ABSTRACT
Membrane-active peptides, components of capsid structural proteins, assist viruses in overcoming the host membrane barrier in the initial stages of infection. Several such peptides have been identified, and their roles in membrane fusion or disruption have been characterized through biophysical studies. In several members of the Picornaviridae family, the role of the VP4 structural peptide in cellular-membrane penetration is well established. However, there is not much information on the membrane-penetrating capsid components of hepatitis A virus (HAV), an unusual member of this family. The VP4 peptide of HAV differs from its analogues in other picornaviruses in being significantly shorter in length and in lacking a signal for myristoylation, thought to be a critical requisite for VP4-mediated membrane penetration. Here we report, for the first time, that the atypical VP4 in HAV contains significant membrane-penetrating activity. Using a combination of biophysical assays and molecular dynamics simulation studies, we show that VP4 integrates into membrane vesicles through its N-terminal region to finally form discrete pores of 5- to 9-nm diameter, which induces leakage in the vesicles without altering their overall size or shape. We further demonstrate that the membrane activity of VP4 is specific toward vesicles mimicking the lipid content of late endosomes at acidic pH. Taken together, our data indicate that VP4 might be essential for the penetration of host endosomal membranes and release of the viral genome during HAV entry.
IMPORTANCE Hepatitis A virus causes acute hepatitis in humans through the fecal-oral route and is particularly prevalent in underdeveloped regions with poor hygienic conditions. Although a vaccine for HAV exists, its high cost makes it unsuitable for universal application in developing countries. Studies on host-virus interaction for HAV have been hampered due to a lack of starting material, since the virus is extremely slow growing in culture. Among the unknown aspects of the HAV life cycle is its manner of host membrane penetration, which is one of the most important initial steps in viral infection. Here, we present data to suggest that a small peptide, VP4, a component of the HAV structural polyprotein, might be essential in helping the viral genome cross cell membranes during entry. It is hoped that this work might help in elucidating the manner of initial host cell interaction by HAV.
INTRODUCTION
Traversing the host membrane barrier is an essential step in the establishment of a viral infection, and viruses typically contain sequestered, hydrophobic, or amphipathic components to execute this step. These components, which are elements of viral glycoproteins or capsid proteins, are instrumental either in negotiating fusion of the viral lipid envelopes with host membranes or in causing disruption of plasma or endosomal membranes (1–3). While membrane fusion proceeds through a mechanistically similar pathway in most viruses, the process of cellular-membrane disruption appears to vary from virus to virus rather than displaying a unified mechanism. The formation of voltage-gated channels, the creation of size-selective pores leading to osmolysis, and the induction of positive curvature on membranes have been proposed as possible mechanisms for the latter process (4–6).
Although virus-mediated membrane penetration has hitherto been neatly classified based on the presence or absence of a lipid envelope, the discovery of viruses at the borderline of enveloped and nonenveloped species has complicated our understanding of this seemingly familiar process (7, 8). The first established example of this phenomenon is hepatitis A virus (HAV), an unusual member of the Picornaviridae family. Recent studies have shown that HAV exists in two forms—a regular, nonenveloped form and an enveloped form (eHAV), which is predominant in the sera of infected individuals (7). Although both forms are equally infectious, their means of cellular-membrane penetration—specifically, whether the two forms of HAV have separate pathways of entry into host cells or whether a unified pathway exists for this purpose—is unclear. While it is possible that eHAV has evolved its own separate mechanism for membrane fusion-based entry, no protein component analogous to glycoproteins of enveloped viruses has been detected in the eHAV lipid envelope so far. Another plausible entry mechanism involves the conversion of eHAV into its nonenveloped counterpart during the initial encounter with host cells. The shedding of the lipid component would allow the viral capsid proteins to interact with the HAV cellular receptor 1 (HAVCR-1) (9, 10) and to disrupt host cell membranes, but there is no experimental support for this pathway of entry either. A study carried out prior to the discovery of eHAV suggests an endosomal route for entry (11); however, the role played by the capsid proteins of HAV in this process has not been addressed.
The HAV capsid, like that of other members of the picornavirus family, is expected to be a pseudo T=3 icosahedral particle constructed from 60 copies each of four structural proteins, VP1, VP2, VP3, and VP4. Whether the small VP4 protein is a component of the HAV capsid is still not clearly known (12). The exact size of VP4 is also ambiguous, since it may be composed of 21 or 23 amino acids based on which one of two alternative start codons is utilized to initiate HAV polyprotein synthesis (12). The absence of a high-resolution structure of the HAV capsid has also precluded an accurate understanding of the positioning of capsid proteins in the capsid shell so far (13). A recently determined, unpublished structure of the HAV capsid could be extremely useful in elucidating the structural details of individual capsid proteins in the future.
For several well-studied picornaviruses, capsid proteins VP1 and VP4 have been implicated in cellular-membrane penetration during virus entry. For poliovirus, membrane disruption during virus entry is negotiated by the N-terminal 31 residues of VP1 and by VP4, a myristoylated, hydrophobic peptide which remains in the interior of the capsid prior to entry (5, 14). The release of the poliovirus genome in the host cell cytosol requires concerted membrane interaction by these components; while VP4 forms voltage-gated channels in the plasma membrane (5), thus allowing the RNA genome to pass through, the amphipathic N terminus of VP1 anchors the virus particle safely onto the membrane during this process through the formation of long, extended networks or channels (15). The VP1 N termini of other picornaviruses, like human rhinovirus type 2 (HRV2), have a more active role in cellular entry, since they probably participate directly in cellular-membrane penetration, as indicated by their ability to disrupt liposomes in vitro (16, 17). The VP4 peptide, which is often myristoylated at the N terminus, has also been found to be directly involved in membrane disruption by picornaviruses other than poliovirus (18). Thus, the necessity for VP4 and the N terminus of VP1 in the entry pathway of picornaviruses has been adequately demonstrated by experiments in which mutation in these regions has severely affected virus infectivity.
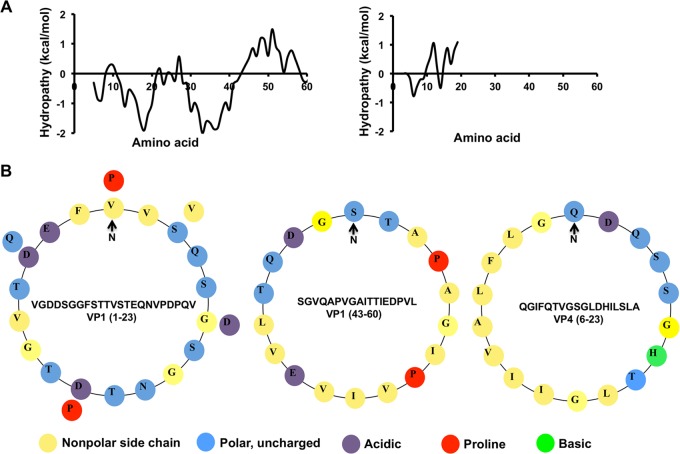
Since it is conceivable that the mechanism of membrane penetration by the nonenveloped form of HAV is similar to that of other picornaviruses, we attempted to identify amphipathic or hydrophobic regions in the VP1 N terminus or VP4 of HAV which could potentially be involved in virus entry. Using hydrophobicity plots, we detected two hydrophobic stretches (residues 1 to 23 and 43 to 60) at the N terminus of VP1, while residues 6 to 23 of the short, 23-amino-acid polypeptide VP4 was found to be almost perfectly amphipathic. Interestingly, it is known that unlike the VP4 peptide in most other members of the picornaviruses, the VP4 peptide of HAV is not myristoylated (19). We tested the ability of synthetic peptides corresponding to either unmyristoylated VP4 or the N-terminal regions of VP1 to interact with fluorescent-dye-filled liposomes in vitro and found that VP4 was extremely efficient in causing disruption of membrane vesicles, although no membrane-penetrating activity was displayed by the N-terminal region of VP1. We also found that the activity of VP4 is highly specific for vesicles mimicking the lipid composition of late endosomal compartments at low-pH conditions. Based on the results of a combination of biochemical methods and molecular dynamics (MD) simulation studies, we propose a mechanism for VP4-orchestrated membrane penetration whereby initial penetration of the N terminus of VP4 into the outer leaflet of membranes eventually results in the induction of small pores in lipid vesicles. Our studies indicate that VP4 might be involved in allowing the viral genome to escape endosomal compartments during entry of the envelope-less form of HAV in host cells.
