Abstract
Rex, a transcriptional repressor that modulates its DNA-binding activity in response to NADH/NAD+ ratio, has recently been found to play a role in the solventogenic shift of Clostridium acetobutylicum. Here, we combined a comparative genomic reconstruction of Rex regulons in 11 diverse clostridial species with detailed experimental characterization of Rex-mediated regulation in C. acetobutylicum. The reconstructed Rex regulons in clostridia included the genes involved in fermentation, hydrogen production, the tricarboxylic acid cycle, NAD biosynthesis, nitrate and sulfite reduction, and CO2/CO fixation. The predicted Rex-binding sites in the genomes of Clostridium spp. were verified by in vitro binding assays with purified Rex protein. Novel members of the C. acetobutylicum Rex regulon were identified and experimentally validated by comparing the transcript levels between the wild-type and rex-inactivated mutant strains. Furthermore, the effects of exposure to methyl viologen or H2O2 on intracellular NADH and NAD+ concentrations, expression of Rex regulon genes, and physiology of the wild type and rex-inactivated mutant were comparatively analyzed. Our results indicate that Rex responds to NADH/NAD+ ratio in vivo to regulate gene expression and modulates fermentation product formation and oxidative stress tolerance in C. acetobutylicum. It is suggested that Rex plays an important role in maintaining NADH/NAD+ homeostasis in clostridia.
INTRODUCTION
Organisms of the genus Clostridium are Gram-positive obligate anaerobes important in human health and physiology, the carbon cycle, and biotechnological applications (1). As anaerobes, clostridia maintain the cellular redox balance mainly through the reactions of central metabolism. The reducing equivalents are generated through the glycolytic pathway and reoxidized through alcohol synthesis, hydrogen production, and other NADH-consuming reactions (Fig. 1) (2). To sustain growth and metabolism, the metabolic network must be operated to maintain the redox balance in the cell.
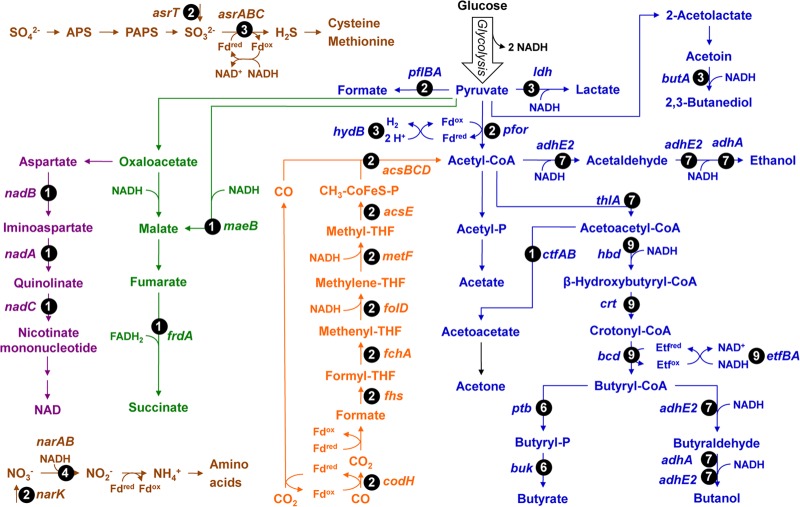
FIG 1.
Metabolic context of the reconstructed Rex regulons in clostridia. The metabolic pathways are color coded as follows: fermentation, blue; Wood-Ljungdahl pathway, orange; (incomplete) TCA cycle, green; NAD biosynthesis, purple; nitrate and sulfate reduction, brown. Numbers in circles indicate the numbers of genomes where the target gene is preceded by a candidate Rex-binding site.
Among Clostridium species, C. acetobutylicum is one of the best-studied species and has been used to develop an industrial acetone, butanol, and ethanol (ABE) fermentation process (3, 4). The redox balance in C. acetobutylicum has been manipulated by using several approaches to push the metabolism toward butanol synthesis. These approaches include the addition of artificial electron carriers such as methyl viologen (MV) or neutral red, increasing the hydrogen partial pressure or gassing with carbon monoxide, and the utilization of reduced substrates like glycerol (2, 5). All these approaches are based on reducing hydrogen formation to provide a surplus of electron, i.e., NAD(P)H, for butanol synthesis. A recent transcriptomic study has gained first insights on the molecular level into the effect of MV addition to cultures of C. acetobutylicum (6). Although some interesting results have been obtained from these studies, the molecular regulatory mechanisms remain to be elucidated.
The strictly anaerobic clostridia have evolved mechanisms to survive limited exposure to air (7). To cope with the oxidative stress, clostridia express genes encoding the components of the detoxification system, which essentially include flavodiiron proteins, desulfoferrodoxin, and rubrerythrins (8). Clostridia use their reducing equivalents to reduce the toxic reactive oxygen species (ROS) and molecular O2, thereby protecting crucial oxygen-sensitive metabolic enzymes (9). To generate the required reducing equivalents, the cellular redox balance needs to be shifted accordingly.
Recently, the redox-sensing transcriptional repressor Rex has been found to play a role in the solventogenic shift of C. acetobutylicum (10). Rex was first discovered in Streptomyces coelicolor and is widely distributed among Gram-positive bacteria. In S. coelicolor and Bacillus subtilis, Rex controls expression of cytochrome bd terminal oxidase and NADH dehydrogenase of the respiratory chain (11, 12). The Rex ortholog in Staphylococcus aureus regulates genes involved in anaerobic respiration and fermentation, such as lactate, formate, and ethanol formation and nitrate respiration (13). In Streptococcus mutans and Enterococcus faecalis, Rex has been shown to be involved in regulation of oxidative stress responses and to influence H2O2 accumulation, respectively (14, 15). The DNA-binding activity of Rex proteins is modulated by the ratio of NADH to NAD+ concentrations (11, 16). The crystal structures of Rex proteins from Thermus aquaticus and B. subtilis in complex with NADH, NAD+, and/or DNA operator have been determined (17, 18). Rex is composed of two domains, an N-terminal winged-helix DNA-binding domain and a C-terminal Rossmann-like domain involved in NADH binding and subunit dimerization.
Although the relative levels of NADH and NAD+ have been shown to influence the DNA-binding activity of Rex based on in vitro binding assays, it remains unclear whether Rex monitors the NADH/NAD+ ratio in vivo to control gene expression. Several genes associated with fermentation pathways have been identified as Rex targets in C. acetobutylicum (10). However, whether Rex also regulates transcription of other genes is not known. The role of Rex-dependent regulation in C. acetobutylicum in response to an altered cellular redox balance such as increased NAD(P)H availability or oxidative stress has not been studied. Moreover, although Rex seems to be widely distributed in clostridia, little is known about its targets and function in the species other than C. acetobutylicum.
In this study, we used a comparative genomic approach to reconstruct Rex regulons in 11 diverse clostridial species. These Clostridium species included the solvent-producing C. acetobutylicum and C. beijerinckii; the organic acid-producing C. butyricum and C. kluyveri; the acetogens that grow on CO2/CO/H2, including C. carboxidivorans and C. ljungdahlii; and the cellulolytic C. cellulovorans. The important human pathogens Clostridium botulinum, C. perfringens, and C. tetani, as well as C. novyi, having potential therapeutic uses in cancers, were also included. The reconstructed clostridial Rex regulons contain the genes associated with important metabolic processes, including fermentation, hydrogen production, NAD biosynthesis, nitrate and sulfite reduction, and CO2/CO fixation. Comparative analysis of reconstructed Rex regulons revealed considerable variations in the regulon content between the analyzed clostridia. The predicted Rex-binding sites in the genomes of Clostridium spp. were verified by in vitro binding assays. Novel members of the Rex regulon in C. acetobutylicum were identified and experimentally validated. Furthermore, the effects of exposure to MV or H2O2 on intracellular NADH and NAD+ concentrations, expression of Rex regulon genes, and physiology were compared between the wild-type and rex-inactivated mutant strains. Our results indicate that Rex monitors NADH/NAD+ ratio in vivo to regulate gene expression and modulates fermentation product formation and oxidative stress response in C. acetobutylicum.
MATERIALS AND METHODS
Bioinformatics tools and resources.
Genome sequences of clostridia analyzed in this study were obtained from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/). Identification of orthologs was performed using the BLAST tool provided by NCBI (19). Orthologs of the Rex protein from C. acetobutylicum ATCC 824 were identified with a 50% protein sequence identity threshold. The ClustalX (version 2.1) program was used for protein sequence alignments (20). Reconstruction of Rex regulons was performed using an established comparative genomics method (21) implemented in the RegPredict webserver (http://regpredict.lbl.gov) (22) and the Genome Explorer software (23). The previously identified Rex recognition DNA motif in Clostridiaceae (24) was used to scan the Clostridium genomes and identify candidate Rex-binding sites. Scores of candidate sites were calculated as the sum of positional nucleotide weights. The score threshold was defined as the lowest score observed in the training set. Genes with candidate upstream binding sites that have high scores and/or are conserved in two or more genomes were included in the Rex regulon. Candidate sites associated with new regulon members were added to the training set, and the respective position weight matrices describing the clostridial Rex-binding DNA motif were rebuilt to improve search accuracy. Functional annotations of the predicted regulon members were based on the SEED database (http://theseed.uchicago.edu/FIG/index.cgi) (25).
Bacterial strains and growth conditions.
C. acetobutylicum strain ATCC 824, its mutant with the rex gene inactivation (rex::intron), and the rex-complemented strain (rex::intron pSY9-rex) were used in this study. C. acetobutylicum strains were precultured anaerobically on clostridial growth medium (CGM) (26) to exponential growth phase. The cultures were started with the same optical density at 600 nm (OD600; ∼0.02) and performed at 37°C in triplicate in 60 ml of P2 minimal medium (27), which contains (per liter) 0.5 g of K2HPO4, 0.5 g of KH2PO4, 2.2 g of CH3COONH4, 0.2 g of MgSO4 · 7H2O, 0.01 g of MnSO4 · H2O, 0.01 g of NaCl, 0.01 g of FeSO4 · 7H2O, 1 mg of p-aminobenzoic acid, 1 mg of vitamin B1, 0.01 mg of biotin, and 60 g of glucose. For methyl viologen-exposed cultures, MV was added to a final concentration of 1 mM when cells were grown in P2 minimal medium to an OD600 of about 0.15. For hydrogen peroxide challenge experiments, cells were grown in P2 minimal medium to an OD600 of about 2.0. Then, cells were exposed to 50, 100, or 200 μM H2O2 or the equal volume of H2O. Because the sensitivity of C. acetobutylicum toward H2O2 was largely dependent on Fenton chemistry (28), 1 mM iron chelator 2,2′-dipyridyl (Sigma-Aldrich) was added to attenuate peroxide-dependent killing of cells. After incubation at 37°C for 30 min, the number of surviving cells was determined as described previously (29). Briefly, aliquots of appropriate dilutions were plated on CGM and incubated anaerobically for 36 h at 37°C. The CFU for each sample were determined and normalized to the number obtained for the nonstressed wild type (100%).
