Abstract
SUMMARY
Chemoreceptors sense environmental signals and drive chemotactic responses in Bacteria and Archaea. There are two main classes of chemoreceptors: integral inner membrane and soluble cytoplasmic proteins. The latter were identified more recently than integral membrane chemoreceptors and have been studied much less thoroughly. These cytoplasmic chemoreceptors are the subject of this review. Our analysis determined that 14% of bacterial and 43% of archaeal chemoreceptors are cytoplasmic, based on currently sequenced genomes. Cytoplasmic chemoreceptors appear to share the same key structural features as integral membrane chemoreceptors, including the formations of homodimers, trimers of dimers, and 12-nm hexagonal arrays within the cell. Cytoplasmic chemoreceptors exhibit varied subcellular locations, with some localizing to the poles and others appearing both cytoplasmic and polar. Some cytoplasmic chemoreceptors adopt more exotic locations, including the formations of exclusively internal clusters or moving dynamic clusters that coalesce at points of contact with other cells. Cytoplasmic chemoreceptors presumably sense signals within the cytoplasm and bear diverse signal input domains that are mostly N terminal to the domain that defines chemoreceptors, the so-called MA domain. Similar to the case for transmembrane receptors, our analysis suggests that the most common signal input domain is the PAS (Per-Arnt-Sim) domain, but a variety of other N-terminal domains exist. It is also common, however, for cytoplasmic chemoreceptors to have C-terminal domains that may function for signal input. The most common of these is the recently identified chemoreceptor zinc binding (CZB) domain, found in 8% of all cytoplasmic chemoreceptors. The widespread nature and diverse signal input domains suggest that these chemoreceptors can monitor a variety of cytoplasmically based signals, most of which remain to be determined.
INTRODUCTION
Chemotaxis is a motility-based response that biases cell movement toward beneficial molecules, called attractants, and away from harmful molecules, also known as repellents. Chemotaxis is initiated through the recognition of attractants and repellents by chemoreceptors, which are the signal-sensing proteins of the bacterial chemotaxis system. The chemoreceptors transduce this information to the central regulator of bacterial chemotaxis, the CheA kinase (Fig. 1) (reviewed in references 1 and 2), which in turn leads to the regulation of flagellar rotation. This paradigm has been well studied in integral membrane chemoreceptors of Escherichia coli and has led to many key insights into signal transduction. There are, however, entire classes of bacterial chemoreceptors that are fundamentally different from those of E. coli (3, 4). Studying these chemoreceptors will likely generate even more insights into the fundamental properties of signal recognition and transduction. We focus here on one type of distinct bacterial chemoreceptor: those that lack transmembrane domains and operate strictly cytoplasmically.
FIG 1.
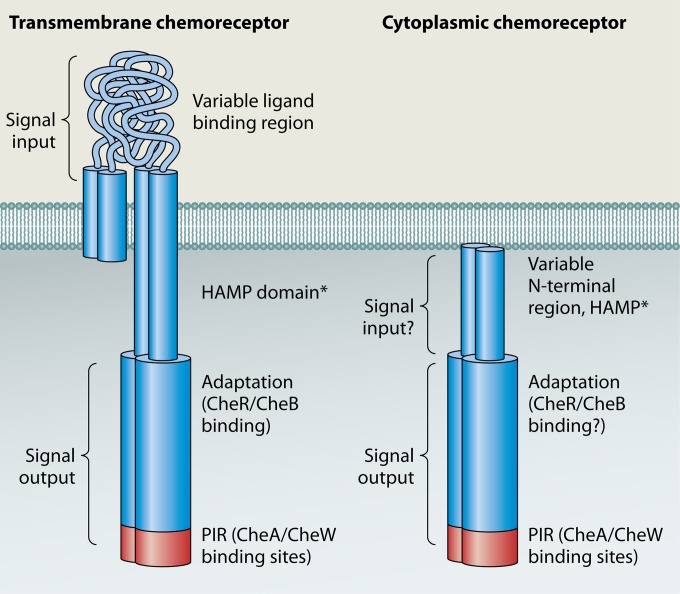
Domain structure of transmembrane (left) and soluble (right) chemoreceptors. Only one type of transmembrane receptor is shown, the so-called class I receptor, with two transmembrane regions and a periplasmic ligand binding region, as these are the best-studied types of transmembrane receptor. Different transmembrane receptor topologies have been described (6, 7). Asterisks indicate a domain that is not always present. Signal output coincides with MA domain in chemorecptors shown. Cytoplasmic chemoreceptors are shown positioned in proximity to the inner membrane but in most cases do not interact with it directly.
Chemoreceptors can be classed as either integral inner membrane proteins or soluble cytoplasmic ones (Fig. 1). All chemoreceptors are identified by the presence of a highly conserved cytoplasmic signaling domain, called the “MA” or “MCP signal” domain, which interacts with the CheW coupling protein and the CheA histidine kinase. Often, chemoreceptors are called methyl-accepting chemotaxis proteins (MCPs) because of their ability to be methylated, but we use the term chemoreceptor because most so-called MCPs have not been experimentally tested for methylation. A substantial fraction of chemoreceptors are cytoplasmic. Specifically, our analysis of 8,384 chemoreceptor proteins in the SMART protein database (5) found that 14.5% lack transmembrane regions and are predicted to reside in the cytoplasm. These cytoplasmic chemoreceptors presumably function similarly to transmembrane chemoreceptors but detect intracellular ligands, although there is little known regarding how they actually sense and transmit signals (6, 7). Furthermore, chemoreceptors can function in processes other than motility, e.g., gene regulation (8). Cytoplasmic chemoreceptors, however, are known to play important roles in many microbial processes, including pathogenesis (9, 10), fruiting body formation (8), as well as mediating taxis in response to cellular energy stores (11, 12), redox (13, 14), and metabolites (15). In this review, we first start with a discussion of basic chemoreceptor attributes and then summarize the current state of understanding about signal recognition, signal transduction, and the subcellular localization of cytoplasmic chemoreceptors.
CHEMORECEPTORS SIGNAL VIA INTERACTIONS WITH THE KEY SIGNALING PROTEINS CheW AND CheA
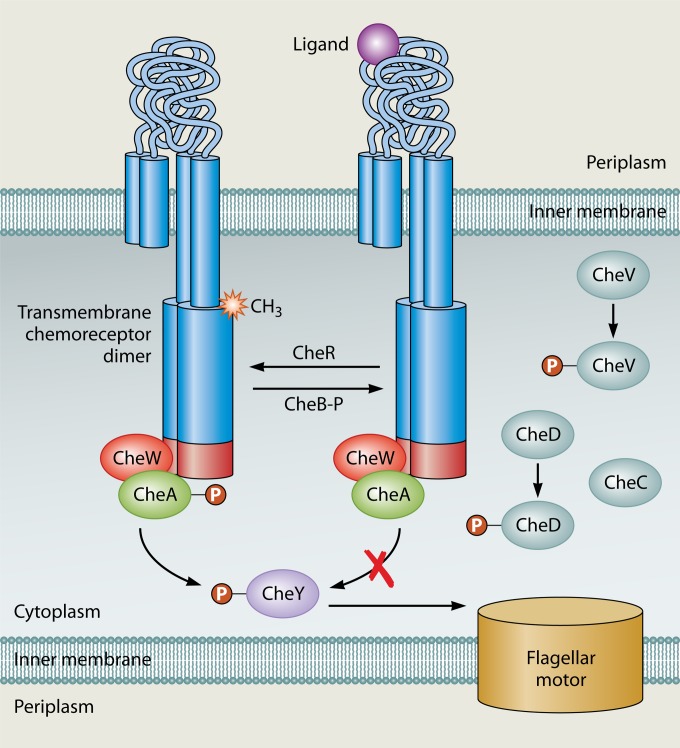
Bacterial chemoreceptors contain signal output regions as well as signal input regions that are, in some cases, clearly defined (4, 6). In all types of chemoreceptors, the signal output region is cytoplasmic and highly conserved because it interacts with the downstream signal transduction proteins CheW and CheA (Fig. 2). Chemoreceptors mediate chemotaxis by controlling CheA's kinase activity (2). CheA phosphorylates CheY, which in turn interacts with the flagellar motor and affects the frequency at which the motor changes the direction of rotation.
FIG 2.
Schematic of chemotaxis signal transduction. Chemoreceptors exist in a ternary complex with the CheW coupling protein and the CheA kinase. The chemoreceptor in the absence of ligand activates CheA, which in turn phosphorylates the response regulator CheY. Phosphorylated CheY interacts with the flagellar motor and affects the direction of motor rotation. CheR is a methyltransferase that acts upon conserved glutamates in the adaptation region of the receptors; methylation at these sites enhances CheA activation by chemoreceptors. CheB is a methylesterase that catalyzes the removal of methyl groups. Its activity is enhanced by phosphorylation via CheA. Additional adaptations proteins, CheV, CheC, and CheD, are described in the text.
Chemoreceptors interact with both CheW and CheA. CheW is a coupling or scaffold protein that is essential to form connections between chemoreceptors and CheA (Fig. 2). Interactions between chemoreceptors and CheW/CheA occur in a subregion of the MA domain called the protein interaction region (PIR) (2, 16). In transmembrane chemoreceptors, the PIR is the portion of the chemoreceptor most distal from the membrane (Fig. 1). The PIR is a four-stranded coiled coil, with two strands originating from each chemoreceptor monomer. Evidence suggests that the PIR adopts the same structure in all receptors, whether cytoplasmic or transmembrane (17). The interaction of CheW with the PIR has been known for a long time and verified by numerous methods, including nuclear magnetic resonance (NMR) chemical shifts (18), targeted disulfide cross-linking (19), and genetic suppressor mutations (20, 21). The interaction of CheA and the PIR, in contrast, has been appreciated only recently but has been documented by several methods, including targeted disulfide cross-linking and mutagenesis (22–24). Interactions between chemoreceptors and CheW, as well as those between chemoreceptors and CheA, are important for controlling CheA kinase activity (23, 25, 26). Cytoplasmic chemoreceptors appear to participate in similar interactions based on observations of interactions between CheW, CheA, and the PIR within the Tm14 cytoplasmic chemoreceptor from Thermotoga maritima (26). Based on these similar interactions, the output of ligand binding in all receptors is likely to trigger conformational changes at the PIR and in turn to affect CheA activation.
