Highlights
-
•
We characterise Sas3p and Gcn5p active HAT complexes in WT and deleted TAP-strains.
-
•
We confirm that Pdp3p interacts with NuA3, histones and chromatin regulators.
-
•
Pdp3p MS-analysis reveals its phosphorylation, ubiquitination and methylation.
-
•
Sas3p can substitute Gcn5p in acetylation of histone H3K14 but not of H3K9.
-
•
Genome-wide profiling of Sas3p supports its involvement in transcriptional elongation.
Abbreviations: ChIP-on-chip, chromatin immunoprecipitation with genome-wide location arrays; HAT, histone acetyltransferase; NuA3, nucleosomal acetyltransferase of histone H3; nt, nucleotide; PTM, post-translational modification; RNAPII, RNA polymerase II; SAGA, Spt-Ada-Gcn acetyltransferase; TAP, tandem affinity purification; TSS, transcription start site; WCE, whole cell extract; WT, wild-type
Keywords: ChIP-on-chip, Chromatin, Histones, Pdp3, TAP–MS strategy, Yeast
Abstract
Histone acetylation affects several aspects of gene regulation, from chromatin remodelling to gene expression, by modulating the interplay between chromatin and key transcriptional regulators. The exact molecular mechanism underlying acetylation patterns and crosstalk with other epigenetic modifications requires further investigation. In budding yeast, these epigenetic markers are produced partly by histone acetyltransferase enzymes, which act as multi-protein complexes. The Sas3-dependent NuA3 complex has received less attention than other histone acetyltransferases (HAT), such as Gcn5-dependent complexes. Here, we report our analysis of Sas3p-interacting proteins using tandem affinity purification (TAP), coupled with mass spectrometry. This analysis revealed Pdp3p, a recently described component of NuA3, to be one of the most abundant Sas3p-interacting proteins. The PDP3 gene, was TAP-tagged and protein complex purification confirmed that Pdp3p co-purified with the NuA3 protein complex, histones, and several transcription-related and chromatin remodelling proteins. Our results also revealed that the protein complexes associated with Sas3p presented HAT activity even in the absence of Gcn5p and vice versa. We also provide evidence that Sas3p cannot substitute Gcn5p in acetylation of lysine 9 in histone H3 in vivo. Genome-wide occupancy of Sas3p using ChIP-on-chip tiled microarrays showed that Sas3p was located preferentially within the 5′-half of the coding regions of target genes, indicating its probable involvement in the transcriptional elongation process. Hence, this work further characterises the function and regulation of the NuA3 complex by identifying novel post-translational modifications in Pdp3p, additional Pdp3p-co-purifying chromatin regulatory proteins involved in chromatin-modifying complex dynamics and gene regulation, and a subset of genes whose transcriptional elongation is controlled by this complex.
1. Introduction
Many regulatory processes of eukaryotic organisms are determined not only by the genome sequence, but also by epigenetic information. Epigenetic modifications generate heritable alterations in gene expression, although the primary nucleotide sequence remains unaltered [1–3]. Major epigenetic events include the methylation of DNA bases and post-translational modifications (PTMs) of histones. DNA methylation has been frequently described as an epigenetic modification related to gene silencing [4]. Meanwhile, histone PTMs typically contribute to the regulation of chromatin-related processes, modulating either directly the chromatin packing by altering the net charge of the histones [5] or indirectly, by regulating the DNA accessibility through nucleosome remodelling. The presence or absence of particular PTMs are likely to determine the sequential recruitment of the PTM enzymes or protein complexes implicated in gene regulation [6–9]. Therefore histone modifications are important in coordinating epigenetic crosstalk. Several types of enzymatic activities are involved in histone modifications [10], of which histone acetyltransferases (HATs) are the best-studied group [11,12]. HAT enzymes acetylate specific lysine residues located mainly on the amino terminal tail of histones [13–16] thereby participating in the recruitment of trans-acting regulatory factors [6–9].
Eleven different proteins exhibiting HAT activity, at least in vitro, have been described to date in the Saccharomyces Genome Database (SGD, www.yeastgenome.org): Elp3p/Hpa1p, Esa1p, Gcn5p, Hat1p, Hpa2p, Hpa3p, Nut1p, Rtt109, Sas2p, Sas3p and Taf1p. Most of these enzymes have physical and functional connections with transcriptional regulation and belong to complexes composed of several subunits, which can modulate their activities. The fact that some of these proteins work as a catalytic subunit of more than one HAT complex, such as Gcn5p, which is a component of SAGA, SLIK/SALSA, ADA and HAT-A2 HAT yeast complexes [11], might explain the diversity of their functions. Histone H3 acetylation at lysines 9 (H3K9) and 14 (H3K14) strongly correlates with transcriptional activity and peaks beyond the transcription start site (TSS) of active genes [17]. Studies suggest that Gcn5p is the HAT responsible for most of these acetylation events in vivo [18–20], which is in accordance with observations that Gcn5p is generally recruited to the promoters of active genes, as previously described for H3K9 and H3K14 acetylation marks [17,21]. Gcn5p also regulates replication-coupled nucleosome activity partly by promoting histone H3 association with CAF-1 (Chromatin Association Factor) via H3 acetylation [22]. Two other HATs also specifically acetylate H3K9 and H3K14 in vivo: the MYST family HAT Sas3p (Something About Silencing protein) and Elp3p (Elongator protein) [18,20]. Sas3p is the catalytic subunit of the NuA3-HAT complex [23] which also contains Eaf6p, Taf14p, Nto1p and Yng1p. Recruitment of NuA3 to nucleosomes in vivo requires the methylation of H3K4 and H3K36, which are catalysed by the Set1p and the Set2p histone methyltransferases, respectively [24]. The function of Sas3p appears to be critical in the absence of Gcn5p [19]. More specifically, deletion of SAS3 in WT cells does not elicit obvious phenotypes, however diminished Sas3p activity in a gcn5Δ null mutant results in cell cycle progression defects, and the complete loss of SAS3 function in gcn5Δ cells leads to cell death [19]. In addition, genome-wide mapping studies have established that Sas3p and Gcn5p are recruited to many of the same actively transcribed genes and that the binding sites of both HATs correlate with H3K14 acetylation [25]. Furthermore, genome-wide mapping approaches suggest that the SAGA-component Gcn5p [26–28] and Yng1p, the histone-binding subunit of NuA3 complex [29], are involved in transcriptional elongation, since their as both has been shown to bind to the coding region of target genes. These observations strongly suggest that the Sas3p and Gcn5p histone acetyltransferases are critical for active transcription, although the molecular mechanisms underlying their regulation have not yet been fully elucidated. In particular, since mutations of the above-mentioned histone H3 lysines result in only mild phenotypes [30,31], the essential function revealed by gcn5Δsas3Δ mutant lethality most likely goes beyond H3 acetylation. Interestingly, in the gcn5 and sas3 histone acetyltransferase double mutant, the inactivation of the chromatin remodelling complex Isw1a triggers the restoration of the RNAPII recruitment to active genes, which emphasise the importance of a fine-tuned balanced between chromatin modifying and remodelling activities for optimal transcription and cell growth [32]. Moreover, previously unanticipated differential roles for Sas3p and Gcn5p in DNA damage response and cell cycle regulation have been recently shown in relation to the β-1,3-glucanosyltransferase Gas1p, which regulates transcriptional silencing independently of its activity on the cell wall [33].
