Abstract
Gram-negative bacteria have evolved several highly dedicated pathways for extracellular protein secretion, including the type II secretion (T2S) system. Since substrates secreted via the T2S system include both virulence factors and degradative enzymes, this secretion system is considered a major survival mechanism for pathogenic and environmental species. Previous analyses revealed that the T2S system mediates the export of ≥20 proteins in Vibrio cholerae, a human pathogen that is indigenous to the marine environment. Here we demonstrate a new role in biofilm formation for the V. cholerae T2S system, since wild-type V. cholerae was found to secrete the biofilm matrix proteins RbmC, RbmA, and Bap1 into the culture supernatant, while an isogenic T2S mutant could not. In agreement with this finding, the level of biofilm formation in a static microtiter assay was diminished in T2S mutants. Moreover, inactivation of the T2S system in a rugose V. cholerae strain prevented the development of colony corrugation and pellicle formation at the air-liquid interface. In contrast, extracellular secretion of the exopolysaccharide VPS, an essential component of the biofilm matrix, remained unaffected in the T2S mutants. Our results indicate that the T2S system provides a mechanism for the delivery of extracellular matrix proteins known to be important for biofilm formation by V. cholerae. Because the T2S system contributes to the pathogenicity of V. cholerae by secreting proteins such as cholera toxin and biofilm matrix proteins, elucidation of the molecular mechanism of T2S has the potential to lead to the development of novel preventions and therapies.
INTRODUCTION
The type II secretion (T2S) system directs the transport of a variety of proteins from Gram-negative bacteria. In Vibrio cholerae, the causative agent of cholera, the T2S system supports the outer membrane translocation of the major virulence factor, cholera toxin, and several other proteins, many of which are associated with the degradation of macromolecules. Known or predicted T2S substrates include chitinases, proteases, lipases/esterases, phospholipases, and nucleases (reviewed in references 1 and 2). While some T2S substrates have been shown to function primarily in either the environmental reservoir or the host, multiple T2S substrates act in both settings (3–6). These enzymes likely support nutrient acquisition and thereby the adaptation and survival of bacteria as they encounter new environmental niches (7, 8). In many of these environments, bacteria are living predominantly as biofilms (9).
Biofilms are microbial communities that are embedded in a matrix comprising polysaccharides, nucleic acids, and associated extracellular proteins. The matrix enables the biofilm to adhere to surfaces better than planktonic cells, forms the scaffold for the construction of the biofilm, and provides mechanical stability once the biofilm is established (reviewed in references 10 to 13). When grown in biofilms, bacteria have access to nutrients accumulating at interfaces. Biofilms also protect the resident bacteria from predators, the effects of extracellular stress, the action of antibiotics, and clearance by the immune system.
Although biofilm formation has been studied extensively in many pathogens, much of the focus has been directed toward extracellular polysaccharides and fimbriae, and only recently has the role of extracellular matrix proteins begun to be addressed (reviewed in reference 14). Proteomic analyses of the biofilm matrix in V. cholerae have revealed several specialized proteins involved in biofilm formation and stabilization (15, 16). These included the RbmA and RbmC proteins, known to be critical for biofilm formation, as well as three proteins not previously associated with biofilms: V. cholerae cytolysin (VCC; VCA0219), a chitinase (VCA0027), and a hemagglutinin-protease (HA/P; VCA0865) (16). We reported previously that RbmC, VCC, HA/P, and chitinase are likely dependent on the T2S system for extracellular secretion, since proteomic analysis identified these proteins in the supernatants isolated from wild-type (WT) V. cholerae, while they were either absent or present at reduced levels in a T2S mutant (17). RbmA is known to be a secreted protein (15), although the mechanism of secretion is not understood. In mature V. cholerae biofilms, RbmA is localized around cell perimeters, is involved predominantly in intercellular interactions, and facilitates the aggregation of bacteria into clusters (16, 18). RbmC encases cell clusters within the biofilm (18), while Bap1, a homologue of RbmC originally identified through transcriptomic analysis of V. cholerae biofilm stages (19), shares limited functional redundancy with RbmC (20) and plays a role in biofilm-surface interactions and in the encasement of cell clusters (16, 18).
Additional extracellular enzymes with degradative function have been detected in the biofilms of other bacteria (for a review, see reference 14). Substrates for these enzymes may consist of matrix components and/or cellular debris and may include polysaccharides, proteins, and lipids. Enzymes that break down these components transform the biofilm into a space where large molecules can be degraded and subsequently taken up and utilized as energy sources (21), since biofilm stability has been shown to be energy dependent (22). Additionally, some extracellular enzymes can degrade structural matrix components to promote the detachment of resident bacteria, thereby allowing new biofilms to be formed (23–26). Given the importance of extracellular proteins in biofilms, protein secretion likely plays a central role in biofilm development and maintenance. The mechanism of delivery of these proteins to the biofilm matrix is generally unknown, however. Here we demonstrate that several proteins with established roles in V. cholerae biofilm formation rely on the T2S system for extracellular release.
MATERIALS AND METHODS
Bacterial growth conditions.
