Abstract
Salmonella encounters various stresses in the environment and in the host during infection. The effects of cold (5°C, 48 h), peroxide (5 mM H2O2, 5 h) and acid stress (pH 4.0, 90 min) were tested on pathogenicity of Salmonella. Prior exposure of Salmonella to cold stress significantly (P < 0.05) increased adhesion and invasion of cultured intestinal epithelial (Caco-2) cells. This increased Salmonella-host cell association was also correlated with significant induction of several virulence-associated genes, implying an increased potential of cold-stressed Salmonella to cause an infection. In Caco-2 cells infected with cold-stressed Salmonella, genes involved in the electron transfer chain were significantly induced, but no simultaneous significant increase in expression of antioxidant genes that neutralize the effect of superoxide radicals or reactive oxygen species was observed. Increased production of caspase 9 and caspase 3/7 was confirmed during host cell infection with cold-stressed Salmonella. Further, a prophage gene, STM2699, induced in cold-stressed Salmonella and a spectrin gene, SPTAN1, induced in Salmonella-infected intestinal epithelial cells were found to have a significant contribution in increased adhesion and invasion of cold-stressed Salmonella in epithelial cells.
INTRODUCTION
Salmonella is an important food-borne pathogen throughout the world. It is transmitted by the fecal-oral route, and most infections occur due to ingestion of contaminated food. Salmonella encounters and survives various stresses, such as cold, acid, and oxidative stress, during its journey from the environment to food to infection in the animal host. The most common stressors include (i) cold stress, because refrigeration (5°C) is commonly used for long-term storage of food; (ii) oxidative stress, because peroxide is commonly used as a food sanitizer during food processing and it is also produced by macrophages and neutrophils as a result of inflammation and oxidative burst during infection; and (iii) acid stress, because acids are commonly used in food processing and, more importantly, gastric acidity is the first line of defense against pathogens within the host gut. Salmonella modulates its gene expression for survival upon exposure to the above stresses, and this can also simultaneously alter the expression of virulence factors and the surface structures of bacteria.
One of the major steps in the successful infection is the ability of Salmonella to adhere to host surfaces. Salmonella expresses two major groups of bacterial adhesins, namely, pilus (fimbrial) and nonpilus (afimbrial) adhesins. The role of many fimbrial operons, such as csg, bcf, fim, lpf, pef, saf, stb, stc, std, stf, sth, sti, and stj, in adhesion of Salmonella to host cells has been investigated (1). Salmonella can either bind directly to host cell surfaces or bind to components of the extracellular matrix (ECM) (2), such as fibronectin, laminin, and plasminogen (3, 4). However, only a small number of afimbrial adhesin factors, such as misL, ratB, shdA, sinH (1), and siiE (5), have been functionally characterized, and in most cases, their binding partners on host cells are not known. Additionally, alterations of the host cell surface due to inflammation can provide alternate adhesin receptors for pathogen binding. Thus, investigating interactions of afimbrial adhesin factors with non-ECM components is equally important. Upon adhesion, Salmonella induces a vast array of cytoplasmic and nuclear responses in epithelial cells, leading to cytoskeletal rearrangement, membrane ruffling and macropinocytosis, induction of transmembrane fluids and electrolyte fluxes, and synthesis of cytokines and mediators of inflammation (6). These functions are shown to be carried out by type 3 secretion system (T3SS) proteins which are encoded on two Salmonella pathogenicity islands (SPIs), SPI1 and SPI2. Expression of SPI1 is required for epithelial cell invasion, and SPI2 is required for intracellular survival and replication within phagocytic cells (6, 7).
Our hypothesis in this study was that prior exposure of Salmonella enterica serovar Typhimurium to abiotic stresses will result in modulation of gene expression and increased host cell adherence and invasion. The specific objective of this study was to investigate the effect of individual stresses on S. Typhimurium. This study tested the effect of cold stress (5°C, 48 h), peroxide stress (5 mM H2O2, 5 h), and acid stress (pH 4.0, 90 min) on the ability of S. Typhimurium to (i) adhere to and invade intestinal epithelial (Caco-2) cells, (ii) differentially regulate gene expression, and (iii) maintain differential gene regulation during infection. We found that prior exposure to cold stress significantly increased the association of S. Typhimurium with intestinal epithelial cells and induced the expression of many virulence-associated genes. Furthermore, we identified a novel receptor-ligand pair involved in the association of cold-stressed S. Typhimurium with intestinal epithelial cells.
MATERIALS AND METHODS
Cell culture, bacterial strain, and growth conditions.
Intestinal epithelial cells (Caco-2; ATCC HTB-37) were obtained from the American Type Culture Collection (Manassas, VA). Cells were seeded to a density of 105 cells/cm2 using Dulbecco's modified Eagle medium (high-glucose modified medium) (DMEM-HMM; catalog no. SH30285; Thermo Scientific, Rockford, IL), nonessential amino acids (Thermo Scientific), 10 mM MOPS (morpholinepropanesulfonic acid) (Sigma, St. Louis, MO), 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] (Sigma), 15 mM HEPES (Sigma) and 2 mM NaH2PO4 (Sigma). Additionally, 16.6% fetal bovine serum (FBS; HyClone Laboratories, Logan, UT) was added for feeding and maintaining cells. Cells were incubated at 37°C with 5% CO2 for 14 days postconfluence to allow differentiation. The cells were serum starved for 24 h prior to use by feeding the cells with cell culture medium described above but without FBS.
S. Typhimurium LT2 ATCC 700720 was used as a wild-type strain in this study. Cell culture medium was used to grow the bacteria at 37°C with shaking at 220 rpm. Bacterial gene knockout procedures for STM2699 and invA were performed as described previously (8).
Stress treatments for bacteria.
A log-phase culture of S. Typhimurium was centrifuged at 7,200 × g for 5 min. The pellets were resuspended at equal density in DMEM-HMM (i) maintained at 5°C for inducing cold stress, (ii) with 5 mM H2O2 added and maintained at 37°C for peroxide stress, and (iii) with preadjusted pH 4.0 and maintained at 37°C for acid stress. These stress treatments were given for 48 h, 5 h, and 90 min, respectively, in two biological replicates. The control for these treatments was a stationary-phase (∼15 h) culture resuspended in DMEM-HMM and maintained at 37°C.
Bacterium-host cell association assay.