MATERIALS AND METHODS
Peptides.
Peptides corresponding to amino acids 1 to 23 and 43 to 60 of HAV VP1, 1 to 23 of HAV VP4, and 364 to 381 of the capsid protein of an insect virus, Flock House virus (FHV), were obtained from GenPro Biotech. The latter is designated the γ1 peptide.
Preparation of liposomes.
Liposomes composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC; Avanti Polar Lipids) and encapsulating the fluorescent dye sulforhodamine B (SulfoB; Sigma-Aldrich) were prepared using the protocol described in reference 20. Briefly, DOPC was dried under N2 gas and rehydrated to a final concentration of 2 mM in 10 mM sodium phosphate buffer (pH 7.0) containing 50 mM SulfoB. After multiple freeze-thaw cycles, the lipid suspension was extruded through a miniextruder (Avanti Polar Lipids) equipped with a polycarbonate filter containing 100-nm-size pores. The liposomes were purified by passing them through a PD-10 column (GE Healthcare) and were utilized for assays within 3 h of preparation. DOPC liposomes containing 8-aminonaphthalene-1,2,3-trisulfonic acid (ANTS) and p-xylene-bis(pyridinium) bromide (DPX) were similarly prepared, with rehydration being carried out in phosphate buffer containing 100 mM ANTS, 200 mM DPX, or a combination of 50 mM ANTS and 100 mM DPX.
Liposomes mimicking cellular membranes/compartments and composed of various molar ratios of cholesterol, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), phosphatidyl ethanolamine, sphingomyelin, phosphatidyl serine, and bis(monoacylglycero) phosphate were prepared similarly, based on the molar ratios mentioned in reference 21 and Table S1 in the supplemental material. The total lipid concentration in the rehydration buffer was maintained at 10 mM.
Liposomes incorporating dextran molecules were prepared similarly but with minor modifications. Three different types of fluorescent-dye-labeled dextrans were utilized: fluorescein isothiocyanate (FITC)-conjugated dextran with a molecular mass of 3,000 to 5,000 Da (FD 3-5), FITC-conjugated dextran with a molecular mass of 10,000 Da (FD 10), and tetramethylrhodamine-conjugated dextran with a molecular mass of 40,000 Da (FD 40) (Sigma-Aldrich). The total concentration of lipid or lipid mixture in rehydration buffer (10 mM sodium phosphate, pH 7.0) was maintained at 10 mM. The rehydration buffer also contained 100 mg/ml of FD 3-5, FD 10, or FD 40. After several freeze-thaw cycles, the mixture was extruded through a polycarbonate filter with 200-nm-size pores. To separate liposomes from unincorporated dextrans, we subjected the mixtures to size exclusion chromatography on a Superdex S200 column (GE Healthcare). The fractions in the void volume of the column were collected, and the extent of fluorescence dequenching upon the addition of 1% Triton X-100 to individual fractions was utilized to confirm the presence of liposomes. The liposomes were utilized for assays within 3 h of preparation.
Liposome disruption assay.
For the assay, 1 μl of purified liposomes was incubated at room temperature with HAV VP4 or VP1 or FHV γ1 peptide at concentrations anging from 300 nM to 50 μM for 20 min. Disruption of SulfoB-containing liposomes was measured with fluorescence spectroscopy (PerkinElmer) based on dequenching of SulfoB fluorescence at 585 nm. The percentage of SulfoB released was calculated as 100[(F1 − F0)/(Ftx100 − F0)], where F1 is fluorescence intensity measured in the presence of peptide, F0 is fluorescence intensity of liposome only, and Ftx100 is fluorescence intensity in the presence of 1% Triton X-100. Similarly, disruption of liposomes incorporating fluorescent-dye-labeled dextrans was measured by the dequenching of FITC (for FD 3-5 and FD 10) and tetramethylrhodamine (for FD 40) at emission wavelengths of 520 nm and 575 nm, respectively.
Dynamic light scattering.
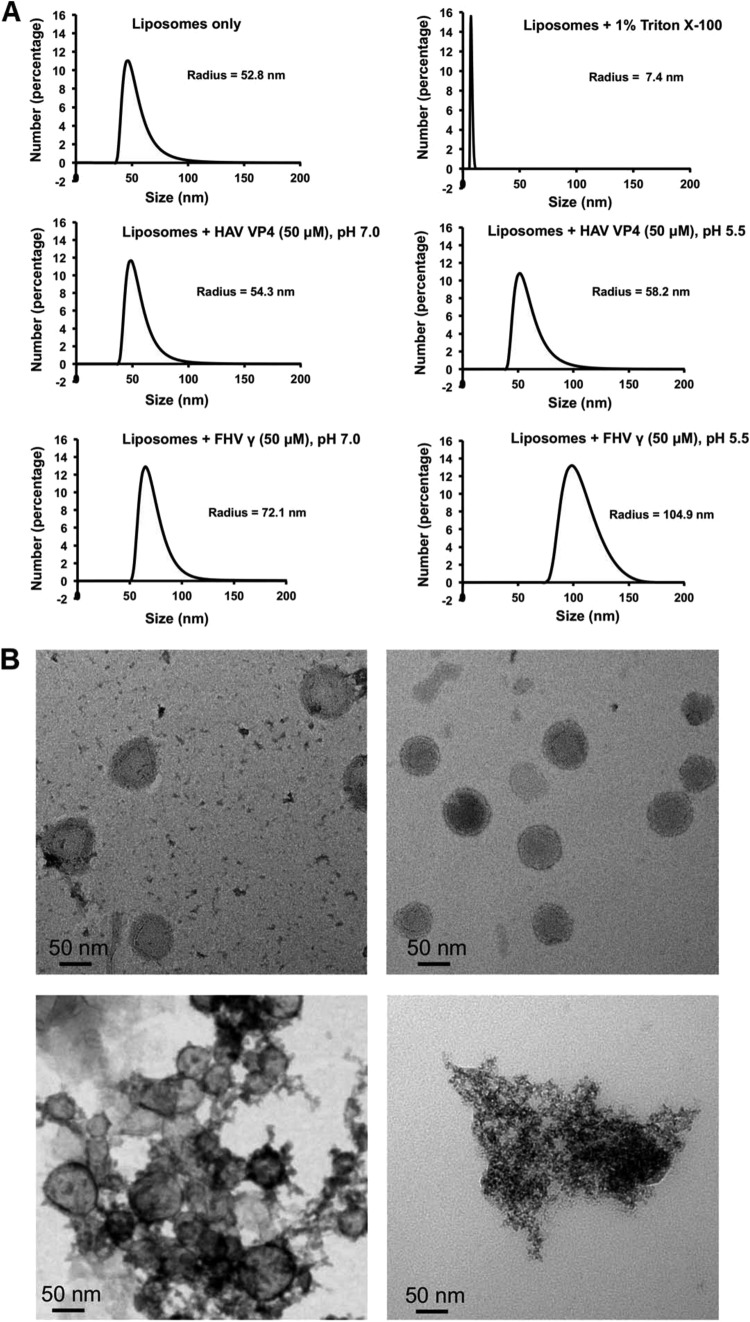
One microliter of purified liposomes, untreated and in buffer or treated with either 50 μM HAV VP4 or FHV γ1 peptide or 1% Triton X-100 at room temperature, was subjected to dynamic light scattering (DLS) in a Zetasizer instrument (Malvern). Data corresponding to each sample were collected in triplicate. The mean hydrodynamic radius, Rh(mean), of representative samples is shown in Fig. 4A.
FIG 4.
(A) Dynamic light-scattering studies showing the mean hydrodynamic radius [Rh(mean)] of DOPC liposomes in buffer or after 20 min of incubation with 1% Triton X-100, 50 μM HAV VP4, or FHV γ1 at pH values of 7.0 and 5.5. (B) Images obtained upon negative staining and electron microscopy of DOPC liposomes, either untreated (top left) or after 20 min of incubation with 50 μM HAV VP4 (top right) at pH 5.5, FHV γ1 (bottom left) at pH 5.5, and 1% Triton X-100 (bottom right).
Circular dichroism.
