Abstract
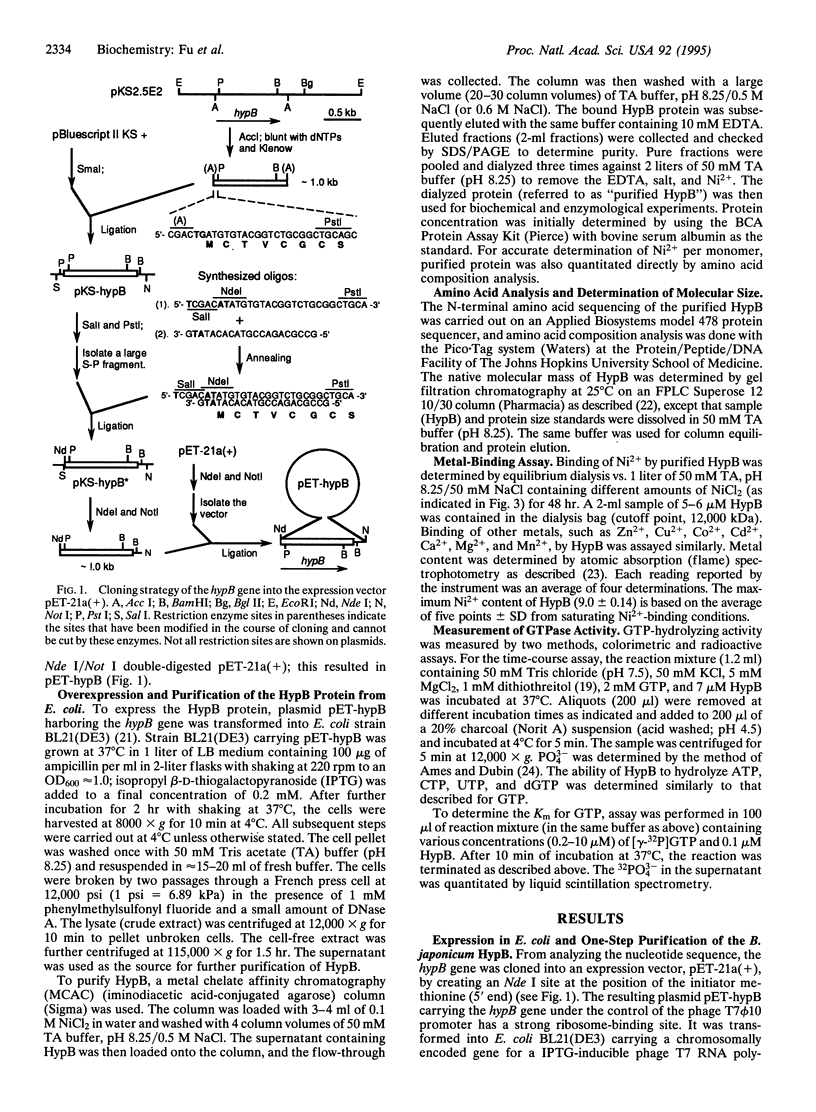
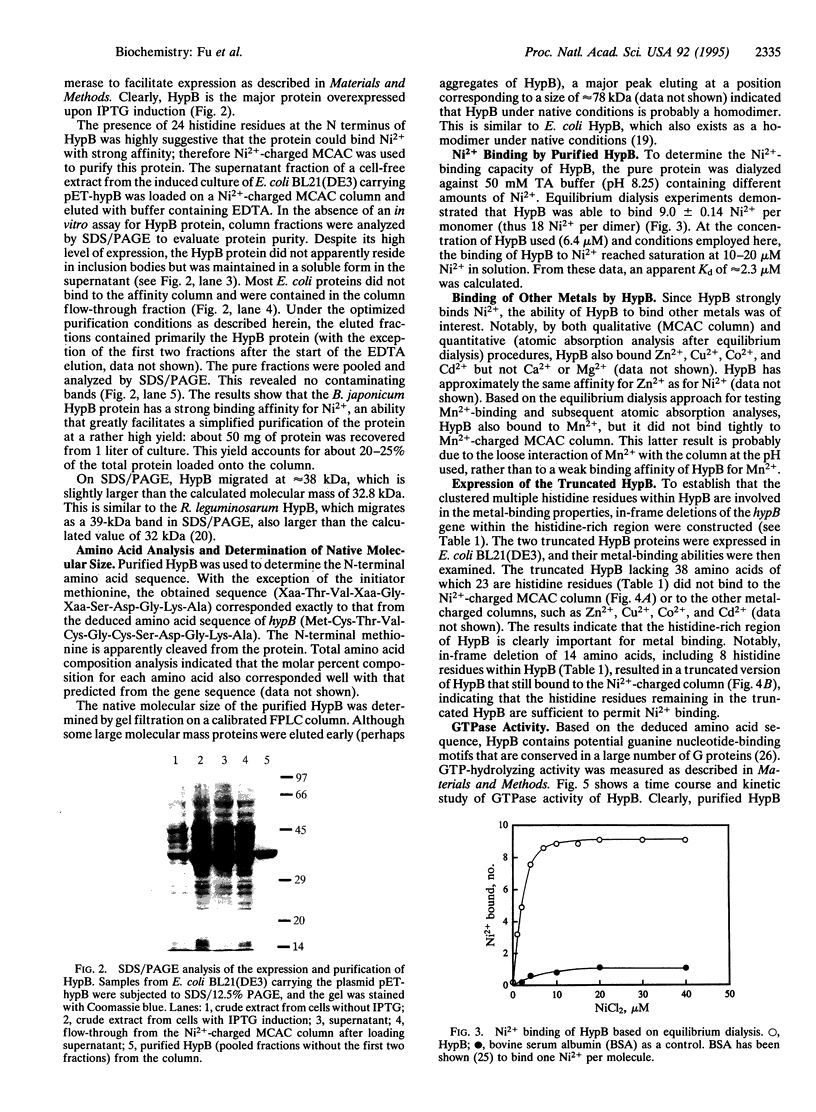
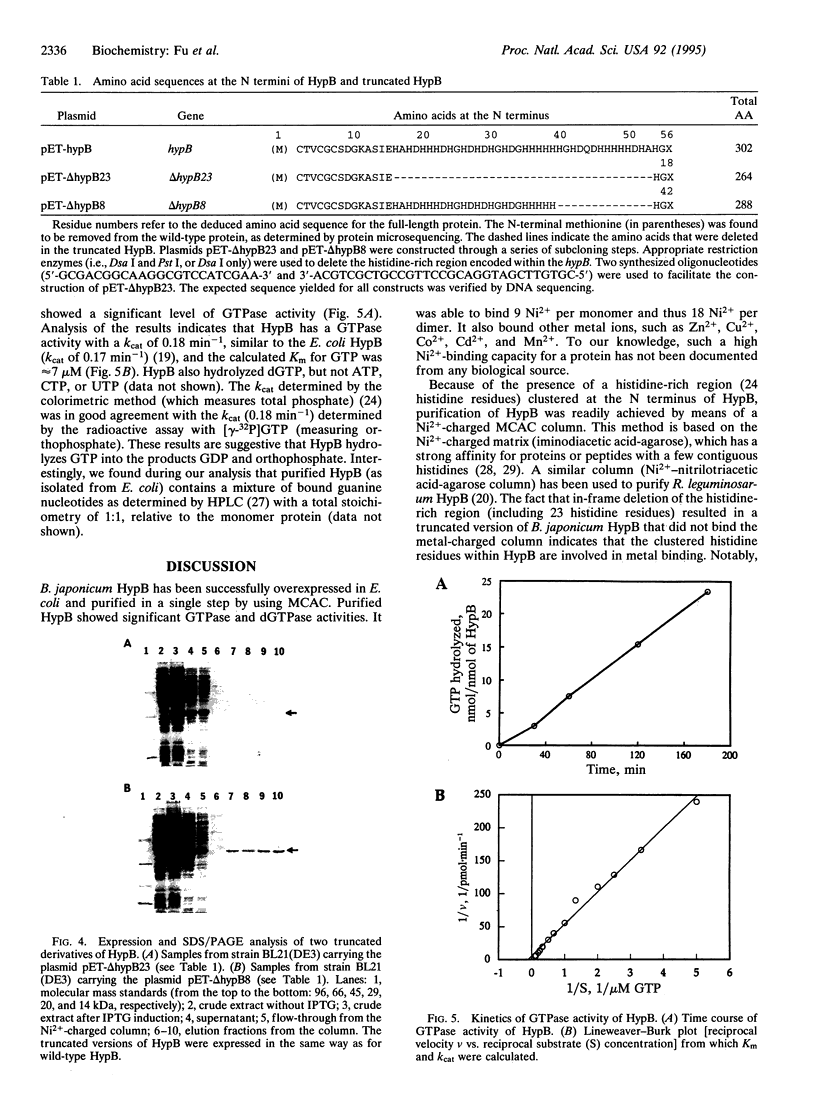
Bradyrhizobium japonicum hypB encodes a protein containing an extremely histidine-rich region (24 histidine residues within a 39-amino-acid stretch) and guanine nucleotide-binding domains. The product of the hypB gene was overexpressed in Escherichia coli and purified by Ni(2+)-charged metal chelate affinity chromatography (MCAC) in a single step. In SDS/PAGE, HypB migrated at 38 kDa--slightly larger than the calculated molecular mass (32.8 kDa). Purified HypB has GTPase activity with a kcat of 0.18 min-1 and a Km for GTP of 7 microM, and it has dGTPase activity as well. HypB exists as a dimer of molecular mass 78 kDa in native solution as determined by fast protein liquid chromatography on Superose 12. It binds 9.0 +/- 0.14 divalent nickel ions per monomer (18 Ni2+ per dimer) with a Kd of 2.3 microM; it also binds Zn2+, Cu2+, Co2+, Cd2+, and Mn2+. In-frame deletion of the histidine-rich region (deletion of 38 amino acids including 23 histidine residues) resulted in a truncated HypB that did not bind to the MCAC column, whereas in-frame deletion of 14 amino acids including 8 histidine residues within HypB resulted in a truncated HypB that still bound to the column. The results indicate that the histidine residues within the histidine-rich region of HypB are involved in metal binding.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AMES B. N., DUBIN D. T. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J Biol Chem. 1960 Mar;235:769–775. [PubMed] [Google Scholar]
- Arp D. J. Rhizobium japonicum hydrogenase: purification to homogeneity from soybean nodules, and molecular characterization. Arch Biochem Biophys. 1985 Mar;237(2):504–512. doi: 10.1016/0003-9861(85)90303-0. [DOI] [PubMed] [Google Scholar]