Mutant construction.
Gene disruption in C. acetobutylicum ATCC 824 was performed by using group II intron-based targetron technology as described previously (30). Briefly, a 350-bp fragment for retargeting an intron to insert within the rex gene (CAC2713) was generated by one-step assembly PCR using the primers shown in Table S1 in the supplemental material according to the protocol of the TargeTron gene knockout system (Sigma). The PCR product was digested and ligated to targetron vector pWJ1 (31), yielding the plasmid pWJ1-rex. The plasmid was methylated in vivo in Escherichia coli ER2275(pAN1) (32) and electroporated into C. acetobutylicum ATCC 824. The transformants were selected on a CGM plate supplemented with erythromycin. The resulting mutant with an intron insertion in the rex gene was confirmed by PCR.
For genetic complementation experiments, the rex gene from C. acetobutylicum was cloned into the pSY9 vector (33) under the control of the constitutive Pptb promoter (34). PCR was carried out using the C. acetobutylicum ATCC 824 genomic DNA and the primers shown in Table S1 in the supplemental material. The obtained plasmid pSY9-rex was electroporated into the rex-inactivated mutant, generating the rex-complemented strain.
RNA isolation and real-time PCR analysis.
Total RNA was isolated from C. acetobutylicum ATCC 824 grown in the P2 minimal medium with or without addition of MV or H2O2. Cells were harvested at mid-exponential growth phase (OD600 of about 2.0), frozen immediately in liquid nitrogen, and ground into powder. RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. Contaminant DNA was removed by DNase I (TaKaRa) digestion. RNA (1 μg) was transcribed into cDNA with random primers using the ReverTra-Plus kit from Toyobo. The product was quantified via real-time PCR using the CFX96 thermal cycler (Bio-Rad). The reaction mixture (20 μl) contained Power SYBR green PCR master mix (Bio-Rad) and 0.4 μm gene-specific primers (as shown in Table S1 in the supplemental material). The PCR parameters were 1 cycle of 95°C for 2 min, followed by 40 cycles of 95°C for 20 s, 60°C for 20 s, and 72°C for 15 s. The accuracy of the PCR product was checked by melting curve analysis. The expression level of each gene was normalized with the value for the CAC2679 gene encoding a pullulanase, which was used as a reference gene with constitutive expression (35). Data were presented as the average of six measurements from two biological replicates, with the corresponding standard deviation.
Protein overexpression and purification.
The rex (CAC2713) gene was PCR amplified from C. acetobutylicum ATCC 824 genomic DNA using the primers shown in Table S1 in the supplemental material. The PCR fragment was ligated into the expression vector pET28a cleaved by BamHI and SalI. The resulting plasmid pET28a-rex was used to produce Rex protein with an N-terminal hexahistidine tag. The plasmid pET28a-rex-Q51K coding for a Rex mutant where the glutamine residue Gln51 was replaced by a lysine residue was constructed with two steps of PCR using pET28a-rex as the template and the mutagenic primers and flanking primers (see Table S1). All recombinant plasmids were sequenced to exclude unwanted mutations in the rex gene. For overproduction of Rex protein and its mutated derivative, E. coli BL21(DE3)pLysS (Novagen) was transformed with expression plasmid pET28a-rex or pET28a-rex-Q51K and cultivated in LB medium at 37°C to an OD600 of 0.8. Protein expression was induced by the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside, and the culture was incubated for another 12 h at 16°C. After the cells were harvested, purification of Rex by nickel-nitrilotriacetic acid affinity chromatography was performed as described previously (36). The purified protein was run on a 12% sodium dodecyl sulfate-polyacrylamide gel to monitor its size and purity.
Electrophoretic mobility shift assay (EMSA).
The 180-bp DNA fragments in the promoter region of individual genes were PCR amplified using the primers shown in Table S1 in the supplemental material. Both forward and reverse primers were Cy5 fluorescence labeled at the 5′ end (Sangong Corp., Shanghai, China), and the PCR products were purified with a PCR purification kit (Axygen). Purified Rex protein or its mutated derivative was incubated with the fluorescence-labeled DNA fragment (1 nM) in 20 μl of binding buffer containing 20 mM Tris (pH 7.5), 0.25 mM dithiothreitol (DTT), 10 mM MgCl2, 5% glycerol, 0.8 μg bovine serum albumin (BSA), and 1 μg salmon sperm DNA (nonspecific competitor). Promoter fragments lacking a putative Rex-binding site were used as negative controls. As potential effectors of Rex-DNA binding, NADH and/or NAD+ was added as indicated. After incubation at 30°C for 20 min, the reaction mixture was electrophoresed at 4°C on a 6% native polyacrylamide gel in 0.5× Tris-borate-EDTA for 1.5 h at 100 V. Fluorescence-labeled DNA on the gel was then detected with the Starion FLA-9000 scanner (FujiFilm, Japan). For the determination of apparent dissociation constants (Kd), the bands were quantified using Quantity One software, and the percentage of shifted DNA was calculated. These values were plotted against the Rex concentration, and Kd values were obtained using the GraphPad Prism software. All determinations were performed at least in triplicate.
Metabolite analysis.
For analysis of extracellular metabolites, culture samples were centrifuged for 10 min at 4°C and 15,000 × g to remove the cells. Acetone, ethanol, and butanol were detected by a gas chromatograph (GC) (Agilent model 7890A) equipped with a capillary column (Alltech EC-Wax; 30 m by 0.32 mm) and a flame ionization detector (Agilent).
The intracellular NADH and NAD+ were extracted and assayed by using the fluorescent NAD+/NADH detection kit (Cell Technology Inc., CA), which utilizes a nonfluorescent detection reagent that is reduced in the presence of NADH to produce its fluorescent analog. Briefly, cells were harvested at mid-exponential growth phase (OD600 of about 2.0) by centrifuging 2 ml of culture broth at 9,000 × g and 4°C for 10 min. Intracellular NADH and NAD+ were extracted using respective extraction buffers by following the manufacturer's instructions. NADH reacted with nonfluorescent detection reagent to form NAD+ and the fluorescent analog. The concentration of the formed fluorescent analog was then determined at 550-nm excitation and 595-nm emission wavelengths by using a spectrofluorometer (Varioskan Flash; Thermo Scientific Co.). NAD+ is further converted to NADH via an enzyme-coupled reaction. The enzyme reaction specifically reacts with NAD+/NADH and not with NADP+/NADPH. A series of NADH and NAD+ standards were used to obtain a calibration curve for determining the concentrations of these compounds in the cell extracts. The intracellular NADH and NAD+ concentrations were then calculated by normalization to cell volume. A predetermined correlation factor of 0.26 g (dry weight) of cells per OD600 and a previously reported intracellular aqueous volume of 1.67 μl per mg (dry weight) of cells (37) were used for calculation. Data were presented as the average of nine measurements from three biological replicates, with the corresponding standard deviation.
RESULTS
Comparative genomic reconstruction of Rex regulons in Clostridium spp.
To reconstruct the Rex regulons in Clostridium species, we applied the integrative comparative genomics approach that combines identification of candidate transcription factor-binding sites with cross-genomic comparison of regulons and with the functional context analysis of candidate target genes. The analyzed clostridia include C. acetobutylicum, C. beijerinckii, C. botulinum, C. butyricum, C. kluyveri, C. novyi, C. perfringens, C. tetani, C. cellulovorans, C. carboxidivorans, and C. ljungdahlii. These 11 species with complete genome sequences belong to Clostridium cluster I (38), whereas they exhibit markedly different phenotypes (for example, they include saccharolytic and proteolytic species as well as solventogenic and acetogenic species). Rex proteins in these clostridia share close sequence homology (>68% identity), and particularly the sequences of the N-terminal DNA-binding domain are highly conserved (see Fig. S1 in the supplemental material). The previously identified Rex-binding DNA motif in Clostridiaceae (24), which has consensus TTGTTAANNNNTTAACAA, was used to search for Rex-binding sites in the genomes of Clostridium species. Finally, we performed a cross-species comparison of the predicted sets of potentially coregulated genes to define the Rex regulon for each species. The candidate members and metabolic context of the Rex regulons in the 11 Clostridium species are shown in Table 1 and Fig. 1, respectively. Detailed information about the predicted DNA-binding sites, candidate Rex target genes, and their known or predicted transcriptional start sites is provided in Table S2 in the supplemental material.
TABLE 1.
