A genetic screen in the green alga Chlamydomonas reinhardtii has molecularly identified >50 temperature-sensitive mutations in genes required for cell division. Most have homologs in land plants. The cyclin-dependent kinase CDKA and the plant-specific CDKB have differentiated cell cycle functions. Potential plant-kingdom-specific cell cycle regulators, and genes conserved across eukaryotes, were identified.
Abstract
Research in yeast and animals has resulted in a well-supported consensus model for eukaryotic cell cycle control. The fit of this model to early diverging eukaryotes, such as the plant kingdom, remains unclear. Using the green alga Chlamydomonas reinhardtii, we developed an efficient pipeline, incorporating robotics, semiautomated image analysis, and deep sequencing, to molecularly identify >50 genes, mostly conserved in higher plants, specifically required for cell division but not cell growth. Mutated genes include the cyclin-dependent kinases CDKA (resembling yeast and animal Cdk1) and the plant-specific CDKB. The Chlamydomonas cell cycle consists of a long G1 during which cells can grow >10-fold, followed by multiple rapid cycles of DNA replication and segregation. CDKA and CDKB execute nonoverlapping functions: CDKA promotes transition between G1 and entry into the division cycle, while CDKB is essential specifically for spindle formation and nuclear division, but not for DNA replication, once CDKA-dependent initiation has occurred. The anaphase-promoting complex is required for similar steps in the Chlamydomonas cell cycle as in Opisthokonts; however, the spindle assembly checkpoint, which targets the APC in Opisthokonts, appears severely attenuated in Chlamydomonas, based on analysis of mutants affecting microtubule function. This approach allows unbiased integration of the consensus cell cycle control model with innovations specific to the plant lineage.
INTRODUCTION
Oscillations of cyclin-dependent kinase (CDK activity), and differently phased oscillations of the anaphase-promoting complex (APC), are central to regulation of the eukaryotic cell cycle (Morgan, 2007). Newborn G1 cells have low CDK activity and high APC activity. Increased CDK activity inactivates the APC and induces DNA replication and then mitosis. In mid-mitosis, the APC gets activated, promoting abrupt cyclin degradation, mitotic exit, and entry into the following G1.
This core CDK-APC oscillator directly regulates many multiprotein complexes involved in DNA replication, spindle assembly and function, and cytokinesis. Surveillance mechanisms (“checkpoints”) impose dependency of a late event (e.g., mitosis) on successful completion of an earlier event (e.g., S-phase), stalling the oscillator until the errors are corrected (Morgan, 2007).
How Universal Is the “Universal Cell Cycle Control” Model?
The model summarized above is heavily based on results in yeast and animals (Opisthokonts). All eukaryotes, including Viridiplantae (land plants and green algae), have homologs of cyclins, CDKs, and the APC. However, these proteins act in similar but not identical ways in plants compared with Opisthokonts (Dissmeyer et al., 2009; Zhao et al., 2012). Furthermore, unbiased genetic searches in plants have revealed cell cycle control components (e.g., Siamese CDK repressors; APC regulators Uvi4 and Osd1) not found in Opisthokonts (Walker et al., 2000; Iwata et al., 2011). Thus, plants have evolved cell cycle control components not found in Opisthokonts and may use shared components differently.
Research in yeast was central to elucidating Opisthokont cell cycle control mechanisms. We have taken a parallel microbial line of attack to Viridiplantae cell cycle control using the single-celled, haploid green alga Chlamydomonas reinhardtii. Chlamydomonas has a generally plant-like genome (Merchant et al., 2007) that diverged from land plants before the series of whole genome duplications took place (Adams and Wendel, 2005), so loss-of-function mutations in single genes can have immediate strong phenotypic consequences.
The Chlamydomonas Cell Cycle
Chlamydomonas grows photosynthetically during the day and can increase cell size >10-fold without DNA replication or cell division. At night, cells undergo rapid cycles of alternating DNA replication, mitosis, and cell division, returning daughters to the normal starting size (Coleman, 1982; Craigie and Cavalier-Smith, 1982; Donnan and John, 1983; Bisova et al., 2005). Daughter cells remain within the mother cell wall after division and then hatch simultaneously as small G1 cells.
In mid-G1, when cells attain sufficient size, and after a sufficient time after the last division, cell cycle progression becomes light independent (Spudich and Sager, 1980). This transition, called “commitment,” is dependent on cell size and time since the last division (Donnan and John, 1983). MAT3 is a homolog of the retinoblastoma tumor suppressor gene (Umen and Goodenough, 2001) that couples the commitment event to cell size. MAT3 interacts genetically and physically with E2F and DP transcription factors (Fang et al., 2006; Olson et al., 2010).
Eleven candidate cell cycle control mutants were previously isolated in Chlamydomonas (Harper et al., 1995). The mutant phenotypes suggested that following commitment, independent “functional sequences” were initiated, one leading to nuclear division and another to cytokinesis. The mutated genes were not molecularly identified.
RESULTS
High-Throughput Isolation of Temperature-Sensitive Lethal Mutations
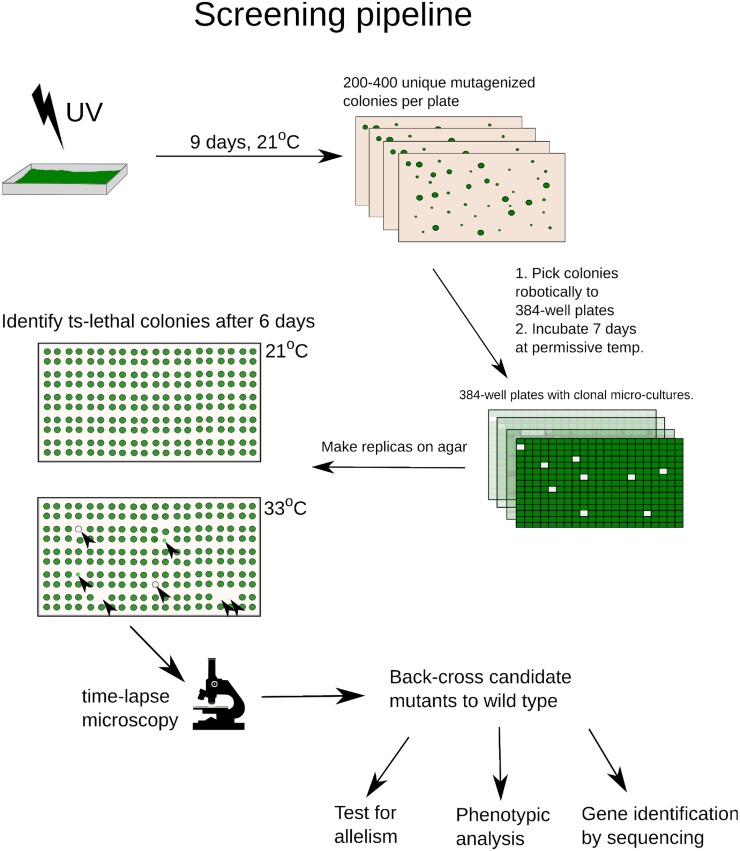
We mutagenized Chlamydomonas with UV to ∼5% survival and robotically picked mutant colonies grown at 21°C, to 384-well microplates. After growth at 21°C, two agar plate replicates were pinned (768 colonies per plate) and incubated at 21 or 33°C (permissive or restrictive temperatures; Harper, 1999). Temperature-sensitive (ts) colonies, with reduced growth at 33°C, were identified by image analysis and picked robotically for further analysis (Figure 1).
Figure 1.
Screening Pipeline.
UV-mutagenized cells were deposited on agar to form colonies and picked robotically into 384-well plates. After replica pinning, ts mutants were identified on the 33°C plate (black arrowheads) based on reduction of biomass compared with 21°C. All ts mutants were screened by time-lapse microscopy to identify potential cell cycle mutants (div/gex; see text). Candidate div and gex mutants were backcrossed to the wild-type parent and analyzed genetically and phenotypically.
[See online article for color version of this figure.]
Characterization of ts Lethal Mutants by Time-Lapse Microscopy Yielded Two Classes of Candidate Cell-Cycle-Specific Mutants
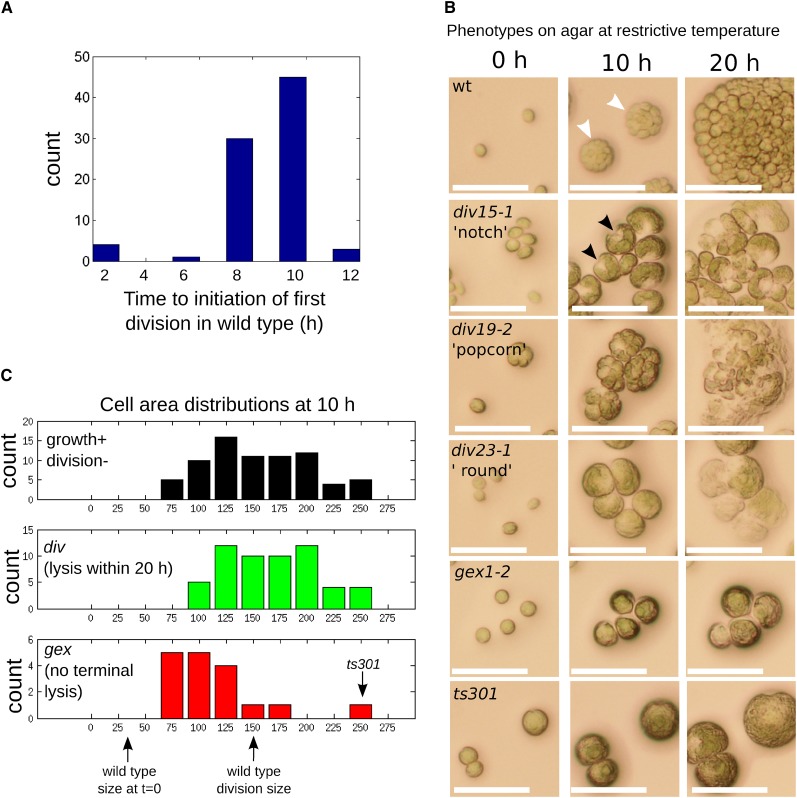
Each ts lethal likely is due to conditional inactivation of some essential gene. To identify candidates for mutations in cell cycle control genes, we employed time-lapse imaging. Cells were pregrown in liquid medium for 2 to 3 d, and agar plates spotted with aliquots in an 8 × 12 array were incubated under constant illumination at restrictive temperature. Conveniently, these conditions resulted in partial cell cycle synchronization: wild-type cells started at approximately the size of newborn cells, enlarged ∼10-fold in size over ∼8 to 10 h, then uniformly divided over the next few hours to form division clusters of 8 to 16 cells (Figures 2A and 2B). The acquired images, taken at 0, 10, 20, and 40 h after the shift to 33°C, allowed a quantitative “cell growth without division” criterion (Nurse et al., 1976), as well as assessment of morphological uniformity of arrest (Hartwell et al., 1970): two classic criteria used to specifically identify cell division cycle mutants.
Figure 2.
Characterization of div and gex Mutants by Time-Lapse Microscopy.
(A) Wild-type cells pregrown at 21°C were spotted on agar at 33°C at time t = 0 h. Most wild-type cells initiated cell division between 8 and 10 h based on the appearance of cleavage structures.
(B) Cellular morphologies of wild-type and representative mutants. Wild-type cells exhibit cleavage into multiple cells (white arrowheads) by 10 h. div mutants grow at a near-normal rate (cf. size of single undivided div mutant cells to size of wild-type cell cluster at 10 h). Many div mutants develop a characteristic notch (a triangular clear area at one point on the perimeter; black arrowheads) or popcorn (multiple aberrant septations, frequently irregular in size and shape) morphologies at 10 h, and all undergo complete cell lysis within 20 h. gex mutant cells grow more slowly and arrest before the first cell division without undergoing terminal cell lysis. The ts301 mutant had a unique phenotype of normal cell growth without any signs of initiating cell division. Bars = 50 μm.
(C) Quantification of cell areas after 10 h in all candidate (growth+ division−) mutants (top panel) and segmented into div mutants (middle panel) and gex mutants (bottom panel). Mean sizes of wild-type cells at t = 0 h and at division are indicated.