All chemoreceptors contain the MA domain, which acts to transduce signal input information to CheA. Only one copy of the MA domain is present in transmembrane chemoreceptors, while more than one copy of the MA domain can be found in cytoplasmic chemoreceptors. For instance, we identified 20 cytoplasmic chemoreceptors with two MA domains. All 20 chemoreceptors consist of an N-terminal region followed by two consecutive MA domains. Eighteen of these chemoreceptors have only small N-terminal regions (class IVb), described in more detail below, while two have longer N-terminal regions (class IVa). None of these chemoreceptors has an annotated sensing domain, and their functions are not yet known. Interestingly, while the MA domains are similar for all these receptors, their N-terminal regions are not, suggesting that they are not strict orthologs. These receptors were found only in Bacteria, within the Actinobacteria, Clostridia, Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria. The genome of Caldicellulosiruptor bescii DSM 6725 is the only one containing more than one (two) of these receptors. On the other hand, we found that the four Vibrio cholerae strains have the same cytosolic chemoreceptor containing two MA domains (see Table S1 in the supplemental material). The relevance of a chemoreceptor having more than one MA domain is not yet known.
SIGNAL TRANSDUCTION IN CHEMORECEPTORS
The signal input region of chemoreceptors is extremely variable, in contrast to the highly conserved signal output region. Transmembrane chemoreceptors typically present an easily identifiable region for signal input outside the cytoplasmic membrane. In contrast, it is often difficult to identify the signal input domain of cytoplasmic chemoreceptors, and it is not yet clear if it always lies N terminal to the MA domain. Lacal and colleagues reported that some cytoplasmic chemoreceptors have a large N-terminal region, with a portion containing known or predicted sensing domains, while other cytosolic chemoreceptors have only a very short N-terminal domain that has no identifiable sensing domains (7).
In transmembrane chemoreceptors, several different organizations have been described (6, 7). The most common organization is typified by the E. coli receptors Tar and Tsr and consists of (i) a short N-terminal sequence that acts as a transmembrane region, (ii) a poorly conserved periplasmic region, (iii) a second transmembrane region, and (iv) a cytoplasmic region containing at least the highly conserved MA domain (Fig. 1). The periplasmic region is therefore flanked by two transmembrane regions, is very diverse in length and sequence, and is responsible for sensing ligands directly or through the interaction with ligand binding proteins (1). Ligand binding is thought to trigger a conformational change in the chemoreceptor that consists of an ∼1.4-Å downward slide of the long transmembrane helix that connects the periplasmic and cytoplasmic regions (16, 27, 28). There have been no studies of ligand-driven conformational changes in cytoplasmic chemoreceptors. Numerous cytoplasmic chemoreceptors contain putative N-terminal ligand binding domains, but C-terminal ones also exist, as we describe below in this review. It is unknown whether cytoplasmic receptors detect ligands by direct binding, operate through protein partners, or both.
A frequently found signal transduction domain in E. coli chemoreceptors is the HAMP domain, which lies between the signal input and output domains (Fig. 1) (29). HAMP domains are thought to function to transmit the transmembrane ligand binding signal to the MA domain, although the exact mechanism is not yet clear. HAMP domains are small 50-amino-acid homodimeric folds that adopt a four-helix bundle structure of two amphipathic helices joined by a linker segment (29). HAMP domains have been observed in crystal structures to adopt multiple conformations, varying by helix rotation, helix translation, and helix-helix crossing angles (30, 31). These different conformations are proposed to underlie signal transduction. About 80% of transmembrane chemoreceptors contain a membrane-proximal HAMP domain, based on our analysis of MA domain-containing proteins in the SMART database (5). Sometimes, more than one copy of the HAMP domain is found in chemoreceptors. However, the HAMP domain may not be essential, since we found that almost 20% of the transmembrane chemoreceptors do not have an annotated HAMP domain.
Given that HAMP domains are canonically thought to function in transmembrane signaling, it was not clear or known whether cytoplasmic chemoreceptors would possess them. To fill this gap, we determined the abundance of the HAMP domain in cytoplasmic chemoreceptors using proteins obtained from the SMART database that lack transmembrane domains (5). Five percent of the 1,217 cytoplasmic chemoreceptors contained a HAMP domain. In these chemoreceptors, the HAMP domain was always N terminal to the MA domain and was frequently accompanied by the PAS domain (∼30% of cytoplasmic chemoreceptors) and more rarely with the CHASE, hemerythrin, NIT, and Cache_1 domains (these domains are discussed below). Generally, HAMP domains were prevalent in cytoplasmic chemoreceptors with long N-terminal domains, so-called class IVa chemoreceptors (7). One possibility is that cytoplasmic chemoreceptors with long N-terminal regions are more likely to contain a signal-sensing domain in this region and thus utilize a HAMP domain, similarly to transmembrane chemoreceptors, in coupling conformational changes from a ligand binding domain to alter CheA kinase activity (29). This possibility, however, remains to be confirmed.
ADAPTATION IN CHEMORECEPTORS
Chemoreceptors are capable of responding to a broad range of chemoeffector concentrations through a process known as adaptation (1, 16). Adaptation typically occurs via posttranslational modifications affecting a chemoreceptor's ability to activate CheA under conditions of prolonged stimulation. In the case of prolonged attractant stimulation, a bacterium's swimming behavior will return to the prestimulation direction change frequency after a period of smooth swimming triggered by attractant addition. The best-understood mechanism of adaptation occurs via the reversible methylation of glutamate residues located in the signal output region of the chemoreceptors. Methyl groups are added by the methyltransferase CheR and removed by the methylesterase CheB (Fig. 2). Methylation of the chemoreceptors increases their ability to activate CheA. There is complex feedback between the CheA kinase and the CheB portion of the methylation system. Specifically, CheA phosphorylates CheB but on a slower time scale than it does CheY. Phosphorylation of CheB activates its esterase activity and leads to demethylation of chemoreceptors, diminishing their capacity to activate CheA (16, 32, 33). A similar system appears to operate in at least some cytoplasmic chemoreceptors, based on the observation that these cytoplasmic chemoreceptors possess a methylation consensus in the adaptation region (15, 34–36). In most cases, however, methylation has not been confirmed, nor is the specific role of methylation known.
Although methylation is the best-understood adaptation method, there are at least two additional adaptation systems utilized by chemoreceptors. These systems have been well characterized only for Bacillus subtilis, so these proteins may function differently outside that system (37). Nonetheless, it is useful to keep in mind that there are additional nonmethylation adaptation systems that may be used by cytoplasmic chemoreceptors. The first system utilizes the CheV proteins, which are chimeras of CheW plus a phosphorylatable receiver (REC) domain (38). CheV proteins allow additional chemoreceptor-kinase control, dictated by the variable phosphorylation state of the CheV REC domain. The second alternative system relies on the CheD and CheC proteins. Both of these proteins have enzymatic activity but additionally perform adaptation via protein-protein interactions (39). CheD interacts directly with chemoreceptors at the methylation region and can deamidate them, thereby increasing their ability to activate CheA (39). CheD availability is modulated by CheC (40). A model has been proposed whereby phosphorylated CheY causes CheC to sequester CheD from the chemoreceptors, inducing adaptation of the chemotaxis system (41). Many species that possess CheD lack CheC, which supports the possibility of variation and diversity in these adaptation systems. To date, no studies about the role(s) of CheV or CheD/C have been done with cytoplasmic chemoreceptors, so it is not yet known whether these types of receptors utilize these diverse adaptation systems.
HIGHER-ORDER STRUCTURES OF CYTOPLASMIC CHEMORECEPTORS
Chemoreceptors exist as homodimers, clustered into trimers of dimers. Homodimers form extensive antiparallel four-helix-bundle coiled-coil interactions that have been seen in structures of cytoplasmic chemoreceptors (42) as well as in soluble cytoplasmic fragments of the Tsr and Tm1143c chemoreceptors (21, 43). These features are shared between different chemoreceptors, supporting that all chemoreceptors are likely to form homodimers (3).
Trimers of dimers are a higher-order form of chemoreceptors that seem to be a universal state of chemoreceptors. Briegel et al. reported that Tm14, a cytoplasmic chemoreceptor of T. maritima, forms trimers-of-dimers in crystals (17). Recently, this work was extended to show that cytoplasmic chemoreceptors from two different species formed trimer-of-dimer structures in cells (44). Similar to transmembrane chemoreceptor arrays, cytoplasmic clusters contain trimers of receptor dimers organized in 12-nm hexagonal arrays. In contrast to transmembrane arrays, however, cytoplasmic clusters comprise two CheA/CheW baseplates sandwiching two opposed receptor arrays. Those authors furthermore showed that cytoplasmic fragments of normally transmembrane E. coli chemoreceptors form similar sandwiched structures in the presence of molecular crowding agents. Together, these results suggest that the 12-nm hexagonal architecture is fundamentally conserved and that sandwiching and crowding can replace the stabilizing effect of the membrane.