Rex regulons in 11 species of clostridiaa
| Operon and metabolism | Presence of gene in Clostridium species: |
Functional role | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. acetobutylicum ATCC 824 | C. beijerinckii NCIMB 8052 | C. botulinum ATCC 3502 | C. butyricum 5521 | C. kluyveri DSM 555 | C. novyi NT | C. perfringens ATCC 13124 | C. tetani E88 | C. cellulovorans 743B | C. carboxidivorans P7 | C. ljungdahlii DSM 13528 | ||
| Fermentation | ||||||||||||
| ldh | +* | − | + | − | − | − | + | − | 0 | 0 | 0 | l-Lactate dehydrogenase |
| pflBA | − | +* | − | − | − | − | − | 0 | + | 0 | 0 | Pyruvate formate-lyase |
| adhA | 0 | +* | + | + | − | + | − | + | − | + | + | Alcohol dehydrogenase [Fe] |
| adhE2 | +* | − | + | + | − | 0 | + | + | + | + | − | Alcohol/acetaldehyde dehydrogenase |
| thlA | +* | + | + | + | − | + | + | + | − | − | 0 | Acetyl-CoA acetyltransferase |
| crt-bcd-etfBA-hbd | +* | + | + | + | + | + | + | + | + | − | 0 | Butyryl-CoA synthesis enzymes |
| ctfAB | − | − | − | 0 | 0 | +* | 0 | 0 | 0 | 0 | 0 | CoA-transferase |
| ptb-buk | +* | + | − | + | 0 | 0 | + | + | − | + | 0 | Phosphotransbutyrylase, butyrate kinase |
| butA | 0 | +* | + | + | 0 | − | 0 | 0 | 0 | − | − | 2,3-Butanediol dehydrogenase |
| hydB | 0 | +* | − | − | 0 | + | − | + | 0 | − | − | Fe-hydrogenase |
| fld-Cbei_4318 | − | +* | − | + | − | − | − | − | 0 | 0 | − | Flavodoxin, pyruvate flavodoxin/ferredoxin oxidoreductase |
| TCA cycle | ||||||||||||
| frdA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | +* | 0 | 0 | 0 | Fumarate reductase flavoprotein subunit |
| maeB | 0 | − | +* | + | 0 | 0 | 0 | 0 | 0 | − | − | Malic enzyme |
| NAD biosynthesis | ||||||||||||
| nadABC | +* | − | − | − | − | − | − | 0 | − | − | − | NAD biosynthesis enzymes |
| Nitrate and sulfite reduction | ||||||||||||
| narAB | 0 | 0 | 0 | + | 0 | 0 | + | 0 | 0 | +* | + | Nitrate reductase |
| narK | 0 | 0 | 0 | 0 | 0 | 0 | − | 0 | 0 | +* | + | Nitrate/nitrite transporter |
| asrABC | +* | − | − | − | 0 | − | − | − | − | + | + | Sulfite reductase |
| asrT | +* | − | − | 0 | 0 | 0 | − | − | − | + | 0 | Predicted sulfite/sulfate transporter |
| Wood-Ljungdahl pathway | ||||||||||||
| codH-cooC-fhs-fchA-folD-metF-lpdA-cooC-acsDCEB | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | +* | +* | Wood-Ljungdahl pathway enzymes |
| Other | ||||||||||||
| grdIH | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | +* | + | Betaine/glycine reductase |
| noxE | 0 | − | + | − | 0 | + | +* | + | 0 | 0 | 0 | NADH oxidase |
| bcd2-fldA-bcd-etfBA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | +* | Acyl-CoA dehydrogenase, acyl-CoA transferase, flavoprotein |
The genes preceded by a conserved Rex-binding site are indicated by +, and the predicted Rex-binding sites verified by targeted experiments are marked by bold type and an asterisk. Genes without a candidate Rex-binding site are indicated by −. The absence of an orthologous gene(s) in the analyzed genomes is indicated by 0.
The reconstructed Rex regulons control the fermentation in all analyzed clostridia (Fig. 1 and Table 1). Most of the predicted Rex targets encode enzymes that consume NADH or other reducing equivalents (e.g., reduced ferredoxin). However, the size and the specific content of reconstructed Rex regulons are highly variable between different clostridial species. For instance, the Rex regulon in C. beijerinckii constitutes 11 operons, whereas in C. kluyveri Rex is predicted to control only one operon. Based on distribution of predicted Rex-regulated genes in Clostridium species, we classified them into the conserved and variable parts of Rex regulons. The conserved part of the Rex regulons includes 5 operons that are potentially regulated by Rex in at least 6 species. They are the adhA gene encoding alcohol dehydrogenase, the adhE2 gene encoding bifunctional alcohol/acetaldehyde dehydrogenase, the thlA gene and crt-bcd-etfBA-hbd operon responsible for the conversion of acetyl coenzyme A (acetyl-CoA) to butyryl-CoA, and the ptb-buk operon for butyrate synthesis. On the other hand, 15 target operons form a group of species-specific regulon members that are preceded by candidate Rex-binding sites in at most 3 genomes analyzed. This group includes the genes involved in fermentation (ldh, pflBA, ctfAB, butA, and Cbei_4318), hydrogen production (hydB), tricarboxylic acid (TCA) cycle (frd and maeB), and nitrate and sulfite reduction (narK, asrABC, and asrT). In addition, the NAD biosynthetic genes nadABC were identified as candidate members of the Rex regulon in C. acetobutylicum. Regulation of the Wood-Ljungdahl pathway (codH-cooC-fhs-fchA-folD-metF-lpdA-cooC-acsDCEB) by Rex, which is used to fix CO2 or CO, was predicted for C. carboxidivorans and C. ljungdahlii.
In summary, the comparative genomics analysis allowed us to reconstruct the Rex regulons in 11 diverse clostridial species. Among these species, C. acetobutylicum has one of the largest sets of Rex targets, including 17 genes organized in 7 operons that contain not only the known targets (ldh, adhE2, thlA, and crt-bcd-etfBA-hbd) but also the newly identified members (ptb-buk, nadABC, and asrTABC). These Rex targets are involved in fermentation, NAD biosynthesis, and sulfite reduction. We then performed experimental characterization of the clostridial Rex-binding motif and the Rex-mediated regulation in C. acetobutylicum as described below.
Rex binds to the promoter regions of predicted target genes in vitro.
To validate the predicted clostridial Rex regulons, electrophoretic mobility shift assays (EMSAs) were performed using the recombinant Rex from C. acetobutylicum, which was overexpressed in E. coli with the N-terminal His6 tag and purified with a nickel-chelating affinity column. For all predicted Rex target operons in C. acetobutylicum, DNA fragments (180 bp) in the promoter regions containing candidate Rex-binding sites were tested in EMSAs (Fig. 2A). A shifted band was observed upon incubation of Rex protein with each promoter fragment, and its intensity was enhanced in the presence of increasing amounts of Rex protein. As a negative control, the promoter fragment of pflBA operon in C. acetobutylicum, which lacks a predicted Rex-binding site, was used, and no binding was observed even at 3,000 nM Rex protein (Fig. 2B). The formation of Rex-DNA complex was suppressed in the presence of 400-fold excess unlabeled DNA fragments but not in the presence of nonspecific competitor, salmon sperm DNA (data not shown). These results confirm that Rex binds specifically to the promoter regions of the predicted Rex target operons in C. acetobutylicum.
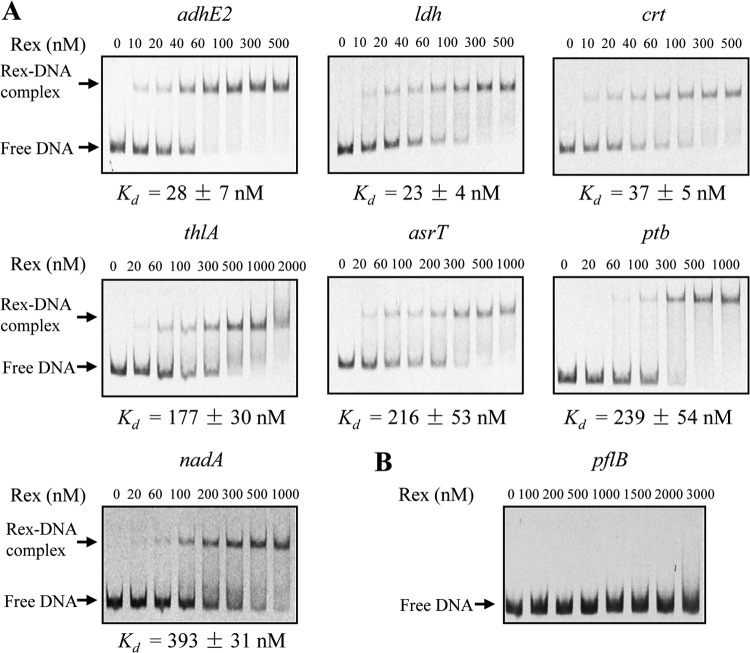
FIG 2.
EMSAs with purified Rex protein and DNA fragments from the promoter regions of predicted target genes in C. acetobutylicum. (A) DNA fragments (1 nM) from the promoter regions of C. acetobutylicum adhE2, ldh, crt, thlA, asrT, ptb, and nadA genes were fluorescence labeled and incubated with the indicated concentrations of Rex protein for 20 min at 30°C. Then, the protein-DNA complexes were resolved by electrophoresis on native 6% polyacrylamide gels. Quantification of the bands allowed the determination of the apparent Kd values (see Materials and Methods). The values shown represent the average and standard deviation of at least three independent assays. (B) As a negative control, the promoter region of the C. acetobutylicum pflBA operon, which lacks a putative Rex-binding site, was used.
The apparent dissociation constant (Kd) values of Rex protein interacting with the tested C. acetobutylicum DNA fragments were determined, and they varied in a wide range from 23 nM to 393 nM (Fig. 2). According to the Kd values, the tested DNA fragments can be divided into two groups. For the first group including the promoter fragments of adhE2, ldh genes, and the crt-bcd-etfBA-hbd operon, Rex protein exhibited a high affinity and the Kd values were in the range of 23 to 37 nM. The second group includes the fragments from the promoter regions of the thlA gene and the asrTABC, ptb-buk, and nadABC operons. The Kd values for this group were in the range of 177 to 393 nM, indicating a lower affinity of Rex to these target fragments.
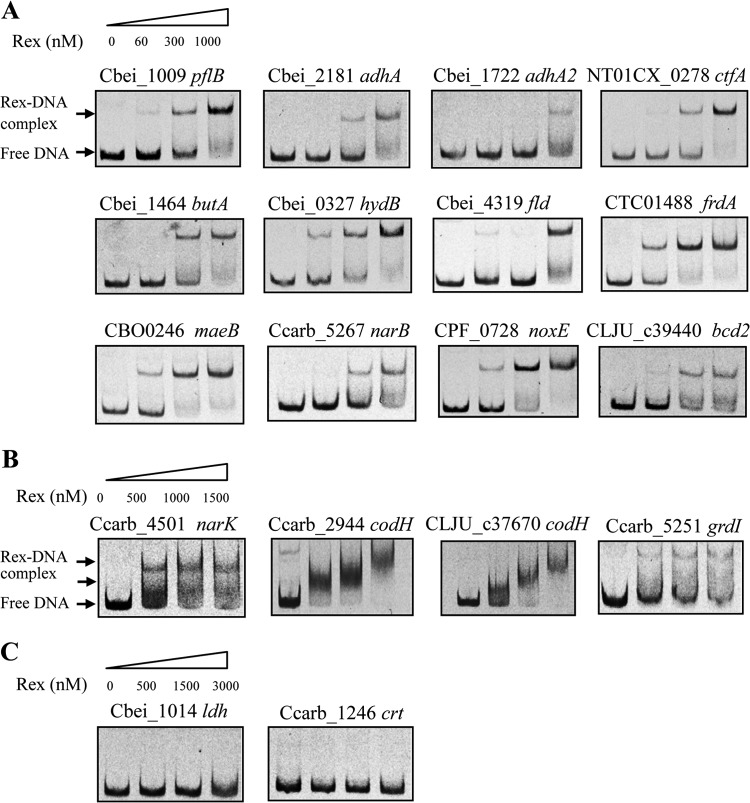
EMSAs were also performed to assess the predicted Rex-binding sites in other analyzed clostridia. For each predicted Rex target operon, the upstream candidate Rex-binding site in one or two genomes was tested (Table 1). Thus, 16 DNA fragments were amplified from the promoter regions of C. beijerinckii pflBA, adhA, adhA2, butA, hydB, and fld-Cbei_4318; C. botulinum maeB; C. novyi ctfAB; C. perfringens noxE; C. tetani frdA; C. carboxidivorans narAB, narK, codH-cooC-fhs-fchA-folD-metF-lpdA-cooC-acsDCEB, and grdIH; and C. ljungdahlii codH and bcd2, respectively. These DNA fragments were tested for binding of C. acetobutylicum Rex protein that is well conserved in the analyzed clostridia. A shift in the presence of purified Rex was observed for all the 16 fragments (Fig. 3A and B). For the C. carboxidivorans narK fragment, two shifted Rex-DNA complexes were detected, supporting our prediction that two DNA-binding sites are present (see Table S2 in the supplemental material). Most of the promoter fragments were completely shifted with 1,000 nM Rex (Fig. 3A and B). In contrast, the promoter fragments of the C. beijerinckii ldh gene and the C. carboxidivorans crt-hbd-thlA-bcd-etfBA operon, which do not contain predicted Rex-binding sites, were not shifted even with 3,000 nM Rex protein (Fig. 3C).