Although Gcn5p has been extensively studied, much less is known about Sas3p. To explore Sas3p functions, we designed a proteomic approach based on the tandem affinity purification (TAP) strategy coupled to a mass spectrometry (MS) analysis and additional genomic studies by ChIP-on-chip assays. TAP-tagged wild-type (WT) and deletion mutant strains were constructed, and in this work, we particularly focused on the purification and characterisation of the active HAT complexes associated with Sas3p and Gcn5p. Notably, we found novel physical associations involving NuA3, nucleosomal histones, Pdp3p, and a variety of proteins involved in chromatin dynamics. At the genomic level, the ChIP-on-chip analyses suggest a role for Sas3p in transcriptional elongation. Altogether, these results significantly improve our knowledge regarding the functionality and dynamic interactions of Sas3p in relation to the presence of Gcn5p. Finally, we also report the likelihood of a functionally relevant crosstalk between NuA3 and Pdp3p.
2. Results and discussion
2.1. Purification of Sas3p reveals its association with histones and Pdp3p
Sas3p is the HAT catalytic subunit of the NuA3 complex. Sas3p, as well as the extensively studied Gcn5p HAT, act on nucleosomal H3, specifically at K9 and K14 [34], as primary sites of acetylation [35]. To study how the absence of one of these two HATs might affect the composition and/or the acetylation activity of the HAT complexes, new strains were constructed. Firstly, we assessed whether the integration of the TAP tag into the genome affects the growth of yeast strains. As shown in Fig. 1, TAP-tagging or SAS3 gene deletion did not produce defective growth at either 30 °C or 37 °C when compared to the corresponding WT cells (BMA64-1A and BY4742, Fig. 1 upper and bottom panels, respectively). On the contrary, the growth defects observed for deleted or TAP-tagged GCN5 (Fig. 1) can be partially explained by previous observations that gcn5Δ mutant cells accumulate in the G2/M phase of the cell cycle [31]. Phenotypic growth defects in the absence of GCN5 depended on the strain background, and no drastic visual defects were observed in the BY4742 background if compared with BMA64-1A. As a control, HAT2-TAP hif1Δ was used as a temperature-sensitive strain (Fig. 1).
Fig. 1.
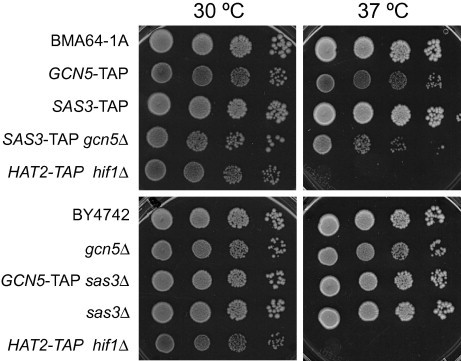
Deletion or TAP-tagging of SAS3, unlike GCN5, does not induce growth defects. Aliquots of 5 μL and 10-fold serial dilutions of the overnight cultures at OD600 ≈ 0.6 were spotted onto YPDA plates. Cells were cultured at 30 °C and 37 °C for 3 days. The temperature-sensitive HAT2-TAP hif1Δ strain, was used as a control.
Sas3p is involved not only in chromatin modification through histone acetylation, but also in chromatin silencing in the silent mating-type cassettes and telomeres. By means of different technical approaches, it has been proposed that 28 proteins physically interact with Sas3p (BIOGRID database). In the present study, SAS3 was TAP-tagged and purified to further explore these interactions. The cell extract preparation and purification for TAP assays were carried out under sufficiently gentle conditions to maintain the integrity of the protein complex. Purification of a non-TAP-tagged strain was used as a negative control to rule out non-specific interactions [36]. Moreover, the typically observed non-specific purifying proteins were also ruled out, such as abundant housekeeping enzymes, translational factors, ribosomal, and heat-shock proteins [37]. The MS identification of the SAS3-TAP co-purified proteins using a nano LC–MS/MS analysis, which was performed using the QSTAR-XL system, is shown in Table 1. Notably, besides the components of the NuA3 complex (Sas3p, Nto1p, Taf30/14p, Yng1p and Eaf6p) and core histones, one of the best represented proteins was Pdp3p (UniProtKB: Q06188), with a 66.8% protein sequence coverage and an emPAI score of 4.56 (Table 1). In this context, it should also be mentioned that the only minor differences were observed among the proteins identified in the extracts from the SAS3-TAP WT or gcn5Δ mutant cells. Thus, in the GCN5-deleted strain, we identified Rim1p, Sec39p and Sas2p (data not shown) instead of Phb2p, Yhz9p and Ubi3p, which were identified in WT SAS3-TAP cells (Table 1). To confirm the physical interactions of Pdp3p with the NuA3 complex components and histones, the PDP3 gene was TAP-tagged and the purified proteins were analysed by MS. First, a portion of the chromatography eluate was analysed by nano LC MALDI-TOF–TOF with the 4700 PA system, which enabled the identification of the four core nucleosomal histones, and only a few peptides corresponding to Taf14p, Yng1p and Eaf6p components of NuA3 (Table 1). Secondly, cell extracts were prepared from a larger number of harvested cells. Approximately 36 μg of TAP-purified proteins were obtained and resolved by SDS–polyacrylamide gel electrophoresis (Fig. 2A). Protein bands were gel-excised and analysed by LC–MS/MS, performed using a 5600 Triple TOF system (Fig. 2B). Protein identification of the major band (slice 1) was carried out by the Protein Pilot software using Paragon Algorithm (AB Sciex). The analysis revealed new PTMs in Pdp3p (methylation at K79 and K126, ubiquitination at K14 and K63, and phosphorylation at S148, S150, S161 and S173; Fig. 2C). Our results corroborated one of the two phosphorylation events described in the BIOGRID database (i.e. at S148). Additionally, analyses of slice 2 identified Eaf6p and the four core histones (Fig. 2B). A thorough PTM search identified several modifications in H3: K23ac, K56ac, K36me, K79me and tri-methylated K79 (Fig. 2C). It must be noted that the histone H3 modifications we identified have been previously linked to active chromatin and genome stability (BIOGRID data). A total of 381 proteins were identified in slices 3 + 4 + 5, including common background proteins such as abundant housekeeping enzymes, ribosomal and heat-shock proteins, and translational factors (data not shown). As shown in Fig. 2B, Pdp3p co-purified not only with the core histones and all the components of the NuA3 complex, but also with several proteins involved in chromatin modification or remodelling.
Table 1.
