Abstract
• Background and Aims Passiflora actinia and P. elegans, two markedly parapatric species, have their southern and northern distribution limits, respectively, in the most southern part of the Brazilian Atlantic Rain Forest. Despite the fact that they are classified in different taxonomic series, previous phylogenetic studies of this genus revealed a high genetic similarity between them. The aim of the present work was to analyse in more detail their geographical range in this region of overlap, to investigate intraspecific genetic variability and phylogeographic structure, and to search for possible hybrids.
• Methods Eighty-two localities were searched for these species, and nuclear internal transcribed spacer (ITS) sequences were investigated for 32 individuals of P. actinia, 20 of P. elegans and one putative interspecific hybrid. Plastid trnL-trnF and psbA-trnH were examined for 12 plants of each species and the putative hybrid.
• Key Results Both species showed a high level of intraspecific and intra-individual ITS variability. Network analysis revealed a north–south geographic gradient in their intra and interspecific relationships. Mismatch analyses suggested a recent population expansion of P. elegans. The plastid markers showed restricted variability but, together with the nuclear data, they contributed to the identification of an interspecific hybrid of intermediate morphology at the border of the distribution of these two species. Both genetic and morphological data indicate the absence of an extensive hybridization zone between these species.
• Conclusions Gene flow between lineages is the possible cause for the presence of different ITS sequences within a given plant, the absence of homogenization being due to the high degree of vegetative reproduction in the two species. Differentiation of P. actinia into geographic groups and the origin of P. elegans may have been influenced by the Atlantic Forest migration towards southern Brazil. The genetic pattern of the interspecific hybrid indicates that plastid inheritance in these species is at least sometimes paternal.
Keywords: Passiflora actinia, Passiflora elegans, Passifloraceae, ITS, trnL-trnF, psbA-trnH, phylogeography, hybridization, quaternary climatic changes, Atlantic Forest, migration
INTRODUCTION
The southern portion of the Atlantic Forest is thought to have been formed by the migration of northern tropical elements (Rambo, 1951), the main event probably having occurred at the beginning of the Holocene (11 000–10 000 years bp) due to increasing temperature and humidity (Roth and Lorscheitter, 1993; Neves and Lorscheitter, 1995). The geographical range of animal and plant species that occur along the Brazilian coast was highly influenced by climatic oscillations in the Quaternary period, mainly due to several direct and indirect effects of changes in sea level that occurred during that period (Villwock and Tomazelli, 1995). These processes had a marked effect on the Atlantic Forest in the last 20 000 years and resulted in periods of expansion and contraction that may have led to fragmentation. The isolation of populations associated with forest fragments probably reduced the gene flow among them, but reproductive isolation does not always happen in such cases and hybrid zones may occur in situations of secondary contact. The analysis of nuclear and plastid DNA is being widely used to evaluate such cases, allowing determination of the direction of the gene flow (Schaal et al., 1998), etc.
The nuclear ribosomal DNA (nrDNA) of higher plants is organized in blocks in one or more chromosomal regions, each block consisting of hundreds to thousands of copies or paralogues (Buckler et al., 1997). Mutations in these tandem repeats within an individual are generally homogenized through concerted evolution (Arnhein, 1983), but the mode and timing of this concerted evolution vary markedly among different groups of plants (Baldwin et al., 1995; Mayer and Soltis, 1999). Generally the nrDNA intragenomic diversity is low, but some species show high heterogeneity in this region (Buckler et al., 1997; Denduangboripant and Cronk, 2000). This can occur if the process is not sufficiently rapid to achieve homogenization of different copies (Campbell et al., 1997; Zhang and Sang, 1999). After intraspecific or interspecific hybridization the different ITS copies (ribotypes) can evolve in different ways (Koch et al., 2003). With more recent hybrids, direct sequencing can reveal an additive pattern (signal of two different nucleotides) in the material studied (Fuertes Aguilar and Nieto Feliner, 2003).
Passiflora is a highly diversified genus of Passifloraceae (passion-flowers), with many of the species being found in tropical America. About 130 Passiflora species occur in Brazil, with the highest number in the Amazon basin (Killip, 1938). The Brazilian state of Rio Grande do Sul, with its subtropical climate, harbours only 15 of these species (Mondin, 2001); among these, P. elegans is notable for being restricted to the state and adjacent regions. Phylogenetic analyses in Passiflora using the ITS region and the plastid trnL-trnF intergenic spacer revealed that P. actinia is closely related to P. elegans (Muschner et al., 2003). Despite their high similarity, however, they had been previously classified in different taxonomic series (P. actinia in Simplicifoliae, P. elegans in Lobatae; Killip, 1938). This classification was based on leaf form, a highly variable trait that is subject to environmental influences in Passiflora (Benson et al., 1976; MacDougal, 1994). These two species are parapatric: the northern limit of P. elegans is the southern limit of P. actinia, and there are no records of sympatry of these species, although both live in the same type of forested habitat. Unlike P. elegans, P. actinia is widely distributed, occurring from 18°S to 30°S along the eastern border of the Atlantic Forest from Espírito Santo to Rio Grande do Sul.
This study is a survey of the intraspecific variation in P. actinia and P. elegans, especially the contact area between them, but also in other regions of their distribution in southern Brazil. Nuclear ITS and plastid trnL-trnF and psbA-trnH markers were used to investigate their relationships and possible hybridization events. The patterns observed were examined taking into consideration the processes of separation and retraction of the Atlantic Forest in the region, and their eventual connections with the evolutionary relationships of these two taxa.
MATERIALS AND METHODS
Samples and DNA isolation
Leaf material from 32 plants of P. actinia, 20 from P. elegans and one from a putative hybrid between P. actinia and P. elegans were obtained at 41 collecting sites (Table 1 and Fig. 1). For the plastid studies, 12 P. actinia, 11 P. elegans and the hybrid were chosen in a way that would adequately cover the study area. Voucher specimens were deposited in the ICN Herbarium, Botany Department, Biosciences Institute, Federal University of Rio Grande do Sul. Total DNA was extracted from young leaves dried in silica gel, using the method of Roy et al. (1992) with a few adaptations.
