Abstract
Upon physical contact with sibling cells, myxobacteria transiently fuse their outer membranes (OMs) and exchange OM proteins and lipids. From previous work, TraA and TraB were identified to be essential factors for OM exchange (OME) in donor and recipient cells. To define the genetic complexity of OME, we carried out a comprehensive forward genetic screen. The screen was based on the observation that Myxococcus xanthus nonmotile cells, by a Tra-dependent mechanism, block swarm expansion of motile cells when mixed. Thus, mutants defective in OME or a downstream responsive pathway were readily identified as escape flares from mixed inocula seeded on agar. This screen was surprisingly powerful, as we found >50 mutants defective in OME. Importantly, all of the mutations mapped to the traAB operon, suggesting that there may be few, if any, proteins besides TraA and TraB directly required for OME. We also found a second and phenotypically different class of mutants that exhibited wild-type OME but were defective in a responsive pathway. This pathway is postulated to control inner membrane homeostasis by covalently attaching amino acids to phospholipids. The identified proteins are homologous to the Staphylococcus aureus MprF protein, which is involved in membrane adaptation and antibiotic resistance. Interestingly, we also found that a small number of nonmotile cells were sufficient to block the swarming behavior of a large gliding-proficient population. This result suggests that an OME-derived signal could be amplified from a few nonmotile producers to act on many responder cells.
INTRODUCTION
Myxobacteria are Gram-negative gliding bacteria that exhibit complex social behaviors. One of these interesting behaviors is their ability to transiently fuse their outer membranes (OMs) and exchange contents (1). The exchange of OM material can be visualized in live cells with fluorescent protein reporters fused to an OM lipoprotein signal sequence or by use of fluorescent OM lipids (2, 3). Phenotypically, OM exchange (OME) can be monitored by extracellular complementation between certain motility mutants. In this assay, a small subset of gliding mutants is transiently rescued or stimulated by the transfer of missing motility proteins from a donor strain that contains the corresponding wild-type protein (4, 5). For stimulation to occur, the cargo protein must reside in the OM, as cytoplasmic and inner membrane proteins are not transferred (2). Because Myxococcus xanthus expresses >400 lipoproteins (6), OME between cells probably involves hundreds of different OM proteins and, consequently, is likely to affect a variety of cellular processes (2). Although the function of OME in wild-type cells is not fully understood, it may play a role in establishing envelope homeostasis and cell-cell communication and may generally assist in the transition from individual cells to coherent multicellular populations (7).
To investigate the mechanism and biological function of OME, we sought to identify cellular factors required for OME. To do this, in a prior study, we characterized a historic mutant derived from the Kaiser Laboratory strain collection (3). This heretofore uncharacterized nonmotile mutant (DK396) was of interest because it universally failed to stimulate all six classes of cgl and tgl motility mutants (4, 8, 9), whose protein products function in adventurous (A)- and social (S)-gliding motility, respectively (for reviews, see references 10 to 12). The genome of this strain was sequenced, and the causative mutation was identified in a gene that we named traA. Interestingly, TraA function is required in both the donor and recipient strains, suggesting that OM transfer is bidirectional. Subsequently, we found that the downstream gene in the bicistronic operon, traB, is also required for OME (3). Immunolocalization studies showed that TraA localizes on the cell surface, with a tendency to reside near the cell poles (13). Phylogenetic and molecular analyses indicated that TraA functions as a homotypic cell surface receptor/adhesin that recognizes other cells bearing identical or similar TraA alleles (13). Thus, the intimate cellular process of sharing OM contents occurs only between siblings or cells that bear a nearly identical traA allele. Molecular recognition within and between species is determined by a polymorphic domain (PA14-like) within TraA. The specific function of TraB is unknown, but it is likely to interact with TraA (3).
In this study, we sought to conduct the first forward genetic screen to define the total complement of proteins involved in OME. This screen was based on the finding that nonmotile mutants block swarm expansion of motile strains by a Tra-dependent mechanism when mixed together (3, 13). To understand the basis of the screen, it is important to recognize that swarm inhibition is distinct from stimulation; the former blocks wild-type motility, and the latter rescues the motility of certain mutants (4). Although the mechanism of swarm inhibition is not well understood, it is thought to involve a signal that is generated from the physiological state of the nonmotile cells, as swarm inhibition initiates after a 1-day incubation on agar surfaces (3, 14). In contrast, stimulation primarily occurs during the first day of incubation and subsides in subsequent days; thus, there appears to be a temporal switch for when stimulation or swarm inhibition can occur. Stimulation and swarm inhibition are similar in the sense that both require cell-cell contact, a hard agar surface, and TraAB function. Here, we exploited the swarm inhibition phenotype as a means to identify mutants defective in OME. In this regard, mutants were easily identified because they resulted in the production of escape flares derived from the motile strain (Fig. 1). The second goal of this work was to identify downstream factors or a pathway(s) that responds to cell-cell interactions mediated by OME. Mutants that affect these factors or this pathway(s) should undergo normal OME but exhibit a relief of swarm inhibition phenotype, because they are defective in perceiving or responding to OME.
FIG 1.
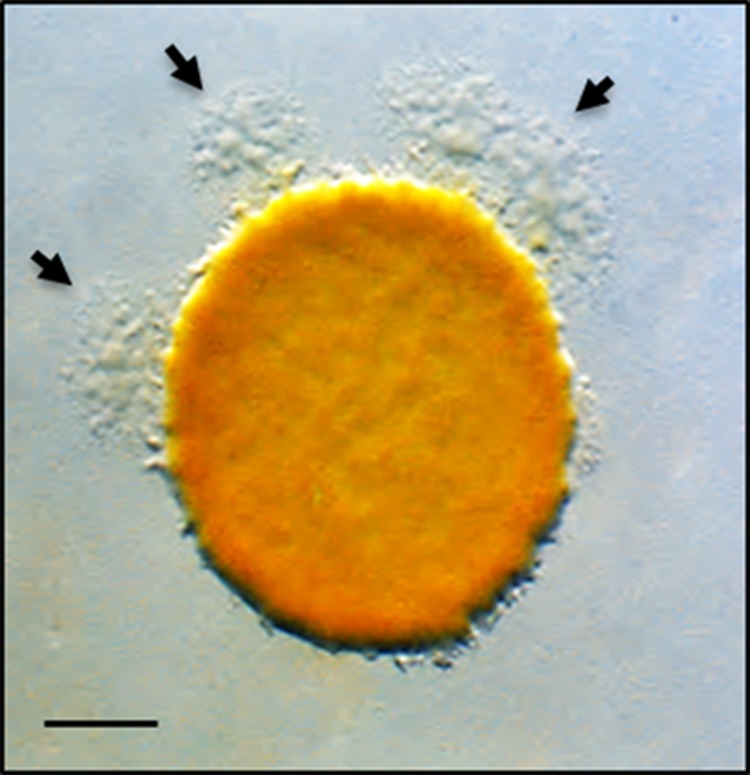
Micrograph showing how escape flare mutants were identified. A nonmotile strain was mixed at a 1:1 ratio with an A-motile strain that was mutagenized with a transposon (representing a pool of mutagenized genomes). After 5 days of incubation, escape flares (arrows) are readily seen. Bar, 1 mm.
