ABSTRACT
Rotavirus (RV) nonstructural protein 4 (NSP4) is a virulence factor that disrupts cellular Ca2+ homeostasis and plays multiple roles regulating RV replication and the pathophysiology of RV-induced diarrhea. Although its native oligomeric state is unclear, crystallographic studies of the coiled-coil domain (CCD) of NSP4 from two different strains suggest that it functions as a tetramer or a pentamer. While the CCD of simian strain SA11 NSP4 forms a tetramer that binds Ca2+ at its core, the CCD of human strain ST3 forms a pentamer lacking the bound Ca2+ despite the residues (E120 and Q123) that coordinate Ca2+ binding being conserved. In these previous studies, while the tetramer crystallized at neutral pH, the pentamer crystallized at low pH, suggesting that preference for a particular oligomeric state is pH dependent and that pH could influence Ca2+ binding. Here, we sought to examine if the CCD of NSP4 from a single RV strain can exist in two oligomeric states regulated by Ca2+ or pH. Biochemical, biophysical, and crystallographic studies show that while the CCD of SA11 NSP4 exhibits high-affinity binding to Ca2+ at neutral pH and forms a tetramer, it does not bind Ca2+ at low pH and forms a pentamer, and the transition from tetramer to pentamer is reversible with pH. Mutational analysis shows that Ca2+ binding is necessary for the tetramer formation, as an E120A mutant forms a pentamer. We propose that the structural plasticity of NSP4 regulated by pH and Ca2+ may form a basis for its pleiotropic functions during RV replication.
IMPORTANCE The nonstructural protein NSP4 of rotavirus is a multifunctional protein that plays an important role in virus replication, morphogenesis, and pathogenesis. Previous crystallography studies of the coiled-coil domain (CCD) of NSP4 from two different rotavirus strains showed two distinct oligomeric states, a Ca2+-bound tetrameric state and a Ca2+-free pentameric state. Whether NSP4 CCD from the same strain can exist in different oligomeric states and what factors might regulate its oligomeric preferences are not known. This study used a combination of biochemical, biophysical, and crystallography techniques and found that the NSP4 CCD can undergo a reversible transition from a Ca2+-bound tetramer to a Ca2+-free pentamer in response to changes in pH. From these studies, we hypothesize that this remarkable structural adaptability of the CCD forms a basis for the pleiotropic functional properties of NSP4.
INTRODUCTION
Ca2+ is an important regulatory molecule involved in many cellular processes. Numerous viral pathogens exploit Ca2+ signaling pathways to establish an environment in the host conducive for robust infection and replication (1). In addition to disrupting the Ca2+ homeostasis of host cells, many virus proteins utilize Ca2+-binding sites to modulate protein structure-function and facilitate virus assembly and capsid integrity.
Rotaviruses (RV) belong to the Reoviridae family and are a major cause of severe diarrheal disease in children throughout the world. RV replication causes numerous changes to cellular Ca2+ homeostasis, and Ca2+ is necessary for infectious virus assembly. Several studies have characterized different cellular proteins and signaling pathways disrupted or exploited by RV-induced elevation in cytosolic Ca2+ levels (2–8). Elevated cytoplasmic Ca2+ also may regulate RV replication by direct binding to virus-encoded Ca2+-binding proteins. Three RV proteins bind or are predicted to bind Ca2+, and this is important to their function. First, the outer capsid protein VP7 binds Ca2+, stabilizing VP7 trimers that are necessary for triple-layered particle assembly and stability (9–11). Second, the nonstructural protein 5 (NSP5) has two DXDXD pseudo EF-hand Ca2+-binding motifs. Although direct evidence that these motifs bind Ca2+ has not been shown, these sites influence the formation of viroplasm-like structures required for RNA replication in a Ca2+-dependent manner (12). Finally, multiple crystal structures of the NSP4 coiled-coil domain (CCD; amino acids 95 to 137) show a Ca2+ ion bound at the core of a tetrameric coiled-coil; however, the biological relevance of this putative Ca2+-binding site remains unknown (12–15).
NSP4 is a viral glycoprotein initially synthesized in the endoplasmic reticulum (ER) that acts as an intracellular receptor that binds to nascent double-layered particles (DLPs) and plays a key role in virus maturation (16). NSP4 exhibits pleiotropic properties, associates with different cell compartments (17), and plays a regulatory role in RV replication through activation of autophagy (4). NSP4 is the only RV-encoded protein that triggers the release of Ca2+ from the ER (18–20) by functioning as a viroporin in infected cells (21). NSP4 also is an oligomeric protein that can adopt multiple forms, which depend on the fragment of NSP4 being analyzed and whether NSP4 is expressed with other RV proteins (22–25). Predictions indicate NSP4 has multiple putative Ca2+-binding sites (26); however, available structural information provides evidence only for a Ca2+-binding site positioned in the core of the tetrameric coiled-coil structure formed by the NSP4 CCD, which folds into an α-helical conformation. The NSP4 Ca2+-binding site is coordinated by residues glutamate 120 (E120) and glutamine 123 (Q123) contributed by each of the four subunits in the coiled-coil structure. The Ca2+-binding residues E120 and Q123 are conserved in all group A RV NSP4 sequences, suggesting that Ca2+ binding to this site is a highly conserved property of NSP4. Structurally, the bound Ca2+ serves to neutralize the like-charge repulsion of the E120 residues in the core of the tetrameric structure. Despite the available structural data for Ca2+ binding to the NSP4 CCD, biochemical and biophysical experimental data confirming Ca2+ binding independent of crystallization conditions are lacking.
Crystallographic structures of the NSP4 CCD from three different RV strains have been determined. Structures of the simian rotavirus strain SA11 NSP4 CCD (synthetic peptide; amino acids 95 to 137) (13) and that of the human strain I321 NSP4 CCD (bacterially expressed; amino acids 95 to 146) (15) both show a tetrameric coiled-coil with a single Ca2+ ion coordinated by residues E120 and Q123 at the core of the oligomer. In contrast, the structure of the NSP4 CCD from human strain ST3 is a pentameric coiled-coil that lacks a Ca2+ ion at its core (14). The authors speculated that the Ca2+-free pentamer structure arose due to the purification and crystallization of the NSP4 CCD at acidic pH, which led to protonation of E120 and stabilization of the Ca2+-free state. The existence of distinct Ca2+-bound tetramer and Ca2+-free pentamer conformations suggests that the E120/Q123 Ca2+-binding site regulates the NSP4 CCD oligomeric form. This present study sought to determine whether Ca2+ or pH regulates the oligomer plasticity of the CCD using biochemical and structural analyses of NSP4 from the SA11 strain.
MATERIALS AND METHODS
Cloning, mutagenesis, and purification of His-TEV NSP4 residues 95 to 146.
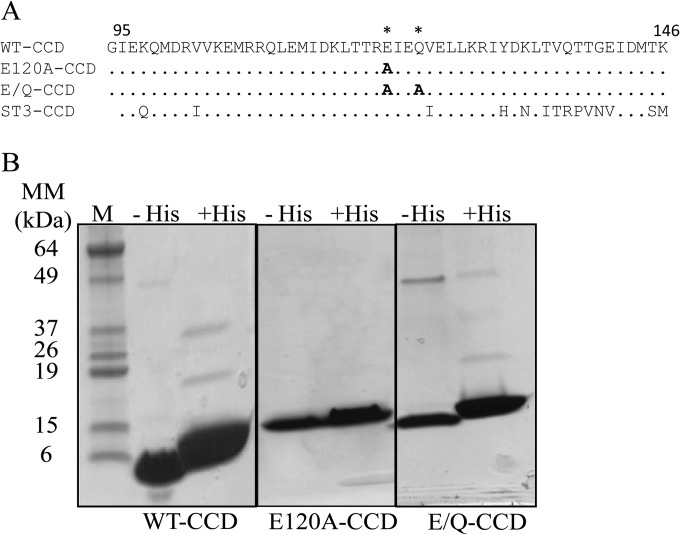
The SA11 NSP4 corresponding to residues 95 to 146 was cloned into the bacterial expression vector pET46Ek/LIC (EMD Biosciences, Inc., San Diego, CA, USA). The tobacco etch virus (TEV) protease cleavage site was positioned between the hexahistidine tag (6×His) and the NSP4 coding region to allow tag removal with TEV protease, which resulted in a nearly native protein containing only a single glycine from the linker at the N terminus. Mutations E120A and E120A/Q123A were introduced into the wild-type (WT) sequence using site-directed mutagenesis (Fig. 1A). The recombinant protein was produced in Escherichia coli BL21(DE3) (Novagen, Madison, WI, USA) cells by the addition of 300 μM isopropyl thiogalactoside (IPTG) at an optical density at 600 nm (OD600) of 0.6 for 3 h. Proteins were purified by lysing the cells using Tris-buffered saline (TBS; neutral pH; 20 mM Tris-HCl and 150 mM NaCl, pH 7.5). Affinity chromatography with Ni-nitrilotriacetic acid (NTA) resin (Qiagen, Valencia, CA, USA) was used for binding of the His-tagged proteins and eluted using the buffer with pH 7.5 (20 mM Tris, 150 mM NaCl, and 500 mM imidazole) or pH 5.6 (low pH) (20 mM sodium acetate [pH 5.6], 150 mM NaCl, and 500 mM imidazole). Further purification of the proteins was carried out using gel filtration through a Superdex S-200 preparatory-grade column attached to an AKTA purifier (GE Biosciences, Piscataway, NJ, USA) using TBS or 20 mM sodium acetate (pH 5.6) and 150 mM NaCl as the mobile phase buffer. The 6×His tag was removed by cleaving of the linker using TEV protease (27) and rebinding to Ni-NTA beads. The 6×His tag was removed for all of the experiments, and protein purity was confirmed by SDS-PAGE (Fig. 1B).
