Abstract
• Background and Aims Species' 2C-values (mass of DNA in G1 phase 2n nuclei) vary by at least four orders of magnitude among seed plants. The 2C-value has been shown to be co-ordinated with a number of other species traits, and with environmental variables. A prediction that species 2C-values are negatively related to leaf life span (LL) and leaf mass per area (LMA) is tested. These leaf traits are components of a major dimension of ecological variation among plant species.
• Methods Flow cytometry was used to measure the 2C-values for 41 Australian seed plant species, 40 of which were new to the literature. Where possible, LL and LMA data from the global literature were combined with 2C-values from our data set and online C-value databases.
• Key Results Across all species, weak positive relationships were found between 2C-values and both LL and LMA; however, these did not reflect the relationships within either angiosperms or gymnosperms. Across 59 angiosperm species, there were weak negative relationships between 2C-values and both LL (r2 = 0·13, P = 0·005) and LMA (r2 = 0·15, P = 0·002). These relationships were the result of shifts to longer LL and greater LMA in woody compared with herbaceous growth forms, with no relationships present within growth forms. It was not possible to explain a positive relationship between 2C-values and LMA (r2 = 0·30, P = 0·024) across 17 gymnosperm species. The 2C-value was not related to LL or LMA either across species within orders (except for LMA among Pinales), or as radiation divergences in a model phylogeny.
• Conclusions Gymnosperms appear to vary along a spectrum different from angiosperms. Among angiosperms, weak negative cross-species relationships were associated with growth form differences, and traced to a few divergences deep in the model phylogeny. These results suggest that among angiosperms, nuclear DNA content and leaf strategy are unrelated.
Keywords: Genome size, C-value, leaf life span, leaf mass per area, LMA, SLA, angiosperms, gymnosperms, correlated divergence analysis, standardized major axis
INTRODUCTION
Nuclear DNA (2C) values
The amount of DNA in G1 phase 2n nuclei is referred to as the 2C-value (2C), and is measured as the mean mass (pg) or mean number of base pairs (gigabase pairs; Gbp) of DNA in the nucleus. Currently, within-species variation in 2C (e.g. Price and Johnston, 1996a; Reeves et al., 1998) is poorly understood and controversial (e.g. Greilhuber, 2005). On the other hand, it is well understood that 2C varies widely across species; by at least four orders of magnitude within angiosperms, in the range of 0·01–100 pg (Bennett et al., 2000), and at least 5-fold within gymnosperms, from 12·96 to 63·5 pg (Murray, 1998). There are some known consequences of having a particular 2C. Species with greater 2C generally have nuclei (and cells) of greater volume (Stebbins, 1971; Price et al., 1973; Edwards and Endrizzi, 1975; Lawrence, 1985; Bennett, 1987; Cavalier-Smith, 2005), which take longer to divide (Van't Hof and Sparrow, 1963; Bennett, 1971; Price, 1976; Cavalier-Smith, 1982). These relationships seem to be principally biophysical; a species of greater 2C requires a nucleus of greater volume, and more time to unravel and duplicate during meiosis and mitosis, compared with a species of lower 2C (Bennett, 1987).
Variation in 2C is associated with variation in a number of other functional traits and environmental variables, and it has been suggested that 2C could be useful as a general predictor of species ecology (Grime and Mowforth, 1982; Bennett, 1987; Grime, 1998; Leitch et al., 1998; Reeves et al., 1998; Knight and Ackerly, 2002). Among 24 species from the Sheffield region, Grime and Mowforth (1982) showed that those with greater 2C tended to expand shoots earlier in the growing season than those with lower 2C. Also, species with greater 2C generally produce seeds of greater mass (Thompson, 1990; Leishman et al., 2000; Knight and Ackerly, 2002). Less well supported are relationships with mean height (e.g. in Senecio; Lawrence, 1985), and fruit and flower size (Stebbins, 1971), both increasing with 2C across the species studied. Environmentally, 2C has been reported to vary with latitude (both positively and negatively; see Knight et al., 2005), precipitation (e.g. Sims and Price, 1985; Wakamiya et al., 1993), altitude (see summary in Knight and Ackerly, 2002) and temperature (Wakamiya et al., 1993). Knight and Ackerly (2002) recently assembled a large regional data set for California, and reported that while the mean 2C of species at any given site in the region did not vary with latitude, the largest were constrained by mean minimum and maximum temperatures of the coldest and hottest months, and mean annual precipitation.
Leaf life span and leaf mass per area
A species' leaf life span (LL) measures how long typical leaves survive until death or loss, averaged across branches and individuals for the species (Chabot and Hicks, 1982). In economic terms, LL measures the duration of return of photosynthetic revenue (Wright et al., 2003). Longer-lived leaves are generally of more substantial construction (thicker or more dense, or both), having greater leaf mass per area (LMA; Reich et al., 1992, 1997; Wright et al., 2002, 2004). The relationship between LL and LMA is maintained across species from diverse habitats and phylogenies, and is a major dimension of variation among plant species (Westoby et al., 2002; Wright et al., 2004). This dimension describes a spectrum of variation in carbon-gain strategy of species; from the quick return on investment end (species with short LL and low LMA), to the slow return on investment end (long LL and high LMA) (Wright et al., 2004). To a large extent, the position along this spectrum describes various life history strategies; fast growing herbaceous species have short LL and low LMA, while woody perennials, particularly evergreens, have longer LL and high LMA.
Why 2C might be related to LL and LMA
We reasoned that there could exist a general relationship between 2C and LL and LMA; specifically, that species with greater 2C might produce shorter lived leaves of lower LMA. Considering that greater 2C is associated with production of larger volume cells, it follows that such tissues may be lower density by having fewer cell walls per unit volume (setting aside between-species variation in other cellular traits). In leaves, variation in tissue density among species is a prime factor driving variation in LMA, the other being lamina depth (Castro-Díez et al., 2000; Niinemets, 2001). Given the robust relationship between LMA and LL (Wright et al., 2004), we might therefore expect leaves of large genome species also to be shorter lived.
Knight et al. (2005) recently compiled specific leaf area (SLA; the inverse of LMA) and 2C data for 67 mainly herbaceous and gymnosperm species from two previous studies (Grime et al., 1997, herbs; Reich et al., 1998, gymnosperms). They found a weak negative relationship between SLA and 2C (weakly positive if expressed as LMA), apparently driven by strong differences in both LMA and 2C between these growth forms in their data set. Strong variation in leaf traits is known to occur between woody and herbaceous growth forms (Wright et al., 2004), and between angiosperms and the largely evergreen gymnosperms (e.g. Reich, 1998), so it is important to understand how variation between these groups might shape a general relationship with 2C. Moreover, gymnosperms are generally known to have larger genomes than angiosperms (Leitch et al., 1998; Murray, 1998), thus we wanted to know whether gymnosperms lie along the same spectrum of variation as angiosperms (i.e. similar LL and LMA at a given 2C).
