Abstract
The 3′ end processing of animal replication-dependent histone mRNAs is activated during G1/S-phase transition. The processing site is recognized by stem-loop binding protein and the U7 snRNP, but cleavage additionally requires a heat-labile factor (HLF), composed of cleavage/polyadenylation specificity factor, symplekin, and cleavage stimulation factor 64 (CstF64). Although HLF has been shown to be cell cycle regulated, the mechanism of this regulation is unknown. Here we show that levels of CstF64 increase toward the S phase and its depletion affects histone RNA processing, S-phase progression, and cell proliferation. Moreover, analyses of the interactions between CstF64, symplekin, and the U7 snRNP-associated proteins FLASH and Lsm11 indicate that CstF64 is important for recruiting HLF to histone precursor mRNA (pre-mRNA)-resident proteins. Thus, CstF64 is central to the function of HLF and appears to be at least partly responsible for its cell cycle regulation. Additionally, we show that misprocessed histone transcripts generated upon CstF64 depletion mainly accumulate in the nucleus, where they are targets of the exosome machinery, while a small cytoplasmic fraction is partly associated with polysomes.
INTRODUCTION
Most eukaryotic mRNAs have poly(A) tails, which are produced in a two-step reaction consisting of an endonucleolytic cleavage of a longer precursor mRNA (pre-mRNA) and the subsequent addition of the poly(A) tail to the upstream fragment (cleavage and polyadenylation [CPA]) (1, 2). The only major class of protein-coding genes whose transcripts are not processed by CPA are the replication-dependent histone genes (3, 4). Their expression is cell cycle regulated to meet the need for histones to package the newly synthesized DNA during the S phase. An ∼40-fold increment in histone mRNA levels during the G1/S-phase transition involves upregulation of both transcription (∼5-fold) and 3′ end processing (∼8-fold) (5). Later, during the G2 phase, the histone mRNAs are destabilized and rapidly degraded.
Histone RNA 3′ end processing consists of a single cleavage and leaves a conserved hairpin structure at the end of the mRNA (3, 4). The cleavage site, usually after a CA dinucleotide, is defined by the upstream hairpin, recognized and bound by stem-loop- or hairpin-binding protein (SLBP or HBP, respectively), and a histone downstream element (HDE), bound through RNA:RNA base pairing by the RNA moiety of the U7 small nuclear ribonucleoprotein (U7 snRNP). These two components are bridged by a 100-kDa zinc finger protein (ZFP100). The U7 snRNP contains a unique Sm core, composed of five canonical Sm proteins and two special Sm-like proteins named Lsm10 and Lsm11 (6, 7). Importantly, the N-terminal domain of Lsm11 is essential for histone RNA processing (6). An interaction of this domain with another histone processing protein, FLASH, is required for further assembly of the cleavage complex (8–10). However, the cleavage activity is associated with a separate moiety, originally termed heat-labile factor (HLF) (11). HLF consists of a subset of the proteins involved in CPA: cleavage/polyadenylation specificity factor (CPSF), of which the 73-kDa subunit catalyzes the cleavage reaction, symplekin, and part of the cleavage stimulation factor (CstF) (12). More recently, our own studies indicated that the 68-kDa subunit of mammalian cleavage factor I (CFIm68) interacts with Lsm11 and plays a role in histone RNA processing as well (13). However, CFIm68 does not appear to be part of the HLF (10, 12).
Both SLBP and HLF have been shown to be cell cycle regulated (14, 15). While the upregulation of SLBP has been studied in some extent and involves changes in SLBP translation, posttranscriptional modifications, and turnover (15–18), no details are known about the cell cycle regulation of HLF.
As mentioned above, only part of CstF is present in the HLF. A first analysis identified CstF64 and CstF77, but not CstF50, as HLF members (12). However, the presence of CstF77 has recently been questioned (10). In CPA, the three CstF components are assumed to form a hexamer composed of two copies of each subunit (19). CstF77 is important for nuclear localization of the other subunits (20) and mediates some interactions with other components of the CPA machinery (1, 2). CstF50 contains WD40 repeats and may form a core for assembly of the other two CstF members. Importantly, CstF64 has an RNA recognition motif (RRM) through which it binds to the U/GU-rich downstream element and thereby helps to define the polyadenylation site (2). However, concerning the role of CstF64 in histone 3′ end processing, it is not known whether it also acts by binding to RNA or what its exact role may be.
The CstF64 gene (CSTF2) is located on the X chromosome but has an autosomal paralogue (CSTF2T) coding for CstF64τ (21). CstF64τ is most predominant in spermatogenic precursor cells where the X chromosome becomes silenced. Nevertheless, it is expressed ubiquitously, albeit at low levels. We previously showed that due to a lower affinity for CstF77 compared to CstF64, it is not efficiently incorporated into CstF complexes, and its free form is relatively unstable (20). However, in case of loss of CstF64, CstF64τ is more efficiently incorporated into CstF and thereby becomes upregulated and can compensate for the loss of CstF64 in CPA. However, our study also indicated that CstF64τ may not be able to contribute to histone RNA processing.
In summary, it is not known how the HLF gains activity or is assembled when cells enter the S phase, how it is distinguished from the polyadenylation machinery, and how it is recruited to histone pre-mRNAs. Additionally, it was shown that if normal processing is prevented, histone transcripts can become polyadenylated at downstream poly(A) sites (22–25). However, neither a possible function nor the metabolism of these transcripts has been characterized.
Here, we address some of the open questions outlined above. Our results indicate that CstF64 is cell cycle regulated and that its depletion slows down S-phase progression. We also show that CstF64 interacts with FLASH:Lsm11, suggesting that it may be the main link that recruits the HLF to the factors bound on histone pre-mRNA. We further confirm that CstF64τ does not efficiently contribute to histone RNA processing and demonstrate that it is deficient in the interaction with FLASH:Lsm11. Finally, we show that most of the misprocessed transcripts generated upon CstF64 depletion are polyadenylated, display a nuclear localization, and are targets of the exosome machinery. However, a small fraction of these read-through transcripts can be found in the cytoplasm, and some of these appear to be associated with polysomes, suggesting that they may direct the synthesis of histone proteins.
MATERIALS AND METHODS
Cell culture, transfection, and synchronization.
HeLa or HEK293T cell lines were grown in Dulbecco's modified Eagle medium (DMEM) (12 g/liter DMEM–F-12 powder [AMIMED BioConcept, Allschwil, Switzerland], 2.44 g/liter NaHCO3 [Merck, Zug, Switzerland]) supplemented with 10% fetal calf serum (Sigma-Aldrich, Buchs SG, Switzerland), 100 U/ml penicillin (AMIMED BioConcept), and 100 μg/ml streptomycin (AMIMED BioConcept) at 37°C in a moist atmosphere containing 5% CO2. For plasmid transfection, cells were grown to 60% confluence in cell culture dishes of the appropriate size. Either FuGene (Promega, Mannheim, Germany) or polyethylenimine (PEI) (Polysciences, Cham, Switzerland) was used as the transfection reagent. RNA interference was achieved by pSUPuro-based depletion (26), followed by selection of transiently transfected cells with 1.5 μg/ml puromycin (Sigma-Aldrich). Synchronization in G2/M was done by addition of 200 ng/ml nocodazole (Sigma-Aldrich) for 18 to 20 h to normal medium. For S-phase synchronization, complete medium was complemented with 4 μg/ml aphidicolin (Sigma-Aldrich) for 24 h; the cells were collected 4 h after release.
Doxycycline (Clontech, TaKaRa Bio Europe SAS, Saint-Germain-en-Laye, France) was added to complete medium at a final concentration of 5 μg/ml to activate HeLa inducible cell lines. The cell lines were created by lentiviral vector transduction according to reference 27 and sorted by fluorescence-activated cell sorter (FACS). The resulting inducible cell lines were always grown in DMEM containing 10% tetracycline-free fetal calf serum (AMIMED BioConcept) until induced. For sequences of the short hairpin RNA (shRNA) constructs, see Table S1 in the supplemental material.
Cell cycle analysis.
Cells were trypsinized, washed with phosphate-buffered saline (PBS), and fixed by dropwise addition to ice-cold 70% ethanol. After removal of the ethanol by centrifugation, the cells were washed once more with PBS, and DNA was stained with propidium iodide (PI) solution (0.1% Triton X-100 in PBS, 0.2 mg/ml RNase A, 0.2 mg/ml PI) for 1 h. Cell cycle profiles were analyzed by cytofluorometry with a BD FACSCalibur (Becton Dickinson, San Jose, CA) and processed with FlowJo software (Tree Star, Ashland, OR).
RNA extraction, RT, and real-time qPCR.