We eliminated from consideration mutants that performed multiple cell division cycles. We also eliminated mutants with severe defects in cell growth that never reached the normal wild-type division size or that grew slowly, followed by morphologically normal cell division in most cells upon attaining normal division size, since these phenotypes suggested a primary defect in cell growth rather than in cell cycle. We also excluded mutants that lost cell integrity promptly upon exposure to high temperature. These criteria eliminated ∼95% of ts lethal mutants (sample images in Supplemental Figure 1).
The remaining mutants all exhibited growth to approximately the wild-type division size by 10 h (Figure 2C). The morphology of arrested cells provided useful empirical criteria for differentiating the mutants.
One class of mutants exhibited formation of surface structures (“notch” or “popcorn”; Figure 2B) at 10 h. Similar structures were noted in Chlamydomonas blocked for DNA replication: Electron microscopy suggested these were incipient cytokinetic structures (Harper and John, 1986). Therefore, we hypothesized that these mutants initiated the cell division cycle but were defective in specific processes. This idea was supported by gene identification: For example, multiple genes required for DNA replication were in this category (see below). For unknown reasons, mutants in this class invariably underwent near-quantitative cell lysis by ∼20 h incubation, as noted previously (Harper et al., 1995).
A second, much smaller class (“round”) did not exhibit surface structures, but shared with the structure-formers the property of cell lysis by ∼20 h (Figure 2B). Characterization of DNA replication in these mutants by flow cytometry showed that all mutants in this class underwent DNA replication despite complete failure of cell division (see below; Supplemental Figure 2). These mutants therefore did initiate the cell division cycle but may have failed specifically in initiating the cytokinetic program. Gene identification supported this idea, since the mutated genes were tubulin folding chaperones required for microtubule function (see below), and microtubules are required for cytokinesis in Chlamydomonas (Ehler and Dutcher, 1998). In a div13-1 div24-1 strain doubly mutant for DNA polymerase α and the tubulin folding chaperone TFC-E (see below), the round phenotype of div24-1 was epistatic to the notch phenotype of div13-1 (Supplemental Figure 3), showing that the notch morphology required functional microtubules. Further, in one notch-forming mutant tested (div48-1), antimicrotubule staining revealed abundant microtubule structures in the vicinity of the notch (Supplemental Figure 4; see below). Therefore, we interpret these structures as incipient cytokinesis.
Thus, we interpret these mutants (notch, popcorn, and round) as initiating the cell division cycle, but failing in specific aspects of its execution. We therefore named these div mutants (“division-defective”). div mutants generally displayed cell growth rates similar to wild type (Figure 2C).
In contrast, mutants that retained cell integrity were generally slower growing than the div mutants (with one clear exception; see below). We tested many for initiation of DNA replication, and all were negative. Since these mutants fail to exhibit any sign of cell cycle initiation (no DNA replication and no candidate incipient cytokinetic structures), we call them gex (“G1 exit-defective”; Figures 2B and 2C).
Phenotypic classification of most mutants into these classes was quite clear; we do note that classification as notch or popcorn was sometimes ambiguous, and both the degree of cell growth defect (Figure 2C) and speed and extent of cell lysis were somewhat variable among the mutants. Therefore, for mutants lacking further phenotypic classification, the div/gex differentiation should be considered preliminary. It is a clear distinction for all mutants discussed in detail below.
The DNA Replication Cycle and CDK Activation in the Wild Type and in gex/div Mutants
We assessed DNA replication at the restrictive temperature by fluorescence-activated cell sorting (FACS). In addition, as a biochemical indicator of cell cycle progression, we measured Cks1-precipitable kinase activity. Cks1 is a conserved protein that can be used as an affinity reagent for active cyclin-CDK complexes in many organisms, including Chlamydomonas (Bisova et al., 2005). We used recombinant Chlamydomonas Cks1 on magnetic beads to purify kinase activity toward the canonical CDK substrate histone H1 from whole-cell lysates. Chlamydomonas, like all Viridiplantae, has two CDKs suspected of involvement in cell cycle control, CDKA and the plant-specific CDKB (Porceddu et al., 2001; Vandepoele et al., 2002; De Veylder et al., 2007) (see below). Cks1 precipitates active cyclin-bound forms of both (De Veylder et al., 1997; Corellou et al., 2005), thus providing a profile of global CDK activity.
Cells were partially presynchronized by growth in 14:10 light-dark cycles at 21°C. At the conclusion of the dark periods, most cells were small and recently hatched, with G1 DNA content. At t = 0, cultures were transferred to 33°C in light for 14 h, then kept at 33°C in the dark for the duration of the experiment.
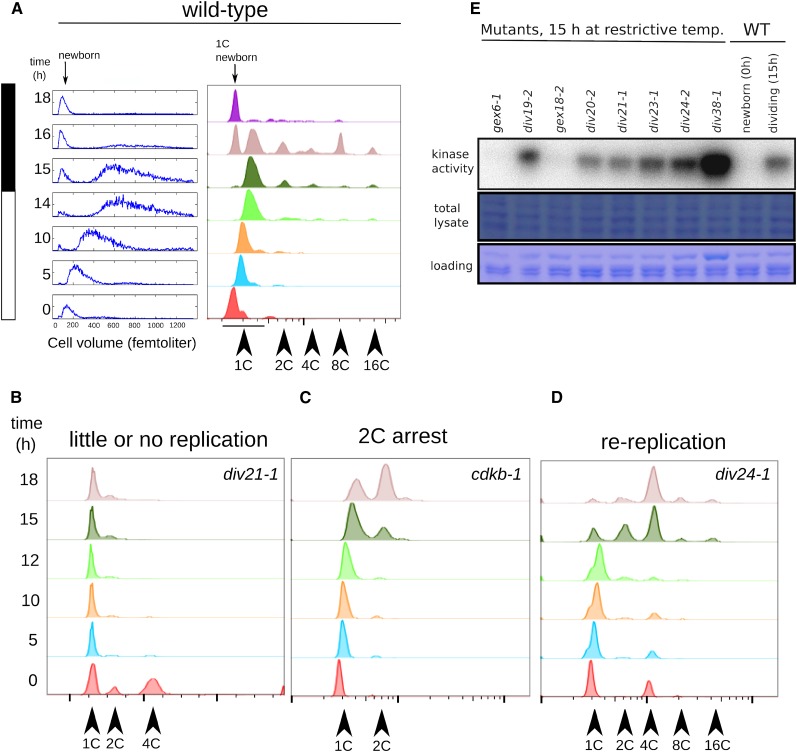
Under these conditions, wild-type cells uniformly increased in cell volume over 14 h from a newborn median size of ∼100 fL to a predivision size of ∼600 fL without initiating DNA replication or division (Figure 3A). (For unknown reasons, cell cycle progression was significantly slower in liquid medium than in the agar plate assay.) Between 14 and 16 h, most cells initiated replication as seen by the appearance of higher order peaks by FACS analysis (Figure 3A). The complex FACS profile at these times represents cells that have undergone 0 to 4 cell cycles. Since the daughters remain inside the mother cell until they hatch, these clusters appear in the FACS profile as 1, 2, 4, 8, and 16C peaks. By 18 h, most cells have completed division and hatched. This is evident from the return to a uniformly 1C population by FACS and a drop in cell size back to the newborn cell size of 100 fL (Figure 3A). The newborn 1C cells had a reproducibly lower FACS signal than the large G1 cells about to enter DNA replication, perhaps due to chloroplast DNA accumulation (Supplemental Figure 5). This shift allows us to distinguish between early and late G1 cells. Synchrony is partial, and accumulation of hatched newborn cells overlaps with entry of late G1 cells into replication. Cell counts and conversion to the newborn FACS signal indicate that DNA replication and cell division are near-quantitative for the wild type. As reported (Bisova et al., 2005), Cks1-precipitable kinase activity rose from a negligible level in newborn cells to a high level around the time of division, then dropped concomitantly with the release of daughter cells (Figure 5C).
Figure 3.
Cell Cycle Analysis of div and gex Mutants.
(A) Cell growth (Coulter counter, left) and DNA content (FACS, right) in a partially synchronized wild-type culture. Cells increase in volume over 14 h, at which point cell division begins as seen by reappearance of small daughter cells (newborn). Initiation of DNA replication occurs at around 14 h. Higher order FACS peaks at the 16-h time point (4C, 8C, and 16C) represent cells undergoing division. Daughter cell release is seen by FACS as a return to a predominantly 1C population at 18 h.
(B) to (D) Representative examples of div mutant replication phenotypes.
(B) Arrest with unreplicated or partially replicated DNA.
(C) Initiation of DNA replication with normal timing by 15 h, followed by arrest with once-replicated DNA in most cells.
(D) Initiation of DNA replication with normal timing, followed by reinitiation and arrest with FACS peaks representing rereplicated DNA (4C, 8C, and 16C peaks). Supplemental Table 1 shows replication phenotypes of all div and gex mutants tested.
(E) Cks1-precipitable kinase activity in wild-type and mutant lysates was measured against histone H1. In all cases tested, gex mutants arrested with background levels, similar to newborn wild-type cells. div mutants arrested with Cks1-kinase levels comparable to or higher than wild-type S/M-phase cells. The extremely high activity in div38-1 might be due to overloading of the kinase reaction (see loading control image).
[See online article for color version of this figure.]
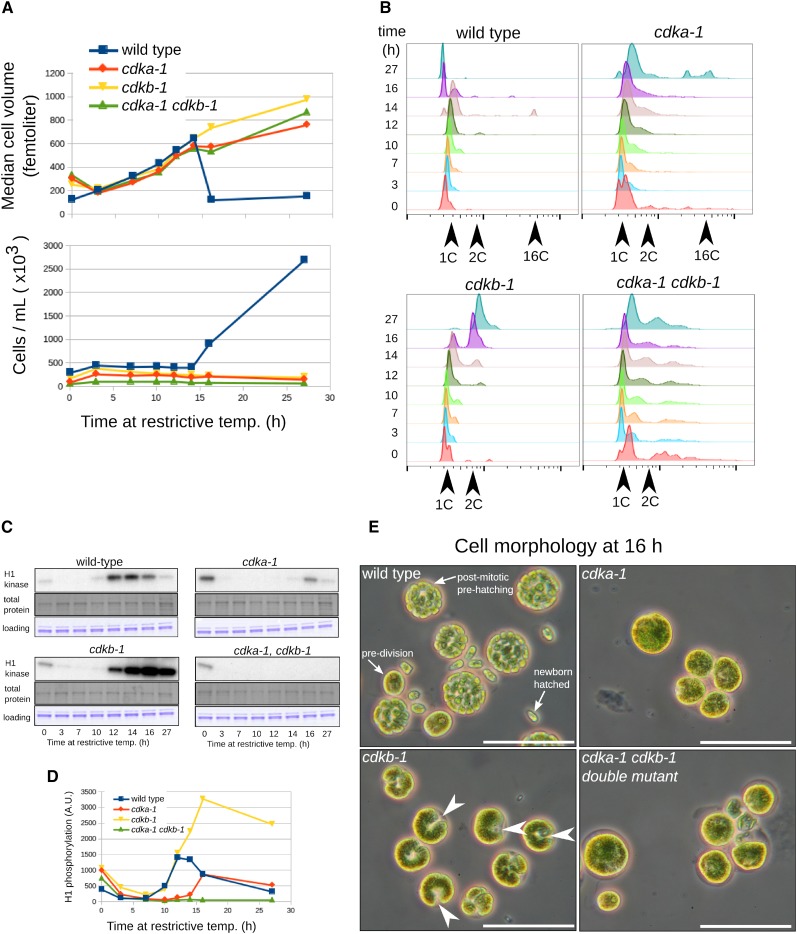
Figure 5.
Differential Functions of CDKA and CDKB.
(A) Cell volume and number (Coulter counter). Near-identical growth rate is observed in all strains during the first 14 h. Wild-type cells release progeny daughter cells between 14 and 16 h as seen by a sharp drop in median cell volume and a corresponding increase in cell number. Mutant cells grow normally but do not undergo division.
(B) Analysis of DNA replication by FACS. Wild-type cells begin replication around 12 to 14 h. cdka-1 cells do not replicate DNA for at least 16 h, though some replication is evident at 27 h. cdkb-1 cells initiate DNA replication approximately on schedule and arrest after one round of replication. The cdka-1 cdkb-1 double mutant has a complex FACS profile, probably due to slow exit from the previous mitosis; there is little or no evidence for DNA replication occurring at restrictive temperature.