Table 1.
Characterization of the P. actinia and P. elegans specimens studied
| Geographical location* |
GenBank accessions |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species and specimen code |
Locality |
Geographical coordinates |
Collector's name or code |
ITS sequences |
ITS 1 |
ITS 2 |
trnL-trnF |
psbA-trnH |
||||||||
| Passiflora actinia | ||||||||||||||||
| A3 | Três Cachoeiras, RS | 29°24′S, 50°06′W | C. Mondin | H8A/H8A | AY219247 | AY219271 | – | – | ||||||||
| A8 | São Francisco de Paula, RS | 29°26′S, 50°36′W | APL 014 | H7A/H7A | AY219248 | AY219272 | AY219312 | AY219288 | ||||||||
| A10 | Serra do Pinto, RS | 29°21′S, 50°10′W | APL 016 | H9A/H9A | AY219249 | AY219273 | AY219313 | AY219289 | ||||||||
| A12 | Praia Grande, SC | 29°10′S, 49°56′W | APL 019 | H9A/H13A | AY219250 | AY219274 | AY219314 | AY219290 | ||||||||
| A15 | Praia Grande, SC | 29°10′S, 49°56′W | APL 022 | H10A/H16A | AY219246 | AY219270 | – | – | ||||||||
| A16 | Jd. América, Curitiba, PR | 25°30′S, 49°18′W | A. Cervi | H18A/H18A | AY219240 | AY219264 | – | – | ||||||||
| A17 | Centro Politécnico, Curitiba, PR | 25°26′S, 49°14′W | A. Cervi | H18A/H18A | AY542629 | AY542658 | AY219315 | AY219291 | ||||||||
| A18 | Centro Politécnico, Curitiba, PR | 25°26′S, 49°14′W | A. Cervi | H18A/H18A | AY542630 | AY542659 | – | – | ||||||||
| A19 | Centro Politécnico, Curitiba, PR | 25°26′S, 49°14′W | A. Cervi | H11A/H17A | AY219244 | AY219268 | – | – | ||||||||
| A20 | Jd. Botânico, Curitiba, PR | 25°26′S, 49°14′W | A. Cervi | H21A/H21A | AY219243 | AY219267 | – | – | ||||||||
| A22 | FLONA, São Francisco de Paula, RS | 29°23′S, 50°22′W | APL 030 | H10A/H16A | AY542640 | AY542669 | – | – | ||||||||
| A23 | Serra do Umbu, RS | 29°29′S, 50°19′W | APL 032 | H7A/H14A | AY219255 | AY219279 | AY219316 | AY219292 | ||||||||
| A24 | J. Velho, São Francisco de Paula, RS | 29°28′S, 50°40′W | PASS 037 | H10A/H16A | AY542641 | AY542670 | – | – | ||||||||
| A32 | D. Pedro de Alcântara, RS | 29°22′S, 49°50′W | PASS 059 | H7A/H15A | AY542642 | AY542671 | AY219317 | AY219293 | ||||||||
| A37 | Serra da Boa Vista, RS | 29°34′S, 50°20′W | APL 055 | H7A/H15A | AY219252 | AY219276 | AY219318 | AY219294 | ||||||||
| A39 | Barra do Ouro, RS | 29°30′S, 50°16′W | APL 062 | H7A/H9A | AY219254 | AY219278 | AY219319 | AY219295 | ||||||||
| A42 | Osório, RS | 29°48′S, 50°18′W | PASS 069 | H7A/H13A | AY219253 | AY219277 | AY219320 | AY219296 | ||||||||
| A47 | Taquara, RS | 29°43′S, 50°42′W | PASS 074 | H7A/H15A | AY542643 | AY542672 | AY219321 | AY219297 | ||||||||
| A50 | Canela, RS | 29°21′S, 50°48′W | APL 105 | H9A/H14A | AY219251 | AY219275 | AY219322 | AY219298 | ||||||||
| A56 | Reserva Biol. da Serra Geral, RS | 29°34′S, 50°12′W | APL 115 | H7A/H15A | AY542644 | AY542673 | AY219323 | AY219299 | ||||||||
| A70 | Urubici, SC | 28°06′S, 49°36′W | A. Cervi | H12A/H18A | AY219245 | AY219269 | – | – | ||||||||
| A88 | Centro Politécnico, Curitiba, PR | 25°26′S, 49°14′W | APL 183 | H18A/H18A | AY542631 | AY542660 | – | – | ||||||||
| A89 | B. Gutierrez, Curitiba, PR | 25°24′S, 49°17′W | APL 184 | H18A/H18A | AY542632 | AY542661 | – | – | ||||||||
| A90 | B. Gutierrez, Curitiba, PR | 25°24′S, 49°17′W | APL 185 | H18A/H20A | AY219241 | AY219265 | – | – | ||||||||
| A92 | B. Alemão, Curitiba, PR | 25°24′S, 49°17′W | APL 187 | H18A/H18A | AY542633 | AY542662 | – | – | ||||||||
| A93 | B. Alemão, Curitiba, PR | 25°24′S, 49°17′W | APL 188 | H18A/H18A | AY542634 | AY542663 | – | – | ||||||||
| A98 | São José dos Pinhais, PR | 25°32′S, 49°13′W | APL 195 | H18A/H18A | AY542635 | AY542664 | – | – | ||||||||
| A110 | São José dos Pinhais, PR | 25°32′S, 49°13′W | APL 208 | H18A/H18A | AY542636 | AY542665 | – | – | ||||||||
| A113 | B. Cascatinha, Curitiba, PR | 25°24′S, 49°18′W | APL 212 | H18A/H18A | AY542637 | AY542666 | – | – | ||||||||
