Significance
Multicellular organisms, by necessity, form highly organized structures. The mechanisms required to construct these often dynamic structures are a challenge to understand. Myxococcus xanthus, a soil bacterium, builds two large structures: growing swarms and fruiting bodies. Because the cells are genetically identical, they rely on regulating protein activity and the levels of gene expression. Moreover, the long, flexible, rod-shaped cells modify each others’ behavior when they collide. By examining development of a Myxococcus swarm, testable rules can be proposed that rely only on cell behavior and cell–cell contact signaling. The mechanisms used by this prokaryote to form complex, dynamic multicellular structures might have been adapted for Hedgehog and Wnt morphogenetic signaling in animals.
Keywords: timer, pattern formation, cell polarity, synchrony, polysaccharide
Abstract
We offer evidence for a signal that synchronizes the behavior of hundreds of Myxococcus xanthus cells in a growing swarm. Swarms are driven to expand by the periodic reversing of direction by members. By using time-lapse photomicroscopy, two organized multicellular elements of the swarm were analyzed: single-layered, rectangular rafts and round, multilayered mounds. Rafts of hundreds of cells with their long axes aligned in parallel enlarge as individual cells from the neighborhood join them from either side. Rafts can also add a second layer piece by piece. By repeating layer additions to a raft and rounding each layer, a regular multilayered mound can be formed. About an hour after a five-layered mound had formed, all of the cells from its top layer descended to the periphery of the fourth layer, both rapidly and synchronously. Following the first synchronized descent and spaced at constant time intervals, a new fifth layer was (re)constructed from fourth-layer cells, in very close proximity to its old position and with a number of cells similar to that before the “explosive” descent. This unexpected series of changes in mound structure can be explained by the spread of a signal that synchronizes the reversals of large groups of individual cells.
Reichenbach documented the species similarities and differences in swarming and fruiting body development in a remarkable series of annotated time-lapse movies (1-4). Due to the way Myxococcus xanthus cells move and to the way cells interact with each other, their swarms spread outward (5). The capacity to spread arises from their ability to build different types of organized, dynamic, multicellular structures: planar, rectangular rafts of cells with their long axes aligned; and round multilayered mounds. Although swarms are round like colonies, they differ from colonies in two ways. (i) Swarms have flat tops; they are not heaped up like a colony. (ii) The rate at which a steady state swarm expands is directly proportional to the rate of individual cell movement, not to its rate of growth, and the increase in cell numbers contribute only 10% of the swarm expansion rate (5). All stages of growth can be found in a swarm. Because M. xanthus swarm cells are moving all of the time, except when they pause briefly to reverse their direction, they offer an experimentally tractable system to investigate how multicellular structures can be constructed according to an inherited plan.
Swarm cells are self-propelled, and their movement enhances growth by giving cells at the top and bottom of a thick swarm equal access to oxygen from above and dissolved solid nutrients from below. Rapidly moving (and growing) cells are found in the 0.5-mm–wide annular ring of moving cells that includes the outermost edge of the swarm, illustrated in Fig. 1. In many respects, an M. xanthus swarm resembles a large school of fish or flock of birds that has no leader. Rather, each swarm cell acts as both leader and follower, giving and taking directions from the movements of neighboring cells. To learn how M. xanthus builds rafts and mounds, we have recorded and studied individual cell behavior in a series of time-lapse movies.
Fig. 1.
A phase contrast image of cells at the edge of a DK1622 swarm on 1% CTT, 1% agar. The swarm is expanding in the radial direction, which is to the right in this image of a small section of the swarm. (Scale bar: 50 µm.) Photographed with a 20× phase contrast objective. A side-by-side cluster of five cells, a slime trail, a multicellular mound with five layers, and a large multicellular raft are identified. The inner edge of the annulus of exponential growth reaches 0.517 µm from the outer edge of the swarm.