MATERIALS AND METHODS
Strains and media.
The bacterial strains used in the study are listed in Table 1 (15, 16). M. xanthus was grown in the dark at 33°C in CTT medium (1% Casitone, 10 mM Tris-HCl [pH 7.6], 8 mM MgSO4, 1 mM KH2PO4) without or with kanamycin (Km; 50 μg/ml), as needed. In the genetic screen and in stimulation assays, 1/2 CTT plates, where the Casitone concentration was reduced to 0.5% and 2 mM CaCl2 was added with 1.5% agar, were used. To generate time-lapse movies of motility, 1/4 CTT 0.8% agarose pads (no CaCl2) were made on glass slides. To make the media, either tap or filter purified water was used. For routine cloning, Escherichia coli DH5α pir+ was grown at 37°C in LB medium with 50 μg/ml Km.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant features or sequence | Source or reference |
|---|---|---|
| Strains | ||
| DH5α pir+ | E. coli cloning strain | Lab collection |
| DK1217 | A− S+ aglB1 | 13 |
| DK8601 | A− S− aglB1 ΔpilA::Tc Tcr | 12 |
| DK8606 | A− S− aglB1 ΔpilA PpilA-gfp Kmr | 12 |
| DK8615 | A+ S− ΔpilQ | 13 |
| DK10410 | A+ S− ΔpilA | 12 |
| DW709 | DK10410 ΔpilA PpilA-gfp Kmr | 28 |
| DW1056 | DK8606 PpilA-SSOM-mCherry(pXW6) PpilA-gfp Smr Tcr Kmr | 2 |
| DW1415 | DK8615 traA::pDP2 Kmr | 3 |
| DW1466 | A− S− ΔcglC Δtgl::Tc Tcr | 3 |
| DW1467 | DK8601 aglB1 ΔtraA ΔpilA::Tc Tcr | 11 |
| DW1478 | DK8615 PpilA-SSOM-mCherry(pXW6) Smr | This study |
| DW1479 | DW1478 traA::pDP2 Kmr Smr | This study |
| DW1601 | DK8615 omrA::EZ-Tn5 Kmr | This study |
| DW1602 | DK8615 omrA::mini-Himar-lacZ Kmr | This study |
| DW1603 | DK8615 omrA::mini-Tn5 Kmr | This study |
| DW1604 | DK8601 aglB1 ΔpilA::Tc omrA::mini-Tn5 Tcr Kmr | This study |
| DW1605 | DK1217 omrA::mini-Tn5 Kmr | This study |
| DW1606 | DK8615 ΔphpA (markerless) | This study |
| DW1607 | DK1217 ΔphpA (markerless) | This study |
| DW1608 | DK8615 ΔMXAN_4428 (markerless) | This study |
| DW1609 | DK8615 omrB::pAD3 Kmr Zmr | This study |
| DW1610 | DK8601 omrB::pAD3 Kmr Zmr | This study |
| DW1611 | DW1603 PpilA-SSOM-mCherry(pXW6) Smr Kmr | This study |
| DW1612 | DW1466 omrA::mini-Tn5 Kmr Tcr | This study |
| Plasmids | ||
| pMini-Himar-lacZ | mariner-derived transposon with lacZ oriR6K, Kmr | Heidi Kaplan |
| pRL27 | Mini-Tn5 oriR6K hyperactive transposase, Kmr | 14 |
| pCR2.1 TOPO XL | Cloning vector, Kmr Zmr | Life Technologies |
| pCR2.1 TOPO TA | Cloning vector, Kmr | Life Technologies |
| pDP2 | traA (insertion cassette) in pCR2.1 TOPO TA, Kmr | 3 |
| pDP21 | PpilA-RBSsyn-traAB cloned in pSWU1, Kmr | 3 |
| pXW6 | PpilA-SSOM-mCherry cassette cloned in pKSAT, Smr | 2 |
| pAD1 | ΔphpA cassette in pBJ114, Kmr | This study |
| pAD2 | ΔMXAN_4428 cassette in pBJ114, Kmr | This study |
| pAD3 | omrB (insertion cassette) in pCR2.1 TOPO XL, Kmr Zmr | This study |
| Primers | ||
| Mariner primer | TGTGTTTTTCTTTGTTAGACCG | |
| EZ-Tn5 KAN-2 Fwd | ACCTACAACAAAGCTCTCATCAACC | |
| EZ-Tn5 KAN-2 Rev | GCAATGTAACATCAGAGATTTTGAG | |
| Tpn RL13 (mini-Tn5) | CAGCAACACCTTCTTCACGA | |
| Tpn RL17 (mini-Tn5) | AACAAGCCAGGGATGTAACG | |
| MXAN_4428delupstrfwd | TATGACCATGATTACGCCAAGCTTAGGTTCACCTCGGTGGC | |
| MXAN_4428delupstrrev | ATCCCAGCGCTGTGAAGCTGTGTGGATGGGACACGTCAGGGT | |
| MXAN_4428deldwnstrfwd | CACACAGCTTCACAGCGCTGGGATGCACGCCAGCACGGCGAC | |
| MXAN_4428deldwnstrrev | CGTTGTAAAACGACGGCCAGTGAATTCCCTCATGAGACGGGCGAAGC | |
| MXAN_4427delupstrfwd | TATGACCATGATTACGCCAAGCTTCTCTCGCTCGTGGGTGAC | |
| MXAN_4427delupstrrev | CCAGCGCTGTGAAGCTGTGTGGCAATGCAGGTCGACAAAG | |
| MXAN_4427deldwnstrfwd | CACACAGCTTCACAGCGCTGGAATCCCGCGACTTTGTTG | |
| MXAN_4427deldwnstrrev | CGTTGTAAAACGACGGCCAGTGAATTCAGTACAGCGCCGTGTAGAGG | |
| MXAN_6818 KO FWD | GGACAGACGGTGTTCTCGTT | |
| MXAN_6818 KO REV | ACAGGAAGCCCACCACCT |
Genetic screen.
