Abstract
• Background and Aims Little information is available on DNA C-values for the New Zealand flora. Nearly 85 % of the named species of the native vascular flora are endemic, including 157 species of Poaceae, the second most species-rich plant family in New Zealand. Few C-values have been published for New Zealand native grasses, and chromosome numbers have previously been reported for fewer than half of the species. The aim of this research was to determine C-values and chromosome numbers for most of the endemic and indigenous Poaceae from New Zealand.
• Scope To analyse DNA C-values from 155 species and chromosome numbers from 55 species of the endemic and indigenous grass flora of New Zealand.
• Key Results The new C-values increase significantly the number of such measurements for Poaceae worldwide. New chromosome numbers were determined from 55 species. Variation in C-value and percentage polyploidy were analysed in relation to plant distribution. No clear relationship could be demonstrated between these variables.
• Conclusions A wide range of C-values was found in the New Zealand endemic and indigenous grasses. This variation can be related to the phylogenetic position of the genera, plants in the BOP (Bambusoideae, Oryzoideae, Pooideae) clade in general having higher C-values than those in the PACC (Panicoideae, Arundinoideae, Chloridoideae + Centothecoideae) clade. Within genera, polyploids typically have smaller genome sizes (C-value divided by ploidy level) than diploids and there is commonly a progressive decrease with increasing ploidy level. The high frequency of polyploidy in the New Zealand grasses was confirmed by our additional counts, with only approximately 10 % being diploid. No clear relationship between C-value, polyploidy and rarity was evident.
Keywords: Chromosome number, C-value, distribution, New Zealand, Poaceae, polyploidy, rarity, taxonomy
INTRODUCTION
Variation in the amount of nuclear DNA of the entire chromosome complement or holoploid genome size, here called C-value following Greilhuber et al. (2005), together with changes in chromosome number and structure, has been implicated in the diversification and speciation of flowering plants (Levin, 2002). This variation is clearly an important component of adaptation, as variation in C-value affects key parameters of plant growth such as the duration of the cell size, cell cycle and life form (Bennett, 1972, 1987; Gregory, 2005). Correlations have also been made between C-value and plant distribution, with the common observation that plants of more tropical latitudes appear to have lower values than those of temperate latitudes (Bennett, 1976; Levin and Funderburg, 1979). However, other studies, e.g. Grime and Mowforth (1982), report the reverse correlation between C-value and latitude. In a large-scale study on the Californian flora, Knight and Ackerly (2002) demonstrated that species with low C-values predominate in all environments and that in extreme environments species with the highest C-values are poorly represented. There has also recently been the suggestion that high C-values may be maladaptive and lead to plant extinction (Vinogradov, 2003). There also appears to be a clear phylogenetic pattern to C-value variation in the Poaceae, with higher values being most common among the members of the Bambusoideae, Oryzoideae, Pooideae (BOP) clade (Kellogg and Bennetzen, 2004).
Polyploidy is clearly one possible contributor to C-value variation, but the relationship between C-value and ploidy is far from clear-cut (Leitch and Bennett, 2004). In their ‘all angiosperms’ and monocot samples Leitch and Bennett (2004) have shown that the mean amount of 1C DNA does not increase in direct proportion to ploidy and that mean genome size (C-value divided by ploidy level) shows a clear decrease. Nevertheless, polyploidy affects many other genetic and phenotypic characters, and in New Zealand, where 63 % of angiosperms are reported to be polyploid (Hair, 1966), it appears to have played an important role in the evolution of the flora. Polyploids also frequently, though not always, have different distributions to their diploid progenitors, and polyploids are often over-represented among colonizer species (Stebbins, 1971; Thompson and Lumaret, 1992; Levin, 2002; Brochmann et al., 2004).
One of the key recommendations that arose from the first Angiosperm Genome Size workshop and conference held at the Royal Botanic Gardens, Kew, in September, 1997 [Annals of Botany 82 (Supplement A)] was the need to obtain an improved representation of the world's flora in the DNA C-value database (http://www.rbgkew.org.uk/cval/homepage.html). One under-represented area is Australasia. There have been few studies of Australasian plants, with only 19 out of 465 first authors of papers listed in the database coming from the region (Bennett and Leitch, 2005). Only three studies have been published that include New Zealand native angiosperms (Murray et al., 1992, 2003; Hanson et al., 2003). Thus, it is timely to produce a survey of a large family, the Poaceae, from New Zealand.
Grasses are a significant component of the New Zealand endemic and indigenous flora. Poaceae are the second largest family in terms of species and in the recent flora (Edgar and Connor, 2000) 157 species are described as endemic and 31 as indigenous. The New Zealand flora is significant for its high level of species endemicity, in that approximately 85 % of the approximately 2300 vascular plant species are endemic (Cockayne, 1967; Wardle, 1991; de Lange and Norton, 1997; Wilton and Breitwieser, 2000); in the grasses 87·6 % are considered to be endemic (Wilton and Breitwieser, 2000). Recent studies have shown that many of the species-rich genera are the results of recent speciation following long-distance dispersal, mainly from Australia, Malesia and South America (Wardle, 1978; Pole, 1994, 2001; McGlone et al., 2001; Winkworth et al., 2002).
Grasses occupy a wide variety of habitats in a landmass composed of islands of varying sizes that span almost 25° of latitude and rise to over 3000 m. These plants also show a variety of distribution patterns from species that are widespread to those that are restricted to a small local area. Two recent publications have provided an excellent framework for further investigation of the New Zealand grasses, a new grass flora (Edgar and Connor, 2000) and an updated catalogue of threatened indigenous New Zealand plants (de Lange et al., 2004). A recent review (Murray, 2005) has suggested that intraspecific C-value variation can be an indicator of taxonomic heterogeneity. With a number of grass genera for which species delimitation is unclear (Edgar and Connor, 2000), C-value data could provide useful indicators.
In this paper we report on the C-values of 155 species (plus a further six subspecific taxa) and new chromosome numbers for 55 taxa of grasses from the New Zealand endemic and indigenous flora.
MATERIALS AND METHODS
Plant material
The plant material used in this study is listed in Table 1. The majority of species have been studied from single individuals as many of them are rare, confined to restricted areas or found in remote parts of the New Zealand botanical region. Figure 1 shows the botanical provincial boundaries of the North and South Islands of New Zealand and the principal offshore islands mentioned in Table 1 so that the origin of the plant samples can be located. All plants were collected from natural populations and grown either in the experimental garden or in a glasshouse at the University of Auckland. The plants studied are listed in Table 1. The recent grass flora (Edgar and Connor, 2000) includes 157 endemic and 31 indigenous species. Here we include a further two species, Paspalum orbiculare and Bromus arenarius, as indigenous although they were treated as naturalized by Edgar and Connor (2000). Voucher specimens of all of the plants used in this study have been deposited either in the herbarium of Auckland Museum (AK), the Allan Herbarium, Landcare Research (CHR) or the Otago University Herbarium (OTA). Chromosome numbers were determined either from root tip meristems or from pollen mother cells using standard preparation techniques. In a small number of species we did not determine the chromosome numbers ourselves but for completeness when determining various statistics additional chromosome numbers were obtained from Dawson (2000) and de Lange and Murray (2002).
Table 1.