To power its gliding movements, M. xanthus bears type IV pili at the leading pole of each cell that retract, pulling it forward (6–8). Such movement is known as S-motility. Three conceptually different motors have been proposed for a second type of movement, A-motility. Focal adhesions—discovered by Mignot et al. (9, 10) and revealed by fluorescently labeled clusters of AglZ, a protein necessary but not sufficient for A-motility (11)—are one proposal. The focal adhesion complexes, found along the sides of cells, are proposed to be connected to cytoskeletal proteins via motor proteins (10, 11). A second proposal considers the deformation of the cell envelope generated by the proton motive force to propel cells in the direction of their long axis (12). Finally, the secretion of polysaccharide slime from nozzles located at the trailing pole of each cell has been proposed to push each cell forward (13–15). In addition to the proposed motors, there is evidence that several lipoproteins (CglB, CglC, CglD, CglE, and CglF) essential for A-motility are localized to the outer surface of the cells’ outer membrane. Mutants that lack any one of the Cgl proteins can be rescued, regaining their A-motility when wild-type cells are mixed with the mutants and allowed to swarm together over an agar surface (16–18). Only the normal A-motility of the mutants is rescued; their genotype remains mutant. These observations showed that Cgl proteins (but not cgl genes) can be transferred efficiently from one cell to another by direct contact (17). Recently, it was demonstrated that two host proteins, TraA and TraB, must be present in both the donor and the recipient for such contact-mediated, outer-membrane lipoprotein transfer (19).
Wu et al. observed that a periodic reversal of gliding direction is necessary in order for M. xanthus to organize a swarm (20). Reversals are controlled by proteins of the Frz chemosensory system (21). Mutants deleted for either the FrzCD protein, the FrzE protein, or the FrzF protein exhibit the same low rate of swarm expansion whereas a mutant deleted for the FrzG protein had a significantly higher expansion rate, closer to the wild-type rate (5). The higher rate in a FrzG deletion could simply reflect a higher level of FrzCD methylation, resulting in unconstrained oscillation. We suggest that FrzCD, FrzE, and FrzF proteins form an oscillator that we call the pacemaker. The pacemaker is understood to drive the MglA small G protein that acts as a switch to reverse the gliding direction (5). We propose that the CglB protein, on the cell surface, forms protein to protein contacts that are the signal required to build multicellular rafts and multilayered mounds in M. xanthus swarms.
Results
Swarm cells are able to grow at their maximum possible rate even as the cell density rises and cells compete with each other for oxygen and other nutrients. Rapid growth of the cells within a mound is facilitated by the circulation of cells from one layer to another. Both dynamic rafts and dynamic mounds are found at the edge of the swarm. The changing form and distribution of cells in a 20× microscopic field is recorded in Table 1.
Table 1.
Multicellular structures found in an A+S+ swarm field
| Day | Multicellular structures found | Movie |
| 1 | Many rafts that were partially covered with patches of second-layer cells. Some rafts emerged as peninsulas extending out from the swarm edge. | S1 |
| 2 | Several small, three-layered, steep-sided dynamic mounds were found. They can elongate, often toward another mound, which they join. Elongated mounds often recondensed to a single round mound. | S2 |
| 4 | Two round, five-layered mounds were evident some distance behind the edge of the swarm. A rapid and synchronous dispersal of all cells from the fifth layer down to the periphery of the fourth layer was observed in both mounds. Dispersal was followed periodically by reconstruction of the fifth layer, three times. | S3 |
| 5 | The whole field all of the way to the outer edge was covered with long ridges that are about five layers high. The ridges move about whereas there are patches of vacant agar. | S4 |
The Pattern of Cell Movement Within a Five-Layered Mound.
Any population of biological oscillators, such as a population of pacemakers, each of whose output is periodic, needs two numbers to describe it: first, its repeat-time or period, which has a stable average value (5); and second, a phase angle between 0° and 360° that designates the stage within one cycle that its pacemaker occupies at each instant of time (5, 22). As long as the pacemakers of two cells are independent of each other, their pacemakers would be expected to be found in different phases. Indeed, they are never found in the same phase before the first explosive dispersion (recorded in Table 2). At that time, several hundred neighboring cells in the same mound share the same phase as they “explosively” disperse. Mounds, like the one identified in Fig. 1, have multiple layers of cells nested one on top of the other. Fig. 2 shows that, in the swarm’s steady state, the mound has exactly five layers. Layers are more striking when the specimen is viewed with oblique illumination as in Fig. 2B. Phase contrast shows the same five layers in Fig. 2A.
Table 2.
Repeated explosive dispersions of the mound top layer
| Fifth layer | Time of the explosion, min | x coord. of mound center, µm | y coord. of mound center, µm | Area of fifth layer before explosion, µm2 |
| First expl. | 7.0* | 56.7 ± 1.27 | 90.72 ± 1.27 | 25.0 ± 2.5 |
| Second expl. | 21 ± 1 | 59.2 ± 1.27 | 88.62 ± 1.27 | 20.8 ± 1.7 |
| Third expl. | 36.7 ± 0.5 | 65.1 ± 1.27 | 91.98 ± 1.27 | 22.5 ± 2.7 |
Fig. 2.