FIG 3.
EMSAs with purified Rex protein and the promoter regions of predicted target genes in Clostridium species other than C. acetobutylicum. (A) EMSAs were performed in the absence (lanes 1) and in the presence of 60, 300, and 1,000 nM Rex protein (lanes 2 to 4, respectively). (B) EMSAs were performed in the absence (lanes 1) and in the presence of 500, 1,000, and 1,500 nM Rex protein (lanes 2 to 4, respectively). (C) The negative controls included the ldh promoter of C. beijerinckii and the crt promoter of C. carboxidivorans, which do not contain the predicted Rex-binding site.
Characterization of the Rex-binding motif in clostridia.
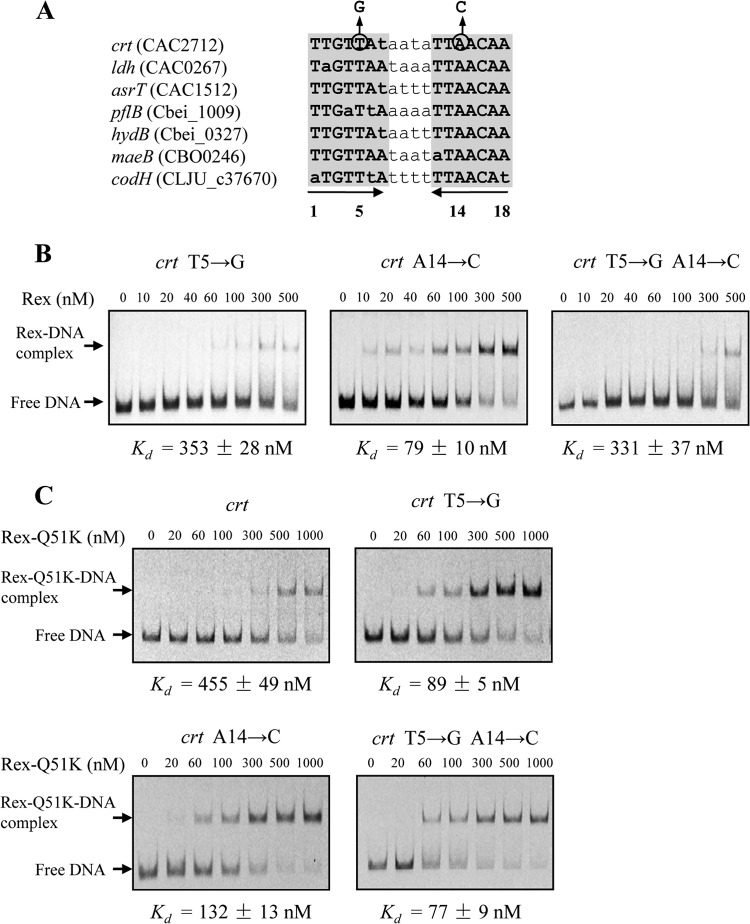
The identified clostridial Rex-binding motif has the consensus TTGTTAANNNNTTAACAA, which deviates in two positions (i.e., positions 5 and 14) from the common consensus TTGTGAANNNNTTCACAA of the Rex-binding motifs in most Gram-positive bacteria such as B. subtilis and S. coelicolor (24). Among the confirmed Rex-binding sites in clostridia, the thymine at position 5 and adenine at position 14 are highly conserved (Fig. 4A). For characterization of the Rex-binding motif in clostridia, mutational analysis was performed on the promoter fragment of the C. acetobutylicum crt-bcd-etfBA-hbd operon. This operon is a conserved Rex regulon member in clostridia, and its promoter fragment showed a substantial shift in the presence of 40 nM Rex (Fig. 2A). We changed the thymine at position 5 to guanine or/and the adenine at position 14 to cytosine to match the common Rex consensus sequence in Gram-positive bacteria (Fig. 4A). The mutated fragments were amplified by PCR and tested in EMSAs for binding of C. acetobutylicum Rex (Fig. 4B). Substitution of the thymine 5 or adenine 14 in the Rex-binding site increased the apparent Kd value of Rex about 10-fold and 2-fold, respectively. The reduced binding affinity of Rex to the mutated Rex-binding sites indicates that the thymine 5 and adenine 14 in the operator are important for Rex binding in clostridia.
FIG 4.
Characterization of the Rex-binding motif in clostridia. (A) Alignment of the Rex-binding sites in the promoter regions of the C. acetobutylicum crt-bcd-etfBA-hbd operon, ldh gene, and asrTABC operon; the C. beijerinckii pflB and hydB genes; the C. botulinum maeB gene; and the C. ljungdahlii codH-cooC-fhs-fchA-folD-metF-lpdA-cooC-acsDCEB operon. The palindromic sequences are shaded. The conserved nucleotides are shown in bold capitals. Bases substituted in the crt promoter for EMSAs are indicated, and the new base is shown above. (B) Mutational analysis of the Rex-binding site in the crt promoter of C. acetobutylicum. The mutations were introduced by PCR, and the corresponding DNA fragments were analyzed by EMSAs with purified Rex protein. The apparent Kd values were determined as described in Materials and Methods. (C) Effect of mutagenesis of Rex on Rex-DNA interactions. A Rex derivative (Rex-Q51K) obtained by site-directed mutagenesis was used in EMSAs to test for binding to the crt promoter and mutated fragments.
In the Rex proteins, Lys47 is a relatively conserved residue in the DNA recognition helix, and it forms a hydrogen bond with guanine 5 of the DNA operator according to a structural study of T. aquaticus Rex (17). However, the Lys47 residue was mutated to glutamine (Gln51) in the Rex proteins from Clostridium spp. (see Fig. S1 in the supplemental material). To assess if this residue substitution influences the Rex-DNA contacts, the Gln51 residue of C. acetobutylicum Rex was changed to a lysine residue by site-directed mutagenesis. The resulting Rex variant was overproduced in E. coli, purified, and used for EMSAs. As shown in Fig. 4C, the mutated protein Rex-Q51K exhibited a 12-fold-increased apparent Kd value for the promoter fragment of the crt-bcd-etfBA-hbd operon. Nevertheless, when the thymine 5 or adenine 14 in the binding sequence was changed to guanine and cytosine, respectively, the binding affinity of the mutated protein Rex-Q51K was significantly increased. Therefore, these results indicate a correlation between a key amino acid residue in the DNA-binding domain of Rex proteins and two nucleotides at symmetrical positions of the palindromic Rex-binding motifs. For clostridial Rex proteins, the Gln51 residue in the recognition helix might position within the major groove of DNA and contact the thymine 5 and adenine 14 of DNA operators (see Fig. S2 in the supplemental material).
Effect of NADH and NAD+ on Rex-DNA interactions.
To test if NADH and NAD+ affect the interaction between the Rex from C. acetobutylicum and its cognate operators, EMSAs were performed using the promoter fragment of the adhE2 gene. As shown in Fig. 5A, the presence of only 5 μM NADH drastically decreased the formation of Rex-DNA complex, whereas the addition of 1 mM NAD+ results in a noticeable enhancement of Rex binding to the DNA fragment (Fig. 5A, lanes 3 and 4). This effect is specific for NADH and NAD+, as it was not found for 10-fold-higher concentrations of NADPH and NADP+ (Fig. 5A, lanes 7 and 8). Furthermore, interaction between the same DNA fragment and C. acetobutylicum Rex protein was assessed in the presence of physiological concentrations of NADH and NAD+. The intracellular NADH and NAD+ pool sizes in C. acetobutylicum are decreased from 0.3 mM and 1.6 mM, respectively, during the exponential growth phase, to 0.1 mM and 1.2 mM, respectively, during the subsequent solventogenic phase (39). Thus, various concentrations of NADH and NAD+, which cover the physiological concentration ranges, were used in EMSAs. As shown in Fig. 5B, the DNA-binding activity of Rex was particularly susceptible to changes in the NADH concentration, but NAD+ clearly influenced the inhibitory effect of NADH on Rex-DNA complex formation. These results strongly suggest that C. acetobutylicum Rex senses and responds to the intracellular ratio of NADH to NAD+ to modulate its DNA-binding activities under physiological conditions.
FIG 5.
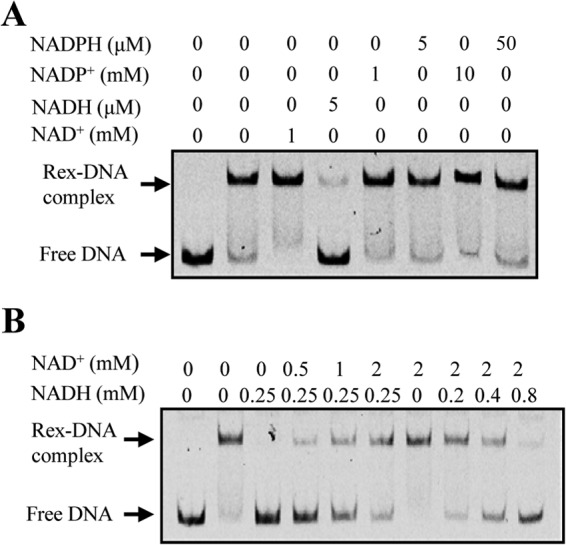
Effect of NADH and NAD+ on the DNA-binding activity of Rex. (A) EMSAs were performed using C. acetobutylicum adhE2 promoter fragment (1 nM), Rex protein (60 nM), and the indicated concentrations of pyridine nucleotides. No protein was added to the first lane. (B) EMSAs were performed as in panel A but with a range of physiological concentrations of NADH and NAD+.