Circular dichroism (CD) spectra of the HAV VP4 and FHV γ1 peptide were measured in the far UV range (190 to 250 nm) using a J-815 CD spectrophotometer (Jasco) with a 1-mm-path-length cuvette. The peptides were at a concentration of 50 μM in the presence of 10 mM sodium phosphate buffer or 50% tetrafluoroethylene (TFE) and at 100 μM in the presence of lipid vesicles. Representative data from three separate studies are shown.
ANTS/DPX assay.
ANTS/DPX liposome leakage/fusion assays were carried out with HAV VP4 and γ1 peptides at a concentration of 50 μM. Peptides were incubated with DOPC liposomes loaded separately with ANTS and DPX for 20 min at room temperature, and leakage of ANTS was assayed by fluorescence spectroscopy (PerkinElmer) (excitation wavelength [λex] = 355 nm; emission wavelength [λem] = 520 nm).
Electron microscopy.
Five microliters of diluted DOPC liposomes, either untreated or incubated with 50 μM HAV VP4 peptide or FHV γ1 peptide at pH 5.5, was applied to 300-square-mesh carbon-coated copper grids (Electron Microscopy Sciences). The excess material was removed by filter paper, followed by the addition of 5 μl of 1% (wt/vol) aqueous solution of uranyl acetate as a negative staining agent. After being washed with distilled water, the grid was dried at room temperature, and the morphology of the liposomes was examined using a Tecnai TF20 transmission electron microscope (FEI Company) at an acceleration voltage of 200 kV.
Molecular dynamics simulation. (i) Structure prediction of HAV VP4.
The three-dimensional structure of HAV VP4 (1MNMSRQGIFQTVGSGLDHILSLA23) was predicted using Bhageerath-H (a homology ab initio hybrid web server for protein tertiary structure prediction) (22–26). Of the five candidate predicted structures (see Fig. S1 in the supplemental material), the lowest-energy structure was used as the starting point for further studies. The predicted structure displayed a helix-turn-helix conformation, which correlated well with structures predicted from other prediction servers.
(ii) System setup.
Three different starting orientations of the HAV VP4 peptide with respect to the membrane surface were used for simulations and analyses. The peptide was placed on top of an equilibrated lipid bilayer comprising 238 POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) molecules (a CHARMM36 force field-compatible POPC molecule was made available by A. Ghosh, Centre for Development of Advanced Computing, India). The POPC bilayer system had 238 lipid molecules in total, i.e., 119 lipid molecules in each leaflet. The orientations of the HAV VP4 peptide with respect to the membrane were (i) with the N-terminal residue, Met1, oriented and placed toward the membrane surface, (ii) with the C-terminal residue, Ala23, oriented and placed toward the membrane surface, and (iii) with a parallel orientation of the peptide on the membrane surface. After setups, three independent, all-atom explicit-solvent molecular dynamics (MD) simulations were carried out with these structures. All the HAV VP4 bilayer systems were fully solvated in water. Counterions (Cl− or Na+) were added to neutralize each system.
(iii) Molecular dynamics simulations.
Simulations were initiated using HAV VP4 in three different orientations with respect to the membrane. All MD simulations were performed for 50 ns in the isothermal-isobaric (NPT) ensemble using the GROMACS 4.6.1 software package (27, 28). The simple point charge (SPC) model was used for water molecules in the simulations (29). All simulations were performed according to the standard protocol, consisting of energy minimization followed by gradual heating of the system. Each of the systems was initially minimized, employing 20,000 steps of steepest descent followed by conjugate gradient minimization. Topology and parameter files were generated using the CHARMM36 force field (30, 31). The equilibration of each system was achieved by performing a 5-ns MD run. Positional restraints were applied to each system during equilibration, while the system was slowly heated up from 0 to 300 K at every 2 ps. After equilibration was reached, three independent 50-ns-duration MD simulations were carried out with periodic boundary conditions at a temperature of 300 K. The Berendsen thermostat with a coupling constant of 0.1 ps was employed to keep the temperature constant (300 K) (32). A pressure coupling was applied semi-isotropically with a coupling constant of 1.0 ps, using the Berendsen algorithm, to keep the pressure constant (1 bar). Lennard-Jones interactions were used as a cutoff at 1.2 nm. The long-range electrostatics was handled by the particle mesh Ewald (PME) method (33), and van der Waals (vdW) interactions were calculated using a cutoff of 1.4 nm. The LINear Constraint Solver (LINCS) algorithm was used to constrain all bond lengths (34). Analyses were performed with GROMACS analysis tools and Visual Molecular Dynamics (VMD) software (version 1.9) (35). All MD simulations were performed on a single graphics processing unit (GPU) card, installed at the Supercomputing Facility (http://www.scfbio-iitd.res.in/) of the Indian Institute of Technology Delhi.
(iv) Graphics.
All figures were generated using VMD (35), the UCSF CHIMERA package (36), and the PyMOL molecular graphics system, version 1.5.0.4 (Schrödinger, LLC; http://www.pymol.org), as required.
RESULTS
HAV VP4, and not the N terminus of VP1, disrupts membranes in vitro.
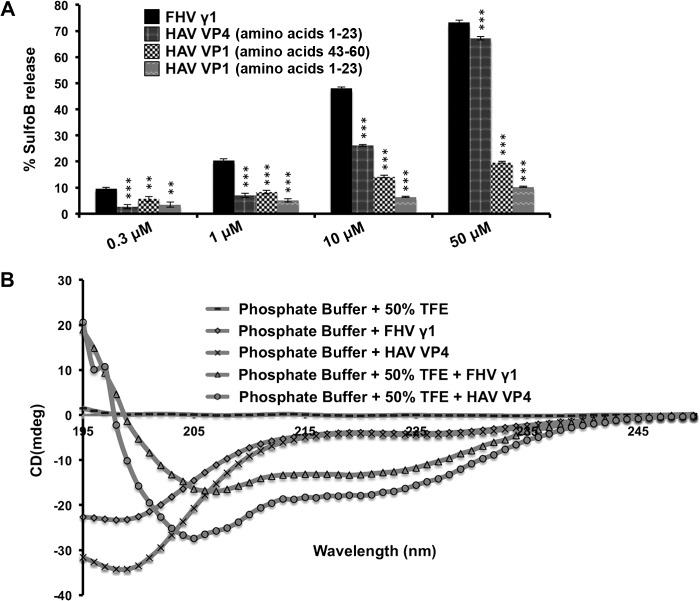
Peptides corresponding to amino acids 1 to 23 and 43 to 60 of VP1 and 1 to 23 of VP4 (Fig. 1A and B) were synthesized, and their ability to interact with liposomes was tested. A peptide corresponding to amino acids 364 to 381 (designated the γ1 region) of the capsid protein of Flock House virus (FHV) was utilized as a positive control for these assays (37). The γ1 region of FHV is well established as being competent for membrane penetration in vitro (37, 38) and required for endosomal-membrane penetration by FHV during entry into host cells (20, 39). The ability of the peptides corresponding to HAV VP4 and the VP1 N terminus to disrupt SulfoB-containing DOPC liposomes was compared with that of the γ1 peptide at concentrations ranging from 300 nM to 50 μM (Fig. 2A). Although the peptides corresponding to the N terminus of HAV VP1 displayed little ability to disrupt liposomes, the peptide corresponding to VP4 was able to disrupt liposomes to almost the same extent as γ1 at a concentration of 50 μM (Fig. 2A). Both γ1 and VP4 were able to cause ∼70% dequenching of SulfoB fluorescence compared to the extent of dequenching (considered 100%) caused by a detergent, Triton X-100. Thus, HAV VP4 has the ability to disrupt liposomes to the same level as a recognized cellular membrane-penetrating peptide from a model nonenveloped virus (39).
FIG 1.
(A) Hydrophobicity plots of the N terminus of VP1 (left) and VP4 (right). (B) Helical wheel representations, from left to right, of VP1 (residues 1 to 23), VP1 (residues 43 to 60), and VP4 (residues 6 to 23). The sequences corresponding to these regions are provided.
FIG 2.
(A) Disruption of DOPC liposomes by VP1 and VP4 peptides at concentrations ranging from 0.3 to 50 μM. The degree of disruption is represented by the extent of dequenching of the fluorescent dye sulforhodamine B (SulfoB), with the extent of dequenching obtained by the addition of 1% Triton X-100 considered to be 100%. Data are represented as the means of the results for triplicate independent samples ± the standard deviation (SD). ***, P < 0.001; **, P < 0.01 (Student's t test in comparison with results for the FHV γ1 peptide). (B) Circular dichroism spectroscopy of HAV VP4 and FHV γ1 in phosphate buffer, pH 7.0, and in the same buffer containing 50% TFE. The concentration of each peptide used was 50 μM. mdeg, millidegrees.