RNA was extracted from cells by Tri-Reagent isolation (1 M guanidine thiocyanate [Sigma-Aldrich], 1 M ammonium thiocyanate [Merck], 0.1 M sodium acetate [pH 5.2], 5% vol/vol glycerin, 38% vol/vol saturated acidic phenol, in diethylpyrocarbonate [DEPC]-treated water [Sigma-Aldrich]). For reverse transcription (RT), usually 1 to 2 μg total RNA was reverse transcribed in 50 μl AffinityScript reverse transcriptase reaction mixture (Stratagene/Agilent Technologies, Basel, Switzerland) complemented with 0.5 mM all four deoxynucleoside triphosphates, 0.4 μM random hexamers, and 8 U RNasin (Promega). For real-time PCR, reverse-transcribed material was amplified in 20 μl by using Mesa Green quantitative PCR (qPCR) MasterMix Plus for SYBR assay ROX (Eurogentec, Seraing, Belgium) with canonical primers or by using qPCR MasterMix Plus with TaqMan primers and probes (see Table S2 in the supplemental material). Data were acquired with Rotor-GeneQ (Qiagen, Hombrechtikon, Switzerland) and analyzed with the associated software. For each primer or probe set, the amplification efficiency was calculated by the standard curve method. The apparent in vivo 3′ end formation efficiency is expressed in molar ratios of precursor over total RNA. The TaqMan assay measures total histone 3C (H3C) mRNA with a primer/probe set spanning the translation initiation codon. The corresponding pre-mRNA is measured with a primer/probe set spanning the 3′ processing site (13).
Selection of polyadenylated RNAs.
Thirty-five microliters (0.175 mg) of oligo(dT)-conjugated magnetic beads [Dynabeads oligo(dT)25 (Invitrogen)] was washed with 500 μl binding buffer (20 mM Tris-HCl [pH 7.5], 1 M LiCl, 2 mM EDTA) and then blocked with 500 μl of the same buffer supplemented with 0.1% yeast tRNA. After a supplemental wash, the beads were resuspended in 100 μl fresh binding buffer.
Ten micrograms of total RNA in 200 μl binding buffer, previously incubated at 65°C for 2 min and quickly cooled in ice, was added to the beads, and the mixture was incubated for 5 min at room temperature. The beads were washed twice with washing buffer (10 mM Tris-HCl [pH 7.5], 0.15 M LiCl, 1 mM EDTA), and then the poly(A)+ RNA fraction was eluted by addition of 20 μl 10 mM Tris-HCl and incubation at 75°C for 2 min. The eluted RNA was then used for reverse transcription.
The nonpolyadenylated RNA contained in the supernatants collected after the annealing step was isolated by phenol-chloroform extraction and ethanol precipitation. The RNA pellet was resuspended in 20 μl DEPC-water and used directly for reverse transcription. For reverse transcription, the same RNA volumes of polyadenylated and nonpolyadenylated RNA were used.
Immunoprecipitation.
Cells were harvested, washed with PBS, and resuspended in hypotonic lysis buffer (10 mM Tris [pH 7.8], 10 mM NaCl, 2 mM EDTA, 0.5% Triton X-100) eventually containing 200 μg/ml RNase A and supplemented with 1× complete EDTA-free protease inhibitor (Roche, Rotkreuz, Switzerland). The cells were lysed for 30 min on ice, followed by the addition of NaCl to 150 mM. The extract was cleared by centrifugation at 4°C at 16,000 × g for 30 min. The cleared extract was then incubated head over tail for 2 h at 4°C with 30 μl anti-Flag M2 magnetic beads (Sigma-Aldrich) or antihemagglutinin (anti-HA) magnetic beads (Pierce-Fisher Scientific, Reinach, Switzerland), followed by five washes with NET-2 HD (50 mM Tris [pH 7.8], 150 mM NaCl, 0.1% NP-40). Elution was achieved by boiling the beads in SDS loading buffer. Immunoprecipitated material was then analyzed by Western blotting, by using the antibodies listed in Table S3 in the supplemental material.
In vitro interaction assay.
HEK293T cells transfected with individual constructs encoding either mFLASH-FLAG or HA-tagged CstF64 (wild-type or mutants) were synchronized in the S phase, harvested, washed in PBS, and then lysed in radioimmunoprecipitation assay (RIPA) buffer without SDS addition (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 50 mM Tris-HCl [pH 8]) supplemented with 1× complete EDTA-free protease inhibitor (Roche) for 15 min on ice. The extracts were cleared by centrifugation at 16,000 × g at 4°C for 30 min. Immunoprecipitation with antibodies directed against the respective tag was performed as described above. Elution was performed with 0.1 M glycine (pH 2.3), neutralized with 1 M Tris-HCl (pH 9.0). The material was then analyzed by SDS-PAGE followed by Coomassie staining. Recombinant proteins were combined in pairs in Tris-buffered saline (TBS) (150 mM NaCl, 50 mM Tris-Cl [pH 7.5]) and incubated at 4°C to test interactions. Samples were then subjected to immunoprecipitation as described above and eluted by boiling the beads in SDS loading buffer. The immunoprecipitated material was analyzed by Western blotting with antibodies directed against the tags listed in Table S3 in the supplemental material.
Nuclear and cytoplasmic fractionation.
Cells were harvested, washed with PBS, resuspended in cytoplasmic buffer (10 mM HEPES [pH 7.9], 60 mM KCl, 1 mM EDTA, 1 mM MgCl2, 1 mM dithiothreitol [DTT], 0.3% NP-40, 1× protease inhibitor, 1× RNase inhibitor, 10% glycerol) and incubated on ice for 2 min. Cytoplasmic proteins were collected in a new tube after centrifugation at 800 × g for 1 min. The pellet was washed once with washing solution (10 mM HEPES [pH 7.9], 60 mM KCl, 1 mM MgCl2, 1 mM DTT, 1× protease inhibitor, 10% glycerol) and then resuspended in nuclear lysis buffer (250 mM Tris-HCl [pH 7.9], 60 mM KCl, 1 mM EDTA, 1 mM MgCl2, 1 mM DTT, 0.3% NP-40, 1× protease inhibitor, 1× RNase inhibitor, 20% glycerol) and shaken for 30 min at 4°C. Nuclear proteins were collected in a new tube after clearing by centrifugation at 16,000 × g for 30 min. The quality of the separations was assessed by Western blotting by detection of α-tubulin and lamin A/C (see Table S3 in the supplemental material for antibody specification). For RNA extraction, Tri-Reagent was added to the obtained fractions, and RNA isolation was continued as described above.
Sucrose gradients.
Usually, 8 to 10 million HeLa cells were pretreated for 30 min with cycloheximide-containing complete medium (100 μg/ml [Sigma-Aldrich]) before being washed in PBS and harvested. Cytoplasmic fractions were obtained by lysis in low-salt buffer (adapted from reference 28 and consisting of 20 mM Tris-HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.3% Triton X-100, 50 mM sucrose) with 1 mM RNAsin (Dundee Cell Products, Dundee, United Kingdom) and 100 μg/ml cycloheximide, by quickly vortexing. Lysates were cleared by centrifugation for 10 min at 10,000 × g. Linear 15 to 45% (wt/wt) sucrose gradients were prepared in a mixture containing 25 mM Tris-HCl (pH 7.5), 25 mM NaCl, 5 mM MgCl2, and 100 μg/ml cycloheximide with a BioComp gradient station (Science Services, Munich, Germany) and then cooled for 30 min at 4°C. Lysates were loaded on gradients and ultracentrifuged at 36,000 rpm at 4°C for 2.5 h in an SW40 Ti swing-out rotor. Fifteen fractions were collected with Fraction collector 2110 (Bio-Rad, Cressier, Switzerland) at a 0.2-mm/s EM1 speed, with a distance of 6.33 mm per fraction, and absorbance was monitored at 254 nm (A254) with an Econo UV monitor (Bio-Rad). Two hundred microliters of each fraction was used for RNA extraction. For EDTA-treated gradients, 20 mM EDTA was directly added to the cleared lysate prior to the gradient centrifugation. As an alternative to prevent polysome formation, cells were incubated for 15 min with 200 nM pactamycin in complete medium before harvesting.
RESULTS
CstF64 is cell cycle regulated, and its depletion affects cell cycle progression, cell growth, and histone RNA processing.
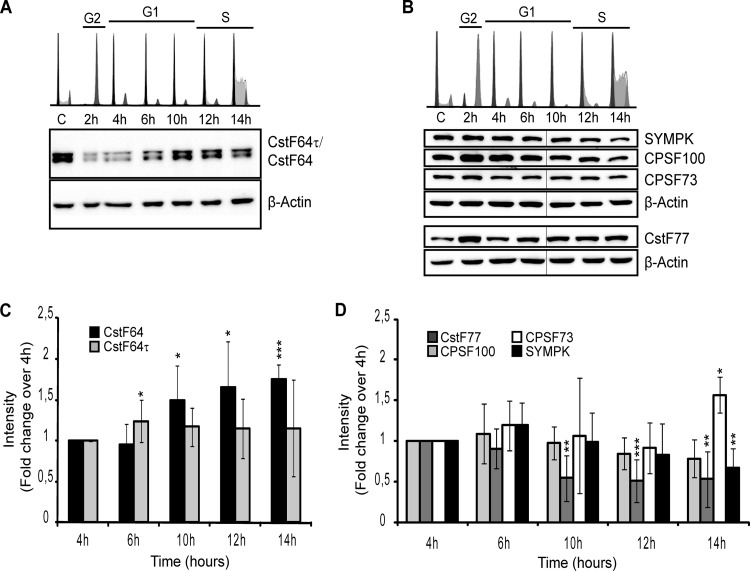
Based on previous indications that CstF64 might be regulated in the cell cycle or in response to the proliferation state of certain cells (29–31) and also based on the presence of CstF64 in the histone processing factor HLF (12), we decided to analyze the levels of CstF64 and CstF64τ as well as those of other proven or potential HLF members during the cell cycle of HeLa cells. For this purpose, HeLa cells were arrested in G2/M by incubation with nocodazole and then released from this block. Cell cycle profiles were analyzed by flow cytometry after DNA staining with propidium iodide. As shown in the top panels of Fig. 1A and B, the cells traversed the G1 phase synchronously and reached the S phase by 12 to 14 h after release. As the cell cycle profiles differed somewhat between individual synchronizations, we chose the 4-h time point as a reference, when the majority of cells were in the early G1 phase in all experiments. The expression of CstF64 and CstF64τ was measured by Western blotting with an antibody recognizing both proteins (Fig. 1A). This suggested that both proteins are upregulated toward the G1/S transition. A quantification of the results from several experiments revealed that this regulation is statistically significant for CstF64 at 10 to 14 h postrelease and amounts to 50 to 75% of the 4-h value (Fig. 1C). In contrast, CstF64τ was only slightly elevated at 6 h (24%) but not at the later time points. Of the other analyzed factors, CstF77 showed no cell cycle regulation (Fig. 1D). Symplekin showed a reduction at 14 h (−33%), and CPSF100 was reduced by 45 to 50% at 10 to 14 h. Finally, CPSF73 showed an elevated level (+57%) at 14 h, but because this protein was measured less often, this result needs to be further investigated. Thus, CstF64 was the only analyzed protein whose expression profile paralleled the upregulation of histone RNA processing.
