Abstract
The nematode worm Caenorhabditis elegans comprises an ancestral immune system. C. elegans recognizes and responds to viral, bacterial, and fungal infections. Components of the RNA interference machinery respond to viral infection, while highly conserved MAPK signaling pathways activate the innate immune response to bacterial infection. C. elegans has been particularly important for exploring the role of innate immunity in organismal stress resistance and the regulation of longevity. Also functions of neuronal sensing of infectious bacteria have recently been uncovered. Studies on nematode immunity can be instructive in exploring innate immune signaling in the absence of specialized immune cells and adaptive immunity.
1. Introduction: The C. elegans model for studying immunity
C. elegans is a small nematode that can be found in the soil, rotting fruit, compost heaps, and snails [1]. The nematode worm has been employed since nearly half a century in the laboratory where it can be maintained in a cost effective manner on agar plates typically containing E. coli OP50 strain as a food source [2]. The worm’s development from embryo to adulthood takes 3 days at a standard temperature of 20°C; within following 3 days of adulthood a single hermaphrodite worm produces approximately 300 genetically identical progeny facilitating the establishment of large animal populations. Males can easily be obtained in order to perform genetic crossings. The average life span of C. elegans at 20°C is approximately 2-3 weeks. The genome of the nematode is sequenced and loss-of-function mutants for the majority of genes are available from public sources. Also gene knock-downs can easily be established by feeding worms on bacteria that contain vectors expressing respective gene-specific dsRNAs. All these factors have to date made C. elegans a model of choice for many genetic studies that produced major breakthroughs such as understanding of cell fate determination during development, the process of apoptosis, mechanisms of RNA interference, the genetic regulation of life span, to only name a few. With the discovery of inducible anti-microbial and anti-viral innate immune responses in the nematode over the past fifteen years, C. elegans has emerged as intriguing model also in the field of immunity.
Several microbes that are known human pathogens also infect the intestine of the nematode. Therefore, their virulence and other aspects of host-pathogen interaction can be studied in the simple metazoan. From an evolutionary perspective, the immune system of C. elegans precedes the highly complex immunity of vertebrates as it contains only some of the most ancestral signaling networks. The reduced complexity can be experimentally advantageous in terms of detailed characterization of the immune signaling cascades. Also systemic interactions between innate immunity and other signaling systems such as stress responses and mechanisms involved in the regulation of longevity have been established by using C. elegans. Recently, a novel function of innate immunity in integrating cell autonomous hazards such as germline DNA damage with systemic stress resistance has been uncovered. Another topic of considerable attention and controversy -the regulation of innate immunity by the nervous system- has also gained some experimental proof from studies in C. elegans. All together, in addition to offering a highly suitable model for classical studies of immunity, the relatively low complexity of the nematode system also provides extended possibilities for dissecting non-conventional aspects of innate immune responses.
2. Anti-viral immune defense in C. elegans
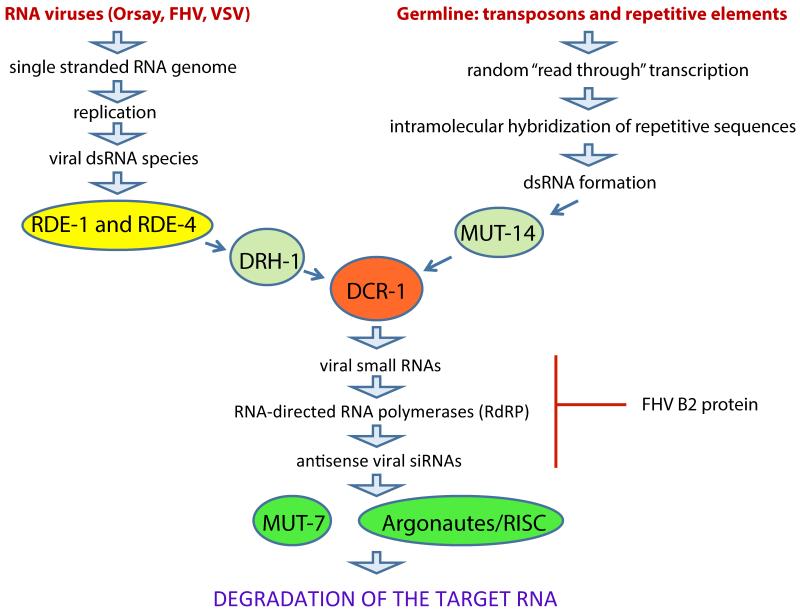
The anti-viral response of the nematode relies on the intracellular RNA interference (RNAi) machinery (Figure 1). Anti-viral surveillance through the RNAi machinery is an ancestral defense mechanism that was first described and characterized in plants [3]. The attempts to dissect the anti-viral immune functions of C. elegans were for a long time complicated by the absence of a natural virus that would infect the nematode. However, studies based on infecting isolated worm cells with the mammalian Vesicular stomatitis virus (VSV) [4] and on transgenic expression of the Flock house virus (FHV) replicon in the worm [5], immediately suggested the key role of the RNAi machinery in the process of antiviral defense with factors like the argonaute protein RDE-1 and the dsRNA binding protein RDE-4 being essential for antiviral silencing.