Our aim in this study was to determine, for all currently available data, whether there existed negative slope relationships between 2C and either LL or LMA. By including new data for woody angiosperms, and all species available from the global literature, we sought to determine whether this relationship existed both within and between growth forms and major seed plant groups. Further, we aimed to test the potential influence of evolutionary history in structuring present-day relationships by conducting phylogenetic analyses: first, whether present-day relationships were consistently found within groups of closely related species; and secondly, whether divergences in 2C were consistently accompanied by divergences in LL and LMA throughout a model phylogeny of the species.
MATERIALS AND METHODS
Direct measurement of LL can take several years, thus published LL data are relatively scarce. Recently, Wright et al. (2004) compiled worldwide leaf trait data, including LL data, for several thousand species from both published and unpublished sources. Where possible, we matched published LL and LMA data from Wright et al. (2004) with 2C data obtained either by our own measurements, or from online angiosperm (Bennett and Leitch, 2003) and gymnosperm (Murray et al., 2003) databases.
2C measurement
Tissues for 2C measurement were obtained either from germinated seeds or from healthy, fully expanded sun leaves collected either from field sites or a garden in Macquarie University (Appendix). Seeds were obtained either from a seed supply company or from personal field collections (Appendix), and they were glasshouse-germinated (24 °C) on moist filter paper in sealed Petri dishes. Pre-germination treatments, including immersion in hot water, soaking or scarification, were applied to seeds of some species to assist germination (Ralph, 1994). We used wheat, Triticum aestivum ‘Chinese Spring’, as the standard for 2C measurement (Bennett and Leitch, 1995). Wheat seeds were germinated alongside study species, and required no pre-germination treatment.
Preparation of tissues for flow cytometry followed methods described by Price and Johnston (1996b), based on the method of Galbraith et al. (1983). We prepared MOPS buffer solution containing, per litre, 4·26 g of magnesium chloride, 8·84 g of sodium citrate, 4·2 g of 3-[N-morpholino]propane sulfonic acid (MOPS), 1 mL of Triton X-100 and 1 mg of boiled RNase, then pH adjusted to 7·0–7·2. We took at least five whole seedlings for each species, or at least one field-collected leaf from at least five individuals, and chopped them in ice-cold buffer using a new scalpel blade. The resulting slurry was filtered through a 53 µm steel mesh filter, and centrifuged at 1000 g for 4 min. After centrifugation, the pellet was resuspended in 250 μL of stain solution (50 ppm propidium iodide in chopping buffer). After a second centrifugation step (1000 g for 4 min), the stain solution was removed and the pellet was resuspended in fresh stain (250 μL). Whenever samples were removed from the centrifuge (i.e. during removal of supernatant and re-suspension), they were placed immediately on ice. Samples were left in a dark refrigerator at 4 °C for 1–2 h, before being analysed using a flow cytometer.
This method was not successful for many species in Myrtaceae and Rutaceae. In these cases, the centrifuge pellet appeared as a mucilaginous matrix, possibly caused by an agglomeration of phenols, tannins or oils (B. Atwell and M. Ludwig, pers. comm.; Grattapaglia and Bradshaw, 1994). Epi-fluorescence microscopy showed a low abundance of nuclei, most enmeshed with various debris. Some limited success (with three garden-grown species from the Myrtaceae) was achieved by washing with 3 % PVPP (an absorbant) in MOPS buffer, before staining, and we include these species in our analyses. Use of seedling root, hypocotyl or leaf tissue did not result in more successful isolation of nuclei, nor did varying the amount of detergent in the chopping buffer, nor using a filter with smaller aperture (20 µm). Further variations based on the method described by Grattapaglia and Bradshaw (1994) were tried, including pulverizing tissues using dry ice and a mortar and pestle, followed by homogenization, but without success.
Relative fluorescences of diploid (2C) nucleus populations of the wheat standard and the study species were determined from simultaneous output histograms from a combined preparation (BD FACSCalibur flow cytometer with BD CellQuest software, BD Biosciences, San Jose, CA, USA). C-values were then calculated using eqn (1):
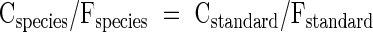 |
where Cspecies and Fspecies are the DNA content and relative fluorescence, respectively, of the subject species, and Cstandard and Fstandard are the DNA content and relative fluorescence, respectively, of the standard (Triticum aestivum ‘Chinese Spring’, 4C = 69·27 pg; Bennett and Leitch, 1995). C-values were estimated in picograms (1 pg = 10−12 g = 0·98 Gbp; Bennett et al., 2000).
Measured 2C varied 20-fold across species, from 0·82 pg (Thryptomene saxicola ‘Payne’, Myrtaceae) to 16·25 pg (Callitris glaucophylla, Cupressaceae) (Appendix). In total, we measured 2C for 41 species, 40 of which are new to the literature.
Compilation of published data
Of the 41 species for which we measured 2C, we matched 24 with LL and LMA data published by Wright et al. (2002) (Appendix). We took the average of two site-based values reported by Wright et al. (2002) for Olearia pimelioides, for each of LL and LMA. Next, we matched previously published LL and LMA data in the global synthesis by Wright et al. (2004) to species' 2C from online databases for angiosperms (Bennett and Leitch, 2003) and gymnosperms (Murray et al., 2003). LL and LMA for a number of species were reported separately for a number of sites, and we took the average of these values. We included only ‘prime’ estimates of 2C (preferred values when multiples were available for a species, as indicated by Bennett and Leitch, 2003) in our data set. When 2C were listed for multiple ploidies of a species, we only included 2C where the correct ploidy was known. In total, we matched LL and 2C data for 52 species. The 2C ranged from 0·4 pg (Betula populifolia, Betulaceae) to 92·0 pg (Trillium grandiflora, Liliaceae). Our final data set contained 2C matched to LL data for 76 species, and to LMA data for 80 species, including LMA data for four grass species, for which LL data were not available (P. Vesk, unpubl. res.).
Statistical analyses
Cross-species analyses
To investigate relationships between 2C and leaf traits across all species (and across species within groups), we utilized linear scaling relationships between mean species trait values plotted on log-scaled axes (Niklas, 1994). Bivariate trait relationships were analysed by fitting standardized major axis (SMA) lines within individual sets, with 95 % slope confidence intervals calculated according to Pitman (1939). For the purposes of these analyses, SMA estimates of the line summarizing the relationship between two variables (i.e. the main axis along which two variables are correlated) are superior to ordinary linear regression estimates because residual variance is minimized in both X and Y dimensions, rather than the Y dimension only (McArdle, 1988). All SMA relationships presented were fitted to data on log base 10 scaled axes.
To compare cross-species relationships between herbaceous/woody and angiosperm/gymnosperm groups, we first tested for differences in the slope and intercept of group SMA relationships. This was done using the program (S)MATR (Falster et al., 2003), which estimated SMA slopes common to both groups following the likelihood ratio method of Warton and Weber (2002). The calculation of common slopes also allowed us to test for elevation (intercept) differences between individual group slopes, as in standard analysis of covariance (ANCOVA). Where significant heterogeneity in group slopes could not be detected, we tested for shifts in elevation and shifts along the common SMA, as reported by Wright et al. (2001). Differences between group means for each trait were then tested for using one-sample analysis of variance (ANOVA).