Summary of the proteins co-purified with Sas3p and Pdp3p identified by mass spectrometry analyses. A portion of eluates of the TAP purifications was analysed by either nano LC–MS/MS Q-TOF (SAS3-TAP) or nano LC MALDI-TOF–TOF (PDP3-TAP). The NuA3 complex components are shown in bold. The proteins associated with Sas3p have been ordered according to the MASCOT protein score; the exponentially modified protein abundance index (emPAI) score is shown in parentheses. (n.d., not determined).
| Protein |
SAS3-TAP |
PDP3-TAP |
||
|---|---|---|---|---|
| Protein sequence coverage (%) | MASCOT protein score (emPAI score) | Protein sequence coverage (%) | MASCOT protein score (emPAI score) | |
| Sas3 | 57.4 | 1633 (1.88) | n.d. | n.d. |
| Nto1 | 54.0 | 981 (1.36) | n.d. | n.d. |
| Pdp3 | 66.8 | 820 (3.56) | 63.8 | 2192 (19.58) |
| Taf30/14 | 87.7 | 555 (2.96) | 69.7 | 156 |
| H2B (HTB1) | 79.4 | 299 (4.58) | 55.7 | 597 (9.64) |
| Yng1 | 57.0 | 256 (1.38) | 15.0 | 26 (0.21) |
| H4 (HHF1) | 78.6 | 215 (2.77) | 49.5 | 157 (1.32) |
| Eaf6 | 56.6 | 208 (3.11) | 28.3 | 34 (0.45) |
| H3 (HHT1) | 73.5 | 144 (0.82) | n.d. | 57 (0.37) |
| H2A (HTA1) | 75.0 | 98 (0.55) | 56.8 | 70 (1.80) |
| Phb2 | 28.1 | 69 (0.20) | n.d. | n.d. |
| Yhz9 | 26.1 | 42 (0.10) | n.d. | n.d. |
| Ubi3 | 11.0 | 31 (0.41) | n.d. | n.d. |
| Rim1 | n.d. | n.d. | 40.0 | 73 (0.87) |
| Taf9 | n.d. | n.d. | n.d. | 28 (0.32) |
Fig. 2.
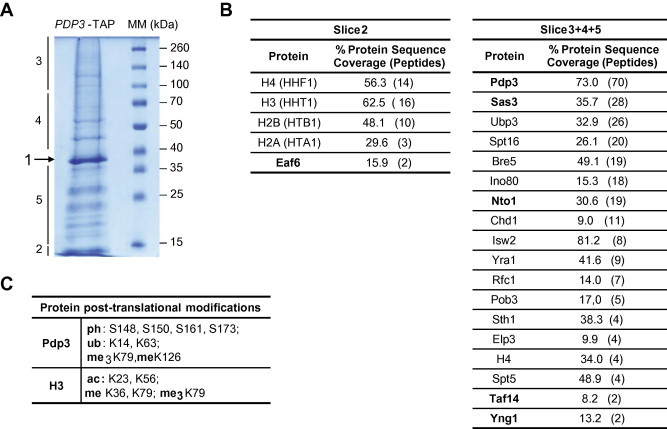
Novel Pdp3p-interacting proteins revealed by tandem affinity purification coupled to mass spectrometry analyses. (A) Proteins co-purifying with PDP3-TAP in yeast cellular extract resolved by SDS-10% PAGE. The excised gel fragments employed for the LC–MS/MS analysis (Triple TOF) are indicated. (B) Summary of the proteins identified in slices 2 and 3 + 4 + 5. The NuA3 complex components are indicated in bold. The percentage of sequence coverage of each protein is shown with the number of unique peptides in parentheses; these values are those at the 95% confidence level. (C) The post-translational modifications shown are those identified in the Protein Pilot search engine at a confidence level ⩾95%. The specific modification and residues are indicated (ph: phosphorylation; ub: ubiquitination; me: mono-methylation; me3: tri-methylation; ac: acetylation).
The network displayed by the STRING representation [38] generated by 22 Pdp3-interacting proteins identified in our analysis (Fig. 3A) shows the increased interacting complexity compared to the network published on BIOGRID (Fig. 3B). Among the transcription-related proteins, we found Ino80p (chromatin remodelling in replication and transcription), Chd1p (a chromatin remodeler, that binds methylated histone), Elp3p (the HAT subunit of elongator complex) and Spt5p (RNAPII core binding, transcription elongation by RNAPII). In particular, the identification of Spt16p and Pob3p, protein components of the yFACT nucleosome assembly complex, and Rfc1p, according to others studies [39], provides insights into the mechanism of NuA3-associated transcription and chromatin regulation.
Fig. 3.
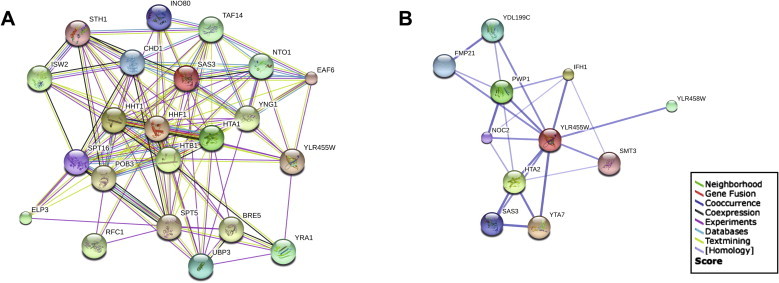
A novel network of Pdp3 interactions identified by TAP–MS. (A) Schematic representation of the interaction network generated from the evidence view of the STRING interactions. The different coloured lines represent the types of experimental evidence available for each of the interacting partners shown. (B) The Pdp3p (YLR455W/PDP3 gene) interaction network previously published on BIOGRID (string-db.org). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Previous studies have reported that Sas3p, core histones, Spt16p and Pdp3p, are co-purified with Yta7p tagged with Protein A [40]. However, an I-DIRT (Isotopic-Differentiation of Interactions as Random or Targeted) analysis applied to YNG1-TAP cells indicated that Pdp3p was a contaminant of NuA3 purification and not a true component of the complex [29]. Pdp3p has recently been described as a nuclear PWWP domain-containing protein [41] that has been physically associated with Sas3p [37]. Additionally, Gilbert and co-workers [41] have recently reported two RNAPII components (Rpb2p and Rpb4p) as Pdp3-interacting proteins, in addition to the NuA3 core-associated proteins and histones. Notably, among the Pdp3-interacting proteins identified in our analyses, we found the RNAPII component Rpb1p (represented by 16 peptides, with a protein sequence coverage of 11.6%).
In order to investigate whether the absence of Sas3p can affect the nature of the proteins associated with Gcn5p and/or its HAT activity, TAP–MS analyses were carried out with complexes purified from the GCN5-TAP WT and GCN5-TAP sas3Δ mutant strains. The LC–MS/MS analyses of a portion of eluates from the TAP purification showed that Gcn5p co-purifies with all the components of the SAGA complex, except Sgf11p, Sus1p (components of the deubiquitination module of SAGA) and Chd1p, a chromatin remodeler (Table 2). These three proteins and Ubp8p were not identified when Sas3p was absent. Interestingly, protein Tra1p, which functions as a recruitment module for SAGA and is also a member of the NuA4 complex, was the most abundant protein in the GCN5-TAP purification from WT cells and one of the minor proteins recovered in the sas3Δ mutant. In both strains, Ahc1p, a component of the Ada2/Gcn5/Ada3 transcription activation complex [42], was found among the less representative proteins.
Table 2.