Caco-2 cells were infected with bacteria at a multiplicity of infection (MOI) of 1:1,000, in a 96-well plate, in three biological replicates. The infected cells were incubated for 60 min, 90 min, and 120 min at 37°C with 5% CO2. Upon incubation, medium was aspirated, cells were washed three times with 200 μl standard Tyrode's buffer and then lysed using 50 μl lysis buffer (AEX Chemunex, France) as described previously (9), and the cell lysate was used to quantify the number of Caco-2 cells and associated bacteria. Quantitative bacterial analysis was done using quantitative PCR (qPCR) with a CFX 96 real-time system (Bio-Rad, Hercules, CA). Reactions were performed with iQ SYBR green Supermix (Bio-Rad) as per the manufacturer's instruction. Briefly, a 25-μl reaction mixture contained 1 μl of cell lysate and 100 nM forward (F) and reverse (R) PCR primers for the 16S rRNA gene (F, 5′-TGT TGT GGT TAA TAA CCG CA-3′; R, 5′-CAC AAA TCC ATC TCT GGA-3′) (10) to quantify S. Typhimurium or the G3PDH gene (F, 5′-ACC ACA GTC CAT GCC ATC AC-3′; R, 5′-TCC ACC ACC CTG TTG CTG TA-3′) to quantify Caco-2 cells (Integrated DNA Technologies, Coralville, IA). The thermocycling parameters for both primer pairs consisted of denaturation at 95°C for 5 min followed by 40 cycles of denaturation, annealing, and extension at 95°C for 15 s, 56°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 1 min. The amplified product was verified using melting curve analysis from 50°C to 95°C with a transition rate of 0.2°C/s. At each time point, one-way analysis of variance (ANOVA) with Tukey's post hoc test was used to find significant differences across treatment and control group means. Differences with P values of <0.05 were considered significant.
Gentamicin protection assay.
Caco-2 cells were infected with bacteria at an MOI of 1:1,000 in a 96-well plate, in three biological replicates. The infected cells were incubated for 60 min at 37°C with 5% CO2. Medium was aspirated, and cells were washed three times with 200 μl standard Tyrode's buffer. To enumerate invading bacteria, cells were incubated with 200 μl of 100 μg/ml gentamicin for 2 h at 37°C with 5% CO2. To enumerate total cell-associated bacteria, cells were incubated with DMEM-HMM (without gentamicin). Cells were washed three times with 200 μl Tyrode's buffer and lysed with 100 μl of 0.01% Triton. The total cell-associated or invading bacteria were enumerated by the pour plate method using nutrient agar (Difco, Detroit, MI). To calculate the number of adherent bacteria, the mean number of invading bacteria was subtracted from the mean of the total number of cell-associated bacteria. The error for adherent bacteria was calculated using the equation (ΔZ)2 = (ΔA)2 + (ΔB)2, where ΔZ is the standard error of mean (SEM) for adherent bacteria, ΔA is the SEM for total host-associated bacteria, and ΔB is the SEM for invading bacteria. An independent two-sample t test was used to find significant differences across treatment and control group means (see Fig. 1B). When appropriate, one-way ANOVA with Tukey's post hoc test was done to find significant differences across treatment and control group means (see Fig. 6 and 7). Differences with P values of <0.05 were considered significant.
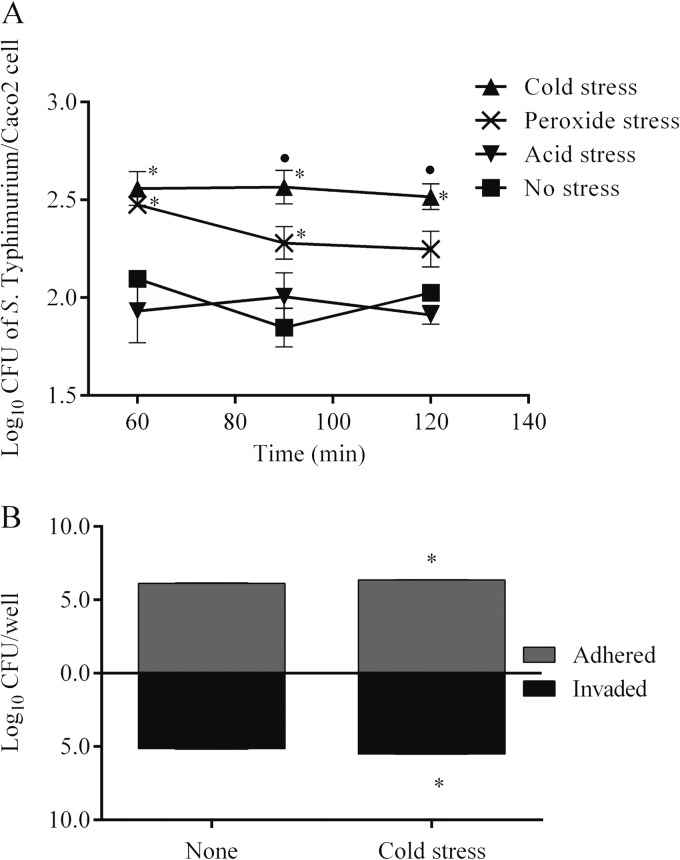
FIG 1.
Association of S. Typhimurium with Caco-2 cells. (A) Total association of cold-, peroxide- and acid-stressed S. Typhimurium with Caco-2 cells measured at 60 min, 90 min, and 120 min postinfection. (B) Adhesion to and invasion of Caco-2 cells by cold-stressed S. Typhimurium measured at 60 min postinfection. Results are means ± SEM (standard errors of the means) for three biological replicates. Asterisks and dots indicate significant differences (P < 0.05) compared to the no-stress control and peroxide-stressed bacteria, respectively.
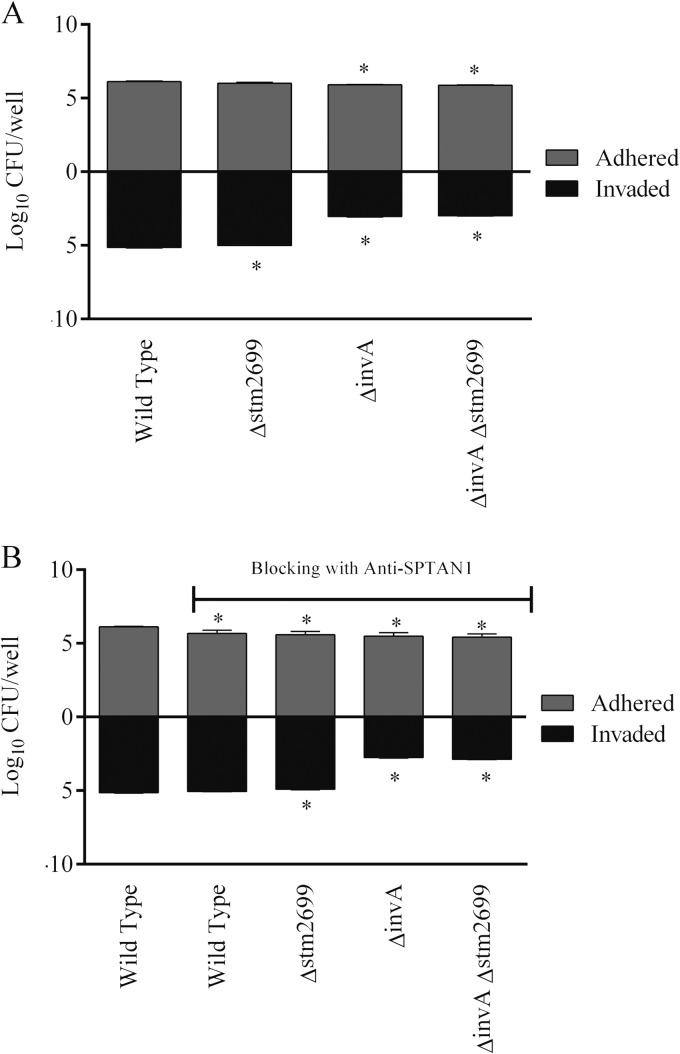
FIG 6.