(C) and (D) Cks1-precipitable kinase activity. The activity observed at t = 0 probably represents cells that have not yet exited the previous mitosis.
(E) Phase microscopy at 16 h. Wild-type cells at various stages of multiple fission are observed: large predivision cells, dividing prehatching cells, and small newborn G1-cells. cdka-1 and cdka-1 cdkb-1 double mutant cells are large, with no evidence of cytokinetic initiation; cdkb-1 cells have developed a uniform notch morphology suggesting cytokinetic initiation (white arrowheads). Bars = 50 μm.
Cell counts and cell size measurements indicated that none of the mutants divided at the restrictive temperature, but continued to grow throughout the time course, as expected from time-lapse microscopy (see above). Also as expected, the gex mutants generally showed a reduced growth rate compared with the wild type, while the div mutants had a growth rate comparable to the wild type over 14 h.
None of the gex mutants tested showed any DNA replication over the course of the experiment. Some div mutants also did not replicate at all. A few replicated once, and others reinitiated DNA replication several times and arrested with FACS peaks at 4C or greater (Figures 3B to 3D; Supplemental Table 1).
Cks1-precipitable kinase activity in cell lysates from several arrested gex mutants was close to background, whereas all div mutants tested arrested with activity at least as high as peak wild-type levels (Figure 3E).
Identification of Causative Mutations
In our collection of ∼200 primary mutants, in all cases except one (ts301, described below), tetrad analysis indicated that the ts mutation segregated as a single locus. We established a combined linkage/complementation test (Methods; Supplemental Figure 6) that yielded 29 complementation groups with more than one member and another ∼30 single-allele complementation groups.
We backcrossed each mutant to the wild type and pooled ∼10 mutant segregants. DNA was extracted from the mutant pool and sequenced at ∼50× coverage (Illumina; see Methods). Reads from such a pool should give uniformly mutant reads only for single nucleotide polymorphisms (SNPs) linked to the causative mutation.
We developed a computational pipeline for detection of uniformly mutant SNPs, with low estimated false-positive and false-negative error rates. In most cases, we detected multiple linked SNPs due to the high density of mutations introduced by UV. Approximately 10% of these SNPs resulted in predicted changes in protein coding sequences based on the genome assembly (Phytozome v.10) (Merchant et al., 2007). Where possible, selection of the most likely causative SNP was based on isolation of multiple independent alleles hitting the same gene model or on the observation that intragenic revertants coreverted a SNP (see Methods). In other cases, we implemented a Bayesian calculation, based on the observation that nearly all definitive mutations (those identified by multiple alleles and/or by reversion analysis) were found to be strongly nonconservative mutations, in conserved positions based on BLAST comparison to Arabidopsis thaliana, while irrelevant passenger mutations generally lacked these attributes. In many cases, this classifier allowed high-probability identification of the causative mutation among several candidates. Linkage analysis mapping the mutations relative to each other (Figure 4; Supplemental Data Set 1) and to known markers was incorporated into this calculation, improving confidence in the assignments. These computational procedures are outlined in Methods. Table 1 and Figure 4 summarize the map locations of the DIV and GEX genes, the most likely gene model harboring the div and gex mutations, and the evidence for this assignment. Complete data for all linked homogeneous functional SNPs, including calculated Bayesian probability, are in Supplemental Data Sets 1 to 3. In some cases, there were multiple candidate targets, none of which attained plausible Bayesian probability; in other cases, there was no candidate at all, indicating the relevant mutation was not readily identifiable by these methods. Such cases are reported in Table 1 and Figure 4 only if tetrad analysis was sufficient to establish a single-gene mutation with an approximate map location, as indicated.
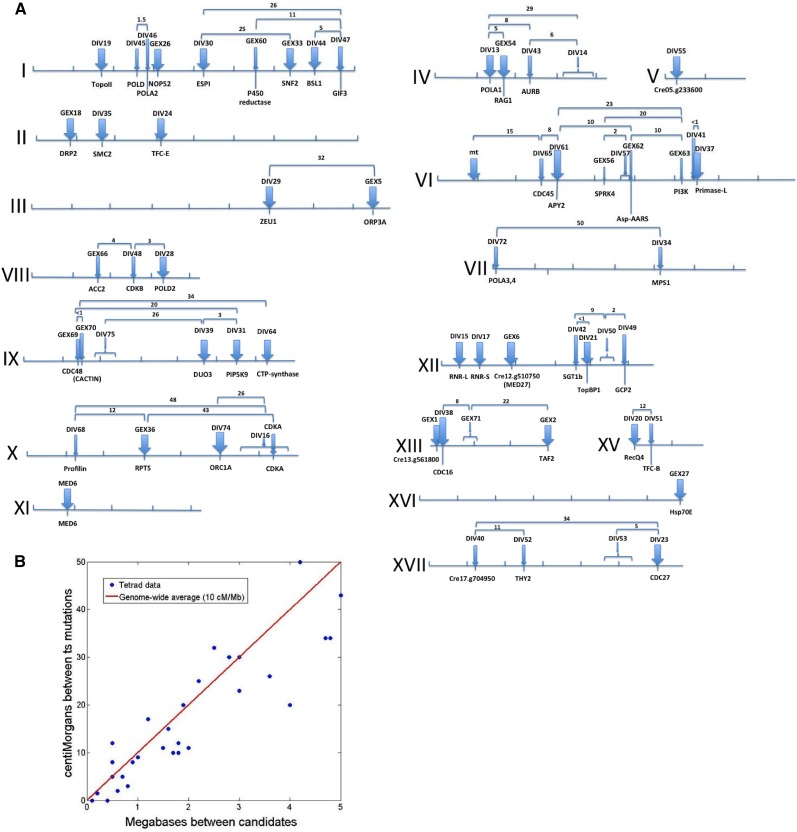
Figure 4.
Genomic Positions of DIV and GEX Genes.
(A) Maps of chromosomes I-XVII. Tick marks, megabases (Mb); thick blue arrows, definitively identified genes (multiple alleles or coreversion; see text); thin blue arrows, most likely identified genes (Tables 1; Supplemental Data Set 1); numbers, measured distance (centimorgans) between the indicated mutations from tetrad analysis (20 to 100 tetrads/cross).
(B) Correlation between physical positions of candidate SNPs and genetic distance (centimorgans [cM]) between mutant loci (data from [A], excluding crosses with predicted distance >4 Mb due to expected higher errors and deviation from linearity at large distances). The genome-wide average of 10 cM/Mb (red line; Merchant et al., 2007) explains 92% of the variance, and the residuals are symmetrical about zero (−1.6 ± 4.7 cM). This close correlation supports the assignments of causative mutations.
[See online article for color version of this figure.]
Table 1. DIV and GEX Gene Map Positions and Best Candidate Genes.
| Genea | Positionb | Cre Locusc | Supportd | No. of Allelese | Ath Best HSPf | Gene Nameg |
|---|---|---|---|---|---|---|
| CDKA | X 6.4 | Cre10.g465900 | L,P | 1 | AT3G48750.1 | CDKA;1 |
| DIV13 | IV 1.4 | Cre04.g214350 | M,L | 3 | AT5G67100.1 | POLA1/ICU2 |
| DIV14 | IV 3.5-4.0 | None | NA | 3 | NA | NA |
| DIV15 | XII 0.1 | Cre12.g492950 | M | 3 | AT2G21790.1 | RNR1 |
| DIV16 | X 5.5-6.5 | None | NA | 4 | NA | NA |
| DIV17 | XII 0.1 | Cre12.g491050 | M | 2 | AT3G23580.1 | RNR2 |
| DIV19 | I 1.7 | Cre01.g009250 | M | 4 | AT3G23890.1 | TOPII |
| DIV20 | XV 0.0 | Cre15.g634701 | M,L | 2 | AT1G27880.1 | RECQ2 |
| DIV21 | XII 3.9 | Cre12.g515900 | M,L | 2 | AT1G77320.1 | TOPBP1 |
| DIV23 | XVII 6.0 | Cre17.g740510 | M,L | 4 | AT2G20000.1 | CDC27b |
| DIV24 | II 3.4 | Cre02.g095113 | R,M | 2 | AT1G71440.1 | TFC-E |
| DIV28 | VIII 3.3 | Cre08.g374050 | M,L | 3 | AT2G42120.2 | POLD2 |
| DIV29 | III 6.0 | Cre03.g190800 | M,L | 5 | AT5G59440.1 | ZEU1 |
| DIV30 | I 4.3 | Cre01.g029200 | M,L,P | 2 | AT4G22970.1 | ESP1 |
| DIV31 | IX 5.5 | Cre09.g400478 | M,L | 2 | AT1G10900.1 | PI4P-5K |
| DIV34 | VII 4.3 | Cre07.g341700 | L,P | 1 | AT1G77720.1 | MPS1 |
| DIV35 | II 1.8 | Cre02.g086650 | L,R | 1 | AT5G62410.1 | SMC2 |
| DIV37 | VI 6.5 | Cre06.g293000 | M,L | 2 | AT1G67320.1 | Primase-L |
| DIV38 | XIII 0.1 | Cre13.g562950 | L,R,P | 1 | AT1G78770.1 | APC6 |
| DIV39 | IX 4.6 | Cre09.g396661 | R,L,P | 1 | AT1G64570.1 | DUO3 |
| DIV40 | XVII 1.2 | (No clear candidates) | NA | 1 | NA | NA |
| DIV41 | VI 6.5 | (No clear candidates) | NA | NA | NA | NA |
| DIV42 | XII 3.4 | Cre12.g513600 | L | 1 | AT4G23570.1 | SGT1A |
| DIV43 | IV 2.3 | Cre04.g220700 | L | 1 | AT2G25880 | AURORA |
| DIV44 | I 7.0 | Cre01.g050850 | R,L | 1 | AT4G03080.1 | BSL1 |
| DIV45 | I 2.6 | Cre01.g015250 | L | 1 | AT5G63960.1 | POLD-cat |
| DIV46 | I 2.9 | Cre01.g017450 | L | 1 | AT1G67630.1 | POLA2 |
| DIV47 | I 7.6 | Cre01.g055200 | L | 1 | AT4G00850.1 | GIF3 |
| DIV48 | VIII 2.6 | Cre08.g372550 | L,P | 1 | AT2G38620.2 | CDKB1;2 |
| DIV49 | XII 4.9 | Cre12.g525500 | L | 1 | AT5G17410 | GCP2 |
| DIV50 | XII 4.4-4.6 | (No clear candidates) | NA | 1 | NA | NA |
| DIV51 | XV 0.4 | Cre15.g636300 | L | 1 | AT3G10220.1 | TFC-B |
| DIV52 | XVII 2.5 | Cre17.g715900 | L | 1 | AT4G34570.1 | THY2 |
| DIV53 | XVII 5.5-6.5 | (No clear candidates) | NA | 1 | NA | NA |
| DIV55 | V 0.3 | Cre05.g233600 | M | 2 | AT1G66750.1 | CDKD1;2 @ |
| DIV57 | VI 4.8 | (No clear candidates) | NA | NA | NA | NA |
| DIV61 | VI 3.0 | Cre06.g273500 | M,L | 2 | AT3G04080.1 | ATAPY1 |
| DIV64 | IX 6.2 | Cre09.g406050 | M,L | 2 | AT1G30820.1 | CTP Synthase |
| DIV65 | VI 2.7 | Cre06.g270250 | L | 1 | AT3G25100.1 | CDC45 |
| DIV68 | X 1.3 | Cre10.g427250 | L | 1 | AT2G19770.1 | PRF5 (profilin) |
| DIV72 | VII 0.5 | Cre07.g312350 | L | 1 | AT5G41880.1 | POLA3,POLA4 |
| DIV74 | X 5.0 | Cre10.g455600 | L,P,R | 1 | AT4G12620.1 | ORC1B |
| DIV75 | IX ∼2.1 | None | NA | NA | NA | NA |
| GEX1 | XIII 0.0 | Cre13.g561800 | M | 4 | None detected | NA |
| GEX2 | XIII 2.9 | Cre13.g583500 | L,M | 2 | AT1G73960.2 | TAF2 |
| GEX5 | III 8.4 | Cre03.g199759 | M,L | 4 | AT5G02100.1 | ORP3A |
| GEX6 | XII 1.9 | Cre12.g510750 | M | 4 | AT3G09180.1 # | MED27 |
| GEX18 | II 0.9 | Cre02.g079550 | M | 2 | AT1G59610.1 | DRP2B |
| GEX26 | I 3.1 | Cre01.g019200 | M | 2 | AT5G20600.1 | NOP52 |
| GEX27 | XVI 5.9 | Cre16.g677000 | M | 2 | AT1G79930.1 | HSP91 |
| GEX33 | I 6.5 | Cre01.g046237 | M,R,L | 2 | AT1G50410.1 | SNF2 |
| GEX36 | X 2.8 | Cre10.g439150 | R,L | 1 | AT3G05530.1 | RPT5A |
| GEX54 | IV 1.7 | Cre04.g212300 | M,L | 2 | AT5G19280.1 | RAG1/KAPP |
| GEX56 | VI 4.3 | Cre06.g278254 | L | 1 | AT3G53030.1 | SRPK4 |
| GEX60 | I 5.5 | Cre01.g039350 | L | 1 | AT4G24520.2 | P450-reductase |
| GEX62 | VI 4.9 | Cre06.g279150 | L | 1 | AT4G31180.1 | Asp-AARS |
| GEX63 | VI 6.2 | Cre06.g290500 | L | 1 | AT4G29380.1 | PI3K-p150 subunit |
| GEX66 | VIII 2.7 | Cre08.g373050 | L | 1 | AT1G36180.1 | ACC2/ACCase |
| GEX69 | IX 1.4 | Cre09.g398550 | L,P | 1 | AT3G01610.1 | CDC48C |
| GEX70 | IX 1.4 | Cre09.g398650 | L | 1 | AT1G03910.1 | CACTIN |
| GEX71 | XIII 1.0 | (No clear candidates) | NA | NA | NA | NA |
DIV/GEX gene.
