Abstract
Tellurium, a metalloid belonging to group 16 of the periodic table, displays very interesting physical and chemical properties and lately has attracted significant attention for its use in nanotechnology. In this context, the use of microorganisms for synthesizing nanostructures emerges as an eco-friendly and exciting approach compared to their chemical synthesis. To generate Te-containing nanostructures, bacteria enzymatically reduce tellurite to elemental tellurium. In this work, using a classic biochemical approach, we looked for a novel tellurite reductase from the Antarctic bacterium Pseudomonas sp. strain BNF22 and used it to generate tellurium-containing nanostructures. A new tellurite reductase was identified as glutathione reductase, which was subsequently overproduced in Escherichia coli. The characterization of this enzyme showed that it is an NADPH-dependent tellurite reductase, with optimum reducing activity at 30°C and pH 9.0. Finally, the enzyme was able to generate Te-containing nanostructures, about 68 nm in size, which exhibit interesting antibacterial properties against E. coli, with no apparent cytotoxicity against eukaryotic cells.
INTRODUCTION
Tellurium (Te), a metalloid belonging to group 16 of the periodic table, can exist in various redox states: telluride (2−), elemental tellurium (0), tellurite (4+), and tellurate (6+). Although the toxicity of the elemental form is not well defined, tellurite (TeO32−) and tellurate (TeO42−) are extremely noxious to most bacteria. The bactericidal activity of tellurite was recognized prior to the use of antibiotics (1), and therefore, Te-based compounds have a long history as antimicrobial and therapeutic agents (2).
Since the early 1900s, it has been recognized that most bacteria generate black deposits when exposed to tellurite (3). Although authors suggested that these deposits were elemental tellurium, this hypothesis was confirmed only in 1962 by X-ray diffraction analysis (4). To generate these deposits, also known as micro- or nanostructures (NS), bacterial cells reduce tellurite to elemental tellurium, at least in part, by the action of oxidoreductases. So far, a number of enzymes have been found to participate in tellurite reduction: nitrate reductases from Escherichia coli, Paracoccus denitrificans, Paracoccus pantotrophus, and Rhodobacter sphaeroides (5, 6); terminal oxidases of the electron transport chain from Gram-negative bacteria (7); dihydrolipoamide dehydrogenase from E. coli, Zymomonas mobilis, Streptococcus pneumoniae, Geobacillus stearothermophilus, and Aeromonas caviae ST (8); catalase from Staphylococcus epidermis CH (9); and NADH dehydrogenase II (NDH-II) (10) and isocitrate dehydrogenase (11) from E. coli. These enzymes display very different cellular functions, suggesting that tellurite reductase (TR) activities may be widely distributed. To date, no tellurite reductase has been used for generating Te-containing nanostructures (TeNSs).
Because of their very interesting physical and chemical properties, TeNSs have attracted significant attention for their use in various fields of nanotechnology. In this context, considerable efforts have been devoted to the chemical synthesis of Te-based NSs, such as hydrothermal or solvothermal methods, refluxing, microwave-assisted synthesis, and chemical vapor and physical vapor deposition (12). Nevertheless, most available protocols require high temperatures, anaerobic conditions (to avoid reagents' oxidation), or toxic chemicals. The use of microbiological systems is thus considered a safe, cost-effective, and environment-friendly process (13).
It was recently reported that chemically synthesized TeNSs display efficient antibacterial properties against E. coli, which seem to be even better than those exhibited by silver NSs (14). Given that Ag nanoparticles are nowadays considered the NSs exhibiting the most effective antimicrobial properties (15), this study opened brand new and promising perspectives for TeNS applications.
In this context, and using a classic biochemical approach, we looked for a novel tellurite reductase with the aim of enzymatically synthesizing TeNSs and assessing their antibacterial properties. In this line, tellurite-reducing activity was assessed in vivo and in cell extracts derived from a collection of Antarctic bacteria that had been previously isolated and characterized in our laboratory (16). Pseudomonas sp. strain BNF22 was selected for TR purification because of the high tellurite reduction shown by whole cells as well as by cell extracts.
This work describes the identification, purification, and use of glutathione reductase (GOR) from Pseudomonas sp. BNF22 for synthesizing TeNSs that exhibit efficient antibacterial properties with no apparent cytotoxicity for eukaryotic MDCK cells. Because of the ever-increasing number of antibiotic-resistant bacteria, TeNSs may represent, in the near future, an attractive alternative to antibiotics.
MATERIALS AND METHODS
Strains and growth conditions.
Pseudomonas sp. BNF22 was routinely grown in LB medium (17) at 25°C (16). Staphylococcus aureus ATCC 6538, E. coli BW25113, E. coli TOP10, and E. coli BL21 were grown in the same medium at 37°C. Growth was initiated by a 1% inoculation of fresh LB medium with a saturated culture. When growth on solid medium was required, LB was supplemented with 2% agar. When required, LB medium was supplemented with 100 μg/ml ampicillin.
For ultrathin section analysis via transmission electronic microscopy (TEM), Pseudomonas sp. BNF22 was grown to an optical density at 600 nm (OD600) of ∼0.4 and exposed to 9 μg/ml tellurite (1/4 MIC [16]) for 14 h. Cells were collected by centrifugation, washed with 10 mM sodium phosphate buffer (pH 6.5), and sent to the Unidad de Microscopía Avanzada (UMA) at Pontificia Universidad Católica de Chile (Santiago, Chile).
TR purification and characterization. (i) Enzymatic assays.
The assay for tellurite reduction was carried out at 30°C in 200 μl of tellurite reductase buffer (TR buffer) which contained 50 mM Tris-HCl (pH 9.0), 1 mM potassium tellurite, 1 mM β-mercaptoethanol (2-ME), and 1 mM NAD(P)H. Tellurite reduction was determined by measuring the increase of A500. One unit of enzymatic activity was defined as the amount of enzyme which caused an increase of 0.001 units of A500 min−1 (18). Specific activity was expressed as units per milligram of protein. Protein concentration was determined according to the method of Bradford (19).
The effect of pH on TR activity was determined at 30°C using the following buffers: 50 mM Na2HPO4-citric acid (pH 3 to 6), 50 mM Tris-HCl (pH 7 to 9), 50 mM NaHCO3-Na2CO3 (pH 10 to 11), and 50 mM KCl-NaOH (pH 12 to 13). The effect of temperature on TR activity was determined in TR buffer at pH 9.0 and at 5, 15, 20, 25, 30, 35, 45, and 55°C. The effect of ionic strength on tellurite reductase activity was determined at 30°C in TR buffer (pH 9.0) supplemented with 50 to 500 mM NaCl.
Glutathione reductase activity was determined by monitoring the decrease of NADPH concentration at 340 nm at 30°C. The reaction mix consisted of 50 mM Tris-HCl (pH 7.0), 2.5 mM glutathione disulfide (GSSG), 0.2 mM NADPH, and 0.6 μg of protein (20).
(ii) Screening for optimal tellurite reductase activity in cell extracts.