To check the conformation of VP4 in a hydrophobic environment, we carried out CD spectroscopy and computed the percentage of secondary-structure elements. While in phosphate buffer, VP4 mostly existed as a random coil, the structure of the peptide altered to a significantly α-helical (47.2%) conformation in the presence of 50% tetrafluoroethylene (TFE) (Fig. 2B). This indicates the possibility of a major structural alteration in VP4 in the hydrophobic environment of cellular membranes. Several tertiary structure prediction softwares indicated that the preferred, energy-minimized conformation for VP4 is predominantly helical, with an elongated, N-terminal helix separated from a diminutive C-terminal one by a loop region (see Fig. S1 in the supplemental material). The γ1 peptide also demonstrated a significantly helical (∼58%) secondary structure in 50% TFE (40) (Fig. 2B).
Liposomes mimicking endosomal vesicles are specifically disrupted by HAV VP4.
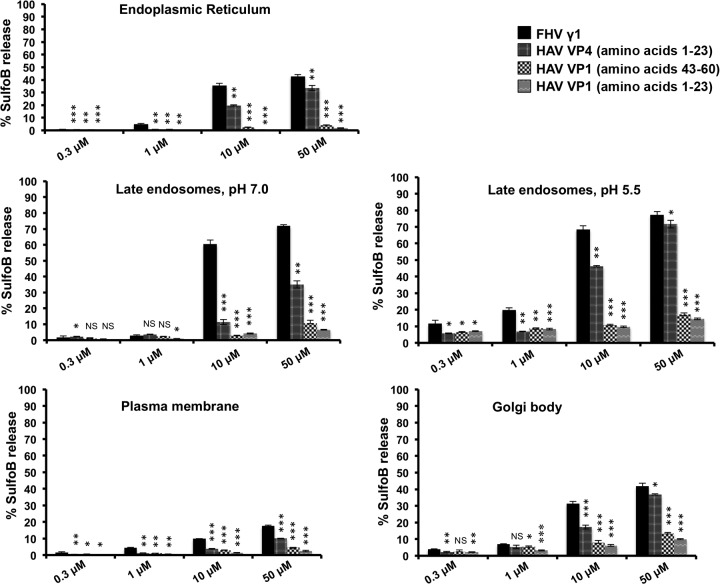
The optimal activity of viral membrane-active peptides is expected to depend on the nature of the limiting membrane. The limiting membrane for HAV entry is not clearly known, with support existing in the literature for both entry through plasma membrane at neutral pH and low-pH-dependent entry through the endosomal route (9–11). It is also possible that different forms of HAV utilize disparate modes of entry (7). In order to check whether there is any lipid composition-specific variation in the membrane penetration activity of HAV VP4, we produced separate batches of liposomes mimicking various cellular organelles. The liposomes were generated from a combination of phosphatidyl choline (POPC), phosphatidyl ethanolamine (POPE), phosphatidyl serine (POPS), sphingomyelin, cholesterol, and bis(monoacylglycero)phosphate (BMP) in order to obtain the closest working approximation of the mammalian plasma membrane as well as the membranes of organelles such as endoplasmic reticulum, Golgi bodies, and late endosomal compartments (21) (see Table S1 in the supplemental material). The extent of disruption of these liposomes by HAV VP4 was quantified at pH 7.0 and additionally at pH 5.5 for liposomes mimicking endosomal compartments. The last condition generated the maximum membrane disruption by HAV VP4, while approximately half the disruption (∼40%) was detected with the same compartments at neutral pH (Fig. 3). This indicates that HAV VP4 is equipped to cause maximum membrane disruption at late endosomal conditions. The 2-fold increase in membrane disruption ability of HAV VP4 at a lower pH may be due to (i) alterations in the conformational state of the peptide or (ii) increased association between peptides and lipid or between multiple peptides as a function of pH change. In order to check the former possibility, we computed the secondary structure of HAV VP4 in lipid vesicles mimicking late endosomal conditions. The α-helical content of VP4 was found to be similar at neutral and low pH conditions (see Fig. S2 in the supplemental material), which indicates that low pH probably promotes association between peptides in a hydrophobic environment rather than increasing the helical content of individual peptides.
FIG 3.
Degree of disruption of liposomes mimicking different cellular organelles by VP4 and VP1 peptides at concentrations ranging from 0.3 to 50 μM. Liposomes mimicking late endosomal vesicles were tested at two pH conditions, 7.0 and 5.5. FHV γ1 was utilized as a control for all assays. Data are represented as the means of the results for triplicate independent samples ± SD. ***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, not significant (P > 0.05; Student's t test in comparison with results for the FHV γ1 peptide).
Interestingly, although the FHV γ1 peptide also caused maximum membrane disruption with liposomes mimicking the lipid content of late endosomal vesicles, it did not demonstrate any pH-specific activity. This is somewhat expected, since the role of low endosomal pH during FHV entry is merely to expose the γ peptide from within the capsid interior, and not to cause conformational changes in γ (20, 40).
Of the rest of the organelle-specific liposomes tested, the ones mimicking the lipid content of Golgi bodies and endoplasmic reticulum membranes were somewhat disrupted by HAV VP4, although there was a minimal effect of the peptide on plasma membrane-specific liposomes (Fig. 3). This indicates the possibility of VP4-mediated entry of HAV into mammalian cells through the endocytic pathway.
HAV VP4 does not cause complete collapse of liposomes.
Unlike hydrophobic fusion proteins of enveloped viruses, which execute the “trimer-of-hairpins” fusion pathway (2), membrane-active peptides of nonenveloped viruses, which damage membranes, have no unifying mechanism for functionality (1). The suggested modes of action for the latter vary widely, with poliovirus VP4 creating voltage-gated channels in membranes (5), reovirus μ1 generating size-selective pores (4, 41), and protein VI of adenovirus collapsing membranes through induction of positive curvature (6). In order to gain an understanding of the mechanism of HAV VP4-mediated disruption of liposomes, we subjected DOPC liposomes to dynamic light-scattering before and after treatment with VP4 peptide and obtained the mean hydrodynamic radius [Rh(mean)] of the liposomes in each case (Fig. 4A). While the addition of 1% Triton X-100 reduced the Rh(mean) of a liposome population from 52.8 nm to 7.4 nm, indicating complete collapse, the addition of 50 μM VP4 hardly altered the mean Rh of liposomes in the observed 20 min. This result indicates that unlike detergents or some nonenveloped viruses like adenovirus, which cause complete collapse of membrane vesicles, HAV VP4 causes membrane damage more discretely, probably through the introduction of small pores or channels reminiscent of the early interaction of reovirus μ1 with model membranes (4). This observation is supported by electron microscopy, which was utilized to visualize liposomes before and after the addition of HAV VP4 (Fig. 4B). In both cases, liposomes were visualized as circular, unilameller vesicles of approximately 100-nm diameter, which indicated that VP4-mediated membrane damage did not alter the essential shape or size of liposomes within 20 min, in spite of causing enough membrane damage within that time period to allow the encapsulated fluorescent dye to escape (Fig. 2A). This manner of membrane disruption is somewhat distinct from that exhibited by the γ1 peptide, which appears to cause substantial disruption and clumping of the vesicles (Fig. 4B). Dynamic light scattering shows that incubation of liposomes with γ1 increases the Rh(mean) of liposomes from 52.8 nm to 72.1 nm, which possibly indicates coalescence of the vesicles (Fig. 4A) in addition to induction of leakage. Similar results were obtained upon testing the effect of HAV VP4 and FHV γ1 on liposomes mimicking the lipid composition of late endosomal vesicles (see Fig. S3 in the supplemental material).
Discrete pores of 5 to 9 nm are formed in endosome-specific vesicles through the action of HAV VP4.