The pMini-Himar-lacZ transposon, pRL27 (17), and EZ-Tn5 (Epicentre) were transformed into M. xanthus (DK8615 or DK10410) using standard protocols (18) (Table 1). After electroporation, the cells were allowed to recover by shaking at 33°C for 5 to 6 h. After ∼6 days of incubation, the colonies from each transformant plate were pooled, and the cell density of each pool was concentrated to ∼1.5 × 109 CFU/ml. A nonmotile M. xanthus strain (DK8601) was also grown and prepared to a similar cell density. Each individual mutant pool and DK8601 were mixed at equal volumes, and about a dozen 5-μl aliquots were pipetted onto 1/2 CTT Omni (Nunc) agar plates with 2 mM CaCl2. The plates were incubated at 33°C and observed for emergent flares after 3 to 5 days. Cells from these flares were collected with a sterile toothpick and transferred onto fresh CTT-Km plates. To verify mutant phenotypes, purified isolates were grown to 3 × 108 CFU/ml and mixed at a 1:1 ratio with DK8601, and aliquots were pipetted onto agar plates. After 2 days of incubation, the spots were visually inspected for emergent flares.
To sequence transposon insertions from M. xanthus, genomic DNA was isolated with an UltraClean microbial DNA isolation kit (MO BIO Laboratories, Inc.) and digested with restriction enzymes (pMini-Himar-lacZ and mini-Tn5, SacII; EZ-Tn5, EcoRI, HindIII, or SalI; New England BioLabs). The digested DNA was ligated with T4 DNA ligase (New England BioLabs) and transformed into E. coli. Candidate clones were then isolated, and in the case of pMini-Himar-lacZ, derived clones were analyzed by diagnostic MluI restriction digestion. Plasmid DNA was sequenced at the University of Wyoming Macromolecular Analysis Core facility with appropriately named primers, which are described in Table 1.
Strain construction.
To backcross transposon insertions into different strains, generalized transduction was done with bacteriophage Mx8. In-frame gene deletion constructs for MXAN_4427 and MXAN_4428 were made using a Gibson cloning strategy (New England BioLabs), wherein PCR products were ligated with the pBJ114 plasmid (19). Restriction-verified plasmids were then electroporated into M. xanthus and plated on CTT-Km plates. Transformants were grown in the absence of Km and then counterselected on CTT plates with 2% galactose. In-frame deletion mutants were confirmed by PCR using primers flanking the deletion (Table 1). Gene disruptions were made using internal gene fragments that were PCR amplified and ligated into the pCR2.1 TOPO XL vector (Table 1). Restriction-verified clones were then transformed into M. xanthus and selected for on CTT-Km plates.
Microbial assays.
To test for protein transfer, cells were grown and concentrated to 3 × 109 CFU/ml in TPM buffer (10 mM Tris-HCl [pH 7.6], 8 mM MgSO4, 1 mM KH2PO4). The cells were then mixed with a nonmotile double-labeled DW1056 strain (in which the cytoplasm was labeled with green fluorescent protein [GFPCyto] and the OM was labeled with an mCherry reporter fused to a signal sequence [SSOM-mCherry]) at a 1:1 ratio. Aliquots of the mixture (5 μl each) were transferred to TPM buffer–0.8% agarose pads and incubated in a humid chamber. After 6 h, the cells were observed by phase-contrast and fluorescence microscopy with a ×20 objective lens for the presence of SSOM-mCherry in the emergent flares (2). In addition, transfer was evaluated with a 100× oil immersion lens. Here mutants that express GFP (recipient) were grown and concentrated as described above. These cells were then mixed with donor M. xanthus cells expressing SSOM-mCherry, and the cell mixture was placed on a TPM buffer plate at a 1:5 ratio (recipient/donor) and incubated in a humid chamber for 2 h. The cells were then collected, washed twice with TPM buffer, and observed on a glass slide. Micrographs were obtained with a phase-contrast/fluorescence microscope that was coupled to digital imaging systems, as described previously (2).
Stimulation tests were done by mixing a strain that is nonmotile but stimulatable (DW1466 ΔcglC tgl::Tc) with various nonmotile donor mutants at a 1:1 ratio and then transferring the cells to 1/2 CTT agar plates with 2 mM CaCl2. To monitor A-gliding motility, the cell density was measured, and cells were concentrated to 1.5 × 109 CFU/ml and placed on 1/2 CTT with 1.5% agar and 2 mM CaCl2. To monitor S-gliding motility, the cells were concentrated as described above and pipetted onto semisolid (0.5% agar) CTT plates with 2 mM MgSO4.
RESULTS
Genetic screen.
To define the OME gene set, we conducted a genetic screen in M. xanthus. Initially, a pilot transposon-based screen was done for donor mutants defective in stimulation. Although this strategy was feasible, we encountered two problems. First, the screen was laborious. Second, mutants which initially appeared to be defective in stimulation based on a cross-streak assay against a stimulatable strain were subsequently found to be false positive, when a more sensitive culture-based assay was used (see below), at a problematically high frequency. The false positives appeared to stem from undefined physiological differences among isolates that arose during screening. On the basis of these outcomes, a different approach was sought.
As described above, in a prior study we discovered that OME can regulate the swarming behavior of M. xanthus cells that are proficient for gliding motility (3). Specifically, when a nonmotile strain, which also could not be stimulated, was mixed at a 1:1 ratio with a motile strain, the ability of the motile strain to swarm was significantly impaired. Importantly, swarm inhibition was relieved when either the motile or nonmotile strain contained a mutation in traA or traB. When the tra mutation was in the motile strain, the swarm rate of the motile strain was fully restored, as if the nonmotile strain elicited no inhibitory effect (see below). In contrast, when the tra mutation was in the nonmotile strain, there was a partial restoration of swarming (3). Additionally, when inhibited swarms were incubated for extended periods, e.g., 7 to 10 days, escape flares were typically found (data not shown). When cells from these flares were isolated and retested, swarm expansion usually ensued when they were mixed with a nonmotile strain, indicating that these isolates contained suppressors that had spontaneously developed. Finally, for reasons that are unknown, OME-dependent swarm inhibition primarily acted on the A-motility system (data not shown).
Based on the above findings, we reasoned that an A-motile strain could be mutagenized and mutant pools could then be mixed with a nonmotile strain to screen for escape flare mutants. Such escape mutants were expected to be defective in OME or in a downstream pathway in the responder cell (an A-motile strain) that no longer responds to OME. To allow facile gene identification and to help ensure a wide distribution of mutations, three different transposons were used: one was mariner based, and two were Tn5 based. Using this strategy, nine separate screens, each consisting of 2,000 to 3,000 mutants, were conducted, resulting in ∼20,000 total transposon mutants being screened. To conduct this screen, the cells in the initial transformation plates, which each contained 200 to 400 transposon mutants, were individually pooled, their cell densities were measured, and the cells were then mixed at a 1:1 ratio with a nonmotile strain. In total, about 80 unique mutant pools were screened. Figure 1 shows a representative example of how escape flares were identified.