FIG 1.
Cell cycle regulation of CstF64. HeLa cells were synchronized in the G2/M phase by nocodazole block. After release, samples were collected at the indicated time points for cell cycle analysis by flow cytofluorometry and for Western blotting. (A) Analysis of CstF64 (64 kDa), CstF64τ (70 kDa), and β-actin (45 kDa [loading control]). (B) Analysis of symplekin (SYMPK [150 kDa]), CPSF100 (100 kDa), CPSF73 (73 kDa), and CstF77 (77 kDa). The upper panels in panels A and B show cytofluorometric analyses of the cell populations at the different time points. DNA was stained with propidium iodide (PI). The first sample (labeled “C”) represents asynchronously cycling cells, G2 corresponds to cells 2 h after block release, early G1 is represented by 4- and 6-h samples, the late G1 phase and S-phase transition are shown in 10- and 12-h samples, and the majority of the cells are in the S phase at 14 h after release. The lower panels contain representative Western blots of extracts from synchronized cells. Thin vertical lines indicate where a lane has been omitted from the figure. (C) Western blot quantifications by densitometry analysis using AIDA software reveal a significant upregulation of CstF64 in the late G1 and S phases (samples from 10 to 14 h). CstF64τ shows a less pronounced increase that is only significant at 6 h postrelease. (D) Quantification and statistical analysis of the other detected proteins, which are nearly constant if not downregulated. The only exception is a small but significant upregulation of CPSF73 in the S phase (14 h). Asterisks in panels C and D indicate significance levels for the Welch t test: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. The data were collected from four independent experiments (biological replicates).
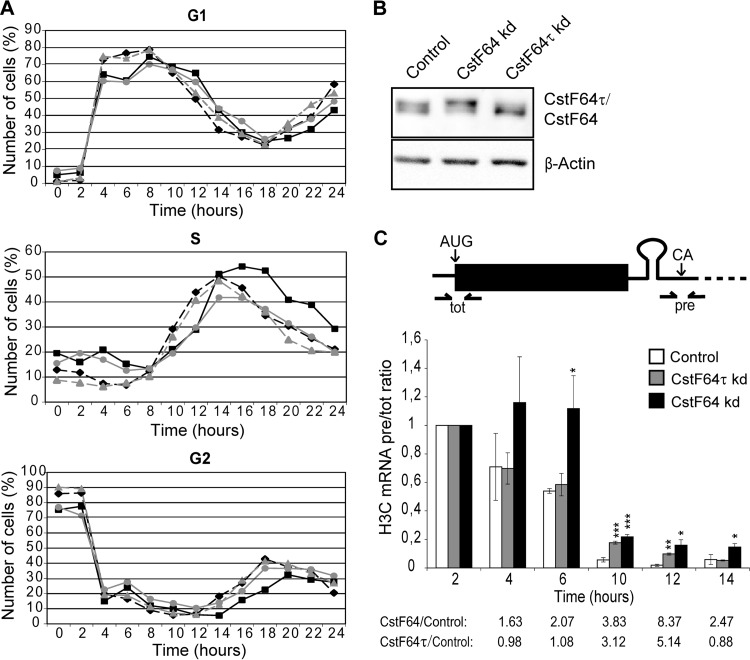
To further analyze the relationship of CstF64 with the cell cycle and with the expression of replication-dependent histone genes, we created HeLa cell lines in which a depletion of either CstF64 or CstF64τ can be induced with doxycycline. With these cells, we performed identical nocodazole synchronization experiments after depletion of either CstF64 or CstF64τ. Note that CstF64 depletion leads to an upregulation of CstF64τ (Fig. 2B), as described previously (20). Importantly, CstF64-depleted cells tended to accumulate in the S phase and showed a delay in progressing though this phase and entering G2 compared with the uninduced control cells (Fig. 2A). This phenomenon was also seen in three additional experiments, two of which are shown in Fig. S1A and B in the supplemental material, even though the actual cell cycle kinetics varied somewhat between different synchronization experiments. In contrast, CstF64τ depletion did not alter the cell cycle progression. Consistent with these findings, cell proliferation was inhibited by prolonged CstF64 depletion (see Fig. S1C).
FIG 2.
Effects of CstF64 depletion on S-phase progression and histone RNA processing. HeLa cell lines carrying doxycycline-inducible shRNA expression cassettes for the depletion of either CstF64 or CstF64τ were generated by lentiviral transduction. After induction of shRNA expression by doxycycline, the cells were arrested in G2/M by nocodazole treatment. (A) After release, the cell cycle progression was monitored by flow cytometry (PI DNA staining) and compared with identically synchronized doxycycline-free cells. The three panels from top to bottom show the subpopulations of G1-, S-, and G2-phase cells. Shown in gray are cells containing shCstF64τ without (triangles) or with (circles) doxycycline induction. Cells containing shCstF64 without (diamonds) or with (squares) doxycycline induction are shown in black. CstF64 depletion appears to delay passage through the S phase, resulting in an accumulation of cells in the S phase and delayed entry into the G2 phase. (B) Western blot showing the efficiency of depletion. The upper panel shows CstF64 and CstF64τ revealed by the H300 antibody, which recognizes both proteins. The lower panel shows β-actin (the loading control). kd, knockdown. (C) Determination of apparent in vivo processing of histone H3C mRNA. (Top) Scheme showing the positions of qPCR primer/probe sets that were used to measure total (tot) and precursor (pre) RNAs. (Bottom) Quantifications of H3C pre-RNA/total RNA ratios for normal, nondepleted cells (control) as well as Cstf64τ-or CstF64-depleted (kd) cells. The pre-RNA/total RNA ratios have been set as 1 for each cell line for cells at 2 h postrelease (which express very little histone RNA) and are shown as multiples of these values for the other time points. Error bars represent standard deviations from three independent experiments (biological replicates). Asterisks indicate significance levels for the Welch t test as described in the legend to Fig. 1. Unless labeled explicitly, the variations are not significant. Fold changes of H3C pre-RNA/total RNA ratios between CstF64-depleted or Cstf64τ-depleted cells and the controls are listed below to illustrate the precursor accumulation during the G1/S-phase transition.
These results indicated that the increment in CstF64 expression that occurs as the cells approach the S phase is connected to phase-specific events as is replication-dependent histone RNA processing. Therefore, we measured the apparent in vivo histone RNA processing efficiency for histone 3C (H3C) (13). Briefly, the levels of H3C mRNA precursors (i.e., uncleaved) and of the total mRNA (i.e., uncleaved plus correctly cleaved) were measured by reverse transcription-quantitative PCR (RT-qPCR) (Fig. 2C, top). The processing efficiency was then calculated as the molar ratio of uncleaved to total H3C mRNA. We call this “apparent in vivo processing efficiency” because the levels of the precursor and total RNAs are also affected by turnover, which is likely to differ between the two RNA species. As expected from our previous data (20), histone RNA processing was impaired after CstF64 depletion. Importantly, however, this effect was relatively more pronounced during the G1/S transition and S phase than in other periods of the cell cycle (Fig. 2C). This effect of CstF64 depletion on histone RNA processing is most likely not due to an indirect effect of blocking CPA, because the upregulated CstF64τ functions well in CPA (20) and global polyadenylation is not significantly affected by a CstF64 depletion (32).
Interestingly, different from asynchronously cycling cells (20), CstF64τ depletion also inhibited the apparent histone RNA processing activity at the 10- and 12-h time points. Whether this indicates a direct role of CstF64τ in histone RNA processing is unclear because, despite its upregulation in CstF64-depleted cells, CstF64τ cannot compensate for the loss of histone RNA processing activity caused by CstF64 depletion.
Taken together, these data therefore indicate that CstF64 is a cell-cycle-regulated member of the histone processing complex (more specifically of the HLF) and that it plays an important role in replication-dependent histone RNA processing.
CstF64 needs to interact with symplekin to fulfill its role in histone RNA processing.