TR activity was determined in crude extracts of Pseudomonas sp. BNF22 previously grown under the following growth conditions: (i) 1× or 0.5× LB medium; (ii) presence or absence of tellurite (1/4 MIC); (iii) early-exponential, mid-exponential, or stationary growth phases; and (iv) presence of NAD(P)H. Growth was monitored at 600 nm, and samples were collected during the different growth phases. These were centrifuged at 9,500 × g for 10 min and washed with buffer A (10 mM sodium phosphate [pH 6.5], 50 mM NaCl, 0.5 mM 2-ME). Phenylmethanesulfonyl fluoride (PMSF; 150 μM final concentration) was added, and cells were disrupted by sonic treatment. Cell debris was discarded by centrifugation at 9,500 × g for 20 min, and the specific TR activity was assessed in the supernatant (cell extract) in the presence of 0.25 mM NADH or NADPH. In addition, to authenticate the protein nature of the reducing species, 5-min heat treatment at 95°C or 15-min SDS (1% final concentration) treatment of the cell extracts was conducted prior to TR activity measurements.
(iii) Purifying TR activity from Pseudomonas sp. BNF22.
Pseudomonas sp. BNF22 was grown in LB medium up to stationary growth phase (24 h), collected by centrifugation, and stored at −80°C until use. Cells (10 g [wet weight]) were suspended in 40 ml of buffer A supplemented with 150 μM PMSF and disrupted by sonication on ice. Cell debris was discarded by centrifugation at 9,500 × g and 4°C for 20 min, and the supernatant was amended with streptomycin sulfate (1% final concentration). After 30 min on ice, precipitated nucleic acids were removed by centrifugation as described above. The supernatant (crude extract) was dialyzed against buffer A for 2 h at 4°C. All subsequent steps were carried out at room temperature, except those in which a dialysis took place (4°C). The crude extract (35 ml) was loaded onto a phosphocellulose (Sigma) column (2.4 by 7.5 cm) stabilized with buffer A. The reductase activity did not bind to the resin, and the pass-through was immediately loaded onto a DEAE-cellulose (Pharmacia) column (2.4 by 6.5 cm) also equilibrated with buffer A. After washing, adsorbed proteins were eluted with 500 ml of a linear gradient 0 to 0.5 M NaCl in buffer A. Active fractions were pooled (42.5 ml) and dialyzed overnight against buffer B (10 mM sodium phosphate, pH 6.5).
Ampholytes (Servalyt; pH 4 to 7, 1.5% final concentration) were added to the solution, and isoelectric focusing was carried out at 40 mA, 4°C, for 2 h using a Rotofor cell (Bio-Rad). Precipitated proteins were removed at 12,000 × g for 10 min, and active fractions were pooled (4.7 ml). The solution was diluted 7-fold with buffer B, amended with 1% ampholytes, and loaded into the Rotofor cell. Isoelectric focusing was carried out as described above. Active fractions were pooled (2.8 ml), concentrated by ultrafiltration (UF; 50-kDa cutoff), and washed 3 times with buffer A. A 200-μl fraction containing the tellurite reductase activity was finally obtained. SDS-PAGE was carried out as described previously (18). The overall purification procedure is summarized in Table 1.
TABLE 1.
Purification of tellurite reductase activity from BNF22
| Step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Purification (fold) |
|---|---|---|---|---|
| Crude extract | 662 | 74,200 | 112 | 1 |
| DEAEa | 63 | 60,633 | 960 | 9 |
| Rotofor 1 | 12 | 38,665 | 3,171 | 28 |
| Rotofor 2 | 2 | 13,683 | 6,655 | 59 |
| UF 50 kDa | 1.6 | 16,235 | 9,945 | 89 |
Phosphocellulose P11 column linked in tandem to a DEAE-Sepharose column. See Materials and Methods for details.
Protein bands observed upon fractionation by SDS-PAGE were cut, dehydrated, and sent for matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) identification at the Centro de Estudios para el Desarrollo de la Química (CEPEDEQ) from the Universidad de Chile (Santiago, Chile).
(iv) Cloning, overexpression of the Pseudomonas sp. BNF22 gor gene, and purification of the recombinant enzyme.
Genomic DNA from Pseudomonas sp. BNF22 was purified using the GeneJET genomic DNA purification kit (Thermo Scientific). Based on MALDI-TOF results, two pairs of primers were designed to amplify the gene encoding the glutathione reductase from Pseudomonas sp. BNF22 and for its subsequent cloning into the vector pET101/D-TOPO. The first pair, GM102-22, was designed based on the published gor sequence from Pseudomonas sp. strain GM102 (GenBank: EJM01012) (5′-CACCATGGCCTACGATTTTGAC-3′ [forward] and 5′-ACTCGCGACTGGCGTGC-3′ [reverse]). The second, degenerate primer pair, DEG-22, was designed using the CODEHOP method (21) and the aligned amino acid sequences (Clustal X2) of glutathione reductases from Pseudomonas protegens, Pseudomonas aeruginosa PAO1, Pseudomonas fluorescens, Pseudomonas stutzeri, Pseudomonas syringae, Pseudomonas entomophila, and Pseudomonas sp. GM102 (5′-CACCATGGCCTTCGATTTYGAYCTD-3′ [forward] and 5′-ACTCGCGTGAGGSGTRCGCAT-3′ [reverse]).
Two PCR steps were required to amplify the gor gene from Pseudomonas sp. BNF22. Reaction mixtures (25 μl) consisted of 1× buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.2 μM (each) primer, 1.25 U of Pfu polymerase (Thermo Scientific), and 50 ng of template DNA. The first step consisted of a touchdown PCR (TD-PCR) using GM102-22 primers under the following conditions: (i) 95°C for 3 min; (ii) 95°C for 45 s, 65°C for 45 s, and 72°C for 2.75 min (25 cycles); and (iii) 72°C for 10 min. The annealing temperature was decreased by 1°C during the first 10 cycles and was kept at 55°C during the 15 remaining rounds. The PCR product was diluted 40-fold with MilliQ water, and a second amplification was performed using DEG-22 primers under the following conditions: (i) 95°C for 3 min; (ii) 95°C for 30 s, 64°C for 30 s, and 72°C for 2.5 min (35 cycles); and (iii) 72°C for 10 min.
The amplified ∼1.4-kb fragment was excised from the agarose gel, and the DNA was recovered using the Zymoclean gel DNA recovery kit (Zymo Research). The fragment was then cloned into the pET101/D-TOPO vector (Invitrogen) and transformed into E. coli TOP10 according to the manufacturer's instructions. The clone pGR223, containing the directionally inserted gor gene, was used for further analysis.
E. coli carrying pGR223 was grown at 37°C to an OD600 of ∼0.6 and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 12 h with vigorous shaking. Cells were suspended in 20 mM sodium phosphate buffer (pH 7.4) that contained 0.5 M NaCl and 20 mM imidazole (buffer C) and disrupted by sonication. His-tagged proteins present in the crude extract were purified by affinity chromatography using HisTrap HP (GE Healthcare) columns. After extensive washing with buffer C, bound proteins were eluted with the same buffer but containing 500 instead of 20 mM imidazole.
(v) Using GOR for TeNS synthesis.