Mounds. (A) Top view, phase-contrast illumination. (Scale bar: 100 µm.) (B) Several mounds viewed under oblique light. (Scale bar: 100 µm.) Each layer (Layer 1–Layer 5) is indicated.
Reflecting a virtually continuous motion of cells at the swarm edge, cells move from one layer in the mound to the next layer, either above or below it. This motion appears to use S-motility because a cell must be pulled off the surface of one layer, before it can be moved to another layer and pilus retraction (S-motility) can pull it off. A force of 100 pN can be produced by retraction of a single pilus of Neisseria gonorrheae to which the pili of M. xanthus are closely related (6). Cells also move to eliminate voids between adjacent cells in the same layer. This motion can involve the relatively weaker force of A-motility, estimated by Wolgemuth et al. as about a piconewton (13) because the cell remains on the same surface. The phase of each cell’s pacemaker is manifested, starting about 1 h into Movie S3. At that moment, the dark, round, top (fifth) layer of the mound splits into fragments that separate from each other at a relatively high speed. The entire fifth layer fragments into 50 or more small, black, rod-shaped spots that appear to represent small clusters of cells, as shown in Fig. 3. Complete fragmentation of the fifth layer occurs within a minute (compare 62.5 min with 63.5 min in Fig. 3). The speed of fragmentation, measured on images from Movie S3 was 12.5 ± 0.4 µm/min. This speed is about seven times the 1.7 ± 0.9 µm/min average displacement speed of any of the single cells moving on the agar outside the mound in the same microscopic field. Less than 1 min after the abrupt speedup had been noted, the gliding speed of cells in the mound had returned to their prefragmentation rate. The speed of cells outside the mound, measured at the time of fifth layer fragmentation, also had the same low prefragmentation rate.
Fig. 3.
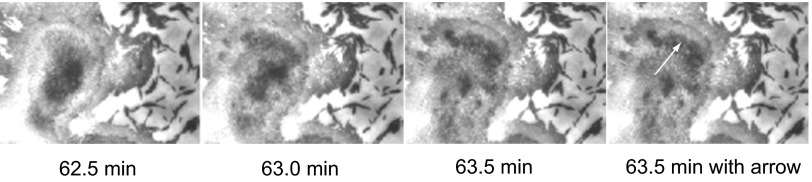
Three consecutive frames from the movie of the mound identified in Fig. 1, showing the explosion of its top layer. Photos were taken every 30 s for 2.5 h. The images shown were exposed at 62.5, 63.0, and 63.5 min. The arrow in the fourth frame shows the lateral displacement of cells from the center of the top layer at 62.5 min to the periphery of the next layer down at 63.5 min, from which the speed of fragmentation was calculated. For the full visual effect, see Movie S1.
Because the fifth layer disappears completely, its descent to the periphery of layer 4 could be explained by the synchronous reversal of all of the cells in the top layer of the mound followed immediately by extension of their type IV pili, by pilus tip attachment to fibrils that envelop layer-4 cells (as sketched in Fig. S1) and by pilus retraction and descent of cells to the periphery of layer 4. Table 3 shows that the area of cells gained by layer 4 is the same as the area of cells lost by layer 5 for the first and second explosions. For the third explosion, however, layer 4 seemed to have gained only 9.1 µm2, significantly less than the 16.8 µm2 loss from layer 5. Note (Table S1) that the areas lost and gained being decreasing with each subsequent explosion (because cells drop one by one from layer 5 to layer 4) could be explained by a wider separation of cells on layer 4 because there are fewer of them. Despite the falloff in number of cells detected, the simultaneous disappearance of cells from layer 5 with the expansion of layer 4 by a comparable area of cells renders the repeated explosion scheme a plausible account of the observations. Moreover, this scheme explains how cells within a mound use their pili to move between any pair of adjacent layers. M. xanthus type IV pili are capable of extending and retracting faster than is necessary for such movement (6, 7).
Table 3.
Comparing the loss from layer 5 with the gain to layer 4
| Fifth Layer | Area | No. of measurements | Mean, µm2 | SD |
| First expl. | Fifth layer loss | 15 | 30.91 | ±3.53 |
| Fourth layer gain | 15 | 31.18 | ±5.14 | |
| Second expl. | Fifth layer loss | 15 | 17.3 | ±2.52 |
| Fourth layer gain | 15 | 16.8 | ±2.61 | |
| Third expl. | Fifth layer loss | 15 | 16.8 | ±2.66 |
| Fourth layer gain | 15 | 9.1 | ±1.94 |
Reversals of Cells in a Raft Are Revealed by Their Gray Level.