Rex negatively regulates expression of its direct target genes in vivo.
To validate the predicted regulation of Rex on gene expression in vivo, the rex gene in C. acetobutylicum was disrupted by insertion of an intron, resulting in the rex-inactivated mutant (confirmed by PCR as shown in Fig. S3 in the supplemental material). The transcript levels of the predicted Rex direct targets in the rex-inactivated mutant were compared with those in the wild type by using quantitative real-time-PCR (qRT-PCR). The two strains were cultivated in minimal medium with 60 g liter−1 of glucose as a carbon source, and no differences in cell growth were observed for them. For comparison of transcript levels, cells were harvested in the exponential growth phase at an OD600 of 2.0 and a growth rate of 0.16 h−1 for both strains, and total RNA was isolated. Six qRT-PCR measurements from two independent cultures were performed. As shown in Table 2, the relative mRNA levels of all the 17 genes were elevated more than 1.5-fold in the rex-inactivated mutant compared with the wild-type strain. The most prominent effect of rex mutation was observed for the adhE2 gene, which showed a ≥160-fold-increased mRNA level in the rex-inactivated mutant. Complementation of the rex-inactivated mutant by using a plasmid construct constitutively expressing rex reduced the adhE2 gene expression (see Fig. S3). The genes with a strongly increased expression in the rex-inactivated mutant also include the ldh and thlA genes (Table 2). The crt-bcd-etfBA-hbd, asrTABC, and nadABC operons showed a 1.5- to 3-fold-elevated transcript level in the rex-inactivated mutant. Expression of the ptb-buk operon was also increased by rex mutation. Therefore, the qRT-PCR results confirm that Rex is a negative regulator of ldh, adhE2, thlA, crt-bcd-etfBA-hbd, ptb-buk, nadABC, and asrTABC operons involved in fermentation, NAD biosynthesis, and sulfite reduction in C. acetobutylicum.
TABLE 2.
Comparison of mRNA levels in C. acetobutylicum wild type and rex-inactivated mutant using qRT-PCR
| Gene name | Gene identifier | mRNA ratioa (rex mutant/WTb) |
|---|---|---|
| adhE2 | CAP0035 | 164.75 ± 6.45 |
| ldh | CAC0267 | 13.75 ± 0.84 |
| thlA | CAC2873 | 12.46 ± 2.34 |
| crt | CAC2712 | 2.43 ± 0.10 |
| bcd | CAC2711 | 2.81 ± 0.78 |
| etfB | CAC2710 | 2.17 ± 0.20 |
| etfA | CAC2709 | 1.55 ± 0.14 |
| hbd | CAC2708 | 2.79 ± 0.37 |
| ptb | CAC3076 | 6.07 ± 1.35 |
| buk | CAC3075 | 1.97 ± 0.12 |
| nadA | CAC1025 | 2.53 ± 0.38 |
| nadB | CAC1024 | 1.79 ± 0.12 |
| nadC | CAC1023 | 1.79 ± 0.26 |
| asrT | CAC1512 | 2.04 ± 0.72 |
| asrA | CAC1513 | 2.41 ± 0.74 |
| asrB | CAC1514 | 2.67 ± 0.36 |
| asrC | CAC1515 | 1.69 ± 0.48 |
Data represent means ± standard deviations of values of mRNA ratios obtained from six measurements starting from two independent cultures. The strains were cultivated in P2 minimal medium, and total RNA was isolated in the exponential growth phase at an OD600 of about 2.0. The P value of the mRNA ratios for all the genes studied is smaller than 0.01.
WT, wild type.
Rex plays a role in maintaining NADH/NAD+ homeostasis in C. acetobutylicum.
To understand the role of Rex-dependent regulation in C. acetobutylicum, we investigated the effects of exposure of the wild-type and rex-inactivated mutant strains to methyl viologen (MV) or hydrogen peroxide (H2O2). First, the effects on intracellular NADH and NAD+ concentrations were determined. The strains were cultivated in minimal medium without or with addition of 1 mM MV or 30 μM H2O2 and harvested in the exponential growth phase at an OD600 of about 2.0. Quantification of intracellular NADH and NAD+ concentrations revealed an increase in the size of the total NAD pool in the rex-inactivated mutant compared to the wild type, which could be due to derepression of NAD biosynthetic genes in the mutant (Table 3). In accordance with previous findings (40), exposure of the wild type to MV caused a 2.5-fold increase in the NADH/NAD+ concentration ratio (Table 3). Although the NADH/NAD+ ratios were similar in the wild type and rex-inactivated mutant grown in cultures without MV addition, MV-exposed rex-inactivated mutant exhibited a 1.5-fold-increased NADH/NAD+ ratio compared to MV-exposed wild type. In contrast, the intracellular NADH/NAD+ ratio was decreased by 21% and 56% in the wild-type and rex-inactivated mutant, respectively, by H2O2 addition (Table 3). Therefore, MV addition resulted in a remarkable increase in intracellular NADH/NAD+ ratio, whereas exposure to H2O2 significantly reduced the NADH/NAD+ ratio. The rex-inactivated mutant showed larger fluctuations in the NADH/NAD+ ratio than did the wild type when exposed to MV or H2O2, suggesting that Rex plays an important role in maintaining NADH/NAD+ homeostasis in C. acetobutylicum.
TABLE 3.
Intracellular NADH and NAD+ concentrations in C. acetobutylicum wild type and rex-inactivated mutanta
| Strain | Addition to cultures | NADH concn (mM) | NAD+ concn (mM) | NADH/NAD+ ratio |
|---|---|---|---|---|
| Wild type | 0.19 ± 0.01 | 0.83 ± 0.01 | 0.23 ± 0.02 | |
| Wild type | MV | 0.27 ± 0.03 | 0.46 ± 0.01 | 0.59 ± 0.05 |
| Wild type | H2O2 | 0.20 ± 0.02 | 1.11 ± 0.11 | 0.18 ± 0.02 |
| rex mutant | 0.26 ± 0.02 | 1.02 ± 0.18 | 0.25 ± 0.02 | |
| rex mutant | MV | 0.34 ± 0.08 | 0.38 ± 0.07 | 0.92 ± 0.11 |
| rex mutant | H2O2 | 0.16 ± 0.02 | 1.44 ± 0.12 | 0.11 ± 0.02 |
Data represent means ± standard deviations of nine measurements from three biological replicates. The strains were grown in the P2 minimal medium without or with addition of 1 mM MV or 30 μM H2O2. The intracellular concentrations of NADH and NAD+ were determined in the exponential growth phase at an OD600 of about 2.0.
Rex monitors NADH/NAD+ ratio in vivo to regulate gene expression.
The effect of exposure to MV or H2O2 on expression of genes in the Rex regulon was compared between C. acetobutylicum wild-type and rex-inactivated mutant strains. The transcript levels of the genes involved in fermentation were determined by using qRT-PCR, because the fermentation genes comprise the major direct targets of clostridial Rex. As shown in Fig. 6A, expression of Rex target genes adhE2, thlA, crt, bcd, and hbd in the wild type was significantly upregulated by MV addition. Most strikingly, the adhE2 gene in the wild type showed an 80-fold-increased mRNA level in the presence of MV, which is consistent with previous reports (6). The transcript levels of adhE2, thlA, crt, bcd, and hbd genes in the rex-inactivated mutant were not significantly affected by MV addition and were higher than the levels measured in MV-exposed wild-type cells (Fig. 6A). These results strongly suggest that Rex responds to the increase in intracellular NADH/NAD+ ratio achieved by MV exposure, leading to derepression of Rex target genes. The ptb, buk, and ldh genes in the wild type were not induced when MV was present in the medium (Fig. 6A), although these genes were identified as Rex direct target genes. This may be explained by possible involvement in their regulation of other, still-unknown regulatory mechanisms in the presence of MV.
FIG 6.
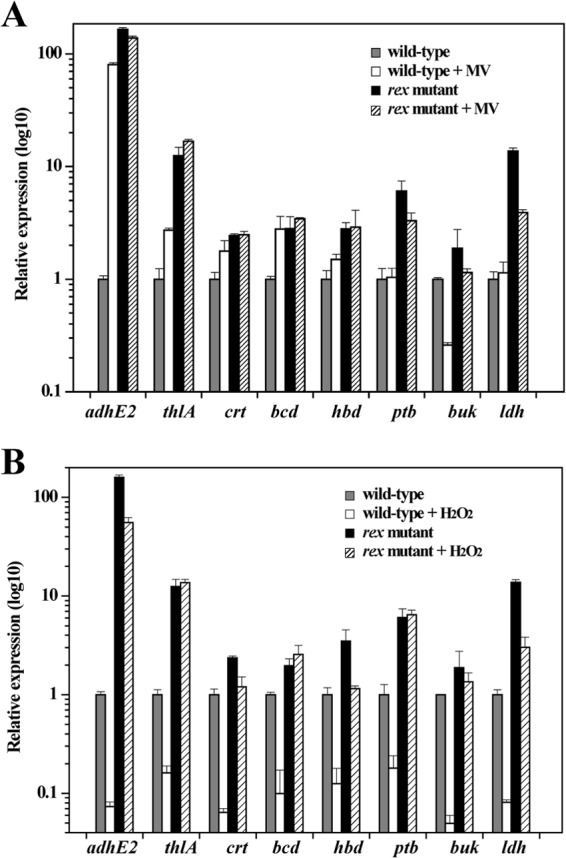
Effect of exposure to MV (A) or H2O2 (B) on transcript levels of the genes involved in fermentation in C. acetobutylicum wild-type and rex-inactivated mutant strains. The strains were grown in P2 minimal medium without or with addition of 1 mM MV or 30 μM H2O2. Total RNA was isolated from cells harvested in the exponential growth phase at an OD600 of about 2.0. The mRNA levels of each gene were determined by qRT-PCR and normalized to the gene expression in the wild-type strain grown in the absence of MV or H2O2. Data represent means ± standard deviations of values from six measurements starting from two independent cultures. Differences in the mRNA levels of adhE2, thlA, crt, bcd, and hbd genes in the wild type between the absence and presence of MV are statistically significant (P < 0.01), while the mRNA levels of all the studied genes in the wild type are significantly different (P < 0.01) upon exposure to H2O2.
