Abstract
Staphylococcus aureus is a major cause of food poisoning outbreaks associated with dairy products, because of the ingestion of preformed enterotoxins. The biocontrol of S. aureus using lactic acid bacteria (LAB) offers a promising opportunity to fight this pathogen while respecting the product ecosystem. We had previously established the ability of Lactococcus lactis, a lactic acid bacterium widely used in the dairy industry, to downregulate a major staphylococcal virulence regulator, the accessory gene regulator (agr) system, and, as a consequence, agr-controlled enterotoxins. In the present paper, we have shown that the oxygen-independent reducing properties of L. lactis contribute to agr downregulation. Neutralizing lactococcal reduction by adding potassium ferricyanide or maintaining the oxygen pressure constant at 50% released agr downregulation in the presence of L. lactis. This downregulation still occurred in an S. aureus srrA mutant, indicating that the staphylococcal respiratory response regulator SrrAB was not the only component in the signaling pathway. Therefore, this study clearly demonstrates the ability of L. lactis reducing properties to interfere with the expression of S. aureus virulence, thus highlighting this general property of LAB as a lever to control the virulence expression of this major pathogen in a food context and beyond.
INTRODUCTION
Staphylococcus aureus is a major cause of food poisoning outbreaks associated with dairy products, because of the ingestion of staphylococcal enterotoxins preformed in foodstuff (1). In fermented milk products, a strategy to combat S. aureus or any other pathogen must respect the product ecosystem, which is crucial to the manufacturing process and the development of the organoleptic properties of the final product. Biocontrol based on the use of nonpathogenic lactic acid bacteria (LAB) appears to be an attractive and sustainable strategy to combat bacterial pathogens. The ability of LAB to inhibit S. aureus growth has been reported in several contexts (2–6) and relies on properties such as acidification, the production of bacteriocins and hydrogen peroxide, or competition for nutrients (3, 6–10). LAB are also reportedly able to interfere with the expression of S. aureus virulence, which includes staphylococcal enterotoxin production (9, 11, 12). However, with just a few exceptions (12), the inhibitory mechanisms underlying this inhibition of virulence are still poorly understood.
We previously investigated the interaction between Lactococcus lactis LD61 and S. aureus MW2 and established that L. lactis downregulated the expression of the accessory gene regulator (agr) system of S. aureus in mixed cultures as well as agr-controlled enterotoxins such as enterotoxin C (13, 14). This agr downregulation by L. lactis was observed under planktonic conditions, in chemically defined medium (CDM) at a constant of pH 6.6 and under microaerobic conditions, and was further validated under sessile growth conditions in a model cheese (15). The agr system is a key virulence regulator in S. aureus, controlling the time course of expression of virulence factors in relation to the growth phase (1, 16, 17). It combines a two-component system and a quorum-sensing system (see Fig. S1 in the supplemental material). The agr system comprises two transcripts, RNAII (which encodes agrBDCA, the structural components of the quorum-sensing system) and RNAIII (which is the effector molecule of agr). The agr system itself is tightly regulated by a complex regulatory network that allows it to respond to several environmental stimuli, which include nutrient availability, pH, and oxygen availability (18–26). Indeed, we previously showed that a drop in the pH partially accounted for the downregulation of agr observed in a cheese matrix in the presence of L. lactis (15).
The present study was undertaken to unravel the mechanisms involved in the downregulation of agr in the presence of L. lactis under planktonic conditions in CDM at a constant pH of 6.6 that we previously reported (13). First, the production of cyclic dipeptide in a Lactobacillus reuteri culture supernatant was previously shown to downregulate the agr system in a mixed culture (12). We thus questioned whether agr downregulation by L. lactis was mediated by the secretion of an effector or whether it required the presence of L. lactis cells, metabolically active or not. Second, compilation of our findings revealed that RNAIII downregulation also occurred with other L. lactis strains (our unpublished results), suggesting that general properties of L. lactis were involved. The contributions of a lower pH and proteolysis were excluded in CDM because the pH was regulated at 6.6 and the nitrogen nutrition of S. aureus was achieved through free amino acids. Nevertheless, we could not totally exclude the contribution of lactic acid production by L. lactis to agr downregulation, even at a constant pH of 6.6. A contribution of the reducing properties of L. lactis to agr downregulation was also postulated. L. lactis is well known for its strong reducing capacities (27, 28). S. aureus senses the redox environment through several regulators, which in turn regulate agr expression (29–37). Among these regulators is SrrAB, a redox sensor that senses low redox potential under conditions of anaerobiosis, rather than low oxygen availability itself, and represses agr expression (34, 36, 37). We thus hypothesized that S. aureus sensed a low redox potential in the presence of L. lactis through SrrAB (36, 37). To sum up, the present study was designed to investigate whether any of the following mechanisms could account for agr downregulation by L. lactis: (i) lactate production, independent of a lower pH; (ii) the release of an effector into the medium; (iii) the necessary presence of L. lactis, alive or not; and (iv) the decrease of the redox potential and the contribution of SrrAB to the signaling pathway.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. lactis subsp. lactis bv. diacetylactis LD61 and S. aureus MW2 were used throughout this work (38, 39).
Mixed cultures of S. aureus and L. lactis and pure cultures (used as controls) were grown in a 2-liter Biostat B Plus fermentor (Sartorius, Melsungen, Germany) in CDM at 30°C with the pH regulated at 6.6 by the automatic addition of 10 N KOH, as previously described (13, 14). The fermentors were equipped with a redox electrode (Pt4805-DPAS-SC K8S/200; Mettler Toledo), a pH electrode (Easyferm Plus K8 200; Hamilton), and a dissolved oxygen probe (InPro 6820/12/220; Mettler Toledo). The redox probes were verified with a standard solution at +300 mV ± 30 mV. The measured redox potential (Em) was corrected with regard to the reference hydrogen electrode to obtain Eh, as previously described (Eh = Em + Er, where Er = 204 mV) (40, 41). Eh was not corrected with regard to pH, as the pH was maintained constant at 6.6 throughout fermentation. Oxygen probes were used to measure the oxygen pressure (pO2). They were standardized in air-sparged water for 100% pO2 and in nitrogen-sparged water for 0% pO2.