Chromosomal position.
Most likely candidate gene model carrying the causative mutation (Phytozome v5.5).
Support for this causative mutation: M, multiple alleles; R, intragenic (pseudo)-reversion; L, linkage between div/gex mutants with linked candidate SNPs (Figure 4); P, cosegregation of ts lethality with the candidate SNP based on sequencing of PCR products from multiple (typically 10 to 20) segregants; NA, not applicable.
Number of alleles isolated based on complementation testing and identification of lesions in the same gene model following sequencing of bulked segregants.
Best Arabidopsis BLAST hit (locus ID: Phytozome v5.5, TAIR10). #, This homology was only detectable through the Volvox carterii homolog to Cre12.g510750 (BLAST score 230), which then detected Arabidopsis Med27 (BLAST score 45). There is no better alignment to Med27 in V. carterii or Chlamydomonas than these, and the alignments have significant mutual overlap.
Name/function of best candidate gene. @, While CDKD1;2 (probable Cdk-activating kinase) is the best Arabidopsis BLAST hit for DIV55, it is unlikely that DIV55 is a true CDKD homolog, since the homology is limited to a part of the kinase domain, and there is a much better Chlamydomonas CDKD homolog than DIV55 (Cre05.g233600; Bisova et al., 2005).
The Bayesian analysis provides identification of likely causative mutations in 21 cases (Table 1; Supplemental Data Set 3); if the Bayesian probability is accurate, then we estimate a greater than 90% chance that at least 18 of these 21 assignments are correct. (On the other hand, it is quite likely that at least one of the 21 assignments is incorrect.) All mutations discussed in detail below have >95% estimated probabilities of correct identification.
DIV Genes
Many of the div mutations fell in conserved genes likely to be involved in controlling or executing diverse cell cycle processes. Many are likely directly related to S-phase execution: deoxynucleotide triphosphate synthesis (RNR and THY2), origin activation (RecQ4 and TopBP1), priming synthesis (Pol-α, four subunits), and elongation (Pol-δ, two subunits). Others are probably involved in microtubule function (tubulin folding chaperones TFC-E and TFC-B), spindle pole organization/nucleation (GCP2), and chromosome dynamics (SMC2, ESP1, and TopoII). Others encode likely coordinators of mitotic progression, such as the APC (CDC16 and CDC27), AuroraB, MPS1, and CDKB.
Most of the DIV genes 37/45 (82%) had strong Arabidopsis BLAST homologs, compared with a random expectation of ∼50% for the annotated Chlamydomonas genome (Merchant et al., 2007) (Phytozome v.10) (P < 0.001). This is presumably due to conservation of core cell cycle functions through Viridiplantae evolution. Interestingly, BLAST searches suggest that some DIV genes might reflect Viridiplantae-specific functions, including the phosphatase BSL1 and the transcription factor GIF3.
GEX Genes
The gex mutations were found mostly in conserved genes related to diverse and generally growth-related processes: gene expression, protein folding, membranes, and lipids. These annotations support the idea that the primary defects of these mutants might be in some aspect of cell growth, consistent with their moderately reduced growth rates. Their total failure of cell division, even when they reach cell sizes above wild-type division size, could reflect some specific growth-monitoring checkpoint. Related ideas have been proposed for the relationship of cell cycle progression to growth-related processes such as ribosome biogenesis in budding yeast (Jorgensen et al., 2004); indeed, GEX26 is a homolog of the rRNA-processing protein Nop52.
Mutations in Cyclin-Dependent Kinases CDKA and CDKB
The mutant ts301 was unique in our screen in having a highly robust growth rate, unlike the bulk of the gex mutants, but failing to initiate DNA replication or cytokinesis and also retaining cell integrity (Figures 2B and 2C). ts301 contained a causative mutation in the sole Chlamydomonas CDKA gene. This mutation (hereafter called cdka-1) changed Phe to Cys in the highly conserved and functionally critical DFG motif (Supplemental Figure 7 and Supplemental Data Set 4) (Endicott and Noble, 2013).
Surprisingly, tetrad analysis and sequence analysis of bulked segregant pools revealed a second causative mutation in ts301: a STOP mutation early in the coding sequence of MED6 (a component of the Mediator transcriptional activator; Malik and Roeder, 2010). The single med6-1 and cdka-1 mutants were viable at restrictive temperature but exhibited distinct phenotypes in time-lapse microscopy (Supplemental Figure 8). med6-1 mutants initiated division on time but displayed a somewhat increased frequency of division failure and altered cell morphology. The cdka-1 single mutant exhibited a long (∼10 h) division delay but ultimately went through morphologically normal division. cdka-1 med6-1 double mutants exhibited tight arrest as large undivided cells. An interesting speculation for this synthetic phenotype is that cdka-1 cells are rescued from lethality by transcription of some genes with strong Mediator dependence.
The div48-1 causative mutation was a Pro-His substitution in the APE motif in the sole Chlamydomonas CDKB gene (hereafter, cdkb-1). This motif is highly conserved (Endicott and Noble, 2013) (Supplemental Figure 7), and the mutation probably resulted in loss of CDKB function at high temperature, since the mutation was recessive (Supplemental Figure 9).
CDK Function in Chlamydomonas Cell Cycle Control
We performed synchronized time courses at restrictive temperature on wild-type, single cdka-1 and cdkb-1, and the double mutant (Figure 5). cdka-1 cells were strongly delayed both in induction of Cks1-precipitable kinase activity and DNA replication. In sharp contrast, cdkb-1 cells exhibited normal initial induction of Cks1-precipitable kinase activity; however, while this activity fell in the wild type as cells divided, in cdkb-1 cells, activity continued to rise, then remained elevated (Figure 5C). cdkb-1 cells initiated DNA replication with at most a slight delay but completed only one round of replication. Also unlike cdka-1 cells, cdkb-1 cells produced a surface notch structure (Figure 5E). The cdka-1 cdkb-1 double mutant exhibited no kinase activity, and little if any DNA replication occurred during the time course. (The latter determination was somewhat complicated by an initially complex FACS profile, presumably due to slow exit from the previous mitosis.) The double mutant exhibited no cytokinetic initiation.
Thus, delayed replication and division in the cdka-1 single mutant are dependent on CDKB; a single cycle of replication and formation of the notch in the cdkb-1 single mutant are dependent on CDKA.
These strongly contrasting phenotypes of cdka-1 and cdkb-1 indicate that CDKA functions early in initiation of DNA replication and initiation of cytokinesis. CDKB is required instead only for later events in division, including spindle formation, nuclear division, and subsequent rounds of DNA replication. CDKB is also required for inactivation of CDK activity at the end of mitosis.
Regulation of Microtubules during Mitosis
In Chlamydomonas, the flagella generally resorb prior to mitosis (Parker et al., 2010), and in mitosis, the basal bodies migrate close to the poles of the microtubules making up the mitotic spindle (O’Toole and Dutcher, 2014). Parallel cortical arrays of microtubules in interphase give way to microtubules lining the cytokinetic cleavage site (Johnson and Porter, 1968; Piasecki et al., 2008; Mittelmeier et al., 2013). We employed antimicrotubule immunofluorescence to assess the status of microtubules in arrested mutants, using deconvolution microscopy (Figure 6). (Note: In this experiment, we employed a plate synchrony method [see Methods] that gave better synchrony and more rapid entry into mitosis for unknown reasons; therefore, times in this experiment are not comparable to times in Figure 5.) Wild-type cells exhibited a peak of dividing cells around 12 h; at this time, we observed a range of microtubule morphologies, including interphase-like arrays, mitotic spindles, and telophase/postdivision rings of microtubules marking cytokinetic cleavage. Arrested cdka-1 and cdkb-1 cells both were large and entirely lacked mitotic spindles. They differed in that cdka-1 mutant cells exhibited poorly organized microtubules in roughly cortical arrays, while cdkb-1 mutant cells arrested with intense microtubule staining at the position of the notch cellular contraction, consistent with this structure representing incipient cytokinesis as suggested above (Johnson and Porter, 1968; Harper and John, 1986).
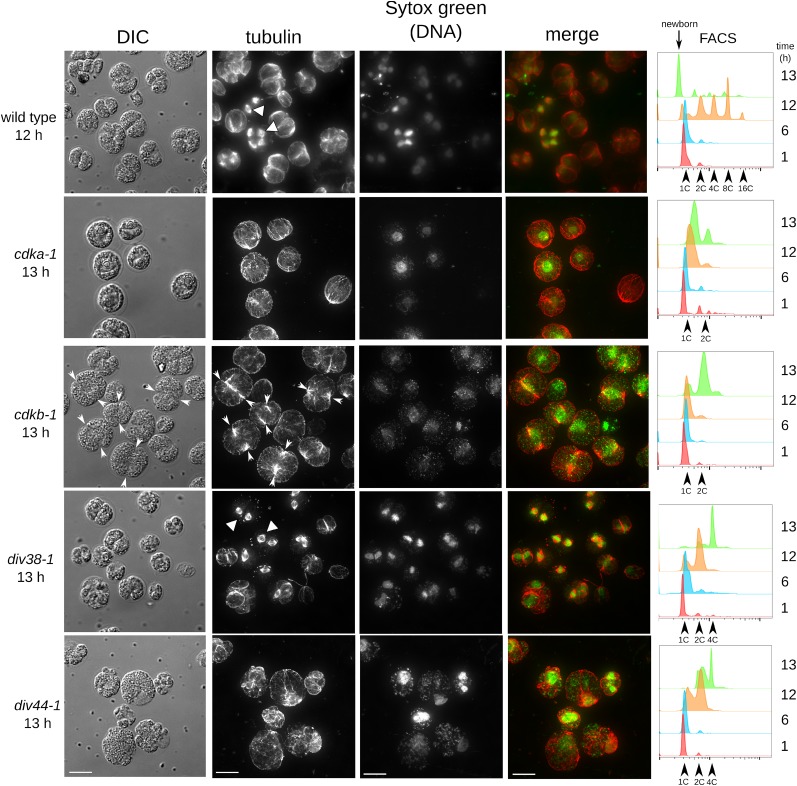
Figure 6.
Distribution of Tubulin and DNA in Arrested Mutants.
Parallel plates of wild-type and mutant strains (cdka-1, cdkb-1, div38-1, and div44-1) were shifted to 33°C at time zero. Tubulin was visualized by anti- α-tubulin indirect immunofluorescence (white triangles: mitotic spindles). DNA was stained with Sytox green. In cdkb-1, white arrowheads indicate location of the notch. DNA content was assayed at 1, 6, 12, and 13 h by FACS (right panel). Images are maximum projections of stacks of ∼30 deconvolved images. Images have been linearly modified to enhance contrast. Bars = 10 μm.