Figure 1. Antiviral responses of C. elegans.
The antiviral response in the nematode C. elegans is mediated by the RNAi machinery. The double stranded RNA intermediates produced during viral replication are first sequestered and retained by the RDE-1/4 dsRNA binding complex. The DExD box RNA helicase DRH-1later unwinds RNA molecules and facilitates their acquisition by the Dicer complex. Small viral RNAs generated by the Dicer homolog DCR-1 are amplified by RNA-directed RNA polymerases (RdRP) generating large numbers of anti-sense viral siRNAs that in turn trigger degradation of the target mRNA in a manner dependent on Argonaut proteins and the Rnase MUT-7. A similar pathway is used for the transcriptional silencing of transposons and repetitive DNA elements in the germline. In this pathway dsRNA species produced through intramolecular hybridization of repetitive sequence regions are sequestered, unwound, and delivered to DCR-1 by the DExD box RNA helicase MUT-14 which acts in a manner similar to DRH-1. RISC - RNA induced silencing complex.
Only recently natural sampling experiments led to the discovery of the first natural viral pathogen of C.elegans: the RNA+ Orsay virus that belongs to the Nodaviridae family [6] and causes easily detectable morphological abnormalities in the intestine of the worm. It was demonstrated that infection load upon treatment with the Orsay virus was elevated in mutants lacking such important RNAi factors as RDE-1, RDE-4, the RNaseD MUT-7 and the dicer-related helicase DRH-1, which also carries a RIG-I-like domain [6-8]. Also the presence of small viral RNAs (both primary sense and secondary effector anti-sense) was detected upon infection. Interestingly, the susceptibility to the viral infection varied between different wild-type C. elegans isolates suggesting natural variation in somatic RNAi efficiency [6].
The opportunity of using bona fide viral infection in combination with previously reported functions of specific RNAi factors significantly extended our mechanistic view of anti-viral surveillance in C. elegans. It is currently believed that upon infection with an RNA virus, the double stranded RNA intermediates produced during replication of the viral genome comprise the initial step of the RNAi response. The intermediates are first sequestered by the dsRNA binding complex RDE-1/RDE-4 whose proposed function is detection and retaining of exogenous dsRNAs [9]. The dsRNA is then passed to the DExD box RNA helicase DRH-1 that interacts with RDE-1/4 [9], unwinds the RNA molecule and facilitates its acquisition by the worm dicing complex [7]. The primary small RNAs produced through processing of the double stranded replication intermediate by the C. elegans Dicer homolog DCR-1 serve as template for RNA-directed RNA polymerases (RdRP) resulting in accumulation of secondary anti-sense viral small RNAs that mediate degradation of the target full length viral RNA in a manner dependent on the Rnase MUT-7 [10] and Argonaut proteins.
Similar mechanisms are also utilized for the silencing of transposons and repetitive elements, such as multiple copy transgenes, in the germline of C. elegans. Thus far, no apparent natural integration of exogenous elements was found in the C. elegans germline, as no vertical transfer of the virus was detected in case of Orsay infection [6]. The germline silencing pathway might be evolutionary derived form the CRISPR mechanisms - an ancestral prokaryotic “immune system” conferring RNAi-like surveillance against exogenous genetic elements (such as phage genomes) integrated into bacterial DNA [11]. In case of germline silencing pathway the initial step that triggers an RNAi response is random “read through” transcription of the DNA region containing repetitive elements. Upon transcription double stranded RNA structures are formed via intramolecular cross-hybridization of repetitive sequences [12]. Such structures are putatively recognized by the DExD box RNA helicase MUT-14 that is proposed to unwind dsRNA and pass it to DCR-1 for processing similar to the somatic DRH-1 helicase [10]. It has been clearly demonstrated that both DCR-1 and MUT-14 are essential for transposon silencing in the germline with MUT-14 playing a role only in the germline and not during somatic RNAi. The final steps of the germline transposon repression pathway are proposed to be the same as the ones described for the somatic anti-viral RNAi.
Different mammalian viruses are known to have developed ways of avoiding their detection by the immune system. For instance, several viruses are capable of blocking MHC class I antigen processing and presentation, thereby escaping recognition by T-killer cells [13]. The most prominent example how viral proteins escape the RNAi surveillance through inhibiting the RNAi machinery in C. elegans is the Flock house virus protein B2 that significantly downregulates exogenous RNAi and enhances susceptibility of worms to the Orsay virus infection without having any effect on the endogenous physiological micro-RNA pathway [14]. Another important connection between C. elegans and mammalian anti-viral mechanisms is the homology between DRH-1 and the RIG-I (Retinoic acid inducible gene I) RNA helicase – a key cytoplasmic sensor of foreign dsRNAs in mammals [7,8]. The strongest homology between the two proteins resides within DExD/H-box helicase domain responsible for RNA binding, while domains involved in protein-protein interactions are different consistent with the fact that DRH-1 targets dsRNAs for Dicer processing whereas RIG-I activates a classical systemic anti-viral immune response mediated by interferones and pro-inflammatory cytokines [15]. It seems probable that the two molecules as well as the two responses to the presence of viral RNA are evolutionary connected with currently existing differences having evolved according to particular needs of respective organismal systems: while cell-autonomous anti-viral surveillance is sufficient in C. elegans a more complex mammalian physiology raises a requirement for a systemic alert and response. It is also worth mentioning that DRH-1 is dispensable for RNAi that targets cellular transcripts [7] making this protein a specific factor in foreign sequence RNAi surveillance.