Both strains were subcultured and inoculated into the fermentor at 106 CFU ml−1, as previously described (14). Bacterial growth was estimated by determining CFU using a micromethod, as previously described (14). The number of CFU of the L. lactis population was determined on M17 (Difco) agar plates supplemented with 0.5% glucose and incubated at 30°C for 24 h. The number of CFU of the S. aureus population was determined after growth on tryptic soy broth (AES, Combourg, France) agar plates supplemented with 6.5% NaCl and incubated at 37°C for 24 h. Bacterial growth was determined in duplicate for each sample (each time point of an independent biological replicate).
Experimental design.
The growth conditions described below were used to test the involvement of different mechanisms in agr downregulation in the presence of L. lactis, as mentioned in the introduction.
(i) Reference conditions.
Experiments were performed in triplicate (independent biological repeats) under microaerobic conditions (air in the headspace of fermentors but no air bubbling). These conditions (CDM at 30°C with a constant pH of 6.6 under microaerobic conditions) are referred to here as the reference conditions.
(ii) Lactate production.
To test the effect of lactate production on agr downregulation, culturing of S. aureus MW2 was performed under reference conditions (CDM with pH regulated at 6.6 under microaerobic conditions) with the addition of a lactate concentration corresponding to the maximum concentration obtained in a mixed culture with L. lactis (i.e., 70 mM).
(iii) Secretion of an effector.
To test whether agr downregulation in the presence of L. lactis was mediated by the secretion of an effector, S. aureus cells were cultured in L. lactis supernatants. Because L. lactis LD61 underwent lysis in CDM under reference conditions at entry into the stationary phase (14), L. lactis culture supernatants were collected both at the end of the exponential phase (SNexp) and in the stationary phase (SNstat) (i.e., before and after the lysis of L. lactis LD61). L. lactis cells were removed by centrifugation (6,500 × g for 10 min at 4°C), and the supernatant was filter sterilized (0.2 μm) prior to inoculation with S. aureus.
(iv) Need for live L. lactis.
In order to determine whether the presence of live L. lactis cells was required, experiments using heat-killed L. lactis cells were performed. L. lactis cells were grown until the end of the exponential phase of growth under reference conditions. The cells were collected by centrifugation, washed, heat killed by incubation for 30 min at 70°C, centrifuged, and suspended in fresh CDM prior to S. aureus inoculation.
(v) Reducing properties of L. lactis.
To test whether the decrease of the redox potential in the presence of L. lactis was involved in agr downregulation, we performed two sets of experiments. First, cells were cultured under reference conditions with the addition of potassium ferricyanide (24 mM), which was previously used to adjust the redox potential and which does not interfere with bacterial growth at this concentration (42). Second, cells were cultured under reference conditions except that pO2 was maintained at 50%.
Construction of the srrA mutant of S. aureus MW2.
The srrA mutant of S. aureus MW2 was obtained essentially as described previously by Hiron et al. (43), with the following modifications. Briefly, a recombination cassette was cloned into Escherichia coli NEB10-beta by using an overlap PCR technique. This recombination cassette led to the insertion of a stop codon and an XhoI restriction site in the srrA coding sequence at position +108, followed by a deletion of 331 bp. To construct this cassette, two fragments (referred to as boxes A2 and B2, corresponding to the upstream and downstream regions of the fragment to be deleted, respectively) were PCR amplified from MW2 genomic DNA by using the primers listed in Table S1 in the supplemental material. Boxes A2 and B2 were mixed at equal concentrations and used as a template for a second PCR using only the two external primers. The internal primers BoxA2-srrAB-R-XhoI and BoxB2-srrAB-F-XhoI contained 20 nucleotides corresponding to boxes B2 and A2, respectively, thus allowing overlap PCR to occur. The hybrid PCR product, referred to as box A2B2, was cloned into the pGEM-T vector (Promega, Lyon, France), resulting in plasmid pGEM-T::BoxA2B2, which was then digested by BamHI and BglII. The BamHI-box A2B2-BglII fragment was then purified after separation by agarose gel electrophoresis and cloned into the temperature-sensitive shuttle vector pMAD (44). The resulting plasmid, pMAD::BoxA2B2, was introduced into S. aureus MW2 following subcloning into S. aureus RN4220, and erythromycin-resistant transformants were selected at 30°C, the permissive temperature for pMAD replication. Deletion of the chromosomal region was subsequently achieved by double-crossover events, as previously described (44). Chromosomal deletions were checked by PCR (see Table S1 in the supplemental material) and reverse transcription-quantitative PCR (RT-qPCR), using primers reported previously (15).
RNA extraction and purification.
RNA samples from S. aureus in pure or mixed cultures were obtained and quantified, and their quality was evaluated as previously described (13).
Reverse transcription-quantitative PCR.
RT-qPCR was performed in triplicate by using oligonucleotides previously designed for gyrB, RNAIII, agrA, and srrA (15). The gene expression level was first reported relative to the expression level of gyrB (internal standard) (15). The expression levels of genes in S. aureus MW2 in the stationary phase (24 h of culture) in a pure culture were used as a reference. The expression levels of genes in the exponential and stationary phases in pure and mixed cultures were then calculated relative to the reference values.
Statistical analysis.
All statistical analyses were performed in triplicate with R software (2013; R Foundation for Statistical Computing, Vienna, Austria). Statistical analysis was applied to normalized gene expression levels by using one-way analysis of variance, considering a P value of <0.05 to identify, for each time point analyzed (exponential phase and stationary phase), genes displaying a significant change in expression levels in pure versus mixed cultures. Multiple comparisons of conditions were achieved by using the Tukey range test.
RESULTS
Lactate is not involved in agr downregulation in the presence of L. lactis.