The strains and plasmids used in this study are listed in Table 1. Bacterial cultures were propagated at 37°C in Luria-Bertani (LB) medium with supplements as specified below. Chromosomal in-frame deletion mutant strains were generated according to previously published protocols (18, 27). The concentrations of antibiotics and inducer used, where appropriate, were as follows: ampicillin, 100 μg/ml; carbenicillin, 200 μg/ml; rifampin, 100 μg/ml; isopropyl-β-d-thiogalactopyranoside (IPTG), 10 to 100 μM.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Properties | Source or reference |
|---|---|---|
| Vibrio cholerae strains | ||
| N16961 | WT El Tor O1 biotype | Laboratory collection |
| NΔepsD | N16961 ΔepsD; Kanr | 54 |
| NΔeps | N16961 ΔepsCDEFGHIJKLMN; Cmr | 54 |
| RA1552 | El Tor O1 biotype, rugose variant; spontaneous Rifr mutant of clinical isolate 92A1552 | 38 |
| RΔABC | Rugose variant; ΔrbmA Δbap1 ΔrbmC | 20 |
| RΔepsF | Rugose variant; ΔepsF | This study |
| RΔVPS | Δvps-I Δvps-II; rugose variant with deletion of vpsA to vpsK and vpsL to vpsQ; Rifr | 30 |
| SA1552 | El Tor O1 biotype, nonrugose | 38 |
| SΔdns | SA1552 Δdns | This study |
| TRH7000 | El Tor O1 biotype; thy Hgr ΔctxAB | Laboratory collection |
| TRHΔepsD | TRH7000 ΔepsC; Kanr | 28 |
| TRHΔepsG | TRH7000 ΔepsG; Kanr | 28 |
| Plasmids | ||
| pEpsD | epsD cloned into pMMB67; Ampr | 28 |
| pEpsG | epsG cloned into pMMB67; Ampr | 28 |
| pHA/P | ha/p cloned into pMMB66; Ampr | 55 |
| pGFP | gfpmut2 cloned into pMMB66; Ampr | 55 |
| pEpsF | epsF cloned into pMMB67; Ampr | This study |
Recombinant DNA techniques.
DNA manipulations were carried out using standard molecular techniques according to the manufacturer's instructions. PCRs were carried out using primers purchased from Bioneer Corporation (Alameda, CA) or Integrated DNA Technologies (Coralville, IA) and the Phusion High-Fidelity PCR kit (New England Biolabs). The sequences of the primers used in the present study are available upon request. Plasmid sequences for epsF expression were verified by sequencing (UC Berkeley DNA Sequencing Facility, Berkeley, CA). Plasmids for the complementation of epsD and epsG have been published previously (28).
SDS-PAGE and immunoblotting.
For all experiments, samples were normalized to equivalent optical densities at 600 nm (OD600) and were resolved using NuPAGE 4-to-12% Bis-Tris gradient gels (Invitrogen). For the immunodetection of RbmC, RbmA, and Bap1 (Fig. 1), samples were concentrated by precipitation utilizing pyrogallol red-molybdate-methanol (PRMM) as described previously (17). Briefly, culture supernatants were mixed with an equal volume of PRMM solution (0.05 mM pyrogallol red, 0.16 mM sodium molybdate, 1.0 mM sodium oxalate, 50 mM succinic acid, 20% methanol [pH 2]), and the pH of the mixture was immediately adjusted to 2.8 to 3.0 for optimal precipitation. Samples were incubated at room temperature for 2 h, followed by overnight incubation at 4°C. Proteins were pelleted by centrifugation at 10,000 × g for 1 h, washed twice with ice-cold acetone, and reconstituted in SDS loading buffer. Immunoblots were blocked in 5% nonfat dry milk in TBS buffer (50 mM Tris-HCl, 200 mM NaCl [pH 7.5]), followed by incubation with anti-Bap1 (1:200), anti-RbmA (1:1,000), and anti-RbmC (1:1,000) antibodies. Secondary detection was performed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG. Immunoblots were imaged using a Typhoon Trio system (Amersham Biosciences).
FIG 1.
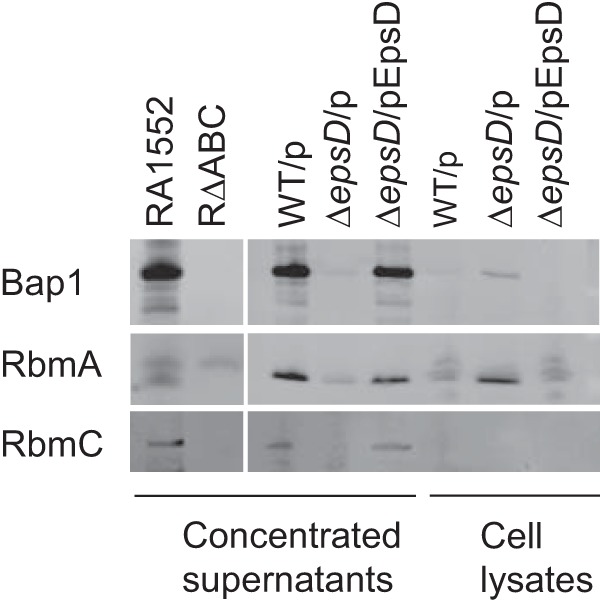
Secretion of the biofilm matrix proteins RbmC, Bap1, and RbmA is dependent on the T2S system. Concentrated culture supernatants of stationary-phase cultures and cell extracts from WT V. cholerae N16961 with vector only (WT/p), an epsD mutant with vector only (ΔepsD/p), and a complemented epsD mutant strain (ΔepsD/pEpsD) were loaded by equivalent optical densities. As controls, a strain with mutations in rbmA, rbmC, and bap1 (RΔABC) and its isogenic rugose parental strain (RA1552) were included. Primary detection was performed with anti-RbmC (1:1,000), anti-Bap1 (1:200), and anti-RbmA (1:1,000) antisera.
Quantification of biofilm formation in V. cholerae.
Biofilm assays were performed as described previously (29) with the following modifications. Briefly, overnight stationary-phase cultures of wild-type and mutant V. cholerae strains were diluted to an OD600 of 0.05 in LB medium, with carbenicillin and 10 μM IPTG if necessary, and 100 μl of each strain was added to 8 wells each of a 96-well polystyrene microtiter plate. Plates were incubated at 30°C for 8, 24, or 48 h. At each time point, culture medium containing nonadherent cells was discarded, plates were rinsed with water, and each well was stained with 125 μl of 0.1% crystal violet for 10 min. The plates were rinsed again and were allowed to dry for 3 h or overnight. Crystal violet was solubilized by the addition of 125 μl of 30% acetic acid for 10 min, and biofilms were quantified by measuring the OD550 using a plate reader. Four biological replicates, each with eight technical replicates, were performed.