- Black L. K., Fu C., Maier R. J. Sequences and characterization of hupU and hupV genes of Bradyrhizobium japonicum encoding a possible nickel-sensing complex involved in hydrogenase expression. J Bacteriol. 1994 Nov;176(22):7102–7106. doi: 10.1128/jb.176.22.7102-7106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Chen J. C., Mortenson L. E. Identification of six open reading frames from a region of the Azotobacter vinelandii genome likely involved in dihydrogen metabolism. Biochim Biophys Acta. 1992 Jun 15;1131(2):199–202. doi: 10.1016/0167-4781(92)90077-d. [DOI] [PubMed] [Google Scholar]
- Colbeau A., Richaud P., Toussaint B., Caballero F. J., Elster C., Delphin C., Smith R. L., Chabert J., Vignais P. M. Organization of the genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis and identification of two hyp regulatory mutants. Mol Microbiol. 1993 Apr;8(1):15–29. doi: 10.1111/j.1365-2958.1993.tb01199.x. [DOI] [PubMed] [Google Scholar]
- Dernedde J., Eitinger M., Friedrich B. Analysis of a pleiotropic gene region involved in formation of catalytically active hydrogenases in Alcaligenes eutrophus H16. Arch Microbiol. 1993;159(6):545–553. doi: 10.1007/BF00249034. [DOI] [PubMed] [Google Scholar]
- Ferber D. M., Maier R. J. Hydrogen-ubiquinone oxidoreductase activity by the Bradyrhizobium japonicum membrane-bound hydrogenase. FEMS Microbiol Lett. 1993 Jul 1;110(3):257–264. doi: 10.1111/j.1574-6968.1993.tb06331.x. [DOI] [PubMed] [Google Scholar]
- Fu C., Javedan S., Moshiri F., Maier R. J. Bacterial genes involved in incorporation of nickel into a hydrogenase enzyme. Proc Natl Acad Sci U S A. 1994 May 24;91(11):5099–5103. doi: 10.1073/pnas.91.11.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Maier R. J. Nucleotide sequences of two hydrogenase-related genes (hypA and hypB) from Bradyrhizobium japonicum, one of which (hypB) encodes an extremely histidine-rich region and guanine nucleotide-binding domains. Biochim Biophys Acta. 1994 Feb 8;1184(1):135–138. doi: 10.1016/0005-2728(94)90163-5. [DOI] [PubMed] [Google Scholar]