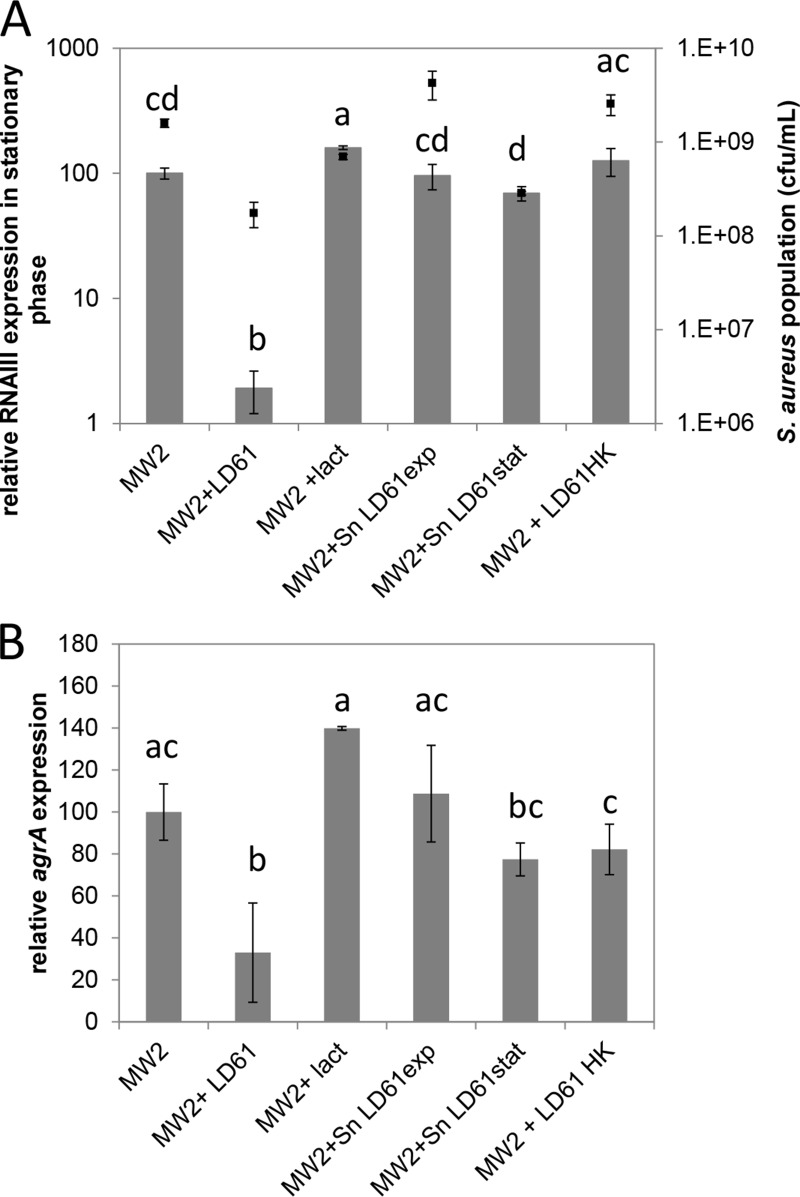
S. aureus MW2 cells were cultured in the presence of lactate. No inhibition of S. aureus RNAIII or agrA expression was observed under these conditions in the stationary phase (Fig. 1). Indeed, the expressions of RNAIII and agrA were slightly favored by the addition of lactate. Therefore, regardless of its acidification potential, lactate is not involved in the agr downregulation observed in mixed cultures.
FIG 1.
Expression of the agr system in S. aureus MW2 in the stationary phase under various conditions. Cells were cultured in CDM at a constant pH of 6.6 under microaerobic conditions. S. aureus was grown either alone (MW2), in the presence of L. lactis LD61 (MW2+LD61), with lactate (MW2+lact), in the supernatant of L. lactis LD61 grown under reference conditions until the exponential phase of growth (MW2+SnLD61exp) or the stationary phase of growth (MW2+SnLD61stat), or in the presence of heat-killed L. lactis LD61 (MW2+LD61HK). The expression levels of RNAIII (A) and agrA (B) were determined by RT-qPCR and are expressed relative to those of RNAIII and agrA in the stationary phase in a pure S. aureus culture (100%), respectively. Multiple comparisons of RNAIII and agrA expression levels under various conditions were achieved by using the Tukey range test and are indicated by letters. The final S. aureus population (black squares) is also plotted in panel A.
Live L. lactis is required to downregulate agr expression in S. aureus.
S. aureus was grown in the presence of L. lactis supernatants. The growth of S. aureus was not altered in SNexp compared to growth in CDM, whereas it was affected when S. aureus was grown in SNstat. Hence, the final S. aureus population in the stationary phase was ∼5-fold smaller in the SNstat supernatant than in a pure culture in CDM. The growth of S. aureus in L. lactis supernatants did not significantly affect RNAIII induction in the stationary phase compared to a pure S. aureus culture on CDM (Fig. 1A). Similar trends were observed for agrA (Fig. 1B).
To investigate whether metabolically active lactococcal cells were required or not, cells were cultured in the presence of heat-killed L. lactis cells. As shown in Fig. 1A, RNAIII induction in the presence of heat-killed L. lactis was similar to that observed in a pure culture of S. aureus, indicating that the mechanism(s) involved in RNAIII downregulation required metabolically active L. lactis. Likewise, agrA expression was not significantly affected in the presence of heat-killed L. lactis (Fig. 1B).
Reduction by L. lactis contributes to RNAIII downregulation.