Phylogenetic analyses
We used different phylogenetic analyses to determine the extent to which 2C and LL were related (a) among groups of closely related present-day species; and (b) when expressed as correlated divergences throughout the evolutionary history of the species.
First, we tested for potential scaling relationships between 2C and both LL and LMA within orders (i.e. among recently diverged species nearer the tips of the phylogenetic tree). We selected only orders containing at least five species: Asterales, Fabales, Fagales, Pinales and Proteales for LL analysis, plus Poales for LMA analysis. We tested for correlations between 2C and each of LL and LMA using Pearson correlation coefficients. Remaining within orders, we next sorted species into woody and herbaceous growth forms. We then constructed growth form contrast graphs to investigate whether comparing woody with herbaceous species within orders resulted in consistent increases in both 2C and leaf trait values.
Secondly, we conducted a correlated divergence analysis (Felsenstein, 1985; Grafen, 1989; Westoby, 1999; Wright and Cannon, 2001) on log-transformed 2C, LL and LMA trait data. Correlated divergence analysis tests whether, throughout successive radiations (nodes) in a clade of organisms, divergences in one trait are consistently co-ordinated with divergences in another. We constructed a model phylogeny for the 80 species (Fig. 1), positioning orders according to APGII (2003). Sub-ordinal species placements were then made according to current phylogenetic models for gymnosperms (Gadek et al., 2000; Rice et al., 1997; Earle, 2002), Poales (GPWG, 2001; E. Kellogg, pers. comm.), Proteales (Douglas, 1995; Wright and Westoby, 2002), Fabales (Doyle et al., 1997; Murphy et al., 2003; M. Crisp, unpubl. res.) and Asterales (Stephens, 2001 onwards; Wright and Westoby, 2002). Divergences in each trait were calculated as differences at each dichotomous node in the phylogeny using mean log-transformed trait values of the two daughter nodes (Felsenstein, 1985), treating all branch lengths as equal. Where unresolved phylogeny resulted in a polytome, we treated the node as a missing datum. However, when a dichotomous node included a polytomous descendant node, we used average log-transformed trait values of the polytome to calculate divergences in the dichotomous ancestor node. The direction of subtraction in calculating divergences is only important to the extent that it must be consistent within each trait. Reversing the direction of subtraction simply reverses the sign of the calculated divergence, producing symmetry about the origin for any two traits. For this reason, regressions on divergence data are fitted through the origin.
Fig. 1.
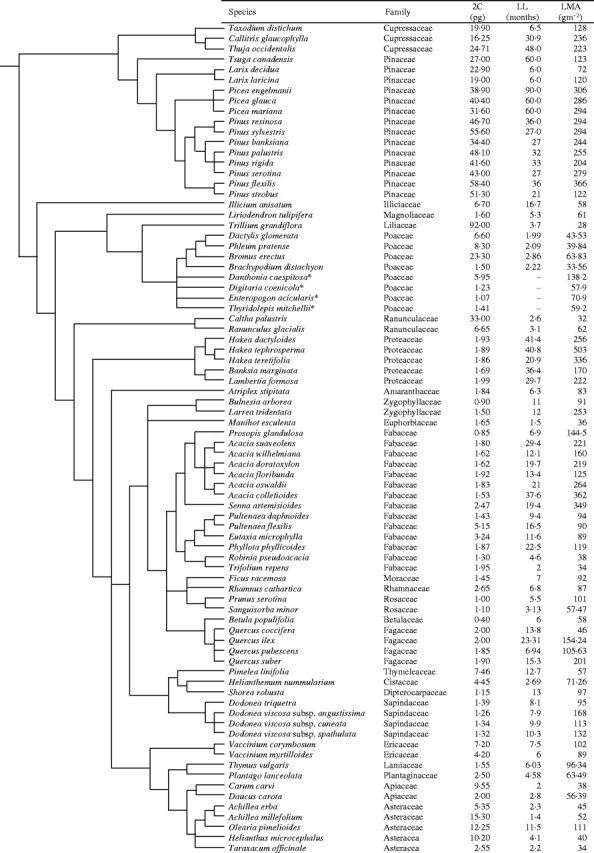
Phylogeny of the 80 study species, including 2C and leaf trait data used in the analyses.
RESULTS
One-way ANOVA confirmed that, in our data set, angiosperms (mean 2C = 5·4 pg) had significantly lower 2C than gymnosperms [mean 2C = 36·5 pg; F(1,92) = 102·01, P < 0·0005], in agreement with published results (e.g. Leitch et al., 1998; Murray et al., 1998).
Cross-species analyses
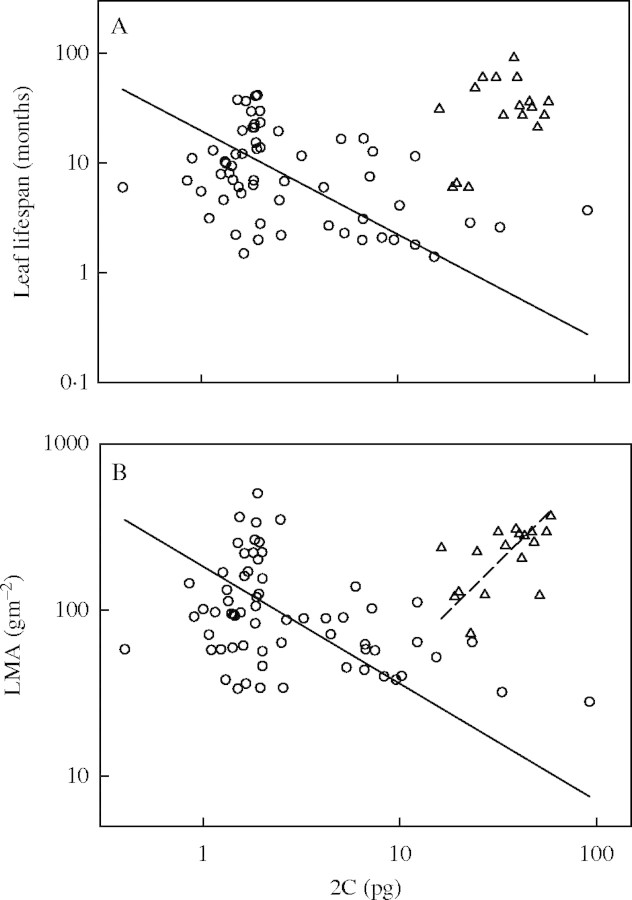
Across 76 species, there was a weak positive relationship between 2C and LL (r2 = 0·061, P = 0·032; Fig. 2A). Similarly, we found no correlation between 2C and LMA across all 80 species (n = 80, r2 = 0·03, P = 0·120; Fig. 2B).
Fig. 2.