| A115 | Pq. Barigüi, Curitiba, PR | 25°25′S, 49°18′W | APL 214 | H18A/H18A | AY542638 | AY542667 | – | – | ||||||||
| A123 | Pq. Passaúna, Curitiba, PR | 25°27′S, 49°22′W | APL 222 | H18A/H18A | AY542639 | AY542668 | – | – | ||||||||
| A126 | B. Zaninelli, Curitiba, PR | 25°23′S, 49°16′W | APL 225 | H18A/H19A | AY219242 | AY219266 | – | – | ||||||||
| Passiflora elegans | ||||||||||||||||
| E5 | Morro Santana, Porto Alegre, RS | 30°04′S, 51°07′W | PASS 010 | H3E/H6E | AY219258 | AY219282 | – | – | ||||||||
| E12 | Caçapava do Sul, RS | 30°33′S, 53°32′W | PASS 026 | H6E/H6E | AY542645 | AY542674 | AY219324 | AY219300 | ||||||||
| E13 | Caçapava do Sul, RS | 30°33′S, 53°32′W | PASS 027 | H4E/H6E | AY219262 | AY219286 | – | – | ||||||||
| E16 | Portão, RS | 29°42′S, 51°14′W | PASS 033 | H1E/H3E | AY219259 | AY219283 | AY219325 | AY219301 | ||||||||
| E20 | Ponta do Cego, Porto Alegre, RS | 30°15′S, 51°06′W | PASS 063 | H1E/H1E | AY219260 | AY219284 | AY219326 | AY219302 | ||||||||
| E22 | Pelotas, RS | 31°48′S, 52°24′W | APL 068 | H1E/H1E | AY542655 | AY542684 | AY219327 | AY219303 | ||||||||
| E26 | Guaíba, RS | 30°04′S, 51°24′W | APL 072 | H3E/H6E | AY542650 | AY542679 | AY219328 | AY219304 | ||||||||
| E29 | Cachoeira do Sul, RS | 30°16′S, 52°57′W | APL 078 | H3E/H6E | AY542651 | AY542680 | AY219329 | AY219305 | ||||||||
| E30 | Santo Antônio da Patrulha, RS | 29°50′S, 50°30′W | PASS 116 | H1E/H3E | AY542654 | AY542683 | AY219330 | AY219306 | ||||||||
| E38 | Lavras do Sul, RS | 30°52′S, 53°43′W | APL 093 | H3E/H6E | AY542652 | AY542681 | AY219331 | AY219307 | ||||||||
| E44 | Paverama, RS | 29°36′S, 51°48′W | PASS 097 | H2E/H6E | AY219261 | AY219285 | AY219332 | AY219308 | ||||||||
| E45 | Paverama, RS | 29°36′S, 51°48′W | PASS 098 | H2E/H6E | AY542657 | AY542686 | – | – | ||||||||
| E48 | Lagoa dos Três Cantos, RS | 28°33′S, 52°51′W | PASS 103 | H3E/H6E | AY542653 | AY542682 | AY219333 | AY219309 | ||||||||
| E49 | São Miguel das Missões, RS | 28°32′S, 54°33′W | PASS 105 | H1E/H1E | AY542656 | AY542685 | AY219334 | AY219310 | ||||||||
| E50 | Espumoso, RS | 28°49′S, 52°40′W | PASS 110 | H6E/H6E | AY542646 | AY542675 | – | – | ||||||||
| E51 | Lageado, RS | 29°20′S, 52°05′W | PASS 111 | H5E/H5E | AY219256 | AY219280 | – | – | ||||||||
| E53 | Lageado, RS | 29°20′S, 52°05′W | PASS 113 | H6E/H6E | AY219257 | AY219281 | – | – | ||||||||
| E55 | Lageado, RS | 29°20′S, 52°05′W | PASS 115 | H6E/H6E | AY542647 | AY542676 | – | – | ||||||||
| E56† | Santo Antônio da Patrulha, RS | 29°50′S, 50°30′W | PASS 118 | H15A/H3E | AY219263 | AY219287 | AY219335 | AY219311 | ||||||||
| E72 | BR290, RS | 30°09′S, 52°08′W | PASS 214 | H6E/H6E | AY542648 | AY542677 | – | – | ||||||||
| E75 | BR386, RS | 29°33′S, 51°53′W | PASS 267 | H6E/H6E | AY542649 | AY542678 | – | – | ||||||||
The abbreviations PR, RS and SC indicate the Brazilian states of Paraná, Rio Grande do Sul and Santa Catarina, respectively.
Interspecific hybrid between Passiflora actinia and P. elegans.
Fig. 1.
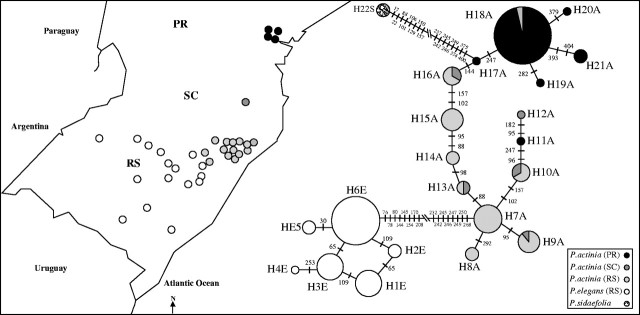
Left, geographical ranges and collecting places of Passiflora actinia (black and light- and dark-shaded circles) and P. elegans (white circles). The abbreviations PR, SC, and RS indicate the Brazilian states of Paraná, Santa Catarina and Rio Grande do Sul, respectively. Right, median-joining network connecting the different ITS sequences. The diameters of the circles indicate their frequencies, and the degree of shading their origin: white, P. elegans, Rio Grande do Sul; light shading, P. actinia, Rio Grande do Sul; dark shading, P. actinia, Santa Catarina; black, P. actinia Paraná; dotted, P. sidaefolia, used as an outgroup. Numbers between the circles indicate the sites where changes occurred, separating the sequences.