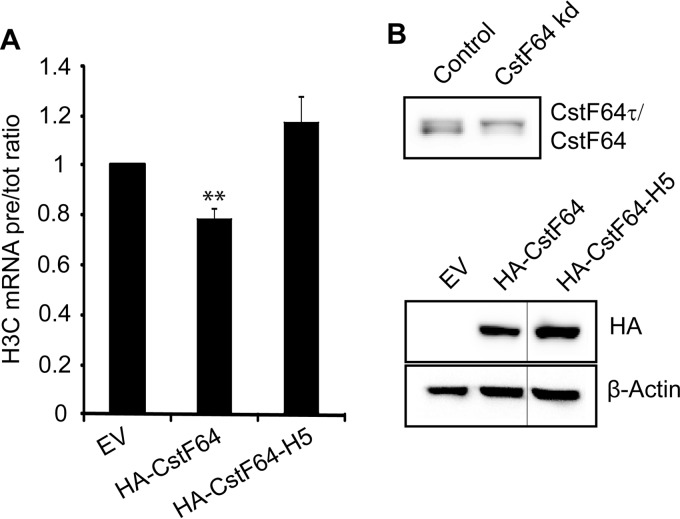
Symplekin has been identified as the heat-labile member of the HLF and has been proposed to act as an assembly platform for the other HLF components (12). Moreover, symplekin binds CstF64 on its hinge domain, which consists of five α-helices (33). This hinge domain of CstF64 mediates mutually exclusive interactions with CstF77 (via helices 1 to 4) and symplekin (via helices 2 to 5) (20). The interaction with CstF77 is essential for the nuclear localization of CstF64. Importantly, we previously showed that a mutant form of symplekin, unable to interact with CstF64, is significantly impaired in its ability to function in histone RNA processing (20). We therefore hypothesized that CstF64 might in turn need to interact with symplekin in order to fulfill its function in histone RNA processing. To address this question, we induced CstF64 depletion in HeLa cells, and then we reintroduced by transient transfection HA-tagged forms of either normal CstF64 or of a helix 5 mutant (HA-CstF64-H5) that is unable to interact with symplekin but still interacts with CstF77 (20). We then performed apparent in vivo processing assays to measure the complementation effect. Full-length HA-CstF64 was able to partly complement the histone processing defect induced by CstF64 depletion (Fig. 3A). Within 24 h posttransfection, it reduced the accumulation of histone precursor transcripts by ∼20%, compared to an empty vector-transfected sample. As CstF64 depletion usually causes a 1.5- to 2-fold increase in the H3C precursor/total RNA ratio in asynchronously dividing cells (20), the reexpression of HA-CstF64 was able to counteract about half of the depletion effect. In contrast, the CstF64-H5 mutant did not rescue the phenotype but rather seemed to worsen it. This indicated that CstF64 must be able to interact with symplekin in order to carry out its function in histone RNA processing.
FIG 3.
Exogenous HA-CstF64 needs to interact with symplekin to complement a histone RNA processing defect caused by CstF64 depletion. CstF64-depleted cells (after 4 days of doxycycline induction) were transiently transfected with HA-tagged cDNAs for either wild-type CstF64 (HA-CstF64) or a CstF64 mutant unable to interact with symplekin (HA-CstF64-H5) (20) or with an empty vector (EV) as a control. Total RNA was isolated 24 h after transfection, and histone H3C precursor (pre-RNA) and total mRNA levels were quantitated by RT-qPCR. (A) H3C pre-RNA/total RNA ratios. HA-CstF64 reduces the precursor accumulation by ∼20% (P = 0.0089, Welch t test). In contrast, the mutant HA-CstF64-H5, unable to bind symplekin, fails to rescue the defect or even aggravates it (P = 0.0962). The error bars represent standard deviations from three biological replicates. (B) Western blots showing the efficiency of CstF64 depletion before transfection (upper panel) and the expression of tagged wild-type and mutant forms of CstF64 24 h posttransfection (lower panel). β-Actin was probed as a loading control. Thin vertical lines indicate where a lane has been omitted from the figure.
CstF64 directly interacts with FLASH and/or Lsm11.
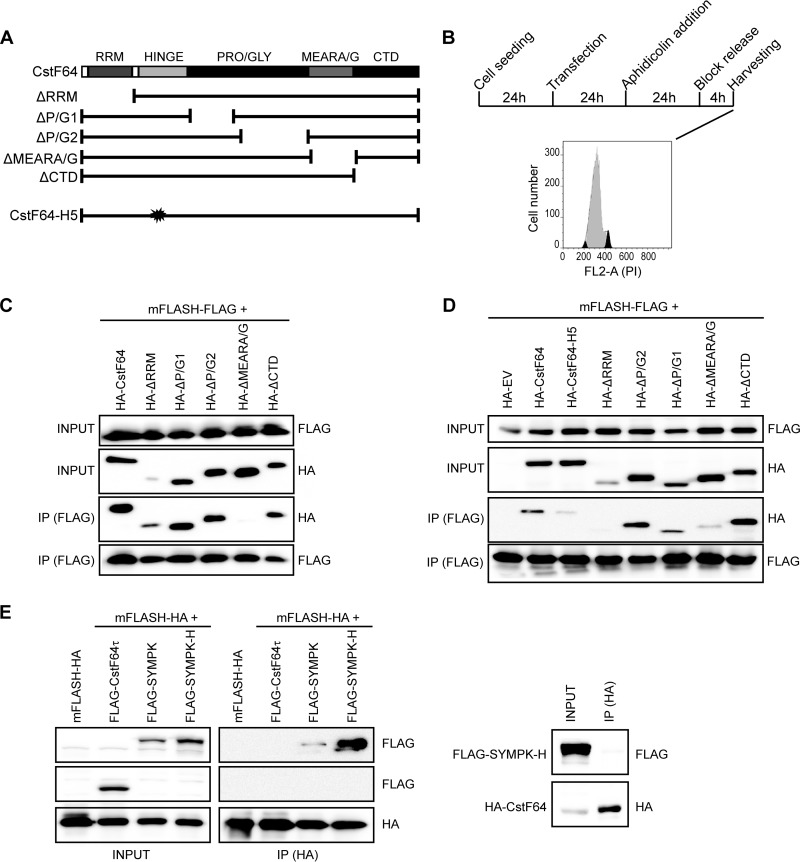
It has recently been suggested that the interaction between FLASH and the amino terminus of Lsm11 is required to form a platform onto which the HLF can bind. Importantly, symplekin and CstF64 were identified as the strongest interactors with FLASH and/or Lsm11 (10). Having shown that CstF64 is cell cycle regulated, we hypothesized that its increased availability during S-phase entry could be required and limiting for the formation or stability of the HLF and/or its recruitment onto histone pre-mRNA. To analyze if CstF64 might indeed help to recruit the HLF to RNA-resident proteins, we aimed to determine whether it directly binds to FLASH or to a combination of FLASH and Lsm11, and which of its domains might be involved. Based on the structure of human CstF64 and literature data, we defined several macrodomains, as depicted in Fig. 4A. We then created a series of HA-tagged CstF64 deletion mutants: ΔRRM lacks the RNA recognition motif (amino acids [aa] 1 to 105); ΔP/G1 and ΔP/G2 have, respectively, lost the first and second halves of a long, unstructured proline/glycine-rich domain. (ΔP/G1 lacks aa 202 to 318, and ΔP/G2 lacks aa 320 to 405.) ΔMEARA/G is missing the 12 repeats of the MEARA/G domain (aa 404 to 519). Finally, ΔCTD lacks the carboxy-terminal domain (aa 509 to 577) (Fig. 4A). Because the endogenous human FLASH protein consists of almost 2,000 aa, we created a FLAG-tagged version of a minimal FLASH domain of 314 amino acids (mFLASH-FLAG) that had previously been shown to be fully functional in histone RNA 3′ end processing (8, 34). Additionally, we produced a FLAG-tagged fusion between mFLASH and the N terminus of Lsm11 (aa 1 to 153; mFLASH:Lsm11-FLAG). First, we showed by immunofluorescence that all recombinant proteins accumulate in the cell nucleus (see Fig. S3A in the supplemental material). Moreover, the ability of endogenous CstF77 to associate with all CstF64 mutants was demonstrated by coimmunoprecipitation (see Fig. S3B).
FIG 4.