Role of STM2699 and SPTAN1 in adhesion and invasion of epithelial cells with nonstressed S. Typhimurium. (A) Role of STM2699 in S. Typhimurium adhesion to and invasion of epithelial cells. (B) Role of STM2699 and SPTAN1 interaction in S. Typhimurium adhesion to and invasion of epithelial cells pretreated with anti-SPTAN1 antibodies. Asterisks indicate significant differences (P < 0.05) compared to the first bar, which represents adhesion and invasion of wild-type S. Typhimurium.
FIG 7.
Role of STM2699 and SPTAN1 in adhesion and invasion of epithelial cells with cold-stressed (5°C, 48 h) S. Typhimurium. (A) Role of STM2699 in adhesion and invasion of cold-stressed S. Typhimurium in epithelial cells. (B) Role of STM2699 and SPTAN1 interaction in adhesion and invasion of cold-stressed S. Typhimurium in epithelial cells pretreated with anti-SPTAN1 antibodies. Asterisks indicate significant differences (P < 0.05) compared to the first bar, representing adhesion and invasion of wild type S. Typhimurium. The dot indicates a significant difference (P < 0.05) compared to the bar representing adhesion and invasion of S. Typhimurium ΔinvA.
Epithelial cell infection assay for determination of gene expression.
Caco-2 cells were cultured in T-75 flasks and were serum starved 24 h before infection. Bacteria (S. Typhimurium that had been cold stressed for 48 h and nonstressed S. Typhimurium), at an MOI of 1:1,000, were used to infect epithelial cells. The controls included cold-stressed and nonstressed S. Typhimurium in DMEM-HMM only (no epithelial cells) and noninfected epithelial cells. The experiment was done in two biological replicates. The infected cells along with the controls were incubated at 37°C with 5% CO2 for 60 min. For infected cells, medium with extracellular bacteria was aspirated after 60 min of infection, and 10 ml of TRIzol LS reagent (Invitrogen, Carlsbad, CA) was added to lyse the cells. Subsequently, the lysate was centrifuged at 7,200 × g for 5 min to pellet the cell-associated bacteria. TRIzol LS supernatant was stored in a clean tube and further processed for extraction of total RNA released from infected Caco-2 cells. The cell-associated bacterial pellet was resuspended in 2 ml of fresh TRIzol LS, gently mixed, and processed for extraction of total RNA. For controls, medium with bacteria was collected and centrifuged at 7,200 × g for 5 min, and 10 ml of TRIzol LS reagent was added, gently mixed, and processed for extraction of total RNA. For the noninfected Caco-2 cells, medium was aspirated, and 10 ml of TRIzol LS reagent was added to the cells, was gently mixed, and processed for extraction of total RNA.
Bacterial RNA extraction, hybridization, and normalization.
Bacterial RNA was prepared from TRIzol LS reagent, and cDNA was generated following procedures described previously (11). Prior to hybridization, the cDNA samples were denatured at 98°C for 10 min followed by snap cooling at 4°C for 5 min. Labeled cDNA was hybridized onto a custom-designed Affymetrix GeneChip covering all the annotated coding sequences of S. Typhimurium LT2 ATCC 700720 (11). Labeled cDNA obtained from pure culture of S. Typhimurium (500 ng) and coculture of S. Typhimurium and Caco-2 cells (2,000 ng) was hybridized on the chips and scanned at the Center for Integrated Biosystems (Utah State University, Logan, UT) following the manufacturer's protocols. Raw data (.cel files) were background corrected, quantile normalized, and summarized using MS-RMA (12).The resultant normalized log2-transformed intensity matrix was used for further statistical analysis.
Caco-2 RNA extraction, hybridization, and normalization.
The TRIzol LS samples (750 μl) containing infected or noninfected Caco-2 cells were frozen (liquid N2) and thawed (70°C) twice followed by addition of 250 μl of water. RNA was extracted using TRIzol LS following manufacturer's instructions. RNA concentration, A260/280 and A260/230 were measured on NanoDrop (Thermo scientific, Waltham, MA) and analyzed for integrity on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Synthesis of cDNA, biotin labeling of cRNA, and fragmentation and purification of cRNA were carried out using one-cycle cDNA synthesis kit (Affymetrix, Santa Clara, CA). Labeled and fragmented cRNA (10 μg) was hybridized onto the Affymetrix U133Plus2 GeneChip as per the manufacturer's recommendations at the Center for Integrated Biosystems (Utah State University, Logan, UT). Raw data (.cel files) were background corrected, quantile normalized, and summarized using RMA (13). RMA-normalized data were then filtered through the PANP algorithm (P. Warren, D. Taylor, P. G. V. Martini, J. Jackson, and J. Bienkowska, presented at the 7th IEEE International Conference on Bioinformatics and Bioengineering, 2007) to make presence-absence calls for each probe set. Probe sets that were called present in at least one of the samples were included for further statistical analysis.
Antibody blocking assay.
The aim of the antibody blocking assay was to determine the role of alpha spectrin in bacterial association. Anti-SPTAN1 antibodies (1:2,000; Novus Biologicals, Littleton, CO) were added to the Caco-2 cells to block exposed alpha spectrin on cell surface. Cells were incubated for 60 min at 37°C with 5% CO2 before bacterial treatments at an MOI of 1:100 were added. The experiment was performed in two biological replicates. Invasion assays were performed as described earlier. One-way ANOVA with Tukey's post hoc test was used to find significant differences across treatment and control group means. The differences with P values of <0.05 were considered significant.
Cell-based caspase assay.
The aim of the cell-based caspase assay was to measure caspase activation upon bacterial infection. Caco-2 cells were washed with phosphate-buffered saline (PBS) before use. Upon addition of bacteria at an MOI of 1:1,000, in three biological replicates, cells were incubated at 37°C with 5% CO2 for 4 h, 6 h, and 8 h. At different time points, caspase activity was measured using Caspase-Glo 8, 9, and 3/7 assay kits (Promega, Madison, WI) following the manufacturer's instructions. Bioluminescence was measured in a DTX 880 multimode-detector plate reader (Beckman Coulter, Brea, CA). Two-way ANOVA with Bonferroni post hoc tests was used to find significant differences between treatment and control group means at different time points. The differences with P values of <0.05 were considered significant.
Statistical analysis for gene expression.
Gene expression profiles for cold-stressed (5°C, 48 h) and nonstressed S. Typhimurium alone and in the presence of epithelial cells were obtained 60 min postinfection. The data were analyzed as two-class unpaired data (cold stress versus no stress), with the T statistic, using significance analysis of microarrays (SAM) (14). All the genes were ranked based on the score (d) from SAM output. This preordered ranked gene list was then used in Gene Set Enrichment Analysis software (GSEA) (15) to detect the coordinate changes in the expression of groups of functionally related genes upon respective treatments. The gene sets with q values of <0.05 were considered significant. The gene sets were defined based on the annotations from the Comprehensive Microbial Resource (CMR) (16), clusters of orthologous groups of proteins (COGs) (17) and the Virulence Factors of Pathogenic Bacteria database (VFDB) (1).