Among the mechanisms that could account for agr downregulation, we hypothesized that the reducing properties of L. lactis contributed to this downregulation. In the pure L. lactis culture, the redox potential decreased markedly (down to −167 mV) after 8 h and then increased subsequently, at the same time that the pO2 increased (Fig. 2B). The pO2 increased at the time when the cultivability of L. lactis declined (Fig. 2A). In the S. aureus pure culture, the redox potential decreased following a three-step kinetic (Fig. 2): first, a drop corresponding to oxygen exhaustion (after 4 h) was observed; next, a slow decline of the redox potential was observed until 10 h of culture, corresponding to the end of the exponential phase of growth; and finally, the redox potential reached its final value (−100 mV) after 20 h, corresponding to the stationary phase. In the mixed culture, a rapid decrease in the redox potential was observed compared to the pure S. aureus culture, reaching −178 mV after 8.5 h of culture. The redox potential then increased and remained constant at −100 mV until 24 h, as in the pure S. aureus culture. The redox potential therefore remained lower in the mixed culture than in the pure S. aureus culture up to 20 h of culture.
FIG 2.
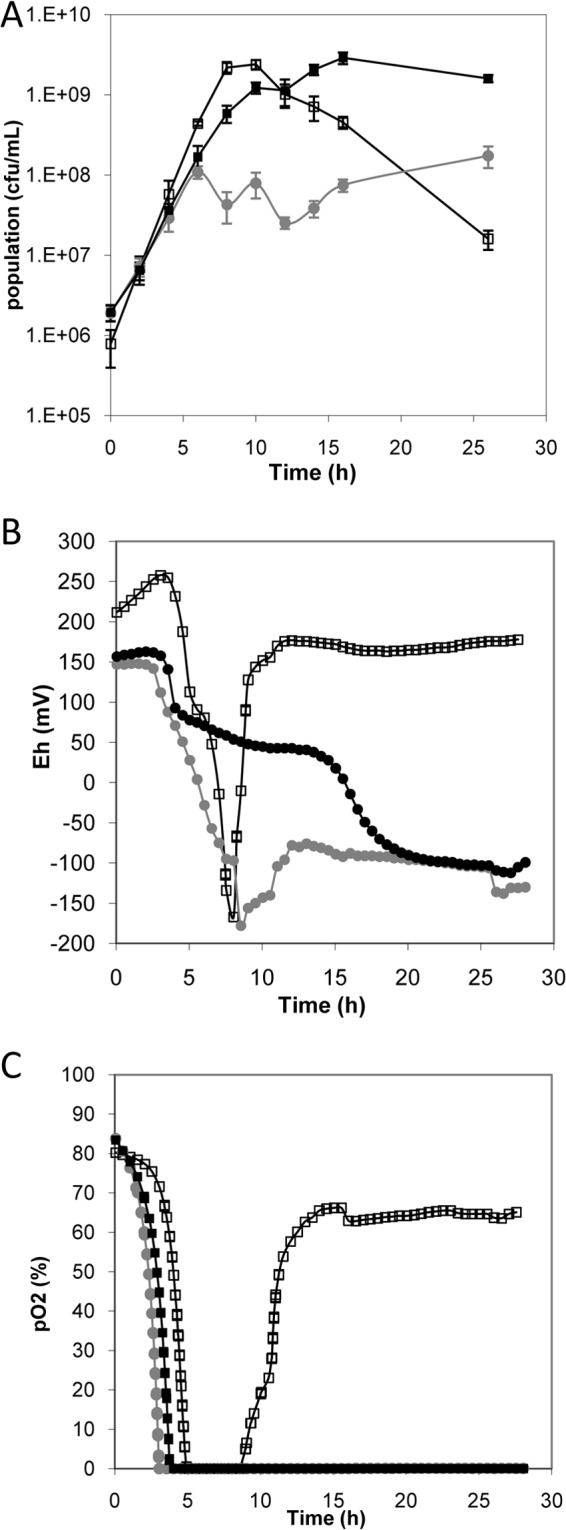
Kinetics of growth (A), redox potential (B), and partial oxygen pressure (C) in cultures of L. lactis LD61 alone (open squares), S. aureus MW2 alone (black squares), or S. aureus MW2 with L. lactis LD61 (gray circles). Cultures were performed under reference conditions, i.e., in CDM at a constant pH of 6.6 under microaerobic conditions. Shown are the kinetics from one representative experiment out of the three performed.
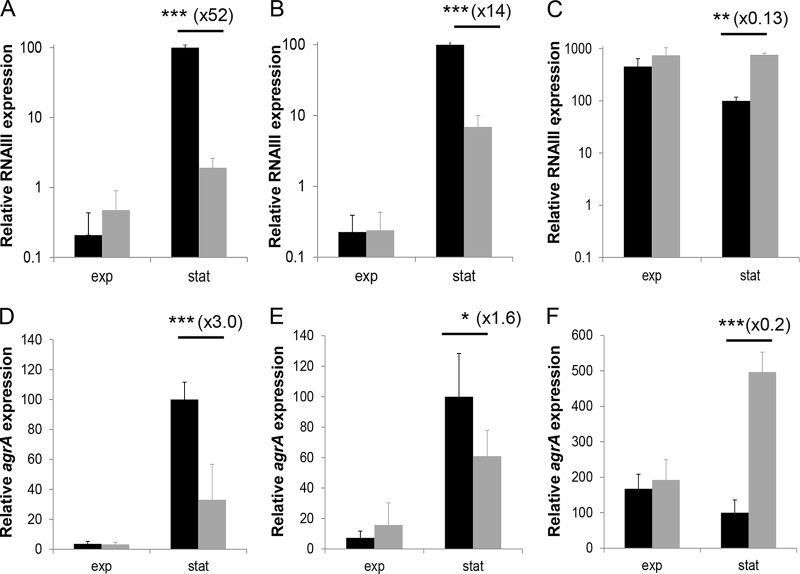
To evaluate the impact of the lower redox potential in the presence of L. lactis on agr expression, we performed two sets of experiments designed to compensate for the reduction in redox potential in a mixed culture (see Materials and Methods). In the presence of potassium ferricyanide, the initial redox potential was higher than that under reference conditions (+500 to 600 mV versus +150 mV) (see Fig. S2 in the supplemental material). The redox potential decreased only moderately until 15 to 16 h, which corresponded to entry into the stationary phase of both pure S. aureus and mixed cultures. A sharp decrease then occurred, reaching final plateaus at −10 and −110 mV in the mixed and pure S. aureus cultures, respectively. As observed under the reference conditions, the induction of RNAIII occurred in pure S. aureus cultures in the presence of potassium ferricyanide (Fig. 3B). Interestingly, in mixed cultures in the presence of potassium ferricyanide, RNAIII downregulation by L. lactis was partially released in the stationary phase. Hence, in the stationary phase, the RNAIII expression level was 52-fold lower in the presence of L. lactis than in a pure S. aureus culture under reference conditions, whereas the repression factor was only 14-fold in the presence of potassium ferricyanide (Fig. 3A and B). A similar trend was observed for agrA expression (Fig. 3D and E).