FIG 1.
Purification of NSP4 WT-CCD and the CCD mutants. (A) Sequence alignment of strain SA11 NSP4 residues 95 to 146 of WT-CCD and the E120A-CCD and E/Q-CCD mutants showing specific amino acid mutations in WT-CCD, indicated with asterisks. The strain ST3 NSP4 CCD is also shown in the alignment to show that the E120 and Q123 residues are conserved. (B) SDS-PAGE gel of the NSP4 WT-CCD, E120A-CCD, and E/Q-CCD purified proteins with His tag (+His) and without His tag (−His). Molecular mass marker bands (lane M) are shown on left of the gel.
BN-PAGE.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) was performed according to the manufacturer's (Life Technologies, Grand Island, NY, USA) protocol. WT NSP4 residues 95 to 146 (WT-CCD), NSP4 residues 95 to 146 with the E120A mutation (E120A-CCD), and NSP4 residues 95 to 146 with the E120A/Q123A mutations (E/Q-CCD) were dialyzed against TBS, with four 1-liter changes each time. Native PAGE sample buffer (Life Technologies) was used for loading the proteins into the gel. Protein samples were separated on native PAGE 4% to 16% Bis-Tris Novex gels (Life Technologies) at 150 V at 4°C. Native Mark unstained protein standards (Life Technologies) were used to estimate the relative molecular masses of the proteins, and proteins were visualized by staining with Coomassie brilliant blue.
Analytical gel filtration.
The molecular masses of WT-CCD and mutants E120A-CCD and E/Q-CCD were analyzed using a Superdex S-200 analytical gel filtration column (GE Biosciences). Gel filtration was carried out in TBS (pH 7.5), sodium acetate (pH 5.6), and 150 mM NaCl buffer (low pH) at a protein concentration of 5 mg/ml. The Superdex S-200 column was calibrated with standard molecular weight marker proteins (Bio-Rad, Hercules, CA), and Blue dextran was used to determine the void volume (Vo), using two different pH buffer systems, and a standard curve was used to calculate the molecular mass of each NSP4 CCD based on the peak elution volume.
Oligomer transition with varying pHs.
To determine whether WT-CCD or the E/Q-CCD mutant could transition from tetramer to pentamer due to changes in pH, we performed size exclusion chromatography experiments on purified protein samples using an analytical Superdex S-200 column. Initially, both WT-CCD and E/Q-CCD proteins were lysed in neutral-pH buffer and purified by affinity chromatography using Ni-NTA resin. Subsequently, the His tag was removed using TEV protease, and the proteins were further purified using gel filtration. First, both WT-CCD and E/Q-CCD proteins were dialyzed in neutral-pH buffer and analyzed by gel filtration with the same buffer. Next, the proteins were dialyzed against low-pH buffer and reanalyzed by gel filtration using the low-pH buffer. Finally, the proteins were dialyzed back into the neutral-pH buffer and analyzed by gel filtration. Proteins were analyzed at 5 mg/ml for all of the gel filtration experiments. Apparent molecular masses were calculated from a standard curve of known proteins (Bio-Rad) run in the respective pH-buffered system.
Isothermal titration calorimetry.
Binding affinities of WT-CCD, E120A-CCD, and E/Q-CCD proteins to Ca2+ were determined by isothermal titration calorimetry (ITC) using a Microcal Auto-iTC200 system (GE Life Sciences). Purified WT-CCD, E120A-CCD, and E/Q-CCD proteins were initially dialyzed in a buffer containing EDTA (20 mM Tris, 150 mM NaCl, and 10 mM EDTA) to remove bulk Ca2+ contamination. Further dialysis continued in the absence of EDTA (20 mM Tris, 150 mM NaCl) and in the presence of Chelex 100 resin (200-400 mesh; Bio-Rad) to remove contaminating divalent ions present in the buffer and protein. All buffers were prepared with OmniSolv water (EMD Bioscience, Billerica, MA, USA), and protein samples were dialyzed against four buffer changes (of 1 liter each) prior to ITC experiments. NSP4 CCD proteins (480 μM) were analyzed by ITC and titrated with 2 mM CaCl2 (prepared with the final dialysis buffer). Titrations were carried out with 5-min-interval injections of 1 μl, for 36 injections at 25°C. For each titration, a control experiment was performed by titrating Ca2+ into buffer alone under identical conditions. Data from the first injection were excluded from the calculation of thermodynamic parameters of Ca2+ binding, and analysis was carried out using Origin7 software and a one-site model. Thermodynamic parameters stoichiometry (N), association constant (Ka), and enthalpy change (ΔH) and entropy were obtained from the titration data. The dissociation constant (Kd) was calculated as 1/Ka.
Crystallization of NSP4 WT-CCD and the E/Q-CCD mutant.
WT-CCD (33 mg/ml) and the E/Q-CCD mutant (41 mg/ml) were crystallized by the hanging-drop vapor diffusion method using the Mosquito crystallization robot (TTP LabTech, Herts, Melbourn, United Kingdom) at 20°C and visualized using Rock Imager (Formulatrix, Waltham, MA, USA). Crystals of WT-CCD were obtained in a condition that contained 25% (wt/vol) PEG 1500 in 0.1 M MIB (sodium malonate, imidazole, and boric acid in a 2:3:3 molar ratio) buffer (pH 7.0). Crystals of the E/Q-CCD mutant were obtained in a condition that contained 0.2 M sodium chloride, 10% (wt/vol) PEG 8000, and 0.1 M NaK phosphate (pH 6.2). All crystals were harvested, placed in the cryoprotectant solution (20% glycerol), and flash-frozen in liquid nitrogen.
Data collection and processing.
Complete data sets were collected for the WT-CCD and E/Q-CCD crystals to a resolution of 2.03 and 1.79 Å, respectively, at the ALS synchrotron facility in Berkeley, CA. The initial data were processed using either the HKL2000 (28) or the IMOSFLM (29) programs. Space groups were confirmed using POINTLESS (30). The previously determined structure of SA11 NSP4 residues 95 to 146 (PDB 1G1J) (13) was used for phasing, using the molecular replacement program PHASER (31). For WT-CCD, the molecular replacement solution was obtained in the space group I222, with two molecules in the asymmetric unit. The molecular replacement solution for E/Q-CCD was found in the space group P 21, with five molecules in the asymmetric unit. Following automated model building and solvent addition using ARP/wARP, iterative cycles of refinement and further model building of the structures were carried out using PHENIX (32) and COOT (33). Simulated annealing difference maps were used to validate the bound ligands. Data collection and refinement statistics are given in Table 1. Protein-Ca2+ and protein-protein interactions were analyzed using COOT (33) and PDBePISA, with donor-to-acceptor distances between 2.6 Å and 3.2 Å for hydrogen-bonding interactions. Figures were prepared using PyMOL and CHIMERA.
TABLE 1.