CstF64 directly interacts with a minimal FLASH domain. (A) Schematic representation of CstF64 mutants. HA-tagged CstF64 mutants were created in which individual structural domains of human CstF64 were deleted. Additionally, we used a mutant with a multiple-point mutation in helix 5 of the hinge domain (CstF64-H5) that is unable to interact with symplekin (20) or an empty vector (EV) control. As a binding partner, we used a FLAG-tagged version of mFLASH (i.e., a minimal 314-aa domain of FLASH that is functional in histone RNA processing) (8, 34). (B) Outline of the experimental strategy. HEK293T cells were transfected either with individual expression constructs (for the experiment shown in panel C) or in pairwise combinations. The transfected cells were synchronized in the S phase with aphidicolin and harvested 4 h after release of the block. The cytofluorometric profile shows the extent of synchronization at the time of harvesting. (C) Association of mFLASH-FLAG with wild-type or mutant CstF64 in vitro. The recombinant proteins expressed in S-phase-synchronized HEK293T cells were purified by antitag immunoprecipitation under stringent conditions. They were then incubated in couples to assay their interaction potential by immunoprecipitation with FLAG-coated magnetic beads. Western blots from input samples (10% of the total) were probed with anti-HA and anti-FLAG antibodies to show the presence of the various proteins. The immunoprecipitated (IP) material was analyzed in the same way. The mutant HA-CstF64 lacking the MEARA/G domain loses the ability to interact with mFLASH-FLAG. (D) Association of mFLASH-FLAG with wild-type or mutant CstF64 in vivo. Lysates from cells transfected with pairwise combinations of expression constructs were subjected to immunoprecipitation with FLAG-coated magnetic beads. Otherwise, the analyses of input and immunoprecipitates (IP) were performed as in panel C. (E) Interaction of FLAG-CstF64τ and wild-type or mutant FLAG-symplekin with HA-tagged mFLASH. Immunoprecipitations from lysates of pairwise cotransfections were performed with HA-coated magnetic beads, and Western blots were probed for the presence of FLAG-tagged symplekin or CstF64τ. The panels on the right show that the symplekin hydro mutant (H) is unable to bind CstF64, as published (20). Nevertheless, it still associates with HA-mFLASH (left panels). The lack of interaction of FLAG-CstF64τ with HA-tagged mFLASH is also shown.
Because direct interaction studies with recombinant proteins produced in bacteria or by coupled in vitro transcription/translation had failed (unpublished results and reference 10), and in vivo coimmunoprecipitation (co-IP) experiments only revealed interactions between CstF64 and FLASH when performed with extracts from S-phase cells (described below), we decided to purify the recombinant proteins from S-phase-synchronized HEK293T cells. Briefly, constructs encoding mFLASH-FLAG, HA-tagged CstF64, or HA-CstF64 mutants were transfected separately into HEK293T cells. The transfected cells were synchronized by aphidicolin block and collected 4 h after release, when the majority of the population was in the S phase (Fig. 4B). The recombinant proteins were purified from whole-cell protein extracts by stringent immunoprecipitation with antibodies against the respective tag, and their amounts and purity were assessed by SDS-PAGE and Coomassie blue staining (see Fig. S2 in the supplemental material).
When these recombinant proteins were used in coimmunoprecipitation experiments, the full-length CstF64 was efficiently precipitated with mFLASH-FLAG, as were the mutants lacking the RRM, P/G1, P/G2, or CTD (Fig. 4C). Importantly, however, the CstF64 mutant lacking the MEARA/G region had a very strongly reduced binding ability. These results were confirmed in multiple in vivo co-IP experiments. For these, the two interaction partners to be tested were cotransfected into HEK293T cells, the cells were similarly synchronized in the S phase, and co-IP experiments were performed with whole-cell protein extracts. As shown in Fig. 4D, wild-type HA-CstF64 and the mutants lacking P/G2 or the CTD could be coprecipitated with mFLASH-FLAG also after in vivo coexpression. The RRM and P/G1 mutants were expressed at lower levels but were nevertheless coprecipitable with mFLASH-FLAG. Again, the MEARA/G deletion mutant showed a strongly reduced interaction. An inverse IP experiment yielded similar results (see Fig. S4A in the supplemental material). Upon precipitation of the HA-tagged ΔMEARA/G mutant, only small amounts of mFLASH-FLAG were coprecipitated, whereas for all other CstF64 mutants, the amount of coprecipitated mFLASH-FLAG was roughly proportional to the amount of precipitated CstF64.
A number of further control experiments validated these findings. We repeated the same experiments with the fusion protein mFLASH:Lsm11-FLAG (see Fig. S4B in the supplemental material) and for both mFLASH-FLAG and mFLASH:Lsm11-FLAG in the presence of 200 μg/ml RNase A (see Fig. S4C). These two controls indicated that these interactions are actually protein mediated. Thus, in conclusion, the CstF64 variant lacking the MEARA/G region exhibited the strongest deficiency in binding to either mFLASH or mFLASH:Lsm11_NTerm in all experiments performed and under all conditions.
In these in vivo co-IP experiments, we also tested the CstF64-H5 (hinge domain helix 5) mutant, which is impaired in its ability to interact with symplekin (20). As expected, this mutant was also significantly impaired in its interactions with mFLASH-FLAG (Fig. 4D) but in this case most likely because of its reduced binding to symplekin. Therefore, we wondered whether symplekin can autonomously bind to FLASH/Lsm11 or whether its recruitment is entirely mediated by CstF64. We thus performed a similar immunoprecipitation from cells that had been cotransfected with HA-tagged mFLASH and FLAG-symplekin (either the wild-type protein or the hydro mutant unable to bind CstF64 [SYMPK-H]). This mutant is altered in four hydrophobic residues of an amphipathic α-helix (residues 442 to 460) in symplekin that mediates the interaction with CstF65 (20). As shown in Fig. 4E, wild-type symplekin and the hydro mutant were both able to bind mFLASH-HA. However, we could confirm that the symplekin hydro mutant cannot be coprecipitated with HA-CstF64 (Fig. 4E, right panel). Finally, we also performed a similar coprecipitation experiment with mFLASH-HA and FLAG-tagged CstF64τ. In this case, the analysis by Western blotting revealed no coimmunoprecipitation (Fig. 4E), indicating that CstF64τ cannot bind to FLASH.
In additional control experiments, we tested all CstF64 mutants for their ability to bind symplekin. As expected, the CstF64-H5 mutant was completely deficient in binding to symplekin (see Fig. S5A in the supplemental material). However, except for a possible slight reduction in the case of the ΔP/G1 mutant, all other CstF64 mutants retained the ability to interact with symplekin (see Fig. S5B). This further supported the view that the observed effect of the ΔMEARA/G mutant may reflect the loss of a direct interaction and is not indirectly caused by an impaired interaction with symplekin.
In summary, the experiments described above demonstrate that CstF64 can directly bind to FLASH and/or Lsm11 and that the MEARA/G region plays an important role in this binding. In contrast, CstF64τ appears to be unable to interact with FLASH, which may explain its lack of activity in histone RNA 3′ end processing. However, our experiments also show that symplekin can bind FLASH independently.
Read-through transcripts of replication-dependent histone genes are predominantly polyadenylated and accumulate in the nucleus.
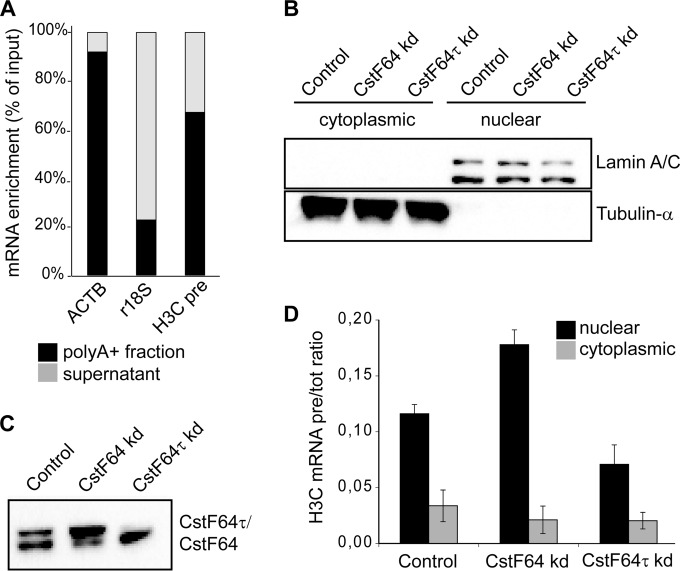
Having shown that unprocessed replication-dependent histone pre-mRNAs exist in HeLa cells throughout the G1 phase and accumulate upon CstF64 depletion (Fig. 2E), we wanted to characterize their nature, metabolism, and potential function. Several reports suggested that upon misprocessing, replication-dependent histone transcripts acquire a poly(A) tail (22–25). Indeed, we found by in silico analysis that several members of the human replication-dependent histone genes contain cryptic polyadenylation signals downstream of the canonical cleavage sites. To identify if and to what extent the unprocessed read-through transcripts are polyadenylated, we selectively purified poly(A)+ cellular RNAs from exponentially growing HeLa cells with oligo(dT)-conjugated magnetic beads and analyzed the enriched RNAs by RT-qPCR. While ∼91% of β-actin transcripts bound to the poly(A) beads, only ∼23% of 18S rRNA was enriched by poly(A) selection (Fig. 5A). In comparison with these positive and negative controls, ∼67% of the replication-dependent histone H3C read-through transcripts were found in the selected material, indicating that a large proportion of the histone precursor transcripts are polyadenylated.
FIG 5.
Polyadenylated histone read-through transcripts accumulate in the nucleus upon CstF64 depletion. (A) The majority of histone read-through transcripts are polyadenylated. Polyadenylated RNAs were selected from total HeLa cell RNA by using oligo(dT)-conjugated magnetic beads. The eluted [poly(A)+] and unbound supernatant fractions were retrotranscribed with random hexamers and subjected to qPCR analysis for the presence of β-actin mRNA (ACTB [positive control]), 18S rRNA (r18S [negative control]), and H3C precursors (H3C pre). (B to D) Untreated HeLa cells as well as CstF64- and CstF64τ-depleted cells were fractionated, and cytoplasmic and nuclear RNAs were extracted. (B) Protein samples from the fractionations were probed with antibodies detecting lamin A/C and α-tubulin as markers of nuclear and cytoplasmic compartments, respectively. kd, knockdown. (C) Nuclear extracts were probed with an antibody recognizing both CstF64 and CstF64τ to reveal the extent of depletion. (D) The levels of unprocessed H3C precursors and total H3C RNA were measured by RT-qPCR experiments. H3C pre-RNA/total RNA ratios were calculated for the nuclear and cytoplasmic fractions, in either the unmodified (Control) or CstF64 or CstF64τ depletion background. The error bars represent standard deviations from three independent experiments (biological replicates).