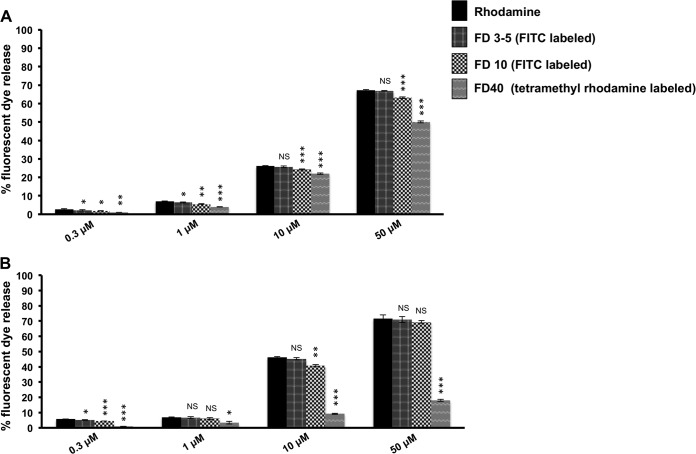
Further studies were carried out to understand the nature of the damage inflicted on host membranes by HAV VP4. DOPC liposomes as well as liposomes mimicking late endosomal compartments, each incorporating fluorescently labeled dextran molecules of various sizes (3 to 5 kDa, corresponding to an average diameter of 2.8 nm, and 10 and 40 kDa, corresponding to diameters of 4.6 and 9 nm, respectively), were generated (41). The ability of HAV VP4 to release each type of dextran from encapsulating vesicles was separately determined by measuring the dequenching of dextran-associated fluorescence (Fig. 5). While HAV VP4 was able to cause almost equivalent release of all three sizes of dextran molecules tested from DOPC liposomes, the release of 40-kDa dextrans from late endosome-mimicking liposomes was significantly reduced, although there was no change in the extent of release of 3- to 5-kDa and 10-kDa dextrans. Thus, although morphological and DLS-based studies of VP4-incubated liposomes indicated formation of discrete pores in the membranes, the size of pores formed was somewhat dependent on the lipid composition of the membrane. While pores generated on DOPC membranes by HAV VP4 probably had a diameter close to or larger than 9 nm, the pores formed on late endosome-specific membrane vesicles essentially were of smaller diameters (5 to 9 nm), which did not allow 40-kDa dextrans to pass through (Fig. 5). It is possible that the presence of a combination of lipids and cholesterol effectively decreased the pore diameters.
FIG 5.
Extent of release of fluorescent-dye-labeled dextrans of various sizes from DOPC liposomes (A) and liposomes mimicking late endosomal vesicles (B) by HAV VP4 at concentrations ranging from 0.3 to 50 μM at pH 5.5. The extent of dequenching upon release of the free or dextran-associated fluorescent dyes rhodamine, FITC, and tetramethyl rhodamine was determined by measuring the increase in fluorescence at emission wavelengths of 585 nm, 520 nm, and 575 nm, respectively, with the extent of dequenching obtained by the addition of 1% Triton X-100 considered to be 100% in each case. The incubation time of peptide with liposome was maintained at 20 min for each assay. Data are represented as the means of the results for triplicate independent samples ± SD. ***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, not significant (P > 0.05; Student's t test in comparison with the results for rhodamine dye release).
HAV VP4 does not induce fusion in membranes.
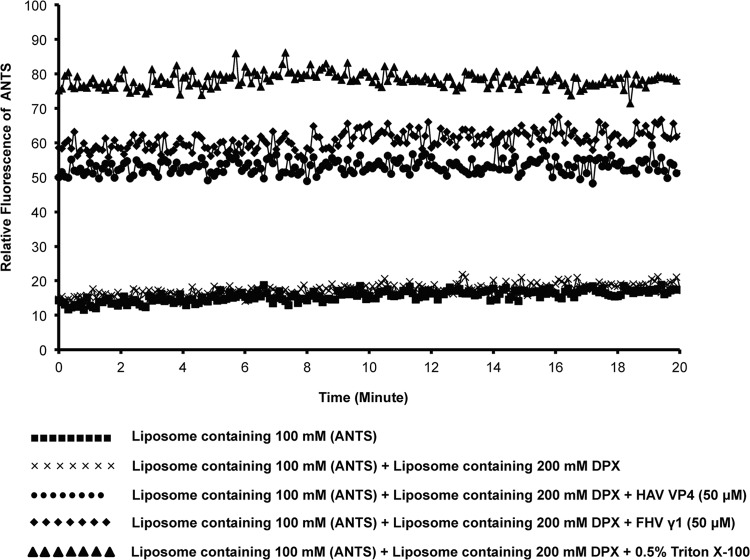
The discovery that HAV exists in both enveloped (eHAV) and nonenveloped forms essentially dictates either (i) two separate modes of entry by the virus and the existence of viable mechanisms for both lipid fusion and membrane disruption or (ii) conversion of one form into the predominant “entry-specific” form. There is no example so far in the literature for the occurrence of both types of entry mechanisms in the same virus capsid. The fusion peptides of enveloped viruses and the membrane penetration peptides of nonenveloped viruses typically have specific and unique ways of interacting with lipid membranes, in spite of occasional structural similarities (40). The HAV VP4 peptide was nonetheless tested for its ability to carry out the mixing of aqueous vesicle content by the ANTS/DPX assay, which is traditionally utilized for detecting vesicle fusion. Separate DOPC liposome mixtures containing either 100 mM the polyanionic fluorescent dye ANTS or 200 mM the cationic quencher DPX or a mixture of 50 mM ANTS and 100 mM DPX were prepared, and ANTS fluorescence in the presence or absence of 50 μM HAV VP4 was monitored over time (Fig. 6). In the event of liposome fusion by the action of VP4, the fluorescence of ANTS would be quenched by DPX. However, the addition of HAV VP4 or FHV γ1 peptide caused an instantaneous increase in ANTS fluorescence, which remained steady over a time period of 20 min, indicating that the peptides cause vesicle leakage instead of fusion (Fig. 6). Similar results were obtained for 0.5% Triton X-100, which is known to cause leakage in liposomes. Thus, HAV VP4 was not a multifunctional peptide capable of inducing membrane fusion, and its functionality appeared limited to causing membrane damage.
FIG 6.
ANTS/DPX fusion assay with HAV VP4 and FHV γ1. A mixture of liposomes containing 100 mM the fluorescent dye ANTS and 200 mM the quencher DPX was either left untreated or treated with 50 μM VP4, FHV γ1, or 0.5% Triton X-100, and the fluorescence of ANTS was monitored continuously for 20 min at an emission wavelength of 520 nm. No decrease in ANTS fluorescence was detected at any point, suggesting leakage, but not fusion, of vesicles.
Molecular dynamics simulations reveal the initial contact of VP4 with membranes.
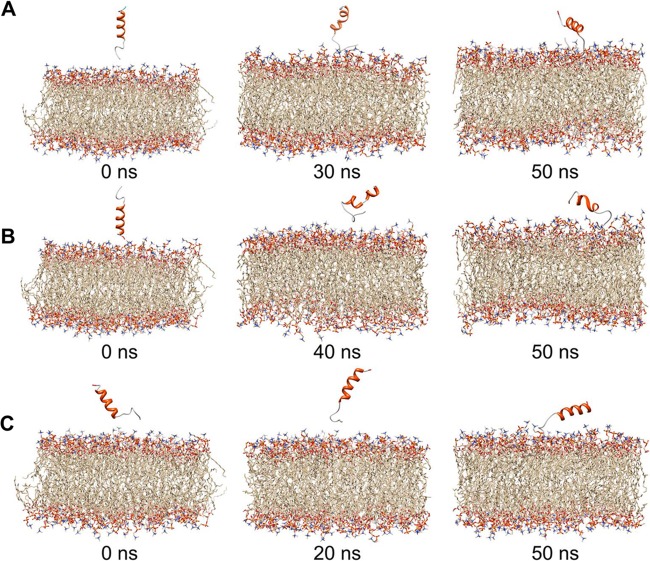
The objective of the simulations of HAV VP4 with preequilibrated POPC membrane was to determine a plausible mode of interaction of HAV VP4 with the membrane and of its penetration into the membrane surface. Three independent 50-ns MD simulations were carried out in order to model the possible modes of interaction of VP4 residues with membrane (Fig. 7A to C). The simulations were carried out with a tertiary structure for VP4 predicted by Bhageerath-H (22–26). Of the five predicted possible structures, the lowest-energy structure, comprised of a helix-turn-helix conformation, was selected as the starting structure for MD simulations (see Fig. S1 in the supplemental material). Three different starting orientations of the peptide with respect to the membrane surface were used for the simulations: (i) the N terminus of VP4 (Met1) oriented and placed toward the membrane, (ii) the C terminus oriented and placed toward the membrane, and (iii) a parallel orientation of the peptide placed on the POPC membrane surface. In the simulation study corresponding to the first orientation of VP4 (Fig. 7A), the Met1 residue rapidly approached the hydrophilic lipid slab of the membrane surface during the initial 5 ns of the simulation. The peptide further attained a parallel orientation with respect to the membrane surface and remained unaltered in this conformation up to ∼25 ns. During this time, the C-terminal helical region remained stable while the loop region at the N terminus appeared to form a hinge-like structure (Fig. 7A). After the 25-ns run, the N-terminal region embedded itself into the upper leaflet of the membrane bilayer and continued in this conformation until the end of the simulation (50 ns) (Fig. 7A). During this period, several hydrogen-bonding interactions among N terminus residues of VP4, such as Met1, Asn2, Ser4, and Arg5, with the head group regions of the POPC membrane were detected (see Table S2 in the supplemental material) and were probably instrumental in allowing the peptide to penetrate into the membrane. The backbone root mean square deviation (RMSD) of VP4 remained stable during the simulation, suggesting that the overall structure of the system remained unaltered and stable during the MD run (see Fig. S4A in the supplemental material).
