): 85–94. [Google Scholar]
- Leitch IJ, Soltis DE, Soltis PS, Bennett MD. 2005. Evolution of DNA amounts across land plants (Embryophyta). Annals of Botany 95: 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingenfelser SF, Dallas CE, Jagoe CH, Smith MH, Brisbin IL, Chesser RK. 1997. Variation in DNA content of blood cells of largemouth bass from contaminated and uncontaminated waters. Environmental Toxicology and Chemistry 16: 2136–2143. [Google Scholar]
- Lockwood SF, Bickham JW. 1991. Genetic stock assessment of spawning Arctic cisco (Coregonus autumnalis) populations by flow cytometric determination of DNA content. Cytometry 12: 260–267. [DOI] [PubMed] [Google Scholar]
- Lockwood SF, Bickham JW. 1992. Genome size in Beaufort Sea coastal assemblages of Arctic ciscoes. Transactions of the American Fisheries Society 121: 13–20. [Google Scholar]
- Lockwood SF, Derr JN. 1992. Intra- and interspecific genome-size variation in the Salmonidae. Cytogenetics and Cell Genetics 59: 303–306. [DOI] [PubMed] [Google Scholar]
- Lockwood SF, Holland BS, Bickham JW, Hanks BG, Bull JJ. 1991. Intraspecific genome size variation in a turtle (Trachemys scripta) exhibiting temperature-dependent sex determination. Canadian Journal of Zoology 69: 2306–2310. [Google Scholar]
- MacCulloch RD, Upton DE, Murphy RW. 1996. Trends in nuclear DNA content among amphibians and reptiles. Comparative Biochemistry and Physiology 113B: 601–605. [Google Scholar]
- MacGillivray CW, Grime JP. 1995. Genome size predicts frost resistance in British herbaceous plants: implications for rates of vegetation response to global warming. Functional Ecology 9: 320–325. [Google Scholar]
- Masterson J. 1994. Stomatal size in fossil plants: evidence for polyploidy in a majority of angiosperms. Science 264: 421–424. [DOI] [PubMed] [Google Scholar]
- Maszewski J, Kolodziejczyk P. 1991. Cell cycle duration in antheridial filaments of Chara spp. (Characeae) with different genome size and heterochromatin content. Plant Systematics and Evolution 175: 23–38. [Google Scholar]
- McLaren IA, Sévigny J-M, Corkett CJ. 1988. Body size, development rates, and genome sizes among Calanus species. Hydrobiologia 167/168: 275–284. [Google Scholar]
- McLaren IA, Sévigny J-M, Frost BW. 1989. Evolutionary and ecological significance of genome sizes in the copepod genus Pseudocalanus Canadian Journal of Zoology 67: 565–569. [Google Scholar]
- McLysaght A, Hokamp K, Wolfe KH. 2002. Extensive genomic duplication during early chordate evolution. Nature Genetics 31: 200–204. [DOI] [PubMed] [Google Scholar]
- Mirsky AE, Ris H. 1951. The desoxyribonucleic acid content of animal cells and its evolutionary significance. Journal of General Physiology 34: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama EN, Petrov DA, Hartl DL. 1998. Genome size and intron size in Drosophila Molecular Biology and Evolution 15: 770–773. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Mowforth MA, Grime JP. 1989. Intra-population variation in nuclear DNA amount, cell size and growth rate in Poa annua L. Functional Ecology 3: 289–295. [Google Scholar]
- Murray BG. 1998. Nuclear DNA amounts in Gymnosperms. Annals of Botany 82 (Suppl. A): 3–15. [Google Scholar]
- Narayan RKJ. 1985. Discontinuous DNA variation in the evolution of plant species. Journal of Genetics 64: 101–109. [Google Scholar]
- Narayan RKJ. 1988. Evolutionary significance of DNA variation in plants. Evolutionary Trends in Plants 2: 121–130. [Google Scholar]
- Narayan RKJ. 1998. The role of genomic constraints upon evolutionary changes in genome size and chromosome organization. Annals of Botany 82 (Suppl. A): 57–66. [Google Scholar]
- Noirot M, Barre P, Louarn J, Duperray C, Hamon S. 2002. Consequences of stoichiometric error on nuclear DNA content evaluation in Coffea liberica var. dewevrei using DAPI and propidium iodide. Annals of Botany 89: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohri D, Fritsch RM, Hanelt P. 1998. Evolution of genome size in Allium (Alliaceae). Plant Systematics and Evolution 210: 57–86. [Google Scholar]
- Orgel LE, Crick FHC. 1980. Selfish DNA: the ultimate parasite. Nature 284: 604–607. [DOI] [PubMed] [Google Scholar]
- Otto SJ, Whitton J. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Ozkan H, Tuna M, Arumuganathan K. 2003. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. Journal of Heredity 94: 260–264. [DOI] [PubMed] [Google Scholar]