X-ray data collection and refinement statistics
| Parameter | Valuea |
|
|---|---|---|
| WT-CCD | E/Q-CCD | |
| Wavelength resolution range (Å) | 44.71–2.03 [2.103–2.03] | 28.24–1.799 [1.863–1.799] |
| Space group | I222 | P 21 |
| Unit cell (Å/°) | 31.97, 35.72, 178.84/90, 90, 90 | 39.86, 53.47, 52.54/90, 95.84, 90 |
| Total no. of reflections | 13,801 | 204,230 |
| Unique no. of reflections | 6,998 | 20,053 |
| Multiplicity | 2.0 [1.9] | 4.6 [4.4] |
| % completeness | 99.3 [94.4] | 97.6 [80.3] |
| Mean I/sigma(I) | 13.4 [2.8] | 15.2 [4.7] |
| Wilson B factor (Å2) | 33.8 | 18.9 |
| R-merge (%) | 3 [25] | 6 [34] |
| R-work (%) | 22.4 [28.1] | 18.7 [20.1] |
| R-free (%) | 27.6 [35.1] | 23.7 [26.1] |
| No. of atoms | ||
| Protein | 626 | 1,638 |
| Ligands | 1 | 7 |
| Water | 39 | 330 |
| RMS bonds (Å) | 0.015 | 0.006 |
| RMS angles (°) | 1.3 | 0.9 |
| Ramachandran favored (%) | 100 | 100 |
| B factors (Å2) | ||
| Avg | 45.0 | 23.5 |
| Protein | 44.7 | 20.7 |
| Ligands | 75.0 | 59.6 |
| Water | 49.7 | 35.2 |
Values in brackets are for the highest-resolution shell.
Protein structure accession numbers.
The coordinates and structure factors for the protein structures determined in this study have been deposited in the Protein Data Bank under accession numbers 4WB4 for WT-CCD and 4WBA for E/Q-CCD mutant.
RESULTS
Ca2+-binding properties of the NSP4 CCD.
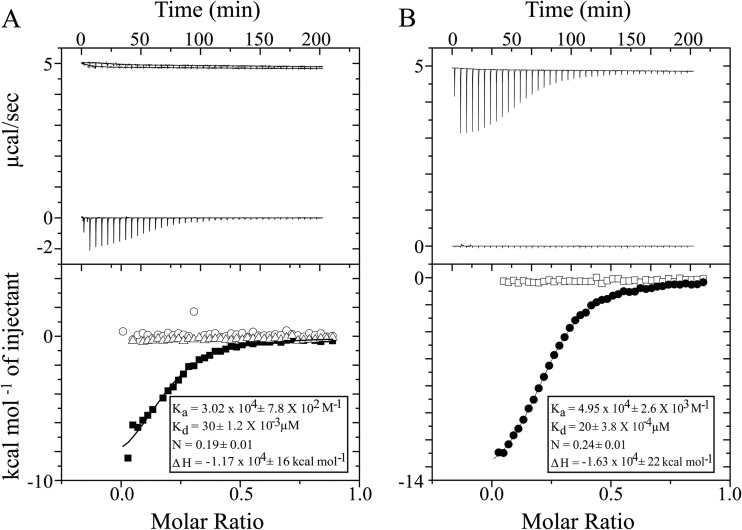
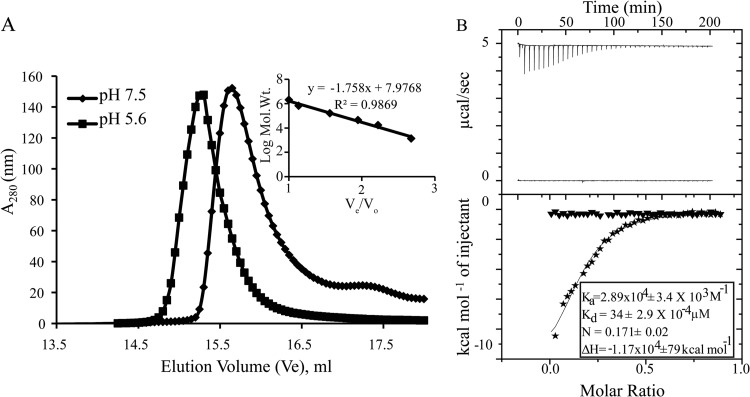
Although Ca2+-binding properties of the NSP4 CCD are characterized by crystallographic studies, which implicate a tetrameric state and conserved E120 and Q123 residues from each subunit in Ca2+ binding, the ion binding of the NSP4 CCD in solution has not been analyzed previously. To analyze the ion-binding properties of NSP4 CCD in solution and the requirement of E120/Q123 in Ca2+ binding, we used ITC to carry out binding analysis of purified WT-CCD and of the E/Q-CCD mutant (Fig. 1B). To prepare the protein for the ITC analysis, residual Ca2+ was removed by a series of dialysis steps with EDTA, and subsequently the EDTA was removed by extensive dialysis in TBS in the presence of Chelex 100 resin. Titration of Ca2+ into WT-CCD resulted in a shallow binding isotherm, with a calculated Ka of 3.2 × 104 M−1 and therefore a Kd of 31.25 μM (Fig. 2A, bottom, inset). The calculated stoichiometry of the Ca2+ ion to WT-CCD was 0.2:1 to 0.25:1, consistent with a tetramer of CCD. Next, we examined the divalent cation-binding specificity of NSP4 by performing titrations identical to those described above but using either MgCl2 or BaCl2 in place of CaCl2. While both Mg2+ and Ba2+ are divalent cations similar to Ca2+, the binding geometry of Mg2+ is octahedral, while the binding geometry of Ca2+ and Ba2+ is pentagonal bipyramidal. Thus, Ba2+ can substitute for Ca2+ but Mg2+ typically cannot. ITC titrations with Mg2+ showed no binding to WT-CCD (Fig. 2B, bottom, inset), but titrations with Ba2+ showed ion binding activity similar to that of Ca2+, although with slightly higher binding affinity (Fig. 2B, bottom, squares). These data indicate that WT-CCD does bind Ca2+, and the binding site requires ions with a pentagonal bipyramidal binding geometry, such as Ca2+ and Ba2+.
FIG 2.
ITC analysis of divalent metal ions binding to NSP4 WT-CCD and CCD mutants. (A) Top, raw data from titrations of a 0.2-ml cell containing 480 μM WT-CCD was titrated with 36 × 1 μl of 2 mM CaCl2 in 20 mM Tris and 150 mM NaCl at pH 7.5; bottom, ITC data showing integrated isotherm and best-associated fit for a one-site model showing Ca2+ binding to WT-CCD (■), E120A-CCD (○), and E/Q-CCD (△) proteins. Average thermodynamic parameters associated with Ca2+ binding to WT-CCD are reported in the inset. E120A-CCD and E/Q-CCD do not show any binding with Ca2+. (B) Top, raw data from titrations of a 0.2-ml cell containing 480 μM WT-CCD was titrated with 36 × 1 μl of 2 mM BaCl2 or MgCl2 in 20 mM Tris and 150 mM NaCl at pH 7.5; bottom, ITC data showing integrated isotherm and best-associated fit for a one-site model showing Ba2+ (●) and Mg2+ (□) binding to WT-CCD at pH 7.5. Average thermodynamic parameters associated with Ba2+ binding to WT-CCD at pH 7.5 are reported in the inset. At pH 7.5, no binding of Mg2+ is observed.
We next examined whether E120 and Q123 are required in Ca2+ binding by mutating either E120 or both E120 and Q123 to alanine (E120A-CCD or E/Q-CCD). Titration of Ca2+ into either E120A-CCD or E/Q-CCD mutants did not show measurable binding (Fig. 2A, bottom [E120A-CCD, circles; E/Q-CCD, triangles]). Taken together, our results show that the NSP4 CCD exhibits specificity for ions such as Ca2+ and Ba2+ and that the Ca2+-binding properties in solution are consistent with crystal structures depicting the NSP4 CCD as a Ca2+-bound tetramer requiring the participation of E120 and Q123. The ITC data indicate that mutation of the Ca2+ ligating residues E120 or E120 and Q123 eliminates the Ca2+-binding site. Thus, E120 and Q123 serve as a functional Ca2+-binding site in the NSP4 CCD.
The NSP4 CCD Ca2+-binding site regulates the oligomeric structure of the CCD.