To help avoid the isolation of spontaneous mutations, escape flares were identified 3 to 5 days after plating. Cells from identified flares were then isolated and retested for a relief of swarm inhibition. This effort yielded 75 mutants that recapitulated the relief of swarm inhibition phenotype after purification. The genomic insertion sites for all 75 transposon mutants were determined and resulted in the identification of 33 genes. Of these, 30 genes were hit only once and 3 genes were hit multiple times. To test if the mutations bred true, a representative mutation from each gene was backcrossed into a parental strain (DK8615). Interestingly, none of the single-hit genes bred true or the phenotype in the resulting transductants could not be confirmed as a bona fide swarm relief. In contrast, all three of the genes that were hit multiple times bred true.
For 2 of the 30 single-hit genes that did not breed true but did exhibit a full swarm relief phenotype in the original mutants (both of which were derived from mariner mutagenesis), we investigated possible explanations for the genetic discrepancies. In these two cases, the mutant strains where found by PCR analysis to have a second transposon insertion in traA, thus accounting for their phenotypes (data not shown). The occurrence of multiple mariner transposon insertions within single mutant strains is in agreement with the findings of a prior study by Hartzell and coworkers (18). For the remaining 28 single-hit mutants, all of which exhibited only a partial swarm expansion phenotype (see below), we concluded that those strains were likely to contain secondary mutations, either spontaneous or transposon based, or the phenotypes were too subtle to be detected after the backcross. In any case, these mutants were not further studied.
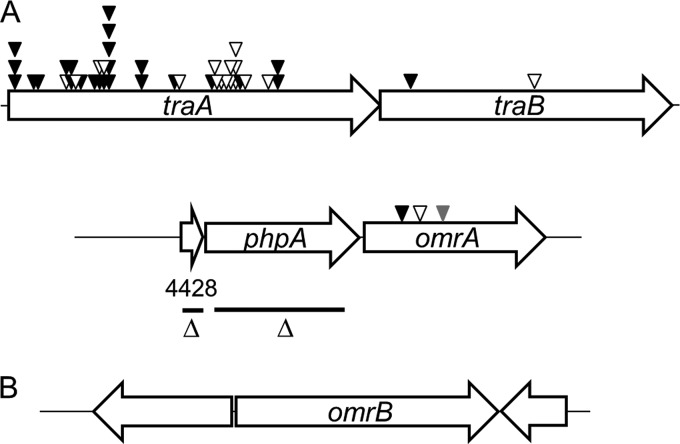
As shown in Fig. 2A, traA was hit 40 times (or 42 times when the 2 mutants described above are included) in this screen, whereas traB was hit twice. These results validate the screen, as over half of the mutations isolated were in genes known to be required for OME and swarm inhibition. Of the 40 sequenced transposon insertions within traA, 31 were clearly from independent isolates (16 from mariner and 15 from Tn5 derived from pRL27), because their insertion sites differed or they were isolated from independent mutagenesis screens. The remaining nine traA insertions could represent sibling clones because the insertions were in the same position and derived from the same mutagenesis screen as the other mutants. The three remaining transposon insertions were in a new gene (MXAN_4426) that we named omrA (OME response A) (Fig. 2A).
FIG 2.
Genetic maps of discovered genes. (A) Transposon insertions isolated in traA (MXAN_6895), traB (MXAN_6898), and omrA (MXAN_4426) are shown. Black, white, and gray triangles, mariner, Tn5, and EZ-Tn5 transposon insertions, respectively. In-frame deletion (Δ) regions in phpA (MXAN_4427) and MXAN_4428 are indicated as black bars. (B) Genetic organization of the omrB (MXAN_6818) locus. Genes are drawn approximately to scale.
Finally, to help ensure that the screen was comprehensive, we isolated spontaneous and UV-induced mutants that similarly produced escape flares. This effort yielded 15 additional mutants that were confirmed upon retesting. From complementation studies, where a traAB-containing plasmid (pDP21) was transformed into these mutant strains, 11 additional mutations were mapped to the traAB locus (data not shown). The mutations in the remaining four strains were not mapped.
The OME response can be amplified by a minority cell population to control behaviors in large populations.
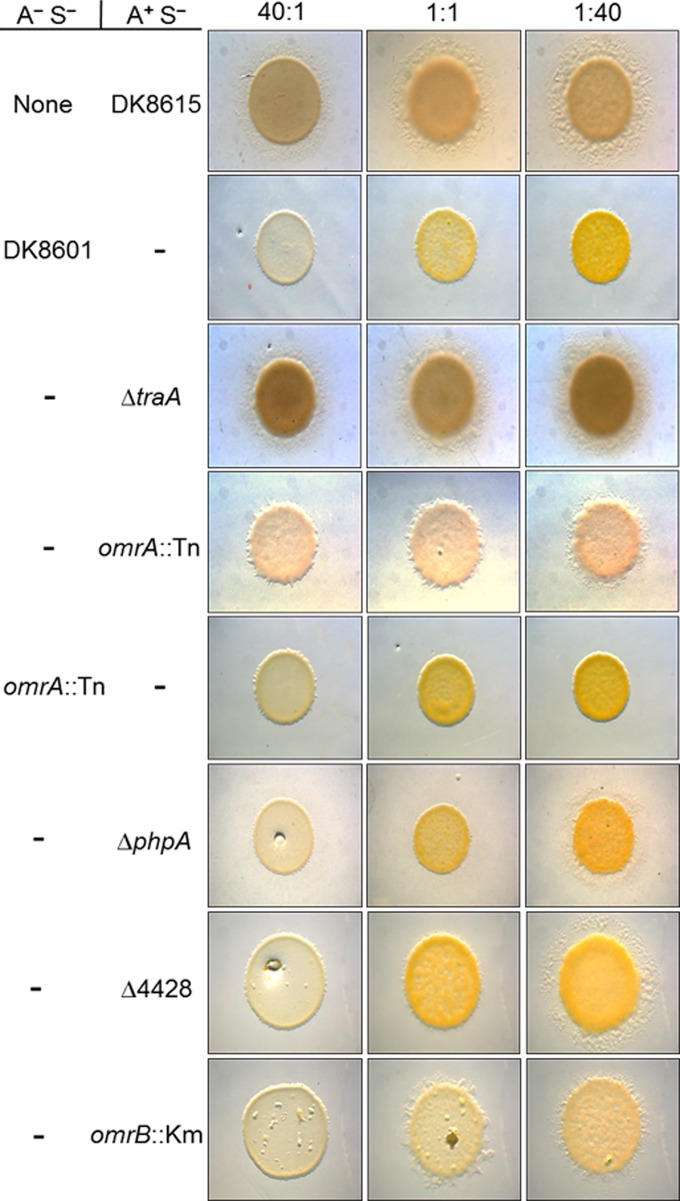
Although the three omrA mutants relieved swarm inhibition, their phenotypes were partial compared to the full relief seen with traA or traB mutants. To help quantify phenotypic differences, the ratio of motile to nonmotile cells was varied. Surprisingly, in the control mixture, a ratio of one nonmotile cell to 40 (or even 50) motile cells resulted in complete swarm inhibition, i.e., inhibition was similar to that at a 1:1 ratio or a reversed 40:1 ratio (Fig. 3, second row, and data not shown). Importantly, this result indicates that a small number of nonmotile cells block the swarm expansion of many cells.