3. Anti-microbial responses in C. elegans
C. elegans is naturally exposed to numerous microbes, some of which can be benign food sources others dangerous pathogens for the worm. The close interaction throughout millions of years of nematode and bacterial evolution led microbes to evolve ways of keeping nematodes at large and worms to shape means of avoiding bacterial infections and toxicity. Most bacterial species that are known to be pathogenic for C. elegans cause deleterious phenotypes by infecting the intestine of the worm. Interestingly, bacterial gut colonization as such is not necessarily detrimental as demonstrated by feeding worms with Enterococcus faecium which propagates in the intestine to large titers without causing significant death of the animals [16]. Microbes only become pathogenic to the worms when they produce active virulence factors. The bacteria causing intestinal infections in C. elegans belong to both Gram-positive (Enterococcus faecalis, Staphylococcus aureus [16,17]) and Gram-negative (Pseudomonas aeruginosa, Salmonella typhimurium, Serratia marcescens, [18-20]) classes. Many of the C. elegans intestinal pathogens are also pathogenic to humans under all or restricted conditions with a number of virulence factors targeting nematodes and mammals, making C. elegans a powerful model system for virulence factor screens [18,21,22].
The most studied C. elegans intestinal pathogens are P. aeruginosa and S. typhimurium. P. aeruginosa is a common soil and water bacterium and an opportunistic human pathogen that causes disease in immune compromised individuals or upon extensive wounding and trauma. Under laboratory conditions the clinical isolate of P. aeruginosa strain PA14 has been demonstrated to kill C. elegans and inflict infection in mice [18,21]. Two types of nematode killing were recognized for P. aeruginosa: the so called “fast killing” occurring within several hours and mediated by bacteria grown on high-osmolarity medium and the “slow-killing” by bacteria cultured on low-nutrient media which takes several days [18,21]. The mechanisms of the two killing processes are distinct: the fast killing is mediated by toxins such as phenazines and does not require living bacteria while slow killing is a bona fide infectious process that involves living microbes. A screen based on using random transposon insertions in PA14 strain led to identification of several virulence factors required for maximum killing of the worms during slow infection [18]. Three of the identified genes - lasR, gacA and lemA- have already been identified as virulence factors in other model systems. GacA and lemA (also known as GacS) are components of GacA/GacS regulatory system that is responsible for bacterial biofilm formation while lasR encodes an essential transcriptional activator. S. typhimurium, a serotype of Salmonella enterica, is lethal to mice by causing typhoid-like disease; C. elegans worms fed with S. typhimurium from adulthood also exhibit reduced life span [19]. Interestingly, infection with Salmonella is accompanied by an increase of programmed cell death in the germline of C. elegans [23] that involves the core apoptotic factors the Apaf1 homolog ced-4 and the caspase ced-3. Mutant worms lacking apoptotic induction are more susceptible to S. typhimurium indicative of a protective role of germline apoptosis during pathogen defense. Bacterial mutants of the phoP/phoQ virulence-signaling pathway and genes involved in lipopolysaccharide synthesis result in reduced killing and lack of programmed cell death during S. typhimurium infection suggesting a link between pathogenicity, germ cell apoptosis, and antimicrobial defense.
Not only the intestine but also the worm’s cuticle can be target of infections. The most well studied cuticle distortion is the one caused by Microbacterium nematophilum. M. nematophilum is a Gram-positive organism that is able to colonize the C. elegans rectum, which is lined with cuticle, as well as the small peri-anal region of the exterior cuticle. During infection the anal region becomes swollen producing a phenotype known as Dar (deformed anal region) [24]. Despite being non-lethal, Microbacterium infection induces an immune response in the nematode [25]. Yersinia pseudotuberculosis a close relative of a bubonic plague pathogen Y. pestis, produces polysaccharide rich biofilms that attach predominantly to the cuticle of the head region of C. elegans preventing the worm from feeding and inhibiting growth [26]. This phenomenon is similar to how Y. pestis distorts the feeding of the flea promoting its transmission to mammals. Feeding blockage in both C. elegans and flea require genes involved in polysaccharide biosynthesis [27].
The mechanisms involved in recognition of microbes by C. elegans are incompletely understood. In mammals and Drosophila, Toll like receptors and Imd/TNF signaling are major driving mechanisms of anti-microbial response. The nematode’s genome encodes a single Toll like receptor homolog TOL-1 that is, however, not implicated in anti-microbial defense unlike its Drosophila and mammalian counterparts [28]; neither are the nematode orthologs of TNF receptor associated factor-1 (TRF-1), Pelle and IL-1R-associated kinases (PIK-1) and inhibitor of NF-κB (IKB-1). Other key components of known anti-microbial recognition cascades such as TLR adaptor MYD88 and NF-κB family of transcription factors have to date not been found in the C. elegans genome. In addition to TOL-1, another gene product containing Toll/IL-1R (TIR) protein-protein interaction domain - a scaffold protein TIR-1, was identified in the nematode. Genetic analysis uncovered that both knock-down and mutations of tir-1 result in severe hypersensitivity to a variety of established C. elegans pathogens implicating TIR-1 in anti-microbial defense [29-31]. However, the receptor(s) that activates TIR-1 in response to microbial presence still remains to be identified.