To further elucidate the metabolism and possible function of these transcripts, we investigated their intracellular localization by nuclear/cytoplasmic fractionation (Fig. 5B) followed by RT-qPCR detection of precursor and total H3C RNA. We performed this experiment in control and in CstF64- or CstF64τ-depleted HeLa cells. As shown in Fig. 5D, the precursor RNA (pre-RNA) transcripts accumulated mostly in the nuclear compartment. Nevertheless, a small fraction was present in the cytoplasm. Based on equal cell equivalents, it appeared that 75 to 80% of U2 snRNA and 80 to 90% of histone read-through transcripts are nuclear under our fractionation conditions. Interestingly, upon CstF64 knockdown (kd), the nuclear accumulation of precursors was increased, whereas CstF64τ depletion led to the opposite result (Fig. 5D). Due to the coregulation of the two paralogues (20), the depletion of CstF64τ results in an overexpression of CstF64 (Fig. 5C), which may explain this effect, as CstF64 but not CstF64τ is active in histone RNA processing.
Nuclear histone read-through transcripts are targets of the exosome machinery.
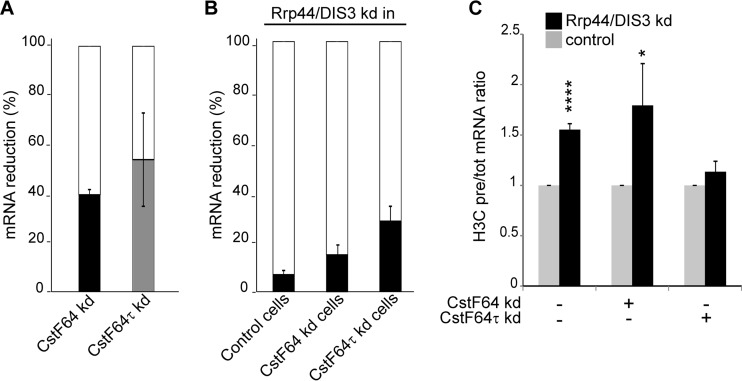
Because most histone precursor transcripts were located in the nucleus (Fig. 5D) and the cell cycle regulation of histone transcripts in budding yeast had been shown to depend in part on the TRAMP-exosome pathway (35), we investigated whether the nuclear histone read-through transcripts are targets of the exosome machinery also in human cells. For this purpose, we created an shRNA construct targeting DIS3, the exonuclease of the human nuclear exosome, and used a scrambled-sequence shRNA as control. The depletions were performed in CstF64-, CstF64τ-depleted, or control HeLa cell backgrounds. The extent of CstF64 or CstF64τ depletion is shown in Fig. 6A, and the extent of Rrp44/DIS3 depletion in each of the three cell populations is shown in Fig. 6B. The RNA from these cells was analyzed for the H3C pre-RNA/total RNA ratio. Consistent with our hypothesis, the unprocessed histone precursor transcripts accumulated upon DIS3 depletion, suggesting that they are physiologically controlled by the nuclear exosome (Fig. 6C). In fact, the fraction of precursor transcripts that were restored by Rrp44/DIS3 depletion was higher in the CstF64-depleted cells than in the other two cell populations, confirming that CstF64 depletion affects histone RNA processing.
FIG 6.
Misprocessed histone transcripts are exosome targets. HeLa cells were depleted of active nuclear exosome by transfection of a DIS3/Rrp44-specific shRNA expression plasmid on the background of normal (control), CstF64- or CstF64τ-depleted cells. A plasmid with a scrambled sequence shRNA was used as control. The cells were selected for the presence of the plasmids by incubation with puromycin for 3 days and harvested. The various depletion efficiencies as well as histone H3C pre-RNA and total RNA levels were analyzed by RT-qPCR. (A) Extent of CstF64 and CstF64τ depletions. kd, knockdown. (B) Extent of DIS3/Rrp44 depletions in control (not depleted for a CstF64 paralogue) as well as CstF64- or CstF64τ-depleted cells. (C) Accumulation of H3C precursors over total transcripts upon impairment of the exosome (black bars). The values in cells not depleted of DIS3/Rrp44 were set as 1 (gray bars) for each tested condition. The error bars in the graphs represent the standard deviations from three independent experiments (biological replicates). Asterisks indicate significance levels for the Welch t test: *, P < 0.05; and ****, P < 0.0001.
Cytoplasmic histone read-through transcripts can associate with polysomes.
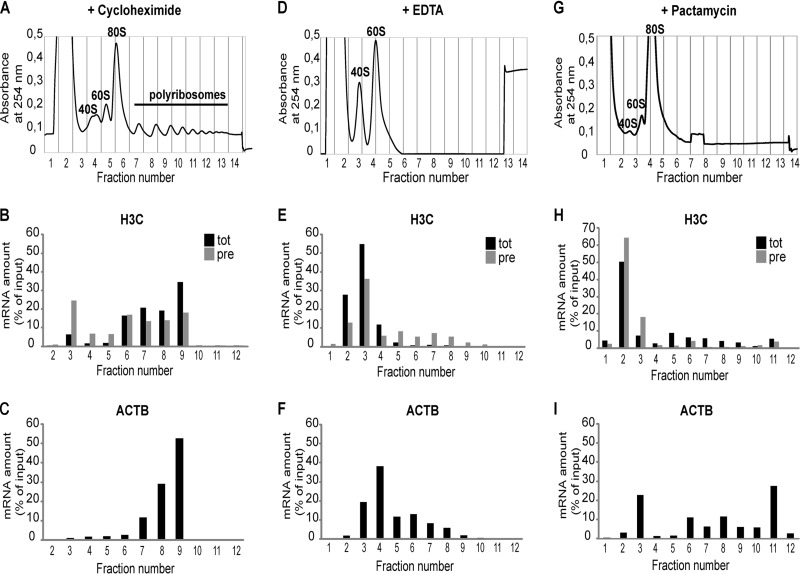
As a large proportion of the histone read-through transcripts appeared to be polyadenylated (Fig. 5A), we asked whether the small fraction of these transcripts that are present in the cytoplasm may associate with polysomes and get translated. Thus, cytoplasmic extracts of HeLa cells were subjected to polysome profiling on 15 to 45% sucrose gradients (Fig. 7A), and the distribution of total and precursor H3C transcripts in the gradient fractions was analyzed by RT-qPCR. As control, we also profiled the polysome association of β-actin mRNA. As the precursor transcripts are present in a very small amount, we plotted the data as a percentage of the input (defined as total signal detected over the entire gradient) for each type of RNA. As expected, β-actin mRNA was found mainly in polysome fractions (Fig. 7C). Total histone RNAs were also mostly present in polysome fractions (Fig. 7B). Importantly, however, ∼2/3 of histone H3C precursors were found in the polysome fractions as well (Fig. 7B, fractions 6 to 10). In Fig. S6 in the supplemental material, we show that this polysome association could be observed in both the G1 and S phases of the cell cycle. Addition of EDTA to the gradients, which dissociates ribosome particles (Fig. 7D), completely shifted the total histone H3C and β-actin RNAs but importantly also the histone H3C pre-RNAs into the free RNA fractions (Fig. 7E and F). Similarly, pactamycin, which prevents polysome formation by blocking translocation (36), led to a shift of all three investigated RNAs into the light fractions of the gradient (Fig. 7H and I). This release was incomplete for β-actin mRNA, which may have been due to some ribosome contaminations of fraction 11 or to alternative complexes containing β-actin mRNA. However, taken together, these experiments indicate that the cytoplasmic histone read-through transcripts are indeed associated with polysomes.
FIG 7.
Polyadenylated histone read-through transcripts associate with polysomes. HeLa cells were treated with cycloheximide to preserve polysomes, and cytoplasmic extracts were subjected to centrifugation on linear 15 to 45% (wt/wt) sucrose gradients. In panels D, E, and F, the gradients contained EDTA to dissociate polysomes. In panels G, H, and I, polysomes were dissociated with pactamycin. Note that pactamycin treatment leads to an accumulation of 80S particles. Moreover, in this experiment, fraction 11 still contained some ribosomal material. This may partly explain the incomplete shift observed for β-actin mRNA. (A, D, and G) Absorption profiles (A254) of the gradient elutions. Fractions 4 and 5 in panel A, 3 to 5 in panel D, and 3 and 4 in panel G contain ribosomal subunits; free RNA is present in earlier fractions and ribosomes and polysomes in later fractions. RNA was extracted from the obtained fractions, and H3C precursor RNA (pre) and total (tot) RNA, as well as β-actin (ACTB) mRNA levels (as a control), were measured by RT-qPCR and plotted as percentage of the calculated input (sum of all fractions). (B, E, and H) Distribution of H3C total (tot) and precursor/read-through (pre) RNAs. (C, F, and I) Distribution of β-actin mRNA. Unprocessed β-actin pre-mRNAs were present in such small amounts that a meaningful distribution could not be determined.
