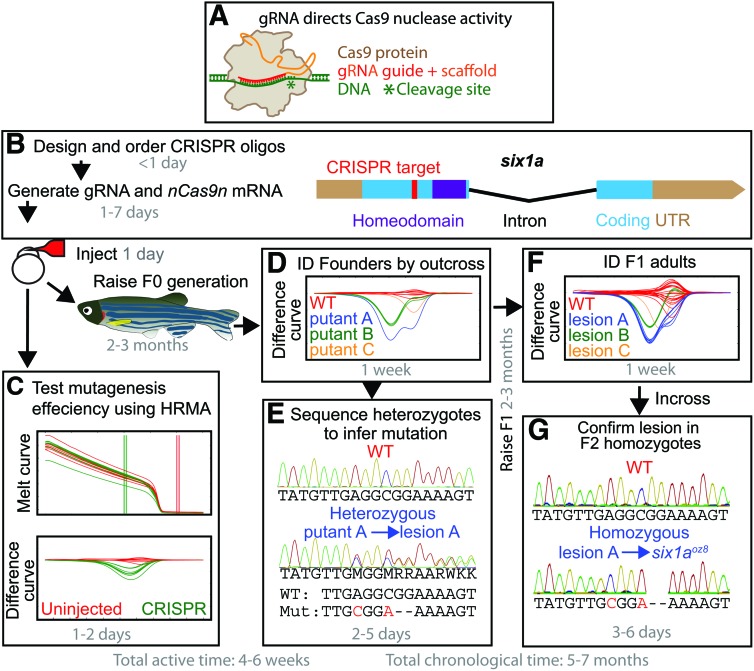
Over the past few years, the cost of directed mutagenesis in zebrafish has plummeted, opening the doors for many researchers to mutate their genes of interest.1 Most recently, CRISPR-Cas-based mutagenesis has taken center stage, with significant advances in zebrafish mutagenesis occurring almost every month. In this system, a short guide RNA (gRNA) directs Cas9 nuclease to cleave specific genomic target sites (Fig. 1A).2 Because it is simple to construct the targeting portion of this system,2 it is possible to target many genes with CRISPR on a limited budget and timescale.1 However, researchers often need to do significant troubleshooting and protocol building before generating mutants with CRISPR, which can slow progress. In this study, we present a detailed start-to-finish guide to CRISPR mutagenesis in zebrafish (Supplementary Data, http://molgen.osu.edu/amacher/resources; Supplementary Data are available online at www.liebertpub.com/zeb), intended to provide a simple method for generating strong loss of function alleles. We have used this pipeline to create mutations in 13 genes to date and provide an example of the pipeline using six1a CRISPR.
FIG. 1.
Mutagenesis pipeline with time estimates. (A) Illustration of a Cas9-gRNA complex bound to target DNA.12 (B) Example of CRISPR target site design for six1a (NCBI Gene ID 494168), with the target sequence downstream of potential alternative start sites and upstream of the homeodomain. (C) Polymerase chain reaction products from a subset of injected embryos (green lines) are subjected to a high-resolution melt curve and normalized to uninjected curves (red lines), revealing the melting difference caused by injection. (D) High-resolution melt analysis (HRMA) difference curves identifying three potentially different lesions in six1a were (E) sequenced from heterozygous F1 generation embryos. HRMA curves identified in this initial analysis are confirmed by (F) genotyping adult F1 generation fish fin DNA. (G) Homozygous mutant F2 generation sequence reads confirm the lesions inferred from heterozygous F1 generation sequences. Once confirmed, lesions are given allele designations. The mutation in six1a lesion A is six1aoz8, a 2 bp deletion that frameshifts Six1a after amino acid 93 of 284 and introduces 37 aberrant aa before terminating. Sequence analysis for lesion B (six1aoz9), which frameshifts Six1a after amino acid 94 and introduces only 12 aberrant aa, is described in the Supplementary Data. Lesion C is another 2 bp deletion (not shown). Color images available online at www.liebertpub.com/zeb
We provide guidelines to identify CRISPR targets sites most likely to cause loss of protein function, two protocols to generate gRNA,2,3 and a pipeline to identify and confirm mutations (Fig. 1B–G). CRISPR target sites are selected at locations that cannot be bypassed by alternative start codon usage or alternative first exon usage and are 5′ to regions predicted to be important for protein function. After injection of gRNA and nuclear localized zebrafish codon-optimized cas9 (nls-zCas9-nls) mRNA,4 mutagenesis is quantitatively assayed using high-resolution melt analysis (HRMA) on polymerase chain reaction (PCR) products spanning the CRISPR target. Successful mutagenesis in the injected generation (F0) results in a mixture of WT and mutant PCR products that melt at lower temperatures than uninjected WT controls (Fig. 1C).5 HRMA can also speed up founder recovery in the first outcrossed generation (F1) (Fig. 1D) because each heterozygous lesion produces a