Chromosome number (numbers in bold taken from Dawson, 2000), ploidy level, DNA C-value (pg per 2C nucleus), place of origin of the plants and location of voucher specimens of plants used in the present study
| Species |
n |
2n |
Ploidy |
2C |
Place of origin |
Voucher |
|---|---|---|---|---|---|---|
| Achnatherum petriei (Buchanan) S.W.L.Jacobs & J.Everett | 42 | 6x | 1·77 | South I., Otago, Awahokomo | AK 286416 | |
| Agrostis dyeri Petrie | 42 | 6x | 10·81 | North I., Wellington, Tararua Ranges | AK 282090 | |
| A. imbecilla Zotov | 42 | 16x | 10·78 | South I., Canterbury, Old Man Range | AK 286444 | |
| A. magellanica Lam. | 84 | 12x | 21·23 | Auckland Is., Enderby I. | AK 281998 | |
| 84 | 12x | 21·77 | Campbell I. | AK 281989 | ||
| A. muelleriana Vickery | 42 | 6x | 11·26 | South I., Canterbury, Crimea Range | AK 281984 | |
| 42 | 6x | – | South I., Southland, Garvie Mountains | AK 282063 | ||
| A. muscosa Kirk | 42 | 6x | 10·58 | South I., Otago, Ohau Downs | AK 286818 | |
| 42 | 6x | – | North I., Taranaki, Egmont National Park | AK 282074 | ||
| 42 | 6x | – | North I., Taranaki, Puketapu Road | AK 282074 | ||
| 42 | 6x | – | South I., Canterbury, Lake Tekapo | AK 286744 | ||
| A. pallescens Cheeseman | 42 | 6x | 10·88 | South I., Southland, Te Anau Downs | AK 286499 | |
| A. personata Edgar | 42 | 6x | 10·64 | South I., Canterbury, Crimea Range | AK 281042 | |
| A. petriei Hack. | 42 | 6x | 10·85 | South I., Otago, Glenmore Station | AK 286748 | |
| Amphibromus fluitans Kirk | 42 | 6x | 7·97 | North I., Wellington, Lake Wairarapa | AK 282157 | |
| Anemanthele lessoniana (Steud.) Veldkamp | 40–44 | 4x | 1·89 | North I., Wellington, Haurangi Range | AK 281149 | |
| Australopyrum calcis Connor & Molloy subsp. calcis | 14 | 2x | 11·27 | South I., Marlborough, Leatham Valley | AK 296501 | |
| A. calcis subsp. optatum | 14 | 2x | 11·60 | South I., Canterbury, Castle Hill | AK 286443 | |
| Connor & Molloy | 14 | 2x | 11·57 | South I., Canterbury, Tengawai | AK 281153 | |
| Austrofestuca littoralis (Labill.) E.B.Alexeev. | 28 | 4x | 7·42 | Great Barrier I., Kaitoke Beach | AK 281576 | |
| Austrostipa stipoides (Hook.f.) S.W.L. Jacobs & J. Everett | 44 | 4x | 3·15 | North I., North Auckland, Ambury Park | AK 280019 | |
| Bromus arenarius Labill. | 28 | 4x | 16·45 | North I., North Auckland, Ponui I. | AK 283726 | |
| Cenchrus caliculatus Cav. | 102 | 6x | 11·12 | Kermadec Is., Raoul I. | AK 282949 | |
| Chionochloa antarctica (Hook.f.) Zotov | 42 | 6x | 5·31 | Campbell I. | AK 281160 | |
| C. australis (Buchanan) Zotov | 42 | 6x | 4·43 | South I., Canterbury, Lake Tennyson | AK 281047 | |
| C. bromoides (Hook.f.) Zotov | 42 | 6x | 6·09 | North I., North Auckland, Mokohinau Is. | AK 283728 | |
| C. cheesemanii (Hack.) Zotov | 42 | 6x | 5·47 | North I., Wellington, Tararua Ranges | AK 281546 | |
| 42 | 6x | – | South I., Marlborough, D'Urville I. | AK 282156 | ||
| C. conspicua subsp. cunninghamii (Hook. f.) Zotov | 42 | 6x | 6·33 | North I., South Auckland, Karangahake Gorge | AK 286731 | |
| C. crassiuscula (Kirk) Zotov subsp. crassiuscula | 42 | 6x | 5·00 | Stewart I., Table Hill | OTA 57968 | |
| C. crassiuscula subsp. directa Connor | 42 | 6x | – | South I., Fiordland, Fiordland National Park | OTA 57599 | |
| C. crassiuscula subsp. torta Connor | 42 | 6x | – | South I., Fiordland, Takahe Valley | OTA 57597 | |
| C. defracta Connor | 42 | 6x | – | South I., Nelson, Mt Dun, Windy Point | OTA 57937 | |
| C. flavescens subsp. brevis Connor | 42 | 6x | 5·17 | South I., Nelson, Mt Maling | AK 281046 | |
| C. flavescens subsp. hirta Connor | 42 | 6x | – | South I., Westland, Mt Ryall | OTA 57964 | |
| C. flavescens subsp. lupeola Connor | 42 | 6x | 5·32 | South I., Nelson, Mt Owen | OTA 57964 | |
| C. flavicans Zotov f. flavicans | 42 | 6x | 7·57 | North I., South Auckland, Hahei | AK 286419 | |
| C. flavicans f. temata Connor | 42 | 6x | 7·77 | North I., Hawke's Bay, Te Mata Peak | AK 286444 | |
| C. juncea Zotov | 42 | 6x | 5·38 | South I., Nelson, Mt Augustus | AK 286772 | |
| C. lanea Connor | 42 | 6x | 5·06 | Stewart I., Table Hill | AK 286646 | |
| C. macra Zotov | 42 | 6x | 5·01 | South I., Otago, Mt St Bathans | AK 286726 | |
| 42 | 6x | – | South I., Otago, Cromwell | AK 286729 | ||
| C. nivifera Connor & K.M.Lloyd | 42 | 6x | – | South I., Southland, Fiordland National Park, Mt Burns | No voucher | |
| C. pallens Zotov subsp. pallens | 42 | 6x | 5·13 | North I., Wellington, Tararua Ranges | AK 281982 | |
| C. pallens subsp. pilosa Connor | 42 | 6x | – | South I., Marlborough, Mt Fyffe | OTA 57595 | |
| C. rigida (Raoul) Zotov subsp. rigida | 42 | 6x | 5·07 | South I., Otago, Lake Mahinerangi | AK 286730 | |
| C. rigida subsp. amara Connor | 42 | 6x | – | South I., Southland, Takahe Valley | OTA 57963 | |
| C. rubra Zotov subsp. rubra var. rubra | 42 | 6x | 5·14 | North I., Wellington, Rangipo Desert | AK 286452 | |
| C. rubra subsp. rubra var. inermis Connor | 42 | 6x | 5·36 | North I., Taranaki, Egmont National Park | AK 286647 | |
| C. rubra subsp. cuprea Connor | 42 | 6x | – | South I., Canterbury, Lake Tennyson | AK 286451 | |
| C. rubra subsp. occulta Connor | 42 | 6x | – | South I., Westland, Paparoa Range | OTA 57962 | |
| 42 | 6x | – | South I., Canterbury, Lake Tennyson | AK 286462 | ||
| C. spiralis Zotov | 42 | 6x | 5·11 | South I., Southland, Mt Luxmore | AK 286781 | |
| C. vireta Connor | 42 | 6x | 4·97 | South I., Westland, Haast Pass | OTA 57585 | |
| Cortaderia fulvida (Buchanan) Zotov | 90 | 10x | 8·30 | North I., South Auckland, Lake Rotorua | AK 281166 | |
| 90 | 10x | 8·36 | North I., North Auckland, Kerikeri River | AK 208542 | ||
| C. richardii (Endl.) Zotov | 90 | 10x | 7·85 | South I., Otago, Awahokomo Creek | AK 256111 | |
| C. splendens Connor | 90 | 10x | 7·89 | North I., North Auckland, Whatipu | AK 283784 | |
| 90 | 10x | 7·82 | North I., South Auckland, Kawhia | AK 281157 | ||
| 90 | 10x | 7·71 | North I., North Auckland, Surville Cliffs | AK 281158 | ||
| 90 | 10x | 8·49 | North I., North Auckland, Kawau I. | AK 287115 | ||
| 90 | 10x | 8·27 | North I., South Auckland, Mercury Is. | AK 287117 | ||
| C. toetoe Zotov | 90 | 10x | 7·94 | North I., South Auckland, Rotorua | AK 281165 | |
| C. turbaria Connor | 90 | 10x | 9·58 | Chatham Is., Rekohu I. | AK 251864 | |
| Deschampsia cespitosa (L.) P.Beauv. | 26 | 2x | 10·43 | Chatham Is., Rekohu I. | AK 282160 | |
| D. chapmanii Petrie | 26 | 2x | 11·05 | South I., Southland, Garvie Mountains | AK 282066 | |
| 26 | 2x | – | Campbell I. | AK 281995 | ||
| D. gracillima Kirk | 26 | 2x | – | Campbell I. | AK 281991 | |
| D. tenella Petrie | 26 | 2x | 10·07 | South I., Southland, Garvie Mountains | AK 282080 | |
| Deyeuxia aucklandica (Hook.f.) Zotov | 56 | 8x | – | North I., Hawke's Bay, Kaweka Range | AK 253147 | |
| 56 | 8x | 14·06 | South I., Southland, Garvie Mountains | AK 282067 | ||
| 56 | 8x | 13·98 | South I., Canterbury, Lake Tennyson | AK 288429 | ||
| D. avenoides (Hook.f.) Buchanan | 70 | 10x | – | North I., North Auckland, Auckland City | AK 281152 | |
| 70 | 10x | 13·53 | South I., Canterbury, Lake Tennyson | AK 255953 | ||
| 70 | 10x | 15·52 | South I., Canterbury, Maitland Forest | AK 282062 | ||
| D. lacustris Edgar & Connor | 28 | 4x | 16·13 | South I., Canterbury, Lake Tennyson | AK 281049 | |
| D. quadriseta (Labill.) Benth. | 56 | 8x | 15·36 | North I., North Auckland, Waikumete Cemetery | AK 250793 | |
| D. youngii (Hook.f.) Buchanan | 28 | 4x | 9·87 | South I., Canterbury, Maitland River | CHR 562376 | |
| D. aff. quadriseta | 56 | 8x | 13·20 | North I., Wellington, Waimarino Plains | AK 286819 | |
| Dichelachne crinita (L.f.) Hook.f. | 70 | 10x | 16·36 | Chatham Is., Rekohu I. | AK 282152 | |
| 70 | 10x | – | South I., Otago, Alexandra | CHR 569774 | ||
| D. inaequiglumis (Hack.) Edgar & Connor | 70 | 10x | 16·40 | North I., North Auckland, Waikumete Cemetery | AK 250805 | |