Proteins that co-purified with Gcn5p from wild-type or SAS3 deleted cells were identified by a mass spectrometry QSTAR XL analysis. The listed proteins have been ordered according to the MASCOT protein score. The emPAI score offers an approximate, label-free, relative protein quantification in a mixture based on protein coverage by peptide matches in a database search (n.i.: not identified in the Protein Pilot search).
|
GCN5-TAP |
GCN5-TAP sas3Δ |
||||
|---|---|---|---|---|---|
| Protein | Protein sequence coverage (%) | MASCOT protein score (emPAI score) | Protein | Protein sequence coverage (%) | MASCOT protein score (emPAI score) |
| Tra1 | 54.97 | 2399 (0.52) | Ngg1/Ada3 | 83.47 | 1163 (1.65) |
| Ngg1/Ada3 | 83.19 | 1817 (3.31) | Spt7 | 55.25 | 558 (0.24) |
| Spt7 | 74.32 | 1768 (1.65) | Ada2 | 74.20 | 544 (1.00) |
| Taf5 | 67.67 | 1021 (1.38) | Gcn5/Ada4 | 70.39 | 490 (1.25) |
| Gcn5/Ada4 | 82.46 | 988 (3.21) | Taf5 | 62.53 | 483 (0.39) |
| Hfi1/Ada1 | 64.34 | 917 (1,88) | Taf12 | 59.00 | 426 (0.44) |
| Ada2 | 80.65 | 899 (3.56) | Hfi1/Ada1 | 63.73 | 400 (0.60) |
| Spt20 | 64.24 | 841 (1.46) | Taf6 | 64.53 | 313 (0.65) |
| Taf6 | 68.41 | 831 (1.86) | Sgf29 | 78.38 | 275 (0.90) |
| Spt8 | 58.64 | 784 (1.07) | Spt20 | 56.95 | 253 (0.33) |
| Taf12 | 50.83 | 769 (1.31) | Spt8 | 50.66 | 246 (0.34) |
| Sgf73 | 48.55 | 541 (0.49) | Tra1 | 40.84 | 223 (0.05) |
| Sgf29 | 79.92 | 425 (1.12) | Sgf73 | 51.14 | 128 (0.09) |
| Ubp8 | 33.76 | 264 (0.27) | Ahc1 | n.i. | 109 (0.16) |
| Spt3 | 51.93 | 253 (0.64) | Spt3 | n.i. | 98 (0.18) |
| Taf10 | 73.79 | 202 (0.72) | Taf10 | 30.10 | 83 (0.31) |
| Taf9 | 63.06 | 181 (1.04) | Taf9 | 44.58 | 49 (0.20) |
| Ahc1 | 48.59 | 174 (0.22) | |||
Overall, our results provide a comprehensive list of physically interacting proteins, thus complementing previous data from low-throughput studies [40,41] and high-throughput studies compiled in the BIOGRID database. The protein network generated in the present work connects Pdp3p with a variety of chromatin-related proteins. These data suggest that novel mechanisms could be involved in the increase in Pdp3p in response to DNA replication stress [43] and its relocation to the cytosol in response to hypoxia [44] reported previously. Therefore, further research is needed to elucidate whether these proteins, Pdp3p and Sas3p, as well as another component of NuA3, together with the core of histones, are functionally linked.
2.2. The TAP-purified Sas3p complex acetylates nucleosomal histone H3
Sas3p and Gcn5p are HAT enzymes that belong to the NuA3 and the SAGA complex, respectively. Both HAT complexes acetylate nucleosomal histones including H3K9 and H3K14 among other residues [34]. The protein complexes associated with Sas3p and purified by tandem affinity chromatography acetylated histones H4, and preferentially H3, when assayed with chicken erythrocyte oligonucleosomes in the presence or absence of Gcn5p and vice versa (Fig. 4). Interestingly, the activity observed for the NuA3 complex purified from the sas3Δ mutant, and also for the SAGA complex purified from the gcn5Δ mutant, was stronger in comparison to the WT. However, we cannot rule out that these differences might be due to a larger quantity of enzyme in the sample. In previous studies, by using the TAP strategy described herein, we were able to purify the HAT B complex that presented Hat1 activity on free histone H4 (results not shown). Thus, we can conclude that the experimental conditions we used to purify HAT complexes are sufficiently gentle to preserve their enzymatic activity.
Fig. 4.
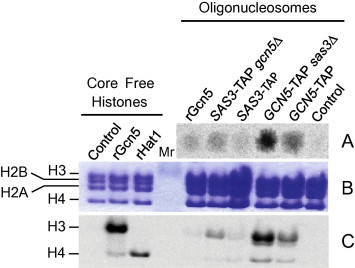
HAT activity of the protein complexes purified by TAP. (A) The HAT assay was performed with a 1/200 portion of the TAP-purified protein samples. 14C-radiolabelled histones were detected with PhosphorImager using 1/4 of the mixture reaction volume. Portions (5 μL) of each sample were spotted across a glass microfiber filter paper and exposed for 24 h. (B) Radiolabelled histones were resolved by SDS–PAGE (16%) and stained with Coomassie Blue. (C) The autoradiography of the gel shown in (B) after 1 week of exposure; rGcn5 and rHat1 correspond to the recombinant HAT proteins used as controls of specificity over H3 [50] and H4 [59], respectively.
2.3. The absence of Sas3p or Gcn5p has different effects on the in vivo acetylation levels of H3 at K9 and K14
It is well-accepted that H3K14 is the main in vitro target for both histone acetyltransferases Sas3p and Gcn5p, although both proteins also acetylate H3K9. It has been reported that these histone modifications play an important role in gene transcriptional activation [24–29]. Fig. 5 shows the results obtained from the Western blot analyses of the total protein extracts employed to assess the in vivo acetylation levels of histone H3 at K14 and K9. Compared to the WT cells, loss of SAS3 or GCN5 did not significantly affect the total histone H3 levels (αH3 C-terminal–antibody). Strikingly, the H3K9 acetylation was practically absent in gcn5Δ cells, but remained virtually unaltered in sas3Δ cells. This shows that Sas3p cannot replace the acetylating function of Gcn5p on H3K9. However, the H3K14 acetylation levels were similar to the WT cells in both mutants. These data suggest that both Sas3p and Gcn5p contribute to reciprocally restore the H3K14ac level, because no change was detected when either of them was absent. Together, these data suggest that Sas3p has a similar but non-redundant function to Gcn5p (Fig. 5).
Fig. 5.
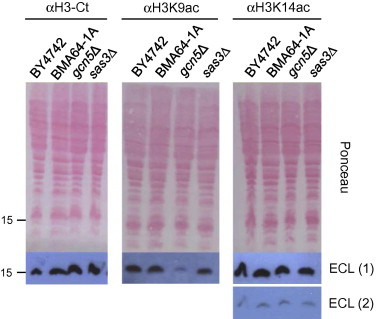
Acetylation levels of histone H3 at lysines 9 and 14 in vivo. The indicated strains were cultured to exponential growth (OD600 ≈ 0.8), alkaline-lysed and pelleted. Proteins contained in the crude extracts (0.3 OD600 units) were resolved by SDS–PAGE (16%) and transferred to nitrocellulose membranes. The amounts of total and modified histones were determined by immunodetection with the antibodies indicated in the figure; different exposure times (ECL 2 vs. ECL 1) demonstrate that the chemiluminescence signal was not saturated. The Ponceau S staining of the membrane is shown as a loading control. The position of the 15 kDa molecular mass marker is indicated on the left.