FIG 7.
Snapshots from molecular dynamics simulation studies of HAV VP4 placed with the N terminus facing the model membrane (A), with the C terminus facing the model membrane (B), and in an orientation parallel to the model membrane (C). Images shown are from the beginning (left), after 20 to 40 ns (middle), and after 50 ns (right) of simulations.
With the second orientation, consisting of the C-terminal residue (Ala23) placed toward the membrane (Fig. 7B), the peptide appeared unstable on the membrane surface for 40 ns. The peptide also exhibited loss of secondary structure during this time. From 40 ns onwards, the peptide managed to position itself in an inclined orientation with respect to the membrane surface, so that the N terminus pointed toward the membrane surface (Fig. 7B). Further, within a span of 2 ns, the peptide acquired an upright position, allowing the N-terminal residues to face the membrane. Eventually, the N terminus region formed a hinge-shaped structure and penetrated the hydrophilic head groups of the upper leaflet of the membrane surface (Fig. 7B). This event was accompanied by H-bonding interactions, forming between Ser4, Arg5 and Ile8 of the peptide, and POPC molecules (see Table S2 in the supplemental material). The backbone RMSD of VP4 in this orientation appeared to be less stable (see Fig. S4B in the supplemental material), indicating that the peptide was not in a favorable orientation to interact with the membrane.
In the third and final orientation, the peptide was placed parallel to the membrane surface (Fig. 7C). During the first 2 ns, the N terminus interacted with the membrane, mainly through H-bonding contacts (see Table S2 in the supplemental material) mediated through Met1, Asn2, and Arg5 of the peptide until the end of the simulation (50 ns). The backbone RMSD of VP4 showed that the peptide and its interactions with the membrane surface were stable throughout the simulation (see Fig. S4C in the supplemental material).
The observation that during the course of the simulations, only the N terminus of HAV VP4 was interacting with and penetrating the membrane bilayer, irrespective of the initial orientation, indicates that this is the perhaps the preferred mode of interaction of VP4 with lipid membranes (Fig. 7A to C). The initial penetration is probably mediated by formation of an H-bonding interaction between Met1, Asn2, and Arg5 residues situated at the N terminus of VP4 with the hydrophilic head groups of the outer leaflet of the lipid bilayer. Also, at several instances during the first and third simulations, a circular ring-like conformation was acquired by the C-terminal region of VP4 (see Fig. S5 in the supplemental material). It is possible that this structural unit observed during simulations might eventually become stable through interaction with lipid molecules and participate in creating a pore through the membrane.
DISCUSSION
HAV is responsible for ∼40% of all cases of viral hepatitis worldwide and is prevalent among populations in developing countries with poor hygienic and sanitary conditions (12). Recent socioeconomic improvements in some countries has created an incipient danger of large-scale outbreaks of HAV due to the collocation of susceptible pockets with generally resistant populations. China has experienced several outbreaks over the last couple of decades, and it has been reported recently that the level of anti-HAV antibodies in Indian newborns has been depleted to 50% to 60% within a span of 15 years (42). Further, economic progress in specific areas has coincided with an epidemiological shift of the disease from usually asymptomatic early childhood infections to symptomatic infections in the 2nd and 3rd decades of life, which carry an increased risk of liver failure (43). Thus, it is essential to develop and implement more effective preventive measures and therapies against HAV, which necessitates a clear understanding of the manner of host cell interaction by HAV. HAV is arguably the least well-studied of all picornaviruses, since the slow-growing nature of the virus precludes production of large quantities of purified material for biochemical and structural studies (13). This has hampered studies on, among other areas, the role of specific capsid proteins in the establishment of infection.
We have identified the capsid structural peptide VP4 of HAV as a potential component for host membrane penetration during entry. The in vitro membrane disruption ability of HAV VP4 is comparable to that of well-established, well-studied membrane penetration peptides, such as the γ1 peptide of the model nonenveloped virus FHV (39). The involvement of VP4 in mediating cellular entry for members of the Picornaviridae family is also well established in literature. Poliovirus VP4 is known to form ion channels in membranes to allow genome escape (5), and a glutathione S-transferase (GST)-tagged recombinant VP4 from human rhinovirus type 16 (HRV16) can cause disruption of model membranes in vitro (18). Although there is no direct evidence for the involvement of the VP4 polypeptides of coxsackievirus, echovirus, and enterovirus in membrane penetration, they are comparable to the VP4 polypeptides of poliovirus and HRV16 in length and degree of hydrophobicity and in having a myristoylation signal (N-terminal G-x-x-x-T/S). These striking similarities suggest possible parallel functionalities during cellular entry. The presence of a hydrophobic myristate has been linked to the efficiency of membrane penetration as well as to the assembly and maturation of viruses in the Picornaviridae and Reoviridae families (44–49). Interestingly, HAV VP4 differs from its analogues in other picornaviruses in a number of fundamental points: it is about a third of the length of other VP4 polypeptides, displays significantly elevated hydrophobicity, and does not contain a myristoylation signal at the N terminus. There is a putative myristoylation signal 5 residues downstream of the VP4 N terminus; however, experimental evidence proves that VP4 does not undergo any modification at this region (19). Thus, the membrane disruption activity displayed by HAV VP4 is entirely a function of its own hydrophobic character and is in no way dependent on myristate-lipid interactions.
By analogy with other members of the Picornaviridae family, we had hypothesized that both VP4 and VP1 might be involved in membrane penetration during HAV entry. The N-terminal region of VP1 in picornaviruses displays various degrees of hydrophobicity and is expected to have essential but somewhat diverse roles during virus entry. The VP1 N terminus from HRV2 has been shown to contain membrane-disrupting activity in vitro and is probably instrumental in allowing the viral genome to escape from endosomes (16, 17). In contrast, the N-terminal residues of poliovirus VP1 are capable of associating with liposomes (14), although there is no experimental evidence for vesicle disruption. The latest data indicate that VP1 probably functions to anchor poliovirus to cellular membranes and to protect the genomic RNA during disassembly, while VP4 primarily creates pores in cellular membranes (5, 15). The two hydrophobic regions identified at the N-terminal domain of HAV VP1 are unable to cause disruption of membrane vesicles in vitro. It is, however, possible that these regions can associate with membranes and assist in virus entry in some fashion.
The membrane activity of HAV VP4 is significantly more pronounced toward liposomes with a lipid profile corresponding to that of the late endosomal compartments, while minimal preference is exhibited for double-membrane vesicles mimicking the mammalian plasma membrane. While early endosomes are generated from plasma membranes and contain a similar lipid composition, upon maturation the membranes of these vesicles become enriched in the anionic lipid bis(monoacylglycero)phosphate (BMP) (21, 50). It has been shown that the presence of BMP is essential for membrane penetration and delivery of cargo by several cell-penetrating peptides (CPPs). CPPs can disrupt late endosomal vesicles containing BMP but are ineffective against the plasma membrane, which lacks BMP while containing a similar proportion of sterols and other lipid components (21, 50). Thus, HAV VP4 might be similar to CPPs in its requirement for the presence of BMP for optimal membrane penetration. VP4 has an additional requirement for low-pH conditions for optimal activity, since only half as much disruption of vesicles mimicking late endosomal compartments is achieved by 50 μM the peptide at pH 7.0. However, we have confirmed that low-pH conditions do not serve to alter the secondary structure of the peptide in any way.