Gene expression profiles were obtained for epithelial cells alone and upon infection with cold-stressed (5°C, 48 h) and nonstressed S. Typhimurium, 60 min postinfection. The data were analyzed as two-class unpaired data (infection with cold-stressed S. Typhimurium versus infection with nonstressed S. Typhimurium), with the T statistic, using SAM (14). All the genes were ranked based on the score (d) from SAM output. This preordered ranked gene list was then used in GSEA (15) to detect the coordinate changes in the expression of groups of functionally related genes upon infection with cold-stressed S. Typhimurium. The gene sets with q values of <0.20 were considered significant. The gene sets were defined based on the annotations in GO gene sets from the Molecular Signatures database (MSigDB) (15).
RESULTS AND DISCUSSION
Effect of stress treatments on bacterial association to epithelial cells.
Among the stresses tested, cold stress of S. Typhimurium significantly (P < 0.05) increased its association with Caco-2 cells from 60 min postinfection (p.i.) until the end of the experiment (i.e., 120 min) compared with the nonstressed control and the peroxide-stressed S. Typhimurium (Fig. 1A). Because the numbers of cold-stressed S. Typhimurium associated with cells remained significantly higher and stable through 120 min, the invasion was measured only at 60 min p.i. Interestingly, the numbers of cold-stressed S. Typhimurium bacteria that invaded Caco-2 cells were significantly (P < 0.05) higher than the numbers of nonstressed S. Typhimurium bacteria (Fig. 1B), suggesting that prior exposure to cold stress significantly increased both adhesion and invasion of S. Typhimurium in epithelial cells. Consequently, for all other follow-up experiments, we focused on cold-stressed Salmonella.
Gene expression profile of cold-stressed S. Typhimurium.
Cold stress of S. Typhimurium modulated expression of several virulence-associated genes and genes associated with protein secretion and trafficking, DNA metabolism and repair, and degradation of RNA (Table 1). Overall, the alteration in global gene expression in cold-stressed S. Typhimurium suggested that both virulence and bacterial metabolism may play a role in pathogenesis. However, in this study our primary focus was on the virulence-associated genes and their potential link to the observed phenotype of increased association of cold-stressed S. Typhimurium to intestinal epithelial cells. To better understand the gene expression profiles and for visualization, the differentially expressed genes from virulence-associated gene categories were further divided into three major functional groups, namely, T3SS-associated genes, plasmid-associated genes, and prophage-associated genes (Fig. 2).
TABLE 1.
Gene categories in S. Typhimurium that were significantly (q < 0.05) regulated due to cold stress (5°C, 48 h)
| Enriched gene categorya | No. of genes regulated | No. of genes in category | Regulation | FDR (q)b |
|---|---|---|---|---|
| SP2-TTSS | 26 | 27 | Induced | 0 |
| TTSS | 27 | 58 | Induced | 0 |
| TTSS effectors | 10 | 22 | Induced | 0 |
| TTSS2 effectors | 9 | 13 | Induced | 0 |
| Cellular processes—pathogenesis | 8 | 32 | Induced | 0.03 |
| Prophage functions | 70 | 143 | Induced | 0.04 |
| Plasmid functions | 26 | 28 | Induced | 0 |
| Protein and peptide secretion and trafficking | 21 | 54 | Induced | 0 |
| Cellular processes—DNA transformation | 28 | 30 | Induced | 0 |
| Purine ribonucleotide biosynthesis | 9 | 18 | Induced | 0.01 |
| CogL—replication recombination and repair | 79 | 157 | Induced | 0.01 |
| Transcription—degradation of RNA | 9 | 15 | Induced | 0.05 |
TTSS, type III secretion system.
FDR, false discovery rate.
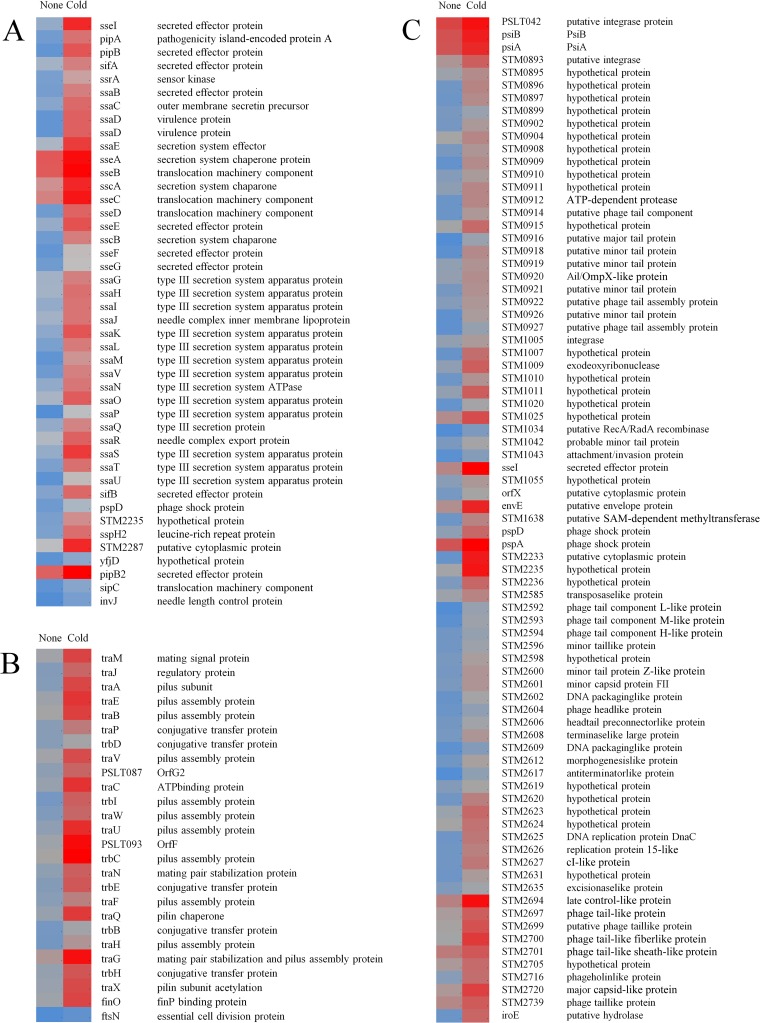
FIG 2.
Heatmap representation of differentially regulated (q < 0.05) genes in S. Typhimurium due to exposure to cold stress (5°C, 48 h). (A) T3SS-associated genes. (B) Plasmid (pSLT)-associated genes. (C) Prophage-associated genes. The blue-gray-red color scale represents low expression intensity (2.4) to high expression intensity (13.5).
(i) T3SS-associated genes.