2.4. Sas3 preferentially binds to the 5′-half of the coding region of target genes
In a previous genome-wide study [25], we demonstrated that Sas3p, like Gcn5p, was recruited to a pool of intensely transcribed genes, and that a considerable overlap was observed between the two cohorts of the Sas3p- and Gcn5p-bound gene pools. However, because the arrays only had a single probe for each gene, it was not possible to determine the location of Sas3p in relation to the ORF. Here we investigated the genome-wide occupancy of Sas3p by ChIP-on-chip using tiled microarrays (244K, Agilent) with an average probe spatial resolution of 50 nt. Use of this technique showed that H3K14 (and H3K9) acetylation is enriched in promoter regions and the TSS of active genes [17,45]. Because Sas3p is a HAT, whose main target in vitro is H3K14, we expected to detect the majority of Sas3p binding to the promoter regions of genes. Surprisingly, we found that Sas3p was associated with the coding regions of genes, with peak enrichment 400 bp from the TSS within the ORF, and that this enrichment dropped substantially towards the 3′ region of the ORF (Fig. 6A). This result is similar to that obtained for Yng1p genome-wide occupancy [29], which also belongs to the NuA3 complex, and suggests that this complex might be involved in transcriptional elongation, at least in an initial step of the process. We employed a graphical representation (real-length representation), where the Sas3p location profiles were centred on the actual TSS [46]. The data for each probe corresponds to their real distance from the TSS for each gene up to the total transcript length, with a maximum of 3000 bp. A similar result was obtained with a graphical representation of the genome-wide data using a metagene profile, where genes were expanded or compressed to fit onto a hypothetical gene of average length (data not shown). Next we analysed whether the positive correlation observed between Sas3p binding and transcriptional activity [25] could be reproduced at a higher level when studying the Sas3p association with the coding regions of the most highly transcribed genes. Fig. 6B shows how the relative Sas3p binding level rises for a set of genes grouped according to the reported presence of RNA polymerase II (RNAPII) [47] (up to 25%, from 25% to 75% and over 75%) in the body of the genes. Moreover, the main correlation between the presence of both RNAPII and Sas3p proteins was observed, not only within the first 1000 bp from the TSS, but also upstream of the coding region. Thus, these results provide new evidence for the involvement of Sas3p in transcriptional elongation, which is in agreement with previous reports [25] showing that Sas3p acts as a general activator of transcription. As both functions have also been described for Gcn5p, the results of the present work provide new evidence to support the hypothesis that Gcn5p and Sas3p share redundant functions [19,25].
Fig. 6.
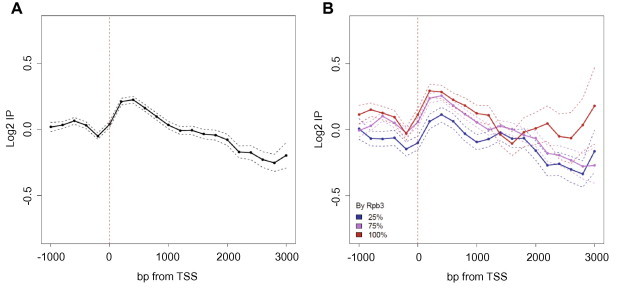
Genome-wide profile of Sas3p location. The Sas3-HA tagged protein was immunoprecipitated with an anti-HA 3F10 antibody. The log2 values of the specific immunoprecipitation of the tagged strain are represented in relation to the immunoprecipitation of a non-tagged strain. For the real-length representation, profiles were centred on the actual transcription start site (TSS) [46] and the data for each probe corresponded to their real distance from the TSS for each gene, from 1000 bp upstream to the total transcript length, to a maximum of 3000 bp downstream. Probes were binned to 200 bp. The mean and confidence intervals for the means (t-test, 95% confidence) were plotted (A). The real-length representation of the profile of the Sas3p location of the genes grouped according to the distribution of RNAPII [47] on the gene (B). The figures show the average of two independent biological experiments.
The genome-wide pattern of Sas3p binding to the genes observed in Fig. 6A might be due to general non-specific Sas3p binding or may result from high Sas3p levels in specific classes of genes, which together form the observed pattern. To investigate this, we analysed the binding pattern of Sas3p to genes. Of the 5603 genes showing probes in the ORF, 5450 had more than three probes inside the ORF. Of these, 2644 genes showed no trend (the type H genes in Fig. 7A). The remaining genes were classified into four clusters according to the Sas3p binding pattern. Cluster 1 included those genes where Sas3p tended to be located at the 5′ of the ORF (936 type L + N genes in Fig. 7B), which, therefore, represents the group that best fits the profile obtained at the genomic level. Cluster 2 contains the genes where Sas3p tended to be located at the 3′ end of the ORF (706 type J + Z genes in Fig. 7C). Cluster 3 included the genes for which Sas3p tended to be located at both the 5′ and 3′ ends of the ORF (405 type U genes in Fig. 7D). Finally, cluster 4 contains the genes for which Sas3p tended to be located preferentially at the central region of the ORF (759 type A + S + X genes in Fig. 7E). Interestingly, a gene ontology (GO) analysis, which was performed separately on the genes belonging to the four clusters, revealed that only the genes included in cluster 1 were statistically grouped into GO categories (Table 3). Among the most statistically significant processes, the over-representation of the intracellular organelle-related categories is noteworthy. Taken together, these findings strongly suggest that Sas3p is present at the 5′ end of the ORF of a set of approximately 1000 genes involved in essential cellular functions, such as organelle biosynthesis and the regulation of biological processes.
Fig. 7.
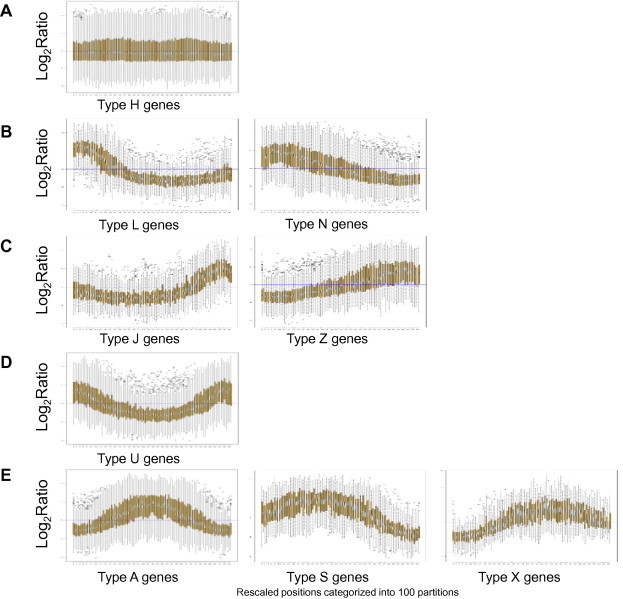
Boxplot representation of the Sas3p binding to different groups of genes. (A) Boxplot of an average type H gene (2644 genes). The chromosomal positions for each gene were categorised into 100 partitions. Log2 ratio values were extracted for the corresponding probes for all these partitions. (B) Boxplot of an average gene of cluster 1 (936 genes, type L or N). Data are represented as in (A). (C) Boxplot of an average gene of cluster 2 (706 genes, type J or Z). Data are represented as in A. (D) Boxplot of an average gene of cluster 3 (405 genes, type U). Data are represented as in A. (E) Boxplot of an average gene of cluster 4 (759 genes, type A, S or X). Data are represented as in A.
Table 3.