The membrane activity of HAV VP4 does not cause any dramatic change in the structure or physical appearance of either DOPC liposomes or liposomes mimicking late endosomes. Our data show that the overall size and shape of liposomes remain constant up to 20 min after incubation with the peptide, while pores of 5- to 9-nm diameter are induced in the membranes, allowing fluorescent-dye-associated dextrans of various sizes to pass through. Thus, the membrane-penetrating mechanism of HAV VP4 appears similar to that of poliovirus and HRV2, both of which form pores in cellular membranes (5, 16, 17). Interestingly, the pores induced by HAV VP4 in DOPC liposomes are somewhat larger in diameter (>9 nm) than those formed in late-endosome-specific liposomes. This slight difference in the end result might reflect a somewhat altered degree or manner of association of VP4 with vesicles containing different lipid compositions. This mode of membrane damage is similar to that carried out by the μ1N peptide of orthoreovirus, which is required for endosomal membrane disruption during virus entry. μ1N has been shown to induce the formation of size-specific pores in membrane vesicles, which eventually rupture due to osmolysis (4, 41).
We utilized all-atom molecular dynamics (MD) simulation studies with a predicted tertiary structure of HAV VP4 in order to understand the mode of interaction of the peptide with model membranes. Although MD simulation is a powerful tool and has been utilized effectively for mapping the manner of membrane interaction by peptides of bacterial and mammalian origin as well as fusion peptides from enveloped viruses like influenza virus and HIV (51–54), there are very few examples of utilization of this technique for understanding the mechanism of action of viral-membrane-disrupting or -associating peptides (51). Among the latter group, only the membrane interaction of the N-terminal region of poliovirus VP1, which is responsible for the association of the virus with the host plasma membrane during genome release, has been mapped using MD simulation (51). Our studies with various orientations of HAV VP4 with respect to a model membrane clearly indicate that the initial encounter with and penetration into the leaflet of cellular membranes is mediated through the N-terminal end of the peptide. Indeed, the relative instability of the backbone RMSD of the system, when the C-terminal end of the peptide is placed close to the model membrane, suggests that this is an unfavorable orientation. Optimal membrane disruption may also require an oligomeric form of VP4. It is possible that VP4 peptides are released from the HAV capsid in an oligomeric form or that association between peptides is promoted during membrane interaction. Significantly longer simulations incorporating multiple copies of VP4 coupled with corroborating functional evidence is required to confirm whether oligomerization is a functional necessity for VP4-mediated membrane disruption.
The recent discovery of an enveloped version of HAV has made understanding the cellular-entry process of this virus a uniquely interesting challenge. Interestingly, both eHAV and nonenveloped HAV are thought to use the same receptor, HAVCR-1, for cellular entry (7). Thus, it is possible that the envelope of eHAV is removed upon initial encounter with host cells, allowing the same set of capsid proteins to mediate receptor interaction and membrane disruption for both the enveloped and the nonenveloped versions. It is difficult to speculate on the possibility of eHAV utilizing membrane fusion to enter host cells, since protein components analogous to lipid-embedded glycoproteins have not yet been detected in eHAV. A fusion-competent peptide was previously discovered in capsid protein VP3 (55, 56); however, utilization of this region for fusion of the eHAV envelope with host cell membranes will necessitate its exposure on the surface of the eHAV lipid coat.
The preferential activity of VP4 toward late-endosome-specific vesicles suggests that this peptide might be involved in endosomal escape of the HAV genome in the initial steps of virus entry. HAV VP4 has previously been shown to have an essential role in virion morphogenesis (57), although detection of this peptide in HAV capsid preparations has been difficult. This is probably due to a combination of limited starting material and the small size of VP4. Additionally, VP4 does not display any fusion activity, which rules out the scenario that it might be a multifunctional membrane-active peptide able to assist in both membrane fusion and disruption activities of the enveloped and nonenveloped versions of the virus, respectively. Thus, it is possible that although HAV spends a major part of its life cycle as an enveloped virus, its mode of entry into host cells is through the traditional route of membrane disruption employed by nonenveloped viruses in general. VP4 may be required for membrane disruption by eHAV after its lipid envelope has been shed during entry.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Department of Biotechnology (DBT), India. A.K.P. was supported by a Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), and M.B. was supported by the Ramalingaswami Fellowship from the Department of Biotechnology (DBT).
We thank John E. Johnson (The Scripps Research Institute, CA, USA) for critical reading of the manuscript and B. Jayaram (Indian Institute of Technology, Delhi, India) for allowing the use of the Supercomputing Facility for Bioinformatics and Computational Biology, IIT-Delhi, for molecular dynamics simulations.
Footnotes
Published ahead of print 13 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01896-14.
REFERENCES
- 1.Banerjee M, Johnson JE. 2008. Activation, exposure and penetration of virally encoded, membrane-active polypeptides during non-enveloped virus entry. Curr. Protein Pept. Sci. 9:16–27. 10.2174/138920308783565732. [DOI] [PubMed] [Google Scholar]
- 2.Kielian M, Rey FA. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67–76. 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai B. 2007. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 23:23–43. 10.1146/annurev.cellbio.23.090506.123454. [DOI] [PubMed] [Google Scholar]
- 4.Agosto MA, Ivanovic T, Nibert ML. 2006. Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc. Natl. Acad. Sci. U. S. A. 103:16496–16501. 10.1073/pnas.0605835103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danthi P, Tosteson M, Li QH, Chow M. 2003. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J. Virol. 77:5266–5274. 10.1128/JVI.77.9.5266-5274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier O, Marvin SA, Wodrich H, Campbell EM, Wiethoff CM. 2012. Spatiotemporal dynamics of adenovirus membrane rupture and endosomal escape. J. Virol. 86:10821–10828. 10.1128/JVI.01428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Z, Lemon SM. 2014. Peek-a-boo: membrane hijacking and the pathogenesis of viral hepatitis. Trends Microbiol. 22:59–64. 10.1016/j.tim.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silberstein E, Konduru K, Kaplan GG. 2009. The interaction of hepatitis A virus (HAV) with soluble forms of its cellular receptor 1 (HAVCR1) share the physiological requirements of infectivity in cell culture. Virology J. 6:175. 10.1186/1743-422X-6-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silberstein E, Xing L, van de Beek W, Lu J, Cheng H, Kaplan GG. 2003. Alteration of hepatitis A virus (HAV) particles by a soluble form of HAV cellular receptor 1 containing the immunoglobin- and mucin-like regions. J. Virol. 77:8765–8774. 10.1128/JVI.77.16.8765-8774.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishop NE. 1998. Examination of potential inhibitors of hepatitis A virus uncoating. Intervirology 41:261–271. 10.1159/000024948. [DOI] [PubMed] [Google Scholar]
- 12.Tesar M, Harmon SA, Summers DF, Ehrenfeld E. 1992. Hepatitis A virus polyprotein synthesis initiates from two alternative AUG codons. Virology 186:609–618. 10.1016/0042-6822(92)90027-M. [DOI] [PubMed] [Google Scholar]
- 13.Martin A, Lemon SM. 2006. Hepatitis A virus: from discovery to vaccines. Hepatology 43:S164–172. 10.1002/hep.21052. [DOI] [PubMed] [Google Scholar]