Cold stress induced expression of several T3SS-associated genes on various SPIs. The role of these genes is to allow bacteria to remain docked at the epithelial cell membrane in order to deliver effectors to the host cell cytosol (18). These genes are also required for later stages of infection and intracellular replication (19). The two-component system ssrAB controls the expression of genes encoding the components of the T3SS and its effectors located within and outside the SPI2 region (20), including ssrA, whose expression was induced 1.8-fold. The genes encoding effectors mainly from the ssa and sse operons, along with sscA, sscB, sifA, sifB, pipA, pipB, and pipB2 from SPI2 and SPI5, were induced. The SPI1 genes invJ, which is responsible for translocation of the effectors (19), and sipC, which is involved in actin bundling (21), were also induced (Fig. 2B).
(ii) Plasmid-associated genes.
Genes of the tra and trb clusters forming a subset of type 4 secretion system (T4SS) (22) were induced (Fig. 2C). These plasmid-borne genes encode the elements of pili required for mating and conjugal DNA transfer. Transcription of the tra operon is activated by TraJ, which is positively regulated by FinO (23), and induction of both traJ and finO was observed (Fig. 2C).
(iii) Prophage-associated genes.
The genes on prophages Fels-1 (22/35), Gifsy-2 (12/52), Gifsy-1 (24/53), and Fels-2 (9/47) were induced (Fig. 2A). The prophages are often identified adjacent to virulence genes and tRNA genes (24) and are known to contribute to pathogenicity, insertion of transferable elements, and lateral spreading of the pathogenicity determinants. The roles of Gifsy-2 and Gifsy-1 prophages have been studied (25), but there is no direct evidence of a direct contribution of Fels-1 and Fels-2 to virulence other than the roles of sodC, nanH, and grvA from Fels-1 (26) and a suspected role of abiU from Fels-2 (26, 27). Other phage genes, such as pspA and pspD, were also induced in response to cold stress (Fig. 2A).
Gene expression profile of cold-stressed S. Typhimurium during infection of epithelial cells.
Infection of epithelial cells with cold-stressed S. Typhimurium resulted in induction of prophage genes, plasmid-associated genes (including the spv operon), stress response and DNA transformation genes, and genes associated with energy metabolism, protein synthesis, polysaccharide metabolism, and chemotaxis (Table 2). In order to determine the link between the gene expression and the observed phenotype of increased epithelial cell adhesion and invasion, we again focused on the virulence-associated gene categories, and the genes from these categories were divided into four major functional groups for visualization, namely, plasmid-associated genes, prophage-associated genes, DNA transposition-associated genes, and spv and other stress response-associated genes.
TABLE 2.
Gene categories in cold-stressed (5°C, 48 h) S. Typhimurium that were significantly (q < 0.05) regulated during infection of epithelial cells
| Enriched gene categorya | No. of genes regulated | No. of genes in category | Regulation | FDR (q)b |
|---|---|---|---|---|
| Prophage functions | 63 | 143 | Induced | 0 |
| Plasmid functions | 26 | 28 | Induced | 0 |
| Transposon functions | 27 | 44 | Induced | 0 |
| SPV locus | 4 | 4 | Induced | 0.04 |
| Adaptations to atypical conditions | 10 | 30 | Induced | 0.04 |
| Cellular processes—DNA transformation | 30 | 30 | Induced | 0 |
| Energy metabolism—anaerobic | 32 | 66 | Induced | 0.04 |
| Synthesis and modification ribosomal proteins | 37 | 61 | Repressed | 0 |
| CogJ—translation | 67 | 170 | Repressed | 0 |
| Cellular processes—chemotaxis and motility | 20 | 40 | Repressed | 0 |
| Energy metabolism—pyruvate dehydrogenase | 3 | 8 | Repressed | 0.02 |
| Metabolism of surface polysaccharides and LPS | 39 | 82 | Repressed | 0.04 |
| Energy metabolism—aerobic | 12 | 24 | Repressed | 0.04 |
SPV, Salmonella plasmid virulence locus.
FDR, false discovery rate.
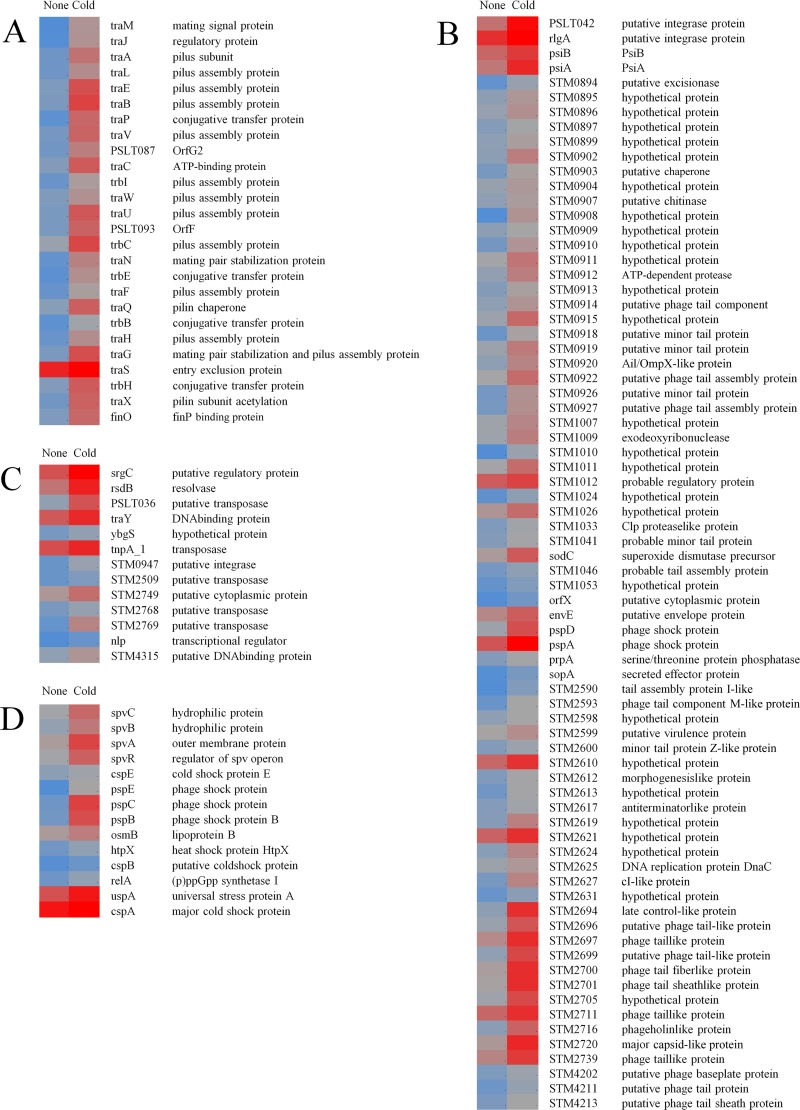
(i) Plasmid-associated genes.