Cross-species relationship between 2C and (A) leaf life span (LL) for 76 species, and (B) leaf mass per area (LMA) for 80 species. Triangles represent gymnosperms and circles are angiosperms. The solid line is the standardized major axis (SMA) for angiosperms; the dashed line is the SMA for gymnosperms. In (A), there was a significant positive relationship across all species (slope not shown). While there was no relationship across gymnosperm species, there was a significant negative relationship across angiosperms (the solid line is the SMA for angiosperms). In (B), there was no relationship across all species. Across gymnosperm species, there was a significant positive relationship, and across angiosperm species there was a negative relationship (the solid line is the SMA for angiosperms). All axes are log base 10 scaled.
Angiosperms/gymnosperms
Although weak, 2C was significantly negatively correlated with LL across all angiosperms (n = 59, r2 = 0·13, P = 0·005; Fig. 2A), but not across gymnosperms (n = 17, r2 = 0·19, P = 0·082). There was a weak negative correlation between 2C and LMA within angiosperms (n = 63, r2 = 0·15, P = 0·002; Fig. 2B), and a stronger, positive correlation within gymnosperms (n = 17, r2 = 0·30, P = 0·024).
Woody/herbaceous
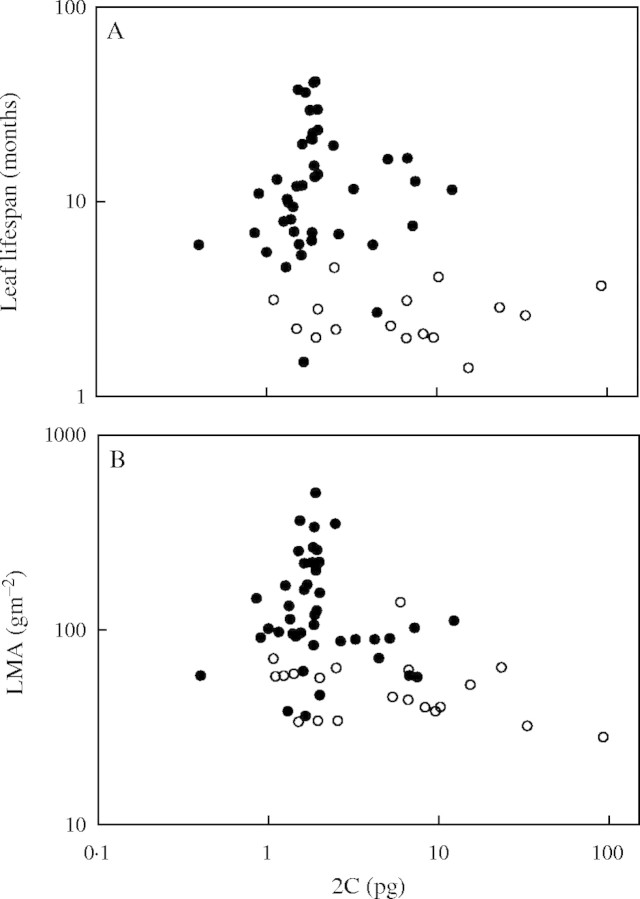
We found significant differences in 2C (t = 3·16, P = 0·004, d.f. = 23·71), LL (t = −11·12, P < 0·0005, d.f. = 55·67) and LMA (t = −7·27, P = 0·025, d.f. = 56·95) between woody and herbaceous angiosperms, using unequal variance t-tests. The negative correlations of LL and LMA with 2C within angiosperms were largely driven by these differences, since these relationships were not retained within angiosperm woodies (LL n = 42, r2 = 0·01, P = 0·624, Fig. 3A; LMA n = 42, r2 = 0·01, P = 0·509, Fig. 3B) or herbs (LL n = 15, r2 < 0·01, P = 0·823, Fig. 3A; LMA n = 19, r2 = 0·10, P = 0·182, Fig. 3B).
Fig. 3.
Scatterplots of (A) LL and (B) LMA against 2C for angiosperms grouped by growth form. Solid circles represent woody species and open circles represent herbaceous species. All axes are log base 10 scaled.
Phylogenetic analyses
Across species within orders
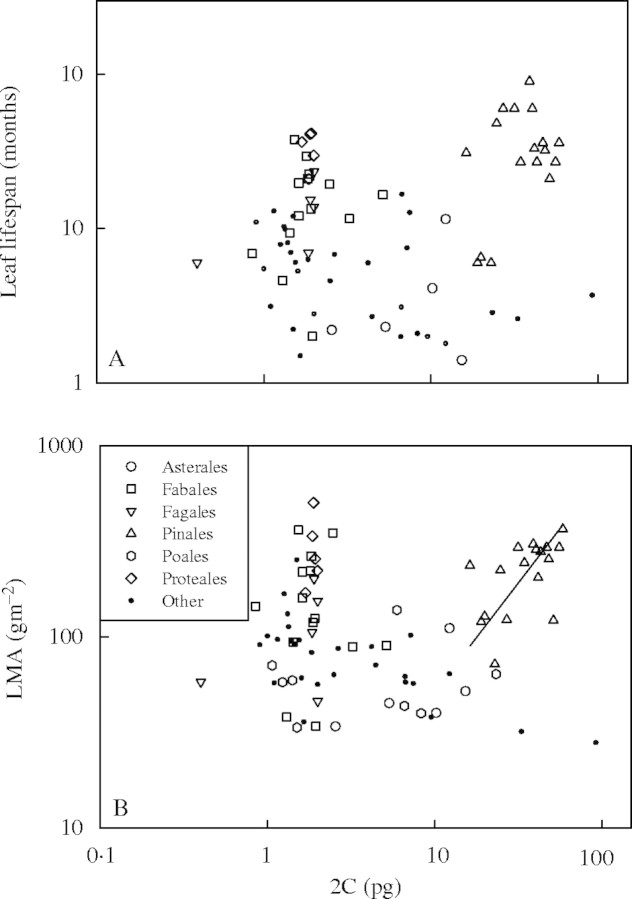
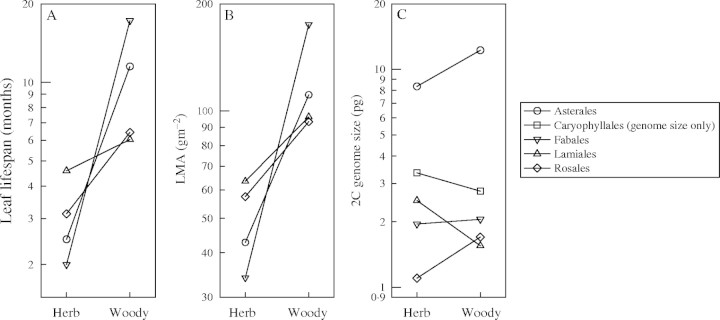
The data set contained species representing 20 seed plant orders. Within angiosperm orders containing at least five species, there was no correlation between 2C and either LL (Fig. 4A; Table 1) or LMA (Fig. 4B; Table 1). As previously reported, there was a weakly significant, positive correlation between LMA and 2C (n = 17, r2 = 0·30, P = 0·024) within the single gymnosperm order, Pinales. Within orders, woody species tended to have higher LL and LMA values than herbaceous species, as expected (Fig. 5A and B). However, woody species did not have consistently larger genomes than herbaceous species within orders (Fig. 5C).
