Abstract
Elephant populations are under intense pressure internationally from habitat destruction and poaching for ivory and meat. They also face pressure from infectious agents, including elephant endotheliotropic herpesvirus 1 (EEHV1), which kills ∼20% of Asian elephants (Elephas maximus) born in zoos and causes disease in the wild. EEHV1 is one of at least six distinct EEHV in a phylogenetic lineage that appears to represent an ancient but newly recognized subfamily (the Deltaherpesvirinae) in the family Herpesviridae.
TEXT
Elephant endotheliotropic herpesvirus 1 (EEHV1) causes a rapidly progressing and usually fatal hemorrhagic disease that occurs in the wild in Asia and affects ∼20% of Asian elephant (Elephas maximus) calves born in zoos in the United States and Europe (1). About 60% of juvenile deaths of captive elephants are attributed to such infections. Development of control measures has been hampered by the lack of systems for culture of the virus in laboratories. Its genetic study has been restricted to analysis of blood, trunk wash fluid, and tissue samples collected during necropsies. Fortunately, methods for amplifying DNA from uncharacterized viruses through the use of degenerate PCR primers that target highly conserved genes (2, 3), plus advances in DNA sequencing, have enabled assembly of complete genome sequences for EEHV1A and EEHV1B (4, 5), as well as numerous subgenomic segments from other EEHV present in clinical specimens. Among other things, this work has led to the discovery of what is now a dozen new herpesviruses of elephants, including five of the gammaherpesvirus lineage (6, 7). In two remarkable papers published in this issue of Journal of Virology (8, 9), L. K. Richman, J.-C. Zong, G. S. Hayward, and colleagues provide evidence to support recognition of five new EEHV species and show that these viruses and EEHV1 belong to a lineage within the family Herpesviridae that is distinct from its three currently recognized herpesvirus subfamilies (the Alpha-, Beta-, and Gammaherpesvirinae), the proposed Deltaherpesvirinae. This work provides a foundation for biological studies of the transmission and pathogenesis of these viruses and will be useful in developing the specific and informative diagnostic tools and therapeutic approaches needed to prevent these viruses from further devastating the already diminished international herd of these magnificent beasts.
A NEW HERPESVIRUS SUBFAMILY?
The current subfamilies within the Herpesviridae are harmonious with relationships readily deduced from genetic information, but they were established on the basis of shared biological properties more than 30 years ago, prior to the availability of biological sequences (10). Subsequent identification of viruses that share virion architecture but little genetic information with other herpesviruses led to the establishment in 2009 of two new virus families, the Alloherpesviridae (viruses of bony fish and frogs) and Malacoherpesviridae (viruses of oysters), and creation of the order Herpesvirales to encompass the three virus families (11) (Fig. 1A). The Herpesvirales stand as one of the seven virologic orders recognized by the International Committee for Taxonomy of Viruses (12).
FIG 1.
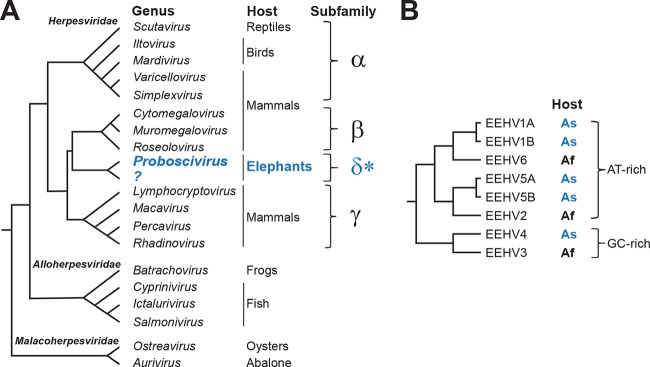
EEHV phylogeny and taxonomy. (A) EEHV lines of descent in the context of established herpesvirus taxonomy. With the exception of the EEHV, the taxonomic relationships shown are based on current herpesvirus taxonomy (http://ictvonline.org/). The proposed (indicated with a blue asterisk) Deltaherpesvirinae (δ) subfamily branches from between the Betaherpesvirinae (β) and Gammaherpesvirinae (γ) lineages. In constructing the diagram, it is assumed that the Proboscivirus genus would be retained in the proposed new subfamily, and the diagram indicates the possibility of establishing a new genus for the GC-rich viruses that are shown in panel B. (B) Relationships among the EEHV. AT- and GC-rich branches are shown, as well as the host species for each virus. The host species were Asian elephants (As) or African elephants (Af). This figure was adapted from reference 15.