DIV38 encodes a homolog of the anaphase-promoting complex subunit CDC16. div38-1 cells frequently arrested with metaphase spindles (Figure 6), consistent with the APC requirement for anaphase in Opisthokonts. div38-1 exhibited a somewhat leaky phenotype, since most cells had apparently passed through one metaphase and then arrested as binucleate cells (frequently with both nuclei associated with a metaphase spindle). Interestingly, the apparent cleavage furrow microtubules were absent specifically from div38-1 mutant cells with spindles.
These observations suggest that CDKA activity is required for all mitotic transitions of microtubules, while CDKB is required specifically for mitotic spindle formation. APC activity may be required specifically for anaphase, after spindle morphogenesis (see below).
We noted also that nuclear DNA staining was generally much tighter and apparently more condensed in cells with mitotic spindles, whether wild-type or in the div38-1 mutant, whereas staining was more diffuse in cdka-1 and cdkb-1 cells (Figure 6). This suggests that CDKs are required for mitotic chromosome condensation in Chlamydomonas, as in Opisthokonts (Hirano, 2005).
CDKA Acts at Cell Cycle Initiation, Upstream of DIV Genes
Many DIV genes are probably required in S-phase and mitosis (Table 1). When grown at restrictive temperature on agar, div mutants frequently display apparent incipient cytokinesis (notch or popcorn phenotypes) at about the time of wild-type cell division at 10 h, followed by complete cell lysis by 20 h (see above). In contrast, cdka-1 does not start dividing until >20 h. We constructed double mutants between cdka-1 and multiple div mutants, and in all cases, we observed that the div notch or popcorn phenotype, and subsequent cell lysis, were suppressed until ∼20 h, the time when cdka-1 cells normally initiate division (Supplemental Figure 10). This observation suggests that CDKA acts upstream of these DIV genes, consistent with the idea that CDKA promotes initiation of entry into the cell division cycle; the DIV genes, in contrast, are required later, and only for specific aspects of the division program.
cdkb-1 differed from other div mutants in that the double mutant with cdka-1 never formed notch structures seen with cdkb-1 CDKA, even after very extended incubation (see above). This suggests some partial overlap of function of CDKA and CDKB for forming these structures.
In contrast, all gex mutants tested were morphologically completely epistatic to cdka-1. This could be a simple consequence of limited cell growth in the gex mutants; genetically, the result shows that the rapid and extensive cell growth in cdka-1 mutant cells is dependent on the GEX genes.
Multiple Rounds of DNA Replication in the Absence of Spindle Microtubules Suggest a Relaxed Spindle Checkpoint in Chlamydomonas
We identified several mutants that reproducibly arrested with rereplicated DNA, as determined by FACS analysis showing a significant fraction of cells with a 4C, or higher, DNA content (as a result of at least two rounds of replication). Three of these rereplication mutants, div24-1, div51-1, and div49-1, were predicted to directly impair or eliminate microtubule function. TFC-E (DIV24) and TFC-B (DIV51) are both tubulin-folding cofactors needed for the production of microtubule filaments (Szymanski, 2002). GCP2 (DIV49) is a conserved member of the γ-tubulin ring complex, which nucleates microtubules (Wiese and Zheng, 2006).
These mutants failed nuclear division and arrested with single nuclei. Mitotic spindles were undetectable in div24-1 or div49-1 mutants, consistent with the idea that absence of nuclear division in the mutants is due to failure to form a spindle (Figure 7), although some aberrant tubulin-containing structures were apparent in the mutants. DNA distribution in some div49-1 cells appeared more fragmented than in div24-1 or the wild type but without the clear signal of separated individual nuclei seen in the wild type.
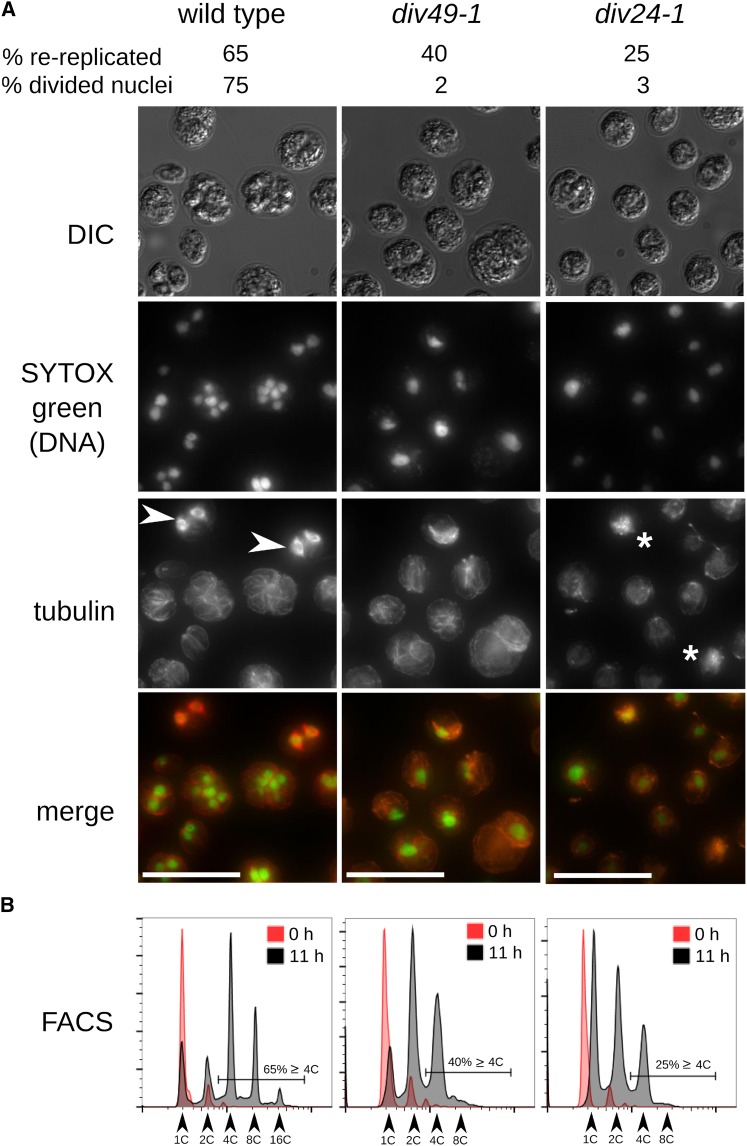
Figure 7.
Rereplication in the Absence of Functional Microtubules Suggests a Weak Spindle Checkpoint.
Parallel agar plates of the wild type, div49-1, and div24-1 were shifted to restrictive temperature at t = 0 h.
(A) Cells imaged after 11 h at restrictive temperature.
(B) DNA content analyzed by FACS at 0 and 13 h. Wild-type cells are in S/M-phase at 11 h (high-ploidy FACS peaks, evident nuclear division, and mitotic spindles [white arrowheads]). In div49-1 and div24-1, 40 and 25%, respectively, of cells have rereplicated DNA, as determined from the proportion of cells with 4C or higher DNA content. Divided nuclei were detected in 2 and 3% of these cells, respectively, and no mitotic spindles were observed. In div24-1, tubulin staining appeared abnormal and frequently aggregated into bright staining patches (white asterisks). Due to uneven SYTOX green staining, exposure times were changed between images. Bars = 25 μm.
Despite lack of nuclear division, div24-1 and div49-1 mutants underwent more than one round of DNA replication (Supplemental Figure 2A). For example, at a time point when only 2 to 3% of div24-1 and div49-1 cells had multiple nuclei, compared with 57% of the wild type, 25, 40, and 65%, respectively, had replicated DNA more than once (Figure 7). Thus, these mutants reinitiate DNA replication without intervening nuclear division.
To confirm independence of DNA rereplication from nuclear division, we treated wild-type cells with the microtubule depolymerizing drug amiprophos-methyl (James et al., 1993). DNA replication started with normal timing and progressed through multiple rounds, without detectable nuclear division. We obtained the same result with an independent wild-type strain S1D2, which is highly polymorphic compared with our standard laboratory strain (Supplemental Figure 2B).
Thus, repeated rounds of DNA replication can occur in Chlamydomonas without intervening nuclear division. In Opisthokonts, the spindle assembly checkpoint monitors kinetochore attachment of microtubules to the chromosomes (Musacchio and Salmon, 2007). Unattached kinetochores lead to inhibition of the Cdc20-APC (Peters, 2006). APC inhibition prevents dissolution of sister chromatid cohesion and independently stabilizes mitotic cyclins, so cyclin-CDK activity remains high during checkpoint-mediated cell cycle arrest. In Opisthokonts, high CDK activity prevents origin loading and, therefore, DNA replication (Blow and Dutta, 2005).
To determine if APC inactivation would block extra cycles of DNA replication in Chlamydomonas, we tested the APC mutants div38-1 and div23-1 (defective in APC subunits CDC16 and CDC27). As noted above, we observed a significantly higher proportion of metaphase cells in arrested div38-1 and div23-1 cells (37 and 29%), respectively, compared with the wild type (∼5%) (Figure 6; Supplemental Figure 11). Analysis of these mutants was somewhat complicated since their arrest was not completely tight; mononucleate and binucleate cells with and without spindles were observed. However, the proportion of 2C and 4C signal in FACS was roughly consistent with the mononucleate and binucleate proportions (Figure 6; Supplemental Figure 11), supporting the idea that APC inactivation coordinately prevented chromosome separation and DNA rereplication. Cks1-precipitable kinase activity was also high in div38-1 and div23-1 (Figure 3E), consistent with a role for the APC in turning off mitotic kinase activity.
DIV30 encodes separase. In Opisthokonts, the spindle checkpoint restrains anaphase by blocking separase activation (Musacchio and Salmon, 2007). Many div30-1 mutant cells underwent two replication cycles without nuclear division. Cytokinesis and septation nevertheless appeared to occur efficiently, resulting in approximately four cell bodies, only one of which had a nucleus (Supplemental Figure 12). Separase inactivation in budding yeast similarly blocks nuclear division, but other aspects of mitotic exit still occur, as does DNA rereplication in the next cell cycle (McGrew et al., 1992). This is presumably because separase is specifically required for sister chromatid separation but not for mitotic cyclin degradation.
In Opisthokonts, the APC is independently required for sister chromatid separation leading to anaphase, and for mitotic kinase inactivation, and is (indirectly) required for subsequent rounds of DNA replication; separase is specifically required for sister chromatid separation. Our results suggest that these APC and separase requirements are conserved in Chlamydomonas. Surprisingly, though, while preventing microtubule function in Opisthokonts results in effective APC inhibition through the spindle assembly checkpoint, this regulation apparently does not occur in Chlamydomonas, resulting in extra rounds of replication without nuclear division in cells lacking a functional mitotic spindle.
Potential Plant Kingdom-Specific Cell Cycle Regulators
In addition to the plant-specific CDKB, other mutations identify regulators either absent from Opisthokont genomes or without clear involvement in Opisthokont cell cycle control. DUO3, a homeobox-containing protein, was identified in Arabidopsis as required for pollen cell division during male gametophyte development. duo3 mutant pollen fail to execute a required second cell division, despite accumulation of cyclin B (Brownfield et al., 2009). DIV39 is the most likely Chlamydomonas DUO3 homolog. Although the homology is restricted to a few segments, the causative mutation in div39-1 (confirmed by isolation of a pseudo-revertant that restored viability; Supplemental Figure 13) alters a conserved residue within one of these segments, near the homeodomain sequence. div39-1 cells arrest with rereplicated DNA and accumulated mitotic spindles and divided nuclei (Supplemental Figure 14). Cortical microtubules normally associated with cytokinesis were seen in div39-1 cells.
DIV44 is the sole Chlamydomonas homolog of the BSU1-like (BSL) family of phosphatases involved in brassinosteroid signaling (Mora-García et al., 2004), a pathway that plays many important roles during plant growth and development. The div44-1 mutation changes a conserved residue in the conserved BSL Kelch domain and resulted in a complex arrest phenotype. Completion of the first round of DNA replication (1C to 2C) occurred with normal timing, indicating that DIV44 is dispensable for S-phase. Subsequent DNA replication was blocked or much delayed, leading to a predominantly 2C arrest with partial leakage to 4C. Apparent cytokinesis was observed in a fraction of div44-1 cells; however, we never observed mitotic spindles in arrested div44-1 cells, and distribution of DNA into the apparent daughter cells was uneven (Figure 6). Thus, DIV44 likely plays an essential if complex role in mitosis in Chlamydomonas, after completion of the first S-phase.