Unless otherwise indicated, the standard conditions for the enzymatic synthesis of TeNSs consisted of 50 mM Tris-HCl (pH 9.0), 1 mM potassium tellurite, 1 mM 2-ME, 1 mM NADPH, and 50 μg/ml of the recombinant GOR. Reactions were carried out at room temperature for 45 min.
Characterizing the TeNSs. (i) Dynamic light scattering (DLS) analysis.
Zeta potential and hydrodynamic diameter of TeNSs were measured at 25°C using a Malvern Zetasizer Nano ZS apparatus. Values were calculated based on at least three separate measurements of 20 runs each.
(ii) UV-vis absorption spectra.
The UV-visible (UV-vis) absorption spectra of TeNSs, GOR (50 μg/ml), potassium tellurite (1 mM), and NADPH (1 mM) were recorded in a Tecan Infinite M200pro reader at room temperature.
(iii) Detection of GOR bound to TeNSs.
Synthesized TeNSs were washed extensively with water to remove all traces of GOR from the supernatant. The NSs were then treated with SDS and 2-ME, and after heating (95°C, 10 min), the suspension was fractionated by SDS-PAGE. Untreated-TeNS supernatant was used as control.
(iv) TEM.
TEM analysis was carried out by placing a drop of the TeNS-containing suspension onto carbon-Formvar-coated copper grids. Electron micrographs were recorded using a Philips Tecnai 12 TEM operating at 80 keV.
(v) EDS.
Energy-dispersive X-ray spectroscopy (EDS) analyses were performed using air-dried TeNSs. The NSs were intensively washed with water and ethanol to remove traces of unreacted reagents. EDS measurements were carried out using an LEO-1420VP scanning electron microscope using the detector system INCAx-sight-7424 (Oxford Instruments).
(vi) ICP-OES analyses.
Inductively coupled plasma-optical emission spectrometry (ICP-OES) analyses were performed using a PerkinElmer Optima 2000 DV apparatus in the axial viewing mode. The wavelength corresponding to tellurium was 214,281 nm. A calibration curve for tellurium (in 2% ultrapure HNO3) ranging from 1 to 200 mg/liter was constructed. Prior to analysis, TeNSs were treated as follows: NS-containing suspensions were centrifuged at 14,000 × g for 90 min. The supernatant was recovered, mixed with 30 μl of ultrapure HNO3 (65%), filtered through a 0.2-μm-pore-size membrane, and analyzed by ICP-OES. On the other hand, TeNSs were washed 2 times with MilliQ water, centrifuged at 14,000 × g for 90 min, suspended in 5% HNO3 (final concentration) until complete dissolution, filtered through an 0.2-μm membrane, and analyzed by ICP-OES.
To determine TeNS stability and the release of Te, synthesized TeNSs were incubated at room temperature for 3 days. Samples were collected at 0, 24, 48, and 72 h.
To assess the relationship between NADPH concentration and the amount of TeNSs generated, the enzymatic synthesis was carried out in the presence of 0.063, 0.125, 0.25, 0.5, and 1 mM NADPH.
TeNS antibacterial properties. (i) Growth curve analysis.
TeNSs synthesized under standard conditions were dialyzed against 50 mM Tris-HCl (pH 9.0) for at least 4 h. Growth curves of E. coli BW25113 and S. aureus ATCC 6538 were monitored in Tecan Infinite M200pro equipment. Dialyzed TeNSs were added when cells reached an OD600 of ∼0.4.
(ii) ROS determination using H2DCFDA.
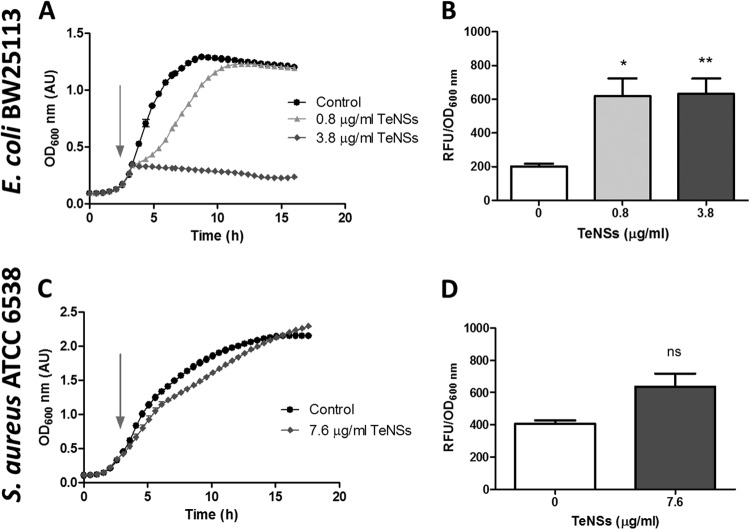
E. coli and S. aureus grown to an OD600 of ∼0.4 were exposed to TeNSs (0.8, 3.8, and 7.6 μg/ml) for 30 min. After being washed two times and suspended in 1 ml of 50 mM phosphate buffer, pH 7.0, cells were incubated for 30 min in the dark with the oxidation-sensitive probe 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA; 0.02 mM). Total reactive oxygen species (ROS) were assessed by monitoring the relative fluorescence units (RFU) (λex, 490 nm; λem, 520 nm) in Tecan Infinite M200pro equipment. RFU were normalized by the optical density of the culture.
(iii) Viability of MDCK cells in the presence of TeNSs.
MDCK cells (20,000/well) were grown in a 96-well plate in RPMI (HyClone) medium supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 2 mM l-glutamine. After 6 h, treatment with 0.8, 3.8, or 7.6 μg/ml TeNSs was carried out and adherent cells were incubated for 16 or 36 h in 5% CO2, in a humidified atmosphere at 37°C. Viability was determined using the reactive alamarBlue (Thermo Scientific) at 570 nm (using 600 nm as a reference wavelength) according to the manufacturer's instructions.
Nucleotide sequence accession number.
The inserted DNA from the Pseudomonas sp. BNF22 gor gene was deposited in GenBank under accession no. KM066115.
RESULTS
Identification, purification, and characterization of a tellurite reductase from Pseudomonas sp. BNF22.
When exposed to tellurite (1/4 MIC), cells of Pseudomonas sp. BNF22, whether stained or not, showed electron-dense nanostructures within the cytoplasm (see Fig. S1A and B in the supplemental material). The nanostructures, which appeared as heterogeneously sized nanotube clusters, most likely resulted from reducing tellurite to elemental tellurium. This reduction is generated, at least in part, by the presence of cytoplasmic tellurite-reducing enzymes (see below).
After screening for the highest TR activity, which occurred in the presence of NADPH, the purification of such enzymes was carried out using crude extracts of cells grown for 24 h (stationary growth phase) in LB medium (see Fig. S2 in the supplemental material). It is worth noting that heating or SDS (1%) treatment greatly decreased TR activity, confirming the protein nature of the reducing activity (Fig. S2). An optimized, reproducible purification procedure was developed, whose different steps are listed in Table 1.