Gene ontology attributes of the genes to which Sas3p binds at the beginning of the ORF. The genes of cluster 1 (936 genes) were classified based on over-represented attributes by using the FuncAssociate 2.0 web tool [http://llama.mshri.on.ca/funcassociate]. The number of genes bound for each category, the number of overall genes within this attribute and the p values are shown. Only the categories with a p value ⩽10−6 are included.
| Go attribute | No. of genes in category | No. of genes | p value |
|---|---|---|---|
| Membrane-bounded organelle/intracellular membrane-bounded organelle | 3725 | 631 | 4.85 × 10−9 |
| Regulation of biological process | 1371 | 264 | 2.06 × 10−7 |
| Organelle | 4061 | 669 | 2.52 × 10−7 |
| Intracellular organelle | 4059 | 668 | 3.35 × 10−7 |
| Regulation of cellular process | 1130 | 255 | 5.51 × 10−7 |
In summary, we have performed proteomic and genomic approaches to provide relevant insight to further extend our knowledge on the NuA3 histone acetyltransferase complex and its role in transcriptional regulation. We studied the crosstalk between Sas3p and Gcn5p, characterised the proteins that co-purified with Sas3p in active HAT complexes and studied its dependency on the presence or absence of Gcn5p. Our work provides experimental-based evidence for the Pdp3 protein–protein interaction in the NuA3 HAT complex, characterising its post-translational modifications, to broaden its protein network among chromatin interacting proteins. Complementarily, the high throughput genomic analyses indicated that Sas3p binding takes place preferentially on the 5′ end of the ORF, mostly in genes involved in intracellular organelle-related processes.
3. Materials and methods
3.1. Yeast strains and culture media
The Saccharomyces cerevisiae yeast strains used in this study are listed in Table 4. Yeast cells were grown at 30 °C in yeast peptone dextrose (YPD) medium containing, or not, supplementary adenine. Epitope tags and gene replacements were introduced into the genome by PCR-mediated one-step gene replacement. The TAP-tagged yeast strains were constructed by gene targeting at the 3′ end of the ORF with a tag variant (CBP-T7-TEV-Protein A-KAN MX6) of the original TAP tag [48], which we constructed in B. Seraphin’s laboratory. Growth was assayed by spotting serial dilutions of fresh yeast cells exponentially cultured on YPDA and incubated at different temperatures.
Table 4.
Yeast strains used in this study.
| Strain | Genotype | Source or reference |
|---|---|---|
| BMA64-1A | MATa ade2-1 his3-11, 15 leu2-3, 112 trp1-Δ2 ura3-1 can1-100 | [57] |
| BY4742 | MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0 | Euroscarf |
| YPH250 | MATa ura 3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 | [58] |
| BQS1033 | BY4742 sas3Δ0::LEU2 | Our laboratory |
| BQS1180 | BY4742 gcn5Δ0::KAN MX4 | Our laboratory |
| BQS1217 | BY4742 SAS3-HA6-HIS3 | [25] |
| BQS1435 | BMA64-1A SAS3::TAP::KAN MX6 | This study |
| BQS1505 | YPH250 hif1Δ::NAT MX4 HAT2::TAP::KAN MX6 | Our laboratory |
| BQS1514 | BMA64-1A GCN5::TAP::KAN MX6 | This study |
| BQS1515 | BQS1033 GCN5::TAP::KAN MX6 | This study |
| BQS1517 | BQS1435 BMA64-1A SAS3::TAP::KAN MX6 gcn5Δ::NAT MX4 | This study |
3.2. Preparation of cell extracts for tandem affinity purification (TAP)
At least 6 L of cultured cells grown to an absorbance of OD600 ≈ 2.0 were harvested, instantly frozen in liquid nitrogen and cryo-ground 6 times during a 3-min period at maximum speed using a MM301 ball mill (Retsch). Frozen powdered cells were transferred into 100-mL containers and were stored at −80 °C until used. Cell extracts were prepared using 2 mL/g of cells of a buffer 40 mM HEPES, pH 7.9, 350 mM NaCl, 10% glycerol and 0.1% Nonidet NP-40, containing protease inhibitors 2 mM benzamidine, 1 mM PMSF, 2 mg/mL leupeptine, 2 mg/mL pepstatin A, 2.4 mg/mL chymostatin and 10 μL/mL Trasylol (Bayer). All the affinity purification steps were carried out at 4 °C, basically as described elsewhere [49]. To increase the SAS3-TAP protein purification yield, instead of commercial resins, we used IgG- (I4506, Sigma) and Calmodulin (P1431, Sigma)-coated Dynabeads (M-280, Invitrogen), prepared following the manufacturer’s instructions. For PDP3-TAP, we used commercial resin IgG-Sepharose (17-0969-01, Amersham) and Calmodulin-coated Dynabeads. Two antibodies were used to detect the TAP-tag: P1291 Sigma and CAB1001 Open Biosystems, which respectively recognise the Protein A or the calmodulin-binding peptide regions of the TAP tag. TAP-purified protein complexes were either directly analysed by MS, or dried, and then solved on sample buffer and resolved in 10% SDS–PAGE. Gel was stained with Colloidal Blue (R0571 Fermentas) prior to the MS analysis.
3.3. In vitro histone acetyltransferase specificity assays
The PhosphorImager HAT assay was performed as previously described [50], with minor modifications. HAT activities were determined over the whole chromatographic TAP-eluates (5 or 10 μL). Sample fractions were incubated for 20 min at 30 °C with chicken erythrocyte oligonucleosomes or free core histones used as a control and prepared as previously described [51] and with 0.005 μCi [1-14C]acetyl-CoA in a final volume of 16 μL. The reactions were finalised by adding 4 μL of 5× SDS sample loading buffer. The histones in the reaction mixtures were resolved by SDS–PAGE (16%). After staining/fixing and drying, gels were exposed to the BAS-SR IP phosphor-storage image plate (FujiFilm) for different times. Phosphor screens were scanned in a FujiFilm FLA-3000 fluorescent imager analyser.
3.4. Histone immunoblotting assays
For the histone Western blots, cells were collected at OD600 ≈ 0.8. WCEs were prepared as described elsewhere [52] and were resolved by SDS–PAGE (16%). Protein gels were transferred to 0.2-μm pore nitrocellulose membranes (Protan, Whatman), as described elsewhere [53]. Membranes, after staining with Ponceau S and subsequent washes, were processed according to the ECL Advance Western Blotting detection system (RNP 2135, GE Healthcare) instructions. The primary antibodies were the following: α-H3Ct (ab1791, Abcam), α-H3K9ac (06-942, Millipore) and α-H3K14ac (07-353, Upstate), and were used at a 10-fold dilution as suggested by the manufacturer. The secondary antibody was horseradish peroxidase-linked anti-rabbit (GE Healthcare), employed at a dilution of 1/40,000.
3.5. ChIP-on-chip experiments
ChIP-on-chip experiments were performed as previously described [45], but with some modifications. Briefly, the antibody used for the genome-wide location analysis was 2 μg of rat anti-HA 3F10 (Roche). The rat antibody was coupled to a suspension of 100 μL of 50% (v/v) Protein G Sepharose 4FF (Amersham Biosciences) equilibrated in lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM Phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 1 pill of protease inhibitor cocktail (Roche) was dissolved in every 25 mL of buffer) supplemented with 5 mg/mL of BSA. Next, 2000 ng of DNA immunoprecipitated from the BQS1217 strain (SAS3-HA tagged) and DNA immunoprecipitated from the BY4742 strain (WT) were labelled with Cyanine 5-dUTP and Cyanine 3-dUTP, respectively, using the ‘CGH Labelling Kit’ (Invitrogen, p/n 18095-011) following the manufacturer’s instructions. Labelled DNA was hybridised to the Yeast Whole Genome ChIP-on-chip Microarray (Agilent p/n G4491A, AMADID: 014741), specifically designed for location analyses.