In parallel to genomics-based approach, genetic screens for mutants showing enhanced killing by P. aeruginosa identified a p38 MAP kinase-related signaling pathway as an essential player in anti-microbial defense (Figure 2). The cascade consists of neuronal symmetry family member 1 (nsy-1), SAPK/ERK kinase 1 (sek-1) and p38 MAPK family member 1 (pmk-1) [32,33]. NSY-1, SEK-1, and PMK-1 seem to function in a pathogen-activated linear phosphorylation cascade, where PMK-1 is phosphorylated by SEK-1 and SEK-1 by NSY-1. This pathway is orthologous to mammalian apoptosis signal-regulating kinase 1 (ASK1) - MAPK kinase 3/6 (MKK3/6) - p38 MAPK cascade, which is also involved in triggering innate immune responses. Knock-down of tir-1 blocks activation of PMK-1 upon infection suggesting TIR-1 as candidate for the most upstream member of the cascade [29]. The phosphorylation state of PMK-1 is also regulated by heavy metal stress activated MAPK kinase MEK-1 and MAP kinase phosphatase VHP-1 [33].
Figure 2. Antimicrobial responses in C. elegans.
The central signaling modules of the antimicrobial response of the nematode are conserved MAP kinase mediated signaling cascades. Upon exposure to fungi and intestinal infection the p38 homolog PMK-1 is induced via upstream kinases NSY-1 and SEK-1 and the scaffold protein TIR-1. The receptor that activates TIR-1 is still unknown. The PMK-1 induced nuclear response is to a large extent mediated by the transcription factor ATF-7 leading to expression of putatively secreted innate immune effectors such as C-type lectins, CUB domain containing proteins and antimicrobial peptides. Another MAP kinase homolog - MPK-1/ERK, is activated during infection with Microbacterium nematophilum by upstream kinases LIN-45 and MEK-2. The transcriptional response that is triggered by MPK-1 is similar to the one mediated by PMK-1 however the exact nuclear mediator is not known. Also the TGF-beta related pathway consisting of a secreted ligand DBL-1, two membrane anchored receptors (SMA-6 and DAF-4) and three cytoplasmic signal transducers (SMA-2-4), is implicated in antimicrobial defense based on the need of its functionality for the infection survival and on gene expression profiles of respective loss of function mutants.
Microarray studies conducted by several groups revealed common gene expression signatures induced by various pathogens in a MAP kinase dependent manner [34-36]. Three groups of putative secreted proteins have been implicated in mediating the worm’s innate immune response (Figure 2). One large group of pathogen induced factors consists of the C-type lectin domain containing proteins that bind specific carbohydrates on the bacterial surface [37]. A second group includes proteins containing a CUB domain characteristic of a diverse set of mostly extracellular proteins, including proteases. CUB stands for C1s/C1r complement components, the embryonic sea urchin protein (Uegf), and bone morphogenetic protein 1 (Bmp1) [38]. The third group of putative secreted proteins is comprised of antimicrobial peptides that may function similarly to mammalian AMPs, despite the lack of direct structural similarity, which, however, might be difficult to determine for such small polypeptide sequences [39]. The transcription factor ATF-7 was found to transcriptionally control the expression of innate immune genes downstream of PMK-1 [40]. PMK-1 signaling also induces the programmed germ cell death upon infection with S. typhimurium [41].
While PMK-1 appears to be the most commonly evoked MAPK in the nematode’s pathogen defense, also the ERK1/2 MAPK homolog MPK-1 can be activated in response to specific pathogens (Figure 2). For example, the Dar response inflicted by M. nematophilum leads to an induction of immune gene expression through MPK-1 [34,42]. In addition to relatively well-characterized MAPK dependent immune response, the DBL-1 (decapentaplegic/bone morphogenic protein like-1) signaling pathway that is homologous to the mammalian TGF-β cascade was implicated in the C. elegans anti-microbial immunity. This pathway consists of a secreted molecule (DBL-1), two membrane anchored receptors (SMA-6 and DAF-4) and three cytoplasmic signal transducers (SMA-2-4). Animals lacking almost any member of this signaling cascade show enhanced susceptibility to microbial infections [25,43]. Also expression pattern of genes induced upon infection with Serratia marcescens overlapped with those controlled by dbl-1 [43]. Interestingly, unlike the C. elegans counterpart, the mammalian TGF-β pathway has a potent immunosuppressive function.