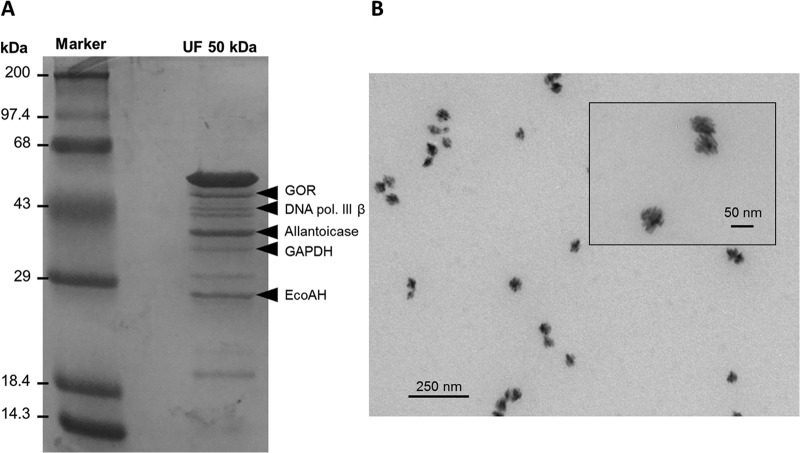
Using this procedure, TR specific activity increased from 112 to 9,945 U/mg (purification, ∼89-fold). SDS-PAGE analysis of the fraction obtained after ultrafiltration (50-kDa molecular mass cutoff) revealed the presence of about 10 bands (Fig. 1A). This fraction generated round, porous nanostructures ∼60 nm in size (Fig. 1B). Because of the reduced number of protein bands in this fraction, and to specifically identify the enzyme responsible for tellurite reduction, SDS-PAGE bands were excised and sent for MALDI-TOF analysis. Five bands were positively identified, which corresponded to glutathione reductase (GOR), DNA polymerase III subunit β (DNA pol. III β), allantoicase, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and enoyl coenzyme A-hydratase (enoyl-CoA-hydratase) (EcoAH) (Fig. 1A). GOR seemed the best candidate to reduce tellurite since it belongs to the flavoprotein disulfide reductase (FDR) family, whose members are characterized by the presence of flavin as cofactor. This family also includes dihydrolipoamide dehydrogenase, previously reported as TR in our laboratory (8).
FIG 1.
Partial purification of the native tellurite reductase and enzymatic synthesis of TeNSs. (A) SDS-PAGE (12%) of the UF 50-kDa fraction. Proteins identified by MALDI-TOF are indicated on the right. GOR, glutathione reductase; DNA pol. III β, DNA polymerase III subunit β; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; EcoAH, enoyl-CoA-hydratase. Fifty micrograms of protein was loaded per well. (B) TEM analysis of the tellurite reduction product generated using the UF 50-kDa fraction.
The Pseudomonas sp. BNF22 gor gene was amplified, cloned, and overexpressed in E. coli. The inserted DNA was sequenced (Macrogen, South Korea), deposited in GenBank (accession number KM066115), and analyzed using BLASTx. Identity ranging from 94 to 97% was observed with the gor gene from other Pseudomonas species (not shown).
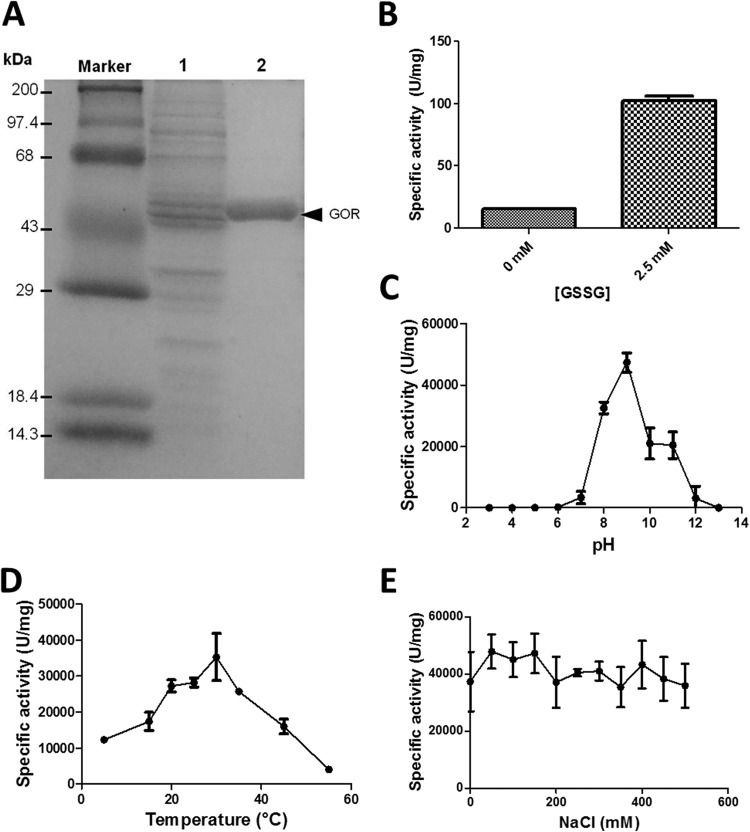
Amplification and cloning were carried out as described in Materials and Methods. The E. coli clone pGR223, containing a correctly inserted gor gene from Pseudomonas sp. BNF22, was used to purify the recombinant protein by HisTrap column chromatography. A band exhibiting an expected molecular mass of ∼50 kDa corresponding to the monomer subunit of GOR was observed (Fig. 2A). The recombinant enzyme was functional, i.e., able to oxidize NADPH efficiently in the presence of GSSG (Fig. 2B).
FIG 2.
Purification, functional expression, and characterization of the recombinant GOR. (A) SDS-PAGE (12%) of the crude extract (lane 1) and HisTrap-purified proteins (lane 2). Twenty-five micrograms of protein was loaded per well. (B) Glutathione reductase activity of the recombinant protein in the presence of 0 or 2.5 mM GSSG. (C to E) Effect of pH (C), temperature (D), and ionic force (E) on the TR activity of the recombinant GOR. Bars represent the averages of three independent trials ± standard deviations.
The effect of pH, temperature, and ionic strength on the TR activity of the recombinant GOR was evaluated. While the optimal conditions for reduction were 30°C and pH 9.0 (Fig. 2C and D), the effect of ionic strength was not significant under the tested conditions (0 to 500 mM NaCl) (Fig. 2E). The recombinant GOR was capable of reducing other chalcogen oxyanions such as selenite and (slightly) tellurate but not sulfite, sulfate, and selenate; it was also unable to reduce copper, gold, or silver (not shown).
GOR-mediated synthesis and characterization of TeNSs.