FIG 3.
OME and the response pathway regulate swarm expansion of mixed strains. Nonmotile (A− S−) and A-motile (A+ S−) strains were mixed at the ratios indicated at the top and incubated for 3 days. The top row is a control where the A-motile strain DK8615 was instead mixed with buffer. All rows below contain strain mixtures; in rows that contain a parental strain (DK8601 or DK8615), the strain is identified by a dash, and in rows that contain derivative strains, the mutations are indicated. The strains are listed in Table 1.
As discussed above, swarm inhibition requires OME, as a tra mutant A-motile strain could swarm equally well whether alone or mixed with nonmotile cells at ratios of 40:1, 1:1, or 1:40 (Fig. 3; compare row 1 to row 3; note that when the motile population was high [1:40 ratio], swarm expansion was the largest). For the omrA mutant, there was a relief of swarm inhibition at all three ratios, although at all ratios the phenotype was partial, meaning that the swarm areas for the traA mutant mixtures or A-motility monocultures were greater (Fig. 3; compare row 4 to rows 3 and 1). From a temporal analysis, the swarm relief phenotypes of the traA and omrA mutants were difficult to visually discern at day 1 or 2 and required a compound microscope to do so; however, by day 3 their phenotypic differences were apparent (Fig. 3 and data not shown).
To examine individual cell motility dynamics, time-lapse movies were made of strain mixtures. As reported previously (3), when a nonmotile strain was mixed with an A-motile strain, the cellular movements of the latter strain significantly decreased or stopped after 1 day of incubation, whereas isogenic traA mutants continued to glide (compare Movies S1 and S2 in the supplemental material; time point, 72 h). Similar to a traA mutant, the omrA mutant retained gliding movements after a 72-h incubation with the nonmotile strain (DK8601), although the movements of the traA mutant were more robust than those of the omrA mutant (compare Movies S2 and S3 in the supplemental material). In addition, by 96 h incubation with DK8601, the relative motility of the omrA mutant stopped, while the traA mutant retained motility (data not shown). Therefore, the full swarm relief phenotype of a mixed traA colony (Fig. 3) can be explained by the sustained and unobstructed cell movements seen by time-lapse microscopy. Similarly, the partial swarm relief phenotype of a mixed omrA colony correlates with reduced cell movements, which subside after a 3-day incubation.
OmrA is not required for OME.
TraA and TraB are required in both donor and recipient cells for OME and swarm inhibition (3). To test whether OmrA functions similarly in donor strains, the omrA mutation was introduced into a nonmotile strain (DK8601). In contrast to what was found in an A-motile strain background, when the nonmotile strain contained the omrA::mini-Tn5 mutation (DW1604), complete swarm inhibition was observed at all strain ratios (Fig. 3, fifth row). Importantly, this result indicates that OmrA plays a role only in responder cells and demonstrates a function distinct from that of TraA and TraB.
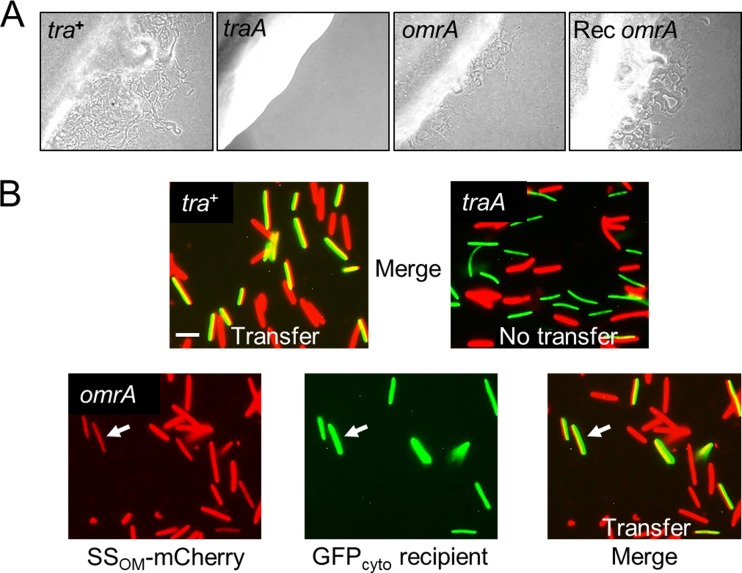
To determine whether OmrA is required for OME, we carried out a stimulation assay. Here, a nonmotile donor strain that contained the omrA::mini-Tn5 allele (DW1604) was mixed with a nonmotile strain (DW1466 ΔcglC Δtgl::Tc) that can be stimulated for both A- and S-motility by OME (3). The nonmotile omrA mutant stimulated DW1466 to glide, whereas the traA mutant did not (Fig. 4A). Compared with the stimulation by the Tra+ parental control, stimulation by the omrA mutant was notably and reproducibly impaired (Fig. 4A); however, the significance of the reduced stimulation is not clear, because we have found that other mutants, as well as medium conditions or the physiological state of cells, can similarly impair stimulation (3, 20) (data not shown). In addition, the omrA::mini-Tn5 allele was also introduced into the DW1466 recipient strain, and in this genetic background, stimulation occurred at wild-type levels (Fig. 4A). These results indicate that unlike the traA or traB mutation, the omrA mutation still allows the CglC and Tgl proteins to be transferred.
FIG 4.
OmrA is not required for stimulation or lipoprotein transfer. (A) A nonmotile but stimulatable recipient (DW1466) was mixed with isogenic nonmotile donors that contain the indicated alleles (DK8601, DW1467, and DW1604 in the first, second, and third panels, respectively). In the far right panel, the stimulatable recipient (Rec) instead contains the omrA mutation (DW1612 mixed with DK8601). Micrographs were taken after 1 day of incubation. (B) (Top) Merged fluorescence micrographs showing that transfer (yellow) of the SSOM-mCherry lipoprotein to a GFP-labeled recipient (DW709) is Tra dependent (DW1478, tra+; DW1479, traA mutant). Bar, 3 μm. (Bottom) Lipoprotein transfer from an isogenic omrA::mini-Tn5 donor (DW1611). To show the transfer unambiguously, the red, green, and merged panels are all shown. Arrows, transfer to a recipient cell.