The rectangular shape of a raft indicates that the rod-shaped cells within it are aligned in the direction of the raft’s long axis. A raft, like the one labeled in Fig. 1, differs from side-by-side clusters of cells also labeled in Fig. 1 in having a lower density of cells. Cells are packed less tightly in a raft than in a cluster, and the surface density (cells per µm2) is lower because the rafts appear gray whereas the side-by-side clusters of similar cells appear black (Fig. 1), like individual cells. With phase-contrast illumination, the gray level is roughly proportional to the surface density. However, the surface density is lower only in the direction of the raft’s long axis whereas the density of packing cells side by side seems to be the same as that observed within a cluster. Within a raft, pairs of adjacent cells are expected to be separated from each other by a layer of the capsular polysaccharide, like a side-by-side cluster of cells, several of which are evident in Fig. 1. Capsular slime protects each cell from autolysis by extracellular lytic enzymes (15). An asymmetric density arises from the way rafts are thought to enlarge: New cells associate with one side or the other of a raft, expanding its width. For example, the raft that is labeled in Fig. 1, when followed from one movie frame to the next, was seen to be joined by cells from its neighborhood, usually one at a time (Figs. S2–S5). The joining process can be observed live in movie S1 in ref. 20. The addition of individual cells to either side of a raft can leave an empty space between the ends of cells that are added to the new course of cells at either edge of the raft. “Course” is understood as a building pattern, a linear formation of cells in a raft. As more courses of cells are added and the raft widens further, courses with space between cell ends develop within the body of the raft. Considering that the space between the ends of two cells in the same course varies from a single point of contact between two hemispherical cell ends to a full cell length, Fig. S4A illustrates possible cell arrangements. This scenario of raft enlargement was observed by tracking the growth of multiple gaps found among several rafts evident in Fig. 1. Gaps enlarged in the direction of the raft’s long axis to a maximum of ∼60% of the raft’s width in 3–7.5 min, as shown in Fig. S4. These observations justify a broad assumption that raft cells are reversing independently of one another. The dynamics of independent reversals help to explain why rafts having many gaps are gray whereas cell clusters and single cells that have no gaps are black whenever a phase-contrast objective lens is used to examine them.
The Role of CglB in Building Rafts and Mounds.
The swarm of an A+S+ strain expands at a constant rate for several hundred hours, almost 100 generations (5). That constancy reflects the remarkable stability of the steady-state organization. Fig. 4 shows that, although a strain lacking the CglB lipoprotein (deletion ASX1) (Materials and Methods) can expand its swarm, after a short initial lag, the instantaneous rate of expansion falls over the 680 h it was followed, and, compared with wild-type cells, a steady expansion rate never was achieved. Instead the expansion rate falls slowly. Up to 285 h, the expansion rate is 0.056 mm/h; from 310 h to 480 h, it is 0.046 mm/h; and, from 495 h to 680 h, it is 0.033 mm/h. Each of these straight-line fitted segments has a correlation coefficient larger than 0.99. The absence of any steady-state behavior coincides with the absence of rectangular rafts and of five-layered mounds at the edge of the mutant swarm. The construction defect is quite particular in that the CglB mutant is still able to build organized structures with small, round second layers, but no mounds with layers. Instead of rectangular rafts whose long axis apparently can be oriented in any direction within the swarm, the CglB mutant arrests morphogenesis having built small “arrowheads” that are shown in Fig. S6 A and B. Unlike the straight sides of a raft that seem to be oriented in any direction, the arrowhead’s tip always points away from the swarm center. Like rafts, arrowheads have a small, stable, round second layer, and, opposite their tip, they often open a single, relatively large gap. Gap opening indicates that cell reversals in each course of the arrowhead are independent of each other, like those observed in rafts. These observations raise several questions: Why do second-layer patches of cells form in particular places and why do they grow to some particular size? We suggest that those places are differentiated by the formation of patches of polysaccharide fibrils (23) assembled on the upper surface of the raft and that the tips of M. xanthus pili can bind the fibrils with a tenacity that resists the large pulling force of pilus retraction (6). Arguments for both propositions are presented in detail in the SI Text. Importantly, we propose that the CglB lipoprotein is needed to build proper rafts and to build layered mounds. CglB is part of a cell–cell signaling process that manages the phase of the pacemaker. Our observed synchronization of fifth-layer cells in the wild-type but not the CglB mutant agrees with this proposition.