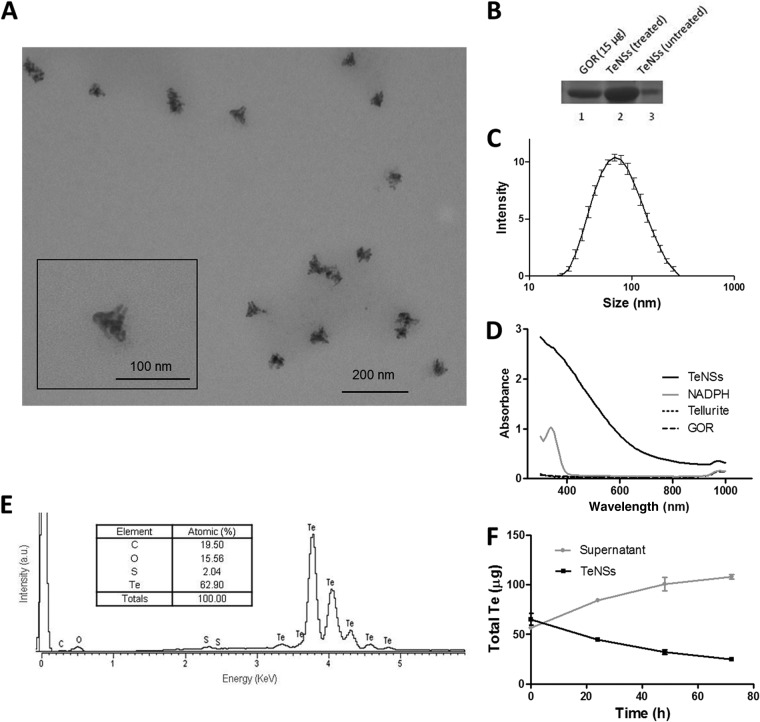
Most of the TeNSs generated under standard conditions (see Materials and Methods) displayed triangular shapes with, most likely, porous surfaces (Fig. 3A). The nanostructures exhibited a size of ∼68 nm and low agglomeration (Fig. 3A and C). The determined zeta potential was −34 mV (not shown). The TeNSs absorbed much more UV-vis light than did GOR or tellurite alone. Nonetheless, no particular absorbance peaks were observed, a common characteristic of tellurium (Fig. 3D) (22). The chemical composition of TeNSs, as determined by EDS, is shown in Fig. 3E. These structures were mainly composed of tellurium (62.9%), and a smaller proportion of carbon, oxygen, and sulfur was also detected. Based on the stoichiometry between Te and O, it may be inferred that tellurium is not present in the TeNSs as tellurium dioxide (TeO2).
FIG 3.
Characterization of the TeNSs generated with the recombinant GOR under standard conditions. (A) TEM micrograph. (B) SDS-PAGE of GOR (lane 1) and supernatants of treated (2-ME, SDS, or heat) (lane 2) and untreated (lane 3) TeNSs. (C) Hydrodynamic diameter distribution (DLS). (D) UV-vis absorption spectra of TeNSs, GOR, tellurite, and NADPH. (E) Representative EDS spectrum of the purified TeNSs. (Inset) Chemical composition of TeNSs. (F) ICP-OES analysis of TeNS stability and release of Te over time. Bars represent the averages of three independent trials ± standard deviations.
The presence of carbon, oxygen, and sulfur suggests that TeNSs are not composed of pure inorganic species of tellurium. Capping with organic compounds, most likely the bound glutathione reductase (GOR), or parts of it, may have been generated. To confirm this hypothesis, the synthesized TeNSs were treated with SDS, 2-ME, and heating, and after centrifuging, the nanostructure-free suspension was fractionated by SDS-PAGE (Fig. 3B). GOR was observed in treated TeNSs, suggesting a strong interaction with the enzyme, probably through covalent bonding. The same amount of untreated TeNSs showed only a light, attenuated band, whose presence could be explained by unspecific GOR binding or by the release of the enzyme from TeNSs due to mechanical stress during the various cycles of washing, centrifugation, and resuspension. Treating TeNSs with 2 M NaCl did not result in GOR release from the TeNSs (not shown), suggesting that no or few ionic interactions exist between the enzyme and TeNSs.
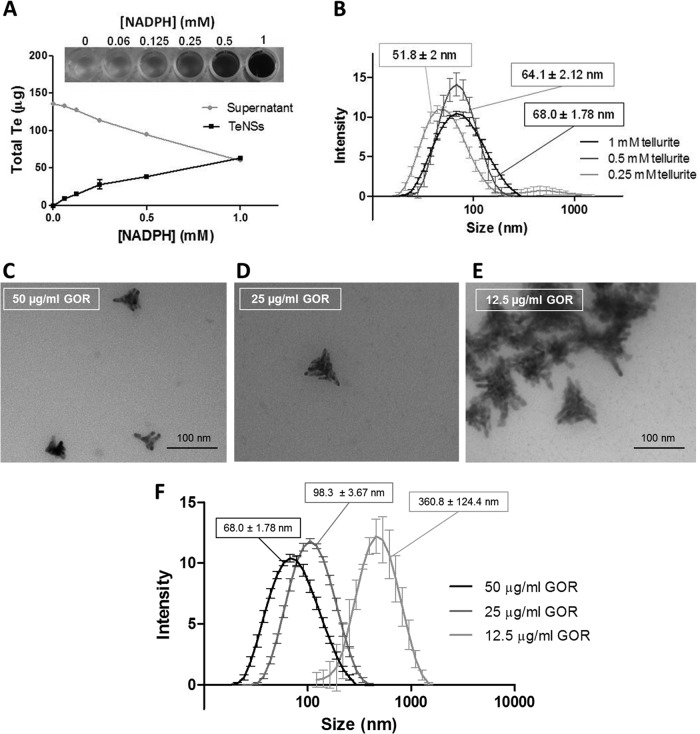
The amount of TeNSs was dependent on NADPH concentration, and under standard conditions, half of tellurite was transformed into TeNSs (Fig. 4A). The effect of protein concentration on the size and shape of TeNSs was determined by DLS and TEM analysis. Lower GOR concentrations resulted in increased hydrodynamic diameters. Indeed, using 50, 25, and 12.5 μg/ml of the purified, recombinant GOR, hydrodynamic diameters of 68, 98.3, and 360.8 nm were observed, respectively (Fig. 4F). These values were confirmed by TEM analysis, where the average size of individual nanostructures increased in the presence of lower GOR concentrations (Fig. 4C to E). Interestingly, in the presence of 12.5 μg/ml GOR, the TeNSs agglomerated, leading to high hydrodynamic diameters (DLS). Treating TeNSs with proteinase K also led to an important increase of hydrodynamic diameters (not shown). Taken together, these results suggest that GOR not only reduces tellurite but also may act as a scaffold that will somehow determine the size and shape of the TeNSs. On the other hand, tellurite concentration seemed to slightly affect the size of the NSs as determined by DLS (Fig. 4B).
FIG 4.
Effect of substrate concentration on TeNS characteristics. (A) Effect of NADPH concentration on the amount of NSs generated as determined by ICP-OES. (B) Hydrodynamic diameter of TeNSs as determined by DLS using 0.25, 0.5, and 1 mM tellurite. (C to E) TEM micrograph of the NSs generated using 50 (C), 25 (D), or 12.5 (E) μg/ml of the recombinant GOR. (F) Hydrodynamic diameter of TeNSs as determined by DLS using the same GOR concentrations. Bars represent the averages of three independent trials ± standard deviations.
Antibacterial properties and cytotoxicity of TeNSs.