Colony morphology and pellicle formation analyses.
Colony corrugation and pellicle formation assays were carried out according to previously published protocols (15, 20, 30). Briefly, for colony morphology studies, cultures grown overnight at 30°C with shaking (200 rpm) were serially diluted with LB medium, and 100 μl of the diluted cultures was plated onto LB agar medium. The cultures were incubated for 4 days (2 days at 30°C, followed by 2 days at room temperature). Assays were repeated with at least three different biological replicates. For the pellicle formation assay, overnight stationary-phase cultures were diluted 1:200 into 5 ml of fresh medium in glass culture tubes (18 by 150 mm). The cultures were incubated at 30°C for 2 days, followed by 1 day at room temperature.
VPS dot blot analyses.
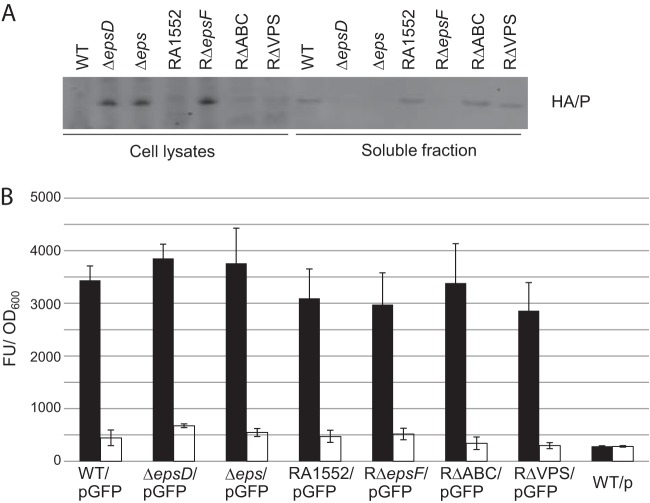
Dot blotting experiments for Vibrio exopolysaccharide (VPS) analysis were carried out according to previously described protocols (18, 30). Briefly, overnight-grown cultures were plated onto LB agar plates and were incubated at 37°C for 1 day. Cells were harvested from the plates by scraping and were resuspended in LB medium. The resuspended cultures were adjusted to the same optical density, and equal volumes of the cell suspensions were shaken gently at 4°C for 1 h. Cells were removed by centrifugation. The supernatants were collected (soluble VPS), and cell pellets were resuspended to the same volume in LB medium (cell-associated VPS). The soluble VPS fraction and intact cell samples (1.0 μl) were serially diluted and were spotted onto nitrocellulose membranes, followed by incubation with anti-VPS antiserum (1:1,000) and HRP-conjugated goat anti-rabbit IgG (1:7,500). Dot blots were imaged using a Typhoon Trio system. Fractions were also analyzed for the presence of the type II secreted protein hemagglutinin-protease (HA/P) by SDS-PAGE and immunoblotting using anti-HA/P antibodies (1:5,000) and for the cytoplasmic marker green fluorescent protein (GFP) by fluorometry (excitation and emission at 485 and 535 nm, respectively).
DNase zymography.
Supernatants from overnight cultures grown in 3-(N-morpholino)propanesulfonic acid (MOPS) minimal medium (1× MOPS salts [40 mM MOPS {pH 7.4}, 4 mM Tricine, 0.1 mM FeSO4·7H2O, 9.5 mM NH4Cl, 0.28 mM KCl, 0.53 mM MgCl2·6H2O, and 50 mM NaCl], 1× trace metals [0.005% MgSO4, 0.0005% MnCl2·4H2O, and 0.0005% FeCl3], and 0.5% glucose; supplemented with 0.1 mM KH2PO4) were subjected to SDS-PAGE without prior boiling using gels that included 200 mg/ml salmon sperm DNA (Invitrogen). To remove the SDS, gels were washed with 2.5% (vol/vol) Triton X-100 followed by 2.5% Triton X-100–50 mM Tris-HCl (pH 7.5) (31). After washing, gels were incubated first at 37°C in 50 mM Tris-HCl (pH 7.5)–50 mM MgCl2 and then at 37°C in 50 mM Tris-HCl (pH 7.5)–50 mM MgCl2–25 mM CaCl2, stained in ethidium bromide, and imaged.
RESULTS
T2S-dependent secretion of biofilm matrix proteins.
RbmA, RbmC, and Bap1 are secreted proteins with established roles in V. cholerae biofilm formation. Previously, proteomic analysis of culture supernatants suggested that RbmC may be a member of the V. cholerae T2 secretome (17). To confirm the role of the T2S system in RbmC secretion and also to determine if the other biofilm matrix components, RbmA and Bap1, are dependent on the T2S system for extracellular secretion, we carried out immunoblot analyses using culture supernatants and whole-cell lysates from cultures of the WT strain N16961, the isogenic ΔepsD T2S mutant, and the complemented mutant strain of V. cholerae. As controls, a V. cholerae strain with mutations in rbmA, rbmC, and bap1 (RΔABC) and its isogenic parental strain (RA1552) were included. Immunoblots showed the presence of polypeptides corresponding to the expected sizes for RbmC (∼102 kDa), RbmA (∼26 kDa), and Bap1 (∼73 kDa) in the supernatant fractions of the WT and the complemented deletion strain (Fig. 1). All three proteins were absent in the supernatant fractions from the T2S mutant. Additionally, RbmA and Bap1 accumulated in the cell fractions of the T2S mutant, suggesting that these proteins are made but cannot be secreted in this strain. We were unable to detect RbmC in cell fractions of the T2S mutant, suggesting that it may be degraded or its expression level may be reduced when it is not secreted from the cell (32). These results indicate that the biofilm matrix proteins are predominantly secreted in WT V. cholerae and require the T2S system for extracellular secretion.