To date, 88 virus species have been formally recognized within the Herpesviridae (the current complete list of approved virus taxons is available at http://ictvonline.org/). In addition, approximately 200 additional viruses detected using methods such as those described above await formal consideration (V. Lacoste, personal communication). With very few exceptions, the amino acid sequence of a small conserved segment of the viral DNA polymerase (∼150 amino acids) is sufficient to not only reliably identify a virus as belonging to the evolutionary lineage represented by the Herpesviridae, but also their subfamily, and in most cases a recognized genus. Early analyses of such sequences from EEHV1 showed that sequences of its DNA polymerase and other highly conserved herpesvirus core genes branched near the base of the betaherpesvirus branch of the herpesvirus tree, leading to the suggestions that the virus might represent a new herpesvirus subfamily (13, 14). EEHV1 is currently formally recognized as the species Elephantid herpesvirus 1, the type species in the Proboscivirus genus within the Betaherpesvirinae. The new papers by Richman et al. (8) and Zong et al. (9) go well beyond this, showing that six distinct EEHV belong to the Proboscivirus lineage; the authors make the case that this lineage represents a novel subfamily within the Herpesviridae.
Evidence for recognition of a new subfamily comes in three main forms: (i) deep branching of conserved herpesvirus core sequences near the points of divergence of the beta- and gammaherpesvirus branches of phylogenetic trees (Fig. 1A); (ii) the presence of genes that are present in alphaherpesvirus and/or gammaherpesvirus genomes and are seldom if ever found in betaherpesviruses, as well as numerous genes that are unique to EEHV (∼60 in the case of EEHV1, or half of its encoded proteins) (Fig. 2); and (iii) an arrangement of herpesvirus core gene blocks different from what has been seen in other herpesvirus lineages (Fig. 2). The latter is important because with few and limited exceptions, herpesvirus core genes are arranged in a colinear manner within subfamilies (15).
FIG 2.
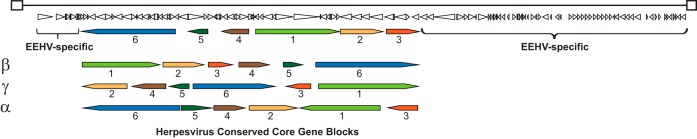
EEHV1 genomic architecture. The 180-kb genome consists of a unique segment bounded by a pair of direct terminal repeats. The locations of the six blocks of core genes conserved among members of the Herpesviridae are shown, along with the orientations of these regions characteristic of the alpha-, beta-, and gammaherpesvirus subfamilies. The conserved core region is flanked by genes not found in other herpesviruses. This figure is based on information in references 4, 8, 9, 12, and 15. The genomic orientation shown is reversed from the depictions in references 8 and 9.
The question of whether the EEHV should be elevated to the level of a virus subfamily is under deliberation by the Herpesvirales Study Group of the International Committee for Taxonomy of Viruses. There are questions and competing arguments that go beyond the question of whether Greek letters will soon be in short supply. Will the other EEHV have genome organizations similar to that of EEHV1? Are there reasons to expect that ongoing searches for new viruses in other organisms will uncover a multitude of analogous new lineages? What unintended consequences might connect to recognition of a new lineage? What depiction of the relationships is most usefully informative? In any respect, the new data are clear that the lineage is deeply rooted within the Herpesviridae.
GENOMIC PLASTICITY AND DIVERSITY
Patterns of sequence variation are fundamental biological properties of individual viruses and groups of viruses. As a group, EEHV display forms and levels of sequence diversity that are collectively unique with respect to forms of variation seen for other groups of herpesviruses. Somewhat analogous to human herpesviruses 6A and 6B (16), most of the 64 genes that span the center of the EEHV1A and EEHV1B genomes are >99% identical, with three interspersed sharply bounded regions of much greater sequence divergence (referred to as chimeric domains, or CD), as well as some sequence rearrangements in the vicinity of the genomic termini (4, 8). Although less sequence is available, EEHV5A and EEHV5B appear to have a similar relationship (9). In contrast to human herpesvirus 6A (HHV-6A) and HHV-6B, the chimeric domains appear to be products of recombination with diverged versions of EEHV species that have not been detected and may no longer be extant. It remains to be determined whether either of these pairs of closely related EEHV represent distinct virus species.