Fig. 4.
Increase in the diameter of a DK1622 A+S+ swarm (squares ☐) as a function of time. The straight line is a least squares best fit, and its slope gives the steady-state rate of swarm expansion. Increase in the swarm diameter of a CglB mutant lacking the lipoprotein (ASX 1 mutant) (Materials and Methods) is represented by open circles (Ο). Each strain was inoculated by toothpick in the center of a 1% CTT agar plate to preserve the cell organization of the inoculum and incubated at 21 °C. Swarm diameters were measured daily. The DK1622 data were reported in ref. 5. Both plots show the means of triplicate swarm measures.
Discussion
Why does a mutant lacking only the CglB protein have the capacity to expand its swarm but not be able to build either proper rafts or layered mounds like the wild type? The focal adhesions discovered in M. xanthus by Mignot et al. (9, 10) that require CglB for their activity may offer an answer. Each adhesion includes a cluster of 15 known proteins that define a particular subgroup of A-motility proteins (11). Although focal adhesions are proposed to be motors, their only experimentally observed activity is the binding of one essential A-motility protein to other A-motility proteins, as demonstrated by GST affinity chromatography (24). At low cell density, the cell’s cluster of focal adhesion proteins was seen to be localized to a series of fluorescent dots spaced roughly 1/2 µm apart along the cell and spanning the forward half of an actively moving cell. (For scale, M. xanthus cells are about 1/2 µm wide and 8–11 µm long.) The dots, recognized by their fluorescence, seemed to assemble at the cell’s leading pole. Those dots vanished as they proceeded into the trailing half of the cell. Also, as cells moved along, some of the fluorescent clusters seemed to attach themselves to the agar substrate because they remained fixed in position for a while, as the cell itself, including its rigid peptidoglycan layer, appeared to move through the attachment to the agar in a culture dish (9, 10). How the rigid peptidoglycan matrix might move through a focal attachment is unknown, but, in any case, we suggest that the transient attachment of a cell to the agar is, in fact, an artifact of the low cell density used to facilitate photomicroscopy of fluorescent cellular proteins in focal adhesions. At higher cell density, Mauriello et al. (11) noticed the transient attachment of pairs of cells moving in opposite directions and bringing their focal adhesions into alignment. We propose that these pairings facilitate the passage of a signal between two cells that involves CglB and other A-motility proteins of a focal adhesion.
Synchronizing Pacemakers.
The proposed signal might explain several observations: the complete synchronization of the pacemakers in adjacent cells in the mound’s fifth layer; the subsequent reconstruction of the fourth and fifth layers of the mound, with new cells at the same position in the microscopic field after each successive explosive dispersion, that can be seen in Movie S5; and finding the reconstructions that follow each other at regular intervals. The numerical data in Table 2 show that it takes 14 ± 1 min to rebuild the fifth layer after an explosive dispersion. Fig. 3 and Table S1 show that all of the cells from the fifth layer descend to the periphery of the fourth layer and that the increase in area of layer 4 equals the original area of layer 5. Equality is clear in the first and second dispersions but is only approached in the third (Table 2). Movie S5 offers a dynamic view of three sequential losses and reconstructions of the fifth layer for comparing the cell arrangements with each other. Apparently, cells within the mound are able to move rapidly from one layer to the next, either up or down. Movement between layers is fast and depends on type IV pili, as explained in the SI Text. After moving rapidly to an adjacent layer, cells, using their A-motility, can move to some new position within the same layer, improving the alignment between the focal adhesions in neighboring cells. Mounds are constructed from the bottom up, adding layer upon layer; thus, cells in the top (fifth) layer have been resident in and communicating with nearest neighbors longer than any other cells in the mound.
Synchronized reversal of all of the cells in the fifth layer can explain why the first explosive dispersion is observed about an hour (63 min) after the movie’s start: time is required for the proposed synchronizing signal to spread over all fifth-layer cells. The data of Table 2 suggest that a reformed fifth layer is composed of many individual cells from the fourth layer that have used their type IV pili to attach to the empty fibril network that defines the fifth layer and that is sketched in Fig. S1. Although it is simplest to imagine that cells drop down layer by layer, our current technique, which gives a large field of view, tends to limit our observations to the fourth and fifth layers of a five-layered mound.