Role of the T2S system in biofilm formation.
The dependency of the biofilm matrix proteins RbmA, RbmC, and Bap1 on the T2S system for extracellular secretion implies that the T2S system contributes to biofilm development and/or maintenance. To test this hypothesis, we analyzed the biofilm formation capacities of different V. cholerae isolates and several isogenic T2S mutants in a quantitative microtiter assay and two qualitative assays that evaluate colony morphology and pellicle formation in liquid cultures. First, WT V. cholerae N16961 and a mutant in which the entire T2S system was deleted (Δeps) were compared using a crystal violet staining-based biofilm assay. Biofilms were allowed to progress for 8, 24, or 48 h and were then stained with crystal violet and quantified; biofilm mass was expressed as the absorbance at 550 nm. V. cholerae lacking the T2S system exhibited a statistically significant defect in biofilm formation relative to the WT strain. Figure 2A shows the dynamics of biofilm formation by these strains over time.
FIG 2.
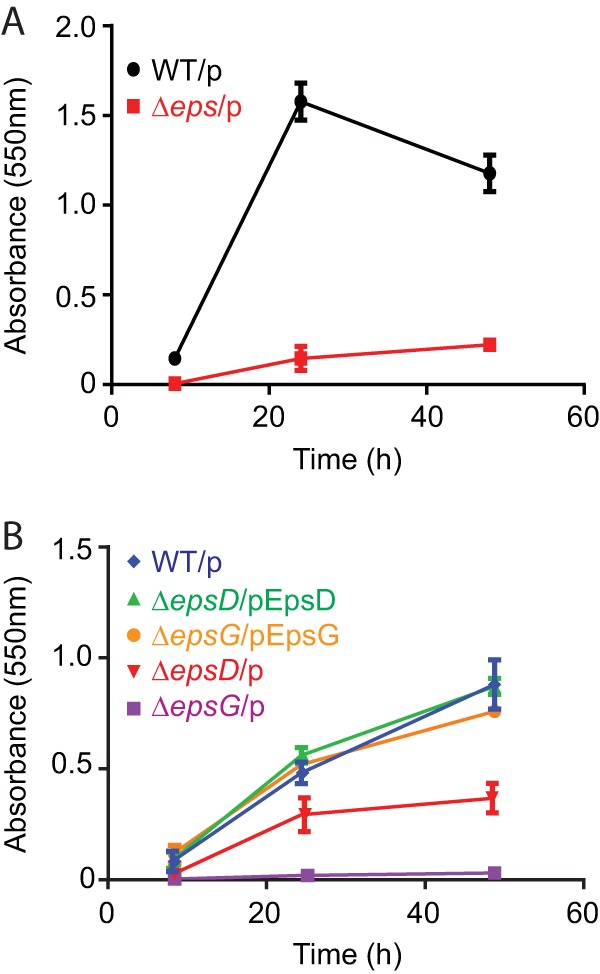
The V. cholerae T2S system supports biofilm formation. Cultures of V. cholerae WT and T2S mutant strains were grown for 8, 24, and 48 h at 30°C in microtiter plates, and biofilm formation by the adherent cells was quantified by crystal violet staining. (A) WT V. cholerae N16961 with vector only (WT/p) and a mutant with the whole eps operon deleted, with vector only (Δeps/p). (B) V. cholerae TRH7000 with vector only (WT/p), epsD and epsG mutants with vector only (ΔepsD/p and ΔepsG/p), and their complemented strains (ΔepsD/pEpsD and ΔepsG/pEpsG). There were statistically significant differences (P ≤ 0.0001) between the WT and mutant strains at all time points tested. The OD600 of the mutant planktonic cultures did not differ significantly from that of the WT culture (data not shown).
We also analyzed the biofilm formation ability of a toxin-negative derivative of N16961, TRH7000, since we have constructed single-gene deletions of all 12 eps genes in this strain background and have shown that each eps gene is required for extracellular secretion (28, 33–36). While we observed some differences between the single T2S gene deletion strains, the mutant strains consistently displayed significantly less biofilm formation than the WT strain. The results for two representative mutants, TRHΔepsD and TRHΔepsG, are shown in Fig. 2B. The reason for the difference between mutants is not known, but it may result from a low level of nonspecific leakage of biofilm matrix proteins in some of the mutants. More importantly, complementation of the T2S mutants restored the ability of these strains to form biofilms. While the T2S mutants had a distinctly smaller amount of stained biofilm mass over the entire 48-h period, the complemented strains closely mimicked the profile of the WT strain. These results indicate that the T2S system is required for biofilm formation in V. cholerae.
The T2S system contributes to rugosity.
To further determine the involvement of T2S in biofilm formation, we analyzed the impact of inactivating the T2S system in a rugose variant of V. cholerae. Rugose strains have an enhanced capacity to produce biofilm matrix materials, and consequently, their biofilm-forming ability can additionally be observed by the development of corrugated colonies and pellicles (37–39). The rugose parental strain RA1552 and an RΔepsF strain were incubated on LB agar plates in order to follow the development of colony corrugation and in LB broth in glass culture tubes in order to analyze pellicle formation at the air-liquid interface. Compared to the parental rugose strain, the T2S mutant formed a smooth-looking colony (Fig. 3, top) and exhibited a drastic reduction in the level of pellicle formation (Fig. 3, bottom). The T2S mutant strain was phenotypically similar to the strain with mutations in rbmA, rbmC, and bap1. Increased colony corrugation and pellicle formation were observed when the T2S secretion mutant was complemented in trans, indicating that extracellular components secreted by the T2S system contribute to colony corrugation and pellicle formation.