Chemoreceptors form extensive multichemoreceptor arrays, composed of many trimers of dimers connected by CheW and CheA at the chemoreceptor PIR (17, 45, 46). In accordance with these observations, CheW and CheA are essential for extensive multichemoreceptor array formation. To gain higher-resolution information on the relative positions of each protein, a crystal structure of a cytoplasmic chemoreceptor, Thermotoga Tm14, was employed (17, 26). The high-resolution structure of Tm14 in complex with CheA and CheW shows an unusual unzipped conformation of the chemoreceptors, into trimers of tetramers, with CheW and CheA engaged in a ring around the PIRs. This unusual unzipped conformation has not been observed in native situations and is believed to be a nonnative feature. However, evolutionary analysis of sequence conservation and mutation patterns suggests that the contacts among the receptor, CheA, and CheW displayed by the structure are relevant to the native chemosensory system (26). This structural model bears strong similarity to interactions predicted and observed for intact transmembrane chemoreceptors by using cryotomography, supporting the accuracy of this idea (17, 44, 47).
CYTOPLASMIC CHEMORECEPTORS ARE PREVALENT IN BACTERIA AND ARCHAEA
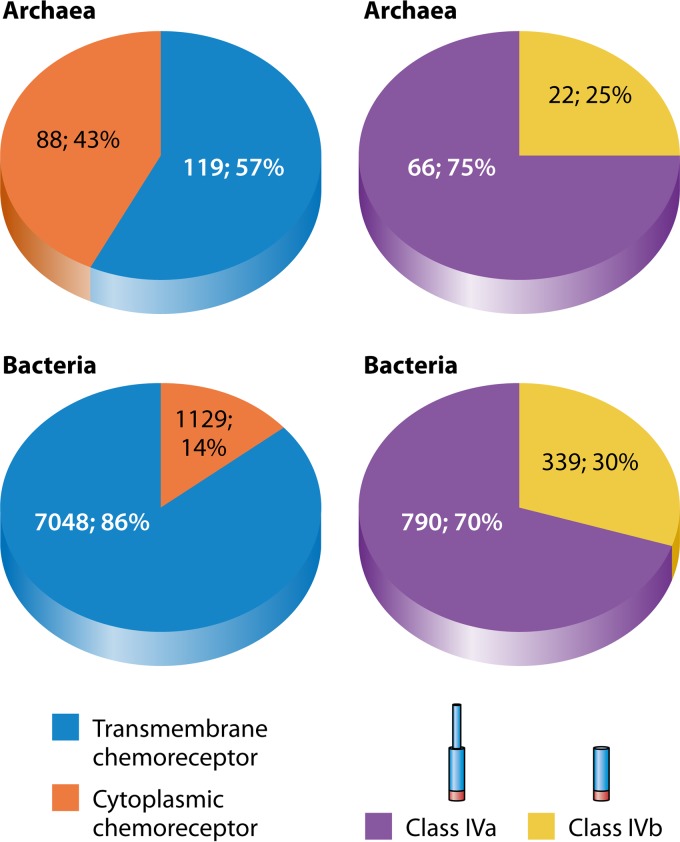
Although there are several examples of cytoplasmic chemoreceptors, their prevalence was unknown. We therefore conducted an analysis of 8,384 chemoreceptors in complete genomes of the SMART database (5). These chemoreceptors were defined by the presence of the MA domain and included 207 from Archaea and 8,177 from Bacteria (Fig. 3). Of the bacterial chemoreceptors, 14% (1,129) were cytoplasmic, based on the lack of transmembrane domains. Cytoplasmic chemoreceptors were more abundant in Archaea, with nearly 43% of all MCPs identified being cytoplasmic (Fig. 3). We found three archaeal species with exclusively cytoplasmic receptors, including Thermofilum pendens Hrk 5 (one chemoreceptor), Archaeoglobus fulgidus DSM 4304 (two chemoreceptors), and “Candidatus Methanoregula boonei” 6A8 (four chemoreceptors). Interestingly, in the archaeon Methanosphaerula palustris E1-9C, 11 out 12 of its chemoreceptors are cytoplasmic. Most genomes, however, have transmembrane chemoreceptors plus some cytoplasmic ones.
FIG 3.
(Left) Abundances of cytoplasmic chemoreceptors in Archaea and Bacteria. Chemoreceptors were identified from complete genomes in the SMART database (5) as proteins with annotated MA domains and further narrowed by identifying those that had no transmembrane domains, to create a set of 1,217 cytoplasmic chemoreceptors. (Right) Results from analyses of only the cytoplasmic chemoreceptors, manually, to determine which chemoreceptors belong to class IVa, those with N-terminal domains of 108 amino acids or longer, or class IVb, those with N-terminal domains of <108 amino acids.
Cytoplasmic chemoreceptors had previously been called class IV chemoreceptors (4) and were subsequently further divided into two different classes according to the length of the polypeptide N terminal to the MA domain (7). Specifically, class IVa chemoreceptors contain an N-terminal domain of at least 108 amino acids, whereas class IVb contains an N-terminal region of <108 amino acids. Many of the class IVa receptors have predicted ligand binding motifs, as described below, but for the most part, we do not yet know the functional significance of long and short N-terminal domains. However, class IVa chemoreceptors are ∼2- to 3-fold more abundant than class IVb chemoreceptors (Fig. 3).
LIGAND BINDING DOMAINS FOUND IN CYTOPLASMIC CHEMORECEPTORS
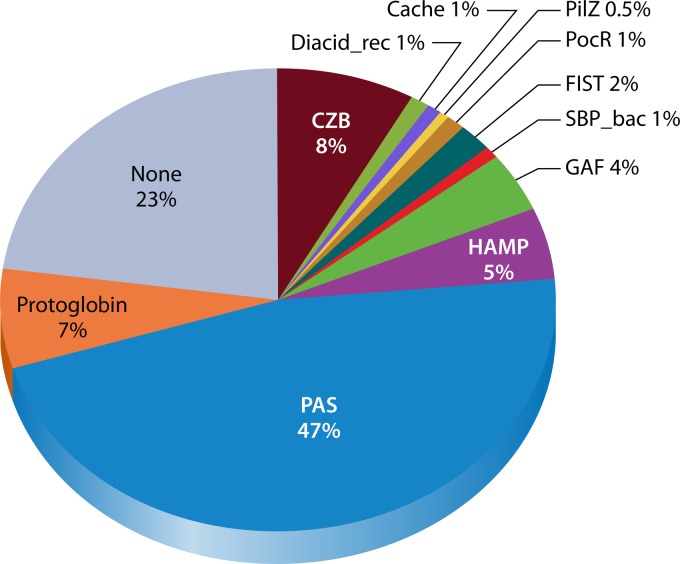
How cytoplasmic chemoreceptors sense their ligands and respond to them is a major remaining question in bacterial chemotaxis. To help fill this gap, we used both the SMART and PFAM databases to analyze the domain content of our set of 1,129 cytoplasmic chemoreceptors obtained from the SMART database. We determined the prevalence of individual domains in cytoplasmic chemoreceptors, the frequency of cooccurrences of domains, and the position of these domains relative to the MA domain. Roughly one-quarter of cytoplasmic chemoreceptors had no identifiable domain other than the MA domain (Fig. 4). Several types of domains were found fairly commonly in cytoplasmic chemoreceptors, including domains that sense small molecules or bind redox-active cofactors and thus are predicted to drive a tactic response to redox or cellular energy levels. Each of these domains is discussed below. Interestingly, some domains were found N terminal to the MA domain, as is typical in transmembrane chemoreceptors, while others were found C terminal to the MA domain. The significance of this positioning, however, is not yet known. Additionally, we do not yet know whether ligand sensing at these domains controls the conformation or availability of the PIR, as has been suggested for transmembrane chemoreceptors (16, 23).
FIG 4.
Domains most commonly found in soluble chemoreceptors. The set of cytoplasmic chemoreceptors (1,217 proteins) was identified as described in the Fig. 3 legend and used as the input for both the SMART (5) and PFAM databases (76) to identify all additional annotated domains. Domains with several subtypes were combined as follows: GAF contains GAF and GAF_2; FIST contains FIST and FIST_C; PAS contains PAS_3, PAS_4, PAS_8, and PAS_9; and Cache contains Cache_1 and Cache_2. Annotated domains with 3 hits or less are not shown here but are included in Table 1. Brief descriptions of domain functions can be found in Table 1.
PAS domains are by far the most abundant sensing domains annotated in cytoplasmic chemoreceptors, occurring in 47% of analyzed proteins, and in most cases, several PAS domains were found per protein (Fig. 4). PAS domains are found in many signaling proteins, from human to bacteria, where they function to bind small molecules (48). Many PAS domain proteins bind a small molecule that in turn detects the signal. For example, heme binding confers oxygen detection, and flavin adenine dinucleotide (FAD) binding allows redox detection. In all cytoplasmic chemoreceptors, the PAS domain was located N terminal to the MA domain (Table 1). A few PAS-containing cytoplasmic chemoreceptors have been implicated in energy taxis, a motility-based response for positioning cells in an optimum microenvironment for energy generation (12, 15) (Table 1).
TABLE 1.