4. Interaction between immunity and longevity pathways
Also mutations in pathways that do not appear to primarily respond to microbial infection can dramatically alter pathogen resistance. Mutations in the insulin-like growth factor receptor-1 homolog daf-2 or inactivation of germline development either by laser ablation or mutations in the Notch receptor glp-1 increase pathogen resistance of C. elegans [44-47] (Figure 3). This phenomenon was shown to depend on the function of the Forkhead family transcription factor DAF-16 mainly in the intestine of the nematode [47]. Interestingly activation of DAF-2 –resulting in inactivation of DAF-16- was proposed as a mechanism of host immunity avoidance by P. aeruginosa [48]. Gene expression studies implicated DAF-16 in inducing putative anti-microbial genes consistent with its role in pathogen tolerance in addition to aging [49,50]. Interestingly despite its requirement for pathogen resistance of daf-2 mutants and germline-depleted animals, lack of daf-16 per se did not sensitize nematodes to pathogen infections unlike pmk-1 deficiency [44]. The letter observation indicated that other cascades could also contribute to pathogen resistance of long-lived animals. Indeed, loss of pmk-1 not only had a significant negative effect on pathogen resistance but also partially suppressed life span extension of daf-2 mutants [35]. Gene expression analysis performed in the same study indicated that distinct genes were positively regulated by PMK-1 and DAF-16 during infection suggesting that these two cascades work in parallel downstream of daf-2 inactivation to mediate pathogen defense. When discussing the negative effect of pmk-1 on lifespan extension one has to however keep in mind additional functions of PMK-1 unrelated to immunity such as for instance activation of SKN-1 transcription factor in response to oxidative stress [51]. It has been demonstrated that inactivation of skn-1 also suppresses long-lived phenotype of daf-2 mutants worms [52]. However, lifespan extension is not always accompanied by enhanced pathogen resistance. For instance, mitochondrial clk-1 mutant animals or animals undergoing caloric restriction become long-lived but not pathogen resistant [53]. Noteworthy, DAF-16 appears to be dispensable for those two longevity assurance pathways [54,55].
Figure 3. Interaction of the C. elegans innate immunity with stress and longevity pathways.
Germline depleted nematodes and nematodes lacking functional insulin-like growth factor 1 receptor homolog DAF-2 are long-lived and pathogen resistant. The extended life span under these conditions is believed to largely depend on the FOXO transcription factor DAF-16, which mediates induction of anti-stress genes. Interestingly, also pathogen resistance is to some extent DAF-16 dependent. The PMK-1/p38 MAP kinase, whose main role is to mediate conventional innate immune responses, was also shown to contribute to enhanced pathogen resistance during reduced insulin-like signaling and lack of the germline. Of note, PMK-1 and DAF-16 seem to regulate innate immune responses by activating non-overlapping sets of genes. In addition to its immune function, PMK-1 also contributes to longevity by mediating responses to oxidative stress in a manner dependent on the transcription factor SKN-1. The induction of conventional innate immune response is accompanied by protein folding stress in the endoplasmic reticulum and intestinal oxidative stress. Stress management mechanisms such as ER unfolded protein response are essential for the animal’s survival during infection.
Similar to humans and other model organisms, immune function of C. elegans appears to decline with age. Adult animals (days1-4 and day 7 of adulthood) showed enhanced susceptibility to a variety of pathogens when compared to worms at the final pre-adulthood larval stage L4 [56]. The OP50 E.coli strain of bacteria, the standard laboratory food of C. elegans, is non-pathogenic when cultured on minimal agar media. While early in life no intact bacteria remain in the nematode’s intestine of the nematode, old intestines become colonized growing to large titers, which is thought to contribute to the death of animals. Indeed worms grown either on heat- or UV-killed OP50 have an extended lifespan [57,58]; and conversely when worms were fed OP50 that was mildly pathogenic due culturing on brain-heart infusions, their life span was shortened [16]. The age related tissue changes were similar in all worms irrespective of the food source indicating that bacteria might not influence the rate of normal aging. Indeed, PMK-1 expression was shown to decline with age [59], which could at least in part explain age-related immune senescence.
5. Interaction between immunity and stress response pathways
While innate immune response is essential for anti-viral and anti-microbial defense, its excessive and uncontrolled induction has proven to be detrimental in humans and various animal models leading to tissue degeneration and immune disorders. Also in C. elegans, the activation of immunity is closely associated with stress. For instance, resistance to infection with Enterococcus faecalis required the generation of reactive oxygen species (ROS) in the intestine, while DAF-16-mediated induction of the ROS-detoxification genes sod-3 and ctl-2 protects the tissues from the ensuing oxidative stress [60]. In addition, PMK-1-dependent anti-oxidant response via activation of SKN-1 might also enable the worms to withstand the dual oxidase Duox1/BLI-3-mediated ROS production during pathogen infection [61].
In addition to coping with ROS, the pathogen defense response also evokes considerable protein folding stress making the functionality of protein refolding cascades an essential pre-requisite of infection tolerance. The protein folding stress might result from a major shift in the proteome towards the production of large amounts of secreted innate immune peptides (see above). The heat stress inducible chaperone system regulated by transcription factor HSF-1 was shown to be required for normal tolerance to a variety of C. elegans pathogens [62]. While inactivation of hsf-1 made worms infection sensitive, heat stress treatment prior to infection enhanced survival. Also life span extension during infection due to inactivation of daf-2 required a functional heat stress response.