Polymerase chain reaction (PCR) amplification and sequencing
The segments chosen for study were: (a) the internal transcribed spacer (ITS) region of nrDNA, using primers and amplification conditions described by Desfeux and Lejeune (1996); and (b) plastid trnL-trnF and psbA-trnH intergenic spacers, employing, respectively, the primers and amplification conditions indicated by Taberlet et al. (1991) (primers e and f) and Sang et al. (1997). Dimethyl sulfoxide (DMSO) at 10 % was used in the ITS PCR, to exclude the presence of silenced alleles (Buckler et al., 1997; Fuertes Aguilar et al., 1999). The amplified material was purified with shrimp alkaline phosphatase and exonuclease I (Amersham Biosciences, Amersham, Bucks., UK) and the strands directly sequenced using the BigDye Terminator Sequencing Kit (PE Applied Biosystems, Foster City, CA, USA) and an ABI Prism 310 (Perkin Elmer, Wellesley, USA) genetic analyser.
Sequence analyses
The GenBank numbers of P. actinia and P. elegans sequences are given in Table 1. For ITS, a site was classified as heterozygous when more than one peak was present in the electropherogram and the weakest signal reached at least 25 % of the strength of the strongest signal (Fuertes Aguilar et al., 1999; Fuertes Aguilar and Nieto Feliner, 2003). To minimize the inclusion of bad reads as polymorphisms, as suggested by these authors, the restriction was added that double peaks had to occur on the same position in both strands. The sequences were aligned in the ClustalX1.81 program (Thompson et al., 1997), with manual corrections using the GeneDoc program (http://www.psc.edu/biomed/genedoc). The variable sites and nucleotide diversity were estimated using the MEGA version 2.1 program (Kumar et al., 2001). For heterozygous individuals, ITS sequence types were estimated using the PHASE program, version 1.0 (Stephens et al., 2001), with manual resolution of the few not determined by the program. Relationships among sequences were inferred using median-joining networks (ε = 0; Bandelt et al., 1999) with the NETWORK program, version 3.1.1.1, available at (http://www.fluxus-engineering.com). Tajima's D (Tajima, 1989), Fu and Li's D* (Fu and Li, 1993) and observed and expected mismatch distribution graphics, were conducted using the DNAsp 3·99 program (Rozas and Rozas, 1999). Passiflora sidaefolia sequences were used as the outgroup.
Seed viability
The seed viability of a possible P. actinia × P. elegans hybrid was investigated by the tetrazolium test (Bonner, 1986). Twenty seeds from three fruits of plant E56 (the putative hybrid) and E30 (which acted as the control) were investigated. The seeds were placed in distilled water at 30 °C for 24 h, longitudinally cut, soaked in a 0·5 % (w/v) 2,3,5 triphenyl tetrazolium chloride solution, and stored at 30 °C until the embryo acquired a reddish coloration (maximum time 48 h).
RESULTS
Geographical range of the species
Field trips were taken to 81 localities in the Brazilian states of Rio Grande do Sul (RS), Santa Catarina (SC) and Paraná (PR). Localities where P. actinia and P. elegans were found are indicated in Fig. 1. Our collections confirmed previous records that the southernmost geographic range of P. actinia is the north-east region of Rio Grande do Sul, being restricted to Atlantic Forest areas, and that P. elegans occurs in the riparian vegetation of the interior of Rio Grande do Sul. Especially in P. elegans, large distances were observed between the sampled populations, and it is hypothesized that this isolation may be related to the high fragmentation of the forest formation with which this species is associated. Populations of both species were composed of a small number of large plants with many branches, making it difficult to ensure that samples came from different individuals.
ITS variation
The ITS 1 and ITS 2 sequences varied between 227 and 228 bp, and 171 and 175 bp in P. actinia and P. elegans, respectively. After alignment the two spacers were analysed together, totalling a matrix of 404 characters (average of 66·7 % GC content). Thirty-two polymorphic sites were found, of which 14 corresponded to fixed interspecies variation, 14 to intraspecific differences in P. actinia and four to intraspecific variability in P. elegans. A large part of this variability was due to heterozygous sites in some individuals (57 % of the polymorphic sites in P. actinia, and 75 % in P. elegans showed this type of variation). Six sequence types were identified in P. elegans (H1E-H6E) and 15 in P. actinia (H7A-H21A; Table 2). Plant E56 was identified as a hybrid (see below) due to its morphological appearance and the occurrence of sequences from both species (H3E/H15A). Nucleotide diversity values were 0·006 for P. actinia and 0·002 for P. elegans.
Table 2.