The omrA mutant was directly tested for transfer of the OM lipoprotein reporter (SSOM-mCherry). As described previously (2), these experiments were conducted by mixing and incubating an SSOM-mCherry donor with a GFP-labeled recipient. Here, the recipient strain contained GFP in the cytoplasm, where it cannot be transferred (2). OME was thus scored as green recipients turning red or yellow in the merged fluorescence micrographs. The omrA mutant efficiently transferred SSOM-mCherry, as the GFP recipient became red (Fig. 4B). Compared with the OME in the positive control, no defect in OME was observed, whereas a traA mutant donor was not able to transfer SSOM-mCherry (Fig. 4B). From these results, we conclude that OmrA is not required for OME but instead plays a role in perceiving or responding to OME (TraAB function).
Genetic characterization of the omrA operon.
omrA resides as the distal gene in an apparent operon with two upstream genes (Fig. 2A), suggesting that these gene products could function together in a biological pathway. To address this possibility, in-frame and markerless deletion mutations were created in the upstream genes (Fig. 2A) in an A-motile (DK8615) background. These mutants, the ΔMXAN_4428 and ΔphpA (ΔMXAN_4427) mutants, were then tested for swarm inhibition. As shown in rows 6 and 7 of Fig. 3, these mutants exhibited a relief of swarm inhibition when mixed at a 1:40 (nonmotile cell-to-motile cell) ratio but not when mixed with higher ratios of nonmotile cells. Additionally, we noted that the swarm expansion was variable even at the 1:40 ratio, as cells often initially emerged as isolated flares, instead of as a continuous swarm front, which subsequently encased the inoculum spot (data not shown). The significance of this modest relief of swarm inhibition is unclear, as the parental strain (DK8615) did expand when the mixed strain ratio was reduced to 1:80 (data not shown). Thus, in this swarm assay, the ΔMXAN_4428 and ΔphpA mutations resulted in a 2-fold increase, at best, in the ratio of nonmotile cells. Finally, we note that these partial or poor swarm relief phenotypes would not have been detected under our stringent screening conditions (1:1 ratio).
The PhpA protein has been biochemically shown to function as a tyrosine phosphatase and, in a physiological context, is involved in exopolysaccharide (EPS) regulation, because a phpA mutant overexpresses EPS (21). Because EPS is required for S-motility (22), we tested the ΔphpA mutant for S-motility defects in a strain that lacks A-motility (A−). The ΔphpA strain had reduced S-motility on semisolid agar, a medium that favors S-motility (23), and the colony center was composed of macroscopic cell aggregates (see Fig. S1A in the supplemental material). Under the same conditions, an omrA mutant also had reduced S-motility; however, its colony morphology was similar to that of the parental strain (see Fig. S1A in the supplemental material). When the ΔphpA mutation was placed in the A+ S− genetic background (DW1606), a reduced-A-motility phenotype was also observed, but no macroscopic aggregates were detected (see Fig. S1B in the supplemental material). In contrast, cells in the same background with omrA or ΔMXAN_4428 mutations exhibited wild-type A-motility (data not shown). Upon microscopic examination, ΔphpA cells were found to clump in solution, and in an S-motility-positive (S+) background, the cell clumps could become large (compare Fig. S1C to D in the supplemental material). In support of these findings, Kimura and colleagues reported that a ΔphpA mutant has a hyperagglutination phenotype (21).
The ΔphpA and ΔMXAN_4428 mutants revealed no growth defect phenotypes, whereas omrA mutants showed several poor growth phenotypes. First, omrA mutants grew slowly, and, second, the cultures stopped growing at a mid-log-phase cell density of ∼2 × 108 CFU/ml (∼70 Klett units). However, when the shaker flasks were incubated for extended periods, the culture turbidity of the omrA mutants eventually increased, suggesting that suppressors had developed. In support of this conclusion, when an omrA mutant culture was subsequently propagated, the cultures exhibited no growth defects; however, the swarm relief phenotype was retained. Last, reviving naive omrA mutants from −80°C freezer stocks proved difficult and in some cases was not possible. Consequently, fresh mutants were constructed by transductions, and transductants were immediately used in the described experiments.
Bioinformatic analysis identifies OmrB.
The OmrA protein has significant sequence homology to Pfam PF03706 (UPF0104) (E value, 1.7e−38). The best-characterized PF03706 family member is MprF from Staphylococcus aureus, where this domain resides in the cytoplasmic membrane and has phospholipid flippase activity (24). Consistent with a PF03706 designation, OmrA, like other family members, was predicted to contain eight transmembrane helices (TMHMM, version 2.0) (25) (Fig. 5). Within the Pfam database (July 2014) (26), PF03706 was most frequently found as a single-domain protein (3,567 homologs); its second most frequent protein form consisted of an architecture where PF03706 was fused to a domain called DUF2156 (PF09924; 962 homologs), as found in MprF (24). To investigate whether other proteins may function in an OmrA pathway, we searched the M. xanthus DK1622 genome for ORFs that contained PF09924 (27). From this search a single gene (MXAN_6818) that contained a domain with significant homology to PF09924 (E value, 1.1e−40) was identified. To test whether this gene functions in an OME response pathway, we created a gene disruption in MXAN_6818. Interestingly, the mutant (DW1609) showed a partial relief of swarm inhibition when mixed at 1:1 and 1:40 ratios of nonmotile to motile cells (Fig. 3, bottom row). Because the MXAN_6818 disruption mutant exhibited a partial swarm relief phenotype and was competent for OME (data not shown), similar to omrA mutants, the gene was designated omrB. However, we emphasize that at a 40:1 ratio the omrB::Km mutant showed complete swarm inhibition, whereas at this ratio the omrA::mini-Tn5 mutant retained partial swarm expansion (Fig. 3), which suggests that the omrA mutant has a more pronounced swarm relief phenotype. As shown in Fig. 2B, omrB is monocistronic in the DK1622 genome.
FIG 5.