Massive production of innate immune peptides during the process of anti-microbial defense generates protein folding stress in the endoplasmic reticulum (ER) and raises a requirement for the functional endoplasmic reticulum unfolded protein response (UPR ER) to ensure optimal infection survival (Figure 3). Worms lacking key regulator of UPR ER, the XBP-1 transcription factor, demonstrate markedly reduced life span during feeding on pathogenic P. aeruginosa [63]. The pathogen sensitivity of xbp-1 mutant worms depends on PMK-1 mediated induction of immune peptides highlighting the initial source of protein folding stress. Also the so-called “non-canonical UPR” mediated by abu (activated in blocked UPR) genes appears to play a role in infection tolerance. CED-1, a phagocytic receptor previously implicated in the engulfment of apoptotic bodies, regulates expression of abu genes during infection [64]. ced-1 loss of function mutants are severely immunocompromised and rapidly killed by pathogenic bacteria.
6. Neuronal regulation of immunity
The modulation of the immune response by the nervous system has been a topic of controversy for many years. While interaction between immunity and neurons always seemed plausible, the complexity of the two systems in most model organisms prevented researches from detailed understanding of this phenomenon. In C. elegans the identity, morphology and synaptic connectivity of its 302 neurons is well described making it a highly suitable model for neural-immune studies. One way for the nematode to respond to the presence of pathogenic bacteria is by activating a conserved pathogen avoidance behavior. By investigating natural variations in C. elegans pathogen resistance a polymorphism in G-protein coupled receptor (GPCR) gene npr-1 was found to impair survival during PA14 infection by inhibiting oxygen dependent pathogen avoidance [65]. An earlier study that relied on systematic inactivation of different GPCRs also identified npr-1 as a factor essential for nematode survival during infection with various bacteria [66]. By placing worms on plates completely covered with bacterial lawn, it became apparent that lack of avoidance behavior could only partially explain the difference in survival between wild type and mutant animals. Indeed, gene expression profiling of npr-1 deficient animals revealed differential regulation of genes known to be expressed in the intestine during infection in a PMK-1 dependent manner [66]. Ablation of AQR, RQR and URX neurons that are involved in integration of environmental signals rescued the pathogen sensitivity phenotype of the npr-1 mutants, suggesting that npr-1 deficiency leads to hyperactivity in these neurons to modulate immunity. Lack of those neurons generally resulted in increased survival of worms during microbial infection, indicating that this neuronal circuit might have an inhibitory function during immune response. Another G-protein coupled receptor OCTR-1 acts in ASH and ASI sensory neurons to suppress not only the infection-driven activation of PMK-1 but also the induction of non-canonical UPR in peripheral tissues. octr-1 deficient animals show enhanced resistance to infection with P. aeruginosa that is independent of pathogen avoidance [67]. Interestingly, OCTR-1 is a catecholamine receptor related to vertebrate adrenergic receptors whose ligand is noradrenalin that mediates responses to acute stress in mammals that are known to be accompanied by immunesuppresion [68]. Altogether the findings discussed above strongly point toward dynamic regulation of immunity by the nervous system.
7. Innate immunity as an integrator of cell-restricted damage and systemic stress resistance
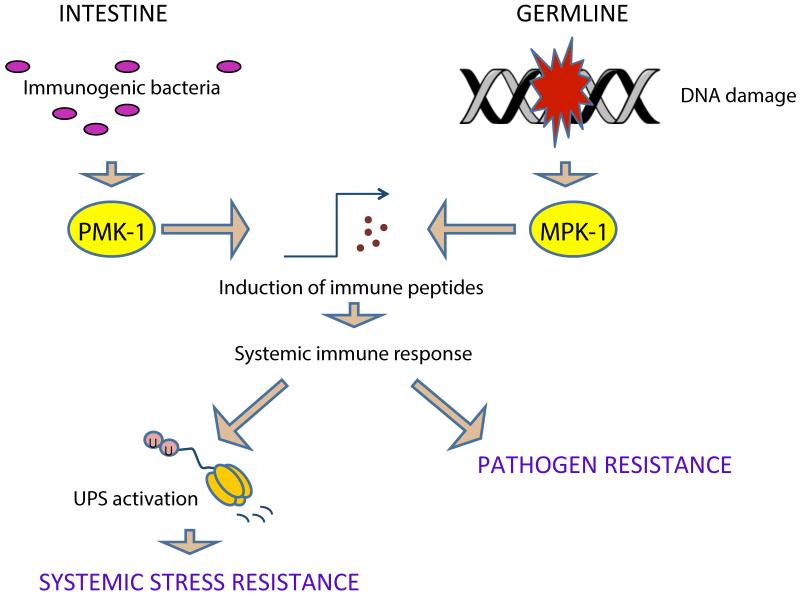
In addition to their conventional role in pathogen clearance and recycling of damaged tissue material, the secreted factors of the innate immune response have recently emerged as a signaling module that connects cell-specific DNA damage to systemic multi-stress resistance (Figure 4). Germline DNA damage-induced systemic stress resistance (GDISR) leads to elevated tolerance of heat and oxidative stress, and pathogen infection [69]. DNA damage in C. elegans germ cells leads to activation of MPK-1 that induces immune peptides in the germline, which in turn lead to enhanced proteostasis in somatic tissues through activation of the ubiquitin proteasome system (UPS). Interestingly, the PMK-1 dependent induction of intestinal immunity by feeding nematodes with immunogenic bacteria had a similar effect on systemic stress tolerance. Transient feeding worms with immunogenic bacteria also led to elevated activity of the ubiquitin proteasome system (UPS). In contrast, prolonged exposure to immunogenic bacteria led to apparent overloading of the UPS indicating an important function for UPS-mediated protein homeostasis during innate immune responses in C. elegans [69]. Intriguingly, upon germ cell DNA damage progeny production is transiently halted due to the activity of cellular DNA damage checkpoints in the germline but resumes later in life, when normally worms have already passed their peak of fertility. GDISR might thus function as an organismal DNA damage checkpoint that allows extension of reproductive lifespan until germ cell have had sufficient time to repair their damaged genomes before offspring production resumes [70,71]. Interestingly systemic effects such as the attenuation of growth factor signaling and induction of innate immunity in response to DNA damage has also been described in Drosophila and mammals [72-74]. The role of innate immunity in integrating cell-specific damage with systemic stress and maintenance responses might thus be highly evolutionary conserved.