On the other hand, exposure to H2O2 resulted in 5- to 20-fold-reduced mRNA levels of Rex regulon members adhE2, ldh, thlA, crt, bcd, hbd, ptb, and buk in the wild type (Fig. 6B). These genes in the rex-inactivated mutant showed unaltered or 2- to 4-fold-decreased expression levels in the presence of H2O2 compared to the culture without H2O2 addition. Thus, the effect of H2O2 addition on expression of these genes in the rex-inactivated mutant was much smaller than that in the wild type. These results indicate that Rex represses its target genes in response to the decrease in intracellular NADH/NAD+ ratio achieved by H2O2 exposure. Therefore, transcriptional analyses of C. acetobutylicum wild type and rex-inactivated mutant exposed to MV or H2O2 reveal that Rex monitors the NADH/NAD+ ratio in vivo to regulate expression of genes in its regulon.
In addition, we studied the effect of rex inactivation on the expression of central metabolic genes that are not members of the predicted Rex regulon (Fig. 7). They include the adhE1 gene encoding bifunctional alcohol/acetaldehyde dehydrogenase; ctfA, ctfB, and adc genes responsible for acetone formation; and bdhA and bdhB genes encoding butanol dehydrogenases. Expression of these genes in the wild type was significantly downregulated by MV addition, which largely coincides with previous reports (6). The transcript levels of adhE1, ctfA, ctfB, and adc genes in the rex-inactivated mutant were about 2-fold lower than those in the wild type, and expression of bdhA and bdhB genes was not significantly affected by rex mutation in the absence of MV. Rex proteins are known to be transcriptional repressors in other bacterial species, and no binding of C. acetobutylicum Rex was observed for the promoter regions of the adhE1-ctfAB operon and adc, bdhA, and bdhB genes in EMSAs (data not shown), suggesting that Rex may indirectly regulate the expression of the adhE1-ctfAB operon and the adc gene in C. acetobutylicum.
FIG 7.
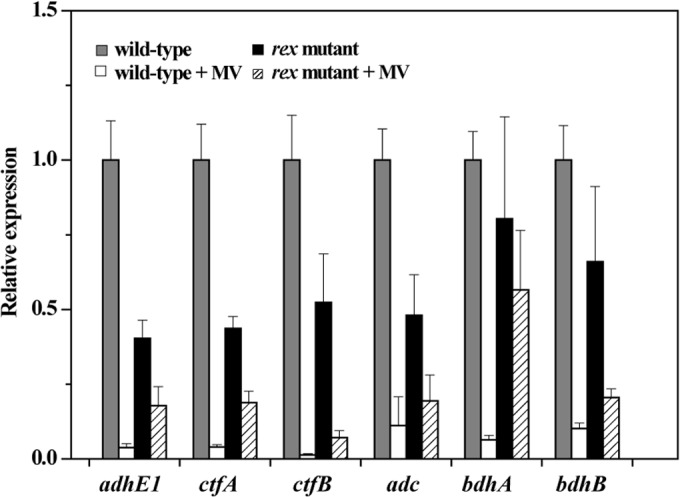
Effect of rex inactivation and MV addition on transcript levels of the fermentation genes that are not members of the predicted Rex regulon. Data represent means ± standard deviations of values from six measurements starting from two independent cultures and are normalized to the expression level in the wild type without MV exposure. Differences in the mRNA levels of adhE1, ctfA, ctfB, and adc genes between the wild type and rex-inactivated mutant are statistically significant (P < 0.01), while the mRNA levels of all the studied genes in the wild type are significantly different (P < 0.01) upon treatment with MV.
Rex modulates fermentation product formation and oxidative stress tolerance in C. acetobutylicum.
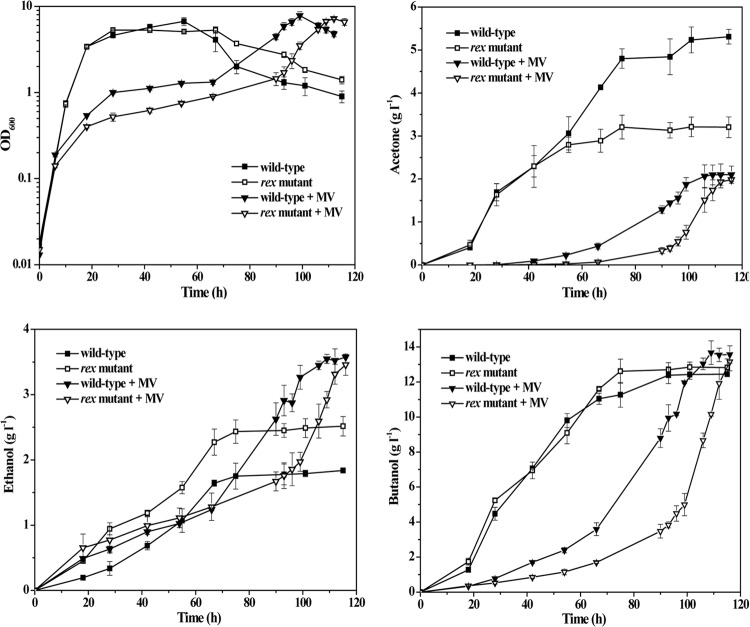
To elucidate the role of Rex in regulation of central metabolism in C. acetobutylicum, we compared the effects of MV exposure on fermentation product formation between the wild-type and rex-inactivated mutant strains. As shown in Fig. 8, the rex-inactivated mutant grew more slowly than the wild type in the presence of MV, whereas the growth rates of the two strains were similar in cultures without MV addition. Determination of fermentation product formation revealed different product spectra between the wild type and rex-inactivated mutant in the absence of MV (Fig. 8), which is in accordance with a recent report (10). Mutation of rex resulted in a significantly increased ethanol and a slightly elevated butanol production, while acetone synthesis was reduced; thus, the alcohol (butanol plus ethanol)-to-acetone ratio was improved from 2.7 to 4.8 (Fig. 8). Complementation of the rex-inactivated mutant by using a plasmid constitutively expressing rex restored a typical wild-type fermentation profile (see Fig. S3 in the supplemental material). Alcohol formation of the wild type was elevated by 22%, whereas acetone production was reduced 2.5-fold, by MV addition. In contrast, exposure of the rex-inactivated mutant to MV resulted in only marginally increased alcohol synthesis and 1.6-fold-decreased acetone production (Fig. 8). Therefore, the effect of MV addition on fermentation production formation in the wild type was more profound than in the rex-inactivated mutant. These results indicate that Rex modulates fermentation product formation and plays an important role in improving the alcohol-to-acetone ratio in C. acetobutylicum cultures.
FIG 8.
Cell growth and fermentation product formation in batch cultures of C. acetobutylicum wild-type and rex-inactivated mutant strains without or with addition of MV. The strains were grown in P2 minimal medium containing 60 g liter−1 of glucose. At an OD600 of about 0.15, MV was added to a final concentration of 1 mM. Cell growth was monitored spectrophotometrically at 600 nm. Formation of acetone, butanol, and ethanol was determined by gas chromatography. The data points and error bars represent means ± standard deviations of values from three independent cultures.
Given that the rex-inactivated mutant exhibited a sharper decrease in intracellular NADH/NAD+ ratio in response to H2O2 exposure than the wild type (Table 3), we wondered whether Rex deficiency would influence the capability of C. acetobutylicum to cope with oxidative stress. Hydrogen peroxide killing assays were used to assess the impact of rex mutation on oxidative stress tolerance of C. acetobutylicum. The wild-type and rex-inactivated mutant strains were incubated with an iron chelator (1 mM dipyridyl) and 50, 100, or 200 μM H2O2 for 30 min, and the survival of cells was determined as CFU. Results showed that the rex-inactivated mutant was more sensitive to H2O2 than the wild type (Fig. 9). The survival rate of the rex-inactivated mutant was approximately 2.5-fold lower than that of the wild type in the presence of 100 and 200 μM H2O2. Expression of a plasmid-encoded rex from a constitutive promoter in the rex-inactivated mutant restored a wild-type tolerance to H2O2. To understand why Rex deficiency increases susceptibility to oxidative stress, we compared between the wild-type and rex-inactivated mutant strains the expression levels of the genes encoding the components involved in detoxification. They include reverse rubrerythrins (rbr3A-rbr3B), desulfoferrodoxin (dfx), rubredoxin (rd), NADH-dependent rubredoxin oxidoreductase (nror), and the oxygen-reducing flavodiiron proteins (fprA1 and fprA2). As shown in Fig. 10, the expression levels of these genes were decreased 2- to 18-fold in the rex-inactivated mutant compared to those in the wild type. Following constitutive expression of a plasmid-borne rex in the rex-inactivated mutant, the transcription of these genes was largely restored. These genes are not preceded by a candidate Rex-binding site, suggesting an indirect effect of Rex on their activation. These results indicate that Rex is involved in regulation of oxidative stress response in C. acetobutylicum.
FIG 9.
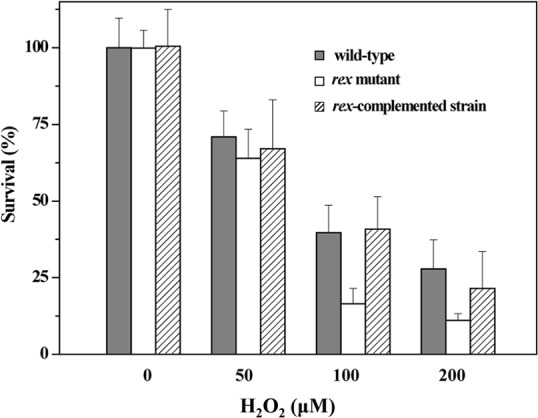
Survival of C. acetobutylicum wild-type, rex-inactivated mutant, and rex-complemented strains after hydrogen peroxide treatment. The strains were grown in P2 minimal medium to an OD600 of about 2.0. Then, cells were exposed to 1 mM iron chelator 2,2′-dipyridyl and the indicated concentrations of H2O2 or the equal volume of H2O. After incubation at 37°C for 30 min, the CFU were determined as the survival of cells and normalized to the number obtained for the nonstressed wild type. Data represent means ± standard deviations of values from three independent experiments.
FIG 10.
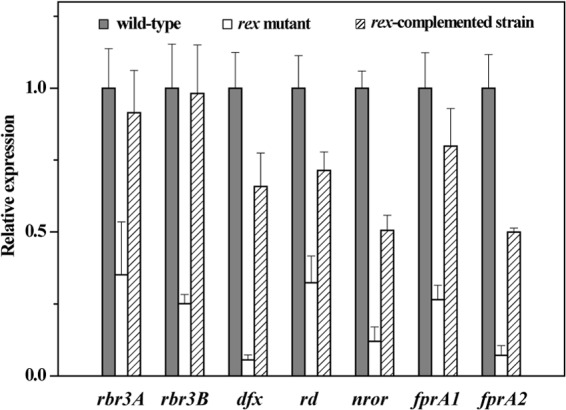
Influence of rex inactivation on transcript levels of the genes involved in detoxification of ROS and molecular O2 in C. acetobutylicum. Total RNA was isolated from the wild-type, rex-inactivated mutant, and rex-complemented strains grown in P2 minimal medium and harvested at an OD600 of about 2.0. The expression levels of each gene were normalized to the gene expression in the wild-type strain. Data represent means ± standard deviations of values from six measurements starting from two independent cultures. Differences in the mRNA levels of all the studied genes between the wild type and rex-inactivated mutant are statistically significant (P < 0.01).