DISCUSSION
Histone production is highly regulated in space and time. As the cell approaches the S phase, histone locus bodies (HLBs) form on the clusters of replication-dependent histone genes. HLBs are devoted to histone gene transcription and RNA processing and contain important factors, such as NPAT (a histone-specific transcription factor), Ars2, and FLASH (37). Moreover, during the G1/S-phase transition, histone gene transcription increases ∼5-fold and 3′ end processing becomes ∼8 times more efficient (5).
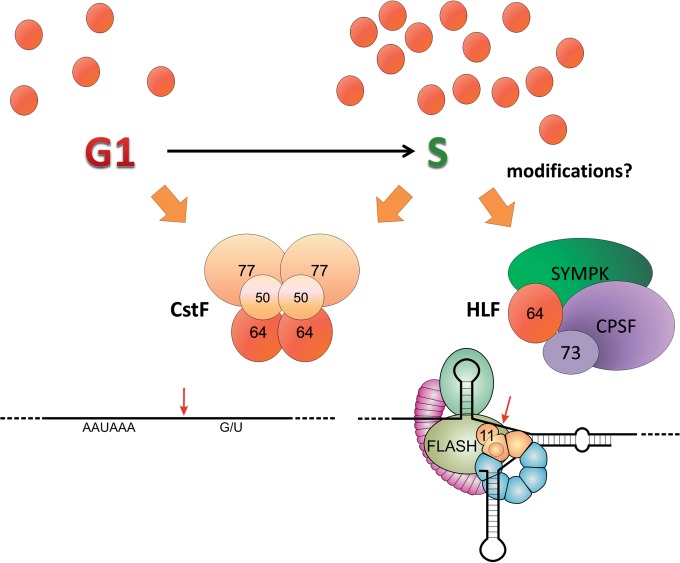
Regarding histone RNA 3′ end processing, two components of the processing machinery have been described to be cell cycle regulated: SLBP (15) and the HLF (14, 38). HLF is known to contain symplekin, CstF64, as well as the entire CPSF complex, including the processing endonuclease CPSF73; evidence concerning the additional presence of CstF77 is controversial (10, 12). However, the mechanisms by which HLF is regulated during the cell cycle and by which its components assemble in a composition distinct from a cleavage/polyadenylation complex have not been elucidated. Furthermore, it is not fully understood how HLF gets recruited to the histone pre-mRNA, even though two of the specific factors interacting with the pre-mRNA (i.e., the U7 snRNP component Lsm11 and FLASH) appear to provide a binding platform for the HLF (10). Our work indicates that CstF64 plays an important role in these processes. The main conclusions, which will be discussed below, are schematically illustrated in Fig. 8.
FIG 8.
Schematic illustration of CstF64's dual roles in cleavage and polyadenylation and histone RNA processing. CstF64 (red brown) can act in cleavage and polyadenylation (CPA) or histone RNA 3′ end processing as a component of, respectively, the hexameric CstF complex or the heat-labile factor (HLF) that additionally contains symplekin (SYMPK) and CPSF (of which the endonuclease CPSF73 is shown as a separate unit). The formation of these alternative complexes depends at least partly on mutually exclusive interactions of the CstF64 hinge domain with CstF77 or SYMPK (20, 33). Low CstF64 levels in the G1 phase may favor CstF formation and hence CPA. During the G1/S transition, increasing CstF64 levels and possibly posttranslational modifications may allow increased HLF formation. Tethering of HLF to the factors bound to a histone pre-mRNA processing site are mediated by symplekin and CstF64, which can both interact with a binding platform provided by FLASH (olive green) and the N terminus of the U7 snRNP component Lsm11 (sand brown). Binding of CstF64 to FLASH depends on the MEARA/G domain, as shown in Fig. 4, and possibly also on posttranslational modifications. Other histone pre-mRNA bound factors shown are stem-loop- or hairpin-binding protein (sea green) and 100-kDa zinc finger protein (violet). Further components of the CPA complex have been omitted for simplicity.
A first conclusion is that CstF64 is regulated not only during a transition from a resting to a proliferative state (29–31) but also during the cell cycle (Fig. 1). Its accumulation during the G1/S-phase transition temporarily mirrors the formation of HLBs (37) and the upregulation of replication-dependent histone mRNA transcription and processing (5). Moreover, CstF64 is important for correct histone RNA processing, since its depletion affects the normal cleavage reaction, so that read-through transcripts accumulate (Fig. 2). Its depletion also affects cell cycle progression, leading to a delayed S phase and inhibition of cell proliferation. These effects are unlikely to be caused by deficient polyadenylation, based on two main arguments. First, CstF64τ gets upregulated upon CstF64 depletion (Fig. 2B) and functions well in CPA (20). Second, a published global transcriptome sequencing (RNA-seq) study did not reveal major CPA changes after CstF64 depletion (32). It rather appears that the upregulation of CstF64 is important for the formation or activation of the HLF during the G1/S transition. In contrast, the other HLF members, as far as they have been analyzed here, do not show a similar upregulation during G1/S transition (Fig. 1D). While the manuscript for this article was under review, the MacDonald group published related findings from mouse embryonic stem cells. When depleted of CstF64, these cells displayed slower growth, loss of pluripotency, and a lengthened G1 phase, correlating with an increase in polyadenylated histone RNAs (39).
A stable interaction between CstF64 and symplekin is very important for histone RNA processing. In fact, both the symplekin “hydro” mutant, unable to interact with CstF64 (20), and the CstF64 “hinge helix 5” mutant, unable to interact with symplekin (Fig. 3), are inefficient in histone RNA processing. Considering that the binding of CstF64 to symplekin and the binding of CstF64 to CstF77 are mutually exclusive (20, 33), it is tempting to speculate that these interactions may dictate in which type of processing reaction the resulting complex gets used (Fig. 8). In G1, a relatively limited amount of CstF64 might be efficiently recruited by CstF77, and this might in turn determine its preferential inclusion in the CPA complex. Inversely, elevated concentrations of CstF64 toward the S phase might kinetically favor the assembly of active HLF through an increased formation of CstF64-symplekin interactions. Another possibility that is not mutually exclusive with the one described above is that posttranslational modifications of CstF64 regulate its differential association with CstF77 or symplekin. That such modifications may occur is supported by the fact that we could only demonstrate and assay CstF64-FLASH interactions with recombinant proteins from S-phase-synchronized cells. Thus, it is quite likely that CstF64 may be the crucial element required to activate HLF either from a precursor composed of symplekin and CPSF or from the individual components.
Additionally, we have shown that besides an indirect binding to FLASH or Lsm11 via symplekin (effect of CstF64 “helix 5” mutant), CstF64 also interacts with FLASH and/or Lsm11 independently of symplekin (Fig. 4). This interaction is strongly affected by deletion of the poorly characterized MEARA/G domain of CstF64. As indicated by circular dichroism spectroscopy and differential scanning calorimetry, this domain appears to adopt a helical conformation (40). Interestingly, a major structural difference between CstF64 and CstF64τ that is unable to bind to FLASH (Fig. 4E) and does not contribute to histone RNA processing (Fig. 2C) resides in this MEARA/G repeat domain. The dissimilarity of this domain in CstF64τ may in part explain the differential functionality of the two paralogues in histone RNA processing. Another reason may be that CstF64τ also appears to interact less efficiently with symplekin than CstF64 (41). Most importantly, however, our results indicate that CstF64 contributes to tethering the HLF to histone pre-mRNA by interactions that are independent of symplekin and that are also not dependent on RNA, as they can be observed in RNase-treated samples (see Fig. S4C in the supplemental material).
In our in vivo binding assays, the ΔP/G1 mutant also seemed to be partly deficient in interaction with mFLASH or mFLASH:Lsm11 (Fig. 4; see Fig. S2D in the supplemental material). This may have been due to its somewhat reduced binding to symplekin (see Fig. S3 in the supplemental material). This explanation would be in line with a recent report suggesting that the proline/glycine-rich domain is involved in strengthening the interaction with symplekin (41).
In exponentially growing cells, read-through transcripts that represent either histone mRNA precursors or molecules that have escaped the canonical processing pathway, are present in extremely small amounts; however, they accumulate to a certain extent in G1-arrested cells (14). Here we have confirmed these findings and extended them to synchronized HeLa cells (Fig. 2C). Importantly, we could also show a higher accumulation of these read-through transcripts after depletion of CstF64 but not CstF64τ. In fact, upon CstF64τ depletion, less read-through, poly(A)+ histone transcripts accumulated (Fig. 5D). This might simply reflect the fact that CstF64τ depletion results in increased CstF64 levels (20). Another possible explanation is that CstF64τ plays a direct role in polyadenylating histone read-through transcripts.