Genes within the tra and trb clusters, forming a subset of T4SS, were induced in response to the cold stress alone and remained induced during the infection. Additionally, traL, which is required for pilus tip formation on cell surfaces (28), and traS, which is involved in entry exclusion during mating pair stabilization (22), were also induced during infection (Fig. 3D).
FIG 3.
Heatmap representation of genes differentially regulated (q < 0.05) in cold-stressed (5°C, 48 h) S. Typhimurium during infection of epithelial cells. (A) Plasmid (pSLT)-associated genes. (B) Prophage-associated genes. (C) DNA transposition-associated genes. (D) spv and other stress response-associated genes. The blue-gray-red color scale represents low expression intensity (2.1) to high expression intensity (11.2).
(ii) Prophage-associated genes.
Several genes located on the prophages Fels-1 (23/35), Gifsy-2 (12/52), Gifsy-1 (15/53), and Fels-2 (11/47) also remained induced during infection (Fig. 3A). Additionally, other phage-associated genes, such as pspABCDE, were induced in cold-stressed S. Typhimurium during infection (Fig. 3A). It is important to note that the psp genes are highly conserved in some pathogens and are involved in infection processes.
(iii) DNA transposition-associated genes.
Genes encoding resolvase, transposase, and integrase, on the chromosome as well as on the plasmid, and involved in DNA transposition were induced during infection (Fig. 3C).
(iv) Stress response and spv operon genes.
Other genes that were induced during infection included the plasmid-associated transcriptional regulator spvR and spvABC, which play a role in intracellular proliferation of Salmonella (29), and stress response-associated genes, such as cspABE, uspA, relA, htpX, and osmB (Fig. 3B).
Additionally, the genes associated with flagellar functions from the fli, flh, and flg operons were found to be repressed in cold-stressed S. Typhimurium during infection. During the infection, flagellar proteins are recognized by Toll-like receptor 5 (TLR5), NLR, and Ipaf on the host cell, leading to induction of the proinflammatory cascade, caspase 1-dependent cell death, and the T-cell-mediated immune response (30). However, downregulation of these genes is considered a means by which Salmonella evades its detection (31). Lipopolysaccharide (LPS), a component of the outer membrane of Salmonella, has been shown to interact with TLR4 (32) and initiate a proinflammatory cascade. The genes encoding surface polysaccharides, including LPS, were also repressed.
Gene expression of epithelial cells in response to infection with cold-stressed S. Typhimurium.
Infection of epithelial cells with cold-stressed S. Typhimurium regulated 18 gene categories related to mitochondrial function along with other gene categories related to ribosomal genes, ribonucleoprotein complex, kinesin complex, RNA splicing, and coenzyme metabolism (Table 3). All the genes regulated in the categories functionally associated with mitochondria were overlaid on the electron transport chain (ETC) for visualization using ingenuity pathway analysis software (IPA; Ingenuity Systems, Redwood City, CA). The genes encoding the subunits of the four complexes of ETC, complex I (NADH dehydrogenase/reductase), complex II (succinate dehydrogenase), complex III (cytochrome bc1 complex), and complex IV (cytochrome c oxidase) were induced. Additionally genes encoding glycerol-3-phosphate dehydrogenase 2 (GPD2) and monoamine oxidase A (MAOA), were also induced. Although ETC is a very efficient system, it is a major site to produce superoxide radicals and reactive oxygen species because there is a high probability of electrons being passed to oxygen directly, instead of the next electron carrier in the chain. An appropriate balance of oxidants and antioxidants is required for cell survival; even a slight perturbation can lead to damage of biological macromolecules and hence cell death. In functionally intact mitochondria, a large number of antioxidant gene products are needed to neutralize the effects of superoxide radicals and reactive oxygen species (33). Consequently, the expression intensities of the components of the antioxidant defense system (33) were further examined. No significant changes in gene expression were observed for any of the genes except SOD2 (superoxide dismutase 2), CAT (catalase), GPX7 (glutathione peroxidase 7), and PRX3 (peroxiredoxin 3), and these genes were induced 1.2-, 1.2-, 1.3-, and 1.5-fold, respectively.
TABLE 3.
Gene categories in epithelial cells that were significantly (q < 0.20) regulated during infection with cold-stressed (5°C, 48 h) S. Typhimurium
| Enriched gene category | No. of genes regulated | No. of genes in category | Regulation | FDR (q)a |
|---|---|---|---|---|
| Ribosome | 18 | 37 | Induced | 0.01 |
| Ribosomal subunit | 11 | 20 | Induced | 0 |
| Organellar ribosome | 12 | 22 | Induced | 0 |
| Mitochondrial ribosome | 12 | 22 | Induced | 0 |
| Mitochondrial small ribosomal subunit | 9 | 11 | Induced | 0 |
| Organellar small ribosomal subunit | 9 | 11 | Induced | 0.01 |
| Small ribosomal subunit | 9 | 11 | Induced | 0.01 |
| Mitochondrial part | 47 | 128 | Induced | 0.01 |
| Mitochondrial envelope | 28 | 85 | Induced | 0.03 |
| Mitochondrial membrane | 26 | 77 | Induced | 0.03 |
| Mitochondrial inner membrane | 24 | 60 | Induced | 0.01 |
| Mitochondrial membrane part | 20 | 50 | Induced | 0 |
| Mitochondrial lumen | 18 | 44 | Induced | 0.03 |
| Mitochondrial matrix | 18 | 44 | Induced | 0.03 |
| Cellular respiration | 9 | 18 | Induced | 0.03 |
| NADH dehydrogenase complex | 7 | 14 | Induced | 0.03 |
| Mitochondrial respiratory chain complex I | 7 | 14 | Induced | 0.03 |
| Respiratory chain complex I | 7 | 14 | Induced | 0.03 |
| Structural constituent of ribosome | 43 | 73 | Induced | 0.00 |
| Ribosome biogenesis and assembly | 6 | 12 | Induced | 0.04 |
| Ribonucleoprotein complex | 41 | 111 | Induced | 0.01 |
| Microtubule motor activity | 9 | 14 | Induced | 0.01 |
| Kinesin complex | 8 | 11 | Induced | 0.03 |
| RNA splicing via transesterification reactions | 10 | 30 | Induced | 0.03 |
| Coenzyme metabolic process | 12 | 31 | Induced | 0.04 |
FDR, false discovery rate.
The other genes induced in epithelial cells during infection with cold-stressed S. Typhimurium were related to microtubule activity and the kinesin complex. The genes located on SPI2 of S. Typhimurium cause accumulation of microtubules around the Salmonella-containing vacuole (SCV) (34), recruitment of kinesin and dynein to regulate vacuolar membrane dynamics (35), and interference with the activity of ubiquitination pathway (36). This, together with the increased adhesion and invasion of cold-stressed Salmonella, explains the induction of genes related to microtubule activity and the kinesin complex.
These observations suggest induction of oxidative stress in epithelial cells infected with cold-stressed S. Typhimurium compared with the infection with nonstressed S. Typhimurium. Oxidative stress has been shown to cause damage leading to apoptosis via caspase activation (37, 38), and hence, activation of caspases 8, 9, and 3/7 was measured.