Sequences identified in ITS regions 1 and 2 of the Passiflora plants investigated
| Nucleotide position |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS 1 |
ITS 2 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sequence type |
30 |
65 |
76 |
78 |
80 |
88 |
95 |
96 |
98 |
102 |
109 |
144 |
145 |
154 |
157 |
170 |
182 |
208 |
232 |
242 |
245 |
246 |
247 |
249 |
250 |
253 |
268 |
282 |
292 |
379 |
393 |
404 |
No. observed |
|||||||||||||||||||||||||||||||
| H1E | – | A | T | C | T | T | A | C | T | C | T | A | G | G | – | T | T | A | G | C | – | G | – | C | C | C | T | G | – | G | G | C | 8 | |||||||||||||||||||||||||||||||
| H2E | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | 2 | |||||||||||||||||||||||||||||||
| H3E | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | 8 | |||||||||||||||||||||||||||||||
| H4E | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | T | · | · | · | · | · | · | 1 | |||||||||||||||||||||||||||||||
| H5E | T | C | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | 2 | |||||||||||||||||||||||||||||||
| H6E | · | C | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | 20 | |||||||||||||||||||||||||||||||
| H8A | · | C | C | T | C | · | · | · | · | · | C | C | A | A | · | C | · | G | A | A | C | C | C | T | T | · | A | · | T | · | · | · | 2 | |||||||||||||||||||||||||||||||
| H7A | · | C | C | T | C | · | · | · | · | · | C | C | A | A | · | C | · | G | A | A | C | C | C | T | T | · | A | · | · | · | · | · | 9 | |||||||||||||||||||||||||||||||
| H9A | · | C | C | T | C | · | C | · | · | · | C | C | A | A | · | C | · | G | A | A | C | C | C | T | T | · | A | · | · | · | · | · | 5 | |||||||||||||||||||||||||||||||
| H10A | · | C | C | T | C | · | · | · | · | – | C | C | A | A | G | C | · | G | A | A | C | C | C | T | T | · | A | · | · | · | · | · | 3 | |||||||||||||||||||||||||||||||
| H11A | · | C | C | T | C | · | · | G | · | – | C | C | A | A | G | C | · | G | A | A | C | C | · | T | T | · | A | · | · | · | · | · | 1 | |||||||||||||||||||||||||||||||
| H12A | · | C | C | T | C | · | C | G | · | – | C | C | A | A | G | C | A | G | A | A | C | C | · | T | T | · | A | · | · | · | · | · | 1 | |||||||||||||||||||||||||||||||
| H13A | · | C | C | T | C | · | · | G | · | · | C | C | A | A | · | C | · | G | A | A | C | C | C | T | T | · | A | · | · | · | · | · | 2 | |||||||||||||||||||||||||||||||
| H14A | · | C | C | T | C | · | · | G | A | · | C | C | A | A | · | C | · | G | A | A | C | C | C | T | T | · | A | · | · | · | · | · | 2 | |||||||||||||||||||||||||||||||
| H15A | · | C | C | T | C | C | C | G | A | · | C | C | A | A | · | C | · | G | A | A | C | C | C | T | T | · | A | · | · | · | · | · | 5 | |||||||||||||||||||||||||||||||
| H16A | · | C | C | T | C | C | C | G | A | – | C | C | A | A | G | C | · | G | A | A | C | C | C | T | T | · | A | · | · | · | · | · | 3 | |||||||||||||||||||||||||||||||
| H17A | · | C | C | T | C | C | C | G | A | – | C | C | A | A | G | C | · | G | A | A | C | C | · | T | T | · | A | · | · | · | · | · | 1 | |||||||||||||||||||||||||||||||
| H18A | · | C | C | T | C | C | C | G | A | – | C | · | A | A | G | C | · | G | A | A | C | C | · | T | T | · | A | · | · | · | · | · | 27 | |||||||||||||||||||||||||||||||
| H19A | · | C | C | T | C | C | C | G | A | – | C | · | A | A | G | C | · | G | A | A | C | C | · | T | T | · | A | A | · | · | · | · | 1 | |||||||||||||||||||||||||||||||
| H20A | · | C | C | T | C | C | C | G | A | – | C | · | A | A | G | C | · | G | A | A | C | C | · | T | T | · | A | · | · | T | · | · | 1 | |||||||||||||||||||||||||||||||
| H21A | · | C | C | T | C | C | C | G | A | – | C | · | A | A | G | C | · | G | A | A | C | C | · | T | T | · | A | · | · | · | – | – | 2 | |||||||||||||||||||||||||||||||
Dots indicate agreement with the reference sequence (H1E) and dashes deletions at the indicated positions.
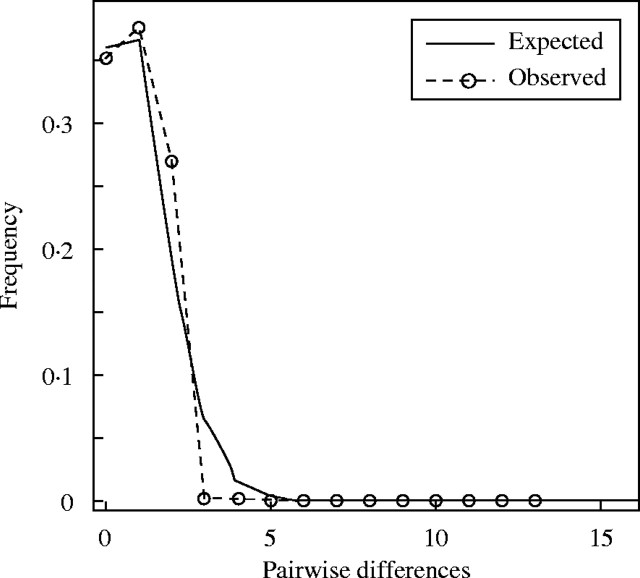
The median-joining network (Fig. 1) clearly showed a north–south direction in the inter- and intraspecific relationships. A clear separation exists between the northern and southern sequences in P. actinia, but there is a series of intermediate sequence types. All P. elegans sequences were consistently associated with those obtained from southern P. actinia plants. In P. elegans, however, it was not possible to identify a relationship between sequences and the geographical origin of the plant. Tajima's D (P. actinia: 1·077; P > 0·10, P. elegans: 0·689; P > 0·10) and Fu and Li's D* (P. actinia: −0·966; P > 0·10, P. elegans: −0·357; P > 0·10) neutrality tests showed non-significant values. A single wave in the mismatch distribution—a histogram of genetic differences between pairs of individuals within a sample—with a peak at about one difference was found among P. elegans plants (Fig. 2) indicating a recent population expansion. The P. sidaefolia (AY102353/AY102373) sequence used as an outgroup connected to the network through the sequences of northern individuals of P. actinia. A reticulation was found between H1E, H2E, H3E and H6E, due to homoplasic substitutions at sites 65 and 109 or to recombination between some copies (Fig. 1).