| D. lautumia Edgar & Connor | 70 | 10x | 17·67 | South I., Marlborough, Flaxbourne River | AK 286907 | |
| D. micrantha (Cav.) Domin | 70 | 10x | 16·85 | North I., Gisborne, Torere Point | AK 286503 | |
| Echinopogon ovatus (G.Forst.) P.Beauv. | 21II | 6x | 10·85 | North I., North Auckland, Hunua Range | AK 286820 | |
| Elymus apricus Á.Löve & Connor | 42 | 6x | 28·20 | South I., Otago, Matangi | AK 286417 | |
| E. enysii (Kirk) Á.Löve & Connor | 28 | 4x | 20·54 | South I., Canterbury, Porters Pass | AK 286502 | |
| E. falcis Connor | 42 | 6x | 27·57 | South I., Canterbury, Waitaki Valley | AK 286908 | |
| E. multiflorus (Hook.f.) Á.Löve & Connor | 42 | 6x | 25·16 | North I., North Auckland, Anawhata | AK 282082 | |
| E. sacandros Connor | 42 | 6x | 27·74 | South I., Marlborough, Flaxbourne River | AK 286643 | |
| E. solandri (Steud.) Connor | 42 | 6x | 28·56 | North I., Wellington, Sinclair Head | AK 281159 | |
| 42 | 6x | 28·40 | South I., Marlborough, Stephens I. | AK 282151 | ||
| 42 | 6x | 26·59 | South I., Marlborough, Upper Wairau River | AK 284756 | ||
| E. tenuis (Buchanan) Á.Löve & Connor | 56 | 8x | 32·09 | South I., Canterbury, Sawdon Run | AK 282069 | |
| Festuca actae Connor | 42 | 6x | 16·52 | South I., Canterbury, Banks Peninsula | AK 281513 | |
| F. coxii (Petrie) Hack. | 56 | 8x | 21·50 | Chatham Is., Rekohu I. | CHR 569775 | |
| F. deflexa Connor | 42 | 6x | 16·02 | South I., Nelson, Mt Owen | OTA 57588 | |
| F. luciarum Connor | 56 | 8x | 19·85 | North I., Hawke's Bay, Mangaharuru Range | OTA 57621 | |
| F. matthewsii (Hack.) Cheeseman subsp. matthewsii | 42 | 6x | 16·26 | South I., Southland, Fiordland National Park | OTA 57938 | |
| F. matthewsii subsp. aquilonia Connor | 42 | 6x | – | South I., Canterbury, Mt Fyffe | OTA 57936 | |
| F. matthewsii subsp. pisamontis Connor | 42 | 6x | – | South I., Otago, Pisa Range | OTA 57945 | |
| F. novae–zelandiae (Hack.) Cockayne | 42 | 6x | 16·83 | South I., Canterbury, Cass | AK 287121 | |
| 42 | 6x | – | South I., Otago, Awahokomo | AK 252541 | ||
| F. ultramafica Connor | 56 | 8x | 20·62 | South I., Nelson, Mt Dun | OTA 57629 | |
| Hierochloe brunonis Hook.f. | 84 | 12x | 27·81 | Campbell I. | AK 281993 | |
| H. equiseta Zotov | 42 | 6x | 18·10 | South I., Canterbury, Porters Pass | CH 562182 | |
| H. fusca Zotov | 84 | 12x | 29·68 | North I., Wellington, Kapiti I. | AK 286448 | |
| 84 | 12x | 27·55 | Chatham Is., Rekohu I. | CHR 562185 | ||
| H. novae–zelandiae Gand. | 28 | 4x | 12·54 | South I., Canterbury, Tennyson Tarns | AK 287053 | |
| H. redolens (Vahl) Roem. & Schult. | 84 | 12x | 29·97 | South I., Canterbury, Rakaia River | CH 562181 | |
| Imperata cheesemanii Hack. | 20 | 2x | 1·45 | Kermadec Is., Raoul I. | AK 253146 | |
| Isachne globosa (Thunb.) Kuntze | 30II | 6x | 3·64 | North I., North Auckland, Western Springs | AK 256108 | |
| Koeleria cheesemanii (Hack.) Petrie | 28 | 4x | 9·95 | South I., Canterbury, Crimea Range | AK 281043 | |
| K. novozelandica Domin sens. str. | 28 | 4x | – | South I., Canterbury, Balmoral | CHR549886 | |
| 28 | 4x | 9·82 | South I., Canterbury, Porters Pass | CHR 569776 | ||
| K. riguorum Edgar & Gibb | 28 | 4x | – | South I., Nelson, Douglas Range | AK 286472 | |
| K. aff. cheesemanii | 28 | 4x | 13·23 | South I., Canterbury, Porters Pass | CHR 569776 | |
| K. aff. novozelandica | 28 | 4x | 10·41 | South I., Otago, Awahokomo | AK 282149 | |
| Lachnagrostis ammobia Edgar | 98 | 14x | 25·48 | Stewart I., Mason Bay | AK 247127 | |
| L. billardierei (R.Br.) Trin. | 56 | 8x | – | Chatham Is., Rekohu I. | AK 282153 | |
| 56 | 8x | – | Chatham Is., Rekohu I. | AK 229942 | ||
| 56 | 8x | – | North I., North Auckland, Kaitarakihi | AK 250913 | ||
| 56 | 8x | – | North I., North Auckland, Taitomo I. | AK 288306 | ||
| 56 | 8x | 18·25 | North I., North Auckland, Kauri Point | AK 288137 | ||
| L. elata Edgar | 98 | 14x | 26·81 | North I., South Auckland, Pureora | AK 283507 | |
| 98 | 14x | – | North I., Wellington, Tongariro National Park | AK 288307 | ||
| L. filiformis (G.Forst.) Trin. | 56 | 8x | 12·57 | North I., North Auckland, Auckland City | AK 281150 | |
| L. leptostachys (Hook.f.) Zotov | 84 | 12x | 25·21 | Auckland Is., Enderby I. | AK 281999 | |
| 84 | 12x | 24·43 | Campbell I. | AK 281988 | ||
| L. littoralis (Hack.) Edgar subsp. littoralis | 56 | 8x | 13·51 | Great Barrier I., Medlands Beach | AK 282162 | |
| 56 | 8x | 15·32 | North I., North Auckland, Cornwallis | AK 288077 | ||
| 56 | 8x | 13·47 | South I., Westland, Punakaiki | AK 253019 | ||
| L. littoralis subsp. salaria Edgar | 56 | 8x | 16·72 | South I., Canterbury, Christchurch | AK 282150 | |
| 56 | 8x | 16·49 | South I., Canterbury, Brooklands | AK 282144 | ||
| L. lyallii (Hook.f.) Zotov | 98 | 14x | 17·09 | South I., Canterbury, Lake Tekapo | AK 288430 | |
| 98 | 14x | 23·47 | South I., Canterbury, Lake Tennyson | AK 286721 | ||
| 98 | 14x | 22·78 | North I., Wellington, Tongariro National Park | AK 252979 | ||
| L. pilosa (Buchanan) Edgar subsp. pilosa | 49II | 98 | 14x | 23·78 | South I., Marlborough, Isolation Creek | AK 256032 |
| 98 | 14x | 24·66 | Chatham Is., Rekohu I. | CHR 562182 | ||
| L. uda Edgar | 98 | 14x | 25·50 | South I., Southland, Garvie Mountains | AK 282068 | |
| Microlaena avenacea (Raoul) Hook.f. | 48 | 4x | 3·40 | North I., North Auckland, Waitakere Ranges | AK 282158 | |
| M. carsei Cheeseman | 48 | 4x | 3·25 | North I., North Auckland, Kerikeri River | AK 281154 | |
| M. polynoda (Hook.f.) Hook.f. | 48 | 4x | 2·37 | North I., North Auckland, Waitakere Ranges | AK 286418 | |
| M. stipoides (Labill.) R.Br. | 48 | 4x | 1·80 | North I., North Auckland, Auckland City | AK 286645 | |
| 48 | 4x | 1·80 | Chatham Is., Rekohu I., Otoi Creek | AK 286445 | ||
| 48 | 4x | 1·83 | North I., Taranaki, New Plymouth | AK 288314 | ||
| Oplismenus hirtellus subsp. | 54 | 6x | 5·21 | North I., North Auckland, Waitakere Ranges | AK 286447 | |
| imbecillis (R.Br.) U.Scholz | 54 | 6x | 5·27 | Kermadec Is., Raoul I. | AK 286724 | |
| Paspalum orbiculare G.Forst | 63 | 6x | 3·13 | North I., North Auckland, Cornwallis Park | AK 252543 | |
| Poa acicularifolia Buchanan subsp. acicularifolia | 28 | 4x | 5·35 | South I., Canterbury, Castle Hill | AK 286743 | |
| Poa acicularifolia subsp. ophitalis Edgar | 28 | 4x | 6·15 | South I., Nelson, Mt Dun | AK 286644 | |
| P. anceps G.Forst subsp. anceps | 28 | 4x | 6·00 | North I., North Auckland, Kaitarakihi Bay | AK 289038 | |
| 28 | 4x | 6·01 | North I., North Auckland, Kauri Point | AK 288074 | ||
| P. anceps subsp. polyphylla (Hack.) Edgar | 28 | 4x | 5·45 | Kermadec Is., Raoul I. | AK 282943 | |
| P. antipoda Petrie | 28 | 4x | – | Antipodes Is. | AK 286714 | |
| P. astonii Petrie | 28 | 4x | 5·98 | South I., Otago, Dunedin | AK 281038 | |
| P. breviglumis Hook.f. | 28 | 4x | 4·24 | North I., Taranaki, Mt Taranaki | AK 288317 | |
| 28 | 4x | 4·38 | South I., Canterbury, Pineleugh | CHR 569777 | ||
| 28 | 4x | 4·55 | North I., Wellington, Tararua Ranges | AK 281888 | ||
| P. buchananii Zotov | 28 | 4x | 5·66 | South I., Canterbury, Crimea Range | AK 281045 | |
| P. chathamica Petrie | 112 | 16x | 21·30 | Chatham Is., Rekohu I. | CHR 562183 | |
| 112 | 16x | 20·96 | Chatham Is., Rekohu I. | AK 286905 | ||
| P. cita Edgar | 84 | 12x | 14·43 | North I., Wellington, Tararua Ranges | AK 282070 | |
| 84 | 12x | 14·99 | South I., Nelson, Charleston | AK 287016 | ||
| P. colensoi Hook.f. | 28 | 4x | 5·61 | North I., Wellington, Tongariro National Park | AK 253027 | |
| 28 | 4x | 5·26 | North I., Taranaki, Egmont National Park | AK 282076 | ||
| 28 | 4x | 5·40 | South I., Canterbury, Lake Tennyson | AK 286711 | ||
| 28 | 4x | 5·16 | South I., Otago, Rastus Burn | CHR 569779 | ||
| 28 | 4x | 5·49 | South I., Otago, Alexandra | AK 256110 | ||
| 28 | 4x | 5·32 | South I., Canterbury, Old Man Range | AK 256156 | ||
| 28 | 4x | 5·47 | South I., Otago, Awahokomo Bluffs | CHR 549885 | ||