FIG 3.
RNAIII expression (A to C) and agrA expression (D to F) of S. aureus MW2 in a pure culture (black) and in mixed cultures with L. lactis LD61 (gray). Cells were cultured under reference conditions (CDM at a constant pH of 6.6 under microaerobic conditions) (A and D), under reference conditions with the addition of potassium ferricyanide (B and E), and in CDM at a constant pH of 6.6 and with pO2 at 50% (C and F). The expression levels of RNAIII and agrA were determined by RT-qPCR during the exponential (exp) and stationary (stat) phases of growth and are expressed relative to those of RNAIII and agrA in the stationary phase in a pure culture (100%), respectively. Significant changes of expression levels in pure versus mixed cultures were determined for each time point by using one-way analysis of variance and are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
A second set of experiments was performed by maintaining the pO2 constant at 50%. Under these conditions, the Eh was kept high (∼220 mV) in both pure and mixed cultures during the entire experiment, and similar growth of S. aureus was obtained in both pure and mixed cultures (data not shown). In a pure S. aureus culture, the RNAIII expression profile was modified compared to that under reference conditions because the agr system was already strongly induced in the exponential phase. Indeed, the expression levels of RNAIII and agrA even decreased in the stationary phase in pure cultures (Fig. 3C and F). In mixed cultures with a pO2 at 50%, RNAIII and agrA downregulation by L. lactis was completely eliminated. Hence, in mixed cultures, the expression levels of RNAIII and agrA were similar to the levels obtained in pure S. aureus cultures in the exponential phase and were even higher in the stationary phase.
Downregulation of the agr system by L. lactis is not mediated by SrrAB in S. aureus.
The involvement of SrrAB in signaling low redox potential was tested by constructing an srrA-null mutant (see Materials and Methods). In experiments with mixed cultures, RNAIII and agrA downregulation still occurred in the srrA mutant and was even more pronounced (see Fig. S3 in the supplemental material), indicating that SrrAB was not involved in agr downregulation by L. lactis.
DISCUSSION
The reducing properties of L. lactis contribute to agr downregulation.
In previous studies, we established the ability of L. lactis LD61 to inhibit the agr system under planktonic conditions in CDM (referred to as reference conditions in the present study) (13). Here, we have clearly demonstrated that the reducing properties of L. lactis contributed to the downregulation of the agr system in mixed cultures. Hence, the redox potential was lower in mixed than in pure cultures of S. aureus until 20 h. This was likely the result of the strong reduction capacity of L. lactis, because it also occurred in the pure L. lactis culture. The contribution of this lower redox potential to agr downregulation was established by the partial release of agr system (RNAIII and agrA) downregulation when reduction of the medium by L. lactis was neutralized by either adding potassium ferricyanide or maintaining the pO2 constant at 50%.
It was observed that the reduction activity of L. lactis and the downregulation of agr were not strictly correlated (Fig. 2B and 3A and D): RNAIII and agrA downregulation occurred in the stationary phase, whereas there were no longer any differences in redox potential at that time (24 h). However, the agr system is an autoinducing system (see Fig. S1 in the supplemental material). The lower redox potential in mixed than in pure S. aureus cultures from the middle of the exponential phase of growth of S. aureus to 20 h of culture probably inhibited autoinduction of the agr system during this period, thus precluding any further induction during the stationary phase.
L. lactis downregulation of agr relies on oxygen-independent mechanisms.
The reducing properties of L. lactis rely on two different mechanisms. The first is oxygen dependent and corresponds to the elimination of oxygen by NADH oxidase (NoxE) (41). The second is oxygen independent and involves the electron transport chain. L. lactis does not possess a complete electron transport chain. However, some components, such as the membrane NADH dehydrogenases NoxA and NoxB as well as menaquinones (MK), have been shown to contribute to these oxygen-independent reducing properties by oxidizing compounds other than oxygen (41). In particular, L. lactis produces short-chain quinone species that are involved in the reduction of O2, Fe3+, and Cu2+ in medium (45). Interestingly, L. lactis can also be a source of quinone for group B streptococci (GBS), which are naturally deficient in MK biosynthesis via bacterial lysis or by direct cell-cell interactions (45). Alternatively, the oxygen-independent reducing capacities of L. lactis may be related to the presence of exofacial thiol groups on the bacterial cell surface, as previously observed (46). These exofacial thiol groups are displayed on proteins anchored to the cell surface, resulting in a reducing microenvironment around the cell.
During our study, the pO2 fell to 0% after 3 to 4 h of growth in both pure cultures of S. aureus and mixed cultures, and differences in redox potentials between these two conditions occurred once oxygen had been eliminated. This indicates that the reducing properties of L. lactis responsible for the lower redox potential in mixed cultures than in pure S. aureus cultures, and hence agr downregulation, rather rely on oxygen-independent mechanisms.