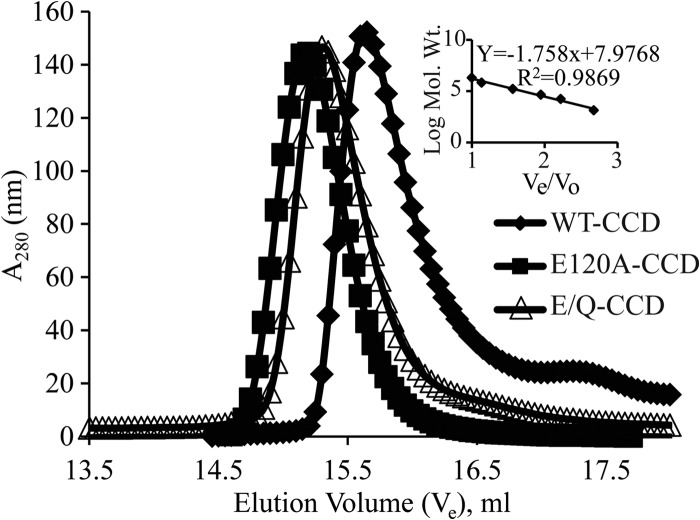
The recently reported crystal structure of the NSP4 CCD of a human strain (14) suggests that lack of Ca2+ binding may alter the oligomeric structure of the CCD. To investigate this further, we analyzed the structures of the SA11 WT-CCD, which has Ca2+-binding activity, and the E/Q-CCD mutants deficient in Ca2+ binding. First, we used BN-PAGE to determine the oligomer sizes of these proteins under native gel electrophoresis conditions. The WT-CCD and E/Q-CCD mutant proteins migrated with very different patterns in BN-PAGE (data not shown). The WT-CCD migrated faster than the mutants, both of which migrated more slowly and in multiple species. The apparent molecular mass of WT-CCD was consistent with a tetramer (∼25.5 kDa). In contrast, both mutants migrated more slowly than WT-CCD, suggesting oligomeric forms larger than a tetramer. However, the resolution of this method was too low to determine a precise oligomeric form. One reason for the poor resolution of the NSP4 CCD mutants is that they have a higher isoelectric point (pI = 8.6), and this would have affected their mobility on the BN-PAGE gel. Next, we used analytical gel filtration to determine more precisely the molecular mass of the WT-CCD and mutant oligomers. Both WT-CCD and the E/Q-CCD mutant proteins were purified as described above, and the 6×His tag was removed. Analytical gel filtration was carried out in normal Tris-buffered saline at room temperature, and the apparent molecular masses of WT-CCD and recombinant proteins were extrapolated from a standard curve created by calibrating the column with standard molecular weight marker proteins. The WT-CCD protein eluted with an apparent molecular mass of 26.4 kDa, consistent with a tetramer (Fig. 3, diamonds), but the E120A-CCD mutant eluted with an apparent molecular mass of 33.6 kDa (Fig. 3, squares), and the E/Q-CCD mutant eluted with an apparent molecular mass of 31 kDa (Fig. 3, triangles). The calculated molecular masses for both mutants are consistent with a pentameric oligomer (Table 2). Together, the BN-PAGE and analytical gel filtration studies show that WT-CCD forms a tetramer, but with loss of Ca2+ binding the CCD mutants form a pentamer. The ITC and gel filtration data presented above suggest that abolishing the NSP4 CCD Ca2+-binding site induced changes in the oligomeric structure of the CCD. To confirm the changes that we observed in solution, we determined the crystal structures of WT-CCD and the E/Q-CCD mutant.
FIG 3.
Effect of mutation of Ca2+ coordinating amino acids on oligomerization of the NSP4 CCD. Molecular mass determinations by size exclusion chromatography. A molecular mass calibration curve was obtained from the elution profiles of the standard proteins (inset). Apparent molecular masses of the proteins were determined from a standard graph. WT-CCD eluted as a tetramer (26.4 kDa) (◆), whereas mutants E120A-CCD (33.6 kDa) (■) and E/Q-CCD (31.1 kDa) (△) eluted as pentamers at pH 7.5. Elution volumes, apparent molecular masses, and numbers of oligomers are reported in Table 2.
TABLE 2.
Elution volume, apparent molecular mass, and number of oligomer species deduced from a standard graph at pH 7.5 and pH 5.6a
| Protein | pH 7.5 |
pH 5.6 |
||||
|---|---|---|---|---|---|---|
| Ve (ml) | Molecular mass (kDa) | No. of monomers | Ve (ml) | Molecular mass (kDa) | No. of monomers | |
| WT-CCD | 15.75 ± 0.05 | 27.1 ± 0.7 | 4.2 ± 0.1 | 15.26 ± 0.01 | 37.35 ± 0.25 | 5.85 ± 0.05 |
| E120A-CCD | 15.17 | 33.6 | 5.3 | |||
| E/Q-CCD | 15.33 ± 15.02 | 30.7 ± 0.4 | 4.9 ± 0.1 | 15.38 | 34.8 | 5.5 |
Ve, elution volume.
Crystal structure of WT-CCD.
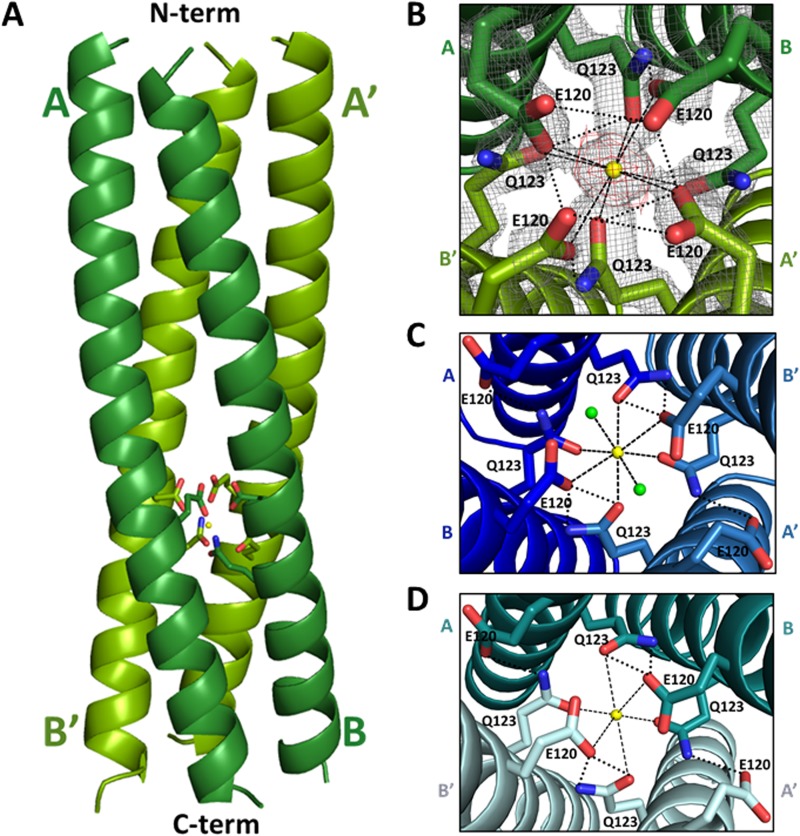
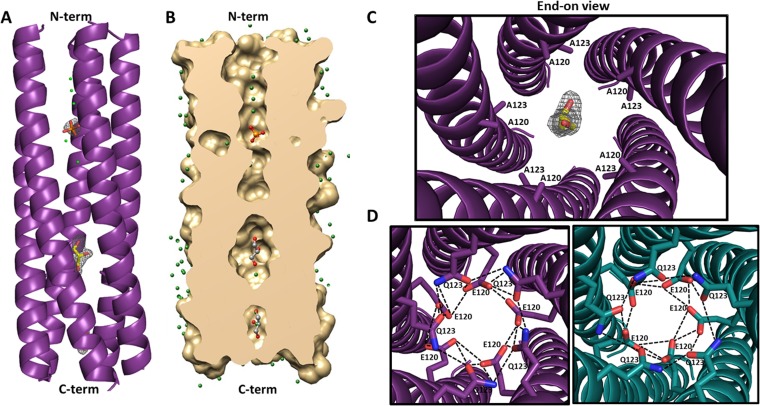
The WT-CCD structure showed the formation of the 4-helix coiled-coil tetramer (Fig. 4A). Detailed analysis revealed that the tetramer structure is similar to the previously reported structures of the strain SA11 NSP4 CCD (13, 15). As in the previous two structures (13, 15), two molecules of WT-CCD in the asymmetric unit (A and B), each with an α-helical conformation, associate with two molecules (A′ and B′) in the neighboring asymmetric unit related by a crystallographic 2-fold symmetry to form a parallel four-stranded coiled-coil structure with a pseudo 4-fold symmetry. We observed a clear density, consistent with earlier structures, corresponding to a single Ca2+ ion bound inside the central channel of the tetramer coordinated by side chain atoms of the E120 and Q123 residues, which form an extensive polar layer within the core (Fig. 4B). Both the strong density in the difference map (>3σ level) and the coordination geometry are highly consistent with the bound Ca2+. The position of the Ca2+ is identical to that in the previous two structures. However, compared to the previous structures, our WT-CCD tetramer showed noticeable variations in the side chain orientations of the Ca2+-binding residues (Fig. 4B to D). In the structures determined by Bowman et al. and Deepa et al. (13, 15), two E120 residues and four Q123 residues coordinate binding of the Ca2+ ion, with the other two E120 residues turned away from the center of the channel forming hydrogen bond interactions with the residues from the neighboring subunits (Fig. 4C and D). In the structure by Bowman et al. (13), in addition to the oxygen atoms of the E120 and Q123 side chains, two water molecules participate in coordinating the Ca2+ ion (Fig. 4C). In our structure and the structure determined by Deepa et al. (15), Ca2+ is coordinated entirely by the side chain oxygen atoms of E120 and Q123; however, our structure shows all four E120 residues oriented toward the center of the core affecting Ca2+ binding (Fig. 4B and D).