| 28 | 4x | 5·22 | South I., Otago, Trotters Gorge | CHR 569778 | ||
| 28 | 4x | 5·52 | South I., Otago, Rastus Burn | AK 286708 | ||
| 28 | 4x | 5·65 | South I., Otago, Blue Creek | AK 286775 | ||
| P. dipsacea Petrie | 28 | 4x | 5·59 | South I., Marlborough, Bert's Creek | AK 285251 | |
| P. foliosa (Hook.f.) Hook.f. | 28 | 4x | 5·99 | Auckland Is., Enderby I. | AK 286739 | |
| P. imbecilla Spreng. | 28 | 4x | 4·26 | South I., Canterbury, Christchurch, Hagley Park | AK 286463 | |
| P. incrassata Petrie | 28 | 4x | 5·60 | South I., Otago, Rastus Burn | AK 286790 | |
| P. intrusa Edgar | 28 | 4x | 6·01 | South I., Southland, Routeburn | AK 286789 | |
| P. kirkii Buchanan | 28 | 4x | 4·89 | South I., Canterbury, Lake Tennyson | AK 282065 | |
| P. lindsayi Hook.f. | 28 | 4x | 4·56 | South I., Canterbury, Tekapo Tarns | AK 282072 | |
| 28 | 4x | 4·51 | South I., Canterbury, Spider Lakes | AK 286497 | ||
| P. litorosa Cheeseman | 263–266 | 38x | 32·24 | Auckland Is., Enderby I. | AK 281151 | |
| 263–266 | 38x | 32·56 | Campbell I. | AK 281987 | ||
| P. maniototo Petrie | 28 | 4x | 5·49 | South I., Otago, Awahokomo Bluffs | AK 259138 | |
| P. matthewsii Petrie | 28 | 4x | 5·49 | South I., Canterbury, Castle Hill | AK 286717 | |
| P. novae–zelandiae Hack. | 28 | 4x | 6·32 | South I., Canterbury, Crimea Range | AK 282179 | |
| 28 | 4x | 6·24 | South I., Otago, Richardson Mountains | AK 281040 | ||
| P. pusilla Berggr. | 28 | 4x | 5·50 | North I., Taranaki, Egmont National Park | AK 286498 | |
| P. ramosissima Hook.f. | 28 | 4x | 5·69 | Auckland Is., Enderby I. | AK 282001 | |
| P. schistacea Edgar & Connor | 28 | 4x | 11·02 | South I., Otago, Hector Mountains, Two-Mile Valley | AK 287133 | |
| P. spania Edgar & Molloy | 28 | 4x | 4·81 | South I., Otago, Awahokomo | AK 282077 | |
| P. sublimis Edgar | 28 | 4x | 5·02 | South I., Canterbury, Arthur's Pass | AK 288308 | |
| P. sudicola Edgar | 28 | 4x | 6·49 | South I., Nelson, Pike Peak | AK 286397 | |
| 28 | 4x | – | South I., Nelson, Pike Peak | AK 282001 | ||
| P. xenica Edgar & Connor | 28 | 4x | 6·67 | South I., Nelson, South Branch Riwaka River | AK 286909 | |
| P. aff. cita(a) | 112 | 16x | 17·44 | South I., Marlborough, Stephens I. | AK 286773 | |
| P. aff. cita(b) | 112 | 16x | 17·40 | South I., Nelson, Golden Bay | AK 286771 | |
| P. aff. colensoi | 28 | 4x | 6·05 | North I., South Auckland, Te Moehau Range | AK 286641 | |
| Puccinellia stricta (Hook.f.) Blom | 14 | 2x | 3·55 | South I., Canterbury, Christchurch | AK 282179 | |
| P. walkeri subsp. chathamica (Cheeseman) Edgar | 42 | 6x | 9·61 | Chatham Is., Rekohu I. | AK 282171 | |
| 42 | 6x | – | Chatham Is., Rekohu I. | AK 287133 | ||
| 42 | 6x | – | Auckland Is., Enderby I. | AK 282000 | ||
| Pyrrhanthera exigua (Kirk) Zotov | approx.156 | 26x | 21·51 | South I., Canterbury, Sawdon Run | AK 253685 | |
| Rytidosperma biannulare (Zotov) Connor & Edgar | 48 | 4x | 8·06 | North I., North Auckland, Waikumete Cemetery | AK 255954 | |
| R. buchananii (Hook.f.) Connor & Edgar | 48 | 4x | 7·07 | South I., Canterbury, Porters Pass | AK 285603 | |
| R. clavatum (Zotov) Connor & Edgar | 24 | 2x | 3·32 | South I., Otago, Ranfurly | AK 256106 | |
| 24 | 2x | – | South I., Canterbury, Waimakariri River | AK 255952 | ||
| R. corinum Connor & Edgar | 48 | 4x | 7·19 | South I., Canterbury, Hakataramea Pass | AK 286053 | |
| R. gracile (Hook.f.) Connor & Edgar | 24 | 2x | 2·48 | South I., Otago, Old Man Range | AK 256105 | |
| R. horrens Connor & Molloy | 24 | 2x | 3·66 | South I., Canterbury, Maitland River | AK 286640 | |
| R. maculatum (Zotov) Connor & Edgar | 48 | 4x | 6·04 | South I., Canterbury, Waimakariri River | AK 256107 | |
| R. petrosum Connor & Edgar | 48 | 4x | 8·26 | North I., Wellington, Cape Palliser | AK 250800 | |
| R. pulchrum (Zotov) Connor & Edgar | 24 | 2x | – | North I., Wellington, Ruahine Range, Toka | AK 286446 | |
| R. pumilum (Kirk) Connor & Edgar | 24 | 2x | 3·24 | South I., Canterbury, Porters Pass | CHR 562374 | |
| R. setifolium (Hook.f.) Connor & Edgar | 24 | 2x | 4·96 | North I., South Auckland, Te Moehau Range | AK 286450 | |
| 24 | 2x | 4·54 | North I., Wellington, Tongariro National Park | AK 253000 | ||
| 24 | 2x | 4·77 | North I., South Auckland, Mt Pirongia | AK 281156 | ||
| R. telmaticum Connor & Molloy | 24 | 2x | 3·43 | South I., Canterbury, Lake Tekapo | CHR 562184 | |
| 24 | 2x | 2·83 | South I., Canterbury, Hakatere | AK 286487 | ||
| R. thomsonii (Buchanan) Connor & Edgar | 48 | 4x | 6·27 | South I., Canterbury, Tekapo Tarns | AK 286745 | |
| D* | ? | 6·63 | South I., Otago, Lake Hawea | AK 286747 | ||
| 24 | 2x | 3·51 | South I., Canterbury, Glenmore | AK 286746 | ||
| R. unarede (Raoul) Connor & Edgar | 48 | 4x | – | North I., Gisborne, Hicks Bay | AK 256109 | |
| 48 | 4x | – | South I., Otago, Flat Top Hill | AK 256013 | ||
| R. viride (Zotov) Connor & Edgar | 24 | 2x | 2·76 | North I., Wellington, Rangipo Desert | AK 286500 | |
| Simplicia buchananii (Zotov) Zotov | 28 | 4x | 11·07 | South I., Nelson, Oparara, Honeycomb Cave | AK 252968 | |
| S. laxa Kirk | 28 | 4x | 10·24 | South I., Otago, Ngatapa | AK 285424 | |
| Spinifex sericeus R.Br. | 18 | 2x | 5·41 | North I., North Auckland, Muriwai Beach | AK 286774 | |
| Stenostachys deceptorix Connor | 28 | 4x | 19·42 | South I., Nelson, Matiri Plateau | AK 286396 | |
| S. gracilis (Hook.f.) Connor | 28 | 4x | 17·91 | South I., Canterbury, Maitland Forest | AK 282084 | |
| S. laevis (Petrie) Connor | 28 | 4x | 18·83 | South I., Canterbury, Lake Tennyson | AK 281985 | |
| Trisetum arduanum Edgar & A.P.Druce | 28 | 4x | 11·07 | North I., South Auckland, Kawhia, Awaroa Reserve | AK 246714 | |
| T. drucei Edgar | 28 | 4x | 11·37 | North I., Wellington, Mangaweka | AK 252495 | |
| T. lepidum Edgar & A.P.Druce | 28 | 4x | 16·70 | South I., Canterbury, Lake Tennyson | AK 281986 | |
| T. serpentinum Edgar & A.P.Druce | 28 | 4x | 10·81 | South I., Nelson, Hackett Creek | AK 252504 | |
| T. spicatum (L.) K.Richt. | 28 | 4x | 9·82 | South I., Canterbury, Crimea Range | AK 282071 | |
| 28 | 4x | – | South I., Canterbury, Porters Pass | CHR 562377 | ||
| D* | ? | 19·48 | Campbell I. | AK 281997 | ||
| T. tenellum (Petrie) A.W.Hill | 28 | 4x | 10·29 | South I., Canterbury, Maitland River | CHR 562375 | |
| T. youngii Hook.f. | 28 | 4x | – | South I., Nelson, Lockett Range | AK 256029 | |
| 28 | 4x | 10·13 | South I., Canterbury, Lake Tennyson | AK 281048 | ||
| T. aff. lepidum | 28 | 4x | 17·02 | South I., Otago, Awahokomo | AK 251835 | |
| Zotovia colensoi (Hook.f.) Edgar & Connor | 48 | 4x | 2·93 | North I., Wellington, Tararua Ranges | AK 281551 | |
| Z. thomsonii (Petrie) Edgar & Connor | approx. 48 | 4x | 2·74 | Stewart I., Mt Rakeahua | AK 283814 | |
| Zoysia minima (Colenso) Zotov | 40 | 4x | 0·99 | North I., North Auckland, Piha Beach | AK 282083 | |
| 40 | 4x | – | South I., Canterbury, Kaitorete Spit | AK 256104 | ||
| Z. pauciflora Mez | 40 | 4x | 0·97 | Great Barrier I., Whangapoua Beach | AK 252969 |
D, plant died before a chromosome count could be made.
Fig. 1.
Map of New Zealand showing the main islands and the traditional botanical provinces.
Plant classification and determination of conservation status
Grasses have been grouped into two major clades, BOP (Bambusoideae, Oryzoideae, Pooideae) and PACC (Panicoideae, Arundinoideae, Chloridoideae + Centothecoideae) by the GPWG (2000). Within this framework we have grouped the New Zealand genera into tribes and subfamilies following Edgar and Connor (2000).
The conservation status of species was obtained from the most recent classification of threatened plants in New Zealand (de Lange et al., 2004). That paper, which uses the New Zealand Threat Classification System (see Molloy et al., 2002), recognizes nine species as ‘Acutely Threatened’, four as ‘Chronically Threatened’ and 58 as ‘At Risk’. Globally all of these taxa fall within the IUCN category of ‘Threatened’ (IUCN, 2000).