Fig. 4.
Plots of (A) LL against 2C for 46 species in Asterales, Fabales, Fagales, Pinales and Proteales, and (B) LMA against 2C for 54 species in Asterales, Fabales, Fagales, Pinales, Poales and Proteales. There were no relationships between 2C and either LL or LMA within individual orders, except for a weakly significant, positive relationship between 2C and LMA within the Pinales. All axes are log base 10 scaled.
Table 1.
Relationships between 2C and LL and LMA within orders containing at least five species
| 2C-LL |
2C-LMA |
|||||
|---|---|---|---|---|---|---|
| Order |
n |
r2 (P) |
n |
r2 (P) |
||
| Asterales | 5 | 0·07 (0·664) | 5 | 0·36 (0·287) | ||
| Fabales | 14 | 0·04 (0·482) | 14 | <0·01 (0·827) | ||
| Fagales | 5 | 0·45 (0·214) | 5 | 0·20 (0·452) | ||
| Pinales | 17 | 0·19 (0·082) | 17 | 0·30 (0·024) | ||
| Poales | – | – | 8 | 0·01 (0·815) | ||
| Proteales | 5 | <0·01 (0·917) | 5 | 0·14 (0·533) | ||
Pearson correlation r2 values are shown for log-transformed leaf traits, with corresponding P-values in parentheses.
Fig. 5.
Within-order comparisons of (A) LL, (B) LMA and (C) 2C between herbaceous and woody growth forms. Data points represent mean trait values for all species of that growth form within any order containing both woody and herbaceous species. Sample sizes for: Asterales (nherb = 4, nwoody = 1), Caryophyllales (nherb = 3, nwoody = 8), Fabales (nherb = 1, nwoody = 13), Lamiales (nherb = 1, nwoody = 1) and Rosales (nherb = 1, nwoody = 3). Trait axes were log base 10 scaled.
Correlated divergence analysis
In total, we calculated divergences in 2C, LL and LMA for 12 gymnosperm ancestor nodes and 41 angiosperm ancestor nodes, not including the root node of the working phylogeny. Since all gymnosperms in our database were within Pinales, we constructed the working phylogeny as a single tree. However, when analysing trait divergences, we omitted the root node from our analyses, since the monophyly of gymnosperms with respect to angiosperms (and therefore the interpretation of trait divergences at the root node in our data set) remains uncertain.
In a divergence graph, a correlation among trait divergences indicates that, throughout the evolutionary history of the species, divergences in a particular trait are consistently accompanied by divergences in the other, evidence for a functional relationship between the traits (Ackerly, 1999; Westoby, 1999). Across all radiations in our model phylogeny (excluding the root node), there was no correlation between divergences in 2C and divergences in either LL (n = 53, r2 < 0·01, P = 0·964; Fig. 6A) or LMA (n = 53, r2 = 0·03, P = 0·189; Fig. 6B). Further, divergences in 2C and LL (Fig. 6A) were not co-ordinated across radiations within either angiosperms (n = 41, r2 < 0·01, P = 0·727) or gymnosperms (n = 12, r2 = 0·17, P = 0·185). Similarly, divergences in 2C and LMA (Fig. 6B) were not co-ordinated across radiations within either gymnosperms (n = 12, r2 = 0·14, P = 0·226) or angiosperms (n = 41, r2 = 0·05, P = 0·149).
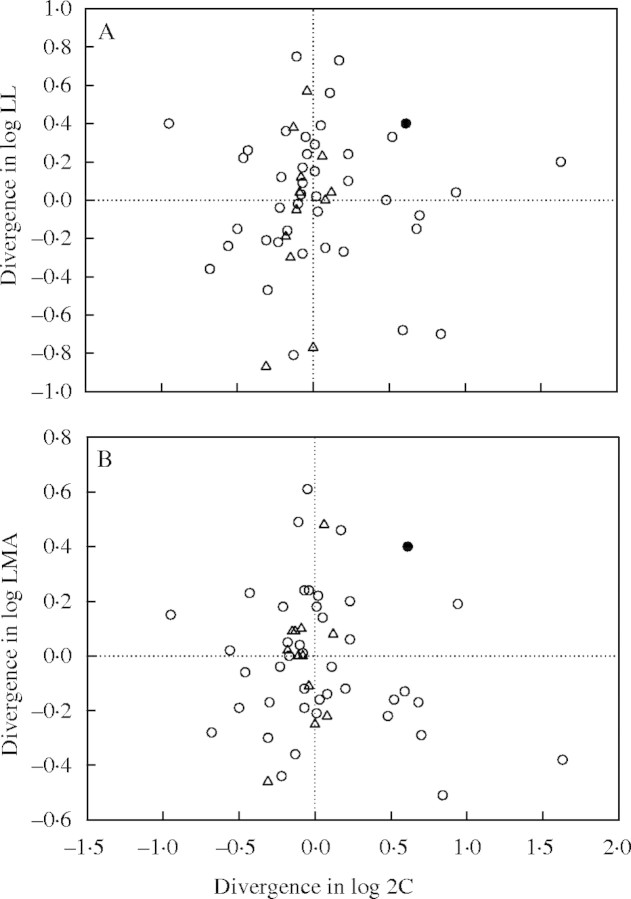
Fig. 6.
Divergences in (A) LL and 2C, and (B) LMA and 2C among radiations in the model phylogeny. Open circles represent nodes within angiosperms (n = 41), triangles nodes within gymnosperms (n = 12), and the solid circle is the root node (connecting angiosperms and gymnosperms in our model phylogeny). Prior to calculating divergences, traits were log base 10 transformed.
Divergence graphs provide further information regarding particular radiations (nodes in the phylogeny) and their effect on cross-species relationships. For example, consider a positive cross-species relationship between two traits. In a divergence graph of the two traits, nodes placed further from the origin, and around the positive 1 : 1 line (i.e. in quadrants two and four), contribute more strongly to the observed cross-species relationship (Westoby, 1999; Wright and Westoby, 2002). For divergences in LL and 2C among angiosperms (Fig. 6A), we identified two nodes in quadrant two and one in quadrant four that contributed strongly to the negative cross-species relationship between 2C and LL (Fig. 2A). These radiations occurred deep in the phylogenetic tree; between Liriodendron and the Trillium–grasses clade, between Thymus–Plantago and the Apiaceae–Asteraceae clade, and between Caltha–Ranunculus and the higher eudicots clade. Each of these radiations contributed a number of species to the cross-species relationship, resulting in a weakly negative relationship, while there was no relationship among correlated divergences. Similar divergences were also important in producing a weakly negative cross-species relationship between LMA and 2C, in the absence of a relationship among phylogenetically independent nodes (Fig. 6B). These were between Liriodendron and the Trillium–grasses clade (and also between Trillium and Poaceae), between Thymus–Plantago and the Apiaceae–Asteraceae clade, and between Caltha–Ranunculus and the higher eudicots clade. Among gymnosperms, the divergence between Larix and Picea–Pinus, at the lower left of quadrant 3 (Fig. 6B), was most important in structuring the positive cross-species relationship between 2C and LMA within gymnosperm species. The root node, representing the radiation of gymnosperms and angiosperms in our phylogeny, occurs in the upper right of Fig. 6. As such, it contributes greatly to the positive cross-species relationships between 2C and each of LL and LMA across all species (angiosperms and gymnosperms combined) in our data set (Fig. 2A and B).