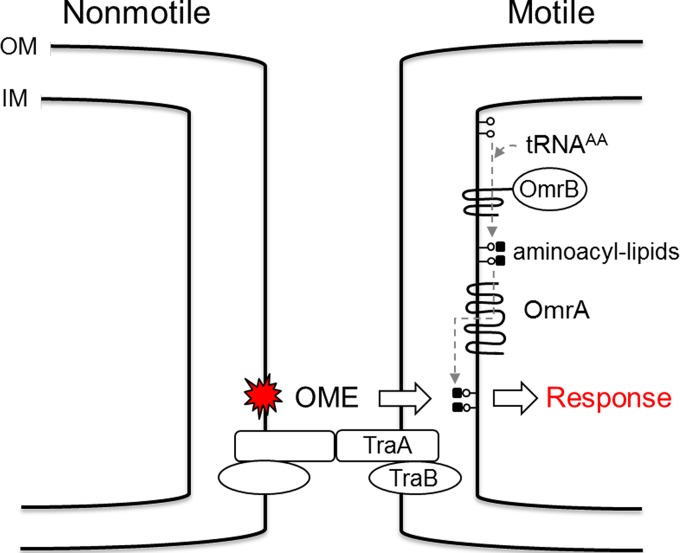
Working model for how OmrA and OmrB modulate the OME response. Here, aminoacylation of phospholipids and transfer to the outer leaflet of the inner membrane allow an undefined OME-derived signal (red star) to be perceived by a responder cell. Lollipops, phospholipids. See the text for additional details. IM, inner membrane; AA, aminoacyl.
The function of MXAN_4428 remains unknown, although the gene product shows significant homology to PF03884 (DUF329; E value, 1e−18), a domain which belongs to the TRASH clan, a family of small proteins that contain conserved cysteine residues thought to bind metal ions (28). In E. coli, YacG, a PF03884 family member, binds GyrB and inhibits DNA gyrase activity (29).
DISCUSSION
In this work, we describe a powerful genetic screen that allowed isolation of M. xanthus mutants defective in OME. In total, 44 transposon mutants with mutations in traA and traB were isolated, and additional point mutations were mapped to this locus. Because no other genes were found to be required for OME, we conclude that the protein machinery directly involved in OM fusion is relatively simple and likely consists of only TraA and TraB in partnering cells. Consistent with this view, in available myxobacterial genomes, the traAB genes are always found in an operon by themselves (30). Furthermore, in other bacterial systems that transport effectors into neighboring cells, namely, type III, IV, V, and VI secretion systems, the genes that encode the transport apparatus are also found to be contained in operons or genetic islands (31–33). In eukaryotes, where membrane fusion systems are better understood, they typically consist of one to a few proteins essential for fusion (34). Thus, on the basis of a priori evidence and our screen, the TraAB proteins may be sufficient to explain OM fusion in myxobacteria. Moreover, as no mutants were found to be defective in the production of OM tubes, which are long narrow outer membrane-enclosed cellular appendages (35), these or other overt cellular structures appear to not be required for OME, as previously suggested (35, 36).
The ease with which traAB mutants were isolated suggests that the screen provided a powerful fitness advantage to A-motile cells when OME was abolished. One obvious fitness gain is the ability of motile cells to escape captivity from nonmotile siblings. Here, escape mutants can seek fresh nutrients and leave crowded areas that contain metabolic wastes. However, given the lopsided outcomes, other fitness gains that are currently not appreciated may be endowed to tra escape mutants.
Our screen yielded a second class of mutants that were fully competent for OME. These mutants were instead defective for the OME response. A working model for the OME response pathway is outlined in Fig. 5. Importantly, because the swarm relief phenotype of the omr mutants was not as robust as that of the tra mutants (Fig. 3), the former mutant class was more difficult to isolate. Consequently, we expect the OME response pathway(s) to consist of additional players that have not yet been identified.
The swarm inhibition phenotype described here cannot be explained simply as a physical obstruction by nonmotile cells, because cells with tra mutations alleviate this block. To explain swarm inhibition, our model proposes that a time-dependent physiological signal or cue is generated in nonmotile cells and is then transmitted to motile cells (Fig. 5). In this model, OME either directly transfers an inhibitory signal or the TraAB function acts as a cue that triggers signal transfer by an undefined mechanism. Consistent with a signaling hypothesis, our findings that a few nonmotile cells can control the gliding behavior of a large population (Fig. 3, 1:40 ratio) can be explained by a mechanism whereby a signal is amplified and propagated from one to many cells. Because our screen pertained to motile responders, signal-generating mutants could not be obtained and the nature of the putative signal(s) remains unknown. Last, our findings from strain mixture experiments suggest that cells within myxobacterial swarms are communicating and cell behaviors are regulated. We hypothesize that the introduction of nonmotile cells within a motile population breaks the harmony of the population and leads to swarm dysfunction. Consistent with this view, Kaiser and Warrick have recently demonstrated that the movement of cells within a swarm shows a surprisingly high degree of synchronization (37). We suggest that this synchronization is disrupted when nonmotile cells are mixed in the population.
Insights into how cells respond to OME can be gleaned from the literature on OmrA and OmrB homologs. In particular, MprF from S. aureus represents a model protein. MprF is a bifunctional enzyme that contains a natural fusion between the domains found in OmrA and OmrB (24). The MprF PF09924 domain (OmrB) catalyzes the synthesis of lysyl-phosphatidylglycerol from lysyl-tRNA and phosphatidylglycerol precursors. The resulting membrane-embedded aminoacyl-phosphatidylglycerol is then flipped from the cytoplasmic membrane leaflet to the outside leaflet by domain PF03706 (OmrA). Within family members, the aminoacyl-phosphatidylglycerol synthases and flippase functions can be promiscuous with respect to substrates containing lysine, alanine, and/or arginine (38–40). Aminoacylation of phosphatidylglycerol in turn results in physicochemical changes in the cytoplasmic membrane and how it interacts with molecules. For example, in S. aureus, gain-of-function mutations in MprF result in antibiotic resistance, including resistance to daptomycin, by altering the membrane surface charge, which in turn repels cationic antibiotics (24). Peschel and colleagues have shown that the two domains in MprF can be genetically separated and yet their functions are retained (41). By analogy to MprF and the PA0920 homolog in Pseudomonas aeruginosa (39), we propose that OmrB similarly functions to aminoacylate phospholipids on the inner leaflet of the cytoplasmic membrane (Fig. 5). Although aminoacylated phosphatidylglycerol has not yet been reported in M. xanthus, this field is an emerging area of research in which bacteria have been found to adapt to their environments by modifying their phospholipids (42). Additionally, it is known that M. xanthus contains chemically diverse phospholipids (43), and MprF homologs in the M. xanthus genome have also been reported by others (44). We propose that, after amino acid addition to phosphatidylglycerol, OmrA flips this substrate to the outer leaflet of the inner membrane (Fig. 5). In support of this model, the finding that both omrA and omrB mutants elicit a swarm relief phenotype (Fig. 3) provides genetic evidence that these proteins function in the same pathway. However, we emphasize that at a high ratio of nonmotile cells, the omrB mutant did not elicit swarm relief, while the omrA mutant did (Fig. 3), suggesting that OmrA plays a more critical role in the responsive pathway. In turn, we suggest that omrA or omrB mutations alter membrane homeostasis, and, consequently, the physiological response of responder cells to OME engagement changes. Possible cellular scenarios to explain an altered response include how effector proteins, small molecules, and/or toxins interact with the inner membrane and/or proteins therein. Currently, we are investigating the molecular details for how cells respond to OME to coordinate multicellular behaviors.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bill Metcalf for pRL27 and Trish Hartzell for pMini-Himar-lacZ.