Phylogenetically, the EEHV form two main clusters, one harboring two closely related pairs of viruses with relatively AT-rich genomes, and another cluster harboring EEHV3 and EEHV4, which have GC-rich genomes (Fig. 1B) (9). Each pair of closely related viruses includes one whose natural host appears to be Asian elephants, and one of African elephants. Thus, Asian and African elephants are both host to at least three distinct, but closely related herpesviruses of the EEHV lineage. A precedent is that humans are hosts to three alphaherpesviruses, herpes simplex viruses 1 and 2, and varicella-zoster virus (formally, Human herpesviruses 1, 2, and 3). Although there were early suggestions that severe hemorrhagic disease associated with EEHV1 was due to infection of Asian elephants with virus from African elephants (13), accumulated evidence shows that these viruses can cause disease as well as persist asymptomatically in their native hosts. While EEHV1 is thus far the major pathogen, the presence of multiple closely related host-specific viruses will complicate efforts to develop vaccines and therapeutic approaches.
Footnotes
Published ahead of print 17 September 2014
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1. Hayward GS. 2012. Conservation: clarifying the risk from herpesvirus to captive Asian elephants. Vet. Rec. 170:202–203. 10.1136/vr.e1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628–1635. 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, Rose TM. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilkie GS, Davison AJ, Watson M, Kerr K, Sanderson S, Bouts T, Steinbach F, Dastjerdi A. 2013. Complete genome sequences of elephant endotheliotropic herpesviruses 1A and 1B determined directly from fatal cases. J. Virol. 87:6700–6712. 10.1128/JVI.00655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ling PD, Reid JG, Qin X, Muzny DM, Gibbs R, Petrosino J, Peng R, Zong JC, Heaggans SY, Hayward GS. 2013. Complete genome sequence of elephant endotheliotropic herpesvirus 1A. Genome Announc. 1(2):e0010613. 10.1128/genomeA.00106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wellehan JF, Johnson AJ, Childress AL, Harr KE, Isaza R. 2008. Six novel gammaherpesviruses of Afrotheria provide insight into the early divergence of the Gammaherpesvirinae. Vet. Microbiol. 127:249–257. 10.1016/j.vetmic.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 7. Latimer E, Zong JC, Heaggans SY, Richman LK, Hayward GS. 2011. Detection and evaluation of novel herpesviruses in routine and pathological samples from Asian and African elephants: identification of two new probosciviruses (EEHV5 and EEHV6) and two new gammaherpesviruses (EGHV3B and EGHV5). Vet. Microbiol. 147:28–41. 10.1016/j.vetmic.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Richman LK, Zong J-C, Latimer EM, Lock J, Fleischer R, Heaggans SY, Hayward GS. 2014. Elephant endotheliotropic herpesviruses EEHV1A, EEHV1B, and EEHV2 from cases of hemorrhagic disease are highly diverged from other mammalian herpesviruses and may form a new subfamily. J. Virol. 88:13523–13546. 10.1128/JVI.01673-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zong J-C, Latimer EM, Long SY, Richman LK, Heaggans SY, Hayward GS. 2014. Comparative genome analysis of four elephant endotheliotropic herpesviruses EEHV3, EEHV4, EEHV5, and EEHV6 from cases of hemorrhagic disease or viremia. J. Virol. 88:13547–13569. 10.1128/JVI.01675-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roizman B, Carmichael LE, Deinhardt F, de The G, Nahmias AJ, Plowright W, Rapp R, Sheldrick P, Takahashi M, Wolf K. 1981. Herpesviridae: definition, provisional nomenclature, and taxonomy. Intervirology 16:201–217. 10.1159/000149269. [DOI] [PubMed] [Google Scholar]
- 11. Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. 2009. The order Herpesvirales. Arch. Virol. 154:171–177. 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pellett PE, Davison AJ, Eberle R, Ehlers B, Hayward GS, Lacoste V, Minson AC, Nicholas J, Roizman B, Studdert MJ, Wang F. 2011. Order Herpesvirales, p 99–107 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 13. Richman LK, Montali RJ, Garber RL, Kennedy MA, Lehnhardt J, Hildebrandt T, Schmitt D, Hardy D, Alcendor DJ, Hayward GS. 1999. Novel endotheliotropic herpesviruses fatal for Asian and African elephants. Science 283:1171–1176. 10.1126/science.283.5405.1171. [DOI] [PubMed] [Google Scholar]
- 14. Ehlers B, Burkhardt S, Goltz M, Bergmann V, Ochs A, Weiler H, Hentschke J. 2001. Genetic and ultrastructural characterization of a European isolate of the fatal endotheliotropic elephant herpesvirus. J. Gen. Virol. 82:475–482. [DOI] [PubMed] [Google Scholar]
- 15. Pellett PE, Roizman B. 2013. Herpesviridae, p 1802–1822 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B. (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 16. Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]