Fig. 2.
Mismatch distribution histogram indicating the observed and expected numbers of pairwise differences in ITS sequences between P. elegans plants. The numbers of pairwise differences are given on the horizontal axis and their frequencies on the vertical axis.
Plastid DNA markers
The results are displayed in Table 1. The trnL-trnF and psbA-trnH spacers ranged in size from 288 to 294 bp and from 309 to 311 bp, respectively (averages, respectively, of 36·1 and 28·9 % GC content). For the trnL-trnF spacer, two differences were found, a 6-bp indel (TAAGAT, sites 265–270) present in P. actinia and absent in P. elegans; and a C (P. actinia)/A (P. elegans) change at site 196. In the psbA-trnH spacer a substitution at position 59, T (P. actinia)/G (P. elegans), and a region characterized by T repetitions (sites 163–172) were found. The latter varied both between species and among plants. All P. actinia plants had eight Ts. In P. elegans plants E16, E29 and E44 had 10 Ts, whereas the others all had nine. Plant E56 of P. elegans showed sequences in both plastid spacers identical to those of P. actinia.
A natural hybrid between P. actinia and P. elegans
Although the two species were not found in sympatry, a population of P. elegans was found only 9 km away from a P. actinia population in Santo Antonio da Patrulha county (29°50′S; 50°30′W). At this site a member of the P. elegans population (E56) was morphologically quite distinct from the others, showing a clearly intermediate phenotype, especially in relation to flower size and leaf shape. The main morphological distinctive characteristics of the two species are that in P. actinia the flowers are larger and the leaves are unlobed, while the leaves of P. elegans are slightly trilobed. In E56 leaves showed various patterns of lobation and the flowers were intermediate in size. All three molecular markers employed support the hypothesis that E56 is a hybrid between P. actinia and P. elegans, as suggested by its intermediate morphology and geographic localization (at the border of the distributions of the two species). The tetrazolium test indicated that seed viability in the hybrid was normal.
DISCUSSION
ITS genetic diversity
Paralogues of nrDNA can become pseudogenes when inactivated or when a single copy is transported to a different genomic region. These pseudogenes can accumulate random substitutions at high rates, reducing the stability of the secondary structure (Buckler and Holtsford, 1996; Koch et al., 2003). It is unlikely, however, that the sequences analysed in this study are pseudogenes, since: (a) DMSO was added to the PCR process, diminishing the possibility of amplification of less stable forms (Buckler et al., 1997, Fuertes Aguilar et al., 1999); (b) the transition/transversion rates and the GC contents are not different from the pattern generally found in the genus (Muschner et al., 2003); and (c) several of the parental copies of the heterozygous individuals were identified.
The high level of ITS polymorphism found here indicates that the process of concerted evolution is not fast enough to homogenize the region. This process acts through recombination or gene conversion (Campbell et al., 1997). Inter-chromosomal exchanges can be facilitated by the terminal location of the nrDNA loci, as occur in Gossypium, Paeonia and Thinopyrum (Wendel et al., 1995; Zhang and Sang, 1999; Li and Zhang, 2002) but in the present case this does not seem to be the case, since Melo and Guerra (2003) mapped the 45S sites, where the ITS region is located, in subterminal positions in P. actinia and P. elegans.
Vegetative reproduction may extend generation time, and this may slow the speed with which concerted evolution/homogenization through recombination can occur (Sang et al., 1995; Buckler IV and Holtsford, 1996). Vegetative reproduction is common in P. actinia and P. elegans. Plants of these species produce many shoots from roots that develop into complete plants; this fact, coupled with recent gene flow between divergent lineages, may explain the ITS polymorphism observed here. The process, however, was not important enough to obscure the interspecific relationships observed in the genus (Muschner et al., 2003).
Relationships of P. actinia and P. elegans and the Quaternary palaeoclimatic history of Rio Grande do Sul
It is clear from the molecular data presented here, as well as other molecular analyses performed by Muschner et al. (2003), that P. actinia and P. elegans are closely related, possible sister species. Additionally, P. actinia and P. elegans have the same chromosome number (2n = 18; Melo and Guerra, 2003), show extremely similar flower morphology and blossom at the same period of the year. In relation to the scenario for their divergence, one hypothesis would postulate that P. elegans diverged from P. actinia before the occurrence of the intraspecific differentiation found today in the latter species. The other scenario would imply that P. elegans diverged from southern populations of P. actinia after the latter separated from the northern groups. However, the phylogenetic connection of P. sidaefolia with the northern clade of P. actinia (Fig. 1) suggests a clear direction on the divergence of these species, and summing up all genetic, geographic and morphological evidence, the proposition that P. elegans diverged from an already differentiated southern group of P. actinia is favoured.
Passiflora actinia shows a high ITS diversity that seems to be geographically structured (Fig. 1), suggesting relatively old populations (Avise, 2000). Conversely, P. elegans shows lower variability and this variation is not geographically structured. The high frequency of sequence H6E, its central position in the network, and its geographic distribution suggest that it may be the plesiomorphic sequence in P. elegans. The left skewed single peak at about one difference in the mismatch distribution (Fig. 2), on the other hand, is an indication that this species went through a severe bottleneck followed by a recent expansion in size and possibly range (Rogers and Harpending, 1992).
Influence of the Pleistocene glaciations on the distribution and genetic diversity of plants has been much studied by molecular markers (Gielly et al., 2001; Zhang et al., 2001), but these studies were mainly limited to the northern hemisphere, with very few relating to tropical forests and even fewer to the species-rich South American Atlantic Forest. Pollen studies performed by Roth and Lorscheitter (1993) and Neves and Lorscheitter (1995) suggest that the central plateau and coastal plains of Rio Grande do Sul had a cold, semiarid climate in the last Pleistocene glacial stage (23 000–11 000 years bp). Open vegetation prevailed and forests were restricted to ecological refuges. The two main events of Atlantic Forest expansion from northern locations into Rio Grande do Sul are related to periods of significant climatic improvement, with rising temperatures and humidity, one at the beginning of the Holocene (11 000–10 000 years bp) and another around 6700 years bp (Lorscheitter, 1997). The data suggest that these migrations may have occurred in a fragmentary way, leading to spatial isolation of populations. The phylogeographic scenarios found here for both P. actinia and P. elegans seems to agree closely with the climatic and biogeographic scenarios described above for the Atlantic Forest in southern Brazil.