Together, these observations lead to the hypothesis that the signal transmitted via CglB and the other A-motility proteins associated with a focal adhesion synchronizes the pacemakers in a pair of cells that are in side-by-side contact. Rapid descent from the fifth to the periphery of the fourth layer suggests that all of the cells in the top layer of the mound would also be reversing their S-engines synchronously. And, because the pacemaker reverses both engines at the same instant (5), every cell in the top layer of the mound is also reversing its A-engines synchronously. Inasmuch as a photo is taken every 30 s for the time-lapse movie, the synchrony of both engines is accurate to at least 0.5 min in 8 min, or to 1 part in 16. In accord with that accuracy, it was observed that the A-engines reverse their polarity within a minute or less (14). Consequently, the population of cells in the fifth layer would be expected to start reversing their engines within one 30-s exposure interval and to have completed the reversal of both engines within the next interval, as observed in Fig. 3. Because explosive dispersions were seen at 14 ± 1-min intervals (Table 2), synchronized cells are understood to have the same clock speed and the same clock phase, and both are obtained from genetically identical pacemakers. The 14-min interval between explosive dispersions includes the time it takes for cells moving at average speed to mount, asynchronously, from the fourth to the fifth layer using their S-motility and then, using their A-motility, to arrange themselves compactly in the new fifth layer.
Signaling Between Cells.
We suggest that clusters of focal adhesion proteins are used to connect the sides of a pair of cells that have stopped moving transiently when their focal adhesions have aligned with each other. This possibility is attractive because those adhesions are restricted to the leading halves of each cell in a pair (10). Using their A-motility, those cells should be able to pair the focal adhesions in their leading halves, slowly and accurately. As noted by Mauriello et al. (25) (and see figure 3 in ref. 25), focal adhesion clusters fluorescently labeled with FrzCD-GFP can align with each other for a short time before dissociating. Rafts, whose cells are aligned with their long axes parallel and continuously moving back and forth at random, should have about half their pairs of laterally adjacent cells moving in opposite directions using A-motility. Our findings and those of others (25) suggest that such signaling could start in rafts. Those conditions would offer a simple explanation why CglB mutants can swarm but can’t build rafts. A-motility, which is responsible for cell movement within a raft, could slowly bring a pair of focal adhesions into perfect alignment with each other. Thus formed, the transiently paired connection is proposed to synchronize the pacemakers of the two interacting cells by resetting both cells to the average value of their preconnection phases. After connecting long enough to average their phases, both cells are expected to reverse, automatically moving away from each other and facilitating the formation of new, transient contacts with other cells, as described in the SI Text. Mauriello et al. (25) observed that pairings between the focal adhesions of two cells were frequently followed by reversal of their gliding direction. Reversal of both partners would help the synchronizing signal spread to new cells by separating cells that had already signaled each other. Separation by reversal from the first signaling partner in a raft would ensure that the next partner would be different from the first. New partners become available in a raft by the opening of gaps (Fig. S4), by the creation of a second layer on the raft, and by creation of multilayered mounds. A mound, which is diagrammed in Fig. S1, places many hundreds of cells in positions able to signal each other.
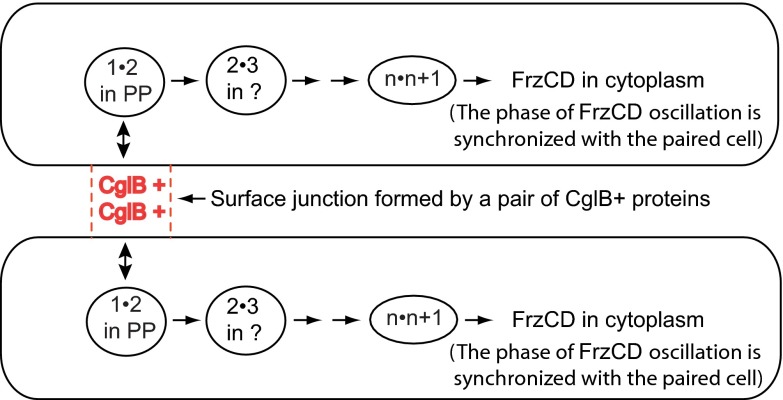
Reversals that follow cell–cell signaling would account for developing some phase correlation between adjacent cells in the same raft, as well as synchronization of all of the cells in the fifth layer of the mound, a process whose steps are laid out in Fig. 5. Every cell in a mound should be available for pairwise signaling, cells in each layer are in contact with one another, and cells in a mound are able to move from one layer to the next. Signaling would be expected to take time to reach all of the cells in the fifth layer, thus accounting for the delay to synchrony in that layer. A delay for spreading the signal for firefly flash synchronization was predicted by Strogatz (26). A-motility proteins that have been reported to be associated with focal adhesions are presented in Table S2. GST affinity chromatography has shown that particular pairs of these proteins are able to bind each other (24). We propose they bind sequentially, to match their particular localization within cells. Focal adhesion binding protein CglB is found in the outer membrane, other focal adhesion binding proteins are found in the periplasm, others are associated with the peptidoglycan sacculus, and still others are found in the cytoplasm where the pacemaker proteins FrzCD, FrzE, and FrzF reside. In the signaling model proposed in Fig. 5, each protein is specific for an upstream and for a downstream binding partner. Binding proteins are also specific for their intracellular compartment(s), as specified in Table S2.