Flow cytometry
Determinations of nuclear DNA C-values were made using flow cytometry. In most cases only a single plant was available for analysis, but where several accessions were available values were measured on different days. All gave consistent results with little day-to-day variation. Nuclei were extracted by chopping fresh young leaves with a pair of single-edged razor blades into a final volume of 10 mL of ice-cold Galbraith's buffer (Galbraith et al., 1983), containing 3 % (w/v) polyvinylpyrrolidone. The chopped material was filtered through a 32-µm steel mesh filter and centrifuged at 300 g for 4 min to obtain a pellet of nuclei. The pellet was resuspended in 300 μL Galbraith's buffer containing100 µg mL−1 propidium iodide. In our laboratory we have found that RNase treatment has no effect on C-value so we routinely omit this step from our procedure. To obtain stable and repeatable results in Cenchrus it was necessary to wash the pellet of nuclei in 15 mL Galbraith's buffer and re-centrifuge before adding the propidium iodide. After staining on ice for at least 60 min, samples were analysed using an EPICS Elite ESP flow cytometer (Beckman-Coulter, Hialeah, FL, USA) using the air-cooled argon laser emitting light at 488 nm. Excitation of the probe propidium iodide was at 488 nm with fluorescence emitted measured using a 610 ± 10-nm bandpass filter. The instrument was aligned daily with flow check beads (Beckman-Coulter) that are labelled with a defined fluorescence intensity. Three replicates of each sample were prepared and at least 5000 nuclei were measured from each replicate.
An initial pilot study to determine the overall range of C-values for the taxa studied used Hordeum vulgare ‘Sultan’ (2C = 11·12 pg DNA/2C nucleus) as an external standard. Once this range was established, we used three different internal standards, H. vulgare ‘Sultan’, Secale cereale ‘Petkus Spring’ (2C = 16·57 pg) and the indigenous Poa anceps subsp. polyphylla (2C = 5·45 pg), which were co-chopped with the taxa to be determined. Poa anceps subsp. polyphylla was calibrated against H. vulgare to provide a grass standard that was closer to the lower values that we obtained in our preliminary study. The flow profiles of two independent runs of P. anceps subsp. polyphylla and H. vulgare are shown in Fig. 2A, B. Neither Zea mays W64A (2C = 5·47 pg) nor Sorghum bicolor ‘Pioneer’ 8695 (2C = 1·74 pg), both grasses and recommended standards (Johnston et al., 1999; Bennett et al., 2000), was available in New Zealand. C-values were reported previously for 26 of the species reported here using Actinidia chinensis as an external standard (Murray et al., 2003). On repeating the analyses with the grass internal standards we have found that these earlier values showed the same ranking but were approximately 30 % lower than those reported here.
Fig. 2.
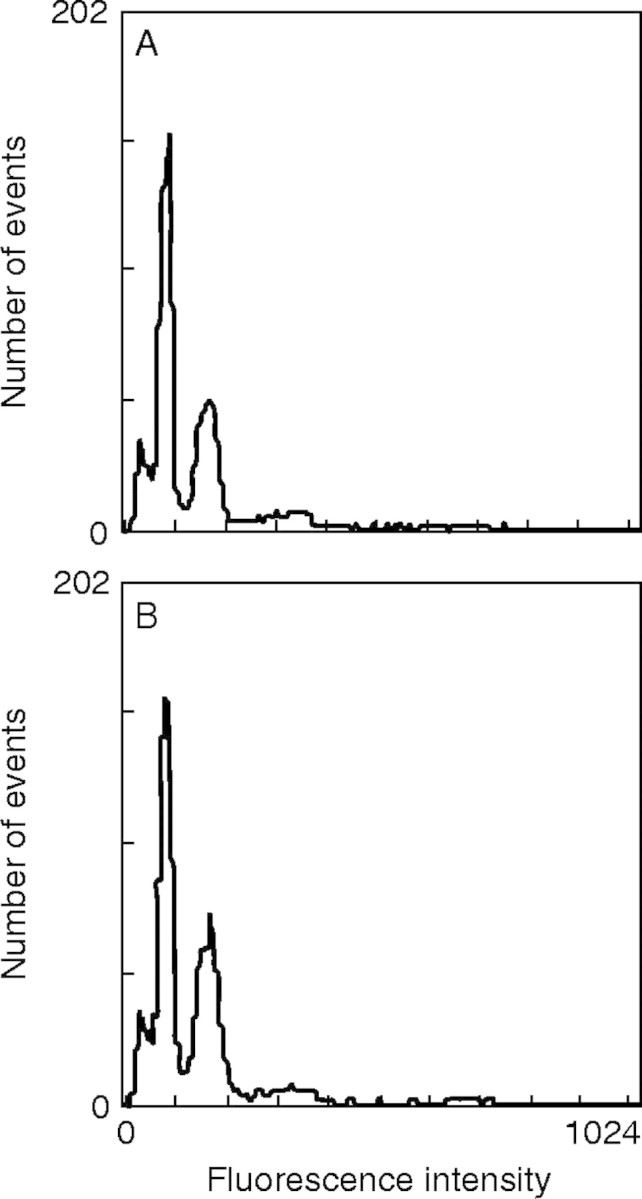
Flow cytometric profiles for two independent runs (A and B) of Poa anceps subsp. polyphylla (left peak) and Hordeum vulgare ‘Sultan’ (right peak) carried out for the calibration of the former as a new standard.
Following Leitch and Bennett (2004) we have calculated mean 2C genome size as the amount of 2C DNA in picograms divided by the ploidy level of the species.
Chromosome number determination
Somatic chromosome numbers were determined from root tips that were pretreated with a saturated solution of paradichlorobenzene for 18 h at 4 °C, fixed in 3 : 1. ethanol/acetic acid and stained with FLP-orcein (Jackson, 1973). Meiotic chromosomes were observed in pollen mother cells that were fixed and stained as outlined above.
RESULTS
C-value variation
C-values were determined for 155 species (161 taxa because in some species two subspecies or forms of a species were measured; Table 1). A wide range of C-values was observed, from 0·97 pg per 2C in Zoysia pauciflora to 32·40 pg per 2C in Poa litorosa (Table 1), representing a 33·4-fold variation. The spread of absolute values within genera varied considerably: some, such as Chionochloa, Cortaderia and Microlaena, showed a narrow range of values whereas others, such as Lachnagrostis and Poa, showed a wide range. When the ranges were expressed in relative terms, the highest over the lowest, Poa with a value of 7·4 was clearly different to the others, such as Chionochloa (1·75), Cortaderia (1·2), Microlaena (1·88) and Lachnagrostis (2·13). There was little evidence of any grouping of values that would result in discontinuities in C-values within genera. At the genus level, measurements were available for ten genera present in both New Zealand and the rest of the world (Table 2). It is difficult to see any clear trends from these comparisons as in some cases, e.g. Elymus, Festuca and Paspalum, the mean genome sizes are remarkably similar in both geographical areas, but in others, e.g. Deschampsia, Imperata and Trisetum, there are large differences. Similarly, comparisons of minimum and maximum C-values for the genera (Table 2) show different patterns. In Agrostis, Festuca, Poa and Trisetum the maximum values for the New Zealand representatives are higher than those from elsewhere but in Bromus, Deschampsia and Imperata they are lower. In Imperata and Koeleria the New Zealand species show lower minimum values than the non-New Zealand samples but the reverse is the case in Deschampsia and Trisetum. However, the limited sizes of the samples must be borne in mind when such comparisons are made.
Table 2.
A comparison of the mean genome size (amount of 2C DNA divided by ploidy level) and minimum and maximum 2C-values (pg) for genera common to both New Zealand and the rest of the world. Data for the rest of the world comparisons were obtained from www.rbgkew.org.uk/cval/homepage.html (C-values) and www.mobot.org (Index to Plant Chromosome Numbers)
| New Zealand |
Rest of World |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genus |
Number of species |
Mean genome size |
Min. 2C-value |
Max. 2C-value |
Number of species |
Mean genome size |
Min. 2C-value |
Max. 2C-value |
||||||
| Agrostis | 8 | 1·80 | 10·58 | 21·50 | 7 | 1·96 | 3·35 | 10·30 | ||||||
| Bromus | 1 | 4·11 | 16·45 | 16·45 | 46 | 3·90 | 3·75 | 32·65 | ||||||
| Deschampsia | 3 | 5·26 | 10·07 | 11·05 | 3 | 4·07 | 9·95 | 18·00 | ||||||
| Elymus | 7 | 4·58 | 20·54 | 32·09 | 12 | 4·55 | 5·85 | 30·30 | ||||||
| Festuca | 7 | 2·67 | 16·02 | 21·50 | 18 | 2·71 | 3·35 | 15·15 | ||||||
| Imperata | 1 | 0·73 | 1·45 | 1·45 | 1 | 2·71 | 10·85 | 10·85 | ||||||
| Koeleria | 4 | 2·72 | 9·82 | 13·23 | 1 | 4·60 | 9·20 | 9·20 | ||||||
| Paspalum | 1 | 0·52 | 3·13 | 3·13 | 3 | 0·57 | 1·20 | 3·05 | ||||||
| Poa | 29 | 1·39 | 4·26 | 32·40 | 6 | 1·52 | 2·35 | 10·75 | ||||||
| Trisetum | 8 | 2·98 | 9·82 | 19·48 | 1 | 1·28 | 5·10 | 5·10 | ||||||
C-value and phylogeny
In Table 3 the 35 genera are grouped into the two major clades, BOP and PACC, and then subfamilies and tribes, and the mean 2C value and mean genome size for each genus has been calculated. Many of the genera in the PACC clade had relatively small mean C-values and mean genome sizes. However, there were some interesting exceptions. Pyrrhanthera in Arundinoideae (PACC) had the third largest C-value observed (21·51 pg per 2C) but the high ploidy level in this monotypic genus (26x) means it had a small mean genome size. The genera in Ehrhartoideae and the tribe Stipeae in Pooideae, in the BOP clade, have low values for both of these measurements.