Activation of caspases.
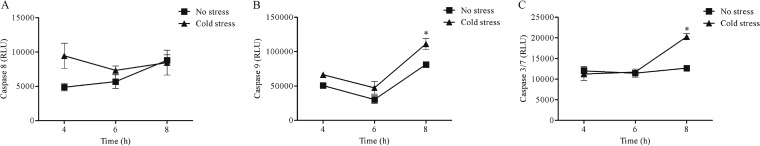
Caspases are intracellular cysteine-containing, aspartic acid-specific proteases implicated in programmed cell death. Caspases are divided into the classifications “initiators” and “executioners” (39, 40). The activation of the executioner caspase, i.e., caspase 3/7, is a committed step in apoptosis and can occur via the extrinsic and/or intrinsic pathway involving the initiator caspase 8 and/or caspase 9, respectively (39, 40). The activation of caspase 3/7 was significantly increased at 8 h p.i. with cold-stressed S. Typhimurium compared to infection with nonstressed S. Typhimurium (Fig. 4C). The activation of caspase 8 did not significantly change upon infection with cold-stressed S. Typhimurium compared to nonstressed S. Typhimurium, at any time points tested during infection (Fig. 4A). On the other hand, the activation of caspase 9 was significantly (P < 0.05) higher at 8 h p.i. with cold-stressed S. Typhimurium compared to infection with nonstressed S. Typhimurium (Fig. 4B). Moreover, activity of caspase 9 increased with time, implying the contribution of caspase 9 in activation of caspase 3/7 via the intrinsic pathway. To summarize, significant activation of caspase 9 and caspase 3/7 was concomitant with the gene regulation observations, suggesting induced apoptosis by the intrinsic (mitochondrial) route, in epithelial cells infected with cold-stressed S. Typhimurium.
FIG 4.
Measurement of caspase 8 (A), caspase 9 (B), and caspase 3/7 (C) activity at 4 h, 6 h, and 8 h postinfection of epithelial cells with nonstressed and cold-stressed (5°C, 48 h) S. Typhimurium. Asterisks indicate significant differences (P < 0.05) between infection with cold-stressed S. Typhimurium and infection with nonstressed S. Typhimurium.
Receptors involved in association of S. Typhimurium with epithelial cells.
Pathogen-host cell association is a function of the regulation of multiple receptor-ligand interactions. A study conducted using a whole-cell cross-linking method revealed a product of STM2699 (a Fels-2 prophage gene) from S. Typhimurium cross-linked to a receptor, SPTAN1 (spectrin), expressed on Caco-2 cells (41). Interestingly, in this study, the expression of STM2699 was induced in response to cold stress alone, and it remained induced during the infection of Caco-2 cells (Fig. 5). As expected, the expression of SPTAN1 in the Caco-2 cells was also induced upon infection with cold-stressed S. Typhimurium (Fig. 5). Consequently, the contribution of STM2699 and SPTAN1 interaction in adhesion and invasion during infection of Caco-2 cells with nonstressed as well as cold-stressed S. Typhimurium was investigated. The gene knockout for STM2699 was created in the wild-type parent as well as the invasion-deficient S. Typhimurium (ΔinvA) strain (42).
FIG 5.
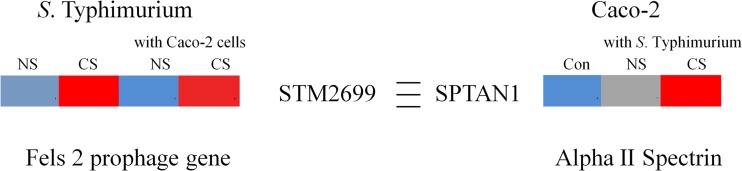
(Left) Gene expression intensities of the S. Typhimurium gene STM2699 in response to no stress and cold stress (5°C, 48 h) alone and during infection of epithelial cells with nonstressed and cold-stressed (5°C, 48 h) S. Typhimurium. (Right) Gene expression intensities of the epithelial cell gene SPTAN1 in epithelial cells alone and during infection with nonstressed and cold-stressed (5°C, 48 h) S. Typhimurium. NS, no stress; CS, cold stress. The blue-gray-red color scale represents low expression intensity (3.7) to high expression intensity (7.5).
Adherence of nonstressed S. Typhimurium ΔSTM2699 to the Caco-2 cells was similar to that of the wild-type strain; however, significantly low numbers of this mutant strain invaded cells (Fig. 6A). Nonstressed S. Typhimurium ΔSTM2699-ΔinvA did not show any further reduction in adhesion or invasion compared with S. Typhimurium ΔinvA (Fig. 6A). These results revealed contribution of STM2699 specifically in cell invasion by nonstressed S. Typhimurium during infection of Caco-2 cells. Pretreatment of Caco-2 cells with anti-SPTANI antibodies showed that the nonstressed wild-type S. Typhimurium adhered in significantly low numbers to the Caco-2 cells, but no alterations in invasion were noted (Fig. 6B). In contrast, S. Typhimurium ΔSTM2699 infection of Caco-2 cells pretreated with anti-SPTANI antibodies resulted in significant reduction in the invasion. Prior treatment of Caco-2 cells with anti-SPTANI antibodies did not show any further reduction in invasiveness of S. Typhimurium ΔSTM2699 ΔinvA compared with the S. Typhimurium ΔinvA strain (Fig. 6B). These results revealed that STM2699-SPTAN1 interaction plays an important role in the invasion of nonstressed S. Typhimurium during infection of Caco-2 cells.
We then tested effect of prior exposure of S. Typhimurium to cold stress on this interaction. Infection of Caco-2 cells with cold-stressed S. Typhimurium ΔSTM2699 reduced adhesion as well as invasion (Fig. 7A). Infection with cold-stressed S. Typhimurium ΔSTM2699 ΔinvA additionally reduced adhesion compared with cold-stressed S. Typhimurium ΔinvA (Fig. 7A). These results revealed the contribution of STM2699 to adhesion and invasion of cold-stressed S. Typhimurium during infection of Caco-2 cells. Pretreatment of Caco-2 cells with anti-SPTANI antibodies did not alter adherence and invasion of cold-stressed wild-type S. Typhimurium; however, infection with cold-stressed S. Typhimurium ΔSTM2699 showed significantly reduced adhesion and invasion (Fig. 7B). Infection with cold-stressed S. Typhimurium ΔSTM2699 ΔinvA of Caco-2 cells pretreated with anti-SPTANI antibodies additionally reduced adhesion compared to infection with cold-stressed S. Typhimurium ΔinvA (Fig. 7B). These results revealed that STM2699-SPTAN1 interaction plays an important role in both adhesion of and invasion by cold-stressed S. Typhimurium during infection of Caco-2 cells.
Concluding remarks.