- Palmer M, Petitpierre E, Pons J. 2003. Test of the correlation between body size and DNA content in Pimelia (Coleoptera: Tenebrionidae) from the Canary Islands. European Journal of Entomology 100: 123–129. [Google Scholar]
- Petitpierre E, Juan C. 1994. Genome size, chromosomes, and egg-chorion ultrastructure in the evolution of Chrysomelidae. In: Jolivet PH, Cox ML, Petitpierre E, eds. Novel aspects of the biology of Chrysomelidae. Dordrecht: Kluwer Academic Publishers, 213–225. [Google Scholar]
- Petrov D. 1997. Slow but steady: reduction of genome size through biased mutation. Plant Cell 9: 1900–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA. 2001. Evolution of genome size: new approaches to an old problem. Trends in Genetics 17: 23–28. [DOI] [PubMed] [Google Scholar]
- Petrov DA. 2002. Mutational equilibrium model of genome size evolution. Theoretical Population Biology 61: 533–546. [DOI] [PubMed] [Google Scholar]
- Poggio L, Rosato M, Chiavarino AM, Naranjo CA. 1998. Genome size and environmental correlations in maize (Zea mays ssp. mays, Poaceae). Annals of Botany 82 (Suppl. A): 107–115. [Google Scholar]
- Price HJ, Hodnett G, Johnston JS. 2000. Sunflower (Helianthus annuus) leaves contain compounds that reduce nuclear propridium iodide fluorescence. Annals of Botany 86: 929–934. [Google Scholar]
- Price HJ, Johnston JS. 1996. Influence of light on DNA content of Helianthus annuus Linnaeus. Proceedings of the National Academy of Sciences of the USA 93: 11264–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price HJ, Sparrow AH, Nauman AF. 1973. Correlations between nuclear volume, cell volume and DNA content in meristematic cells of herbaceous angiosperms. Experientia 29: 1028–1029. [Google Scholar]
- Prokopowich CD, Gregory TR, Crease TJ. 2003. The correlation between rDNA copy number and genome size in eukaryotes. Genome 46: 48–50. [DOI] [PubMed] [Google Scholar]
- Pryer KM, Schneider H, Smith AR, Cranfill R, Wolf PG, Hunt JS, Sipes SD. 2001. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 409: 618–622. [DOI] [PubMed] [Google Scholar]
- Raina SN, Srivastav PK, Rama Rao S. 1986. Nuclear DNA variation in Tephrosia Genetica 69: 27–33. [Google Scholar]
- Rasch EM, Barr HJ, Rasch RW. 1971. The DNA content of sperm of Drosophila melanogaster Chromosoma 33: 1–18. [DOI] [PubMed] [Google Scholar]
- Rayburn AL, Dudley JW, Biradar DP. 1994. Selection for early flowering results in simultaneous selection for reduced nuclear DNA content in maize. Plant Breeding 112: 318–322. [Google Scholar]
- Rodriguez-Juiz AM, Torrado M, Méndez J. 1996. Genome-size variation in bivalve molluscs determined by flow cytometry. Marine Biology 126: 489–497. [Google Scholar]
- SanMiguel P, Bennetzen JL. 1998. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Annals of Botany 82 (Suppl. A): 37–44. [Google Scholar]
- Sella G, Redi GA, Ramella L, Soldi R, Premoli MC. 1993. Genome size and karyotype in some interstitial polychaete species of the genus Ophryotrocha (Dorvilleidae). Genome 36: 652–657. [DOI] [PubMed] [Google Scholar]
- Shahbasov VG, Ganchenko AV. 1990. Nonspecific tolerance and DNA content in genome of amphibians. Doklady Akademii Nauk SSSR 314: 971–975. [PubMed] [Google Scholar]
- Soldi R, Ramella L, Gambi MC, Sordino P, Sella G. 1994. Genome size in Polychaetes: relationship with body length and life habit. In: Dauvin J-C, Laubier L, Reish DJ, eds. Actes de la 4ième Conférence internationale des Polychètes: Mémoires du Muséum National d'Histoire Naturelle 162: 129–135. [Google Scholar]
- Sparrow AH, Nauman AF. 1973. Evolutionary changes in chromosome size and DNA content in the grasses. Brookhaven Symposia in Biology 25: 367–389. [Google Scholar]
- Sparrow AH, Nauman AF. 1976. Evolution of genome size by DNA doublings. Science 192: 524–527. [DOI] [PubMed] [Google Scholar]
- Sparrow AH, Price HJ, Underbink AG. 1972. A survey of DNA content per cell and per chromosome of prokaryotic and eukaryotic organisms: some evolutionary considerations. In: Smith HH, ed. Evolution of genetic systems. New York: Gordon and Breach, 451–494. [PubMed] [Google Scholar]
- Swift H. 1950. The desoxyribose nucleic acid content of animal nuclei. Physiological Zoology 23: 169–198. [DOI] [PubMed] [Google Scholar]