characteristic melt curve.5 We recognize that HRMA machines typically cost $10–25K, but costs can be mitigated by sharing one machine between several laboratories or by pairing an existing quantitative PCR machine with online HRMA software6; alternative approaches have also been developed to identify lesions.7–10 The insertions and deletions of genomic DNA (indels) caused by CRISPR mutagenesis result in overlapping sequence reads in heterozygotes, and the mutant read can easily be extracted from heterozygous sequencing by subtracting WT sequence (Fig. 1E). The HRMA-identified potential mutants (putants) found in F1 generation embryos are confirmed on F1 adults (Fig. 1F), and sequencing from F2 generation homozygotes confirms the F1 heterozygous sequencing (Fig. 1G). This pipeline can be completed using 5 weeks labor, spread over 5–7 months (Fig. 1), and during this time, a single researcher can construct many CRISPR mutants in parallel.
The CRISPR-Cas technology has provided an abundance of options for genome modification.11 In this protocol, we do not explore every possible CRISPR application, but focus on a single purpose: to frameshift target genes at locations where small (<60 bp) indels are likely to cause loss of protein function. Although our protocol is written with simple indel construction in mind, we expect that it will provide zebrafish researchers with a good starting point to explore the vast array of CRISPR-Cas applications.
Supplementary Material
Acknowledgments
The Amacher laboratory fish facility staff provided excellent fish care. April DeLaurier, Ryan Anderson, Joy-El Talbot, and Tom Gallagher provided comments on the article. This work was supported by the Pelotonia Postdoctoral Fellowship Program and by NIH grant R01GM088041.
Disclosure Statement
No competing financial interests exist.
References
- 1.Blackburn PR, Campbell JM, Clark KJ, Ekker SC. The CRISPR system—keeping zebrafish gene targeting fresh. Zebrafish 2013;10:116–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. . Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013;31:227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, et al. . Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 2013;140:4982–4987 [DOI] [PubMed] [Google Scholar]
- 4.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A 2013;110:13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, et al. . Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 2012;8:e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dwight ZL, Palais R, Wittwer CT. uAnalyze: web-based high-resolution DNA melting analysis with comparison to thermodynamic predictions. IEEE/ACM Trans Comput Biol Bioinform 2012;9:1805–1811 [DOI] [PubMed] [Google Scholar]
- 7.Ota S, Hisano Y, Ikawa Y, Kawahara A. Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells 2014;19:555–564 [DOI] [PubMed] [Google Scholar]
- 8.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 2009;19:1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu C, Zhang Y, Yao S, Wei Y. A PCR based protocol for detecting indel mutations induced by TALENs and CRISPR/Cas9 in zebrafish. PLoS One 2014;9:e98282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, et al. . Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 2014;9:e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampson TR, Weiss DS. Exploiting CRISPR/Cas systems for biotechnology. Bioessays 2013;36:34–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, et al. . Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014;156:935–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.