Table 3.
A summary of C-values and mean genome sizes in the 35 endemic or indigenous genera of New Zealand grasses arranged in clades following the most recent grass phylogeny (GPWG, 2000) and then by subfamily and tribe following Edgar and Connor (2000)
| Clade, subfamily, tribe and genus |
Mean 2C DNA amount (pg) |
Mean genome size (pg) |
||
|---|---|---|---|---|
| PACC—Arundinoideae | ||||
| Danthonieae | ||||
| Chionochloa | 5·40 | 0·90 | ||
| Pyrrhanthera | 21·51 | 0·83 | ||
| Rytidosperma | 5·00 | 1·73 | ||
| Cortaderiinae | ||||
| Cortaderia | 8·35 | 0·83 | ||
| PACC—Chloridoideae | ||||
| Chloroideae | ||||
| Zoysia | 0·89 | 0·25 | ||
| PACC—Panicoideae | ||||
| Paniceae | ||||
| Cenchrus | 11·12 | 1·86 | ||
| Oplismenus | 5·24 | 0·87 | ||
| Paspalum | 3·13 | 0·52 | ||
| Spinifex | 5·41 | 2·71 | ||
| Isnachneae | ||||
| Isachne | 3·64 | 0·61 | ||
| Andropogoneae | ||||
| Imperata | 1·45 | 0·73 | ||
| BOP—Ehrhartoideae | ||||
| Ehrharteae | ||||
| Microlaena | 2·71 | 0·68 | ||
| Zotovia | 2·84 | 0·71 | ||
| BOP—Pooideae | ||||
| Stipeae | ||||
| Achnatherum | 1·77 | 0·30 | ||
| Anemanthele | 1·89 | 0·47 | ||
| Austrostipa | 3·15 | 0·79 | ||
| Poeae | ||||
| Austrofestuca | 7·42 | 1·86 | ||
| Festuca | 18·23 | 2·67 | ||
| Poa | 7·84 | 1·39 | ||
| Puccinellia | 6·58 | 1·69 | ||
| Agrostideae | ||||
| Agrostis | 12·16 | 1·80 | ||
| Deyeuxia | 13·95 | 2·09 | ||
| Dichelachne | 16·82 | 1·68 | ||
| Echinopogon | 10·85 | 1·81 | ||
| Lachnagrostis | 20·44 | 1·86 | ||
| Simplicia | 10·66 | 2·67 | ||
| Amphibromus | 7·97 | 1·33 | ||
| Deschampsia | 10·52 | 5·26 | ||
| Koeleria | 10·85 | 2·72 | ||
| Trisetum | 12·97 | 2·98 | ||
| Hierochloe | 23·41 | 2·67 | ||
| Bromeae | ||||
| Bromus | 16·45 | 4·11 | ||
| Hordeae | ||||
| Australopyrum | 11·43 | 5·72 | ||
| Elymus | 27·02 | 4·58 | ||
| Stenostachys | 18·72 | 4·68 | ||
C-value and ploidy level
The mean, standard deviation and range of C-values for different ploidy levels are given in Table 4. There is a progressive increase in mean C-value with increasing ploidy level, with the exception of the 10x category, but it is clear that there is a large range of values at each level and that there no defined incremental increase with increasing ploidy. Nine of the New Zealand genera show a range of different ploidy levels and in seven of these, Deyeuxia, Elymus, Festuca, Hierachloe, Lachnagrostis, Poa and Puccinellia, there is a progressive decrease, to different degrees, in genome size with increasing ploidy (Table 5). In Rytidosperma the tetraploids have a slightly higher genome size than the diploids and in Agrostis the hexaploids and 12-ploids have almost identical genome sizes (Table 5).
Table 4.
Mean, standard deviation and range of C-values at different ploidy levels in New Zealand Poaceae
| Ploidy level |
2x |
4x |
6x |
8x |
10x |
12x |
14x |
16x |
|---|---|---|---|---|---|---|---|---|
| No. of species analysed | 15 | 64 | 41 | 11 | 10 | 6 | 5 | 3 |
| Mean | 5·35 | 8·01 | 10·65 | 18·02 | 12·26 | 24·57 | 24·62 | 18·66 |
| Standard deviation | 3·50 | 4·72 | 7·54 | 5·60 | 4·27 | 5·71 | 2·17 | 2·14 |
| Range | 1·45–11·50 | 0·97–20·54 | 1·77–28·56 | 12·57–32·09 | 7·85–17·67 | 21·50–29·97 | 21·11–26·81 | 17·40–21·13 |
Table 5.
The relationship between mean genome size (amount of 2C DNA divided by ploidy level) and ploidy level in species from nine genera of Poaceae from New Zealand that have a range of ploidy levels
| Ploidy |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genus |
2x |
4x |
6x |
8x |
10x |
12x |
14x |
16x |
38x |
||||||||
| Agrostis | – | – | 1·80 | – | – | 1·79 | – | – | – | ||||||||
| Deyeuxia | – | 3·25 | – | 1·75 | 1·45 | – | – | – | – | ||||||||
| Elymus | – | 5·14 | 4·55 | 4·01 | – | – | – | – | – | ||||||||
| Festuca | – | – | 2·74 | 2·58 | – | – | – | – | – | ||||||||
| Hierochloe | – | 3·14 | 3·02 | – | – | 2·40 | – | – | – | ||||||||
| Lachnagrostis | – | – | – | 1·91 | – | 2·07 | 1·76 | – | – | ||||||||
| Poa | – | 1·44 | – | – | – | 1·23 | – | 1·17 | 0·85 | ||||||||
| Puccinellia | 1·78 | – | 1·60 | – | – | – | – | – | – | ||||||||
| Rytidosperma | 1·68 | 1·80 | – | – | – | – | – | – | – | ||||||||
C-value and rarity
We obtained C-values for a significant proportion of the plants in the various conservation categories and the results are shown in Table 6. We also examined the relationship between rarity and C-value at generic level. Because the numbers of species within genera are much smaller at this level we have grouped the plants classified into the three categories outlined above as a single category ‘Threatened’ and compared their values with those of the non-threatened members of four genera, Chionochloa, Poa, Rytidosperma and Trisetum, for which the chromosome numbers of the plants analysed are the same. In Chionochloa the comparison between threatened and non-threatened is 5·56 (n = 9) to 5·46 (n = 10), in Poa 5·80 (n = 9) to 5·75 (n = 17), in Rytidosperma 8·26 (n = 1) to 6·96 (n = 5) and in Trisetum 11·09 (n = 2) to 11·60 (n = 5).
Table 6.
Mean C-values (± s.d.) and the percentage of polyploids (with the number of species for which data were available in parentheses) in the endemic and indigenous species of New Zealand Poaceae in relation to their conservation status (see text for explanation of conservation categories)
| Conservation status |
Mean C-value |
Percentage polyploids |
|---|---|---|
| Acutely threatened | 8·95 ± 2·36 (n = 8) | 87·5 (n = 8) |
| Chronically threatened | 9·33 ± 2·99 (n = 3) | 75·0 (n = 4) |
| At risk | 12·80 ± 8·98 (n = 40) | 98·0 (n = 49) |
| Not threatened | 11·05 ± 7·38 (n = 113) | 89·7 (n = 117) |
Intraspecific C-value variation
Deyeuxia avenoides, Lachnagrostis littoralis, L. lyallii and Rytidosperma thomsonii all appear to show intraspecific C-value variation. In the first three species, the plants all had the same chromosome number but in R. thomsonii two different chromosome numbers were obtained (Table 1). In D. avenoides, the lower C-value was 87 % of the higher value, in L. littoralis it was 88 % and in L. lyallii it was 74 %. In R. thomsonii diploid and tetraploid plants were counted and the tetraploids had 1·84 times the DNA C-value of the diploid.
Chromosome numbers and ploidy levels
The chromosome numbers for 55 species are published here for the first time and, in addition, we report five new chromosome numbers in species for which chromosome numbers have been reported previously. These latter species are Deyeuxia aucklandica, Lachnagrostis pilosa subsp. pilosa, Rytidosperma buchananii, R. thomsonii and Trisetum tenellum (Table 1). With these new counts, chromosome numbers are now known for 186 species of endemic and indigenous grasses (91·6 % of the total of 203 that we have recognized in this paper; Table 7). If infraspecific ranks are included, there are 214 taxa and of these 193 (90·2 %) have been counted.
Table 7.