FIG 3.
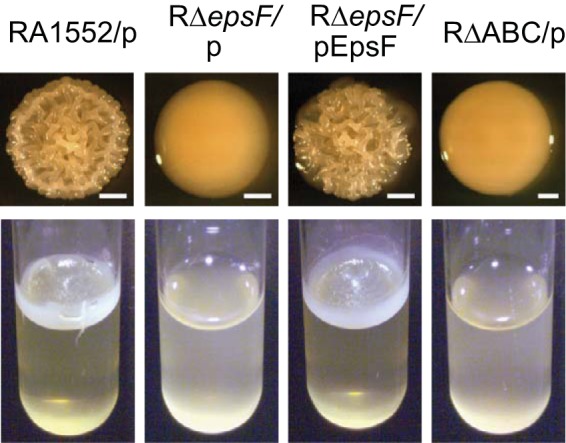
Colony corrugation and pellicle formation are dependent on the T2S system. Shown are colony (top) and pellicle (bottom) phenotypes of rugose strain RA1552 with vector only (RA1552/p), RΔepsF with vector only (RΔepsF/p) or with the complementing plasmid (RΔepsF/pEpsF), and RΔrbmA Δbap1 ΔrbmC with vector only (RΔABC/p). Colonies were incubated for 4 days (bars, 1 mm), while pellicles were incubated for 3 days.
DNase secretion is not T2S dependent.
In addition to the biofilm matrix proteins RbmA, RbmC, and Bap1, other extracellular proteins also have the capacity to control biofilms. V. cholerae has been shown to produce two secreted DNases, Xds and Dns (40–42), and recent work has demonstrated a role for these DNases in the development of normal 3-dimensional (3-D) biofilm architecture and biofilm dispersal on abiotic surfaces (42). In order to determine if the T2S system is also responsible for the secretion of these extracellular proteins, we performed DNase zymography on culture supernatants of N16961 and the ΔepsD mutant. As controls, a strain with a deletion of dns and its isogenic parental strain (SA1552) were included. As seen in Fig. 4, Dns was secreted and active in N16961 and T2S mutant samples, suggesting that Dns is not a T2S substrate. Xds activity, on the other hand, was low and variable, preventing us from determining conclusively whether Xds is secreted by the T2S system (not shown).
FIG 4.
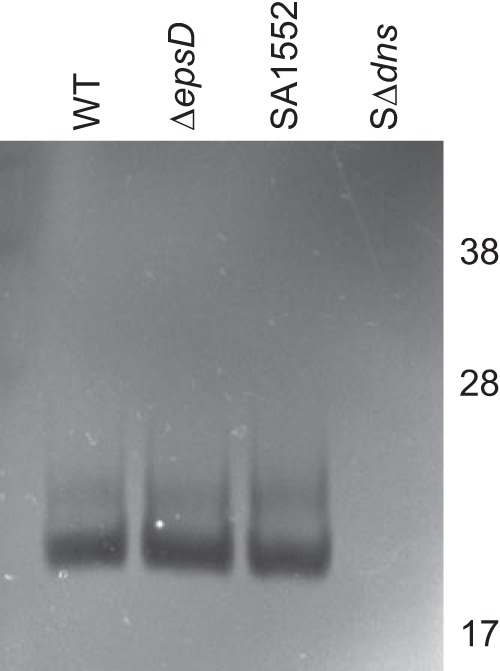
The T2S system does not secrete the DNase Dns. DNA zymography of culture supernatants from the WT V. cholerae strain N16961 and V. cholerae ΔepsD was performed. DNase activity is detected by the clearing of DNA from the SDS-PAGE gel. As controls, a strain with a mutation in dns and its isogenic parental strain (SA1552) were included. Molecular weight markers (in thousands) are indicated.
VPS production is not dependent on T2S.
A hallmark of V. cholerae biofilms is the production of VPS (Vibrio exopolysaccharide), and many of the phenotypic properties of rugose variants have been directly attributed to this exopolysaccharide. It was concluded previously that the T2S system is involved in VPS production, since inactivation of the T2S genes epsD and epsE in a rugose variant of N16961 resulted in a smooth colony morphology (43). In order to evaluate the contribution of the T2S system to VPS secretion, we isolated VPS from cells grown on LB agar and performed VPS dot blot analysis (Fig. 5). A strain lacking VPS (RΔVPS) was included as a control. Soluble and cell-associated VPSs were detected in the nonrugose WT strain N16961 and its isogenic T2S mutants (the ΔepsD and Δeps mutants), as well as in the rugose WT strain RA1552, its isogenic T2S mutant (RΔepsF), and the strain with all three secreted biofilm matrix protein genes deleted (RΔABC). Although more VPS was detected in the rugose strains than in the nonrugose strains, no difference in VPS production was observed between the T2S mutants and their isogenic WT strains in either background, suggesting that VPS does not depend on the T2S system for extracellular transport. The presence of VPS in the released, soluble fraction was not due to lysis of the cells, since fluorescence from the cytoplasmic marker GFP was detected only in the cell pellet, not in the fraction containing soluble VPS (Fig. 6B).
FIG 5.