Domains identified in cytoplasmic chemoreceptorsa
| Domain | % (total no.) of chemoreceptors with domain located: |
No. of chemoreceptors of class IVa/class IVb | Total no. of proteins | Database | |
|---|---|---|---|---|---|
| N-terminal | C-terminal | ||||
| PAS/PAC | 100 (367) | 0 | 362/5 | 367 | SMART |
| CZB | 0 | 100 (114) | 17/97 | 114 | PFAM |
| Protoglobin | 100 (104) | 0 | 10/22 | 104 | PFAM |
| FIST | 100 (14) | 0 | 14/0 | 14 | SMART |
| GAF | 100 (14) | 0 | 14/0 | 14 | SMART |
| Cache_1 | 17 (2) | 83 (10) | 2/10 | 12 | PFAM |
| Diacid_rec | 100 (9) | 0 | 1/8 | 9 | PFAM |
| SBP_bac_5 | 0 | 100 (9) | 0/9 | 9 | PFAM |
| PilZ | 0 | 100 (7) | 6/1 | 7 | PFAM |
| PBPb | 0 | 100 (3) | 0/3 | 3 | SMART |
| Hemerythrin | 0 | 100 (2) | 2/0 | 2 | PFAM |
| Fe_hyd_lg_C | 100 (2) | 0 | 2/0 | 2 | PFAM |
| HNOB | 100 (2) | 0 | 2/0 | 2 | PFAM |
| NMT1 | 0 | 100 (1) | 1/0 | 1 | PFAM |
| CBS | 100 (1) | 0 | 1/0 | 1 | SMART |
| NIT | 100 (1) | 0 | 1/0 | 1 | PFAM |
| CHASE3 | 100 (1) | 0 | 1/0 | 1 | PFAM |
| Bac_globin | 100 (1) | 0 | 1/0 | 1 | PFAM |
| DUF3365 | 100 (1) | 0 | 1/0 | 1 | PFAM |
N-terminal indicates the percentage of chemoreceptors with the indicated domain found on the amino-terminal side of the MA domain, with the total number in parentheses. C-terminal indicates the percentage of chemoreceptors with the indicated domain found on the carboxy-terminal side of the MA domain, with the total number in parentheses. Class IVa chemoreceptors are cytoplasmic receptors with N-terminal domains that are 108 amino acids or longer, while class IVb chemoreceptors have N-terminal regions of <108 amino acids (7). “Database” indicates whether the domains were identified by using the SMART or PFAM database. The following domains were included in this table. PAS domain proteins interact with and respond to small molecules through bound cofactors, including FAD and heme groups (48). CZB domains bind zinc through conserved histidines and cysteines and in some cases are known to sense zinc, although this has not been confirmed for other CZB proteins (49, 50). PilZ domains bind and mediate a response to the secondary messenger c-di-GMP (77). Protoglobin domains coordinate a heme group and can respond to oxygen (78). FIST domains are proposed to bind small ligands, including amino acids (51). GAF domains interact with and respond to 3′,5′-cyclic GMP (cGMP) (53, 79). The Diacid_rec domain is proposed to bind and respond to carbohydrates (54). The PocR domain is a variant of the PAS domain and is predicted to bind hydrocarbons (55). Cache_1 and SBP_bac_5 domains are predicted to have a role in small-molecule recognition (56, 57). PBPb domains are high-affinity small-molecule binding domains characterized in ABC transporters (57). Hemerythrins bind and respond to oxygen through coordinated iron atoms (80, 81) and have also been reported to mediate responses to nitric oxide (82). NMT1 domains have been characterized for their role in the synthesis of the pyrimidine moiety of thiamine and are regulated by thiamine (83, 84). The HNOB domain coordinates heme and is predicted to interact with and respond to gaseous ligands, including nitric oxide (85). CBS domains bind and respond to molecules with adenosyl groups, such as AMP and ATP or S-adenosylmethionine (86). The NIT domain binds and responds to nitrate and nitrite (87). The CHASE3 domain has been characterized as an extracellular sensory domain, although the perceived ligand is unknown (88). The Bac_globin domain coordinates heme as a prosthetic group and binds oxygen reversibly (14, 89). DUF3365 domains are present in bacteria but are functionally uncharacterized.
The second most common domain is the chemoreceptor zinc binding (CZB) domain, found in ∼8% of all cytoplasmic chemoreceptors (Fig. 4). In all cases, the CZB domain was C terminal to the MA domain (Table 1). The CZB domain was first identified in the Helicobacter pylori TlpD (TlpDHP) chemoreceptor (49). It consists of a set of conserved histidines and one cysteine and binds zinc (49). The CZB domain is also found in nonchemoreceptor proteins, including the diguanylate cyclase DgcZ/YdeH. In this protein, it responds to zinc and exhibits zinc-dependent allosteric control on the associated domain (50). The CZB domain was typically the sole identifiable domain in the chemoreceptors probed, although in a few cases, a protoglobin domain was also found.
The third most common sensing domain found in cytoplasmic chemoreceptors is the protoglobin domain, which occurred in 7% of proteins analyzed (Fig. 4). The protoglobin domain is a member of the hemoglobin superfamily and binds heme. The B. subtilis cytoplasmic chemoreceptor HemAT has a heme-bound protoglobin domain that drives aerotaxis (14). The protoglobin domain was found exclusively N terminal to the MA domain (Table 1) and was typically the sole identifiable domain in chemoreceptors, with rare exceptions containing it in addition to the C-terminal CZB or PilZ domain.
Several other domains were found in ∼1% of cytoplasmic chemoreceptors. Several domains were N terminal to the MA domain, including the FIST, GAF, Diacid_rec, and PocR domains. Two domains were C terminal to the MA domain, the SBP_bac_5 and PilZ domains. One domain was found either N- or C-terminally placed, the Cache_1 domain (Fig. 4 and Table 1). The first N-terminal domain, FIST, has been postulated to bind small ligands, such as amino acids, based on the chromosomal proximity of FIST-encoding genes to those coding for proteins involved in amino acid metabolism and transport (51). Interestingly, when this domain is present, the HAMP domain is not, and this domain was detected only in proteobacteria. Another N-terminal domain is GAF. The GAF domain is named based on the proteins in which it was originally found: cyclic GMP (cGMP)-specific phosphodiesterases, adenylyl cyclases, and FhlA (52, 53). The GAF domains were often found in multiple copies per chemoreceptor (Table 1). The Diacid_rec domain is found in several proteins characterized as carbohydrate diacid regulators (54). It is always located N terminal to the MA domain (Table 1). Lastly, the PocR domain is a variant of the PAS domain and is predicted to bind hydrocarbons (55). When present, it was the only annotated domain outside the MA domain and was always N terminal to it (Table 1).
The Cache_1 domain was the only domain found to be located either N or C terminal to the MA domain (Table 1). Cache_1 is typically an extracellular domain that is predicted to have a role in small-molecule recognition in a wide range of proteins, including the animal dihydropyridine-sensitive voltage-gated Ca2+ channel alpha-2δ subunit and various bacterial chemotaxis receptors (56). The name Cache comes from calcium channels and chemotaxis receptors.
Lastly, the SBP_bac_5 and PilZ domains were found exclusively C terminal to the MA domain (Table 1). The SBP_bac_5 domain was originally characterized as an extracellular domain that binds small molecules and participates in high-affinity transport (57). Members include proteins such as the periplasmic oligopeptide binding proteins (OppA), the periplasmic murein peptide binding protein of E. coli (MppA), the periplasmic nickel binding protein (NikA) of E. coli, and the heme binding lipoprotein (HbpA or DppA) of Haemophilus influenzae. In cytoplasmic chemoreceptors, SBP-bac_5 was found only in clostridial species. The PilZ domain, in comparison, binds the bacterial second messenger cyclic di-GMP (c-di-GMP) (58). c-di-GMP is associated with the regulation of biofilm formation, the control of exopolysaccharide synthesis, flagellar- and pilus-based motility, gene expression, and other aspects of bacterial physiology in diverse bacteria (58). The PilZ domain was found in soluble chemoreceptors combined with a variety of other domains, e.g., PAS and protoglobin domains N terminal to the MA domain and the CZB domain C terminal to the MA domain. PilZ domains were found only in cytoplasmic chemoreceptors from alphaproteobacteria. Recently, the Alexandre group showed that the PilZ domain at the C-terminal end of a transmembrane chemoreceptor bound c-di-GMP and modulated chemotactic signaling, supporting that ligand binding at C-terminal domains can influence chemoreceptor function (59).
The diversity of domains in cytoplasmic chemoreceptors suggests that these proteins sense and integrate a wide group of intracellular signals. Furthermore, these domains may be divided presumptively into those that sense redox, such as the PAS domain; those that sense oxygen, such as the protoglobin domain; and those that respond to small molecules, such as the PAS and PilZ domains. It remains unknown how cytoplasmic chemoreceptors convert signals into a tactic response.
SUBCELLULAR LOCALIZATION OF CYTOPLASMIC CHEMORECEPTORS
Transmembrane chemoreceptors localize predominantly at or in the vicinity of the cell pole but can also be found in the lateral membrane (60). In both locations, they form extensive multichemoreceptor arrays, although the polar ones tend to be larger (46). Cytoplasmic chemoreceptors, in contrast, appear to have a wider subcellular distribution, ranging from colocalization with transmembrane chemosensory clusters to a diffuse cytoplasmic distribution and cytoplasmic cluster localization, as described in more detail below. One point to consider in analyzing chemoreceptor localization is whether a particular microbe has only one or more than one set of chemotaxis signaling proteins. Many bacteria have multiple sets of chemotaxis signaling proteins that can act under distinct conditions or even function in processes such as twitching motility or transcription (32) (Table 2). Presumably, if there is only one set of signaling proteins, all chemoreceptors must share these proteins and therefore would be more likely to be colocalized. The localization of cytoplasmic chemoreceptors is by far the best-studied attribute of these proteins, so below, we summarize a significant body of work in this area.
TABLE 2.