DISCUSSION
New 2C-values presented in this study
We have presented new 2C estimates for 41 species of Australian plants, incorporating a range of growth forms, from grasses to evergreen woody shrubs and trees, generally from low nutrient, sclerophyllous vegetation types. The 2C-values ranged from 0·82 to 16·25 pg among these species. Compared with angiosperm trait values obtained from the literature, the species we measured had larger LL and LMA values (e.g. Fig. 2). These species persist in relatively low rainfall, low nutrient habitats, and their greater leaf trait values reflect the slow growing, evergreen growth strategies which predominate in these environments (Westoby et al., 2002; Wright and Westoby, 2002).
Only one of the species for which we measured 2C, Callitris glaucophylla, was previously recorded in the C-value database (as Callitris glauca, a synonym; Australian Plant Name Index, http://www.anbg.gov.au/cpbr/databases/apni.html, accessed June 2004). Our measurement of 16·25 pg is close to the previous determination of 16·5 pg (Bennet and Leitch, 2003). Importantly, we present species 2C from floras that are poorly represented in the current 2C literature.
The relationship between 2C and leaf traits
In our data, the weak positive relationship between 2C and LL across all 76 species (Fig. 2A) was in large part the result of a major divergence (in our model phylogeny) between angiosperms and gymnosperms in both 2C and LL (Fig. 6), and did not reflect the relationships within either group. Separate analysis of gymnosperm data showed a significant positive relationship between 2C and LMA, but not LL. This result is unexplained, possibly due to a small gymnosperm sample size. Across angiosperms, there was a weak negative relationship between 2C and both LL and LMA. The looseness of these relationships (LL r2 = 0·13, P = 0·005; LMA r2 = 0·15, P = 0·002) indicates that it would not be practical to use 2C for predicting leaf strategies of angiosperm species. Among angiosperms, the negative relationship was driven by trait differences between two life history strategies, summarized as woody and herbaceous growth forms. The negative relationship was not apparent within either herbaceous or woody species, nor between herbaceous and woody species within orders. It originated mainly at a few deep phylogenetic divergences, which then propagated into a larger number of species for cross-species analysis (Ackerly, 1999; Westoby, 1999). Correlated divergence analysis indicated no evidence for co-ordinated divergences of 2C and leaf traits throughout the model of evolutionary history of the species, and ordinal analyses indicated that the negative relationship is not reproduced nearer the tips of the phylogeny.
Data compiled by Knight et al. (2005) for a mix of seed plant species indicated a weak negative relationship between SLA (= 1/LMA) and 2C (n = 67, r2 = 0·176, P < 0·001; Knight et al., 2005). This relationship appears on inspection to be produced by the inclusion of gymnosperms with low SLA (high LMA) and greater 2C, as reported in our data.
Previous research has identified 2C as a potential indicator of plant response and growth, albeit in data sets often limited to particular sites or phylogenetic groups (but see Levin and Funderburg, 1979; Knight and Ackerly, 2002). In this study, we investigated the relationship between 2C and two leaf traits which represent a major spectrum of ecological variation among seed plants; the leaf economics spectrum (Westoby et al., 2002; Wright et al., 2004). Investigation of species data assembled from the global literature and spanning diverse evolutionary groups demonstrated that a relationship between 2C and leaf economic strategy is unlikely.
SUPPLEMENTARY INFORMATION
Supplementary information, available from the journal website (http://aob.oxfordjournals.org), provides 2C, LL and LMA data, compiled for 52 species from the literature.
APPENDIX: SPECIES FOR WHICH 2C-VALUES WERE MEASURED IN THIS STUDY
Measurements used tissue from either seedlings (‘seed’) or leaves (‘leaf’). Seedlings were germinated from seed obtained from a commercial supply company (‘commercial’), or field collections (‘field’). Healthy, fully expanded sun leaves were collected either from the field or from a garden at Macquarie University. nsp is the number of G1 phase nuclei measured to determine the relative fluorescence of the species, compared with wheat (Triticum aestivum ‘Chinese Spring’, nwheat).
| Species |
Family |
Growth type |
Material |
Origin |
nsp |
nwheat |
2C (pg) |
|---|---|---|---|---|---|---|---|
| Acacia colletioides | Fabaceae | Shrub | Seed | Commercial | 196 | 503 | 1·53 |
| Acacia doratoxylon | Fabaceae | Shrub | Seed | Commercial | 244 | 709 | 1·62 |
| Acacia floribunda | Fabaceae | Shrub | Seed | Commercial | 411 | 317 | 1·92 |
| Acacia oswaldii | Fabaceae | Shrub | Seed | Commercial | 252 | 73 | 1·83 |
| Acacia suaveolens | Fabaceae | Shrub | Seed | Commercial | 266 | 276 | 1·80 |
| Acacia wilhelmiana | Fabaceae | Shrub | Seed | Commercial | 229 | 258 | 1·62 |
| Atriplex nummularia | Amaranthaceae | Shrub | Seed | Field | 423 | 298 | 5·98 |
| Atriplex semibaccata | Amaranthaceae | Semi-shrub | Seed | Field | 548 | 630 | 1·70 |
| Atriplex stipitata | Amaranthaceae | Semi-shrub | Seed | Field | 409 | 137 | 1·84 |
| Atriplex vesicaria | Amaranthaceae | Semi-shrub | Seed | Field | 133 | 455 | 2·99 |
| Austrodanthonia caespitosa | Poaceae | Grass | Seed | Field | 177 | 158 | 5·95 |
| Banksia marginata | Proteaceae | Tree | Seed | Commercial | 418 | 194 | 1·69 |
| Callitris glaucophylla | Cupressaceae | Tree | Seed | Field | 467 | 152 | 16·25 |
| Calandrinnia polyandra | Portulacaceae | Herb | Seed | Commercial | 120 | 475 | 2·14 |
| Chenopodium desertorum | Amaranthaceae | Forb | Seed | Field | 340 | 179 | 2·20 |
| Digitaria coenicola | Poaceae | Grass | Seed | Field | 338 | 29 | 1·23 |
| Dodonea viscosa subsp. angustissima | Sapindaceae | Shrub | Seed | Field | 557 | 168 | 1·26 |
| Dodonea viscosa subsp. cuneata | Sapindaceae | Shrub | Seed | Field | 256 | 222 | 1·34 |
| Dodonea viscosa subsp. spatulata | Sapindaceae | Shrub | Seed | Field | 289 | 265 | 1·32 |
| Dodonea triquetra | Sapindaceae | Shrub | Seed | Commercial | 243 | 77 | 1·39 |
| Einadia nutans | Amaranthaceae | Forb | Seed | Field | 266 | 336 | 2·01 |