Figure 4. Non-conventional roles of the innate immunity.
Recently, a non-conventional function of the innate immune response as integrator linking cell autonomous damage to the induction of systemic stress resistance was identified in C. elegans. Germline DNA damage led to the activation of immune genes in the germline of the nematode that depended on the MPK-1/ERK MAP kinase activity. The innate immune response resulted in pathogen resistance and activation of the ubiquitin proteasome system (UPS) in peripheral tissues. The UPS activation was responsible for the enhanced tolerance of nematodes to heat and oxidative stress. Interestingly, similar stress resistance could be achieved by transient feeding of worms with immunogenic bacteria. The latter effect was dependent on the PMK-1/p38 kinase activity.
8. Conclusions
C. elegans has become an important metazoan model for studying mechanisms and consequences of innate immune responses. The powerful genetics of the nematode have produced major advances in understanding the interactions between viral RNA and the RNAi machinery. Ancestral pathways through which animals respond to bacterial infections have been established in C. elegans. The worm has been particularly valuable for investigating host-pathogen interactions and revealing links between innate immune responses and mechanisms regulating longevity and stress resistance. In the future, it will be of pivotal interest to further integrate innate immune signaling into physiological alterations throughout the organisms and to better understand the interactions between distinct tissues; for example how the behavioral responses in neurons interact with the immune signaling in the intestine.
Acknowledgements
ME received the EMBO long-term fellowship. BS acknowledges funding from the Deutsche Forschungsgemeinschaft DFG (CECAD, SFB 829, KFO 286), the European Research Council (ERC Starting grant 260383), FP7-PEOPLE-2012-ITN Marie Curie (CodeAge 316354, aDDRess 316390, MARRIAGE 316964 and European Reintegration grant ERG 239330), the German-Israeli Foundation (GIF 1104-68.11/2010), the Deutsche Krebshilfe (109453), and the Bundesministerium für Bildung und Forschung (BMBF-SyBaCol).
References
- [1].Barriere A, Felix MA. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics. 2007;176:999–1011. doi: 10.1534/genetics.106.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aliyari R, Ding S-W. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol Rev. 2009;227:176–88. doi: 10.1111/j.1600-065X.2008.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–7. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- [5].Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, et al. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–3. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Felix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Guo X, Zhang R, Wang J, Ding S-W, Lu R. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16085–90. doi: 10.1073/pnas.1307453110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ashe A, Bélicard T, Le Pen J, Sarkies P, Frézal L, Lehrbach NJ, et al. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife. 2013;2:e00994. doi: 10.7554/eLife.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–71. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- [10].Tijsterman M, Ketting RF, Okihara KL, Sijen T, Plasterk RHA. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science. 2002;295:694–7. doi: 10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- [11].Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sijen T, Plasterk RHA. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–4. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- [13].Horst D, Verweij MC, Davison AJ, Ressing ME, Wiertz EJHJ. Viral evasion of T cell immunity: ancient mechanisms offering new applications. Current Opinion in Immunology. 2011;23:96–103. doi: 10.1016/j.coi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- [14].Guo X, Lu R. Characterization of virus-encoded RNA interference suppressors in Caenorhabditis elegans. J Virol. 2013;87:5414–23. doi: 10.1128/JVI.00148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Loo Y-M, Gale M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–92. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, et al. A simple model host for identifying Gram-positive virulence factors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10892–7. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a Model Host for Staphylococcus aureus Pathogenesis. Infect Immun. 2003;71:2208–17. doi: 10.1128/IAI.71.4.2208-2217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tan M-W, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:715–20. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–42. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- [20].Kurz CL, Ewbank JJ. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat Rev Genet. 2003;4:380–90. doi: 10.1038/nrg1067. [DOI] [PubMed] [Google Scholar]
- [21].Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- [22].Kurz CL, Chauvet S, Andrès E, Aurouze M, Vallet I, Michel GPF, et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. Embo J. 2003;22:1451–60. doi: 10.1093/emboj/cdg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Aballay A, Ausubel FM. Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella typhimurium-mediated killing. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2735–9. doi: 10.1073/pnas.041613098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hodgkin J, Kuwabara PE, Corneliussen B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr Biol. 2000;10:1615–8. doi: 10.1016/s0960-9822(00)00867-8. [DOI] [PubMed] [Google Scholar]
- [25].Nicholas HR, Hodgkin J. Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol Immunol. 2004;41:479–93. doi: 10.1016/j.molimm.2004.03.037. [DOI] [PubMed] [Google Scholar]