Additional DIV genes may operate in plant-specific aspects of cell cycle control, such as GIF3, PI4,5-kinase, apyrase APY2, profilin, and SGT1; further work is required to elucidate their function. The DIV screen is incomplete (many single-allele isolates), so it is likely that many additional plant-kingdom-specific regulators can be discovered by this approach.
DISCUSSION
CDKA Promotes Cell Cycle Initiation
CDKA appears to be specifically required at the transition from cell growth to cell cycle initiation, a natural “switch” in the Chlamydomonas life-cycle (see Introduction), as DNA replication, initiation of cytokinesis, and induction of Cks1-precipitable kinase activity were strongly delayed in the cdka-1 mutant. Cell division was not permanently blocked, however. This could reflect a truly nonessential role for CDKA; alternatively, the cdka-1 allele could have reduced function, perhaps requiring a longer time to build up enough activity to trigger cell cycle entry. In Arabidopsis, CDKA;1 is dispensable, though cdka;1 mutant cells grow very large before dividing (Nowack et al., 2012), providing an interesting parallel to our findings with the cdka-1 mutant.
CDKA-dependent initiation of the cell division cycle could be related to classical commitment, a point at which entry into the division cycle is independent of photosynthesis and biomass accumulation (Spudich and Sager, 1980). Complicating this proposal, commitment appears to be passed several hours before any evident accumulation of Cks1-precipitable kinase activity (Bisova et al., 2005). A related question involves whether CDKA is required for timely initiation of each individual division cycle within the S/M period. Intriguingly, the Rb homolog MAT3 is required in Chlamydomonas to enforce commitment (Umen and Goodenough, 2001), and Rb is thought to be the primary target of CDKA in activating the cell division cycle in Arabidopsis (Nowack et al., 2012), since Rb inactivation largely bypassed the CDKA requirement. Preliminary analysis of Chlamydomonas mat3 cdka-1 double mutants suggests a more complex picture, since the double mutant appeared phenotypically intermediate between the single mutants. This suggests that MAT3 is unlikely to be the sole functional target of CDKA in Chlamydomonas.
The early role for CDKA makes a clear prediction about epistasis between cdka-1 and div mutants blocking later steps during S/M phase: The cdka-1 mutation should delay expression of div phenotypes if they depend on the upstream CDKA-regulated step. This was indeed observed for all cdka-1 div double mutants tested. In the case of the cdka-1 cdkb-1 double mutant, this delay was extended to a near-permanent block. cdka-1 cdkb-1 double mutants arrest with a similar morphology to cdka-1 med6-1 double mutants, suggesting the speculation that CDKB (or genes involved in CDKB activation) are MED6 transcriptional targets, accounting for cell cycle arrest in the cdka-1 med6-1 double mutant.
CDKB Is Required for Mitosis after CDKA-Dependent Cell Cycle Initiation
The cdkb-1 mutant had a completely distinct phenotype from cdka-1: a single round of DNA replication, initiated at about the normal time, combined with cytokinetic initiation, followed by arrest prior to nuclear division, with no mitotic spindle, and apparently with uncondensed nuclear DNA. Cks1-precipitable CDK activity was induced normally in the cdkb-1 mutant (Figure 5C), but while this activity then fell upon mitotic exit in the wild type, it continued to increase in the cdkb-1 mutant, indicating a role for CDKB in inactivating mitotic CDK activity.
CDKA, in addition to promoting DNA replication, may prevent reloading of origins (by analogy to diverse well-established mechanisms in Opisthokonts; Blow and Dutta, 2005). If CDKB is required for inactivating CDKA, then this could explain the occurrence of only a single round of replication in the cdkb-1 mutant.
Involvement of CDKB in plant cell mitosis is a long-standing hypothesis (De Veylder et al., 2011). However, multiple paralogous copies of CDKB in higher plants have prevented a direct test, which is provided by our approach with the single-copy genome of Chlamydomonas.
Regulatory Interactions between CDKA and CDKB in the Plant Lineage
In all plant genomes, CDKA is the closest sequence relative of Opisthokont Cdc2/Cdk1 (Vandepoele et al., 2002), the main mitotic kinase in these organisms (Nurse, 1990). Our findings that CDKA is mainly required for timing of cell cycle initiation, while CDKB is directly required for mitosis, suggest a shift in CDK function in the plant lineage, such that the plant-specific B-type CDKs have taken over as the main mitotic regulators. Careful analysis of a cdka;1 null mutant in Arabidopsis led to a similar conclusion (Nowack et al., 2012). In Arabidopsis, CDKA likely activates CDKB by a transcriptional mechanism (Nowack et al., 2012). As in land plants, CDKB transcription is strongly activated in S/M cells (Bisova et al., 2005); it is unknown whether CDKA is required for this activation.
Conservation and Divergence in Genetic Control over S/M-Phase Events
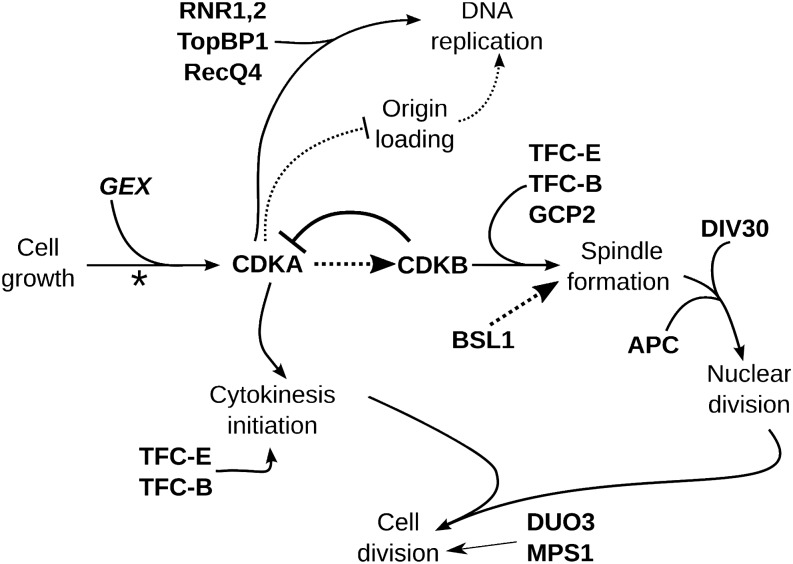
Many DIV genes have clear homologs implicated in cell cycle progression in Opisthokonts (Table 1). However, in addition to CDKA and CDKB, our results point to many other aspects of Chlamydomonas cell cycle control that diverge from the Opisthokont model, either in regulatory relationships of conserved proteins or in the evolution of novel components in the plant lineage. Figure 8 shows a scheme placing CDKA, CDKB, and other genes in an overview of Chlamydomonas cell cycle control.
Figure 8.
Chlamydomonas Cell Cycle Regulation.
In natural conditions, small newborn cells probably hatch at dawn. Photosynthesis supports severalfold (2n-fold) cell growth in G1 during the day (dependent on the GEX genes) and division occurs at night. In mid-G1, the “commitment” point marks a decision to complete cell division (asterisk). Cell division consists of multiple cycles of DNA replication, nuclear division, and cytokinesis. We hypothesize that CDKA is a key initiator of the cell division program, promoting DNA replication and initial steps in cytokinesis. Mitotic progression is then primarily driven by CDKB. CDKA may activate CDKB; CDKB in turn is required to downregulate CDK activity at the end of mitosis. These ideas, and the placement of other genes in this regulatory scheme, are explained in the text.
Mutations in RNR (ribonucleotide reductase), RECQ4, and TOPBP1 prevent DNA replication as expected from the known function of these proteins in Opisthokonts. However, these mutations do not block initiation of cytokinesis (see Results). This observation suggests surprising independence of cytokinetic initiation from the nuclear cycle, compared with the tight linkage observed in Opisthokonts, confirming conclusions of Harper and John (1986).
The Chlamydomonas APC is required at the metaphase-to-anaphase transition (nuclear division; Figure 8), as in Opisthokonts. Also as in Opisthokonts, the APC may be required for resetting DNA replication origins to allow another round of replication (Supplemental Figure 11). The phenotype of separase (DIV30) inactivation was repeated replication without nuclear division but with unimpeded cytokinesis, resulting in multiple aploid cells and a single cell with one polyploid nucleus. This phenotype is consistent with conservation of cohesin cleavage as the core separase function.
The BSL family of Kelch-domain-containing phosphatases is absent from Opisthokont genomes. BSU1 (BRI1 SUPPRESSOR1) is involved in brassinosteroid signaling in Arabidopsis (Mora-García et al., 2004). Other BSLs might have evolutionarily ancient essential functions unrelated to brassinosteroid signaling (Maselli et al., 2014); rice (Oryza sativa) homologs have been associated with a grain length quantitative trait locus (Zhang et al., 2012). In brassinosteroid signaling, the BRI1 (BRASSINOSTEROID INSENSITIVE1) receptor signals to BSU1, which regulates BIN2 (BR INSENSITIVE2); BIN2 in turn regulates transcription factors such as BZR1 (BRASSINAZOLE RESISTANT1) (Kim and Wang, 2010). BIN2 is well conserved in Chlamydomonas, but BRI and BZR1 are undetectable in Chlamydomonas by BLAST. We speculate that the ancestral function of the BSL phosphatases was to promote mitosis, perhaps through BIN2 regulation
DIV44 and CDKB are both required for efficient initiation of the first mitosis after a single round of DNA replication, since div44-1 and cdkb-1 mutant cells both arrest with 2C DNA and no spindles (Figure 6). In div44-1 cells, DNA replication initiated with essentially normal timing, whereas cdkb-1 had a small but detectable S-phase delay. div44-1 and cdkb-1 are the only mutants identified so far with this phenotype. The observed 2C arrest cannot easily be explained by a checkpoint-mediated arrest and block to rereplication, since the spindle checkpoint appears weak or nonexistent. div44-1 mutant cells are distinct from cdkb-1 cells in exhibiting partially penetrant leakage into a morphologically highly abnormal cell division (Figure 6). Further work is required to understand this phenotype.
DIV34 (MPS1) and DIV39 (DUO3) probably act late during S/M-phase; both mutants rereplicated DNA to 4C or more (Supplemental Figures 14 and 15), had apparently segregated nuclei, and appeared to have progressed at least partially through cytokinesis. The relationship of these phenotypes to mps1 phenotypes in Opisthokonts, or to duo3 phenotypes in plants, is unclear.
Weakened Checkpoint Control in Chlamydomonas
Interference with microtubule function triggers spindle assembly checkpoint-mediated cell cycle arrest in mitosis in Opisthokonts (Musacchio and Salmon, 2007). We were unable to detect an efficient spindle assembly checkpoint response in Chlamydomonas, despite the presence in the genome of clear homologs of most spindle checkpoint genes. Interestingly, a similar phenomenon of rereplication of DNA without nuclear division has been observed in Arabidopsis tissues following interference with microtubule function (Grandjean et al., 2004; Castellano and Sablowski, 2008). How Chlamydomonas normally ensures proper ordering, without effective checkpoint control, is unclear; it may be that a simple timing mechanism, as proposed for animal embryos (Murray and Kirschner, 1989) and checkpoint-deficient yeast (Li and Murray, 1991), is sufficient.
Outlook
Our approach allows unbiased identification of genes required for cell cycle progression in the plant superkingdom and provides tools (conditional alleles) for immediate functional analysis. The latter point is central. Almost all the genes identified (Table 1) are single copy in Chlamydomonas but multigene paralogous families in higher plants, so that even a simple loss-of-function analysis in higher plants can require combining many different disruption mutations in one line. These mutants should allow discovery and analysis of plant-lineage-specific innovations in cell cycle control. Innovations will likely comprise both components not found in Opisthokonts and altered network structures for shared components (Cross et al., 2011). Understanding these innovations is of considerable significance given the central importance of plant cell proliferation for maintenance of life on earth.
METHODS
Strains and Media
Chlamydomonas reinhardtii strains cc124 (mt−) and congenic ‘iso10' (mt+) (here ‘M10' and ‘P10') were provided by Susan Dutcher. Medium was prepared as described (Dutcher, 1995; Harris, 2008).