FIG 4.
Comparison of the Ca2+-binding sites in the SA11 NSP4 CCD structures. (A) A side view of the ribbon representation of the NSP4 CCD (residues 95 to 145) tetramer presented in this paper. The bound Ca2+ ion (yellow sphere) along with E120 and Q123 (rendered as sticks, with nitrogen shown in blue and oxygen shown in red) are shown. The tetramer is formed by the association of two parallel helices (A and B, shown in dark green) in the asymmetric unit in the crystal with their crystallographic 2-fold symmetry-related mates (A′ and B′; shown in light green). The helical nature of all four chains extends from residue 95 (labeled as N-term on the top) to 136 (labeled as C-term on the bottom), with the last 10 residues (137 to 145) not clearly resolved in the structure. (B) Closeup of the end-on view of the Ca2+-binding site, with E120 and Q123 residues shown as sticks and the Ca2+ ion shown as a yellow sphere. In this structure, Ca2+ ion binding is coordinated by four E120 and Q123 residues (rendered as sticks, with nitrogen shown in blue and oxygen shown in red). 2Fo-Fc density for the E120 and Q123 residues at a contour level of 1σ is shown in gray, and Fo-Fc difference map density for Ca2+ (3σ contour level) is shown in red. (C) Ca2+-binding site in the SA11 NSP4 CCD tetramer (PDB 1G1I) reported by Bowman et al. (13). A and B subunits in the asymmetric unit (dark blue) and their crystallographic 2-fold symmetry-related mates A′ and B′ (light blue) are shown. In this structure, Ca2+ ion (yellow sphere) binding is coordinated by two water molecules (shown as green spheres), two E120 residues, and four Q123 residues (rendered as sticks). (D) Ca2+-binding site in the SA11 NSP4 CCD tetramer (PDB 2O1K) reported by Deepa et al. (15). Two molecules in the asymmetric unit (A and B [dark teal]) and their crystallographic 2-fold symmetry-related mates (A′ and B′ [light teal]) are shown. Ca2+ ion (yellow sphere) binding is coordinated by two E120 residues and four Q123 residues (rendered as sticks). In panels B, C, and D, the ionic interactions with Ca2+ and hydrogen bond interactions between the side chains are shown as dashed and dotted black lines, respectively.
Crystal structure of the E/Q-CCD mutant.
The E/Q-CCD mutant crystallized in the space group P 21 with five molecules in the asymmetric unit. These molecules in the α-helical conformation interact with one another, with a total buried surface area of 11,140 Å2, to form a pentameric coiled-coil structure consistent with our solution studies (Fig. 5A, B, and C). The pentameric association of the E/Q-CCD subunits is very similar to that observed in the ST3 NSP4 CCD pentamer reported by Chacko et al., which was determined in the absence of Ca2+ (14). The two structures superimpose with a root mean square deviation (RMSD) of ∼0.8 Å for matching Cα atoms. In our E/Q mutant pentamer, inside the channel, a difference map revealed several small but strong density (>3σ level) features in addition to those that could be interpreted as water molecules. The strong piece of density toward the N-terminal region of the pentamer was interpreted as a phosphate molecule based on the tetrahedral shape and the positions of oxygen atoms from the surrounding Q109 residues and two water molecules within hydrogen-binding distances. In addition, two more strong elongated densities were observed toward the C-terminal region, which we have attributed to two glycerol molecules interacting mainly with the neighboring water molecules (Fig. 5A and B). One of these glycerol molecules is localized in close proximity to the mutated E120A and Q123A residues (Fig. 5C). In the ST3 CCD pentamer, however, only two water molecules inside the channel are reported.
FIG 5.
Structure of the E120A/Q123A SA11 NSP4 CCD pentamer. (A) A side view of the ribbon representation of E120A/Q123A SA11 NSP4 CCD (residues 95 to 146) pentamer formed by five parallel helices (purple). The helical nature of all five chains extends from residues 95 (labeled as N-term on the top) to 137 (labeled as C-term on the bottom), with the last nine residues not clearly resolved in the structure. A phosphate molecule (orange sticks with oxygen shown in red) and two glycerol molecules (yellow sticks with oxygen shown in red) are depicted inside the channel. Fo-Fc difference map densities (3σ level) for the phosphate and glycerol molecules are shown in gray. Water molecules are shown as green spheres. (B) A longitudinal slice of the E/Q-CCD pentamer surface model (tan) revealing a cross section of the axial pore. A phosphate molecule is rendered as orange sticks, and two glycerol molecules are rendered as gray sticks, with oxygen shown in red. Water molecules are shown as green spheres. (C) Closeup of the end-on view of the mutated Ca2+-binding site, with A120 and A123 residues shown as purple sticks, a glycerol molecule shown as yellow sticks, and oxygen shown in red. Difference map density for the glycerol molecule is shown in gray. (D) Modeling of the E120 and Q123 residues into the E/Q-CCD pentamer structure and comparison with the ST3 CCD pentamer (PDB 3MIW) reported by Chacko et al. (14). Right, alanine residues at positions 120 and 123 in the E/Q-CCD were mutated back to E and Q (purple sticks, with nitrogen shown in blue and oxygen shown in red), respectively, as in the native SA11 sequence. The diameter of the pentamer accommodates all 10 residues and allows for formation of hydrogen bond interactions (dotted black lines) between the residues that are very similar to the interactions formed in the ST3 pentamer. Left, closeup end-on view of the hydrogen bond interaction (dashed black lines) between E120 and Q123 residues in the ST3 CCD pentamer (dark teal). E120 and Q123 residues are rendered as teal sticks, with nitrogen and oxygen atoms shown in blue and red, respectively.
To examine if the SA11 E/Q-CCD mutant pentamer can accommodate the native E/Q residues, we computationally mutated back the alanine residues in the mutant to their respective E and Q residues. In this simulated native structure, both E and Q residues can be easily accommodated without any steric clashes, making a hydrogen bond network similar to that observed in the ST3 pentamer structure (Fig. 5D). This suggests that the native SA11 CCD can form a pentameric structure. Interestingly, the Q residues in the simulated native pentamer structure can hydrogen bond to the hydroxyl groups of the glycerol, suggesting that the native SA11 CCD could accommodate a glycerol molecule.
Low pH abolishes NSP4 Ca2+-binding activity and affects oligomerization properties.
One notable difference between the NSP4 CCD tetramer structures reported (including our structure, reported here) and the recently reported ST3 NSP4 CCD pentameric structure is that the latter protein was purified and crystalized at low pH, whereas the others were purified and crystalized at neutral pH. Chacko et al. (14) speculated that the lower pH caused protonation of the E120 residue, preventing Ca2+ binding and, therefore, making the Ca2+-free pentameric structure more thermodynamically stable. The crystal structure clearly showed that this NSP4 oligomer did not bind Ca2+, but no studies examined whether acidic pH abrogated NSP4 binding of Ca2+. To examine the effect of pH on the oligomeric state of the CCD, we purified strain SA11 WT-CCD at neutral pH for binding to the Ni2+-NTA beads but eluted the protein at low pH. Analytical gel filtration (Fig. 6A, squares) showed that this protein eluted with an apparent molecular mass of 39 kDa (Table 2), more consistent with a pentamer (based on the molecular weight standards run at low pH), whereas WT-CCD, when eluted from Ni-NTA resin in neutral-pH buffer, eluted as a tetramer (Fig. 6A, diamonds; Table 2). Next, to examine the effect of pH on Ca2+ binding, we prepared WT-CCD in low-pH buffer conditions for ITC experiments using low-pH buffer for dialysis rather than the previously used neutral-pH buffer. Titration of Ca2+ into the WT-CCD protein at low pH showed no Ca2+-binding activity (Fig. 6B, bottom, triangles). However, the same batch of protein, when dialyzed in neutral-pH buffer, showed Ca2+-binding activity similar to that of the WT-CCD protein that was maintained in neutral-pH buffer throughout the preparation (Fig. 6B, bottom, stars and inset). Thus, similar to mutational ablation of the Ca2+-binding site, in the presence of acidic pH, the Ca2+-binding activity was abolished, and this correlates with a change in the oligomeric state of the NSP4 CCD.