Release of Te from the nanostructures to the surrounding milieu was observed over time, and about 40% of the initial TeNSs remained after 72 h (Fig. 3F). Then, the antibacterial effect of these TeNSs was tested against E. coli BW25113 and S. aureus ATCC 6538. As shown in Fig. 5A and C, GOR-generated nanostructures displayed antibacterial properties showing higher growth inhibition against E. coli than against S. aureus. The amount of ROS in E. coli increased significantly in the presence of TeNSs compared to controls, a situation that was not observed in S. aureus (Fig. 5B and D). To improve the action of TeNSs against S. aureus, the combined effect of 2-ME and TeNSs was analyzed. As shown in Fig. S3 in the supplemental material, 2-ME greatly enhanced the antibacterial effect of the TeNSs against this Gram-positive coccus, a phenomenon that was previously reported when tellurite and 2-ME were combined (23).
FIG 5.
Antibacterial properties of TeNSs synthesized using the recombinant GOR under standard conditions. (A and C) Growth curves of E. coli BW25113 (A) and S. aureus ATCC 6538 (C) exposed to Te-based nanostructures when cells reached an OD600 of ∼0.4. S. aureus was not affected by 0.8 and 3.8 μg/ml of NSs, and those data are not shown, for clarity. (B and D) Oxidative status assessment using H2DCFDA in E. coli BW25113 (B) and S. aureus ATCC 6538 (D). Bars represent the averages of three independent trials ± standard deviations. ns, nonsignificant; **, P < 0.01; *, P < 0.05.
To further demonstrate the clinical potential of the TeNSs, a viability assay to evaluate their cytotoxicity was carried out using MDCK cells (Fig. 6). The presence of TeNSs (up to 7.6 μg/ml) showed a very low cytotoxicity, since they did not significantly influence the viability of MDCK cells after 36 h of exposure. Thus, the enzymatically synthesized TeNSs appear promising as novel antibacterial agents.
FIG 6.
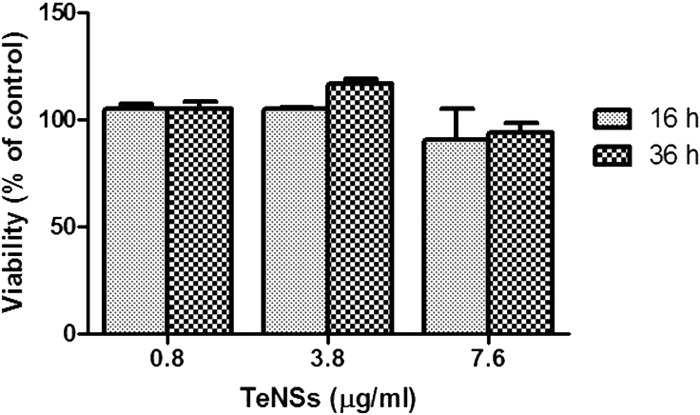
Viability of MDCK cells exposed to 0.8, 3.8, or 7.6 μg/ml of TeNSs for 16 or 36 h as determined by alamarBlue. Untreated cells were used as a control. Bars represent the averages of five independent trials ± standard deviations.
DISCUSSION
The use of microorganisms for synthesizing nanostructures emerges as an eco-friendly and exciting approach and has been investigated in the last few years (13). Many studies indicate that NADH- and NADPH-dependent enzymes are central factors in the microbial synthesis of metal(loid) nanostructures (24–26). Nevertheless, the direct use of a pure enzyme to generate nanostructures is still a rare event. To our knowledge, only Ag0 (27); Au0 (28, 29); CdS (30); and Ni7S6, PbS, and Co3S4 (31) nanostructures have been synthesized that way.
To date, only a few enzymes capable of reducing tellurite using NAD(P)H as electron donors have been identified. These include catalase (9), NADH dehydrogenase II (NDH-II) (10), isocitrate dehydrogenase (11), and lipoamide dehydrogenase (8). However, none of them have been used for generating tellurium nanostructures.
The presence of intracellular TeNSs was observed in Pseudomonas sp. BNF22. These deposits appeared as clusters of nanotubes that could reach 600 nm in length. Because of the low homogeneity of TeNSs and the biotechnological drawbacks inherent to their intracellular synthesis (greater downstream processing), we decided to purify the cytoplasmic enzyme responsible for tellurite reduction and thus get better control of TeNSs' shape and size.
According to previous reports (32), GOR uses preferably NADPH as a cofactor, and it is also probable that there exists a larger enzyme amount in the stationary growth phase, a result which has also been observed in E. coli (33).
Tellurite reductase was partially purified using an optimized and reproducible protocol (Table 1). Of the five bands identified by MALDI-TOF in the UF 50-kDa fraction, only two of them are capable of performing NAD(P)H-dependent redox reactions: glutathione reductase and glyceraldehyde-3-phosphate dehydrogenase. GAPDH preferably uses NAD+ as a cofactor for the oxidation of glyceraldehyde-3-phosphate, while GOR utilizes NADPH to reduce GSSG. As mentioned, GOR belongs to the FDR family (32), along with mercuric reductase (MerA), which reduces mercury at the expense of NADPH oxidation, and dihydrolipoamide dehydrogenase, which displays TR activity (8). Finally, GOR from E. coli exhibits gold-reducing activity that was used to generate gold nanoparticles (29). To our knowledge, no metal(loid)-reducing activity has been reported for GAPDH. Based on these arguments, it was thus hypothesized that GOR was the enzyme responsible for TR activity. It is somehow intriguing that the specific activity of the recombinant GOR is only 4-fold higher than that exhibited by the UF 50-kDa fraction. A number of possibilities might explain this difference, including enzyme activators that are present in the partially purified preparation but not in the final recombinant fraction, different codon usage, lack of proper protein folding because of the His tag, etc. However, and since the recombinant GOR proved very efficient in synthesizing Te-containing nanostructures, we did not attempt to evaluate the referred difference in specific activity.
It was recently demonstrated that flavin adenine dinucleotide (FAD) as well as the catalytic cysteine residues of the E. coli dihydrolipoamide dehydrogenase is essential for tellurite reduction (34). Cysteine residues are very sensitive to pH and deprotonating thiol groups (-SH) generate thiolate (-S−) (pKa of ∼8) (35). This pKa implies that most -SH groups should be protonated at pH 7.0 and therefore inactive (Fig. 2). This would in part explain the strong pH influence on GOR to reduce tellurite, although other effects on the structure of the protein and on other substrates' redox groups should not be ruled out.
The composition of the reaction mix greatly influenced the TeNS characteristics, including amount, size, shape, and agglomeration (Fig. 4). Additional experiments are required to fully optimize the process of TeNS synthesis and to determine the influence of each substrate on its characteristics.
The presence of porous nanostructures is an interesting feature (Fig. 3A). Pores increase the surface area-to-volume ratio of the NSs, since a greater fraction of atoms is located at the surface and leads to increased chemical reactivity of the NSs (36). We decided to test the chemical reactivity of the TeNSs as an antibacterial agent since it was recently shown that chemically synthesized tellurium NSs showed very promising antibacterial properties against E. coli, comparable to or higher than those to silver NSs (14). The same antibacterial properties were then reported using Te nanostructures against S. aureus, P. aeruginosa, Salmonella enterica serovar Typhi, and Klebsiella pneumoniae (37). Moreover, recently an interesting observation was made by Mohanty and colleagues (38), who reported that tellurium nanorods possessed antivirulence properties against P. aeruginosa and were able to effectively inhibit the production of pyoverdine, one of the main virulence factors in this bacterium. The exact mechanism of action of tellurium nanorods remains unclear.