The number of species at different ploidy levels in the 36 genera of Poaceae that contain species endemic or indigenous to New Zealand and the percentage of species at the different ploidy levels. This table includes the chromosome counts made here plus those taken from Dawson (2000) and de Lange and Murray (2002). This list also includes a number of undescribed taxa and chromosome races (see footnotes)
| Ploidy level |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genus |
No. of species in NZ* |
No. of species counted |
2x |
4x |
6x |
8x |
10x |
12x |
>12x |
||||||
| Achnatherum | 1 | 1 | – | – | 1 | – | – | – | – | ||||||
| Agrostis | 10 | 8 | – | – | 7 | – | – | 1 | – | ||||||
| Amphibromus | 1 | 1 | – | – | 1 | – | – | – | – | ||||||
| Anemanthele | 1 | 1 | – | 1 | – | – | – | – | – | ||||||
| Australopyrum | 1 | 1 | 1 | – | – | – | – | – | – | ||||||
| Austrofestuca | 1 | 1 | – | 1 | – | – | – | – | – | ||||||
| Austrostipa | 1 | 1 | – | 1 | – | – | – | – | – | ||||||
| Bromus | 1 | 1 | – | 1 | – | – | – | – | – | ||||||
| Cenchrus | 1 | 1 | – | – | 1 | – | – | – | – | ||||||
| Chionochloa | 23 | 23 | – | – | 23 | – | – | – | – | ||||||
| Cortaderia | 5 | 5 | – | – | – | – | 5 | – | – | ||||||
| Deschampsia | 5 | 4 | 4 | – | – | – | – | – | – | ||||||
| Deyeuxia | 6 | 6 | – | 2 | – | 3 | 1 | – | – | ||||||
| Dichelachne | 4 | 4 | – | – | – | – | 4 | – | – | ||||||
| Echinopogon | 1 | 1 | – | – | 1 | – | – | – | – | ||||||
| Elymus | 7 | 7 | – | 1 | 5 | 1 | – | – | – | ||||||
| Festuca | 10 | 10 | – | 1 | 4 | 4 | – | – | 1 × 24x | ||||||
| Hierochloe | 7 | 5 | – | 1 | 1 | – | – | 3 | – | ||||||
| Imperata | 1 | 1 | 1 | – | – | – | – | – | – | ||||||
| Isachne | 1 | 1 | – | – | 1 | – | – | – | – | ||||||
| Koeleria | 5 | 5 | – | 5 | – | – | – | – | – | ||||||
| Lachnagrostis | 13 | 11 | – | – | – | 4 | – | 2 | 5 × 14x | ||||||
| Lepturus | 1 | 0 | – | – | – | – | – | – | – | ||||||
| Microlaena | 4 | 4 | – | 4 | – | – | – | – | – | ||||||
| Oplismenus | 1 | 1 | – | – | 1 | – | – | – | – | ||||||
| Paspalum | 1 | 1 | – | – | 1 | – | – | – | – | ||||||
| Poa | 41 | 36 | – | 29 | – | 1 | – | 1 | 4 × 16x;1 × 38x | ||||||
| Puccinellia | 4 | 4 | 1 | 1 | 1 | 1 | – | – | – | ||||||
| Pyrrhanthera | 1 | 1 | – | – | – | – | – | – | 1 × 26x | ||||||
| Rytidosperma | 22 | 19 | 11 | 7 | 1 | – | – | – | – | ||||||
| Simplicia | 2 | 2 | – | 2 | – | – | – | – | – | ||||||
| Spinifex | 1 | 1 | 1 | – | – | – | – | – | – | ||||||
| Stenostachys | 3 | 3 | – | 3 | – | – | – | – | – | ||||||
| Trisetum | 11 | 11 | – | 10 | – | 1 | – | – | – | ||||||
| Zotovia | 3 | 2 | – | 2 | – | – | – | – | – | ||||||
| Zoysia | 2 | 2 | – | 2 | – | – | – | – | – | ||||||
| Totals | 203 | 186 | 19 (10·2 %) | 74 (39·8 %) | 49 (26·3 %) | 15 (8·1 %) | 10 (5·4 %) | 7 (3·8 %) | 12 (6·5 %) | ||||||
The total number of species in this table differs from that of Edgar and Connor (2000) for the following reasons. One species of Paspalum and one of Bromus have been treated as indigenous (see Materials and Methods); Deyeuxia includes one undescribed taxa, D. aff. quadriseta, and two taxa within D. avenoides differing in C-value; Koeleria includes two undescribed taxa, K. aff. novozelandica and K. aff. cheesemanii; Poa includes three undescribed taxa, P. aff. cita (a), P. aff. cita (b) and P. aff. colensoi; Rytidosperma includes the 2x and 4x races of R. thomsonii and the 4x and 6x races of R. buchananii; and Trisetum includes one undescribed taxa, T. aff. lepidum, and the 4x and 8x races of T. tenellum.
The large majority (91 %) of species in the 36 endemic and indigenous genera are polyploid; diploids are confined to six genera, Australopyrum, Deschampsia, Imperata, Puccinellia, Rytidosperma and Spinifex (Table 7). Of the species that are polyploid, 39·8 % were tetraploid and 26·3 % were hexaploid, with smaller percentages at the higher ploidy levels (Table 7). High ploidy levels were seen in Poa, four species were 16x and another was 38x, and the endemic, monotypic Pyrrhanthera was 26x. Of the genera with ten or more species, Chionochloa was unusual in that all 23 species were at the same ploidy level (6x) whereas the other large genera had species at a variety of ploidy levels; for example, Festuca, with ten species, had four ploidy levels.
Polyploidy and rarity
Information is available for eight grass taxa that are classified as ‘Acutely Threatened’, four that are ‘Chronically Threatened’, 49 that are ‘At Risk’ and 117 that are ‘Not Threatened’. The percentage of polyploids in each category is given in Table 6. If the three threatened categories are combined then 94·8 % of these species are polyploid, slightly higher than the 89·7 % for the non-threatened category.
DISCUSSION
The present study has increased the representation of grasses in the C-value database by about 30 % and the total number of New Zealand angiosperms to 149. This number includes the first reports for 21 additional genera of Poaceae, five of which, Anemanthele, Pyrrhanthera, Simplicia, Stenostachys and Zotovia, are endemic to New Zealand. The new values show a 33·4-fold variation for the New Zealand plants (0·97–32·40 pg per 2C nucleus), well within the range of values for the family as a whole (0·50–43·25 pg per 2C nucleus). Owing to the high level of endemism in the New Zealand flora, few intraspecific comparisons between New Zealand and non-New Zealand plants can be made, but among indigenous plants some comparisons are possible. An example is Deschampsia cespitosa, a widespread cosmopolitan species that in New Zealand is diploid and has 10·43 pg per 2C. By comparison, Bennett et al. (1982), who did not count the chromosomes of the plant from South Georgia that they studied, but assumed that it was diploid, reported a C-value of 8·55 pg per 2C. Unfortunately, there is no voucher specimen associated with the measurement made by Bennett et al. (1982) so the identity of their plant cannot be confirmed. The value for Imperata cheesemanii from New Zealand is lower than the value for the one other Imperata species so far recorded: otherwise, the minimum values obtained for genera present in New Zealand are higher than the minimum values recorded for the same genera elsewhere in the world. In seven such genera the maximum values for New Zealand species are higher than those found elsewhere. Poa flabellata from South Georgia is 4x and has a C-value of 5·45 pg per 2C (Bennett et al., 1982) similar to the New Zealand 4x Poa species.
Obtaining C-values for this sample of grasses was relatively straightforward and good, symmetrical peaks with low coefficients of variation were generally obtained. Although in most cases only a single sample was available, when we did have more than one there was good agreement between measurements from different plants made on different days, usually less than 5 % difference. There are four exceptions for which there was a greater than 10 % difference between samples and these are discussed briefly below. One other unexpected result was obtained from among the Poa species that had 2n = 4x = 28. Poa schistacea had a C-value of 11·02 pg but 22 other Poa species had C-values between 4·26 and 6·67 pg, with the P. schistacea value being approximately double the mean value of the other species.
C-values, distribution patterns and rarity
Our measurements, although admittedly limited, provide little evidence to support the contention that large C-values are maladaptive and may be a cause of extinction (Vinogradov, 2003). However, our values are relatively low compared with the global sample used by Vinogradov: the highest C-value that we obtained, 32·24 pg per 2C for Poa litorosa, is below the range of his ‘Global concern’ category but higher than the mean of his ‘Local concern’ category. In our sample, there are no clear differences between species that are rare or with restricted distribution and those species that are widespread. When phylogenetic constraints are reduced, by restricting the analysis to species with the same chromosome number within a single genus, there is again no large difference between the restricted and widespread species in the four genera (Chionochloa, Poa, Rytidosperma and Trisetum) for which such a comparison is possible. There also does not appear to be any correlation between polyploidy and rarity, but it must again be borne in mind that the sample sizes are not large and that the majority of New Zealand grasses are polyploid. It is also interesting that in several genera (Agrostis, Festuca, Poa, Puccinellia) the species with the highest C-values and chromosome numbers are found in the most extreme environments such as the sub-Antarctic [Auckland (Campbell) and Enderby Islands] and Chatham Islands. The genus Elymus has the highest mean C-value of all the grass genera in New Zealand (27·02 pg per 2C) yet it is by no means the most uncommon or threatened (de Lange et al., 2004). Many of the least common or seriously threatened species, such as Poa spania, Amphibromus fluitans and Simplicia laxa, are all within the lower half of C-values for New Zealand grasses. Differences in C-value do appear to reflect the geographical origin of the genera, with five that we have identified (following Clayton and Renvoize, 1986) (Imperata, Isachne, Paspalum, Oplismenus, Zoysia) as being of tropical origin all having C-values in the lowest end of the range we observed. This is in line with previous observations that tropical species of plants typically have lower C-values than temperate species (Bennett, 1976; Levin and Funderburg, 1979).
Leitch and Bennett (2004), in a survey of amounts of nuclear DNA, have pointed out that in angiosperms the mean genome size of polyploids was significantly lower than that of diploids. We have performed a similar analysis of the nine genera of New Zealand grasses that contain species with different ploidy levels and have found that most also show smaller genome sizes in polyploids compared with diploids. In some cases the differences are not great and it is possible that this reflects the recent nature of speciation/polyploidization that is commonly found in the New Zealand angiosperm flora (Wagstaff and Garnock-Jones, 1998; Heenan et al., 2002) and that genome diminution in some genera may reflect a longer timescale since speciation.
Taxonomic implications of C-value and chromosome variation
Four examples of putative intraspecific C-value variation have been observed. Three of these species (Deyeuxia avenoides, Lachnagrostis littoralis and L. lyallii) have been long recognized as being highly variable, showing differences in habit and distribution (Edgar, 1995). We also found examples of intraspecific variation in chromosome number in two species of Rytidosperma but were only able to measure C-values in one of them, R. thomsonii. This latter species is reported to have robust and small-statured races that related to the observed differences in chromosome number and C-value (B. P. J. Molloy, personal communication).