Culturing Conditions and Time-Course Experiments
Cells were cultured in liquid TAP medium or on TAP plates with 1.5% agar. For time-course experiments in liquid, cells were precultured in TAP at 21°C under 14-h/10-h light/dark cycles (50 μmol photons s−1 m−2 supplied by cool white compact fluorescent bulbs) in glass flasks. At t = 0, each culture was diluted to OD750 = 0.05 and shifted to a 33°C shaking water bath illuminated at ∼195 μmol photons s−1 m−2. This procedure resulted in partial synchronization of wild-type cells, as indicated by an enrichment of dividing cells between 14 and 16 h (light microscopy) and cells with higher order DNA content (FACS).
Division synchrony could be improved by incubation on TAP agar plates. Cells were pregrown in liquid TAP cultures to a density of 106 cells/mL, then spread evenly on agar plates, one for each time point. After 1 to 2 d of pregrowth at 21°C, the plates were shifted to 33°C at 65 μmol photons s−1 m−2. A majority of wild-type cells (∼90%) were reproducibly dividing after 11 h. Cells were harvested by washing the plate with 10 mL of prewarmed TAP medium. For unknown reasons, this procedure was especially important for APC mutants, which had a significant cell growth defect in liquid medium but not on plates.
Mutagenesis
Twenty-five milliliters of a culture at OD750 ∼0.1 was exposed to a G30T8 UV bulb for 3 to 3.5 min while shaking, then incubated at 21°C in the dark. After 12 h, aliquots of the culture were tested for viable cell number. The estimate of viable cells was based on the ability of a single cell to form a ≥8-cell microcolony on TAP agar in 20 h at 33°C. The culture was diluted so that pinning to agar with a 384-pin tool (V&P Scientific; 150 μL/spot) gave, on average, one viable cell per pin spot (∼300 colonies per plate). Colonies formed after 7 to 9 d at 21°C were picked robotically by the RapidPick colony picker (Hudson Robotics) into 384-well plates with TAP medium and incubated at 21°C for 7 d. Replica plates were made with the pinning tool and incubated for 6 d at 21°C and 3 d at 33°C. ts mutants were scored based on difference in biomass accumulation at 33°C compared with 21°C.
Time-Lapse Microscopy
ts mutants were pregrown in 96-well plates at 21°C for 2 d, and 1-μL aliquots transferred to a TAP agar plate using a 96 pin replica tool (V&P Scientific). The plate was imaged after 0, 10, 20, and 40 h at restrictive temperature on a Leica DMIRE2 microscope with a 10×/0.25-numerical aperture (NA) bright-field objective in a temperature-control chamber (33°C) and with a motorized stage for precise revisiting of positions. Images were collected with a Canon EOS Rebel T2i.
Genetic Analysis and Complementation/Recombination Testing
Genetic crosses were performed essentially as described (Dutcher, 1995). Tetrad dissection was with a Zeiss Axioskop 40 Tetrad microscope.
For the complementation/recombination test, mt− and mt+ gametes were mixed in 96-well plates, using a multi-drop plate filler (Thermo Scientific). After 1.5 h, 10 μL of each cross was spotted on agar using a 96-channel pipette (Rainin) in duplicates. One copy was incubated for 5 d at 33°C under continuous illumination and the other for 5 d at 25°C in the dark. Unmated parents were spotted at restrictive temperature as a control. Mutants were considered allelic if (1) reciprocal crosses failed to produce colonies at 33°C and (2) if zygospores were produced on the dark-plate.
Genome Resequencing
Each mutant was backcrossed to M10 or P10 and 10 to 12 ts segregants isolated, grown out at 21°C, and pooled in equal proportion in 200 mL TAP cultures. Cells were resuspended in 8 mL TEN (10 mM Tris, 10 mM EDTA, and 150 mM NaCl, pH 8) with 0.5% SDS and 0.1 mg/mL proteinase K and heated to 50°C. After 1 h, the mixture was sonicated to break open the cells and then returned to 50°C for another 1 h. The lysate was extracted twice with 16 mL phenol-chloroform and the aqueous phase precipitated with 16 mL 100% ethanol. The precipitate was rinsed with 70% ethanol and resuspended in TE (10 mM Tris and 1 mM EDTA, pH 8). NaCl was added to 1 M, and the solution phenol chloroform extracted and ethanol precipitated. The pellet was washed in 70% ethanol and resuspended in 200 μL TE. Ten micrograms of RNase A was added, 37°C for 1 h, and DNA purified with the Qiaex gel extraction kit (Qiagen). DNA was fragmented by sonication (Covaris S2 Focused Ultrasonicator) to ∼300 bp. Five micrograms of sonicated DNA was end-repaired and dA-tailed. Illumina TruSeq sequencing adapters were ligated to dA-tailed DNA. (All enzymatic reactions were with New England Biolabs kits.) Adaptor-ligated DNA (300 to 600 bp) was purified (Qiagen). DNA concentration was usually 2 to 10 ng/μL. Samples with different indices were pooled with approximately equal representation. Typically seven samples were pooled and sequenced in one lane. Sequencing, 100 bp paired-end, was performed on an Illumina HiSeq2000 instrument by Beijing Genomics Institute.
Sequence Analysis
Illumina reads were aligned to the Chlamydomonas genome (Phytozome v5.3; Merchant et al., 2007) using Bowtie 2.0.6 (Langmead and Salzberg, 2012). Sequence results per position were extracted from the resulting alignment file with mpileup (Li et al., 2009).
Identification of SNPs was based on read counts in the bulked segregant pools compared with Illumina sequence libraries from wild-type unmutagenized parents. A generalized angle (acos(S.W/sqrt(S.S * W.W)), where S and W are the read count vectors at a given position from pooled segregant and wild-type libraries, was calculated between the six-element vectors derived from read counts for each possible call (A,C,G,T,Del,Ins) from the segregant pools and from the parents. High angle (>60 degrees) marked candidate SNPs with significant read counts. We also required low coefficients of variation (i.e., consistent calls in the libraries) of the read count vectors (<1.7). These two criteria gave SNP detection with low false-positive and false-negative rates, based on synthetic data, on sequencing of independent libraries from the same or related material and on the distribution of SNPs compared with expectations from Mendelian segregation with recombination in the segregant pool.
Gene annotations (Phytozome) showed when SNPs altered predicted coding sequence (Supplemental Data Sets 1 to 3). In cases with multiple independent alleles that had cds-changing lesions in the same gene model, we assigned the causative mutations to that model. In other cases, we selected intragenic revertants (see below) and performed sequencing of PCR products (∼200 bp) covering the original SNPs. If a revertant coreverted (or pseudo-reverted) a SNP, we assigned the causative mutation to its gene model.
We used these definitive assignments, and irrelevant passenger mutations from these same libraries, to construct a training set of causative and passenger mutations. The causative mutations were significantly more likely to be deleterious (high negative Blosum62 scores) and also significantly more likely to alter a conserved residue based on BLAST alignment to Arabidopsis. (This was also true comparing the definitive mutations to random mutations generated in silico.) We generated an eight-category classification based on these factors; the likelihoods for each category for causative and passenger mutations allowed Bayesian discrimination when applied to a new set of linked mutations not in the training set. Supplemental Data Sets 2 and 3 show the Blosum and BLAST categories for definitive mutations and for all linked cds-changing SNPs in libraries without a definitive assignment. In almost all of the latter cases, it is notable that one SNP resembled the definitive class in these properties, while the rest did not. Additionally, for all nondefinitive mutations, and for many definitive mutations, we confirmed genetic location by mapping relative to other ts mutations with high-confidence assignment, and the apparent map positions were integrated into the Bayesian calculation as well.
Note that by the nature of the methodology, a region of uniform mutant SNPs detected in bulked segregants analysis strongly suggests a genomic location for the causative mutation, independent of whether one of the SNPs can be clearly assigned as causative. In such cases (Figure 4, Table 1), provided that we have independent confirmation by tetrad analysis of map location, we report the approximate map position of the causative mutation.
Isolation of Revertants
Approximately 5 × 107 cells were collected by centrifugation, plated on TAP agar, exposed to a G30T8 bulb for 2 to 3 min, and incubated in the dark for 12 h at 21°C. Revertant colonies were then selected at 33°C in the light. DNA from revertants was prepared as described (Lin et al., 2013), 1 μL was used for PCR reactions using PuReTaq beads (GE Healthcare) with gene-specific primers (Supplemental Table 2), and products were sequenced (Eurofins Genomics). PCR cycles were as follows: melting 98°C for 45 s, annealing step-down 72 to 65°C over eight cycles 30 s, then 26 cycles at 65°C 30 s, elongation 72°C for 30 s. We assayed between 3 and 20 revertants for mutation (reversion or pseudo-reversion) within a PCR fragment of ∼200 bp, centered on the original mutation. We considered that identification of any true or pseudo-revertants within this fragment provided definitive evidence for gene identification, even if other revertants lacked such changes; we assume that the other revertants that retain the original mutant sequence were intragenic but elsewhere in the sequence or else extragenic. (Initial tetrad analysis suggested that in almost every case, the revertants are intragenic since they do not recombine with the original mutation.) Chance reversion of an irrelevant SNP should have a probability many orders of magnitude lower than 0.05.
FACS and Coulter Counter Analysis
The equivalent of 10 mL culture, OD750 ∼0.05, was spun down and resuspended in 8 mL ethanol:HAc 3:1 and incubated at room temperature for 2 h. Cells were spun down, washed in 5 mL PBS, and resuspended in 2 mL PBS with RNase A (100 μg/mL), 2 h at 37°C, spun down, and resuspended in PBS + 500 nM Sytox Green (Invitrogen). FACS analysis was performed on either a BD FACSCalibur or a BD Accuri C6 instrument (BD Biosciences), with excitation by a 488-nm laser. Assignment of cell populations representing 1C, 2C, 4C, 8C, and 16C DNA content was determined based on wild-type cells. Newborn 1C cells had a lower-valued peak than 1C predivision cells and 1C cells at t = 0 h. We suspect that this reflects differences in growing conditions at 21 versus 33°C and concomitant differences in the amount of chloroplast DNA (Supplemental Figure 5). Frequently, the population of 1C cells was split into two closely spaced peaks at the beginning of the time course. The two peaks corresponded to two cell populations with different side-scatter and likely reflect different-sized daughter cells resulting from the multiple fission cell division mechanisms of Chlamydomonas.
Cell volumes and numbers were determined with 1% glutaraldehyde-fixed cells on the Z2 Beckman Coulter Counter.
Fluorescence Microscopy and Immunostaining
Indirect immunofluorescence was done as described (Mahjoub et al., 2004) except autolysin treatment was omitted and poly-l-lysine (Sigma-Aldrich) was used. The primary antibody was monoclonal anti-α-tubulin (Sigma-Aldrich; clone B512) at a 1:200 dilution in PBS. The secondary antibody was anti-mouse-Alexa 568 (Invitrogen) 1:200. Cover slips were washed three times in PBS, incubated 30 min in PBS + 500 nM Sytox Green, and then washed once in PBS. Imaging was performed on a Zeiss Axioplan 2 microscope with a Zeiss Plan Apochromat 63×/1.40-NA oil differential interference contrast objective, equipped with an AttoArc HBO 100-W mercury arc bulb. Images were collected with a Hamamatsu CCD camera. Filter cubes for FITC and CY3 were used to acquire Sytox Green and Alexa 568 fluorescence, respectively. Stacks of five to seven images were collected and merged using the Openlab acquisition software. The intensity of SYTOX green generally varied across the slide, so exposure times were changed between images. Therefore, this information was used only to determine the distribution of DNA, not to compare intensities. For deconvolution microscopy, images were collected using a DeltaVision microscope (Applied Precision/GE Healthcare Life Sciences) with an Olympus IX-70 microscope stand, a sCMOS camera, and an InsightSSI light source. A UPlanSApo 100×/1.40-NA objective was used. FITC and TRITC filter sets were used to image Sytox green and Alexa-568 fluorescence, respectively. Images were deconvolved using SoftWoRX software and are presented as maximum intensity projections of ∼30 deconvolved images.