DISCUSSION
In this work, we performed comparative genomic reconstruction of Rex regulons in 11 diverse clostridial species by combining the identification of candidate Rex-binding sites with cross-genomic comparison of regulons. Considerable variations were revealed in the sizes and gene contents of reconstructed Rex regulons between different species. The predicted Rex-binding sites in the genomes of Clostridium spp. were experimentally validated. New target genes of Rex in C. acetobutylicum, which are involved in fermentation, NAD biosynthesis, and sulfite reduction, were identified. Moreover, we compared the effects of exposure to methyl viologen or H2O2 on intracellular NADH/NAD+ ratio, expression of Rex targets, and physiology between C. acetobutylicum wild-type and rex-inactivated mutant strains. Our results demonstrate that Rex responds to changes in the NADH/NAD+ ratio in vivo to regulate gene expression and modulates fermentation product formation and oxidative stress response in C. acetobutylicum.
Addition of MV to cultures is one of the approaches that have been widely used to shift the metabolism of C. acetobutylicum away from hydrogen production toward alcohol formation (5). Under this condition, the intracellular NADH/NAD+ ratio increases; thus, Rex dissociates from its operator sites, leading to derepression of adhE2, thlA, crt, bcd, and hbd genes. Among the four known genes encoding alcohol or butanol dehydrogenases (i.e., adhE1, adhE2, bdhA, and bdhB), only the adhE2 gene is a direct target of Rex and upregulated by MV addition (Fig. 6 and 7). Consistently, previous studies have shown that the adhE2-encoded NADH-dependent aldehyde/alcohol dehydrogenase is related to an alcohologenic phenotype (41). Therefore, Rex-mediated regulation of adhE2, thlA, crt, bcd, and hbd genes probably plays a crucial role in enhanced alcohol production in MV-exposed C. acetobutylicum. In fact, we found that although MV addition resulted in a remarkable increase in NADH/NAD+ ratio in the rex-inactivated mutant (Table 3), it did not significantly affect the expression of adhE2, thlA, crt, bcd, and hbd genes and its influence on fermentation product formation in the rex-inactivated mutant was modest compared to that in the wild type (Fig. 6 and 8). The rex-inactivated mutant exhibited a notably reduced acetone production, which is consistent with the significantly decreased transcript levels of ctfA, ctfB, and adc genes (Fig. 7 and 8). This result suggests that Rex may modulate acetone formation in C. acetobutylicum by regulating expression of the acetone synthesis genes, although ctfA, ctfB, and adc genes are not Rex direct targets. Our speculation is that an indirect effect of Rex might occur via additional regulators such as Spo0A, which is a major regulator of sporulation and required for transcription of ctfA, ctfB, and adc genes in C. acetobutylicum (42, 43), which is consistent with the observation of impaired spore formation for the rex-inactivated mutant (data not shown). Therefore, our results reveal that Rex plays an important role in improving the alcohol-to-acetone ratio in C. acetobutylicum cultures.
In addition to MV addition, other approaches such as carbon monoxide sparging or utilization of glycerol as a substrate have also been used to shift the solvent ratio toward butanol in C. acetobutylicum cultures. These approaches aim to inhibit hydrogenase activity, and the reduction of hydrogen formation results in an increased electron flow toward butanol synthesis. We speculate that Rex-dependent regulation in response to intracellular NADH/NAD+ ratio is also involved in these physiological interventions. Genetic manipulations have also been applied to reduce by-product formation of C. acetobutylicum; however, most of these attempts did not result in a desired butanol producer (44, 45). For example, Jiang et al. constructed an adc-inactivated mutant which produced much less acetone, but butanol titers were also reduced and could be restored to the level of the parent strain only with pH control and MV addition to cultures (45). Based on understanding of redox-dependent regulatory mechanisms, alternative engineering targets could be designed to alter the intracellular redox status and improve the butanol production of C. acetobutylicum.
The strictly anaerobic clostridia can withstand limited air exposure upon activation of their reductive machinery for the scavenging of ROS and molecular O2. We found that the C. acetobutylicum rex-inactivated mutant is more susceptible to H2O2 killing than the wild type (Fig. 9), indicating that Rex modulates oxidative stress tolerance in this obligate anaerobe. Although involvement of Rex in regulation of the oxidative stress response has also been reported for the facultative anaerobe S. mutans (15), the mechanisms may be different between S. mutans and clostridia. Our results demonstrate that Rex responds to the decrease in intracellular NADH/NAD+ ratio achieved by H2O2 exposure to repress its target genes, including those encoding NADH-consuming enzymes in central metabolism (e.g., ldh, adhE2, bcd-etfBA, and hbd) (Fig. 6). This may increase the availability of reducing power needed for reduction of H2O2. Moreover, expression of the genes encoding the components involved in detoxification of ROS and oxygen was downregulated in the rex-inactivated mutant (Fig. 10), although these genes are not preceded by Rex-binding sites, suggesting that Rex could indirectly enhance the detoxification system in C. acetobutylicum. Because these genes are primary targets of the transcriptional repressor PerR in C. acetobutylicum according to a previous study (46), we speculate that Rex may regulate expression of these genes via PerR or other transcription factors. However, more work is needed to elucidate the mechanism of the involvement of Rex in regulation of the oxidative stress response. The Rex and PerR regulatory systems that both are widely distributed in Clostridium species seem to play important roles in the oxidative stress defense in C. acetobutylicum, but they sense different signals and possess different direct targets. Whereas Rex senses intracellular NADH/NAD+ ratio to regulate many fermentation genes, PerR is a peroxide sensor that negatively controls expression of the genes involved in the oxygen and ROS detoxification.
An important role of Rex in maintaining NADH/NAD+ homeostasis in C. acetobutylicum was revealed based on our measurements of intracellular NADH and NAD+ concentrations. When exposed to MV or H2O2, the rex-inactivated mutant exhibited larger fluctuations in the NADH/NAD+ ratio than the wild type (Table 3). Further studies are required to identify the mechanism by which Rex functions to prevent large fluctuations in the NADH/NAD+ ratio in C. acetobutylicum. It is hypothesized that Rex regulates the expression of NADH-consuming enzymes (e.g., aldehyde/alcohol dehydrogenase) in response to increased NAD(P)H availability or oxidative stress to help maintain redox homeostasis in the cell. In addition to the direct and indirect targets identified in this study, Rex may also control the expression of many other enzymes involved in the redox balance in C. acetobutylicum, and the transcriptome analysis of the rex-inactivated mutant is now under way. It is worth noting that the influence of exposure to MV or H2O2 on intracellular NAD+ concentration was more profound than that on NADH concentration (Table 3). This suggests that NAD+ has an important role in modulating the DNA-binding activity of Rex in C. acetobutylicum, although in vitro binding assays showed that the binding affinity of Rex for NAD+ is much lower than that for NADH (Fig. 5) (16). NAD+ competes with NADH for binding to Rex, thereby impairing the inhibitory effect of NADH on Rex-DNA complex formation. Allosteric activation for DNA binding by NAD+ has been reported for B. subtilis Rex (16) and may also exist for C. acetobutylicum Rex.
This study gains an insight into the potential regulatory role of Rex in clostridial species other than C. acetobutylicum based on comparative genomic reconstruction of Rex regulons. In C. beijerinckii, another solvent-producing species, the predicted Rex regulon contains genes involved in fermentation (pflBA, adhA, adhA2, thlA, thlA2, crt-bcd-etfBA-hbd, ptb-buk, butA, and fld-Cbei_4318) and hydrogen production (hydB). Experimental evidence that Rex binds upstream of these target genes in C. beijerinckii was provided by EMSAs (Fig. 3). Among these candidate Rex targets, the adhA (Cbei_2181) and adhA2 (Cbei_1722) genes encode two primary alcohol dehydrogenases responsible for production of butanol and ethanol in C. beijerinckii (47). The hydB gene (Cbei_0327) codes for a hydrogenase that uses reduced ferredoxin as the electron donor, and reduced ferredoxin could be generated by pyruvate ferredoxin/flavodoxin oxidoreductase encoded by Cbei_4318. The predicted regulation of both hydB and Cbei_4318 expression by Rex suggests that Rex may be involved in modulation of hydrogen production in C. beijerinckii. Consistently, previous studies have shown that the presence of reduced electron shuttling compounds such as anthrahydroquinone-2,6-disulfonate increased the hydrogen yield of C. beijerinckii, suggesting that hydrogen production is modulated by the redox status in the cell (48). To assess the regulatory role of Rex in C. beijerinckii, we constructed the rex-inactivated mutant of C. beijerinckii. Our results showed that rex inactivation did not significantly alter the fermentation product spectra in C. beijerinckii, although it resulted in derepression of predicted Rex target genes (see Fig. S4 in the supplemental material). One possible explanation is that Rex coordinately regulates alcohol formation and hydrogen production and the distribution of electron flow through these pathways is generally rigid in C. beijerinckii. So far, manipulation of the redox balance to shift the electron flow away from hydrogen production toward alcohol production is limited to C. acetobutylicum, and its successful application in C. beijerinckii has never been reported. Therefore, the redox-dependent regulatory mechanisms in C. beijerinckii probably differ from that in C. acetobutylicum. The different metabolic responses to perturbations in cellular redox balance between the two solventogenic clostridia may be partly attributed to the variability of the Rex regulon members.
The acetogenic C. carboxidivorans, C. ljungdahlii, and Clostridium autoethanogenum are capable of using the Wood-Ljungdahl pathway to fix CO2 or CO and convert it into acetyl-CoA. This feature makes them become promising production strains for industrial syngas fermentations (49). However, regulation of the Wood-Ljungdahl pathway genes in these acetogenic clostridia remains to be explored. Here, we predicted a candidate Rex-binding site located upstream of the Wood-Ljungdahl pathway gene cluster in the genomes of C. carboxidivorans and C. ljungdahlii. Binding of Rex to the promoter region of this gene cluster in both clostridia was verified by EMSAs (Fig. 3). A putative Rex-binding site upstream of the Wood-Ljungdahl pathway gene cluster was also identified in the genome of C. autoethanogenum (data not shown). This suggests that Rex may play a role in regulation of CO or CO2 reduction in C. carboxidivorans, C. ljungdahlii, and C. autoethanogenum. Whether rex inactivation will lead to derepression of Wood-Ljungdahl pathway genes and improvement of syngas fermentation in these acetogenic clostridia needs to be tested. Nevertheless, this study offers an insight into redox-dependent gene regulation in these species, which could be useful for designing sophisticated metabolic engineering approaches to increase the product yields of syngas fermentation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Qing Zhang from the University of Arkansas for Medical Sciences for helpful discussions.