FIG 6.
Effect of pH on Ca2+ binding and oligomerization of WT-CCD. (A) Molecular mass determinations by size exclusion chromatography. A molecular mass calibration curve was obtained from the elution profiles of the standard proteins (inset). Apparent molecular masses of the proteins were determined from a standard graph reported in Table 2. WT-CCD eluted as a 26.4-kDa oligomer at pH 7.5 (◆), consistent with a tetramer, whereas it eluted as 37.6 kDa at pH 5.6 (■), consistent with a larger oligomeric form (pentamer). (B) Top, raw data from titrations of a 0.2-ml cell containing 480 μM WT-CCD was titrated with 36 × 1 μl of 2 mM CaCl2 in 20 mM Tris and 150 mM NaCl at pH 7.5 and 20 mM NaAc and 150 mM NaCl at pH 5.6; bottom, ITC data showing integrated isotherm and best-associated fit for a one-site model showing Ca2+ binding to WT-CCD at pH 7.5 (stars) and at pH 5.6 (▼). Average thermodynamic parameters associated with Ca2+ binding to WT-CCD at pH 7.5 are reported in the inset. At pH 5.6, no binding of Ca2+ is observed.
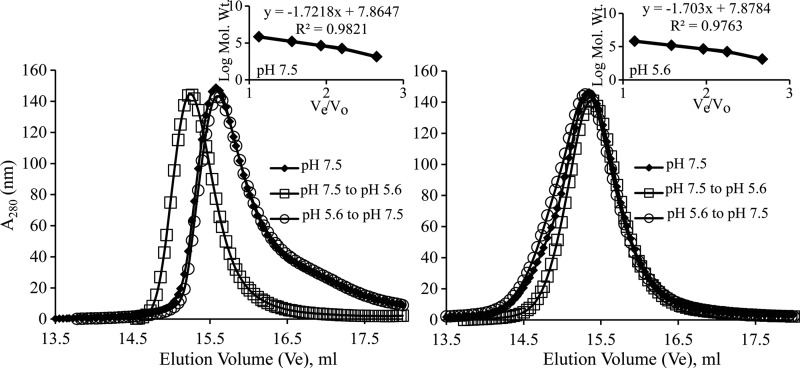
Reversible oligomer transition of NSP4 WT-CCD with varying pHs.
To determine whether the same NSP4 CCD can reversibly transition between tetramer and pentamer states by altering the pH, we first purified WT-CCD and the E/Q-CCD mutant in a neutral-pH buffer and determined their oligomeric states using analytical gel filtration. WT-CCD eluted as a tetramer (Fig. 7A, diamonds; Table 2), whereas the E/Q-CCD mutant eluted as a pentamer (Fig. 7B, diamonds; Table 2). Next, we equilibrated the two protein samples at low-pH buffer by dialysis and carried out analytical gel filtration. In the acidic buffer (pH 5.6), both WT-CCD and the E/Q-CCD mutant eluted as pentamers, suggesting that the WT-CCD protein underwent a change in its oligomeric state (Fig. 7A and B, squares; Table 2). Finally, when both the proteins were dialyzed back into the neutral-pH buffer and reanalyzed, WT-CCD eluted as a tetramer (Fig. 7A, circles; Table 2), while the E/Q-CCD mutant eluted as a pentamer (Fig. 7B, circles; Table 2). These results strongly suggest that the WT-CCD protein undergoes a reversible, pH-driven change in its oligomeric state and abrogation of Ca2+ binding eliminates the pH sensitivity and stabilizes the pentameric state that mimics the Ca2+-free pentameric state of the WT-CCD.
FIG 7.
Effect of pH on oligomer transition of NSP4 WT-CCD and E/Q-CCD. (A) Molecular mass determinations by size exclusion chromatography with varying pHs. WT-CCD eluted as a tetramer at pH 7.5 (◆ and ○), whereas it eluted as a larger oligomeric form as a pentamer at pH 5.6 (□). (B) E/Q-CCD eluted as a pentamer consistently at two different pH buffers (◆, □, and ○). A molecular mass calibration curve was obtained from the elution profiles of the standard proteins (inset). Apparent molecular masses of the proteins were determined from a standard graph (Table 2).
DISCUSSION
The rotavirus enterotoxin NSP4, with its ability to alter intracellular Ca2+ levels, is implicated in multiple functions critical for rotavirus replication and morphogenesis. In the absence of a crystal structure of full-length NSP4, which is prone to severe aggregation, crystallographic studies of the NSP4 CCD have provided valuable structural information on the possible oligomeric states and Ca2+-binding properties of NSP4. These structural studies have shown that the NSP4 CCD from two different RV strains can adopt two distinct oligomeric states, a Ca2+-bound tetramer and a Ca2+-free pentamer, despite conservation of the residues that coordinate Ca2+. These observations raise several interesting questions: Is the preference for a particular oligomeric state strain dependent? What factors influence this preference? Does the NSP4 CCD from a single strain have an intrinsic structural plasticity to transit between the two oligomeric states? Using a combination of biochemical, biophysical, and crystallographic approaches, we show here that the NSP4 CCD of the SA11 strain can adopt both Ca2+-bound tetrameric and Ca2+-free pentameric states and undergo reversible transition between these states in response to variations in pH.
Ca2+ binding is obligatory for the CCD tetrameric state.
Our crystal structure of the NSP4 CCD at a 2-Å resolution with clear densities for the bound Ca2+ and for the side chains of the residues that coordinate Ca2+, together with previous crystallographic studies, affirms the notion that Ca2+ binding is the critical factor in stabilizing the tetrameric state formed by the parallel association of α-helical subunits. The folding of the CCD subunit into an α-helical conformation and formation of the parallel coiled-coil tetrameric structures are consistent with the presence of a discernible heptad repeat in the NSP4 CCD sequence. Peptide sequences with a heptad repeat are known to form such higher-order supercoiled structures (34). However, the tetrameric coiled-coil structure of the NSP4 CCD with a pseudo 4-fold symmetry shows deviation from the coiled-coil tetramer expected from a sequence containing a strict heptad repeat, because the two consecutive polar layers inside the channel are formed by the Q123 and E120 residues, which correspond to the “a” and “d” positions of the heptad repeat. These residues inside the tetramer channel coordinate the bound Ca2+ ion. It is evident from the NSP4 CCD structures that in the absence of Ca2+, electrostatic repulsion between the negatively charged E120 residues inside the channel would lead to destabilization of the tetramer. The narrow space inside the channel is not sufficient for the side chains of these residues to assume different conformations to avoid the electrostatic repulsion. Ca2+ binding neutralizes the charge repulsion and allows the side chains of E120 and Q123 to engage in intersubunit hydrogen bond interactions. Thus, any event that affects Ca2+ binding would lead to destabilization of the tetrameric organization of the NSP4 CCD. Bowman et al. (13) suggested that this region of the tetramer can be susceptible to changes in the oligomeric state of NSP4 and can function as a switch triggered by interactions with other proteins during virus replication.
The CCD exhibits high selectivity for Ca2+.
It is clear from the crystal structures of the Ca2+-bound CCD tetramer that the disposition of the side chains of the E120 and Q123 residues inside the tetramer channel is ideally suited for binding Ca2+. However, comparative analysis of our NSP4 CCD tetramer crystal structure with the previous structures shows that although the position of Ca2+ remains invariant, slight variations are possible in how these side chains orient to chelate Ca2+. In the structure determined by Bowman et al. (13), with the side chains oriented slightly differently, two water molecules complete the coordination with Ca2+. In our structure as well as in the structure determined by Chacko et al. (14), Ca2+ is coordinated entirely by the side chain oxygen atoms of E120 and Q123 despite slightly altered side chain conformations. The studies by Bowman et al. (13), together with our ITC results, show that the CCD tetramer also supports binding of Sr2+ and Ba2+, which have pentagonal bipyramid coordination chemistry similar to that of Ca2+ but not Mg2+. In the context of the in vivo functions of NSP4, Ca2+ binding of course is the most relevant.
The E120 and Q123 residues of the NSP4 CCD that coordinate Ca2+ binding are completely conserved among the NSP4 sequences of group A rotaviruses (35). In addition, the CCD itself, with the essential signatures of a heptad repeat, is the most conserved region of NSP4. From these observations, it can be surmised that the Ca2+-stabilized coiled-coil tetrameric state is a common feature of all group A rotavirus NSP4 CCDs. The intrinsic property of the CCD to exist as a tetramer in the presence of Ca2+, not only in the crystal but also in solution, as shown by our gel filtration studies, strongly suggests that this region dictates a context-dependent oligomeric state of NSP4 during virus replication (see below). However, because of the strong tendency of the recombinant full-length NSP4 to aggregate, a direct demonstration of its oligomeric state either in solution or in the crystal has thus far not been possible.