In this study, we showed that tellurium-based NSs exhibit efficient antibacterial properties against E. coli, whose growth was almost completely inhibited at concentrations as low as 3.8 μg/ml. On the other hand, almost no growth inhibition of S. aureus was observed at concentrations up to 7.6 μg/ml. This difference may be explained by the putative mechanism of action of the nanostructures described below.
ICP analysis of nanostructures clearly showed a decrease of NSs' stability over time, most likely resulting from an oxidation of the nanostructures in suspension. To our knowledge, this is the first report of tellurium oxidation and solubilization in an aqueous system. It was described that the mechanism of action of silver nanoparticles is due, at least in part, to the release of the noxious silver ion (15). A similar mechanism most likely occurs with TeNSs. The soluble tellurium derivative released by Te-containing NSs is probably tellurite, which in turn can damage E. coli cells (39). Tellurite release was suggested by TEM, with the observation of tellurium nanorods inside E. coli after being exposed to Te-based nanostructures (not shown). These nanorods were clearly different in shape from the triangular nanostructures, and thus, it is likely that these nanorods are generated via intracellular reduction of tellurite released from the TeNSs. The significant increase of ROS detected in E. coli cells exposed to tellurium NSs is also a usual response to tellurite exposure. As previously reported, S. aureus is much more resistant to tellurite than E. coli and generates smaller amounts of ROS in the presence of tellurite (23). The same trend is observed in Fig. 5D, where the increase of ROS in the presence of TeNSs is nonsignificant compared to the control. The combined effect of 2-ME and TeNSs against S. aureus was tested. As shown in Fig. S3 in the supplemental material, 2-ME greatly enhanced the antibacterial effect of the tellurium NSs, a phenomenon that was previously reported when both tellurite and 2-ME were simultaneously administered to S. aureus (23), supporting the putative tellurite release from the Te-containing nanostructures.
Finally, it was observed that the synthesized tellurium-based NSs did not exhibit cytotoxicity to MDCK cells (Fig. 6). Similar noncytotoxic properties were observed with tellurium nanorods against human bronchial epithelial cells and murine macrophages (38). Additional experiments are under way to determine the effectiveness of the TeNSs against other, different bacterial species and to confirm their apparent nonnoxious properties against mammalian cells. The antibacterial properties of the TeNSs reported here are very encouraging and could, in the near future, represent an alternative strategy to Ag-containing NSs or antibiotic therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cristian Calderón, Laboratorio de Fisicoquímica, Universidad de Santiago de Chile, Santiago, Chile, for assistance with DLS analyses.
This work was supported by grants from Fondecyt (Fondo Nacional de Ciencia y Tecnología) Regular 1130362 and Postdoctorado 3120049 to C.C.V. and F.A.S., respectively, and from INACH (Instituto Antártico de Chile) DG_02-13 to B.P. B.P. was supported by a doctoral fellowship from MECESUP (Mejoramiento de la Calidad y Equidad de la Educación Superior), Chile.
Footnotes
Published ahead of print 5 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02207-14.
REFERENCES
- 1.Fleming A. 1932. On the specific antibacterial properties of penicillin and potassium tellurite. J. Pathol. Bacteriol. 35:831–842. 10.1002/path.1700350603. [DOI] [Google Scholar]
- 2.Chasteen TG, Fuentes DE, Tantaleán JC, Vásquez CC. 2009. Tellurite: history, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 33:820–832. 10.1111/j.1574-6976.2009.00177.x. [DOI] [PubMed] [Google Scholar]
- 3.King WE, Davis L. 1914. Potassium tellurite as an indicator of microbial life. Am. J. Public Health 4:917–932. 10.2105/AJPH.4.10.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tucker FL, Walper JF, Appleman MD, Donohue J. 1962. Complete reduction of tellurite to pure tellurium metal by microorganisms. J. Bacteriol. 83:1313–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avazeri C, Turner RJ, Pommier J, Weiner JH, Giordano G, Vermeglio A. 1997. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology 143:1181–1189. 10.1099/00221287-143-4-1181. [DOI] [PubMed] [Google Scholar]
- 6.Sabaty M, Avazeri C, Pignol D, Vermeglio A. 2001. Characterization of the reduction of selenate and tellurite by nitrate reductases. Appl. Environ. Microbiol. 67:5122–5126. 10.1128/AEM.67.11.5122-5126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trutko SM, Akimenko VK, Suzina NE, Anisimova LA, Shlyapnikov MG, Baskunov BP, Duda VI, Boronin AM. 2000. Involvement of the respiratory chain of Gram-negative bacteria in the reduction of tellurite. Arch. Microbiol. 173:178–186. 10.1007/s002039900123. [DOI] [PubMed] [Google Scholar]
- 8.Castro ME, Molina R, Díaz W, Pichuantes SE, Vásquez CC. 2008. The dihydrolipoamide dehydrogenase of Aeromonas caviae ST exhibits NADH-dependent tellurite reductase activity. Biochem. Biophys. Res. Commun. 375:91–94. 10.1016/j.bbrc.2008.07.119. [DOI] [PubMed] [Google Scholar]
- 9.Calderón IL, Arenas FA, Pérez JM, Fuentes DE, Araya MA, Saavedra CP, Tantaleán JC, Pichuantes SE, Youderian PA, Vásquez CC. 2006. Catalases are NAD(P)H-dependent tellurite reductases. PLoS One 1:e70. 10.1371/journal.pone.0000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Díaz-Vásquez WA, Abarca-Lagunas MJ, Arenas FA, Pinto CA, Cornejo FA, Wansapura PT, Appuhamillage GA, Chasteen TG, Vásquez CC. 2014. Tellurite reduction by Escherichia coli NDH-II dehydrogenase results in superoxide production in membranes of toxicant-exposed cells. BioMetals 27:237–246. 10.1007/s10534-013-9701-8. [DOI] [PubMed] [Google Scholar]
- 11.Reinoso CA, Appanna VD, Vásquez CC. 2013. α-Ketoglutarate accumulation is not dependent on isocitrate dehydrogenase activity during tellurite detoxification in Escherichia coli. Biomed Res. Int. 2013:784190. 10.1155/2013/784190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim B, Park BK. 2012. Synthesis of self-aligned tellurium nanotubes by a sodium thiosulfate-assisted polyol method. Electron Mater. Lett. 8:33–36. 10.1007/s13391-011-0960-7. [DOI] [Google Scholar]