This work was supported in part by the National Basic Research Program of China (973: 2012CB721101) and the National Natural Science Foundation of China (31121001 and 31470168).
Footnotes
Published ahead of print 2 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02037-14.
REFERENCES
- 1.Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET. 2012. Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr. Opin. Biotechnol. 23:364–381. 10.1016/j.copbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Lütke-Eversloh T, Bahl H. 2011. Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr. Opin. Biotechnol. 22:634–647. 10.1016/j.copbio.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Dürre P. 2007. Biobutanol: an attractive biofuel. J. Biotechnol. 2:1525–1534. 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- 4.Nölling J, Breton G, Omelchenko MV, Makarova KS, Zeng QD, Gibson R, Lee HM, Dubois J, Qiu DY, Hitti J, Wolf YI, Tatusov RL, Sabathe F, Doucette-Stamm L, Soucaille P, Daly MJ, Bennett GN, Koonin EV, Smith DR. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823–4838. 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girbal L, Soucaille P. 1998. Regulation of solvent production in Clostridium acetobutylicum. Trends Biotechnol. 16:11–16. 10.1016/S0167-7799(97)01141-4. [DOI] [Google Scholar]
- 6.Hönicke D, Janssen H, Grimmler C, Ehrenreich A, Lütke-Eversloh T. 2012. Global transcriptional changes of Clostridium acetobutylicum cultures with increased butanol:acetone ratios. N. Biotechnol. 29:485–493. 10.1016/j.nbt.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Hillmann F, Riebe O, Fischer R-J, Mot A, Caranto JD, Kurtz DM, Jr, Bahl H. 2009. Reductive dioxygen scavenging by flavo-diiron proteins of Clostridium acetobutylicum. FEBS Lett. 583:241–245. 10.1016/j.febslet.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riebe O, Fischer R-J, Wampler DA, Kurtz DM, Jr, Bahl H. 2009. Pathway for H2O2 and O2 detoxification in Clostridium acetobutylicum. Microbiology 155:16–24. 10.1099/mic.0.022756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawasaki S, Sakai Y, Takahashi T, Suzuki I, Niimura Y. 2009. O2 and reactive oxygen species detoxification complex, composed of O2-responsive NADH:rubredoxin oxidoreductase-flavoprotein A2-desulfoferrodoxin operon enzymes, rubperoxin, and rubredoxin, in Clostridium acetobutylicum. Appl. Environ. Microbiol. 75:1021–1029. 10.1128/AEM.01425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wietzke M, Bahl H. 2012. The redox-sensing protein Rex, a transcriptional regulator of solventogenesis in Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 96:749–761. 10.1007/s00253-012-4112-2. [DOI] [PubMed] [Google Scholar]
- 11.Brekasis D, Paget MSB. 2003. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J. 22:4856–4865. 10.1093/emboj/cdg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gyan S, Shiohira Y, Sato I, Takeuchi M, Sato T. 2006. Regulatory loop between redox sensing of the NADH/NAD+ ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J. Bacteriol. 188:7062–7071. 10.1128/JB.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagels M, Fuchs S, Pané-Farré J, Kohler C, Menschner L, Hecker M, McNamarra PJ, Bauer MC, von Wachenfeldt C, Liebeke M, Lalk M, Sander G, von Eiff C, Proctor RA, Engelmann S. 2010. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76:1142–1161. 10.1111/j.1365-2958.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vesic D, Kristich CJ. 2013. A Rex family transcriptional repressor influences H2O2 accumulation by Enterococcus faecalis. J. Bacteriol. 195:1815–1824. 10.1128/JB.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bitoun JP, Liao S, Yao X, Xie GG, Wen ZT. 2012. The redox-sensing regulator Rex modulates central carbon metabolism, stress tolerance response and biofilm formation by Streptococcus mutans. PLoS One 7:e44766. 10.1371/journal.pone.0044766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang E, Bauer MC, Rogstam A, Linse S, Logan DT, von Wachenfeldt C. 2008. Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol. Microbiol. 69:466–478. 10.1111/j.1365-2958.2008.06295.x. [DOI] [PubMed] [Google Scholar]
- 17.McLaughlin KJ, Strain-Damere CM, Xie K, Brekasis D, Soares AS, Paget MSB, Kielkopf CL. 2010. Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol. Cell 38:563–575. 10.1016/j.molcel.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Wang E, Ikonen TP, Knaapila M, Svergun D, Logan DT, von Wachenfeld C. 2011. Small-angle X-ray scattering study of a Rex family repressor: conformational response to NADH and NAD(+) binding in solution. J. Mol. Biol. 408:670–683. 10.1016/j.jmb.2011.02.050. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 21.Rodionov DA. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 107:3467–3497. 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, Gelfand MS, Arkin AP, Mironov AA, Dubchak I. 2010. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 38:W299–W307. 10.1093/nar/gkq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mironov AA, Vinokurova NP, Gelfand MS. 2000. Software for analysis of bacterial genomes. Mol. Biol. 34:222–231. 10.1007/BF02759643. [DOI] [PubMed] [Google Scholar]
- 24.Ravcheev DA, Li X, Latif H, Zengler K, Leyn SA, Korostelev YD, Kazakov AE, Novichkov PS, Osterman AL, Rodionov DA. 2012. Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex. J. Bacteriol. 194:1145–1157. 10.1128/JB.06412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702. 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiesenborn DP, Rudolph FB, Papoutsakis ET. 1988. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl. Environ. Microbiol. 54:2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baer SH, Blaschek HP, Smith TL. 1987. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl. Environ. Microbiol. 53:2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642. 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 29.Hillmann F, Fischer R-J, Saint-Prix F, Girbal L, Bahl H. 2008. PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum. Mol. Microbiol. 68:848–860. 10.1111/j.1365-2958.2008.06192.x. [DOI] [PubMed] [Google Scholar]
- 30.Shao L, Hu S, Yang Y, Gu Y, Chen J, Jiang W, Yang S. 2007. Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum. Cell Res. 17:963–965. 10.1038/cr.2007.91. [DOI] [PubMed] [Google Scholar]
- 31.Xiao H, Gu Y, Ning Y, Yang Y, Mitchell WJ, Jiang W, Yang S. 2011. Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate-dependent phosphotransferase system-deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose. Appl. Environ. Microbiol. 77:7886–7895. 10.1128/AEM.00644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mermelstein LD, Papoutsakis ET. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59:1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren C, Gu Y, Hu S, Wu Y, Wang P, Yang Y, Yang C, Yang S, Jiang W. 2010. Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum. Metab. Eng. 12:446–454. 10.1016/j.ymben.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Tummala SB, Welker NE, Papoutsakis ET. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alsaker KV, Paredes CJ, Papoutsakis ET. 2005. Design, optimization and validation of genomic DNA microarrays for examining the Clostridium acetobutylicum transcriptome. Biotechnol. Bioprocess Eng. 10:432–443. 10.1007/BF02989826. [DOI] [Google Scholar]
- 36.Yang C, Rodionov DA, Rodionova IA, Li X, Osterman AL. 2008. Glycerate 2-kinase of Thermotoga maritima and genomic reconstruction of related metabolic pathways. J. Bacteriol. 190:1773–1782. 10.1128/JB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terracciano JS, Kashket ER. 1986. Intracellular conditions required for initiation of solvent production by Clostridium acetobutylicum. Appl. Environ. Microbiol. 52:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JAE. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812–826. 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 39.Amador-Noguez D, Brasg IA, Feng X-J, Roquet N, Rabinowitz JD. 2011. Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum. Appl. Environ. Microbiol. 77:7984–7997. 10.1128/AEM.05374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chauvatcharin S, Siripatana C, Seki T, Takagi M, Yoshida T. 1998. Metabolism analysis and on-line physiological state diagnosis of acetone-butanol fermentation. Biotechnol. Bioeng. 58:561–571. . [DOI] [PubMed] [Google Scholar]
- 41.Fontaine L, Meynial-Salles I, Girbal L, Yang XH, Croux C, Soucaille P. 2002. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:821–830. 10.1128/JB.184.3.821-830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravagnani A, Jennert KCB, Steiner E, Grunberg R, Jefferies JR, Wilkinson SR, Young DI, Tidswell EC, Brown DP, Youngman P, Morris JG, Young M. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37:1172–1185. 10.1046/j.1365-2958.2000.02071.x. [DOI] [PubMed] [Google Scholar]
- 43.Alsaker KV, Spitzer TR, Papoutsakis ET. 2004. Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell's response to butanol stress. J. Bacteriol. 186:1959–1971. 10.1128/JB.186.7.1959-1971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehmann D, Hönicke D, Ehrenreich A, Schmidt M, Weuster-Botz D, Bahl H, Luetke-Eversloh T. 2012. Modifying the product pattern of Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 94:743–754. 10.1007/s00253-011-3852-8. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y, Xu C, Dong F, Yang Y, Jiang W, Yang S. 2009. Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab. Eng. 11:284–291. 10.1016/j.ymben.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Hillmann F, Döring C, Riebe O, Ehrenreich A, Fischer R-J, Bahl H. 2009. The role of PerR in O2-affected gene expression of Clostridium acetobutylicum. J. Bacteriol. 191:6082–6093. 10.1128/JB.00351-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Li X, Mao Y, Blaschek HP. 2012. Genome-wide dynamic transcriptional profiling in Clostridium beijerinckii NCIMB 8052 using single-nucleotide resolution RNA-Seq. BMC Genomics 13:102. 10.1186/1471-2164-13-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatch JL, Finneran KT. 2008. Influence of reduced electron shuttling compounds on biological H2 production in the fermentative pure culture Clostridium beijerinckii. Curr. Microbiol. 56:268–273. 10.1007/s00284-007-9073-9. [DOI] [PubMed] [Google Scholar]
- 49.Bengelsdorf FR, Straub M, Dürre P. 2013. Bacterial synthesis gas (syngas) fermentation. Environ. Technol. 34:1639–1651. 10.1080/09593330.2013.827747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.