Fig. 5.
Proposed pathway of the signal that synchronizes the pacemakers of a pair of cells. Arrows point toward the next pair of sequential A-motility proteins that bind together. Numbers indicate their position in the sequence of pairwise binding steps (proteins are listed in Table S2). Locations of CglB and FrzCD proteins have been established and are indicated. Designation 1•2 is the first pair of proteins to bind, 2•3 is the second pair, and n•n+1 is the next-to-last pair. The last pair is n+1•FrzCD. Strictly, FrzCD is a methylated regulatory protein that, by naming conventions, isn’t considered an A-motility protein although it does control the reversal frequency. The two cells shown are joined for a short time, long enough to complete the whole binding cascade; they are joined by an interaction between an undetermined number of CglB protein molecules plus some number of associated CglC, CglD, CglE, or CglF protein molecules in the outer membranes of both cells, forming the junctional structure indicated in the diagram. Further details can be found in the Discussion.
When the CglB proteins on the surface of those cells pair exactly with each other, then a molecular bridge, colored red in Fig. 5, would be formed that connects the two cells together. Thus connected, the CglB molecular assembly is proposed to synchronize the pacemakers of the connected cells by resetting both their phases to the average value of their preconnection phases. Assuming that all phases, 0–360°, are equally likely in a swarm, the final average phase would be 180°, a reversal. In other words, the pacemakers of layer 5 cells would lead them to reverse direction of both their A-engines and their S-engines, synchronously as observed (Fig. 3).
It is interesting to note that members of the Wnt and Hedgehog families of proteins regulate the timing and expression levels of morphologic regulators in many animal tissues in what could be similar ways (27, 28). Both families of proteins can polarize individual cells and change their behaviors much like the signal that synchronizes cell movements in myxobacteria. Constructing dynamic multicellular structures using contact signaling and polarity reversals to control cell motility may be an evolved function.
Materials and Methods
Bacteria.
Cultures of M. xanthus, of DK1622, and of a Cgl B deletion mutant, ASX1 (29), were routinely grown on 1% agar CTT [1% casitone, 10 mM Tris⋅HCl (pH 8.0), 8 mM MgSO4, 10 mM KPO4 (pH 7.6)] at 20 °C on plates. Cultures were maintained by transfer to a fresh plate at 3-wk intervals. Swarms were propagated by inoculating an agar plate, with bacteria harvested from the edge of a mature swarm using the tip of a sterilized round toothpick, and incubating at 20 °C (room temperature). Most likely, clusters of aligned cells carried on the toothpick helped nucleate the swarm. Bacteria grew within the stab wound and within a day or two glided up the side of the wound and then onto the horizontal surface of the agar, forming a nearly circular disk.
Time-Lapse Photomicroscopy.
To prepare motion pictures for Table 1, the inoculated swarm plates were examined each morning to assess their stage of development. A section of the swarm edge was selected at which individual cells and clusters were visible under the microscope (Nikon Eclipse E800) fitted with a 20× phase contrast objective at 20 °C (room temperature). Pictures were taken at 30-s intervals for 60–150 min. When possible, the microscope was refocused in the 25-s interval between exposures to compensate for drying of the illuminated agar. Images were collected with a SPOT RT SE Monochrome 6 CCD camera (Diagnostic Instruments) controlled through SPOT software, version 3.5.6. Serial images were saved as QuickTime movies that could be examined frame by frame using QT player v7.4.5. Particular cells or multicellular clusters were chosen from one of the time-lapse movies. Using several tools of NIH Image J (v1.40g), outlines of areas covered with cells were traced and measured in terms of number of pixels. There were 9.18647 image pixels per micrometer, established from the photograph of a ruled grid of 1/400 square millimeters. Tracks were measured with the MtrackJ plugin. The measurement were repeated, averaged, and divided by the number of frames spanned by the movement and the time elapsed per frame (30 s) to determine the cell velocity.