The CCD forms pentamers in the absence of Ca2+.
What kind of structure can the NSP4 CCD adopt in the absence of Ca2+? The possibility that the NSP4 CCD can form a coiled-coil pentameric structure in the absence of Ca2+ was first indicated by the crystal structure of NSP4 CCD from human group A rotavirus strain ST3 (14). However, it was not clear from those studies whether this is merely due to a strain difference or whether the CCD from a single strain could adopt a pentameric structure in the absence of Ca2+, as the structure of the ST3 NSP4 CCD in the presence of Ca2+ could not be determined. Our gel filtration studies of the SA11 CCD under conditions that abrogate Ca2+ binding, such as low pH or by mutating the E120 and Q123 residues individually or in tandem, indicate that the SA11 CCD does indeed form pentamers in the absence of Ca2+. An unequivocal demonstration of the pentamer formation in the absence of Ca2+ would have been to crystallize the native SA11 CCD under Ca2+-free conditions and determine its structure. Despite determining eight structures of WT-CCD crystals grown under a variety of conditions, which included denaturation and renaturation protocols and Ca2+-chelating agents, it was not possible to crystallize SA11 CCD without trace Ca2+ as a contaminant binding to the CCD and resulting in tetramer formation. However, it was relatively easy to control for Ca2+ contamination in our solution studies. In lieu of the native CCD structure, our strategy was then to determine the crystal structure of the E120A/Q123A mutant, which clearly showed a coiled-coil pentameric structure similar to that of the ST3 CCD pentamer, substantiating the results from our solution studies.
Although several structures of dimeric, trimeric, and tetrameric coiled-coil structures of peptide sequences bearing heptad repeats have been determined (34, 36), there are only a few structures of coiled-coil pentamers. In addition to the two NSP4 CCD pentamers, the only other pentamer structure is that of the oligomerization domain of the cartilage oligomeric matrix protein (COMP) of the thrombospondin family (37). Despite a lack of sequence similarity with the NSP4 CCD, except for some of the signature elements of a heptad repeat, all three pentamer structures show very similar parallel association of the α-helical subunits, with an average RMSD of ∼1.0 Å. In the case of the COMP pentamer, however, intersubunit disulfide bonds stabilize the pentameric structure, which is not the case with the NSP4 CCD pentameric structures.
pH modulates Ca2+ binding and oligomerization states of the CCD.
In the pentameric structure with an increased pore size of the channel, the E120 and Q123 residues have more space to adopt appropriate conformations to avoid electrostatic repulsions. In the ST3 CCD pentamer structure, these E120 and Q123 residues that point away from the center of the channel engage in intersubunit hydrogen bond interactions lining the interior surface of the channel and contribute to the stability of the pentameric structure. Computational back mutation of the alanine residues to native residues in our E120A/Q123A mutant pentamer structure indicates that identical interactions are possible in the SA11 WT-CCD as well. It is very likely that the low pH, which increases the protonation state of the E120 residues, plays a major role in relieving the electrostatic repulsion and allowing the formation of the pentamer structure, because the pentamer formation of SA11 WT-CCD is observed only at low pH in our gel filtration studies, which also abrogates Ca2+ binding. It is important to note that the ST3 CCD was crystallized under low-pH conditions (14). The effect of pH on Ca2+ binding, as well as the oligomeric state of NSP4 CCD, is further demonstrated by our observation that the SA11 CCD elutes as a tetramer by reversal of the pH back to neutral. A novel inference from these observations is that the NSP4 CCD is a pH sensor, and in response to variations in pH, it can reversibly transit between Ca2+-bound tetrameric and Ca2+-free pentameric states. A similar interplay between a buried polar residue and pH in altering the oligomeric nature has been observed in other proteins that form coiled-coil structures (38).
The NSP4 CCD pentamer allows for binding of different small molecule ligands.
While inside the channel of the Ca2+-bound CCD tetramer, with the exception of Ca2+ and a small number of water molecules, no other small ligands are observed. In contrast, in the pentameric channel, which is relatively more hydrophobic than that of the tetramer, our crystallographic analysis revealed a phosphate ion and two glycerol molecules along with several water molecules. The phosphate ion binds near the N-terminal region of the pentamer hydrogen bonded to side chains of Q109 residues, whereas the glycerol is observed in the close proximity of the E120A and Q123A residues, interacting mainly with the surrounding water molecules. When the alanine residues are computationally reverted back to E and Q, these can hydrogen bond to the hydroxyl groups of the glycerol, suggesting that the native SA11 CCD also could accommodate a glycerol molecule. Binding of these small molecules inside the channel likely contributes to additional stability of the pentameric state. While the binding of these molecules can be viewed as an artifact of the crystallization conditions, our observations point to the possibility that small molecules can indeed bind to the inside of the channel, and the relevance of this observation in the overall functionality of NSP4 during viral replication needs further studies. The relatively hydrophobic interior of the channel may also facilitate passage/penetration of hydrated ions such as Ca2+, as proposed in the case of the COMP pentameric channel (37).
The CCD likely dictates the structure-function of NSP4.
The NSP4 CCD with its unique structural properties, a conserved heptad repeat, the unique combination of E120 and Q123, and a pH-sensitive E120, which not only allows formation of Ca2+-bound tetrameric and Ca2+-free pentameric structures but also a reversible transition between these two states, is likely the principal domain that dictates context-dependent oligomeric states of the full-length NSP4. We suggest that the structural plasticity of NSP4, controlled by the coordinated interplay between Ca2+ binding and variations in pH, constitutes the underlying mechanistic basis for regulating the disparate functions of NSP4, which is known to localize to various cellular sites, including the ER, ER-Golgi intermediate compartment (ERGIC), viroplasms, and plasma membrane, during the replication cycle of rotavirus. Previous data from our laboratory (39) showed that the single amino acid mutation of E120A within the full-length NSP4 abrogated binding to integrin α2β1. This mutation also inhibited C2C12-α2 cell adhesion and spreading. In addition, the E120A mutation abrogated the diarrhea-inducing property of the protein. These results support the idea that mutation of a single amino acid in this domain could affect the functions of full-length NSP4. Although it is currently unclear as to which oligomeric state is selected for a particular function of NSP4, it is possible that the pentameric state of NSP4, for example, is preferred for its viroporin activity, or when functioning as an intracellular receptor to facilitate the budding of icosahedral double-layer particles, or when it is transiting from the ER to the low-pH ERGIC compartment. In contrast, the tetrameric state may be important for interacting with cellular proteins such as LC3, which is exploited by RV for viral morphogenesis (4). Identification of the biochemical factors that stabilize either the tetramer or the pentamer form, as well as mutations that lock the NSP4 CCD into a single conformation, is critical to differentiate between the functions carried out by the NSP4 tetramer and pentamer.
In summary, our studies presented here reveal unique perspectives first to the functional aspects of rotavirus NSP4 and second to the structural aspects of protein sequences with a heptad repeat that form coiled-coil oligomeric structures. In regard to NSP4, our results support novel interpretations, including that unique features of the NSP4 CCD confer the structural plasticity for regulating the structure-function of NSP4 and that the CCD domain is a pH sensor that facilitates reversible transition from a Ca2+-bound tetrameric state to a Ca2+-free pentameric state. Further studies, including raising monoclonal antibodies specific to each of the states, are required to gain further understanding of the role of the suggested structural plasticity of NSP4 for its function during rotavirus replication, morphogenesis, and pathogenesis. Considering that coiled-coil domains are one of the common features implicated in a wide range of functions, including ion channels and protein-protein interactions, the NSP4 CCD is a plausible model system to further our understanding of the sequences and structural properties necessary for nucleating different coiled-coil oligomeric structures and the mechanism of transition between these structures.
ACKNOWLEDGMENTS
We acknowledge the support from NIH grants R01 AI080656 (M.K.E.), R37 AI36040 (B.V.V.P.), K01 DK093657 (J.M.H.), and P30 DK56338 that supports the Texas Medical Center Digestive Diseases Center, and from Robert Welch Foundation grant Q1279 (B.V.V.P.). The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
We acknowledge the use of the synchrotron beam lines at Advanced Light Source (5.0.1), Berkeley, CA, for diffraction data collection and thank their staff for excellent help. We also thank the Auto ITC facility in the Drug Development Core, Baylor College of Medicine.