Characteristics of well-studied cytoplasmic chemoreceptorsa
| Cytoplasmic receptor | Species | Domain(s) |
Input or phenotype | Localization(s) | Methylation |
No. of chemosensory systems (source or database[s]) | Reference(s) | ||
|---|---|---|---|---|---|---|---|---|---|
| NH3 | COOH | Consensus | Verified | ||||||
| TlpD (HP0599) | H. pylori | None | CZB | Electron transport chain blockage | P/C | No | No | 1 | 11, 49 |
| Tm14 (TM0014) | T. maritima | None | None | Unknown | ND | ND | No | 1 (Mist and MicrobesOnline) | 26, 42 |
| McpS (PA1930) | P. aeruginosa | PAS (2) | None | Overexpression disrupts polar localization of transmembrane chemoreceptors | P | Yes | ND | 4 (Porter et al.) | 34 |
| IcpA/McpE (SMc03004) | S. meliloti | Protoglobin | None | Unknown | P | No | No | 2 (Porter et al.) | 61, 90 |
| McpY (SMc03087) | S. meliloti | HAMP, PAS | None | Unknown | P | Yes | ND | 2 (Porter et al.) | 61, 90 |
| HemAT (BSU10380) | B. subtilis | Protoglobin | None | Protoglobin-bound heme senses oxygen | P | Yes | Yes | 1 (Porter et al.) | 14, 64 |
| YfmS (BSU07360) | B. subtilis | None | None | Unknown | P | ND | ND | 1 (Porter et al.) | 14 |
| AerC (AZOBR_70123) | A. brasilense | PAS (2) | None | Cytoplasmic redox/oxygen via FAD bound in PAS domain | P/C | ND | ND | 4 (Russell et al.) | 12 |
| TlpC (RSP_1589) | R. sphaeroides | None | None | Unknown | CC | No | 3 (Porter et al.) | 44, 91 | |
| TlpT (RSP_0044) | R. sphaeroides | None | None | Unknown | CC | No | 3 (Porter et al.) | 36, 67 | |
| FrzCD (MXAN_4141) | M. xanthus | None | None | Senses side-to-side contact from neighboring cells; fruiting body formation | D, F | Yes | Yes | 8 (Porter et al.) | 74, 92, 93 |
| Mcp7 (MXAN_6962) | M. xanthus | None | None | Coupling between sporulation and aggregation | P (sub) | ND | ND | 8 (Porter et al.) | 75 |
| McpB (CC_0428) | C. crescentus | Protoglobin | None | Unknown | C | Yes | ND | 2 (Mist, MicrobesOnline) | 35 |
| Car (OE_5243F) | H. salinarum | PAS | None | Arginine | C | ND | ND | 1 (Mist) | 15 |
Domains are given as either amino terminal (NH3) or carboxy terminal (COOH) relative to the MA domain. Localization is indicated as follows: P, polar; P/C, polar and cytoplasmic; P (sub), subpolar; CC, cytoplasmic clusters; D, cytoplasmic and dynamic; F, filaments; C, cytoplasmic but exact location unknown; ND, not determined. “Methylation consensus” indicates whether methylation consensus sequences have been identified in the protein, while “verified” indicates whether these were experimentally confirmed (Yes) or not yet determined (ND). Numbers of chemotaxis operons and encoded CheA proteins were obtained from data reported previously by Porter et al. (32) and Russell et al. (59) and in the Mist (94) and MicrobesOnline (95) databases, as indicated. This number is usually estimated based on the number of encoded CheA proteins.
Cytoplasmic Chemoreceptors with Nearly Exclusive Localization at the Pole with Transmembrane Chemoreceptor Arrays
Some cytoplasmic chemoreceptors are found predominantly at the pole of the cells. One such protein is Pseudomonas aeruginosa McpS (we use the genus and species abbreviation as a subscript to help differentiate the many similarly named chemoreceptors, e.g., McpSPA) (Table 2 and Fig. 5). The P. aeruginosa PAO1 genome encodes 4 chemotaxis pathways with 26 chemoreceptors, 3 of which lack transmembrane regions and are therefore predicted to be soluble (34). McpSPA contains an N-terminal PAS domain and predicted methylation sites, but it is not yet known what it responds to or whether it is methylated (34). McpSPA localizes to the pole along with several other chemoreceptors, based on immunofluorescence (34). Increasing McpSPA levels disrupted the polar localization of transmembrane chemoreceptors, but McpSPA was still observed at the pole. Increased levels of McpSPA also caused decreased motility in a soft-agar chemotaxis assay. There have been no studies on the proteins required for McpSPA polar localization. Taken together, this cytoplasmic chemoreceptor appears to reside in a polar complex with transmembrane chemoreceptors, and this macromolecular structure is sensitive to the concentration of McpSPA, perhaps through displacing interactions between receptors or signaling proteins (34).
FIG 5.
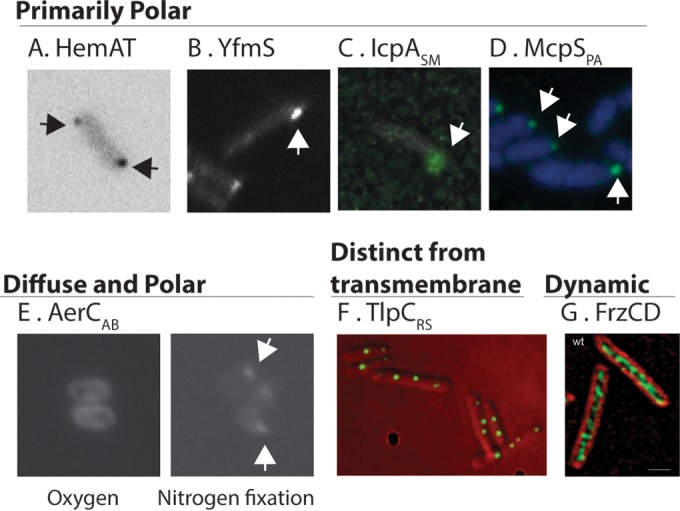
Examples of the diverse subcellular locations of cytoplasmic chemoreceptors. At the top are chemoreceptors that are primarily polar, shown as distinct bright polar spots. (A) B. subtilis HemAT visualized as HemAT-YFP. (Courtesy of George Ordal. Similar images can be found in reference 62.) (B) B. subtilis YfmS visualized as YfmS-YFP. (Courtesy of George Ordal. Similar images can be found in reference 62.) (C) S. meliloti IcpA-enhanced GFP. (Adapted from reference 61 with permission.) (D) P. aeruginosa McpS, visualized by using immunofluorescence with anti-His antibody to recognize His-McpS. (Adapted from reference 34 with permission.) Shown at the bottom are chemoreceptors that (i) are diffuse or polar under different environmental conditions, (ii) reside in an internal cytoplasmic cluster that is distinct from the polar transmembrane chemoreceptors, or (iii) are dynamic. (E) A. brasilense AerC, visualized as AerC-YFP, under different growth conditions. (Courtesy of Gladys Alexandre. Similar images can be found in reference 12.) (F) R. sphaeroides TlpC visualized as TlpC-GFP. (Courtesy of Judith Armitage. Similar images can be found in reference 66.) (G) M. xanthus FrzCD, visualized with anti-FrzCD, and bacterial cells stained with FM4-64. (Reprinted from reference 74 with permission.)
IcpA and McpY of Sinorhizobium meliloti (IcpASM and McpYSM, respectively) are also found nearly exclusively at the pole (Table 2 and Fig. 5). IcpASM and McpYSM are the two cytoplasmic chemoreceptors in the nine-chemoreceptor/two-pathway system of S. meliloti. IcpASM contains a predicted protoglobin domain at its N terminus, and McpYSM contains two PAS domains in tandem at its N terminus; the input signals of these two chemoreceptors, however, are not known (61). IcpASM and McpYSM are both located at the pole, based on analysis of green fluorescent protein (GFP) fusion proteins (61). They furthermore colocalized with the transmembrane chemoreceptor McpX and fluorescently tagged CheA, suggesting that they are part of a large chemoreceptor signaling complex. Interestingly, McpYSM and IcpASM required different sets of proteins to localize to the pole. Specifically, IcpASM was not affected by the loss of other chemoreceptors, CheA or both CheW1 and CheW2, while the polar localization of McpYSM decreased without these proteins (61). Fractionation of S. meliloti cells followed by Western blotting revealed that McpYSM was found in the cytosolic fraction, while IcpASM was present exclusively in the membrane fraction. This membrane association was probed by mild treatment of membrane fractions with detergent, after which IcpASM still showed an association with the membrane fraction (61). The localization of IcpASM is intriguing, as it suggests that cytoplasmic chemoreceptors may interact at the pole with partners apart from chemoreceptor arrays and chemotaxis signaling proteins.
B. subtilis HemAT is yet another example of a polarly localized cytoplasmic chemoreceptor (Table 2 and Fig. 5). B. subtilis encodes one chemotaxis system with 10 chemoreceptors, 2 of which are soluble: HemAT and YfmS (62). HemAT mediates aerotaxis via oxygen binding through a coordinated heme group at its N terminus within a protoglobin domain (63). The ligand of YfmS is unknown; the sole annotated domain in this protein is the MA domain. The localization of yellow fluorescent protein (YFP)-tagged HemAT and YfmS was studied by fluorescence microscopy, and they both localize primarily to the pole, similar to the transmembrane receptor McpB (64). HemAT is also found in Halobacterium salinarum, but its localization in this microbe is not known. It has been suggested that these receptors may be part of a large membrane chemoreceptor array and undergo conformational changes upon ligand binding that propagate to neighboring receptors. This hypothesis could explain the fact that aerotaxis is more efficient when HemAT is not the sole receptor present (62).