In line with previous studies (summarized by Dawson in Edgar and Connor, 2000) the majority of new chromosome counts confirm further examples of polyploidy with diploids confined to six genera, Australopyrum, Deschampsia, Imperata, Puccinellia, Rytidosperma and Spinifex. There may be some debate as to what is a diploid in some of these genera because the basic number (x) is 13 in Deschampsia, 10 in Imperata and 12 in Rytidosperma. We have assumed that 2n = 26, 2n = 20 and 2n = 24 are diploid numbers in these three genera as these are the lowest numbers that are found and the plants are bivalent forming. In addition to the new count of 2n = 48 for Rytidosperma thomsonii, we have obtained new counts for four other species that have been studied previously. In Deyeuxia aucklandica, de Lange and Murray (2002) reported 2n = 42 whereas the new material had 2n = 56. de Lange and Murray (2002) found 2n = 56 for Lachnagrostis pilosa subsp. pilosa compared with 2n = 98 here and they, together with Calder (1937), found 2n = 72 for Rytidosperma buchananii compared with 2n = 48 reported here. The final example is Trisetum tenellum with a count of 2n = 28 obtained here compared with 2n = 56 reported by de Lange and Murray (2002). There are relatively few examples of intraspecific chromosome number variation in the New Zealand flora; Murray et al. (1989) reported that only approximately 2 % of the species for which chromosome numbers were known had different chromosome races. Further investigation of these new examples is needed to ascertain whether the chromosome races and putative C-value variants are sufficiently distinct for them to be recognized as distinct taxa.
Acknowledgments
We thank Dean Baigent-Mercer, Amanda Baird, John Barkla, Steve Benham, Jonathon Boow, John Braggins, Andrea Brandon, Jim Clarkson, Shannel Courtney, Geoff Davidson, Lisa Forester, Rhys Gardner, Bridget Gibb, Terry Hatch, Graham Jane, Peter Johnson, Phil Knightbridge, Kelvin Lloyd, Geoff McCauley, Brian Molloy, David Norton, Colin Ogle, Brian and Chris Rance, Matt Renner, Nick Singers, Mike Thorsen and Matt von Konrat for their help in obtaining plant material for this study, Mei Nee Lee for her help with accessioning the voucher specimens at the Auckland Museum Herbarium, Alison Duffy for assistance in preparing flow cytometry samples, Jingli Zhang for operation of the flow cytometer, and Henry Connor, Peter Heenan and Ilia Leitch for their comments on an earlier draft of the manuscript.
LITERATURE CITED
- Bennett MD. 1972. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London B 181: 109–135. [DOI] [PubMed] [Google Scholar]
- Bennett MD. 1976. DNA amount, latitude, and crop plant distribution. Environmental and Experimental Botany 16: 93–108. [Google Scholar]
- Bennett MD. 1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106 (Suppl.): 177–200. [Google Scholar]
- Bennett MD, Leitch IJ. 2005. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Annals of Botany 95: 45–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Bhandol P, Leitch IJ. 2000. Nuclear DNA amounts in angiosperms and their modern uses—807 new estimates. Annals of Botany 86: 859–909. [Google Scholar]
- Bennett MD, Smith JB, Smith RIL. 1982. DNA amounts of angiosperms from the Antarctic and South Georgia. Environmental and Experimental Botany 22: 307–318. [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, Scheen A-C, Elven R. 2004. Polyploidy in arctic plants. Biological Journal of the Linnean Society 82: 521–536. [Google Scholar]
- Calder JW. 1937. A cytological study of some New Zealand species of Danthonia Journal of the Linnean Society (Botany) 51: 1–9. [Google Scholar]
- Clayton WD, Renvoize SA. 1986.Genera Gramineum: grasses of the world. London: Her Majesty's Stationery Office. [Google Scholar]
- Cockayne L. 1967.New Zealand plants and their story, 4th edn. Wellington: Government Printer. [Google Scholar]
- Dawson MI. 2000. Index of chromosome numbers of indigenous New Zealand spermatophytes. New Zealand Journal of Botany 38: 47–150. [Google Scholar]
- Edgar E. 1995. New Zealand species of Deyeuxia P.Beauv. and Lachnagrostis Trin. (Gramineae: Aveneae). New Zealand Journal of Botany 33: 1–33. [Google Scholar]
- Edgar E, Connor HE. 2000.Flora of New Zealand volume V Gramineae. Lincoln, New Zealand: Manaaki Whenua Press. [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissue. Science 220: 1049–1051. [DOI] [PubMed] [Google Scholar]
- GPWG. 2000. A phylogeny of the grass family (Poaceae) as inferred from eight character sets. In: Jacobs SWL, Everett J, eds. Grasses: systematics and evolution. Collingwood, Victoria: CSIRO Publishing, 3–7. [Google Scholar]
- Gregory TR. 2005. The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Annals of Botany 95: 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Dolezel, J, Lysák MA, Bennett MD. 2005. The origin, evolution and proposed stabilization of the terms ‘genome size’, and ‘C-value’ to describe nuclear DNA content. Annals of Botany 95: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP, Mowforth MA. 1982. Variation in genome size: an ecological interpretation. Nature 299: 151–153. [Google Scholar]
- Hair JB. 1966. Biosystematics of the New Zealand flora 1945–1964. New Zealand Journal of Botany 4: 559–595. [Google Scholar]
- Hanson L, Brown RL, Boyd A, Johnson MAT, Bennett MD. 2003. First nuclear DNA C-values for 28 angiosperm genera. Annals of Botany 91: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heenan PB, Mitchell AD, Koch M. 2002. Molecular systematics of the New Zealand Pachycladon (Brassicaceae) complex: generic circumscription and relationships to Arabidopsis sens. lat. and Arabis sens. lat. New Zealand Journal of Botany 40: 543–562. [Google Scholar]
- IUCN. 2000.IUCN red list categories. Gland: IUCN. [Google Scholar]
- Jackson RC. 1973. Chromosome evolution in Haplopappus gracilis: a centric transposition race. Evolution 27: 243–256. [DOI] [PubMed] [Google Scholar]
- Johnston JS, Bennett MD, Rayburn AL, Galbraith DW, Price HJ. 1999. Reference standards for determination of DNA content of plant nuclei. American Journal of Botany 86: 609–613. [PubMed] [Google Scholar]
- Kellogg EA, Bennetzen JL. 2004. The evolution of nuclear genome structure in seed plants. American Journal of Botany 91: 1709–1725. [DOI] [PubMed] [Google Scholar]
- Knight CA, Ackerly DD. 2002. Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecology Letters 5: 66–76. [Google Scholar]
- de Lange PJ, Murray BG. 2002. Contributions to a chromosome atlas of the New Zealand flora—37. Miscellaneous families. New Zealand Journal of Botany 40: 1–23. [Google Scholar]
- de Lange PJ, Norton DA. 1997. Revisiting rarity: a botanical perspective on the meanings of rarity and the classification of New Zealand's uncommon plants. In: Lynch R, ed. Ecosystems, entomology and plants The Royal Society of New Zealand Miscellaneous series 48: 145–160. [Google Scholar]
- de Lange PJ, Norton DA, Heenan PB, Courtney SP, Molloy BPJ, Ogle CC, et al. 2004. Threatened and uncommon plants of New Zealand. New Zealand Journal of Botany 42: 45–76. [Google Scholar]
- Leitch IJ, Bennett MD. 2004. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society 82: 651–633. [Google Scholar]
- Levin DA. 2002.The role of chromosomal change in plant evolution. Oxford: Oxford University Press. [Google Scholar]
- Levin DA, Funderberg SW. 1979. Genome size in angiosperms: temperate versus tropical species. American Naturalist 114: 784–795. [Google Scholar]
- McGlone MS, Duncan RP, Heenan PB. 2001. Endemism, species selection and the origin and distribution of the vascular plant flora of New Zealand. Journal of Biogeography 28: 199–216. [Google Scholar]
- Molloy J, Bell B, Clout M, de Lange P, Gibbs G, Given D, et al. 2002.Classifying species according to threat of extinction—a system for New Zealand. Wellington: Department of Conservation. [Google Scholar]
- Murray BG. 2005. When does intraspecific C-value variation become taxonomically significant? Annals of Botany 95: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BG, Braggins JE, Newman PD. 1989. Intraspecific polyploidy in Hebe diosmifolia (Cunn.) Cockayne et Allan (Scrophulariaceae). New Zealand Journal of Botany 27: 587–589. [Google Scholar]
- Murray BG, Cameron EK, Standring LS. 1992. Chromosome numbers, karyotypes, and nuclear DNA variation in Pratia Gaudin (Lobeliaceae). New Zealand Journal of Botany 30: 181–187. [Google Scholar]
- Murray BG, Weir IE, Ferguson AR, de Lange PJ. 2003. Variation in C-value and haploid genome size in New Zealand native grasses. New Zealand Journal of Botany 41: 63–69. [Google Scholar]
- Pole M. 1994. The New Zealand flora—entirely long-distance dispersal? Journal of Biogeography 21: 625–635. [Google Scholar]
- Pole MS. 2001. Can long-distance dispersal be inferred from the New Zealand plant fossil record? Australian Journal of Botany 49: 357–366. [Google Scholar]
- Stebbins GL. 1971.Chromosomal evolution in higher plants. London: Edward Arnold. [Google Scholar]
- Thompson JD, Lumaret R. 1992. The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends in Ecology and Evolution 7: 302–307. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. 2003. Selfish DNA is maladaptive: evidence from the plant Red List. Trends in Genetics 19: 609–614. [DOI] [PubMed] [Google Scholar]
- Wagstaff SJ, Garnock-Jones PJ. 1998. Evolution and biogeography of the Hebe complex (Scrophulariaceae) inferred from ITS sequences. New Zealand Journal of Botany 36: 425–437. [Google Scholar]
- Wardle P. 1978. Origin of the New Zealand mountain flora, with special reference to trans-Tasman relationships. New Zealand Journal of Botany 16: 535–550. [Google Scholar]
- Wardle P. 1991.The vegetation of New Zealand. Cambridge: Cambridge University Press. [Google Scholar]
- Wilton AD, Breitwieser I. 2000. Composition of the New Zealand seed plant flora. New Zealand Journal of Botany 38: 537–549. [Google Scholar]
- Winkworth RC, Wagstaff SJ, Glenny D, Lockhart PJ. 2002. Plant dispersal N.E.W.S. from New Zealand. Trends in Ecology and Evolution 17: 514–520. [Google Scholar]