It is generally admitted that extracellular, and consequently intracellular, redox potential can drastically affect bacterial physiology. However, studies that have focused on the role of redox potential are scarce. Hence, it was previously established that the redox potential can influence the growth of bacteria, including starter LAB (47, 48), probiotic bacteria (49, 50), and undesirable bacteria (51). However, to our knowledge, although a direct effect of the redox potential on bacterial growth has been proven through chemical control of the redox potential or a modification of the gas environment (H2 and N2) (47–51), the effects of the reducing properties of LAB on the growth of pathogens or undesirable bacteria have never been clearly addressed. Redox potential is a complex parameter to study: it is not easy to grasp (black box with several inputs and outputs), and it is difficult to control. Increasing the complexity of the system by adding bacterial interactions probably explains the scarcity of reports in this field. Nevertheless, some papers have mentioned that the reducing potential of LAB likely influences the growth of pathogens (27, 52). This assumption was not supported by experiments but rather relied on the previously generally accepted idea that the reducing properties of LAB were related to oxygen consumption only. In other words, the consumption of oxygen by LAB results in an anaerobic environment, which may impair the growth of obligate aerobes. The same assumption can be made for virulence expression instead of growth. Indeed, it could be assumed that the oxygen-dependent reducing properties of L. lactis, i.e., its ability to eliminate oxygen, would influence the expression of S. aureus virulence, in the same way that oxygen exhaustion has been shown to influence the expression of S. aureus virulence in pure cultures (26, 37, 53). However, the effect of L. lactis reducing properties, including oxygen-dependent and -independent mechanisms, on virulence expression in food pathogens such as S. aureus has never been explored. Our study rigorously demonstrates that the reducing properties of L. lactis can downregulate the expression of the agr system, a major staphylococcal virulence regulator. Furthermore, the monitoring of both redox potential and oxygen pressure allowed us to discriminate between oxygen-dependent and oxygen-independent mechanisms.
In an attempt to identify the signaling pathways involved in agr downregulation by L. lactis, an srrA-null mutant was constructed. SrrAB has been shown to sense low redox potential under conditions of anaerobiosis, rather than oxygen availability itself, and to repress agr expression (36, 37). The activation signal is thought to be reduced menaquinones (54). In addition, srrAB is also under the control of Rex, which senses the NADH:NAD+ ratio (33). The expression level of srrA was measured in both pure and mixed S. aureus cultures, which revealed that it was 4-fold higher in mixed than in pure S. aureus cultures in the stationary phase (data not shown). This higher expression level of srrA in mixed cultures was in agreement with the release of Rex repression (33) and the autoinduction of srrAB expression under reduced conditions (37). These findings thus corroborated the hypothesis that SrrAB sensed the lower redox potential in the presence of L. lactis. SrrAB was thus postulated to be the most promising candidate among all the other redox sensors. However, agr downregulation still occurred in an srrA mutant of S. aureus. This suggests that either SrrAB did not mediate agr downregulation by L. lactis reducing properties or SrrAB was not the sole regulator involved in the signaling pathway. Indeed, other transcriptional regulators involved in the regulatory network controlling agr expression have been shown to be redox sensitive. These transcriptional regulators include SarA, SarZ, MgrA, Rex, AirSR, and the agr system itself (29–37, 55), although the role of these regulators at low redox potential has not yet been clearly established. The signaling pathways involved in agr downregulation by L. lactis reducing properties probably rely on several of these redox-sensitive regulators, in view of the redundancy between these regulators in terms of signals and regulons. Surprisingly, agr downregulation was even more pronounced in the S. aureus srrA mutant. The redox status of cells is known to be altered in an srrAB mutant because of the higher levels of tricarboxylic acid (TCA) cycle enzymes found in an srrAB mutant than in the wild type, combined with lower levels of alcohol and lactate dehydrogenases (36). The altered redox status of the cells may have reinforced and/or acted synergistically with the L. lactis effect.
The search for other mechanisms involved in agr downregulation by L. lactis.
The release of agr downregulation by L. lactis in the presence of ferricyanide was partial, suggesting the involvement of other mechanisms. Although we did not identify these other mechanisms, we were able to definitely exclude any contribution of lactate production by L. lactis, at a neutral pH, to agr downregulation. The lack of an effect of L. lactis supernatants on agr expression also revealed that the downregulation of agr by L. lactis was not due to the secretion of an inhibitory compound by L. lactis, unless this compound is highly labile. Furthermore, this result also indicates that agr downregulation was not due to the exhaustion of a nutrient or effector required for agr induction under mixed-culture conditions. The agr system is a quorum-sensing-controlled system and thus directly senses population density (19). A smaller population of S. aureus in a mixed culture could thus also contribute to weaker agr induction under these conditions. However, even if the final S. aureus population obtained in a mixed culture with L. lactis was similar to that obtained in SNstat, RNAIII expression was strongly downregulated in the presence of L. lactis and was hardly affected when S. aureus was grown in SNstat. This indicates that L. lactis downregulation of RNAIII expression was not due to the smaller S. aureus population under mixed-culture conditions. Further experiments will now be necessary to complete the mechanistic scheme of agr downregulation by L. lactis.
In conclusion, it appears that the ability of L. lactis to downregulate the agr system relies on at least two general properties of LAB: their ability to decrease both pH (15) and redox potential via oxygen-independent mechanisms (this study). Taking account of these two properties as both technological and sanitary parameters should contribute to a rational use of LAB starter strains that are able to limit the expression of this major staphylococcal virulence regulator and, as a result, of agr-controlled enterotoxins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tarek Msadek for supplying us with plasmid pMAD.
This work was funded by French National Research Agency (ANR) project Nabab (ANR-08-ALIA-11).
Footnotes
Published ahead of print 5 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02287-14.