- 13.Narayanan KB, Sakthivel N. 2010. Biological synthesis of metal nanoparticles by microbes. Adv. Colloid Interface Sci. 156:1–13. 10.1016/j.cis.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Lin ZH, Lee CH, Chang HY, Chang HT. 2012. Antibacterial activities of tellurium nanomaterials. Chem. Asian J. 7:930–934. 10.1002/asia.201101006. [DOI] [PubMed] [Google Scholar]
- 15.Rai M, Yadav A, Gade A. 2009. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 27:76–83. 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Arenas FA, Pugin B, Henríquez NA, Arenas-Salinas MA, Díaz-Vásquez WA, Pozo MA, Muñoz CM, Pérez-Donoso JM, Vásquez CC. 2014. Isolation, identification and characterization of highly tellurite-resistant, tellurite-reducing bacteria from Antarctica. Polar Sci. 8:40–52. 10.1016/j.polar.2014.01.001. [DOI] [Google Scholar]
- 17.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 18.Chiong M, González E, Barra R, Vásquez C. 1988. Purification and biochemical characterization of tellurite-reducing activities from Thermus thermophilus HB8. J. Bacteriol. 54:610–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Mata A, Pinto CM, López-Barea J. 1984. Purification by affinity chromatography of glutathione reductase (EC 1.6.4.2) from Escherichia coli and characterization of such enzyme. Z. Naturforsch. 39:908–915. [DOI] [PubMed] [Google Scholar]
- 21.Staheli JP, Boyce R, Kovarik D, Rose TM. 2011. CODEHOP PCR and CODEHOP PCR primer design. Methods Mol. Biol. 687:57–73. 10.1007/978-1-60761-944-4_5. [DOI] [PubMed] [Google Scholar]
- 22.Molina RC, Burra R, Pérez-Donoso JM, Elías AO, Muñoz C, Montes RA, Chasteen TG, Vásquez CC. 2010. Simple, fast, and sensitive method for quantification of tellurite in culture media. Appl. Environ. Microbiol. 76:4901–4904. 10.1128/AEM.00598-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugin B, Cornejo FA, García JA, Díaz-Vásquez WA, Arenas FA, Vásquez CC. 2014. Thiol-mediated reduction of Staphylococcus aureus tellurite resistance. Adv. Microbiol. 4:183–190. 10.4236/aim.2014.44024. [DOI] [Google Scholar]
- 24.Ahmad A, Mukherjee P, Mandal D, Senapati S, Khan MI, Kumar R, Sastry M. 2002. Enzyme mediated extracellular synthesis of CdS nanoparticles by the fungus, Fusarium oxysporum. J. Am. Chem. Soc. 124:12108–12109. 10.1021/ja027296o. [DOI] [PubMed] [Google Scholar]
- 25.Senapati S, Ahmad A, Khan MI, Sastry M, Kumar R. 2005. Extracellular biosynthesis of bimetallic Au-Ag alloy nanoparticles. Small 1:517–520. 10.1002/smll.200400053. [DOI] [PubMed] [Google Scholar]
- 26.Correa-Llantén DN, Muñoz-Ibacache SA, Castro ME, Muñoz PA, Blamey JM. 2013. Gold nanoparticles synthesized by Geobacillus sp. strain ID17 a thermophilic bacterium isolated from Deception Island, Antarctica. Microb. Cell Fact. 12:75. 10.1186/1475-2859-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anil Kumar S, Abyaneh MK, Gosavl SW, Kulkarni SK, Pasricha R, Ahmad A, Khan MI. 2007. Nitrate reductase-mediated synthesis of silver nanoparticles from AgNO3. Biotechnol. Lett. 29:439–445. 10.1007/s10529-006-9256-7. [DOI] [PubMed] [Google Scholar]
- 28.Kumar A, Abyaneh MK, Gosavl SW, Kulkarni SK, Ahmad A, Khan MI. 2007. Sulfite reductase-mediated synthesis of gold nanoparticles capped with phytochelatin. Biotechnol. Appl. Biochem. 47:191–195. 10.1042/BA20060205. [DOI] [PubMed] [Google Scholar]
- 29.Scott D, Toney M, Muzikár M. 2008. Harnessing the mechanism of glutathione reductase for synthesis of active site bound metallic nanoparticles and electrical connection to electrodes. J. Am. Chem. Soc. 130:865–874. 10.1021/ja074660g. [DOI] [PubMed] [Google Scholar]
- 30.Ansary AA, Kumar A, Krishnasastry MV, Abyaneh MK, Kulkarni SK, Ahmad A, Khan MI. 2007. CdS quantum dots: enzyme mediated in vitro synthesis, characterization and conjugation with plant lectins. J. Biomed. Nanotechnol. 3:406–413. 10.1166/jbn.2007.045. [DOI] [Google Scholar]
- 31.Ansary AA, Khan MI, Gaikwad SM. 2012. In vitro enzyme mediated synthesis of metal sulfide nanoparticles: control of particle size of CdS, Ni7S6, PbS, Co3S4 nanoparticles using synthetic peptides. Sci. Adv. Mater. 4:179–186. [Google Scholar]
- 32.Argyrou A, Blanchard JS. 2004. Flavoprotein disulfide reductases: advances in chemistry and function. Prog. Nucleic Acid Res. Mol. Biol. 78:89–142. 10.1016/S0079-6603(04)78003-4. [DOI] [PubMed] [Google Scholar]
- 33.Potamitou A, Holmgren A, Vlamis-Gardikas A. 2002. Protein levels of Escherichia coli thioredoxins and glutaredoxins and their relation to null mutants, growth phase, and function. J. Biol. Chem. 277:18561–18567. 10.1074/jbc.M201225200. [DOI] [PubMed] [Google Scholar]
- 34.Arenas FA, Leal CA, Pinto CA, Arenas-Salinas MA, Morales WA, Cornejo FA, Díaz-Vásquez WA, Vásquez CC. 2014. On the mechanism underlying tellurite reduction by Aeromonas caviae ST dihydrolipoamide dehydrogenase. Biochimie 102:174–182. 10.1016/j.biochi.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Vlamis-Gardikas A. 2008. The multiple functions of the thiol-based electron flow pathways of Escherichia coli: eternal concepts revisited. Biochim. Biophys. Acta 1780:1170–1200. 10.1016/j.bbagen.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Teng X, Liang X, Maksimuk S, Yang H. 2006. Synthesis of porous platinum nanoparticles. Small 2:249–253. 10.1002/smll.200500244. [DOI] [PubMed] [Google Scholar]
- 37.Zare B, Faramarzi MA, Sepehrizadeh Z, Shakibaie M, Rezaie S, Shahverdi AR. 2012. Biosynthesis and recovery of rod-shaped tellurium nanoparticles and their bactericidal activities. Mater. Res. Bull. 47:3719–3725. 10.1016/j.materresbull.2012.06.034. [DOI] [Google Scholar]
- 38.Mohanty A, Kathawala MH, Zhang J, Chen WN, Loo JSC, Kjelleberg S, Yang L, Cao B. 2014. Biogenic tellurium nanorods as a novel antivirulence agent inhibiting pyoverdine production in Pseudomonas aeruginosa. Biotechnol. Bioeng. 111:858–865. 10.1002/bit.25147. [DOI] [PubMed] [Google Scholar]
- 39.Pérez JM, Calderón IL, Arenas FA, Fuentes DE, Pradenas GA, Fuentes EL, Sandoval JM, Castro ME, Elías AO, Vásquez CC. 2007. Bacterial toxicity of potassium tellurite: unveiling an ancient enigma. PLoS One 2:e211. 10.1371/journal.pone.0000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.