- Fu C., Maier R. J. Organization of the hydrogenase gene cluster from Bradyrhizobium japonicum: sequences and analysis of five more hydrogenase-related genes. Gene. 1994 Jul 22;145(1):91–96. doi: 10.1016/0378-1119(94)90328-x. [DOI] [PubMed] [Google Scholar]
- Fu C., Maier R. J. Sequence and characterization of three genes within the hydrogenase gene cluster of Bradyrhizobium japonicum. Gene. 1994 Apr 8;141(1):47–52. doi: 10.1016/0378-1119(94)90126-0. [DOI] [PubMed] [Google Scholar]
- Glennon J. D., Sarkar B. Nickel(II) transport in human blood serum. Studies of nickel(II) binding to human albumin and to native-sequence peptide, and ternary-complex formation with L-histidine. Biochem J. 1982 Apr 1;203(1):15–23. doi: 10.1042/bj2030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi A., Rossmann R., Böck A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158(6):444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- Kim H., Maier R. J. Transcriptional regulation of hydrogenase synthesis by nickel in Bradyrhizobium japonicum. J Biol Chem. 1990 Nov 5;265(31):18729–18732. [PubMed] [Google Scholar]
- Kim H., Yu C., Maier R. J. Common cis-acting region responsible for transcriptional regulation of Bradyrhizobium japonicum hydrogenase by nickel, oxygen, and hydrogen. J Bacteriol. 1991 Jul;173(13):3993–3999. doi: 10.1128/jb.173.13.3993-3999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Pankratz H. S., Wang S., Scott R. A., Finnegan M. G., Johnson M. K., Ippolito J. A., Christianson D. W., Hausinger R. P. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993 Jun;2(6):1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S., Jacobi A., Schlensog V., Böhm R., Sawers G., Böck A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- Maier T., Jacobi A., Sauter M., Böck A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993 Feb;175(3):630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porath J., Carlsson J., Olsson I., Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature. 