The presence of a viable hybrid at the place where the two species are geographically closest indicates secondary contact but absence of a hybrid zone, probably due to their spatial segregation, possibly influenced by external environmental agents, such as humidity, soil type, altitude, rainfall and other ecological factors (as described for other species by Fritsche and Kaltz, 2000; Gielly et al., 2001).
Genetic characterization of the hybrid
Interspecific hybridization is a widespread phenomenon in plants (Rieseberg, 1997). Natural hybrids are frequent in Passiflora (Vanderplank, 1996), but had not been previously described between P. actinia and P. elegans. In the present study, a single hybrid plant was identified due to its intermediate floral and vegetative morphology, a finding that was afterwards confirmed by the genetic markers, with the nuclear nucleotide sites that differentiate P. actinia and P. elegans showing an additive pattern in the hybrid. The plastid markers indicated that P. actinia was the donor species of the plastid DNA in this hybrid. It was found in a P. elegans population, the closest P. actinia group occurring 9 km apart. Therefore, it is more likely that this hybrid occurred as a result of a single pollen dispersion and that plastid transmission in these species may be paternal, in contrast to the normal maternal inheritance in flowering plants. The only species of Passiflora considered (P. edulis) in a previous study of organelle inheritance in plants (Harris and Ingram, 1991) showed evidence, through epifluorescence microscopic studies, of paternal plastid inheritance. In addition, molecular markers have indicated that Turnera ulmifolia (also Passifloraceae; Angiosperm Phylogeny Group, 2003) shows a mixed, but paternally biased pattern of plastid inheritance (Shore and Triassi, 1998), indicating with the present results that paternal plastid inheritance may be of more widespread occurrence in this group of plants. Mitochondrial DNA in the hybrid documented here is, however, of maternal (P. elegans) origin (V. C. Muschner, unpubl. res.).
CONCLUSIONS
The present work confirmed previous records that the southernmost geographical range of P. actinia is the Atlantic Forest area situated in the north-eastern region of the Brazilian state of Rio Grande do Sul, whereas P. elegans occurs in the riparian vegetation of the interior of that state. The variability found in P. actinia is structured along a north–south gradient, whereas P. elegans has experienced a recent population bottleneck followed by an expansion. The recent demographic histories of both species were strongly influenced by the climatic changes that occurred in this area during the Quaternary period. A natural hybrid was found at the border of the distribution of these two species, and its genetic constitution suggests that plastid inheritance is paternal in these species.
Acknowledgments
Thanks are due to Prof. Maria Luiza Lorscheitter, Botany Department, Biosciences Institute of our University, for help with the interpretation of the geological past of Rio Grande do Sul, and to the personnel of the Geoprocessing Laboratory, Department of Ecology of the same institute, who helped in the evaluation of the soil types of our collecting sites. Financial help was provided by Programa de Apoio a Núcleos de Excelência (PRONEX), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) and Pró-Reitoria de Pesquisa da Universidade Federal do Rio Grande do Sul (PROPESQ-UFRGS).
LITERATURE CITED
- Angiosperm Phylogeny Group. 2003. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society 141: 399–436. [Google Scholar]
- Arnhein N. 1983. Concerted evolution of multigene families. In: Nei M, Koehn RK, eds. Evolution of genes and proteins. Sunderland: Sinauer, 38–61. [Google Scholar]
- Avise JC. 2000.Phylogeography: the history and formation of species. Cambridge: Harvard University Press. [Google Scholar]
- Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. 1995. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=journals&list_uids=20275&dopt=full 82: 247–277. [Google Scholar]
- Bandelt H-J, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Benson WW, Brown Jr KS, Gilbert LE. 1976. Coevolution of plants and herbivores: passion flower butterflies. Evolution 29: 659–680. [DOI] [PubMed] [Google Scholar]
- Bonner FT. 1986. Measurement of seed vigor for loblolly (Pinus taeda) and slash pines (Pinus elliottii). Forest Science 32: 170–178. [Google Scholar]
- Buckler ES IV, Holtsford TP. 1996.Zea ribosomal repeat evolution and substitution patterns. Molecular Biology and Evolution 13: 623–632. [DOI] [PubMed] [Google Scholar]
- Buckler ES IV, Ippolito A, Holtsford TP. 1997. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics 145: 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CS, Wojciechowski MF, Baldwin BG, Lawrence AA, Donoghue MJ. 1997. Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex (Rosaceae). Molecular Biology and Evolution 14: 81–90. [DOI] [PubMed] [Google Scholar]
- Denduangboripant J, Cronk QCB. 2000. High intraindividual variation in internal transcribed spacer sequences in Aeschynanthus (Gesneriaceae): implications for phylogenetics. Proceedings of the Royal Society of London 267: 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfeux C, Lejeune, B. 1996. Systematics of Euromediterranean Silene (Caryophyllaceae): evidence from a phylogenetic analysis using ITS sequences. Comptes Rendus de l'Academie des Sciences de Paris 319: 351–358. [PubMed] [Google Scholar]
- Fritsche F, Kaltz O. 2000. Is the Prunella (Lamiaceae) hybrid zone structured by an environmental gradient? Evidence from a reciprocal transplant experiment. American Journal of Botany 87: 995–1003. [PubMed] [Google Scholar]