REFERENCES
- 1.Hennekinne JA, De Buyser ML, Dragacci S. 2011. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 36:815–836. 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard DS, Rault L, Berkova N, Le Loir Y, Even S. 2013. Inhibition of Staphylococcus aureus invasion into bovine mammary epithelial cells by contact with live Lactobacillus casei. Appl. Environ. Microbiol. 79:877–885. 10.1128/AEM.03323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlier C, Cretenet M, Even S, Le Loir Y. 2009. Interactions between Staphylococcus aureus and lactic acid bacteria: an old story with new perspectives. Int. J. Food Microbiol. 131:30–39. 10.1016/j.ijfoodmicro.2008.06.032. [DOI] [PubMed] [Google Scholar]
- 4.Klostermann K, Crispie F, Flynn J, Ross RP, Hill C, Meaney W. 2008. Intramammary infusion of a live culture of Lactococcus lactis for treatment of bovine mastitis: comparison with antibiotic treatment in field trials. J. Dairy Res. 75:365–373. 10.1017/S0022029908003373. [DOI] [PubMed] [Google Scholar]
- 5.Nader-Macias ME, Otero MC, Espeche MC, Maldonado NC. 2008. Advances in the design of probiotic products for the prevention of major diseases in dairy cattle. J. Ind. Microbiol. Biotechnol. 35:1387–1395. 10.1007/s10295-008-0438-2. [DOI] [PubMed] [Google Scholar]
- 6.Elkins CA, Muñoz ME, Mullis LB, Stingley RL, Hart ME. 2008. Lactobacillus-mediated inhibition of clinical toxic shock syndrome Staphylococcus aureus strains and its relation to acid and peroxide production. Anaerobe 14:261–267. 10.1016/j.anaerobe.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Ammor S, Tauveron G, Dufour E, Chevallier I. 2006. Antibacterial activity of lactic acid bacteria against spoilage and pathogenic bacteria isolated from the same meat small-scale facility: 2—behaviour of pathogenic and spoilage bacteria in dual species biofilms including a bacteriocin-like-producing lactic acid bacteria. Food Control 17:462–468. 10.1016/j.foodcont.2005.02.007. [DOI] [Google Scholar]
- 8.Barber LE, Deibel RH. 1972. Effect of pH and oxygen tension on staphylococcal growth and enterotoxin formation in fermented sausage. Appl. Microbiol. 24:891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haines WC, Harmon LG. 1973. Effect of variations in conditions of incubation upon inhibition of Staphylococcus aureus by Pediococcus cerevisiae and Streptococcus lactis. Appl. Microbiol. 25:169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otero MC, Nader-Macias ME. 2006. Inhibition of Staphylococcus aureus by H2O2-producing Lactobacillus gasseri isolated from the vaginal tract of cattle. Anim. Reprod. Sci. 96:35–46. 10.1016/j.anireprosci.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Laughton JM, Devillard E, Heinrichs DE, Reid G, McCormick JK. 2006. Inhibition of expression of a staphylococcal superantigen-like protein by a soluble factor from Lactobacillus reuteri. Microbiology 152:1155–1167. 10.1099/mic.0.28654-0. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Wang W, Xu SX, Magarvey NA, McCormick JK. 2011. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. U. S. A. 108:3360–3365. 10.1073/pnas.1017431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Even S, Charlier C, Nouaille S, Ben Zakour NL, Cretenet M, Cousin FJ, Gautier M, Cocaign-Bousquet M, Loubière P, Le Loir Y. 2009. Staphylococcus aureus virulence expression is impaired by Lactococcus lactis in mixed cultures. Appl. Environ. Microbiol. 75:4459–4472. 10.1128/AEM.02388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nouaille S, Even S, Charlier C, Le Loir Y, Cocaign-Bousquet M, Loubière P. 2009. Transcriptomic response of Lactococcus lactis in mixed culture with Staphylococcus aureus. Appl. Environ. Microbiol. 75:4473–4482. 10.1128/AEM.02653-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cretenet M, Nouaille S, Thouin J, Rault L, Stenz L, François P, Hennekinne J-A, Piot M, Maillard MB, Fauquant J, Loubière P, Le Loir Y, Even S. 2011. Staphylococcus aureus virulence and metabolism are dramatically affected by Lactococcus lactis in cheese matrix. Environ. Microbiol. Rep. 3:340–351. 10.1111/j.1758-2229.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 16.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yarwood JM, Schlievert PM. 2003. Quorum sensing in Staphylococcus infections. J. Clin. Invest. 112:1620–1625. 10.1172/JCI20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bronner S, Monteil H, Prevost G. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183–200. 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449. 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 20.Bischoff M, Entenza JM, Giachino P. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171–5179. 10.1128/JB.183.17.5171-5179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimoto DF, Higginbotham RH, Sterba KM, Maleki SJ, Segall AM, Smeltzer MS, Hurlburt BK. 2009. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol. Microbiol. 74:1445–1458. 10.1111/j.1365-2958.2009.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somerville GA, Proctor RA. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 73:233–248. 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bore E, Langsrud S, Langsrud O, Rode TM, Holck A. 2007. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153:2289–2303. 10.1099/mic.0.2007/005942-0. [DOI] [PubMed] [Google Scholar]
- 24.Regassa LB, Novick RP, Betley MJ. 1992. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 60:3381–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinrick B, Dunman PM, McAleese F, Murphy E, Projan SJ, Fang Y, Novick RP. 2004. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186:8407–8423. 10.1128/JB.186.24.8407-8423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ster C, Gilbert FB, Cochard T, Poutrel B. 2005. Transcriptional profiles of regulatory and virulence factors of Staphylococcus aureus of bovine origin: oxygen impact and strain-to-strain variations. Mol. Cell. Probes 19:227–235. 10.1016/j.mcp.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Brasca M, Morandi S, Lodi R, Tamburini A. 2007. Redox potential to discriminate among species of lactic acid bacteria. J. Appl. Microbiol. 103:1516–1524. 10.1111/j.1365-2672.2007.03392.x. [DOI] [PubMed] [Google Scholar]