| Enteropogon acicularis | Poaceae | Grass | Seed | Field | 149 | 80 | 1·07 |
| Eucalyptus elata | Myrtaceae | Tree | Leaf | Garden | 59 | 57 | 1·22 |
| Eutaxia microphylla | Fabaceae | Semi-shrub | Seed | Commercial | 192 | 281 | 3·24 |
| Hakea dactyloides | Proteaceae | Shrub | Seed | Commercial | 625 | 180 | 1·93 |
| Hakea tephrosperma | Proteaceae | Shrub | Seed | Commercial | 661 | 284 | 1·89 |
| Hakea teretifolia | Proteaceae | Shrub | Seed | Commercial | 258 | 112 | 1·86 |
| Lambertia formosa | Proteaceae | Shrub | Seed | Commercial | 347 | 148 | 1·99 |
| Maireana decalvans | Amaranthaceae | Shrub | Seed | Field | 578 | 243 | 2·85 |
| Melaleuca quinquinervia | Myrtaceae | Tree | Leaf | Garden | 81 | 55 | 1·94 |
| Olearia pimelioides | Asteraceae | Semi-shrub | Seed | Field | 186 | 148 | 12·25 |
| Phyllota phyllicoides | Fabaceae | Shrub | Leaf | Field | 190 | 70 | 1·87 |
| Pimelea linifolia | Thymeleaceae | Shrub | Seed | Commercial | 186 | 421 | 7·46 |
| Ptilotus atriplicifolius | Amaranthaceae | Forb | Seed | Field | 207 | 88 | 6·69 |
| Pultanaea daphnoides | Fabaceae | Shrub | Seed | Commercial | 221 | 517 | 1·43 |
| Pultanaea flexilis | Fabaceae | Shrub | Seed | Commercial | 162 | 303 | 5·15 |
| Salsola kali | Amaranthaceae | Semi-shrub | Seed | Field | 86 | 241 | 1·22 |
| Sclerolaena diacantha | Amaranthaceae | Semi-shrub | Seed | Field | 253 | 291 | 2·47 |
| Senna artemisioides | Fabaceae | Shrub | Seed | Field | 231 | 93 | 2·47 |
| Thryptomene saxicola ‘Payne’ | Myrtaceae | Shrub | Leaf | Garden | 61 | 45 | 0·82 |
| Thyridolepis mitchelliana | Poaceae | Grass | Seed | Field | 173 | 134 | 1·41 |
Supplementary Material
Acknowledgments
We warmly thank Duncan Veal for the generous provision of laboratory resources, and Thusitha Gunasekera, Martin Slade and Belinda Ferrari for laboratory assistance. Martha Ludwig and Brian Atwell suggested innovations to the flow cytometric method. We are grateful to Michael Bennett, Bill Taylor and the New South Wales Department of Agriculture, who supplied various seeds as standards for flow cytometry. The authors thank Angela Moles, Dan Falster and two reviewers for their comments on a draft.
LITERATURE CITED
- Ackerly DD. 1999. Comparative plant ecology and the role of phylogenetic information. In: Press, MC, Scholes, TD, Barker, MG, eds. Physiological plant ecology. Oxford: Blackwell Science, 391–413. [Google Scholar]
- APGII. 2003. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society 141: 399–436. [Google Scholar]
- Bennett MD. 1971. The duration of meiosis. Proceedings of the Royal Society B: Biological Sciences 178: 277–299. [Google Scholar]
- Bennett MD. 1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106: 177–200. [Google Scholar]
- Bennett MD, Leitch IJ. 1995. Nuclear DNA amounts in angiosperms. Annals of Botany 76: 113–176. [Google Scholar]
- Bennett MD, Leitch IJ. 2003.Angiosperm DNA C-values database http://www.rbgkew.org.uk/cval/homepage.html. Accessed July 2003. [Google Scholar]
- Bennett MD, Bhandol P, Leitch IJ. 2000. Nuclear DNA amounts in angiosperms and their modern uses—807 new estimates. Annals of Botany 86: 859–909. [Google Scholar]
- Castro-Díez P, Puyravaud JP, Cornelissen JHC. 2000. Leaf structure and anatomy as related to leaf mass per area variation in seedlings of a wide range of woody plant species and types. Oecologia 124: 476–486. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1982. Skeletal DNA and the evolution of genome size. Annual Review of Biophysics and Bioengineering 11: 273–302. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2005. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Annals of Botany 95: 147–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot BF, Hicks DJ. 1982. The ecology of leaf life spans. Annual Review of Ecology and Systematics 13: 229–259. [Google Scholar]
- Douglas AW. 1995. Affinities. Flora of Australia 16: 6–14. [Google Scholar]
- Doyle JJ, Doyle JL, Ballenger JA, Dickson EE, Kajita T, Ohashi H. 1997. A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. American Journal of Botany 84: 541–554. [PubMed] [Google Scholar]
- Earle CJ. 2002.Gymnosperm database http://www.conifers.org/taxa.htm. Accessed July, 2003. [Google Scholar]
- Edwards GA, Endrizzi JL. 1975. Cell size, nuclear size and DNA content relationships in Gossypium. Canadian Journal of Genetics and Cytology 17: 181–186. [Google Scholar]
- Falster DS, Warton DI, Wright IJ. 2003. (S)MATR: standardised major axis tests and routines, Version 1·0 Available at http://www.bio.mq.edu.au/ecology/SMATR. [Google Scholar]
- Felsenstein J. 1985. Phylogenies and the comparative method. American Naturalist 125: 1–15. [Google Scholar]
- Gadek PA, Alpers DL, Heslewood MM, Quinn CJ. 2000. Relationships within Cuppressaceae sensu lato: a combined morphological and molecular approach. American Journal of Botany 87: 1044–1057. [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051. [DOI] [PubMed] [Google Scholar]
- Grafen A. 1989. The phylogenetic regression. Philosophical Transactions of the Royal Society B: Biological Sciences 326: 119–157. [DOI] [PubMed] [Google Scholar]
- GPWG. 2001. Phylogeny and subfamilial classification of the grasses (Poaceae). Annals of the Missouri Botanic Garden 88: 373–457. [Google Scholar]
- Grattapaglia D, Bradshaw HD. 1994. Nuclear DNA content of commercially important Eucalyptus species and hybrids. Canadian Journal of Forest Research 24: 1074–1078. [Google Scholar]
- Greilhuber J. 2005. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany 95: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP. 1998. Plant classification for ecological purposes: is there a role for genome size? Annals of Botany 82: 117–120. [Google Scholar]
- Grime JP, Mowforth MA. 1982. Variation in genome size—an ecological interpretation. Nature 299: 151–153. [Google Scholar]
- Grime JP, Thompson K, Hunt R, Hodgson JG, Cornelissen JHC, Rorison IH, et al. 1997. Integrated screening validates primary axes of specialisation in plants. Oikos 79: 259–281. [Google Scholar]
- Knight CA, Ackerly DD. 2002. Variation in nuclear DNA content across envrionmental gradients: a quantile regression analysis. Ecology Letters 5: 66–76. [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. 2005. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany 95: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence ME. 1985.Senecio L. (Asteraceae) in Australia: nuclear DNA amounts. Australian Journal of Botany 33: 221–232. [Google Scholar]