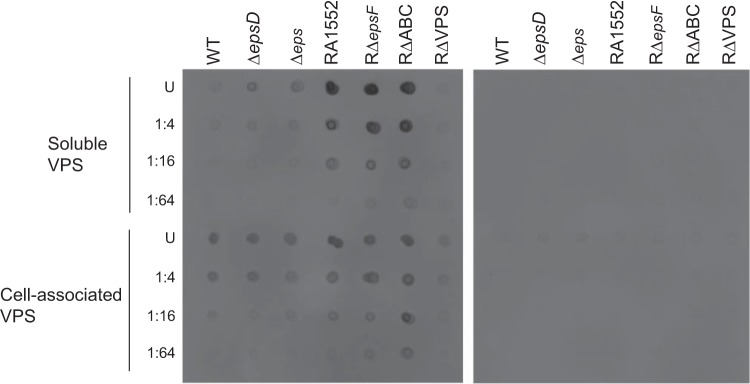
VPS is produced and secreted independently of the T2S system. VPS immunodot blot analysis was conducted with the WT strain N16961, the ΔepsD and Δeps mutants, the WT rugose strain RA1552, RΔepsF, RΔrbmA Δbap1 ΔrbmC (RΔABC), and a strain lacking VPS (RΔVPS). Dot blotting was carried out by spotting the soluble VPS fraction and cells onto nitrocellulose membranes. Undiluted samples (U) and dilutions are indicated. (Left) Incubation with anti-VPS (1:1,000) and HRP-conjugated goat anti-rabbit IgG (1:7,500). (Right) Secondary-antibody control.
FIG 6.
Growth on LB agar does not interfere with the function of the T2S system, and no leakage of a cytoplasmic marker is observed. Shown are data for soluble VPS fractions and cells from the WT V. cholerae strain N16961, the ΔepsD and Δeps mutants, the WT rugose strain RA1552, and the RΔepsF, RΔABC, and RΔVPS strains, with plasmids encoding either HA/P (pHA/P) or GFP (pGFP), grown on LB agar plates. (A) All samples were normalized to equivalent optical densities. Primary detection was performed with anti-HA/P (1:5,000) antiserum. Secondary detection was performed with HRP-conjugated goat anti-rabbit IgG (1:15,000). (B) GFP fluorescence was measured using excitation and emission wavelengths of 485 nm and 530 nm, respectively. Filled and open bars indicate fluorescence detected in cells and soluble VPS fractions, respectively. Results from three independent experiments are presented as average fluorescence units (FU)/OD600 unit ± standard errors.
As a control to ensure that the T2S system is functional in cells grown on agar, we performed immunoblot analyses of the same fractions using antibodies against a well-characterized T2S-dependent protein, hemagglutinin-protease (HA/P). HA/P was detected in the soluble fractions only for strains containing a functional T2S system, while the protein accumulated in the cells of T2S mutants (Fig. 6A). Collectively, these data suggest that the biofilm defects observed in the T2S mutants are not due to interference with VPS production or secretion.
DISCUSSION
The T2S system supports bacterial persistence in many environments by secreting enzymes and toxins. Here we demonstrate that proteins with well-known roles in V. cholerae biofilm formation rely on the T2S system for extracellular secretion. We reported previously that RbmC likely relies on the T2S apparatus for extracellular transport (17), so we tested the hypothesis that like RbmC, Bap1 and RbmA would also require the T2S system for export into the biofilm matrix. Indeed, immunoblot analyses using PRMM-precipitated V. cholerae culture supernatants and whole-cell lysates showed the presence of RbmC, RbmA, and Bap1 in the supernatant fractions of the WT and complemented deletion strains, while they were absent in the supernatant of the T2S mutant (Fig. 1). Cell-associated RbmA and Bap1 instead accumulated in the T2S mutant. RbmC, on the other hand, was not detected in T2S mutant cells, suggesting that it may be degraded or its expression level may be reduced when it is not secreted via the T2S system.
Previously, a P. aeruginosa mini-Tn5-lux mutant library screen identified a T2S mutant with reduced biofilm-forming ability (44). Similarly, Baldi et al. (45) observed less biofilm in a T2S mutant of enteropathogenic Escherichia coli after 96 h in a continuous-culture flow cell system than in the wild type. Thus, we compared the biofilm formation capacities of WT V. cholerae and T2S mutants, including the ΔepsG, ΔepsD, and Δeps mutants. Comparisons of the relative densities of the adherent cells by crystal violet staining showed that V. cholerae lacking a functional T2S system had a statistically significant defect in biofilm formation relative to that by the WT at all time points tested (Fig. 2). Complementation of the T2S genes restored the abilities of these strains to form biofilms to near-WT levels, indicating that the T2S system is required for biofilm production by V. cholerae.
The biofilm-forming capacities of strains with single mutations in rbmA, rbmC, or bap1 are not significantly different from that of the rugose variant (15, 20). However, mutations in rbmA do cause cells to form biofilms that are less structured, more fragile, and more sensitive to sodium dodecyl sulfate than WT biofilms (15), suggesting that changing the profile of extracellular proteins in the biofilms can alter their integrity. A double deletion mutant (ΔrbmC Δbap1) and a triple deletion mutant (ΔrbmA ΔrbmC Δbap1) have significantly reduced biofilm-forming capabilities (15, 20). Here we show that T2S mutants, unable to secrete RbmA, RbmC, Bap1, and possibly additional contributing proteins, are severely deficient in biofilm formation (Fig. 2).
In response to stress, V. cholerae undergoes phenotypic variation that results in a morphologically distinct variant termed the rugose variant (37, 38, 46, 47). Rugose variants form rough/wrinkled colonies, show increased resistance to stresses, and have an enhanced ability to form biofilms and pellicles (30, 37, 38). Genomic expression profiling of rugose variants indicated that the regulation of ≥100 genes differs from that for smooth variants and, in particular, that several genes encoding T2S system proteins are upregulated (48). These findings are supported by our observation that a deletion of the T2S gene epsF results in both loss of rugosity and loss of pellicle formation (Fig. 3), and they agree with the report by Ali et al. (43) showing that two other T2S proteins, EpsD and EpsE, are needed for the formation of the rugose phenotype.