Footnotes
Published ahead of print 17 September 2014
REFERENCES
- 1. Zhou Y, Frey TK, Yang JJ. 2009. Viral calciomics: interplays between Ca2+ and virus. Cell Calcium 46:1–17. 10.1016/j.ceca.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhowmick R, Halder UC, Chattopadhyay S, Chanda S, Nandi S, Bagchi P, Nayak MK, Chakrabarti O, Kobayashi N, Chawla-Sarkar M. 2012. Rotaviral enterotoxin nonstructural protein 4 targets mitochondria for activation of apoptosis during infection. J. Biol. Chem. 287:35004–35020. 10.1074/jbc.M112.369595. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3. Chattopadhyay S, Basak T, Nayak MK, Bhardwaj G, Mukherjee A, Bhowmick R, Sengupta S, Chakrabarti O, Chatterjee NS, Chawla-Sarkar M. 2013. Identification of cellular calcium binding protein calmodulin as a regulator of rotavirus A infection during comparative proteomic study. PLoS One 8:e56655. 10.1371/journal.pone.0056655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crawford SE, Hyser JM, Utama B, Estes MK. 2012. Autophagy hijacked through viroporin-activated calcium/calmodulin-dependent kinase kinase-beta signaling is required for rotavirus replication. Proc. Natl. Acad. Sci. U. S. A. 109:E3405–E3413. 10.1073/pnas.1216539109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diaz Y, Pena F, Aristimuno OC, Matteo L, De Agrela M, Chemello ME, Michelangeli F, Ruiz MC. 2012. Dissecting the Ca(2)(+) entry pathways induced by rotavirus infection and NSP4-EGFP expression in Cos-7 cells. Virus Res. 167:285–296. 10.1016/j.virusres.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 6. Hyser JM, Utama B, Crawford SE, Broughman JR, Estes MK. 2013. Activation of the endoplasmic reticulum calcium sensor STIM1 and store-operated calcium entry by rotavirus requires NSP4 viroporin activity. J. Virol. 87:13579–13588. 10.1128/JVI.02629-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ruiz MC, Cohen J, Michelangeli F. 2000. Role of Ca2+ in the replication and pathogenesis of rotavirus and other viral infections. Cell Calcium 28:137–149. 10.1054/ceca.2000.0142. [DOI] [PubMed] [Google Scholar]
- 8. Zambrano JL, Sorondo O, Alcala A, Vizzi E, Diaz Y, Ruiz MC, Michelangeli F, Liprandi F, Ludert JE. 2012. Rotavirus infection of cells in culture induces activation of RhoA and changes in the actin and tubulin cytoskeleton. PLoS One 7:e47612. 10.1371/journal.pone.0047612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aoki ST, Settembre EC, Trask SD, Greenberg HB, Harrison SC, Dormitzer PR. 2009. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 324:1444–1447. 10.1126/science.1170481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aoki ST, Trask SD, Coulson BS, Greenberg HB, Dormitzer PR, Harrison SC. 2011. Cross-linking of rotavirus outer capsid protein VP7 by antibodies or disulfides inhibits viral entry. J. Virol. 85:10509–10517. 10.1128/JVI.00234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dormitzer PR, Greenberg HB, Harrison SC. 2000. Purified recombinant rotavirus VP7 forms soluble, calcium-dependent trimers. Virology 277:420–428. 10.1006/viro.2000.0625. [DOI] [PubMed] [Google Scholar]
- 12. Sen A, Sen N, Mackow ER. 2007. The formation of viroplasm-like structures by the rotavirus NSP5 protein is calcium regulated and directed by a C-terminal helical domain. J. Virol. 81:11758–11767. 10.1128/JVI.01124-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bowman GD, Nodelman IM, Levy O, Lin SL, Tian P, Zamb TJ, Udem SA, Venkataraghavan B, Schutt CE. 2000. Crystal structure of the oligomerization domain of NSP4 from rotavirus reveals a core metal-binding site. J. Mol. Biol. 304:861–871. 10.1006/jmbi.2000.4250. [DOI] [PubMed] [Google Scholar]
- 14. Chacko AR, Arifullah M, Sastri NP, Jeyakanthan J, Ueno G, Sekar K, Read RJ, Dodson EJ, Rao DC, Suguna K. 2011. Novel pentameric structure of the diarrhea-inducing region of the rotavirus enterotoxigenic protein NSP4. J. Virol. 85:12721–12732. 10.1128/JVI.00349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deepa R, Durga Rao C, Suguna K. 2007. Structure of the extended diarrhea-inducing domain of rotavirus enterotoxigenic protein NSP4. Arch. Virol. 152:847–859. 10.1007/s00705-006-0921-x. [DOI] [PubMed] [Google Scholar]
- 16. Estes MK, Greenberg HB. 2013. Rotaviruses, p 1347–1401 In Knipe DM, Howley PM. (ed), Fields virology, 6th ed, vol 2 Lippincott, William and Wilkins, Philadelphia, PA. [Google Scholar]
- 17. Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. 2006. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J. Virol. 80:6061–6071. 10.1128/JVI.02167-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diaz Y, Chemello ME, Pena F, Aristimuno OC, Zambrano JL, Rojas H, Bartoli F, Salazar L, Chwetzoff S, Sapin C, Trugnan G, Michelangeli F, Ruiz MC. 2008. Expression of nonstructural rotavirus protein NSP4 mimics Ca2+ homeostasis changes induced by rotavirus infection in cultured cells. J. Virol. 82:11331–11343. 10.1128/JVI.00577-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tian P, Ball JM, Zeng CQ, Estes MK. 1996. The rotavirus nonstructural glycoprotein NSP4 possesses membrane destabilization activity. J. Virol. 70:6973–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tian P, Estes MK, Hu Y, Ball JM, Zeng CQ, Schilling WP. 1995. The rotavirus nonstructural glycoprotein NSP4 mobilizes Ca2+ from the endoplasmic reticulum. J. Virol. 69:5763–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hyser JM, Collinson-Pautz MR, Utama B, Estes MK. 2010. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. mBio 1(5):e00265–10. 10.1128/mBio.00265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hyser JM, Zeng CQ, Beharry Z, Palzkill T, Estes MK. 2008. Epitope mapping and use of epitope-specific antisera to characterize the VP5* binding site in rotavirus SA11 NSP4. Virology 373:211–228. 10.1016/j.virol.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jagannath MR, Kesavulu MM, Deepa R, Sastri PN, Kumar SS, Suguna K, Rao CD. 2006. N- and C-terminal cooperation in rotavirus enterotoxin: novel mechanism of modulation of the properties of a multifunctional protein by a structurally and functionally overlapping conformational domain. J. Virol. 80:412–425. 10.1128/JVI.80.1.412-425.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poruchynsky MS, Atkinson PH. 1991. Rotavirus protein rearrangements in purified membrane-enveloped intermediate particles. J. Virol. 65:4720–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sastri NP, Pamidimukkala K, Marathahalli JR, Kaza S, Rao CD. 2011. Conformational differences unfold a wide range of enterotoxigenic abilities exhibited by rNSP4 peptides from different rotavirus strains. Open Virol. J. 5:124–135. 10.2174/1874357901105010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Au KS, Chan WK, Burns JW, Estes MK. 1989. Receptor activity of rotavirus nonstructural glycoprotein NS28. J. Virol. 63:4553–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14:993–1000. 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 28. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30. Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72–82. 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 31. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34. Burkhard P, Stetefeld J, Strelkov SV. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11:82–88. 10.1016/S0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 35. Lin SL, Tian P. 2003. Detailed computational analysis of a comprehensive set of group A rotavirus NSP4 proteins. Virus Genes 26:271–282. 10.1023/A:1024451314534. [DOI] [PubMed] [Google Scholar]
- 36. Lupas AN, Gruber M. 2005. The structure of alpha-helical coiled coils. Adv. Protein Chem. 70:37–78. 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 37. Malashkevich VN, Kammerer RA, Efimov VP, Schulthess T, Engel J. 1996. The crystal structure of a five-stranded coiled coil in COMP: a prototype ion channel? Science 274:761–765. 10.1126/science.274.5288.761. [DOI] [PubMed] [Google Scholar]
- 38. Akey DL, Malashkevich VN, Kim PS. 2001. Buried polar residues in coiled-coil interfaces. Biochemistry 40:6352–6360. 10.1021/bi002829w. [DOI] [PubMed] [Google Scholar]
- 39. Seo NS, Zeng CQ, Hyser JM, Utama B, Crawford SE, Kim KJ, Hook M, Estes MK. 2008. Integrins alpha1beta1 and alpha2beta1 are receptors for the rotavirus enterotoxin. Proc. Natl. Acad. Sci. U. S. A. 105:8811–8818. 10.1073/pnas.0803934105. [DOI] [PMC free article] [PubMed] [Google Scholar]