- Leishman MR, Wright IJ, Moles AT, Westoby M. 2000. The evolutionary ecology of seed size. In: Fenner M, ed. Seeds: the ecology of regeneration in plant communities. Wallingford: CABI, 31–57. [Google Scholar]
- Leitch IJ, Chase MW, Bennett MD. 1998. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Annals of Botany 82 (Suppl A): 85–94. [Google Scholar]
- Levin DA, Funderburg SW. 1979. Genome size in Angiosperms: temperate versus tropical species. American Naturalist 114: 784–795. [Google Scholar]
- McArdle BH. 1988. The structural relationship—regression in biology. Canadian Journal of Zoology 66:2329–2339. [Google Scholar]
- Murphy DJ, Miller JT, Bayer RJ, Ladiges PY. 2003. Molecular phylogeny of Acacia subgenus Phyllodineae (Mimosoideae: Leguminosae) based on DNA sequences of the internal transcribed spacer region. Australian Systematic Botany 16: 19–26. [Google Scholar]
- Murray BG. 1998. Nuclear DNA amounts in gymnosperms. Annals of Botany 82: 3–15. [Google Scholar]
- Niinemets U. 2001. Global-scale climatic controls of leaf dry mass per area, density, and thickness in trees and shrubs. Ecology 82: 453–469. [Google Scholar]
- Niklas KJ. 1994.Plant allometry: the scaling of form and process. Chicago: Chicago University Press. [Google Scholar]
- Pitman ETG. 1939. A note on normal correlation. Biometrika 31: 9–12. [Google Scholar]
- Price, HJ, Sparrow AH, Nauman AF. 1973. Correlations between nuclear volume, cell volume and DNA content in mersitematic cells of herbaceous angiosperms. Experientia 29: 1028–1029. [Google Scholar]
- Price HJ. 1976. Evolution of DNA content in higher plants. Botanical Review 42: 27–52. [Google Scholar]
- Price HJ, Johnston JS. 1996. Influence of light on DNA content of Helianthus annuus Linnaeus. Proceedings of the National Academy of Sciences of the USA 93: 11264–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price HJ, Johnston JS. 1996. Analysis of plant DNA content by Feulgen microspectrophotometry and flow cytometry. In: Jauhar PP, ed. Methods of genome analysis in plants. Boca Raton: CRC Press, 115–132. [Google Scholar]
- Ralph M. 1994.Germination of local native plant seed. Fitzroy: Murray Ralph. [Google Scholar]
- Reeves G, Francis D, Davies MS, Rogers HJ, Hodkinson TR. 1998. Genome size is negatively correlated with altitude in natural populations of Dactylis glomerata Annals of Botany 82: 99–105. [Google Scholar]
- Reich PB. 1998. Variation among plant species in leaf turnover rates and associated traits: implications for growth at all life stages. In: Lambers H, Poorter H, Van Vuuren MMI, eds. Inherent variation in plant growth. Physiological mechanisms and ecological consequences. Leiden: Backhuys Publishers, 467–487. [Google Scholar]
- Reich PB, Walters MB, Ellsworth DS. 1992. Leaf life-span in relation to leaf, plant and stand characteristics among diverse ecosystems. Ecological Monographs 62: 365–392. [Google Scholar]
- Reich PB, Walters MB, Ellsworth DS. 1997. From tropics to tundra: global convergence in plant functioning. Proceedings of the National Academy of Sciences of the USA 94: 13730–13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich PB, Walters MB, Ellsworth DS, Vose JM, Volin JC, Gresham C, et al.1998. Relationships of leaf dark respiration to leaf nitrogen, specific leaf area and leaf life-span: a test across biomes and functional groups. Oecologia 114: 471–482. [DOI] [PubMed] [Google Scholar]
- Rice KA, Donoghue MJ, Olmsted RG. 1997. Analyzing large data sets: rbcL revisited. Systematic Biology 46: 554–563. [DOI] [PubMed] [Google Scholar]
- Sims LE, Price HJ. 1985. Nuclear DNA content variation in Helianthus (Asteraceae). American Journal of Botany 72: 1213–1219. [Google Scholar]
- Stebbins GL. 1971.Chromosome evolution in higher plants. London: Edward Arnold. [Google Scholar]
- Stephens PF. 2001 onwards.Angiosperm phylogeny website http://www.mobot.org/MOBOT/research/APweb/. Accessed July 2003. [Google Scholar]
- Thompson K. 1990. Genome size, seed size and germination temperature in herbaceous angiosperms. Evolutionary Trends in Plants 4: 113–116. [Google Scholar]
- Van't Hof J, Sparrow AH. 1963. A relationship between DNA content, nuclear volume and minimum mitotic cycle time. Proceedings of the National Academy of Sciences of the USA 49: 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamiya I, Newton RJ, Johnston JS, Price HJ. 1993. Genome size and environmental factors in the genus Pinus American Journal of Botany 80: 1235–1241. [Google Scholar]
- Warton D, Weber NC. 2002. Common slope tests for bivariate errors-in-variables models. Biometrical Journal 44: 161–174. [Google Scholar]
- Westoby M. 1999. Generalization in functional plant ecology: the species sampling problem, plant ecology strategy schemes, and phylogeny. In: Pugnaire FI, Valladares F, eds. Handbook of functional ecology. New York: Marcel Dekker, 847–872. [Google Scholar]
- Westoby M, Falster D, Moles AT, Vesk P, Wright IJ. 2002. Plant ecological strategies: some leading dimensions of variation between species. Annual Review of Ecology and Systematics 33: 125–159. [Google Scholar]
- Wright IJ, Cannon K. 2001. Relationships between leaf lifespan and structural defences in a low nutrient, sclerophyll flora. Functional Ecology 15: 351–359. [Google Scholar]
- Wright IJ, Westoby M. 2002. Leaves at low versus high rainfall: coordination of structure, lifespan and physiology. New Phytologist 155: 403–416. [DOI] [PubMed] [Google Scholar]
- Wright IJ, Reich PB, Westoby M. 2001. Strategy-shifts in leaf physiology, structure and nutrient content between species of high and low rainfall, and high and low nutrient habitats. Functional Ecology 15: 423–434. [Google Scholar]
- Wright IJ, Westoby M, Reich PB. 2002. Convergence towards higher leaf mass per area in dry and nutrient-poor habitats has different consequences for leaf life span. Journal of Ecology 90: 534–543. [Google Scholar]
- Wright IJ, Reich PB, Westoby M. 2003. Least-cost input mixtures of water and nitrogen for photosynthesis. The American Naturalist 161: 98–111. [DOI] [PubMed] [Google Scholar]
- Wright IJ, Reich PB, Westoby M, Ackerly DD, Baruch Z, Bongers F, et al. 2004. The worldwide leaf economics spectrum. Nature 428: 821–827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.