The strong connection between VPS and biofilm formation led us to compare the VPS levels of the T2S mutants. We observed no difference in VPS levels between the WT and a T2S mutant (Fig. 4). Nor did we observe a deficiency in VPS secretion in the strain in which all three biofilm matrix proteins were deleted. It is therefore unlikely that the loss of rugosity and/or biofilm production in T2S mutants is due to involvement of the T2S pathway in VPS secretion. It is also unlikely that any of the proteins required for VPS synthesis are dependent on the T2S system for localization. It was concluded previously that the T2S system is involved in VPS production (43); however, this conclusion was made prior to the knowledge of the important role of matrix proteins in biofilm formation. While VPS may make up the majority of the mass of the V. cholerae biofilm matrix (49), it is clear that the presence of VPS alone is not enough to form a functional, productive biofilm.
While it is possible that the primary step of biofilm formation affected by the T2S system in V. cholerae is the building of a biofilm matrix, it is conceivable that other stages of biofilm maturation in V. cholerae and/or other organisms could also be dependent on this secretion system. For instance, extracellular enzymes, such as proteases and lipases, that are secreted by the T2S system may increase the longevity of the biofilm by digesting lysed cells or biofilm matrix components for nutrient acquisition and recycling. Additionally, extracellular enzymes can degrade structural matrix components to promote the detachment of resident bacteria, thereby allowing new biofilms to be formed. It is clear, however, that the T2S system does not support the extracellular transport of all proteins that participate in the formation, stability, and dispersion of biofilms, since our data suggest that the secretion of the DNase Dns does not require the T2S system. While the mechanism by which Dns translocates the outer membrane of V. cholerae has yet to be determined, it is apparent that biofilms are affected by a variety of extracellular components, and these components are transported by different secretion pathways, some of which may be coregulated.
In many bacteria, signaling by the second messenger 3′,5′-cyclic diguanylic acid (c-di-GMP) determines the timing of complex biological processes such as biofilm formation and maturation. High intracellular levels of c-di-GMP lead to an increase in the production of matrix components, including exopolysaccharides and extracellular DNA, and an increase in the level of biofilm formation (50–52). Interestingly, the level of transcription of T2S genes increased 2-fold after c-di-GMP levels were elevated by overexpression of the diguanylate cyclase VCA0956 (53). The correlation between the expression of T2S components and that of genes involved in biofilm formation, including the vps genes, suggests that these systems may be coregulated by c-di-GMP signaling; however, the mechanism through which c-di-GMP activates T2S gene expression remains to be determined.
Since biofilm formation is a key step in environmental persistence and a source of infections, understanding the mechanisms by which the widely distributed, highly conserved T2S system is regulated and contributes to biofilm formation and maintenance is of great medical importance and may identify ways to eradicate and prevent the development of biofilms.
ACKNOWLEDGMENTS
This work was supported by awards R01AI049294 (to M.S.) and R01AI055987 (to F.H.Y.) from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print 29 September 2014
REFERENCES
- 1.Korotkov KV, Sandkvist M, Hol WG. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10:336–351. 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandkvist M, Morales V, Bagdasarian M. 1993. A protein required for secretion of cholera toxin through the outer membrane of Vibrio cholerae. Gene 123:81–86. 10.1016/0378-1119(93)90543-C. [DOI] [PubMed] [Google Scholar]
- 3.Cianciotto NP. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13:581–588. 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Sikora AE. 2013. Proteins secreted via the type II secretion system: smart strategies of Vibrio cholerae to maintain fitness in different ecological niches. PLoS Pathog. 9:e1003126. 10.1371/journal.ppat.1003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirn TJ, Jude BA, Taylor RK. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863–866. 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]
- 6.Evans FF, Egan S, Kjelleberg S. 2008. Ecology of type II secretion in marine gammaproteobacteria. Environ. Microbiol. 10:1101–1107. 10.1111/j.1462-2920.2007.01545.x. [DOI] [PubMed] [Google Scholar]
- 7.Putker F, Tommassen-van Boxtel R, Stork M, Rodriguez-Herva JJ, Koster M, Tommassen J. 2013. The type II secretion system (Xcp) of Pseudomonas putida is active and involved in the secretion of phosphatases. Environ. Microbiol. 15:2658–2671. 10.1111/1462-2920.12115. [DOI] [PubMed] [Google Scholar]
- 8.Mulcahy H, Charron-Mazenod L, Lewenza S. 2010. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 12:1621–1629. 10.1111/j.1462-2920.2010.02208.x. [DOI] [PubMed] [Google Scholar]
- 9.Kolter R, Greenberg EP. 2006. Microbial sciences: the superficial life of microbes. Nature 441:300–302. 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland IW. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222–227. 10.1016/S0966-842X(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 11.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187–209. 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 12.Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17:109–118. 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 36:893–916. 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633. 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 15.Fong JCN, Karplus K, Schoolnik GK, Yildiz FH. 2006. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J. Bacteriol. 188:1049–1059. 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Absalon C, Van Dellen K, Watnick PI. 2011. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 7:e1002210. 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. 2011. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J. Biol. Chem. 286:16555–16566. 10.1074/jbc.M110.211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berk V, Fong JCN, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. 2012. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337:236–239. 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moorthy S, Watnick PI. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623–1635. 10.1111/j.1365-2958.2005.04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong JCN, Yildiz FH. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol. 189:2319–2330. 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allison DG, Ruiz B, SanJose C, Jaspe A, Gilbert P. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179–184. 10.1111/j.1574-6968.1998.tb13225.x. [DOI] [PubMed] [Google Scholar]