Cytoplasmic Chemoreceptors with both Polar Colocalization with Transmembrane Receptor Clusters and Diffuse Cytoplasmic Localization
The localization of some cytoplasmic chemoreceptors seems to be multifaceted, present in both soluble and membrane fractions. This is the case for Azospirillum brasilense AerC (AerCAB) and TlpDHP (Table 2 and Fig. 5). A. brasilense codes for four chemotaxis pathways, with 51 chemoreceptors. AerCAB is a cytoplasmic chemoreceptor that possesses two N-terminal PAS domains and a C-terminal MA domain (12). AerCAB binds FAD at each of two N-terminal PAS domains and is postulated to sense intracellular redox, via the bound FAD, and direct an oxygen-repellent response to support nitrogen fixation (12). AerCAB-YFP localization was determined by fluorescence microscopy and was found to be both diffuse in the cytoplasm and localized in foci at the pole, depending on growth conditions (Fig. 5) (12). Under conditions promoting nitrogen fixation, as opposed to growth in the presence of ammonia, AerCAB was found predominantly at the pole (12). In contrast, growth in the presence of ammonia created dimmer polar foci. Polar localization required proteins of the Che1 operon (CheA1, CheW1, CheB1, and CheY1) (12, 65) as well as specific nitrogen-fixing growth conditions. FAD binding was not required for AerCAB polar localization but did alter migration in a soft-agar assay and abolished aerotaxis in oxygen gradients (12). Nitrogen fixation is an energetically intensive process, and the enzyme responsible for splitting N2, nitrogenase, is inhibited by oxygen. In this manner, AerCAB would help A. brasilense restrict nitrogen fixation to low-oxygen environments.
TlpDHP is found both at the pole with the other chemoreceptors and throughout the cytoplasm (11). H. pylori possesses three transmembrane chemoreceptors in addition to TlpDHP, all part of one chemotaxis pathway. TlpDHP binds zinc at its conserved C-terminal CZB domain (49). The direct signal of TlpDHP is unknown, but the receptor has been reported to mediate a repellent response to inhibitors of the electron transport chain (11). TlpDHP has been detected in cytoplasmic and membrane portions of fractionated cells in approximately equivalent amounts (11). Membrane localization of TlpDHP depends on the transmembrane chemoreceptors CheA, CheW, and CheV1 when assayed by subcellular fractionation and Western blotting (K. D. Collins and K. M. Ottemann, unpublished data). This finding suggests that TlpD may exist in a complex with the other chemotaxis proteins but is also able to reside in the cytoplasm.
Cytoplasmic Chemoreceptors That Form Clusters Distinct from the Transmembrane Polar Receptors
Studies of Rhodobacter sphaeroides chemotaxis have provided an intriguing case of cytoplasmic chemoreceptors that localize exclusively internal to the cytoplasm, away from the polar transmembrane chemoreceptors (Table 2 and Fig. 5). R. sphaeroides possesses three chemotaxis pathways with 13 chemoreceptors, including 4 that lack predicted transmembrane domains. Two of these, R. sphaeroides TlpC (TlpCRS) and TlpTRS, have been studied. TlpCRS and TlpTRS localize to a cytoplasmic cluster (36) that contains a repertoire of chemotaxis signaling proteins (66). Localization of the cytoplasmic cluster is cell cycle dependent (36), and duplication of the cluster occurs prior to cell division, allowing segregation of each duplicated cluster to a daughter cells. Mechanisms controlling the duplication of the cluster remain unknown. Clusters contain TlpCRS, TlpTRS, CheA3, CheA4, CheW4, CheR3, and a ParA homolog called PpfA (all encoded in the same operon) (66). Cluster formation relies on the presence of TlpTRS and CheW4 (67), while the positioning and segregation of the cluster rely on the presence of the N terminus of TlpTRS and the activity of PpfA (68, 69). Cells lacking ppfA do not form a second cytoplasmic cluster during cell division, and only one daughter inherits a cluster as a result. Daughter cells lacking a cluster are nonchemotactic until they synthesize new cluster components (68). PpfA interacts with TlpTRS via its N terminus, which is thought to stimulate PpfA ATPase activity. Additional PpfA interactions with chromosomal DNA may fix the cluster localization and allow the cluster to be segregated during cell division with chromosomal DNA (69). This localization mechanism may be widespread, given that many cytoplasmic chemoreceptors are encoded in the same operon as a protein with PpfA homology (46), and positioning of chemosensory arrays in Vibrio also seems to depend on the presence and activity of a ParA-like protein, ParC (70, 71).
Cytoplasmic Chemoreceptors with Dynamic Cytoplasmic Localization
The bacterium Myxococcus xanthus presents an interesting case for soluble chemoreceptors in social behaviors and developmentally regulated processes. This Gram-negative soil bacterium demonstrates social behavior in coordinating movement during predatory hunting and also undergoes fruiting body formation during nutrient limitation, both of which are controlled in part by chemosensory systems (8, 72). M. xanthus contains eight chemotaxis pathways that perform a variety of functions. It possesses two soluble chemoreceptors, FrzCD and Mcp7 (Table 2). FrzCD controls vegetative swarming and starvation-induced aggregation prior to fruiting body formation (8), while Mcp7 regulates coupling between aggregation and sporulation (73). FrzCD shows a dynamic localization that appears to be sensitive to cell-cell contacts and is organized in helical filaments spanning the length of the cell (Table 2 and Fig. 5). M. xanthus employs two forms of gliding motility, both of which rely on cellular reversals but are not flagellar based (8). The Frz pathway, including FrzCD, along with chemotaxis signaling proteins, including the FrzE histidine kinase, control the frequency of reversals. Mutations in the Frz pathway alter the frequency of reversals. Fluorescence microscopy revealed that FrzCD localizes in helical filaments that cover the cell length and that FrzCD is dynamic within these structures, continuously changing its position, number, and intensity (74, 75). FrzCD in one cell transiently aligns with that in another as cells make side-to-side contacts. An intact Frz signaling pathway is required for FrzCD localization, as an frzE mutant showed more diffuse FrzCD clusters that were also less abundant than in the wild type. Clusters within the frzE mutant were still dynamic, however, constantly changing their number, position, and intensity. FrzCD is proposed to operate as a regulator of reversal frequency through its tracking along a cytoskeletal filament from one pole of the cell to the other, and cell-cell contacts increase the frequency with which FrzCD stimulates reversals (74). FrzCD appears to respond to the presence of attractants and repellents through levels of methylation, and the modification appears to have consequences on fruiting body formation (8). The second M. xanthus cytoplasmic chemoreceptor, Mcp7, displays subpolar localization at either one or both poles and was the most mobile of the M. xanthus chemoreceptors (75). This chemoreceptor is encoded in the che7 locus with cognate copies of CheA, CheW, CheY, CheB, CheR, and the accessory protein Cpc7. Mcp7 mutants prematurely sporulate before cell populations aggregate. Mcp7 activity, specifically the phosphorylation of CheY encoded in the che7 locus (CheY7), negatively regulates sporulation. Phosphorylated CheY7 physically interacts with Cpc7, and this complex negatively regulates sporulation until cell aggregation is complete (73). These two soluble chemoreceptors provide an important reminder that not all chemosensory modules are strictly involved in motility and may instead provide a connection between sensing environmental signals and developmentally controlled behaviors or processes.
Cytoplasmic Chemoreceptors with Unknown Cytoplasmic Localization
Several cytoplasmic receptors have been studied only by subcellular fractionation and clearly localize to the cytoplasmic fraction of cells. It is not known, however, where within the cell (e.g., polar or clustered) these chemoreceptors reside. Caulobacter crescentus McpB (McpBCC) and Halobacterium salinarum Car are examples of this class of cytoplasmic chemoreceptors (Table 2). The C. crescentus chemotaxis system consists of two chemosensory pathways of 19 predicted chemoreceptors, with McpBCC being one of five cytoplasmic chemoreceptors. McpBCC contains a predicted N-terminal protoglobin domain and appears to be methylated based on the migration of the protein in strains lacking either CheB or CheR (35). The localization of McpBCC was determined by cellular fractionation to be exclusively in the cytoplasmic fraction (35). The presence of an N-terminal protoglobin domain, similarly to HemAT, suggests that McpBCC may play a role in aerotaxis, although this role has not been validated (35).
H. salinarum is a halophilic archaeon encoding one chemosensory system with 18 chemoreceptors, 6 of which are predicted to be cytoplasmic. Car is one of these cytoplasmic chemoreceptors and possesses a PAS domain at its N terminus. Strains lacking Car lose the ability to respond to arginine in a soft agar chemotaxis assay but retain the ability to respond to other amino acids, suggesting that this cytoplasmic chemoreceptor mediates a tactic response to arginine (15). Car localizes to the cytoplasmic fraction, based on subcellular fractionation (15). Car is believed to monitor the concentrations of arginine in the cytoplasm, possibly as part of a metabolism-dependent motility response (15).