- 14.Fricks CE, Hogle JM. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. 2013. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J. Virol. 87:3903–3914. 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prchla E, Kuechler E, Blaas D, Fuchs R. 1994. Uncoating of human rhinovirus serotype 2 from late endosomes. J. Virol. 68:3713–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prchla E, Plank C, Wagner E, Blaas D, Fuchs R. 1995. Virus-mediated release of endosomal content in vitro: different behavior of adenovirus and rhinovirus serotype 2. J. Cell Biol. 131:111–123. 10.1083/jcb.131.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis MP, Bottley G, Beales LP, Killington RA, Rowlands DJ, Tuthill TJ. 2008. Recombinant VP4 of human rhinovirus induces permeability in model membranes. J. Virol. 82:4169–4174. 10.1128/JVI.01070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tesar M, Jia XY, Summers DF, Ehrenfeld E. 1993. Analysis of a potential myristoylation site in hepatitis A virus capsid protein VP4. Virology 194:616–626. 10.1006/viro.1993.1301. [DOI] [PubMed] [Google Scholar]
- 20.Odegard AL, Kwan MH, Walukiewicz HE, Banerjee M, Schneemann A, Johnson JE. 2009. Low endocytic pH and capsid protein autocleavage are critical components of Flock House virus cell entry. J. Virol. 83:8628–8637. 10.1128/JVI.00873-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Meer G, Voelker DR, Feigenson GW. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhingra P, Jayaram B. 2013. A homology/ab initio hybrid algorithm for sampling near-native protein conformations. J. Comput. Chem. 34:1925–1936. 10.1002/jcc.23339. [DOI] [PubMed] [Google Scholar]
- 23.Mishra A, Rao S, Mittal A, Jayaram B. 2013. Capturing native/native like structures with a physico-chemical metric (pcSM) in protein folding. Biochim. Biophys. Acta 1834:1520–1531. 10.1016/j.bbapap.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Jayaram B, Bhushan K, Shenoy SR, Narang P, Bose S, Agrawal P, Sahu D, Pandey V. 2006. Bhageerath: an energy based web enabled computer software suite for limiting the search space of tertiary structures of small globular proteins. Nucleic Acids Res. 34:6195–6204. 10.1093/nar/gkl789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishra A, Rana SP, Mittal A, Jayaram B. 2014. D2N: distance to the native. Biochim. Biophys. Acta 1844:1798–1807. 10.1016/j.bbapap.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Jayaram B, Dhingra P, Mishra A, Kaushik R, Mukherjee G, Singh A, Shekhar S. Bhageerath-H: a homology/ab initio hybrid server for predicting tertiary structures of monomeric soluble proteins. BMC Bioinformatics, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. 2005. GROMACS: fast, flexible, and free. J. Comput. Chem. 26:1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 28.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E. 2013. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berendsen HJC, Postma JPM, Gunsteren WFV, Hermans J. 1981. Interaction models for water in relation to protein hydration, p 331–342 D. Reidel Publishing Co., Dordrecht, The Netherlands. [Google Scholar]
- 30.Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD., Jr 2012. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theor. Comput. 8:3257–3273. 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klauda JB, Monje V, Kim T, Im W. 2012. Improving the CHARMM force field for polyunsaturated fatty acid chains. J. Phys. Chem. B 116:9424–9431. 10.1021/jp304056p. [DOI] [PubMed] [Google Scholar]
- 32.Berendsen HJC, Postma JPM, Van Gunsteren WF, DiNola A, Haak JR. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]
- 33.Essmann U, Perera L, Berkowitch ML, Darden T, Hsing L, Pedersen LG. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. 10.1063/1.470117. [DOI] [Google Scholar]
- 34.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. . [DOI] [Google Scholar]
- 35.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38, 27, 28. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 36.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 37.Bong DT, Janshoff A, Steinem C, Ghadiri MR. 2000. Membrane partitioning of the cleavage peptide in flock house virus. Biophys. J. 78:839–845. 10.1016/S0006-3495(00)76641-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janshoff A, Bong DT, Steinem C, Johnson JE, Ghadiri MR. 1999. An animal virus-derived peptide switches membrane morphology: possible relevance to nodaviral transfection processes. Biochemistry 38:5328–5336. 10.1021/bi982976i. [DOI] [PubMed] [Google Scholar]
- 39.Odegard A, Banerjee M, Johnson JE. 2010. Flock House virus: a model system for understanding non-enveloped virus entry and membrane penetration. Curr. Top. Microbiol. Immunol. 343:1–22. 10.1007/82_2010_35. [DOI] [PubMed] [Google Scholar]
- 40.Maia LF, Soares MR, Valente AP, Almeida FC, Oliveira AC, Gomes AM, Freitas MS, Schneemann A, Johnson JE, Silva JL. 2006. Structure of a membrane-binding domain from a non-enveloped animal virus: insights into the mechanism of membrane permeability and cellular entry. J. Biol. Chem. 281:29278–29286. 10.1074/jbc.M604689200. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Agosto MA, Ivanovic T, King DS, Nibert ML, Harrison SC. 2009. Requirements for the formation of membrane pores by the reovirus myristoylated μ1N peptide. J. Virol. 83:7004–7014. 10.1128/JVI.00377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franco E, Meleleo C, Serino L, Sorbara D, Zaratti L. 2012. Hepatitis A: epidemiology and prevention in developing countries. World J. Hepatol. 4:68–73. 10.4254/wjh.v4.i3.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathur P, Arora NK. 2008. Epidemiological transition of hepatitis A in India: issues for vaccination in developing countries. Indian J. Med. Res. 128:699–704. [PubMed] [Google Scholar]
- 44.Chow M, Moscufo N. 1995. Myristoyl modification of viral proteins: assays to assess functional roles. Methods Enzymol. 250:495–509. 10.1016/0076-6879(95)50093-6. [DOI] [PubMed] [Google Scholar]
- 45.Moscufo N, Simons J, Chow M. 1991. Myristoylation is important at multiple stages in poliovirus assembly. J. Virol. 65:2372–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marc D, Drugeon G, Haenni AL, Girard M, van der Werf S. 1989. Role of myristoylation of poliovirus capsid protein VP4 as determined by site-directed mutagenesis of its N-terminal sequence. EMBO J. 8:2661–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marc D, Girard M, van der Werf S. 1991. A Gly1 to Ala substitution in poliovirus capsid protein VP0 blocks its myristoylation and prevents viral assembly. J. Gen. Virol. 72:1151–1157. 10.1099/0022-1317-72-5-1151. [DOI] [PubMed] [Google Scholar]
- 48.Marc D, Masson G, Girard M, van der Werf S. 1990. Lack of myristoylation of poliovirus capsid polypeptide VP0 prevents the formation of virions or results in the assembly of noninfectious virus particles. J. Virol. 64:4099–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tillotson L, Shatkin AJ. 1992. Reovirus polypeptide σ3 and N-terminal myristoylation of polypeptide μ1 are required for site-specific cleavage to μ1C in transfected cells. J. Virol. 66:2180–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang ST, Zaitseva E, Chernomordik LV, Melikov K. 2010. Cell-penetrating peptide induces leaky fusion of liposomes containing late endosome-specific anionic lipid. Biophys. J. 99:2525–2533. 10.1016/j.bpj.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong GS, Chen CP, Lin MH, Kruger J, Becker CF, Fink RH, Fischer WB. 2012. Molecular dynamics simulations and conductance studies of the interaction of VP1 N-terminus from polio virus and gp41 fusion peptide from HIV-1 with lipid membranes. Mol. Membr. Biol. 29:9–25. 10.3109/09687688.2011.644589. [DOI] [PubMed] [Google Scholar]
- 52.Cirac AD, Moiset G, Mika JT, Kocer A, Salvador P, Poolman B, Marrink SJ, Sengupta D. 2011. The molecular basis for antimicrobial activity of pore-forming cyclic peptides. Biophys. J. 100:2422–2431. 10.1016/j.bpj.2011.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haria NR, Monticelli L, Fraternelli F, Lorenz CD. 2014. Plasticity and conformational equilibria of influenza fusion peptides in model lipid bilayers. Biochim. Biophys. Acta 1838:1169–1179. 10.1016/j.bbamem.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 54.Poojari C, Xiao D, Batista VS, Strodel B. 2013. Membrane penetration induced by aggregates of human islet amyloid polypeptides. Biophys. J. 105:2323–2332. 10.1016/j.bpj.2013.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chávez A, Busquets MA, Pujol M, Alsina MA, Cajal Y. 1998. pH-induced destabilization of lipid bilayers by a peptide from the VP3 protein of the capsid of hepatitis A virus. Analyst 123:2251–2256. 10.1039/a804562c. [DOI] [PubMed] [Google Scholar]
- 56.Chávez A, Pujol M, Haro I, Alsina MA, Cajal Y. 2001. Membrane fusion by an RGD-containing sequence from the core protein VP3 of hepatitis A virus and the RGA-analogue: implications for viral infection. Biopolymers 58:63–77. . [DOI] [PubMed] [Google Scholar]
- 57.Probst C, Jecht M, Gauss-Muller V. 1999. Intrinsic signals for the assembly of hepatitis A virus particles. Role of structural proteins VP4 and 2A. J. Biol. Chem. 274:4527–4531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.