Data were processed by ChIP Analytics, 1.3 (Agilent). Blank subtraction normalisation, and inter-array and intra-array median normalisation were performed. Probes were lifted over to the sacCer3 genome version (April 2011). Transcript boundaries [46] were downloaded from SGD (R64-1-1, http://www.yeastgenome.org). Raw and processed data were deposited in GEO: GSE56889. Plots were performed using R and Bioconductor [54]. Probes were binned to 200 bp, and the mean and its confident intervals (t-test, 95% confidence) were plotted. Experiments were carried out in duplicate.
3.6. Mass spectrometry analysis
The TAP-tagged co-purified proteins were treated and identified by LC–MS/MS at the 95% confidence level, as previously described [36,55]. Briefly, the proteins from the TAP-eluates were precipitated with TCA/acetone, solved and digested with trypsin. The resulting peptides were loaded into LC Packings Pep Map C18 columns and chromatographed by elution with a linear gradient of 5% to 45–50% of acetonitrile in 0.1% formic acid for 120 min. The peptides from the Sas3p sample were analysed in a nano ESI Q-TOF mass spectrometer (QSTAR XL, ABI) and those from the Pdp3p sample in a MALDI-TOF–TOF (4700 Proteomic Analyser, ABI). Searches were made in the Swiss-Prot database using the MASCOT and Protein Pilot search engines.
Slices from the gel corresponding to PDP3-TAP were excised and digested with sequencing grade trypsin (Promega), as described elsewhere [56]. The digestion mixture was dried in a vacuum centrifuge, and dissolved in 2% ACN, 0.1% TFA (samples 1 and 2 in 10 μL; samples 4,5 and 6 in 10 μL). Then 5 μL of each sample were loaded into a trap column (Nano LC Column, 3 μ C18-CL, 75 um × 15 cm; Eksigen) and desalted with 0.1% TFA at 2 μL/min for 10 min. Peptides were loaded into an analytical column (LC Column, 3 μ C18-CL, 75 um × 25 cm, Eksigen), equilibrated in 5% acetonitrile 0.1% FA (formic acid). Peptide elution was carried out with a linear gradient of 5–40% B in 30 min (A: 0.1% FA; B: ACN, 0.1% FA) at a flow rate of 300 nL/min. Peptides were analysed in a nano ESI qQTOF mass spectrometer (5600 TripleTOF, AB Sciex). TripleTOF was operated in the information-dependent acquisition mode, in which a 0.25-s TOF MS scan from 350 to 1250 m/z, was performed, followed by 0.05-s production scans from 100 to 1500 m/z on the 50 most intense 2–5 charged ions. MS/MS information was sent to MASCOT v 2.3.02 or to PARAGON via v 4.5 (AB Sciex). Samples 3, 4 and 5 were searched combined for customer convenience.
3.6.1. MASCOT search engine (matrix–science)
A database search was conducted in Swiss-Prot. Searches were made with tryptic specificity, and by allowing one missed cleavage and tolerance on the mass measurement of 50 ppm in the MS mode and of 0.6 Da for the MS/MS ions. The carbamidomethylation of Cys was used as a fixed modification, and oxidation of Met and deamidation of Asn and Gln were employed as variable modifications. The proteins with a score higher than homology or the significance threshold were identified with confidence ⩾95%. The protein score in the results reported from an MS/MS search was derived from the ions scores.
3.6.2. Protein Pilot v4.5 search engine (AB Sciex)
Protein Pilot default parameters were used to generate a peak list directly from the 5600 Triple TOF wiff files. The Paragon algorithm of Protein Pilot was used to search the Expasy protein database (538585 sequences; 191240774 residues) with the following parameters: trypsin specificity, cys-alkylation, taxonomy restriction to yeast, and search effort set at rapid. To avoid using the same spectral evidence in more than one protein, the identified proteins were grouped based on the MS/MS spectra by the Protein Pilot Progroup algorithm. Thus the proteins sharing MS/MS spectra were clustered, regardless of the peptide sequence assigned. The protein within each group that explained more spectral data with confidence was shown as the primary protein of the group. Only the proteins of the group for which individual evidence existed (unique peptides with enough confidence) were also listed, usually toward the end of the protein list. The proteins with an unused score >1.3 were identified with confidence ⩾95%.
3.7. Evidence of databank submission
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with dataset identifiers PXD001089 and DOI http://dx.doi.org/10.6019/PXD001089 and PXD001090 with DOI http://dx.doi.org/10.6019/PXD001090. The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org) consortium through IntAct (PMID: 22121220) and, assigned the identifier IM-23292. The data deposited in GEO (GSE56889) are accessible to reviewers through this link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=kryfcgkmrdsfpab&acc=GSE56889).
Acknowledgements
We wish to specially thank L. Valero who carried out the proteomic analyses in the CIPF and in the SCSIE Proteomics laboratory, a member of the ISCIII Carlos III Networked Proteomics Platform. The authors are also indebted to V. Pelechano for the data analysis of the genome-wide location, to R. Sendra and L. Yenush for critically reading and making useful corrections in the manuscript. This work has been supported by research grant BFU2008-01976 BMC to V.T. from the Spanish Ministry of Science and Innovation and by the Generalitat Valenciana grant (Prometeo 2011/088, Spain). S.V.-M. designed and performed most of the experiments; P.R. constructed materials and performed the preliminary TAP experiments; L.M.-P. carried out the ChIP-on-chip experiments; C.P.M.-J. contributed to the final manuscript preparation and discussion. V.T. conceived, designed and performed the ChIP-on-chip experiments, and wrote the manuscript; M.P. conceived and performed experiments, and wrote the manuscript. All the authors read, corrected and approved the paper.
Contributor Information
Sara Vicente-Muñoz, Email: svicente@cipf.es.
Paco Romero, Email: francisco.romero-gascon@uv.es.
Lorena Magraner-Pardo, Email: lomapar@alumni.uv.es.
Celia P. Martinez-Jimenez, Email: celia.martinez@uv.es.
Vicente Tordera, Email: vicente.tordera@uv.es.
Mercè Pamblanco, Email: merce.pamblanco@uv.es.
Appendix A. Supplementary data
Supplementary material.
Supplementary material.