CONCLUSIONS
Cytoplasmic chemoreceptors are common among microbes of the bacterial and archaeal domains, comprising 14% and 43% of chemoreceptors, respectively. Existing studies, although limited, suggest that cytoplasmic chemoreceptors share the same key structural features as transmembrane chemoreceptors: a homodimeric trimer-of-dimer configuration, organized into 12-nm hexagonal arrays. However, the locations of the cytosolic chemoreceptor arrays vary. Certain cytoplasmic chemoreceptors have been observed to localize to the pole with transmembrane chemoreceptors, and signaling could occur through interactions with other receptors in that array. Other chemoreceptors show diffuse cytoplasmic and polar localizations, suggesting trafficking between these two locations. The localization of R. sphaeroides cytoplasmic chemoreceptors in internal clusters offers the most well-studied example of solely cytoplasmic chemoreceptors, and these receptors seem to be engaged in a complex that is capable of signaling independently of transmembrane signaling centers. The case of FrzCD offers an interesting alternative to other cytoplasmic chemoreceptor signaling as part of a dynamic structure that operates along the length of Myxococcus. This paradigm of signaling may not be unique to this bacterium and instead may offer insight into cytoplasmic chemoreceptors in contexts outside flagellar-driven chemotaxis. The driving force for distinct cellular locations is not yet known. It may be the location of the signal, some aspect of signal transduction, another factor, or some combination of these that determines whether a receptor is cytoplasmic or polar, static or dynamic. Cytoplasmic chemoreceptors bear diverse signal input domains, which are mostly N terminal to the MA domain. Similar to transmembrane receptors, the most common signal input domain is the PAS domain, and a variety of other N-terminal domains exist. It is also common, however, for cytoplasmic receptors to have C-terminal domains. The most common of these is the recently identified CZB domain, found in 8% of all cytoplasmic chemoreceptors. The reason why some domains are N terminal while others are C terminal to the MA domain is not yet known. Some cytosolic chemoreceptors do not seem to contain an input signal region in their sequence, suggesting that these chemoreceptors might play some sort of structural role instead of receiving signals or that there are as-yet-unidentified input domains. There are many open questions about this interesting group of chemoreceptor proteins. Future studies of these chemoreceptors will undoubtedly generate significant insights into the fundamental properties of signal recognition and transduction.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Daniela Keilberg and the reviewers for insightful comments on the manuscript and to George Ordal, Birgit Scharf, Janine Maddock, Gladys Alexandre, Judy Armitage, and David Zusman for providing permission and/or images to use in Fig. 5.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MMBR.00033-14.
REFERENCES
- 1.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024–1037. 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Lai W-C. 2010. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr. Opin. Microbiol. 13:124–132. 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander RP, Zhulin IB. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 104:2885–2890. 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhulin IB. 2001. The superfamily of chemotaxis transducers: from physiology to genomics and back. Adv. Microb. Physiol. 45:157–198. 10.1016/S0065-2911(01)45004-1. [DOI] [PubMed] [Google Scholar]
- 5.Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864. 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wuichet K, Alexander RP, Zhulin IB. 2007. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 422:1–31. 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacal J, García-Fontana C, Muñoz-Martínez F, Ramos J-L, Krell T. 2010. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 12:2873–2884. 10.1111/j.1462-2920.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 8.Zusman DR, Scott AE, Yang Z, Kirby JR. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 5:862–872. 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 9.Rolig AS, Shanks J, Carter JE, Ottemann KM. 2012. Helicobacter pylori requires TlpD-driven chemotaxis to proliferate in the antrum. Infect. Immun. 80:3713–3720. 10.1128/IAI.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behrens W, Schweinitzer T, Bal J, Dorsch M, Bleich A, Kops F, Brenneke B, Didelot X, Suerbaum S, Josenhans C. 2013. Role of energy sensor TlpD of Helicobacter pylori in gerbil colonization and genome analyses after adaptation in the gerbil. Infect. Immun. 81:3534–3551. 10.1128/IAI.00750-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schweinitzer T, Mizote T, Ishikawa N, Dudnik A, Inatsu S, Schreiber S, Suerbaum S, Aizawa SI, Josenhans C. 2008. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J. Bacteriol. 190:3244–3255. 10.1128/JB.01940-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Z, Ulrich LE, Zhulin IB, Alexandre G. 2010. PAS domain containing chemoreceptor couples dynamic changes in metabolism with chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 107:2235–2240. 10.1073/pnas.0910055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong CS, Shitashiro M, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. 2004. Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 231:247–252. 10.1016/S0378-1097(04)00009-6. [DOI] [PubMed] [Google Scholar]
- 14.Hou S, Larsen RW, Boudko D, Riley CW, Karatan E, Zimmer M, Ordal GW, Alam M. 2000. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature 403:540–543. 10.1038/35000570. [DOI] [PubMed] [Google Scholar]
- 15.Storch KF, Rudolph J, Oesterhelt D. 1999. Car: a cytoplasmic sensor responsible for arginine chemotaxis in the archaeon Halobacterium salinarum. EMBO J. 18:1146–1158. 10.1093/emboj/18.5.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9–19. 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. 2012. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. U. S. A. 109:3766–3771. 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vu A, Wang X, Zhou H, Dahlquist FW. 2012. The receptor-CheW binding interface in bacterial chemotaxis. J. Mol. Biol. 415:759–767. 10.1016/j.jmb.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatnagar J, Borbat PP, Pollard AM, Bilwes AM, Freed JH, Crane BR. 2010. Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy. Biochemistry 49:3824–3841. 10.1021/bi100055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu JD, Parkinson JS. 1991. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J. Bacteriol. 173:4941–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S-Y, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. 2006. Reconstruction of the chemotaxis receptor-kinase assembly. Nat. Struct. Mol. Biol. 13:400–407. 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 22.Asinas AE, Weis RM. 2006. Competitive and cooperative interactions in receptor signaling complexes. J. Biol. Chem. 281:30512–30523. 10.1074/jbc.M606267200. [DOI] [PubMed] [Google Scholar]
- 23.Piasta KN, Ulliman CJ, Slivka PF, Crane BR, Falke JJ. 2013. Defining a key receptor-CheA kinase contact and elucidating its function in the membrane-bound bacterial chemosensory array: a disulfide mapping and TAM-IDS study. Biochemistry 52:3866–3880. 10.1021/bi400385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Vu A, Lee K, Dahlquist FW. 2012. CheA-receptor interaction sites in bacterial chemotaxis. J. Mol. Biol. 422:282–290. 10.1016/j.jmb.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cardozo MJ, Massazza DA, Parkinson JS, Studdert CA. 2010. Disruption of chemoreceptor signalling arrays by high levels of CheW, the receptor-kinase coupling protein. Mol. Microbiol. 75:1171–1181. 10.1111/j.1365-2958.2009.07032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Fleetwood AD, Bayas C, Bilwes AM, Ortega DR, Falke JJ, Zhulin IB, Crane BR. 2013. The 3.2 A resolution structure of a receptor:CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays. Biochemistry 52:3852–3865. 10.1021/bi400383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chervitz SA, Falke JJ. 1996. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. U. S. A. 93:2545–2550. 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ottemann KM, Xiao W, Shin Y-K, Koshland DE. 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 285:1751–1754. 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 29.Parkinson JS. 2010. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 64:101–122. 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 30.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929–940. 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 31.Airola MV, Watts KJ, Bilwes AM, Crane BR. 2010. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure 18:436–448. 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9:153–165. 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 33.Anand GS, Stock AM. 2002. Kinetic basis for the stimulatory effect of phosphorylation on the methylesterase activity of CheB. Biochemistry 41:6752–6760. 10.1021/bi012102n. [DOI] [PubMed] [Google Scholar]
- 34.Bardy SL, Maddock JR. 2005. Polar localization of a soluble methyl-accepting protein of Pseudomonas aeruginosa. J. Bacteriol. 187:7840–7844. 10.1128/JB.187.22.7840-7844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potocka I, Thein M, Osteras M, Jenal U, Alley MRK. 2002. Degradation of a Caulobacter soluble cytoplasmic chemoreceptor is ClpX dependent. J. Bacteriol. 184:6635–6641. 10.1128/JB.184.23.6635-6642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wadhams GH, Martin AC, Porter SL, Maddock JR, Mantotta JC, King HM, Armitage JP. 2002. TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol. Microbiol. 46:1211–1221. 10.1046/j.1365-2958.2002.03252.x. [DOI] [PubMed] [Google Scholar]
- 37.Rao CV, Glekas GD, Ordal GW. 2008. The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol. 16:480–487. 10.1016/j.tim.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM. 2010. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol. 18:494–503. 10.1016/j.tim.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glekas GD, Plutz MJ, Walukiewicz HE, Allen GM, Rao CV, Ordal GW. 2012. Elucidation of the multiple roles of CheD in Bacillus subtilis chemotaxis. Mol. Microbiol. 86:743–756. 10.1111/mmi.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao X, Muff TJ, Park S-Y, Zhang S, Pollard AM, Ordal GW, Bilwes AM, Crane BR. 2006. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 124:561–571. 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 41.Muff TJ, Ordal GW. 2007. The CheC phosphatase regulates chemotactic adaptation through CheD. J. Biol. Chem. 282:34120–34128. 10.1074/jbc.M706432200. [DOI] [PubMed] [Google Scholar]