A similar accumulation of read-through transcripts has previously been described after the depletion of several other factors important for histone RNA processing (22–25). In agreement with these previous studies, we could show that the majority of these read-through transcripts are polyadenylated (Fig. 5). Importantly, our fractionation studies showed that these read-through transcripts are mostly present in the nuclear compartment (Fig. 5), where they are targeted for degradation by the exosome complex (Fig. 6). As many human histone genes contain cryptic polyadenylation sites downstream of the canonical cleavage site, this might represent a safety mechanism to avoid read-through into neighboring genes in the tightly packed histone gene clusters, as previously suggested (22–25). In case of misprocessing during pervasive transcription in the S phase, read-through might otherwise result in fusion transcripts that could be deleterious, especially if they would be exported to the cytoplasm and translated. Polyadenylation of histone read-through transcripts might therefore be a signal to trigger their degradation. In this context, it is interesting that in budding yeast, the TRAMP-exosome complex regulates histone metabolism (35). Moreover, it has recently been shown that polyadenylation induces exosome-mediated decay of promoter upstream transcripts (PROMPTs) to control promoter directionality (42, 43). The same authors revealed that exosome components are associated with the 3′ ends of replication-dependent histone genes (42).
Here we also showed that a small fraction of the histone read-through transcripts are present in the cytoplasm (Fig. 5). Moreover, these transcripts are potentially capable of being translated, as they can associate with polysomes (Fig. 7). This might allow cells to replace decaying histones outside the S phase or during quiescence and might be particularly important for those histones for which there are no replication-independent gene variants. In this context, a recent study showed that a subset of histone H2B-encoding polyadenylated transcripts can associate with polysomes under certain cellular stress conditions, suggesting that this pathway may be regulated (44).
Finally, one can speculate that a low level of transcription over the entire cell cycle may help to keep the histone loci in a euchromatic state, in order to allow a rapid transcriptional response at the onset of the S phase of every replication cycle. A low level of read-through transcripts might thus represent a trade-off to allow such a quick response. Alternatively, the histone read-through transcripts might also play a hitherto unsuspected regulatory role.
Most importantly, however, our study indicates that CstF64 is an important cell-cycle-regulated member of the HLF and that it determines the formation of the active HLF and contributes to histone RNA 3′ end processing by helping to tether the HLF and its catalytic endonuclease CPSF73 to the factors already bound to histone pre-mRNA—i.e., FLASH and the U7 component Lsm11.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Judith Trüb for excellent technical support, Gian-Reto Lohrer for performing statistical analyses, and Marc Ruepp and Pamela Nicholson (all associated with the University of Bern) for various materials and discussions. Moreover, we thank Silvia Barabino (University of Milano Bicocca) and Marc Ruepp for helpful comments on the manuscript.
This work was supported by the Swiss National Science Foundation (grants 31003A-120064 and 31003A-135644 to D.S.) and the Kanton Bern.
We have no conflicts of interest.
V.R. performed experimental procedures under the guidance of D.S. E.G. performed experimental procedures under the guidance of D.S. and V.R. V.R. and D.S. conducted the project, designed experiments, and wrote the manuscript.
Footnotes
Published ahead of print 29 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00791-14.
REFERENCES
- 1.Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 65:1099–1122. 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller B, Schümperli D. 1997. The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin. Cell Dev. Biol. 8:567–576. 10.1006/scdb.1997.0182. [DOI] [PubMed] [Google Scholar]
- 4.Dominski Z, Marzluff WF. 2007. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene 396:373–390. 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris ME, Böhni R, Schneiderman MH, Ramamurthy L, Schümperli D, Marzluff WF. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillai RS, Grimmler M, Meister G, Will CL, Lührmann R, Fischer U, Schümperli D. 2003. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 17:2321–2333. 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai RS, Will CL, Lührmann R, Schümperli D, Müller B. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20:5470–5479. 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang XC, Xu B, Sabath I, Kunduru L, Burch BD, Marzluff WF, Dominski Z. 2011. FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol. Cell. Biol. 31:1492–1502. 10.1128/MCB.00979-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell 36:267–278. 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X-C, Sabath I, Dêbski J, Kaus-Drobek M, Dadlez M, Marzluff WF, Dominski Z. 2013. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol. Cell. Biol. 33:28–37. 10.1128/MCB.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gick O, Krämer A, Vasserot A, Birnstiel ML. 1987. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc. Natl. Acad. Sci. U. S. A. 84:8937–8940. 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolev NG, Steitz JA. 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 19:2583–2592. 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruepp MD, Vivarelli S, Pillai R, Kleinschmidt N, Azzouz TN, Barabino SM, Schümperli D. 2010. The 68 kDa subunit of mammalian cleavage factor I interacts with the U7 small nuclear ribonucleoprotein and participates in 3′ end processing of animal histone mRNAs. Nucleic Acids Res. 38:7637–7650. 10.1093/nar/gkq613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lüscher B, Schümperli D. 1987. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 6:1721–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188–4198. 10.1128/MCB.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez R, Marzluff WF. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 22:7093–7104. 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan N, Lam TT, Fritz A, Rempinski D, O'Loughlin K, Minderman H, Berezney R, Marzluff WF, Thapar R. 2012. The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol. Cell. Biol. 32:4306–4322. 10.1128/MCB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M, Lam TT, Tonelli M, Marzluff WF, Thapar R. 2012. Interaction of the histone mRNA hairpin with stem-loop binding protein (SLBP) and regulation of the SLBP-RNA complex by phosphorylation and proline isomerization. Biochemistry 51:3215–3231. 10.1021/bi2018255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno-Morcillo M, Minvielle-Sébastia L, Mackereth C, Fribourg S. 2011. Hexameric architecture of CstF supported by CstF-50 homodimerization domain structure. RNA 17:412–418. 10.1261/rna.2481011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruepp MD, Schweingruber C, Kleinschmidt N, Schümperli D. 2011. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol. Biol. Cell 22:91–104. 10.1091/mbc.E10-06-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dass B, McMahon KW, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. 2001. The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. J. Biol. Chem. 276:8044–8050. 10.1074/jbc.M009091200. [DOI] [PubMed] [Google Scholar]
- 22.Narita T, Yung TMC, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell 26:349–365. 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Sullivan KD, Steiniger M, Marzluff WF. 2009. A core complex of CPSF73, CPSF100, and symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell 34:322–332. 10.1016/j.molcel.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirngruber J, Johnsen SA. 2010. Induced G1 cell-cycle arrest controls replication-dependent histone mRNA 3′ end processing through p21, NPAT and CDK9. Oncogene 29:2853–2863. 10.1038/onc.2010.42. [DOI] [PubMed] [Google Scholar]
- 25.Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. 2009. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 10:894–900. 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brummelkamp TR, Bernards R, Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553. 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 27.Wiznerowicz M, Trono D. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77:8957–8961. 10.1128/JVI.77.16.8957-8951.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krichevsky AM, Kosik KS. 2001. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32:683–696. 10.1016/S0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 29.Martincic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze MT, Milcarek C. 1998. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc. Natl. Acad. Sci. U. S. A. 95:11095–11100. 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takagaki Y, Manley JL. 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2:761–771. 10.1016/S1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 31.Shell SA, Hesse C, Morris SM, Milcarek C. 2005. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J. Biol. Chem. 280:39950–39961. 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- 32.Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 1:753–763. 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Takagaki Y, Manley JL. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20:1515–1525. 10.1128/MCB.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff WF, Dominski Z. 2011. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA 17:1132–1147. 10.1261/rna.2566811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beggs S, James TC, Bond U. 2012. The polyA tail length of yeast histone mRNAs varies during the cell cycle and is influenced by Sen1p and Rrp6p. Nucleic Acids Res. 40:2700–2711. 10.1093/nar/gkr1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis D, Nierhaus KH. 2004. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin. Mol. Cell 13:113–124. 10.1016/S1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
- 37.White AE, Burch BD, Yang X-C, Gasdaska PY, Dominski Z, Marzluff WF, Duronio RJ. 2011. Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 193:677–694. 10.1083/jcb.201012077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stauber C, Schümperli D. 1988. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 16:9399–9414. 10.1093/nar/16.20.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Youngblood BA, Grozdanov PN, MacDonald CC. 2014. CstF-64 supports pluripotency and regulates cell cycle progression in embryonic stem cells through histone 3′ end processing. Nucleic Acids Res. 42:8330–8342. 10.1093/nar/gku551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson JM, McMahon KW, MacDonald CC, Makhatadze GI. 1999. MEARA sequence repeat of human CstF-64 polyadenylation factor is helical in solution. A spectroscopic and calorimetric study. Biochemistry 38:12869–12875. [DOI] [PubMed] [Google Scholar]
- 41.Yao C, Choi E-A, Weng L, Xie X, Wan J, Xing Y, Moresco JJ, Tu PG, Yates JR, Shi Y. 2013. Overlapping and distinct functions of CstF64 and CstF64τ in mammalian mRNA 3′ processing. RNA 19:1781–1790. 10.1261/rna.042317.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, Sandberg R, Hyman A, LaCava J, Rout MP, Andersen JS, Bertrand E, Jensen TH. 2013. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 20:1367–1376. 10.1038/nsmb.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ntini E, Jarvelin AI, Bornholdt J, Chen Y, Boyd M, Jorgensen M, Andersson R, Hoof I, Schein A, Andersen PR, Andersen PK, Preker P, Valen E, Zhao X, Pelechano V, Steinmetz LM, Sandelin A, Jensen TH. 2013. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 20:923–928. 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- 44.Kari V, Karpiuk O, Tieg B, Kriegs M, Dikomey E, Krebber H, Begus-Nahrmann Y, Johnsen SA. 2013. A subset of histone H2B genes produces polyadenylated mRNAs under a variety of cellular conditions. PLoS One 8:e63745. 10.1371/journal.pone.0063745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.