This work was supported by NIH grant GM101449 to D.W.
Footnotes
Published ahead of print 29 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02217-14.
REFERENCES
- 1.Nudleman E, Wall D, Kaiser D. 2005. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309:125–127. 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 2.Wei X, Pathak DT, Wall D. 2011. Heterologous protein transfer within structured myxobacteria biofilms. Mol. Microbiol. 81:315–326. 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- 3.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. 2012. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS Genet. 8:e1002626. 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pathak DT, Wall D. 2012. Identification of the cglC, cglD, cglE, and cglF genes and their role in cell contact-dependent gliding motility in Myxococcus xanthus. J. Bacteriol. 194:1940–1949. 10.1128/JB.00055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodgkin J, Kaiser D. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U. S. A. 74:2938–2942. 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhat S, Zhu X, Patel RP, Orlando R, Shimkets LJ. 2011. Identification and localization of Myxococcus xanthus porins and lipoproteins. PLoS One 6:e27475. 10.1371/journal.pone.0027475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wall D. 2014. Molecular recognition in myxobacterial outer membrane exchange: functional, social and evolutionary implications. Mol. Microbiol. 91:209–220. 10.1111/mmi.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Soto JP, Kaiser D. 1997. The tgl gene: social motility and stimulation in Myxococcus xanthus. J. Bacteriol. 179:4361–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez AM, Spormann AM. 1999. Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J. Bacteriol. 181:4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nan B, Zusman DR. 2011. Uncovering the mystery of gliding motility in the myxobacteria. Annu. Rev. Genet. 45:21–39. 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nan B, McBride MJ, Chen J, Zusman DR, Oster G. 2014. Bacteria that glide with helical tracks. Curr. Biol. 24:R169–R173. 10.1016/j.cub.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wall D, Kaiser D. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1–10. 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 13.Pathak DT, Wei X, Dey A, Wall D. 2013. Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS Genet. 9:e1003891. 10.1371/journal.pgen.1003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pathak DT, Wei X, Wall D. 2012. Myxobacterial tools for social interactions. Res. Microbiol. 163:579–591. 10.1016/j.resmic.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wall D, Kaiser D. 1998. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc. Natl. Acad. Sci. U. S. A. 95:3054–3058. 10.1073/pnas.95.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wall D, Kolenbrander PE, Kaiser D. 1999. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J. Bacteriol. 181:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193–201. 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 18.Youderian P, Burke N, White DJ, Hartzell PL. 2003. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol. 49:555–570. 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- 19.Julien B, Kaiser AD, Garza A. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 97:9098–9103. 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nudleman E, Wall D, Kaiser D. 2006. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 60:16–29. 10.1111/j.1365-2958.2006.05095.x. [DOI] [PubMed] [Google Scholar]
- 21.Mori Y, Maeda M, Takegawa K, Kimura Y. 2012. PhpA, a tyrosine phosphatase of Myxococcus xanthus, is involved in the production of exopolysaccharide. Microbiology 158:2546–2555. 10.1099/mic.0.059824-0. [DOI] [PubMed] [Google Scholar]
- 22.Lu A, Cho K, Black WP, Duan XY, Lux R, Yang Z, Kaplan HB, Zusman DR, Shi W. 2005. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol. Microbiol. 55:206–220. 10.1111/j.1365-2958.2004.04369.x. [DOI] [PubMed] [Google Scholar]
- 23.Shi W, Zusman DR. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. U. S. A. 90:3378–3382. 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst CM, Peschel A. 2011. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol. Microbiol. 80:290–299. 10.1111/j.1365-2958.2011.07576.x. [DOI] [PubMed] [Google Scholar]
- 25.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 26.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, Wiegand R, Kaplan HB. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200–15205. 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamsheer K M, Laxmi A. 2014. DUF581 is plant specific FCS-like zinc finger involved in protein-protein interaction. PLoS One 9:e99074. 10.1371/journal.pone.0099074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sengupta S, Nagaraja V. 2008. YacG from Escherichia coli is a specific endogenous inhibitor of DNA gyrase. Nucleic Acids Res. 36:4310–4316. 10.1093/nar/gkn355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42:D560–D567. 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149. 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konovalova A, Petters T, Sogaard-Andersen L. 2010. Extracellular biology of Myxococcus xanthus. FEMS Microbiol. Rev. 34:89–106. 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- 33.Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ, Low DA. 2014. Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb. Perspect. Med. 4:a010025. 10.1101/cshperspect.a010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens S, McMahon HT. 2008. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9:543–556. 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 35.Wei X, Vassallo CN, Pathak DT, Wall D. 2014. Myxobacteria produce outer membrane-enclosed tubes in unstructured environments. J. Bacteriol. 196:1807–1814. 10.1128/JB.00850-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartzell T. 2014. All in the family: kin contact leads to outer membrane exchange. J. Bacteriol. 196:1789–1792. 10.1128/JB.01617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaiser D, Warrick H. 2014. Transmission of a signal that synchronizes cell movements in swarms of Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 111:13105–13110. 10.1073/pnas.1411925111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy H, Ibba M. 2009. Broad range amino acid specificity of RNA-dependent lipid remodeling by multiple peptide resistance factors. J. Biol. Chem. 284:29677–29683. 10.1074/jbc.M109.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arendt W, Hebecker S, Jager S, Nimtz M, Moser J. 2012. Resistance phenotypes mediated by aminoacyl-phosphatidylglycerol synthases. J. Bacteriol. 194:1401–1416. 10.1128/JB.06576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slavetinsky CJ, Peschel A, Ernst CM. 2012. Alanyl-phosphatidylglycerol and lysyl-phosphatidylglycerol are translocated by the same MprF flippases and have similar capacities to protect against the antibiotic daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 56:3492–3497. 10.1128/AAC.00370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660. 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy H, Dare K, Ibba M. 2009. Adaptation of the bacterial membrane to changing environments using aminoacylated phospholipids. Mol. Microbiol. 71:547–550. 10.1111/j.1365-2958.2008.06563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curtis PD, Geyer R, White DC, Shimkets LJ. 2006. Novel lipids in Myxococcus xanthus and their role in chemotaxis. Environ. Microbiol. 8:1935–1949. 10.1111/j.1462-2920.2006.01073.x. [DOI] [PubMed] [Google Scholar]
- 44.Roy H. 2009. Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol. IUBMB Life 61:940–953. 10.1002/iub.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.