1975 Dec 18;258(5536):598–599. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- Rey L., Imperial J., Palacios J. M., Ruiz-Argüeso T. Purification of Rhizobium leguminosarum HypB, a nickel-binding protein required for hydrogenase synthesis. J Bacteriol. 1994 Oct;176(19):6066–6073. doi: 10.1128/jb.176.19.6066-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey L., Murillo J., Hernando Y., Hidalgo E., Cabrera E., Imperial J., Ruiz-Argüeso T. Molecular analysis of a microaerobically induced operon required for hydrogenase synthesis in Rhizobium leguminosarum biovar viciae. Mol Microbiol. 1993 May;8(3):471–481. doi: 10.1111/j.1365-2958.1993.tb01591.x. [DOI] [PubMed] [Google Scholar]
- Sayavedra-Soto L. A., Powell G. K., Evans H. J., Morris R. O. Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8395–8399. doi: 10.1073/pnas.85.22.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Stults L. W., Moshiri F., Maier R. J. Aerobic purification of hydrogenase from Rhizobium japonicum by affinity chromatography. J Bacteriol. 1986 Jun;166(3):795–800. doi: 10.1128/jb.166.3.795-800.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibelius K. H., Du L., Tito D., Stejskal F. The Azotobacter chroococcum hydrogenase gene cluster: sequences and genetic analysis of four accessory genes, hupA, hupB, hupY and hupC. Gene. 1993 May 15;127(1):53–61. doi: 10.1016/0378-1119(93)90616-b. [DOI] [PubMed] [Google Scholar]
- Van Soom C., Verreth C., Sampaio M. J., Vanderleyden J. Identification of a potential transcriptional regulator of hydrogenase activity in free-living Bradyrhizobium japonicum strains. Mol Gen Genet. 1993 May;239(1-2):235–240. doi: 10.1007/BF00281623. [DOI] [PubMed] [Google Scholar]