- 22.Saville RM, Rakshe S, Haagensen JAJ, Shukla S, Spormann AM. 2011. Energy-dependent stability of Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 193:3257–3264. 10.1128/JB.00251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasa I, Penades JR. 2006. Bap: a family of surface proteins involved in biofilm formation. Res. Microbiol. 157:99–107. 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Tielker D, Hacker S, Loris R, Strathmann M, Wingender J, Wilhelm S, Rosenau F, Jaeger KE. 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151:1313–1323. 10.1099/mic.0.27701-0. [DOI] [PubMed] [Google Scholar]
- 25.Diggle SP, Stacey RE, Dodd C, Camara M, Williams P, Winzer K. 2006. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ. Microbiol. 8:1095–1104. 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- 26.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59:1229–1238. 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 27.de Lorenzo V. 1994. Designing microbial systems for gene expression in the field. Trends Biotechnol. 12:365–371. 10.1016/0167-7799(94)90037-X. [DOI] [PubMed] [Google Scholar]
- 28.Lybarger SR, Johnson TL, Gray MD, Sikora AE, Sandkvist M. 2009. Docking and assembly of the type II secretion complex of Vibrio cholerae. J. Bacteriol. 191:3149–3161. 10.1128/JB.01701-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461. 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 30.Fong JCN, Syed KA, Klose KE, Yildiz FH. 2010. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology 156:2757–2769. 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714. 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song T, Sabharwal D, Gurung JM, Cheng AT, Sjostrom AE, Yildiz FH, Uhlin BE, Wai SN. 2014. Vibrio cholerae utilizes direct sRNA regulation in expression of a biofilm matrix protein. PLoS One 9:e101280. 10.1371/journal.pone.0101280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overbye LJ, Sandkvist M, Bagdasarian M. 1993. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene 132:101–106. 10.1016/0378-1119(93)90520-D. [DOI] [PubMed] [Google Scholar]
- 34.Sandkvist M, Bagdasarian M, Howard SP, DiRita VJ. 1995. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 14:1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandkvist M, Michel LO, Hough LP, Morales VM, Bagdasarian M, Koomey M, DiRita VJ. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray MD, Bagdasarian M, Hol WG, Sandkvist M. 2011. In vivo cross-linking of EpsG to EpsL suggests a role for EpsL as an ATPase-pseudopilin coupling protein in the type II secretion system of Vibrio cholerae. Mol. Microbiol. 79:786–798. 10.1111/j.1365-2958.2010.07487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wai SN, Mizunoe Y, Takade A, Kawabata SI, Yoshida SI. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96:4028–4033. 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yildiz FH, Dolganov NA, Schoolnik GK. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716–1726. 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newland JW, Green BA, Foulds J, Holmes RK. 1985. Cloning of extracellular DNase and construction of a DNase-negative strain of Vibrio cholerae. Infect. Immun. 47:691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Focareta T, Manning PA. 1987. Extracellular proteins of Vibrio cholerae: molecular cloning, nucleotide sequence and characterization of the deoxyribonuclease (DNase) together with its periplasmic localization in Escherichia coli K-12. Gene 53:31–40. 10.1016/0378-1119(87)90090-4. [DOI] [PubMed] [Google Scholar]
- 42.Seper A, Fengler VHI, Roier S, Wolinski H, Kohlwein SD, Bishop AL, Camilli A, Reidl J, Schild S. 2011. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol. 82:1015–1037. 10.1111/j.1365-2958.2011.07867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali A, Johnson JA, Franco AA, Metzger DJ, Connell TD, Morris JG, Jr, Sozhamannan S. 2000. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68:1967–1974. 10.1128/IAI.68.4.1967-1974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overhage J, Lewenza S, Marr AK, Hancock REW. 2007. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J. Bacteriol. 189:2164–2169. 10.1128/JB.01623-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldi DL, Higginson EE, Hocking DM, Praszkier J, Cavaliere R, James CE, Bennett-Wood V, Azzopardi KI, Turnbull L, Lithgow T, Robins-Browne RM, Whitchurch CB, Tauschek M. 2012. The type II secretion system and Its ubiquitous lipoprotein substrate, SslE, are required for biofilm formation and virulence of enteropathogenic Escherichia coli. Infect. Immun. 80:2042–2052. 10.1128/IAI.06160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White BP. 1938. The rugose variant of vibrios. J. Pathol. 46:1–6. 10.1002/path.1700460102. [DOI] [Google Scholar]
- 47.Morris JG, Jr, Sztein MB, Rice EW, Nataro JP, Losonsky GA, Panigrahi P, Tacket CO, Johnson JA. 1996. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 174:1364–1368. 10.1093/infdis/174.6.1364. [DOI] [PubMed] [Google Scholar]
- 48.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497–515. 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 49.Yildiz F, Fong J, Sadovskaya I, Grard T, Vinogradov E. 2014. Structural characterization of the extracellular polysaccharide from Vibrio cholerae O1 El-Tor. PLoS One 9:e86751. 10.1371/journal.pone.0086751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857–869. 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Römling U, Gomelsky M, Galperin MY. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629–639. 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 52.Römling U. 2013. Microbiology: bacterial communities as capitalist economies. Nature 497:321–322. 10.1038/nature12103. [DOI] [PubMed] [Google Scholar]
- 53.Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600–3613. 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sikora AE, Lybarger SR, Sandkvist M. 2007. Compromised outer membrane integrity in Vibrio cholerae type II secretion mutants. J. Bacteriol. 189:8484–8495. 10.1128/JB.00583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott ME, Dossani ZY, Sandkvist M. 2001. Directed polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc. Natl. Acad. Sci. U. S. A. 98:13978–13983. 10.1073/pnas.241411198. [DOI] [PMC free article] [PubMed] [Google Scholar]