- Swift H. 1950. The constancy of desoxyribose nucleic acid in plant nuclei. Proceedings of the National Academy of Sciences of the USA 36: 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarski H. 1983. Cell size and the concept of wasteful and frugal evolutionary strategies. Journal of Theoretical Biology 105: 201–209. [DOI] [PubMed] [Google Scholar]
- Tate JA, Soltis DE, Soltis PS. 2005. Polyploidy in plants. In: Gregory TR ed. The evolution of the genome. San Diego: Elsevier, 371–426. [Google Scholar]
- Thindwa HP, Teetes GL, Johnston JS. 1994. Greenbug DNA content. Southwestern Entomologist 19: 371–378. [Google Scholar]
- Thomas CA. 1971. The genetic organization of chromosomes. Annual Review of Genetics 5: 237–256. [DOI] [PubMed] [Google Scholar]
- Thompson K. 1990. Genome size, seed size and germination temperature in herbaceous angiosperms. Evolutionary Trends in Plants 4: 113–116. [Google Scholar]
- Thorgaard GH. 1983. Chromosomal differences among rainbow trout populations. Copeia 1983: 650–662. [Google Scholar]
- Tiersch TR, Simco BA, Davis KB, Chandler RW, Wachtel SS, Carmichael GJ. 1990. Stability of genome size among stocks of the channel catfish. Aquaculture 87: 15–22. [Google Scholar]
- van de Peer Y, Taylor JS, Meyer A. 2003. Are all fishes ancient polyploids? Journal of Structural and Functional Genomics 2: 65–73. [PubMed] [Google Scholar]
- van Munster EB, Stap J, Hoebe RA, Te Meerman GJ, Aten JA. 1999. Difference in volume of X- and Y-chromosome-bearing bovine sperm heads matches difference in DNA content. Cytometry 35: 125–128. [DOI] [PubMed] [Google Scholar]
- Vendrely R, Vendrely C. 1948. La teneur du noyau cellulaire en acide désoxyribonucléique à travers les organes, les individus et les espèces animales: techniques et premiers résultats. Experientia 4: 434–436. [DOI] [PubMed] [Google Scholar]
- Vignali R, Nardi I. 1996. Unusual features of the urodele genome: do they have a role in evolution and development? International Journal of Developmental Biology 40: 637–643. [PubMed] [Google Scholar]
- Vilhar B, Dermastia M. 2002. Standardisation of instrumentation in plant DNA image cytometry. Acta Botanica Croatia 61: 11–26. [Google Scholar]
- Vilhar B, Greilhuber J, Koce JD, Temsch EM, Dermastia M. 2001. Plant genome size measurement with DNA image cytometry. Annals of Botany 87: 719–728. [Google Scholar]
- Vinogradov AE. 1995. Nucleotypic effect in homeotherms: body mass-corrected basal metabolic rate of mammals is related to genome size. Evolution 49: 1249–1259. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. 1999. Intron-genome size relationship on a large evolutionary scale. Journal of Molecular Evolution 49: 376–384. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE, Chubinishvili AT. 1999. Genome reduction in a hemiclonal frog Rana esculenta from radioactively contaminated areas. Genetics 151: 1123–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H. 2000. Nuclear DNA amounts in mosses (Musci). Annals of Botany 85: 531–546. [Google Scholar]
- Wakamiya I, Newton RJ, Johnston JS, Price HJ. 1993. Genome size and environmental factors in Pinus American Journal of Botany 80: 1235–1241. [Google Scholar]
- Wakamiya I, Price HJ, Messina MG, Newton RJ. 1996. Pine genome size diversity and water relations. Physiologia Plantarum 96: 13–20. [Google Scholar]
- Watanabe K, Yahara T, Denda T, Kosuge K. 1999. Chromosomal evolution in the genus Brachyscome (Asteraceae, Astereae): statistical tests regarding correlation between changes in karyotype and habit using phylogenetic information. Journal of Plant Research 112: 145–161. [Google Scholar]
- Wendel JF. 2000. Genome evolution in polyploids. Plant Molecular Biology 42: 225–249. [PubMed] [Google Scholar]
- Wendel JF, Cronn RC, Alvarez I, Liu B, Small RL, Senchina DS. 2002. Intron size and genome size in plants. Molecular Biology and Evolution 19: 2346–2352. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Cronn RC, Johnston JS, Price HJ. 2002. Feast and famine in plant genomes. Genetica 115: 37–47. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Wessler SR. 2000. Retrotransposon-mediated genome evolution on a local ecological scale. Proceedings of the National Academy of Sciences of the USA 97: 6250–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GK-S, Passey DA, Huang Y-Z, Yang Z, Yu J. 2000. Is ‘junk’ DNA mostly intron DNA? Genome Research 10: 1672–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyngaard GA, Rasch EM. 2000. Patterns of genome size in the copepoda. Hydrobiologia 417: 43–56. [Google Scholar]