- 42.Pollard AM, Bilwes AM, Crane BR. 2009. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry 48:1936–1944. 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim KK, Yokota H, Kim S-H. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787–792. 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 44.Briegel A, Ladinsky MS, Oikonomou C, Jones CW, Harris MJ, Fowler DJ, Chang YW, Thompson LK, Armitage JP, Jensen GJ. 2014. Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. eLife 3:e02151. 10.7554/eLife.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khursigara CM, Lan G, Neumann S, Wu X, Ravindran S, Borgnia MJ, Sourjik V, Milne J, Tu Y, Subramaniam S. 2011. Lateral density of receptor arrays in the membrane plane influences sensitivity of the E. coli chemotaxis response. EMBO J. 30:1719–1729. 10.1038/emboj.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sourjik V, Armitage JP. 2010. Spatial organization in bacterial chemotaxis. EMBO J. 29:2724–2733. 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. U. S. A. 106:17181–17186. 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Draper J, Karplus K, Ottemann KM. 2011. Identification of a chemoreceptor zinc-binding domain common to cytoplasmic bacterial chemoreceptors. J. Bacteriol. 193:4338–4345. 10.1128/JB.05140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zähringer F, Lacanna E, Jenal U, Schirmer T, Boehm A. 2013. Structure and signaling mechanism of a zinc-sensory diguanylate cyclase. Structure 21:1149–1157. 10.1016/j.str.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 51.Borziak K, Zhulin IB. 2007. FIST: a sensory domain for diverse signal transduction pathways in prokaryotes and ubiquitin signaling in eukaryotes. Bioinformatics 23:2518–2521. 10.1093/bioinformatics/btm384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aravind L, Ponting CP. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22:458–459. 10.1016/S0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 53.Heikaus CC, Pandit J, Klevit RE. 2009. Cyclic nucleotide binding GAF domains from phosphodiesterases: structural and mechanistic insights. Structure 17:1551–1557. 10.1016/j.str.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monterrubio R, Baldoma L, Obradors N, Aguilar J, Badia J. 2000. A common regulator for the operons encoding the enzymes involved in D-galactarate, D-glucarate, and D-glycerate utilization in Escherichia coli. J. Bacteriol. 182:2672–2674. 10.1128/JB.182.9.2672-2674.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anantharaman V, Aravind L. 2005. MEDS and PocR are novel domains with a predicted role in sensing simple hydrocarbon derivatives in prokaryotic signal transduction systems. Bioinformatics 21:2805–2811. 10.1093/bioinformatics/bti418. [DOI] [PubMed] [Google Scholar]
- 56.Anantharamana V, Aravind L. 2000. Cache—a signaling domain common to animal Ca2+-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25:535–537. 10.1016/S0968-0004(00)01672-8. [DOI] [PubMed] [Google Scholar]
- 57.Tam R, Saier MH., Jr 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 59.Russell MH, Bible AN, Fang X, Gooding JR, Campagna SR, Gomelsky M, Alexandre G. 2013. Integration of the second messenger c-di-GMP into the chemotactic signaling pathway. mBio 4(2):e00001-13. 10.1128/mBio.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. 2009. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 7:e1000137. 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meier VM, Scharf BE. 2009. Cellular localization of predicted transmembrane and soluble chemoreceptors in Sinorhizobium meliloti. J. Bacteriol. 191:5724–5733. 10.1128/JB.01286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cannistraro VJ, Glekas GD, Rao CV, Ordal GW. 2011. Cellular stoichiometry of the chemotaxis proteins in Bacillus subtilis. J. Bacteriol. 193:3220–3227. 10.1128/JB.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.El-Mashtoly SF, Gu Y, Yoshimura H, Yoshioka S, Aono S, Kitagawa T. 2008. Protein conformation changes of HemAT-Bs upon ligand binding probed by ultraviolet resonance Raman spectroscopy. J. Biol. Chem. 283:6942–6949. 10.1074/jbc.M709209200. [DOI] [PubMed] [Google Scholar]
- 64.Lamanna AC, Ordal GW, Kiessling LL. 2005. Large increases in attractant concentration disrupt the polar localization of bacterial chemoreceptors. Mol. Microbiol. 57:774–785. 10.1111/j.1365-2958.2005.04728.x. [DOI] [PubMed] [Google Scholar]
- 65.Bible AN, Stephens BB, Ortega DR, Xie Z, Alexandre G. 2008. Function of a chemotaxis-like signal transduction pathway in modulating motility, cell clumping, and cell length in the alphaproteobacterium Azospirillum brasilense. J. Bacteriol. 190:6365–6375. 10.1128/JB.00734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wadhams GH, Warren AV, Martin AC, Armitage JP. 2003. Targeting of two signal transduction pathways to different regions of the bacterial cell. Mol. Microbiol. 50:763–770. 10.1046/j.1365-2958.2003.03716.x. [DOI] [PubMed] [Google Scholar]
- 67.Wadhams GH, Martin AC, Warren AV, Armitage JP. 2005. Requirements for chemotaxis protein localization in Rhodobacter sphaeroides. Mol. Microbiol. 58:895–902. 10.1111/j.1365-2958.2005.04880.x. [DOI] [PubMed] [Google Scholar]
- 68.Thompson SR, Wadhams GH, Armitage JP. 2006. The positioning of cytoplasmic protein clusters in bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:8209–8214. 10.1073/pnas.0600919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberts MAJ, Wadhams GH, Hadfield KA, Tickner S, Armitage JP. 2012. ParA-like protein uses nonspecific chromosomal DNA binding to partition protein complexes. Proc. Natl. Acad. Sci. U. S. A. 109:6698–6703. 10.1073/pnas.1114000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ringgaard S, Schirner K, Davis BM, Waldor MK. 2011. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev. 25:1544–1555. 10.1101/gad.2061811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ringgaard S, Zepeda-Rivera M, Wu X, Schirner K, Davis BM, Waldor MK. 2014. ParP prevents dissociation of CheA from chemotactic signaling arrays and tethers them to a polar anchor. Proc. Natl. Acad. Sci. U. S. A. 111:E255–E264. 10.1073/pnas.1315722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirby JR. 2009. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu. Rev. Microbiol. 63:45–59. 10.1146/annurev.micro.091208.073221. [DOI] [PubMed] [Google Scholar]
- 73.Darnell CL, Wilson JM, Tiwari N, Fuentes EJ, Kirby JR. 2014. Chemosensory regulation of a HEAT-repeat protein couples aggregation and sporulation in Myxococcus xanthus. J. Bacteriol. 196:3160–3168. 10.1128/JB.01866-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mauriello EM, Astling DP, Sliusarenko O, Zusman DR. 2009. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc. Natl. Acad. Sci. U. S. A. 106:4852–4857. 10.1073/pnas.0810583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moine A, Agrebi R, Espinosa L, Kirby JR, Zusman DR, Mignot T, Mauriello EMF. 2014. Functional organization of a multimodular bacterial chemosensory apparatus. PLoS Genet. 10:e1004164. 10.1371/journal.pgen.1004164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. 2007. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 26:5153–5166. 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang W, Phillips GN., Jr 2003. Structure of the oxygen sensor in Bacillus subtilis. Structure 11:1097–1110. 10.1016/S0969-2126(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 79.Ho Y-SJ, Burden LM, Hurley JH. 2000. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19:5288–5299. 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martins LJ, Hill CP, Ellis WR. 1997. Structures of wild-type chloromet and L103N hydroxomet Themiste zostericola myohemerythrins at 1.8 A resolution. Biochemistry 36:7044–7049. 10.1021/bi9630422. [DOI] [PubMed] [Google Scholar]
- 81.Throup JP, Zappacosta F, Lunsford RD, Annan RS, Carr SA, Lonsdale JT, Bryant AP, McDevitt D, Rosenberg M, Burnham MK. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392–10401. 10.1021/bi0102959. [DOI] [PubMed] [Google Scholar]
- 82.Strube K, de Vries S, Cramm R. 2007. Formation of a dinitrosyl iron complex by NorA, a nitric oxide-binding di-iron protein from Ralstonia eutropha H16. J. Biol. Chem. 282:20292–20300. 10.1074/jbc.M702003200. [DOI] [PubMed] [Google Scholar]
- 83.Rodriguez-Navarro S, Llorente B, Reyes-Lamothe R, Ramne A, Uber G, Marchesan D, Dujon B, Herrero E, Sunnerhagen P, Perez-Ortin JE. 2002. Functional analysis of yeast gene families involved in metabolism of vitamins B1 and B6. Yeast 19:1261–1276. 10.1002/yea.916. [DOI] [PubMed] [Google Scholar]
- 84.Maundrell K. 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265:10857–10864. [PubMed] [Google Scholar]
- 85.Iyer LM, Anantharaman V, Aravind L. 2003. Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics 4:5. 10.1186/1471-2164-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. 2004. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Invest. 113:274–284. 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shu CJ, Ulrich LE, Zhulin IB. 2003. The NIT domain: a predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem. Sci. 28:121–124. 10.1016/S0968-0004(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 88.Zhulin IB, Nikolskaya AN, Galperin MY. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in Bacteria and Archaea. J. Bacteriol. 185:285–294. 10.1128/JB.185.1.285-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hou S, Freitas T, Larsen RW, Piatibratov M, Sivozhelezov V, Yamamoto A, Meleshkevitch EA, Zimmer M, Ordal GW, Alam M. 2001. Globin-coupled sensors: a class of heme-containing sensors in Archaea and Bacteria. Proc. Natl. Acad. Sci. U. S. A. 98:9353–9358. 10.1073/pnas.161185598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meier VM, Muschler P, Scharf BE. 2007. Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti. J. Bacteriol. 189:1816–1826. 10.1128/JB.00883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Porter SL, Warren AV, Martin AC, Armitage JP. 2002. The third chemotaxis locus of Rhodobacter sphaeroides is essential for chemotaxis. Mol. Microbiol. 46:1081–1094. 10.1046/j.1365-2958.2002.03218.x. [DOI] [PubMed] [Google Scholar]
- 92.McCleary WR, McBride MJ, Zusman DR. 1990. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J. Bacteriol. 172:4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scott AE, Simon E, Park SK, Andrews P, Zusman DR. 2008. Site-specific receptor methylation of FrzCD in Myxococcus xanthusis controlled by a tetra-trico peptide repeat (TPR) containing regulatory domain of the FrzF methyltransferase. Mol. Microbiol. 69:724–735. 10.1111/j.1365-2958.2008.06323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ulrich LE, Zhulin IB. 2007. MiST: a microbial signal transduction database. Nucleic Acids Res. 35:D386–D390. 10.1093/nar/gkl932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38:D396–D400. 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.