Cks1-Precipitable Kinase Assay
Cks1 pull-down and kinase activity measurements were done essentially as described (Levine et al., 1996) using 20 mL of OD750 = 0.05 Chlamydomonas culture and Chlamydomonas Cks1 fused to glutathione S-transferase, expressed in Escherichia coli, and covalently linked to epoxy-functionalized Dynabeads (Invitrogen). Quantification was by transfer to polyvinyl difluoride membrane, exposure to phosphor screen (Molecular Dynamics), and imaging with a Biosciences Typhoon 9400 Variable Imager (Amersham).
Accession Numbers
Accession numbers (Phytozome v.10; http://phytozome.jgi.doe.gov/pz/portal.html) of Chlamydomonas genes in which we isolated mutations are found in Table 1. Arabidopsis accession numbers in Table 1 refer to numbers provided in Phytozome v.10.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ts Lethal Mutant Phenotypes Excluded from Further Characterization.
Supplemental Figure 2. DNA Rereplication Occurs in Cells Probably Lacking Normal Microtubule Function.
Supplemental Figure 3. The “Round” Phenotype of div24-1 Is Epistatic over the “Notch” Phenotype of div21-1.
Supplemental Figure 4. Distribution of DNA and Tubulin in Arrested cdkb-1 Cells.
Supplemental Figure 5. Assigning FACS Peaks.
Supplemental Figure 6. A Test for Allelism among div and gex Mutants.
Supplemental Figure 7. ts Mutations in cdka-1 and cdkb-1 Change Conserved Residues in the Activation Loop of Chlamydomonas CDKA and CDKB Genes.
Supplemental Figure 8. cdka-1 Is Synthetically Lethal with med6-1.
Supplemental Figure 9. The cdkb-1 and div44-1 Alleles Are Recessive: Doubly Heterozygous Vegetative Diploids Are Viable at High Temperature.
Supplemental Figure 10. Epistasis between cdka-1 and Various div Mutants Indicates That (1) the Notch/Popcorn Phenotype Is CDKA Dependent and (2) That CDKA and CDKB Functionally Overlap.
Supplemental Figure 11. Arrest Phenotypes of div38-1 and div23-1 Suggest a Role for the APC at the Metaphase-to-Anaphase Transition.
Supplemental Figure 12. DIV30 Is Required for Nuclear Division.
Supplemental Figure 13. Causative Mutation in div44-1 and div39-1 Confirmed by Reversion Analysis.
Supplemental Figure 14. DIV39 Acts Late in S/M Phase.
Supplemental Figure 15. DIV34 Acts Late during S/M Phase.
Supplemental Table 1. Replication Phenotypes in div and gex Mutants.
Supplemental Table 2. Primers Used in PCR Reactions.
Supplemental Data Set 1. Complete DIV/GEX Evidence Table.
Supplemental Data Set 2. Definitive DIV/GEX Detailed Information.
Supplemental Data Set 3. Nondefinitive DIV/GEX Detailed Information.
Supplemental Data Set 4. CDK Sequence Alignment.
Supplementary Material
Acknowledgments
We thank S. Dutcher and J. Umen for strains, advice, and useful discussion. We also thank K. Pecani for essential assistance with sequencing library preparation and for isolation of the div68-1 mutant as well as C. Atkins for initial work on revertant isolation and screening. We thank Alison North and The Rockefeller University Bio-Imaging Resource Center for key help with deconvolution microscopy. This work was supported by National Institutes of Health Grant 5R01GM078153-07 and by The Rockefeller University.
AUTHOR CONTRIBUTIONS
F.T. and F.R.C. designed and executed the experiments and wrote the article.
Glossary
- CDK
cyclin-dependent kinase
- APC
anaphase-promoting complex
- ts
temperature-sensitive
- FACS
fluorescence-activated cell sorting
- SNP
single nucleotide polymorphism
- NA
numerical aperture
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adams K.L., Wendel J.F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Bisova K., Krylov D.M., Umen J.G. (2005). Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol. 137: 475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J., Dutta A. (2005). Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6: 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L., Hafidh S., Durbarry A., Khatab H., Sidorova A., Doerner P., Twell D. (2009). Arabidopsis DUO POLLEN3 is a key regulator of male germline development and embryogenesis. Plant Cell 21: 1940–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano M.M., Sablowski R. (2008). Phosducin-Like Protein 3 is required for microtubule-dependent steps of cell division but not for meristem growth in Arabidopsis. Plant Cell 20: 969–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman A. (1982). The nuclear cell cycle in Chlamydomonas (Chlorophyceae). J. Phycol. 18: 192–195. [Google Scholar]
- Corellou F., Camasses A., Ligat L., Peaucellier G., Bouget F.Y. (2005). Atypical regulation of a green lineage-specific B-type cyclin-dependent kinase. Plant Physiol. 138: 1627–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Cavalier-Smith T. (1982). Cell volume and the control of the Chlamydomonas cell cycle. J. Cell Sci. 191: 173–191. [Google Scholar]
- Cross F.R., Buchler N.E., Skotheim J.M. (2011). Evolution of networks and sequences in eukaryotic cell cycle control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3532–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Inzé D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8: 655–665. [DOI] [PubMed] [Google Scholar]
- De Veylder L., Larkin J.C., Schnittger A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16: 624–634. [DOI] [PubMed] [Google Scholar]
- De Veylder L., Segers G., Glab N., Casteels P., Van Montagu M., Inzé D. (1997). The Arabidopsis Cks1At protein binds the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett. 412: 446–452. [DOI] [PubMed] [Google Scholar]
- Dissmeyer N., Weimer A.K., Pusch S., De Schutter K., Alvim Kamei C.L., Nowack M.K., Novak B., Duan G.-L., Zhu Y.-G., De Veylder L., Schnittger A. (2009). Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21: 3641–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnan L., John P.C. (1983). Cell cycle control by timer and sizer in Chlamydomonas. Nature 304: 630–633. [DOI] [PubMed] [Google Scholar]
- Dutcher S.K. (1995). Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 47: 531–540. [DOI] [PubMed] [Google Scholar]
- Ehler L.L., Dutcher S.K. (1998). Pharmacological and genetic evidence for a role of rootlet and phycoplast microtubules in the positioning and assembly of cleavage furrows in Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton 40: 193–207. [DOI] [PubMed] [Google Scholar]
- Endicott J.A., Noble M.E. (2013). Structural characterization of the cyclin-dependent protein kinase family. Biochem. Soc. Trans. 41: 1008–1016. [DOI] [PubMed] [Google Scholar]
- Fang S.-C., de los Reyes C., Umen J.G. (2006). Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean O., Vernoux T., Laufs P., Belcram K., Mizukami Y., Traas J. (2004). In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16: 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. (1999). Chlamydomonas cell cycle mutants. Int. Rev. Cytol. 189: 131–176. [Google Scholar]
- Harper J., John P. (1986). Coordination of division events in the Chlamydomonas cell cycle. Protoplasma 131: 118–130. [Google Scholar]
- Harper J., Wu L., Sakuanrungsirikul S., John P. (1995). Isolation and partial characterization of conditional cell division cycle mutants in Chlamydomonas. Protoplasma 186: 149–162. [Google Scholar]
- Harris E. (2008). The Chlamydomonas Sourcebook: Introduction into Chlamydomonas and Its Laboratory Use. (San Diego, CA: Elsevier Academic Press; ). [Google Scholar]
- Hartwell L.H., Culotti J., Reid B. (1970). Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 66: 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. (2005). Condensins: organizing and segregating the genome. Curr. Biol. 15: R265–R275. [DOI] [PubMed] [Google Scholar]
- Iwata E., Ikeda S., Matsunaga S., Kurata M., Yoshioka Y., Criqui M.-C., Genschik P., Ito M. (2011). GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 23: 4382–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S.W., Silflow C.D., Stroom P., Lefebvre P.A. (1993). A mutation in the alpha 1-tubulin gene of Chlamydomonas reinhardtii confers resistance to anti-microtubule herbicides. J. Cell Sci. 106: 209–218. [DOI] [PubMed] [Google Scholar]
- Johnson U.G., Porter K.R. (1968). Fine structure of cell division in Chlamydomonas reinhardtii. Basal bodies and microtubules. J. Cell Biol. 38: 403–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J.R., Schneper L., Broach J.R., Tyers M. (2004). A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18: 2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-W., Wang Z.-Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine K., Huang K., Cross F.R. (1996). Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol. Cell. Biol. 16: 6794–6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Murray A.W. (1991). Feedback control of mitosis in budding yeast. Cell 66: 519–531. [DOI] [PubMed] [Google Scholar]
- Lin H., Miller M.L., Granas D.M., Dutcher S.K. (2013). Whole genome sequencing identifies a deletion in protein phosphatase 2A that affects its stability and localization in Chlamydomonas reinhardtii. PLoS Genet. 9: e1003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub M.R., Qasim Rasi M., Quarmby L.M. (2004). A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol. Biol. Cell 15: 5172–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Roeder R.G. (2010). The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maselli G.A., Slamovits C.H., Bianchi J.I., Vilarrasa-Blasi J., Caño-Delgado A.I., Mora-García S. (2014). Revisiting the evolutionary history and roles of protein phosphatases with Kelch-like domains in plants. Plant Physiol. 164: 1527–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew J.T., Goetsch L., Byers B., Baum P. (1992). Requirement for ESP1 in the nuclear division of Saccharomyces cerevisiae. Mol. Biol. Cell 3: 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelmeier T.M., Thompson M.D., Öztürk E., Dieckmann C.L. (2013). Independent localization of plasma membrane and chloroplast components during eyespot assembly. Eukaryot. Cell 12: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. (2007). The Cell Cycle: Principles of Control. (London: New Science Press. [Google Scholar]
- Murray A.W., Kirschner M.W. (1989). Dominoes and clocks: the union of two views of the cell cycle. Science 246: 614–621. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. (2007). The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8: 379–393. [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Harashima H., Dissmeyer N., Zhao X., Bouyer D., Weimer A.K., De Winter F., Yang F., Schnittger A. (2012). Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 22: 1030–1040. [DOI] [PubMed] [Google Scholar]
- Nurse P. (1990). Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux P., Nasmyth K. (1976). Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146: 167–178. [DOI] [PubMed] [Google Scholar]
- Olson B.J.S.C., Oberholzer M., Li Y., Zones J.M., Kohli H.S., Bisova K., Fang S.-C., Meisenhelder J., Hunter T., Umen J.G. (2010). Regulation of the Chlamydomonas cell cycle by a stable, chromatin-associated retinoblastoma tumor suppressor complex. Plant Cell 22: 3331–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E.T., Dutcher S.K. (2014). Site-specific basal body duplication in Chlamydomonas. Cytoskeleton (Hoboken) 71: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.D.K., Hilton L.K., Diener D.R., Rasi M.Q., Mahjoub M.R., Rosenbaum J.L., Quarmby L.M. (2010). Centrioles are freed from cilia by severing prior to mitosis. Cytoskeleton (Hoboken) 67: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.-M. (2006). The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7: 644–656. [DOI] [PubMed] [Google Scholar]
- Piasecki B.P., LaVoie M., Tam L.W., Lefebvre P.A., Silflow C.D. (2008). The Uni2 phosphoprotein is a cell cycle regulated component of the basal body maturation pathway in Chlamydomonas reinhardtii. Mol. Biol. Cell 19: 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porceddu A., Stals H., Reichheld J.P., Segers G., De Veylder L., Barroco R.P., Casteels P., Van Montagu M., Inzé D., Mironov V. (2001). A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J. Biol. Chem. 276: 36354–36360. [DOI] [PubMed] [Google Scholar]
- Spudich J.L., Sager R. (1980). Regulation of the Chlamydomonas cell cycle by light and dark. J. Cell Biol. 85: 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski D. (2002). Tubulin folding cofactors: half a dozen for a dimer. Curr. Biol. 12: R767–R769. [DOI] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15: 1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.D., Oppenheimer D.G., Concienne J., Larkin J.C. (2000). SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127: 3931–3940. [DOI] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. (2006). Microtubule nucleation: gamma-tubulin and beyond. J. Cell Sci. 119: 4143–4153. [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang J., Huang J., Lan H., Wang C., Yin C., Wu Y., Tang H., Qian Q., Li J., Zhang H. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 109: 21534–21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Harashima H., Dissmeyer N., Pusch S., Weimer A.K., Bramsiepe J., Bouyer D., Rademacher S., Nowack M.K., Novak B., Sprunck S., Schnittger A. (2012). A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet. 8: e1002847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.