In this study, we found that prior exposure of S. Typhimurium to cold stress (5°C, 48 h) significantly increased adhesion and invasion in Caco-2 cells compared to nonstressed S. Typhimurium. The gene expression data further revealed significant induction of virulence-associated genes which remained induced during infection of the host cells, indicating increased pathogenicity of cold-stressed S. Typhimurium. Simultaneous gene expression profiling of Caco-2-cells indicated induction of mitochondrial dysfunction, induction of oxidative stress response, and imbalance of oxidants and antioxidants upon infection with cold-stressed S. Typhimurium. Induced damage of epithelial cells through intrinsic (mitochondrial) pathway was confirmed by induction of caspase 9 and caspase 3/7 activity during infection with cold-stressed S. Typhimurium. Furthermore, we found that the STM2699-SPTAN1 (protein-protein) interaction increased the adhesion and invasion during the infection of Caco-2 cells with cold-stressed S. Typhimurium. These observations together indicate that exposure to cold stress (5°C, 48 h) may potentially increase the pathogenicity of S. Typhimurium. We showed previously that preadaptation to cold stress increases the survival during subsequent acid stress exposure, which mimics the conditions of the gastric transit (11). Follow-up studies to measure the transcriptional response and host-cell interaction of S. Typhimurium upon exposure to cold stress followed by acid stress will provide indispensable targets needed at each step for successful infection.
ACKNOWLEDGMENT
Funding for this project was provided by USDA CSREES grant 2006-34526-433 17001 to B.C.W.
Footnotes
Published ahead of print 5 September 2014
Present address: Jigna Shah, Veterinary Biomedical Sciences, University of Minnesota, Saint Paul, Minnesota, USA; Prerak T. Desai, Department of Pathology and Laboratory, University of California, Irvine, California, USA.
REFERENCES
- 1.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33:D325–D328. 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patti JM, Allen BL, McGavin MJ, Hook M. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585–617. 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 3.Olsen A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655. 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 4.Sjobring U, Pohl G, Olsen A. 1994. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA). Mol. Microbiol. 14:443–452. 10.1111/j.1365-2958.1994.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 5.Gerlach RG, Jäckel D, Stecher B, Wagner C, Lupas A, Hardt WD, Hensel M. 2007. Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell. Microbiol. 9:1834–1850. 10.1111/j.1462-5822.2007.00919.x. [DOI] [PubMed] [Google Scholar]
- 6.Galan JE. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263–271. 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 7.Marcus SL, Brumell JH, Pfeifer CG, Finlay BB. 2000. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2:145–156. 10.1016/S1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai PT, Walsh MK, Weimer BC. 2008. Solid-phase capture of pathogenic bacteria by using gangliosides and detection with real-time PCR. Appl. Environ. Microbiol. 74:2254–2258. 10.1128/AEM.02601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziemer C, Steadham S. 2003. Evaluation of the specificity of Salmonella PCR primers using various intestinal bacterial species. Lett. Appl. Microbiol. 37:463–469. 10.1046/j.1472-765X.2003.01430.x. [DOI] [PubMed] [Google Scholar]
- 11.Shah J, Desai PT, Chen D, Stevens JR, Weimer BC. 2013. Preadaptation to cold stress in Salmonella enterica serovar Typhimurium increases survival during subsequent acid stress exposure. Appl. Environ. Microbiol. 79:7281–7289. 10.1128/AEM.02621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens JR, Ganesan B, Desai P, Rajan S, Weimer BC. 2008. Statistical issues in the normalization of multi-species microarray data, p 47–62 In Proceedings of Conference on Applied Statistics in Agriculture. [Google Scholar]
- 13.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121. 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. 2001. The Comprehensive Microbial Resource. Nucleic Acids Res. 29:123–125. 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 18.Cornelis GR. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811–825. 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 19.Hansen-Wester I, Hensel M. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549–559. 10.1016/S1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- 20.Garmendia J, Beuzon CR, Ruiz-Albert J, Holden DW. 2003. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 149:2385–2396. 10.1099/mic.0.26397-0. [DOI] [PubMed] [Google Scholar]
- 21.Hayward RD, Koronakis V. 1999. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 18:4926–4934. 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1–15. 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 23.Camacho EM, Casadesus J. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44:1589–1598. 10.1046/j.1365-2958.2002.02981.x. [DOI] [PubMed] [Google Scholar]
- 24.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 25.Figueroa-Bossi N, Bossi L. 1999. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33:167–176. 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- 26.Brussow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602. 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyd EF, Brussow H. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521–529. 10.1016/S0966-842X(02)02459-9. [DOI] [PubMed] [Google Scholar]
- 28.Anthony KG, Klimke WA, Manchak J, Frost LS. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J. Bacteriol. 181:5149–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulig PA, Danbara H, Guiney DG, Lax AJ, Norel F, Rhen M. 1993. Molecular analysis of spv virulence genes of the salmonella virulence plasmids. Mol. Microbiol. 7:825–830. 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 30.Miao EA, Andersen-Nissen E, Warren SE, Aderem A. 2007. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 29:275–288. 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- 31.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. 2006. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol. Microbiol. 61:795–809. 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 32.Du X, Poltorak A, Silva M, Beutler B. 1999. Analysis of Tlr4-mediated LPS signal transduction in macrophages by mutational modification of the receptor. Blood Cells Mol. Dis. 25:328–338. 10.1006/bcmd.1999.0262. [DOI] [PubMed] [Google Scholar]
- 33.Lin MT, Beal MF. 2006. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–795. 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 34.Kuhle V, Jackel D, Hensel M. 2004. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic 5:356–370. 10.1111/j.1398-9219.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 35.Guignot J, Caron E, Beuzon C, Bucci C, Kagan J, Roy C, Holden DW. 2004. Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J. Cell Sci. 117:1033–1045. 10.1242/jcs.00949. [DOI] [PubMed] [Google Scholar]
- 36.Rytkonen A, Poh J, Garmendia J, Boyle C, Thompson A, Liu M, Freemont P, Hinton JC, Holden DW. 2007. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl. Acad. Sci. U. S. A. 104:3502–3507. 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hampton MB, Orrenius S. 1997. Dual regulation of caspase activity by hydrogen peroxide: implications for apoptosis. FEBS Lett. 414:552–556. 10.1016/S0014-5793(97)01068-5. [DOI] [PubMed] [Google Scholar]
- 38.Zamzami N, Marchetti P, Castedo M, Decaudin D, Macho A, Hirsch T, Susin SA, Petit PX, Mignotte B, Kroemer G. 1995. Sequential reduction of mitochondrial transmembrane potential and generation of reactive oxygen species in early programmed cell death. J. Exp. Med. 182:367–377. 10.1084/jem.182.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandra J, Samali A, Orrenius S. 2000. Triggering and modulation of apoptosis by oxidative stress. Free Radic. Biol. Med. 29:323–333. 10.1016/S0891-5849(00)00302-6. [DOI] [PubMed] [Google Scholar]
- 40.Pop C, Salvesen GS. 2009. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 284:21777–21781. 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desai PT. 2011. Molecular Interactions of Salmonella with the host epithelium in presence of commensals. Utah State University, Logan, UT. [Google Scholar]
- 42.Galan JE, Curtiss R., III 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 86:6383–6387. 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]