- [26].Tan L, Darby C. A movable surface: formation of Yersinia sp. biofilms on motile Caenorhabditis elegans. J Bacteriol. 2004;186:5087–92. doi: 10.1128/JB.186.15.5087-5092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature. 2002;417:243–4. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- [28].Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–21. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- [29].Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6593–8. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Couillault C, Pujol N, Reboul J, Sabatier L, Guichou J-F, Kohara Y, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–94. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- [31].Shivers RP, Kooistra T, Chu SW, Pagano DJ, Kim DH. Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe. 2009;6:321–30. doi: 10.1016/j.chom.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–6. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- [33].Kim DH, Liberati NT, Mizuno T, Inoue H, Hisamoto N, Matsumoto K, et al. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10990–4. doi: 10.1073/pnas.0403546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–16. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schulenburg H, Hoeppner MP, Weiner J, Bornberg-Bauer E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology. 2008;213:237–50. doi: 10.1016/j.imbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- [38].Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol. 1993;231:539–45. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- [39].Thomas JH. Concerted evolution of two novel protein families in Caenorhabditis species. Genetics. 2006;172:2269–81. doi: 10.1534/genetics.105.052746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, Whitney JK, et al. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 2010;6:e1000892. doi: 10.1371/journal.pgen.1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Aballay A, Drenkard E, Hilbun LR, Ausubel FM. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol. 2003;13:47–52. doi: 10.1016/s0960-9822(02)01396-9. [DOI] [PubMed] [Google Scholar]
- [42].Nicholas HR, Hodgkin J. The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr Biol. 2004;14:1256–61. doi: 10.1016/j.cub.2004.07.022. [DOI] [PubMed] [Google Scholar]
- [43].Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–14. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- [44].Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- [45].Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–6. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- [46].Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- [47].Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–45. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- [48].Evans EA, Kawli T, Tan M-W. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 2008;4:e1000175. doi: 10.1371/journal.ppat.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–83. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- [50].McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–21. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- [51].Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–83. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tullet JMA, Hertweck M, An JH, Baker J, Hwang JY, Liu S, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–38. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Evans EA, Chen WC, Tan M-W. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell. 2008;7:879–93. doi: 10.1111/j.1474-9726.2008.00435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–3. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- [55].Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13091–6. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Laws TR, Harding SV, Smith MP, Atkins TP, Titball RW. Age influences resistance of Caenorhabditis elegans to killing by pathogenic bacteria. FEMS Microbiol Lett. 2004;234:281–7. doi: 10.1016/j.femsle.2004.03.034. [DOI] [PubMed] [Google Scholar]
- [57].Garigan D, Hsu A-L, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–12. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Youngman MJ, Rogers ZN, Kim DH. A decline in p38 MAPK signaling underlies immunosenescence in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002082. doi: 10.1371/journal.pgen.1002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chávez V, Mohri-Shiomi A, Maadani A, Vega LA, Garsin DA. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 2007;176:1567–77. doi: 10.1534/genetics.107.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hoeven RVD, McCallum KC, Cruz MR, Garsin DA. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 2011;7:e1002453. doi: 10.1371/journal.ppat.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13092–7. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–5. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Haskins KA, Russell JF, Gaddis N, Dressman HK, Aballay A. Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev Cell. 2008;15:87–97. doi: 10.1016/j.devcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–4. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. 2008;322:460–4. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science. 2011;332:729–32. doi: 10.1126/science.1203411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Aballay A. Role of the nervous system in the control of proteostasis during innate immune activation: insights from C. elegans. PLoS Pathog. 2013;9:e1003433. doi: 10.1371/journal.ppat.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ermolaeva MA, Segref A, Dakhovnik A, Ou H-L, Schneider JI, Utermöhlen O, et al. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 2013;501:416–20. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ermolaeva MA, Schumacher B. Systemic DNA damage responses: organismal adaptations to genome instability. Trends Genet. 2014 doi: 10.1016/j.tig.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ermolaeva MA, Schumacher B. The innate immune system as mediator of systemic DNA damage responses. Communicative & Integrative Biology. 2013 doi: 10.4161/cib.26926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Karpac J, Younger A, Jasper H. Dynamic coordination of innate immune signaling and insulin signaling regulates systemic responses to localized DNA damage. Dev Cell. 2011;20:841–54. doi: 10.1016/j.devcel.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Garinis GA, Uittenboogaard LM, Stachelscheid H, Fousteri M, van Ijcken W, Breit TM, et al. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol. 2009;11:604–15. doi: 10.1038/ncb1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Schumacher B, van der Pluijm I, Moorhouse MJ, Kosteas T, Robinson AR, Suh Y, et al. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008;4:e1000161. doi: 10.1371/journal.pgen.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]