- 28.Cachon R, Jeanson S, Aldarf M, Divies C. 2002. Characterization of lactic acid starters based on acidification and reduction activities. Lait 82:281–288. 10.1051/lait:2002010. [DOI] [Google Scholar]
- 29.Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, He C. 2006. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2:591–595. 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 30.Chen PR, Nishida S, Poor CB, Cheng A, Bae T, Kuechenmeister L, Dunman PM, Missiakas D, He C. 2009. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. 71:198–211. 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall JW, Ji Y. 2013. Sensing and adapting to anaerobic conditions by Staphylococcus aureus. Adv. Appl. Microbiol. 84:1–25. 10.1016/B978-0-12-407673-0.00001-1. [DOI] [PubMed] [Google Scholar]
- 32.Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188:1899–1910. 10.1128/JB.188.5.1899-1910.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pagels M, Fuchs S, Pane-Farre J, Kohler C, Menschner L, Hecker M, McNamarra PJ, Bauer MC, von Wachenfeldt C, Liebeke M, Lalk M, Sander G, von Eiff C, Proctor RA, Engelmann S. 2010. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76:1142–1161. 10.1111/j.1365-2958.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430–2438. 10.1128/JB.186.8.2430-2438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun F, Ji Q, Jones MB, Deng X, Liang H, Frank B, Telser J, Peterson SN, Bae T, He C. 2012. AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J. Am. Chem. Soc. 134:305–314. 10.1021/ja2071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Throup JP, Zappacosta F, Lunsford RD, Annan RS, Carr SA, Lonsdale JT, Bryant AP, McDevitt D, Rosenberg M, Burnham MK. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392–10401. 10.1021/bi0102959. [DOI] [PubMed] [Google Scholar]
- 37.Yarwood JM, McCormick JK, Schlievert PM. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113–1123. 10.1128/JB.183.4.1113-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 39.Falentin H, Naquin D, Loux V, Barloy-Hubler F, Loubière P, Nouaille S, Lavenier D, Le Bourgeois P, François P, Schrenzel J, Hernandez D, Even S, Le Loir Y. 2014. Genome sequence of Lactococcus lactis subsp. lactis bv. diacetylactis LD61. Genome Announc. 2(1):e01176-13. 10.1128/genomeA.01176-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abraham S, Cachon R, Jeanson S, Ebel B, Michelon D, Aubert C, Rojas C, Feron G, Beuvier E, Gervais P, Coninck J. 2013. A procedure for reproducible measurement of redox potential (E h) in dairy processes. Dairy Sci. Technol. 93:675–690. 10.1007/s13594-013-0134-5. [DOI] [Google Scholar]
- 41.Tachon S, Brandsma JB, Yvon M. 2010. NoxE NADH oxidase and the electron transport chain are responsible for the ability of Lactococcus lactis to decrease the redox potential of milk. Appl. Environ. Microbiol. 76:1311–1319. 10.1128/AEM.02120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kieronczyk A, Cachon R, Feron G, Yvon M. 2006. Addition of oxidizing or reducing agents to the reaction medium influences amino acid conversion to aroma compounds by Lactococcus lactis. J. Appl. Microbiol. 101:1114–1122. 10.1111/j.1365-2672.2006.02999.x. [DOI] [PubMed] [Google Scholar]
- 43.Hiron A, Borezee-Durant E, Piard JC, Juillard V. 2007. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 189:5119–5129. 10.1128/JB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891. 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rezaiki L, Lamberet G, Derre A, Gruss A, Gaudu P. 2008. Lactococcus lactis produces short-chain quinones that cross-feed group B Streptococcus to activate respiration growth. Mol. Microbiol. 67:947–957. 10.1111/j.1365-2958.2007.06083.x. [DOI] [PubMed] [Google Scholar]
- 46.Michelon D, Abraham S, Ebel B, De Coninck J, Husson F, Feron G, Gervais P, Cachon R. 2010. Contribution of exofacial thiol groups in the reducing activity of Lactococcus lactis. FEBS J. 277:2282–2290. 10.1111/j.1742-4658.2010.07644.x. [DOI] [PubMed] [Google Scholar]
- 47.Jeanson S, Hilgert N, Coquillard M-O, Seukpanya C, Faiveley M, Neveu P, Abraham C, Georgescu V, Fourcassié P, Beuvier E. 2009. Milk acidification by Lactococcus lactis is improved by decreasing the level of dissolved oxygen rather than decreasing redox potential in the milk prior to inoculation. Int. J. Food Microbiol. 131:75–81. 10.1016/j.ijfoodmicro.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 48.Ouvry A, Waché Y, Tourdot-Maréchal R, Diviès C, Cachon R. 2002. Effects of oxidoreduction potential combined with acetic acid, NaCl and temperature on the growth, acidification, and membrane properties of Lactobacillus plantarum. FEMS Microbiol. Lett. 214:257–261. 10.1111/j.1574-6968.2002.tb11356.x. [DOI] [PubMed] [Google Scholar]
- 49.Dave RI, Shah NP. 1997. Effect of cysteine on the viability of yoghurt and probiotic bacteria in yoghurts made with commercial starter cultures. Int. Dairy J. 7:537–545. 10.1016/S0958-6946(97)00053-8. [DOI] [Google Scholar]
- 50.Dave RI, Shah NP. 1997. Effectiveness of ascorbic acid as an oxygen scavenger in improving viability of probiotic bacteria in yoghurts made with commercial starter cultures. Int. Dairy J. 7:435–443. 10.1016/S0958-6946(97)00026-5. [DOI] [Google Scholar]
- 51.Alwazeer D, Delbeau C, Divies C, Cachon R. 2003. Use of redox potential modification by gas improves microbial quality, color retention, and ascorbic acid stability of pasteurized orange juice. Int. J. Food Microbiol. 89:21–29. 10.1016/S0168-1605(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 52.Beresford TP, Fitzsimons NA, Brennan NL, Cogan TM. 2001. Recent advances in cheese microbiology. Int. Dairy J. 11:259–274. 10.1016/S0958-6946(01)00056-5. [DOI] [Google Scholar]
- 53.Fuchs S, Pane-Farre J, Kohler C, Hecker M, Engelmann S. 2007. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189:4275–4289. 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakano MM, Zuber P. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165–190. 10.1146/annurev.micro.52.1.165. [DOI] [PubMed] [Google Scholar]
- 55.Sun F, Liang H, Kong X, Xie S, Cho H, Deng X, Ji Q, Zhang H, Alvarez S, Hicks LM, Bae T, Luo C, Jiang H, He C. 2012. Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. Proc. Natl. Acad. Sci. U. S. A. 109:9095–9100. 10.1073/pnas.1200603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.