To measure the gap widths, sequential images in the movie of the swarm edge (published as movie S1 in ref. 20) were measured using the Image J line-selection tool. To measure the area of a patch of cells, the polygonal selection tool was used. Both measurements were repeated 10–20 times and averaged, and the SE was computed.
Supplementary Material
Acknowledgments
We thank Dr. Marianne Powell for her discussion and careful editing of the manuscript. The Nikon Eclipse E800 microscope and the SPOT RT SE CCD camera were purchased in 2003 with support from Public Health Service Grant GM23441 from the National Institute of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411925111/-/DCSupplemental.
References
- 1.Reichenbach H. 1966. Myxococcus spp (Myxobacterales): Swarming and fruiting body development (Institut fur den Wissenschaftlichen Film, Göttingen, Germany), film E 778/1965.
- 2.Reichenbach H. 1968. Archangium violaceum (Myxobacteriales): Swarming and fruiting body development (Institut fur den Wissenschaftlichen Film, Göttingen, Germany), film E 777/1965.
- 3.Reichenbach H. 1974. Chondromyces apiculatus (Myxobacteriales): Swarming and morphogenesis (Institut fur den Wissenschaftlichen Film, Göttingen, Germany), film E 779/1965.
- 4.Reichenbach H, Galle HK, Heunert HH. 1975/1976. Stigmatella aurantiaca (Myxobacterales): Swarming and morphogenesis (Institut fur den Wissenschaftlichen Film, Göttingen, Germany), film E2421.
- 5.Kaiser D, Warrick H. Myxococcus xanthus swarms are driven by growth and regulated by a pacemaker. J Bacteriol. 2011;193(21):5898–5904. doi: 10.1128/JB.00168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier B, et al. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci USA. 2002;99(25):16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satyshur KA, et al. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15(3):363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76(11):5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mignot T, Merlie JP, Jr, Zusman DR. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science. 2005;310(5749):855–857. doi: 10.1126/science.1119052. [DOI] [PubMed] [Google Scholar]
- 10.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315(5813):853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mauriello EM, Nan B, Zusman DR. AglZ regulates adventurous (A-) motility in Myxococcus xanthus through its interaction with the cytoplasmic receptor, FrzCD. Mol Microbiol. 2009;72(4):964–977. doi: 10.1111/j.1365-2958.2009.06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nan B, et al. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108(6):2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. How myxobacteria glide. Curr Biol. 2002;12(5):369–377. doi: 10.1016/s0960-9822(02)00716-9. [DOI] [PubMed] [Google Scholar]
- 14.Yu R, Kaiser D. Gliding motility and polarized slime secretion. Mol Microbiol. 2007;63(2):454–467. doi: 10.1111/j.1365-2958.2006.05536.x. [DOI] [PubMed] [Google Scholar]
- 15.Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol Mol Biol Rev. 2009;73(1):155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci USA. 1977;74(7):2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309(5731):125–127. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 18.Wall D, Kaiser D. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc Natl Acad Sci USA. 1998;95(6):3054–3058. doi: 10.1073/pnas.95.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol. 2011;81(2):315–326. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y, Kaiser AD, Jiang Y, Alber MS. Periodic reversal of direction allows Myxobacteria to swarm. Proc Natl Acad Sci USA. 2009;106(4):1222–1227. doi: 10.1073/pnas.0811662106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5(11):862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 22.Winfree AT. Biological rhythms and the behavior of populations of coupled oscillators. J Theor Biol. 1967;16(1):15–42. doi: 10.1016/0022-5193(67)90051-3. [DOI] [PubMed] [Google Scholar]
- 23.Behmlander RM, Dworkin M. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J Bacteriol. 1994;176(20):6295–6303. doi: 10.1128/jb.176.20.6295-6303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nan B, Mauriello EM, Sun I-H, Wong A, Zusman DR. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol. 2010;76(6):1539–1554. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mauriello EM, Astling DP, Sliusarenko O, Zusman DR. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc Natl Acad Sci USA. 2009;106(12):4852–4857. doi: 10.1073/pnas.0810583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strogatz SH. Sync: The Emerging Science of Spontaneous Order. New York: Hyperion; 2003. [Google Scholar]
- 27.Nusse R. Wnt signaling. Cold Spring Harb Perspect Biol. 2012;4(5, a011163):1–3. doi: 10.1101/cshperspect.a011163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12(6):393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez AM, Spormann AM. Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J Bacteriol. 1999;181(14):4381–4390. doi: 10.1128/jb.181.14.4381-4390.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.