References
- 1.Deal R.B., Henikoff S. Capturing the dynamic epigenome. Genome Biol. 2010;11:218. doi: 10.1186/gb-2010-11-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Martin C., Zhang Y. Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 5.Henikoff S., Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Tordera V., Sendra R., Perez-Ortin J.E. The role of histones and their modifications in the informative content of chromatin. Experientia. 1993;49:780–788. doi: 10.1007/BF01923548. [DOI] [PubMed] [Google Scholar]
- 9.Turner B.M. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 10.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee K.K., Workman J.L. Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 12.Shahbazian M.D., Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 13.Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 14.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Krebs J.E. Moving marks: dynamic histone modifications in yeast. Mol. BioSyst. 2007;3:590–597. doi: 10.1039/b703923a. [DOI] [PubMed] [Google Scholar]
- 16.Waterborg J.H. Dynamics of histone acetylation in Saccharomyces cerevisiae. Biochemistry. 2001;40:2599–2605. doi: 10.1021/bi002480c. [DOI] [PubMed] [Google Scholar]
- 17.Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Tong I.L., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Durant M., Pugh B.F. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:2791–2802. doi: 10.1128/MCB.26.7.2791-2802.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe L., Auston D., Grant P., John S., Cook R.G., Workman J.L., Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristjuhan A., Walker J., Suka N., Grunstein M., Roberts D., Cairns B.R., Svejstrup J.Q. Transcriptional inhibition of genes with severe histone H3 hypoacetylation in the coding region. Mol. Cell. 2002;10:925–933. doi: 10.1016/s1097-2765(02)00647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert F., Pokholok D.K., Hannett N.M., Rinaldi N.J., Chandy M., Rolfe A., Workman J.L., Gifford D.K., Young R.A. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess R.J., Zhou H., Han J., Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol. Cell. 2010;37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John S., Howe L., Tafrov S.T., Grant P.A., Sternglanz R., Workman J.L. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 2000;14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- 24.Martin D.G.E., Grimes D.E., Baetz K., Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol. Cell. Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosaleny L.E., Ruiz-García A.B., García-Martínez J., Pérez-Ortín J.E., Tordera V. The Sas3p and Gcn5p histone acetyltransferases are recruited to similar genes. Genome Biol. 2007;8:R119. doi: 10.1186/gb-2007-8-6-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govind C.K., Zhang F., Qiu H., Hofmeyer K., Hinnebusch A.G. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 27.Wyce A., Xiao T., Whelan K.A., Kosman C., Walter W., Eick D., Hughes T.R., Krogan N.J., Strahl B.D., Berger S.L. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 28.Zapater M., Sohrmann M., Peter M., Posas F., De Nadal E. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol. Cell. Biol. 2007;27:3900–3910. doi: 10.1128/MCB.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taverna S.D., Ilin S., Rogers R.S., Tanny J.C., Lavender H., Li H., Baker L., Boyle J., Blair L.P., Chait B. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi J.K., Grimes D.E., Rowe K.M., Howe L.J. Acetylation of Rsc4p by Gcn5p is essential in the absence of histone H3 acetylation. Mol. Cell. Biol. 2008;28:6967–6972. doi: 10.1128/MCB.00570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W., Bone J.R., Edmondson D.G., Turner B.M., Roth S.Y. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lafon A., Petty E., Pillus L. Functional antagonism between Sas3 and Gcn5 acetyltransferases and ISWI chromatin remodelers. PLoS Genet. 2012;8:e1002994. doi: 10.1371/journal.pgen.1002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eustice M., Pillus L. Unexpected function of the glucanosyltransferase Gas1 in the DNA damage response linked to histone H3 acetyltransferases in Saccharomyces cerevisiae. Genetics. 2014;196:1029–1039. doi: 10.1534/genetics.113.158824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lafon A., Chang C.S., Scott E.M., Jacobson S.J., Pillus L. MYST opportunities for growth control: yeast genes illuminate human cancer gene functions. Oncogene. 2007;26:5373–5384. doi: 10.1038/sj.onc.1210606. [DOI] [PubMed] [Google Scholar]
- 35.Kuo Y.M., Andrews A.J. Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PLoS ONE. 2013;8:e54896. doi: 10.1371/journal.pone.0054896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pamblanco M., Oliete-Calvo P., García-Oliver E., Luz Valero M., Sanchez del Pino M.M., Rodríguez-Navarro S. Unveiling novel interactions of histone chaperone Asf1 linked to TREX-2 factors Sus1 and Thp1. Nucleus. 2014;5:26–38. doi: 10.4161/nucl.29155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 38.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., Von Mering C. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smart S.K., Mackintosh S.G., Edmondson R.D., Taverna S.D., Tackett A.J. Mapping the local protein interactome of the NuA3 histone acetyltransferase. Protein Sci. 2009;18:1987–1997. doi: 10.1002/pro.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tackett A.J., Dilworth D.J., Davey M.J., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert T.M., McDaniel S.L., Byrum S.D., Cades J.A., Dancy B.C., Wade H., Tackett A.J., Strahl B.D., Taverna S.D. An H3K36me3 binding PWWP protein targets the NuA3 acetyltransferase complex to coordinate transcriptional elongation at coding regions. Mol. Cell. Proteomics. 2014;13:2883–2895. doi: 10.1074/mcp.M114.038224. mcp.M114.038224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eberharter A., Sterner D.E., Schieltz D., Hassan A., Yates J.R., 3rd, Berger S.L., Workman J.L. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tkach J.M., Yimit A., Lee A.Y., Riffle M., Costanzo M., Jaschob D., Hendry J.A., Ou J., Moffat J., Boone C. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dastidar R., Hooda J., Shah A., Cao T., Henke R., Zhang L. The nuclear localization of SWI/SNF proteins is subjected to oxygen regulation. Cell Biosci. 2012;2:30. doi: 10.1186/2045-3701-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magraner-Pardo L., Pelechano V., Coloma M.D., Tordera V. Dynamic remodeling of histone modifications in response to osmotic stress in Saccharomyces cerevisiae. BMC Genomics. 2014;15:247. doi: 10.1186/1471-2164-15-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jasiak A.J., Hartmann H., Karakasili E., Kalocsay M., Flatley A., Kremmer E., Strässer K., Martin D.E., Söding J., Cramer P. Genome-associated RNA polymerase II includes the dissociable Rpb4/7 subcomplex. J. Biol. Chem. 2008;283:26423–26427. doi: 10.1074/jbc.M803237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 49.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 50.Poveda A., Sendra R. An easy assay for histone acetyltransferase activity using a phosphorimager. Anal. Biochem. 2008;383:296–300. doi: 10.1016/j.ab.2008.08.029. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz-García A.B., Sendra R., Pamblanco M., Tordera V. Gcn5p is involved in the acetylation of histone H3 in nucleosomes. FEBS Lett. 1997;403:186–190. doi: 10.1016/s0014-5793(97)00049-5. [DOI] [PubMed] [Google Scholar]
- 52.Kushnirov V.V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 53.Thiriet C., Albert P. Rapid and effective Western blotting of histones from acid–urea–triton and sodium dodecyl sulfate polyacrylamide gels: two different approaches depending on the subsequent qualitative or quantitative analysis. Electrophoresis. 1995;16:357–361. doi: 10.1002/elps.1150160161. [DOI] [PubMed] [Google Scholar]
- 54.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oleaga A., Pérez-Sánchez R., Pagés E., Marcos-Atxutegi C., Simón F. Identification of immunoreactive proteins from the dog heartworm (Dirofilaria immitis) differentially recognized by the sera from dogs with patent or occult infections. Mol. Biochem. Parasitol. 2009;166:134–141. doi: 10.1016/j.molbiopara.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Shevchenko A., Jensen O.N., Podtelejnikov A.V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Boucherie H., Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baudin-Baillieu A., Guillemet E., Cullin C., Lacroute F. Construction of a yeast strain deleted for the TRP1 promoter and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast. 1997;13:353–356. doi: 10.1002/(SICI)1097-0061(19970330)13:4<353::AID-YEA86>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 58.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poveda A., Sendra R. Site specificity of yeast histone acetyltransferase B complex in vivo. FEBS J. 2008;275:2122–2136. doi: 10.1111/j.1742-4658.2008.06367.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.