- Fu YX, Li WH. 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes Aguilar J, Nieto Feliner G. 2003. Additive polymorphisms and reticulation in an ITS phylogeny of thrifts (Armeria, Plumbaginaceae). Molecular Phylogenetics and Evolution 28: 430–447. [DOI] [PubMed] [Google Scholar]
- Fuertes Aguilar J, Rosselló JA, Nieto Feliner G. 1999. Nuclear ribosomal DNA (nrDNA) concerted evolution in natural and artificial hybrids of Armeria (Plumbaginaceae). Molecular Ecology 8: 1341–1346. [DOI] [PubMed] [Google Scholar]
- Gielly L, Debussche M, Thompson JD. 2001. Geographic isolation and evolution of Mediterranean endemic Cyclamen: insights from chloroplast trnL (UAA) intron sequence variation. Plant Systematics and Evolution 230: 75–88. [Google Scholar]
- Harris SA, Ingram R. 1991. Chloroplast DNA and biosystematics: the effects of intraspecific diversity and plastid transmission. Taxon 40: 393–412. [Google Scholar]
- Killip EP. 1938. The American species of Passifloraceae. Botanical Series, Field Museum of Natural History 19: 1–613. [Google Scholar]
- Koch MA, Dobes C, Mitchell-Olds T. 2003. Multiple hybrid formation in natural populations: concerted evolution of the internal transcribed spacer of nuclear ribosomal DNA (ITS) in North American Arabis divaricarpa (Brassicaceae). Molecular Biology and Evolution 20: 338–350. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software, Arizona State University, Tempe, Arizona, USA. Program available on the internet (http://www.megasoftware.net). [DOI] [PubMed] [Google Scholar]
- Li D, Zhang X. 2002. Physical localization of the 18S-5·8S-26S rDNA and sequence analysis of ITS regions in Thinopyrum ponticum (Poaceae: Triticeae): implications for concerted evolution. Annals of Botany 90: 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorscheitter ML. 1997. Paleoambientes do sul do Brasil no Quaternário através da palinologia: revisão dos resultados obtidos. Revista Universidade de Guarulhos, Geociências 2: 197–199. [Google Scholar]
- MacDougal JM. 1994. Revision of Passiflora, subgenus Decaloba, Section Pseudodysosmia (Passifloraceae). Systematic Botany Monographs 41: 1–146. [Google Scholar]
- Mayer MS, Soltis PS. 1999. Intraspecific phylogeny analysis using ITS sequences: insights from studies of the Streptanthus glandulosus complex (Cruciferae). Systematic Botany 24: 47–61. [Google Scholar]
- Melo NF de, Guerra M. 2003. Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Annals of Botany 92: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondin CA. 2001.Passiflora organensis Gardner (Passifloraceae), primeira citação de ocorrência para o Rio Grande do Sul. Pesquisas-Botânica 51: 147–150. [Google Scholar]
- Muschner VC, Lorenz AP, Cervi AC, Bonatto SL, Souza-Chies TT, Salzano FM, Freitas LB. 2003. A first molecular phylogenetic analysis in Passiflora (Passifloraceae). American Journal of Botany 90: 1229–1238. [DOI] [PubMed] [Google Scholar]
- Neves PCF, Lorscheitter ML. 1995. Upper Quarternary paleoenvironments in the northern coastal plain of Rio Grande do Sul, Brazil. Quarternary South American Antarctic Peninsula 9: 39–67. [Google Scholar]
- Rambo B. 1951. A imigração da selva higrófila no Rio Grande do Sul. Sellowia 3: 55–91. [Google Scholar]
- Rieseberg LH. 1997. Hybrid origins of plant species. Annual Review of Ecology and Systematics 28: 359–389. [Google Scholar]
- Rogers AR, Harpending H. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution 9: 552–569. [DOI] [PubMed] [Google Scholar]
- Roth L, Lorscheitter ML. 1993. Palynology of a bog in Parque Nacional de Aparados da Serra, east plateau of Rio Grande do Sul, Brazil. Quarternary South American Antarctic Peninsula 8: 39–69. [Google Scholar]
- Roy A, Frascaria N, Mackay J, Bousquet J. 1992. Segregating random amplified polymorphic DNAs (RAPDs) in Betula alleghaniensis Theoretical and Applied Genetics 85: 173–180. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. 1999. DnaSP, version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. 1995. Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implications for biogeography and concerted evolution. Proceedings of the National Academy of Sciences of the USA 92: 6813–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany 84: 1120–1136. [PubMed] [Google Scholar]
- Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA. 1998. Phylogeographic studies in plants: problems and prospects. Molecular Ecology 7: 465–474. [Google Scholar]
- Shore JS, Triassi M. 1998. Paternally biased cpDNA inheritance in Turnera ulmifolia (Turneraceae). American Journal of Botany 85: 328–332. [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: 4876–4882. Program available on the internet (ftp://ftp-igbmc.u-strasbg.fr/pub/clustalx). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderplank J. 1996.Passion flowers, 2nd edn. Cambridge: Massachusetts Institute of Technology Press. [Google Scholar]
- Villwock JA, Tomazelli LJ. 1995. Geologia costeira do Rio Grande do Sul. Universidade Federal do Rio Grande do Sul, Notas Técnicas 8: 1–45. [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T. 1995. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proceedings of the National Academy of Sciences of the USA 92: 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Sang T. 1999. Physical mapping of ribosomal RNA genes in paeonies (Paeonia, Paeoniaceae) by fluorescent in situ hybridization: implications for phylogeny and concerted evolution. American Journal of Botany 86: 735–740. [PubMed] [Google Scholar]
- Zhang L-B, Comes HP, Kadereit JW. 2001. Phylogeny and quaternary history of the European montane/alpine endemic Soldanella (Primulaceae) based on ITS and AFLP variation. American Journal of Botany 88: 2331–2345. [PubMed] [Google Scholar]