ABSTRACT
Adenovirus type 5 E4orf4 is a multifunctional protein that regulates viral gene expression. The activities of E4orf4 are mainly mediated through binding to protein phosphatase 2A (PP2A). E4orf4 recruits target phosphoproteins into complexes with PP2A, resulting in dephosphorylation of host factors, such as SR splicing factors. In the current study, we utilized immunoprecipitation followed by mass spectrometry to identify novel E4orf4-interacting proteins. In this manner we identified Nup205, a component of the nuclear pore complex (NPC) as an E4orf4 interacting partner. The arginine-rich motif (ARM) of E4orf4 was required for interaction with Nup205 and for nuclear localization of E4orf4. ARMs are commonly found on viral nuclear proteins, and we observed that Nup205 interacts with three different nuclear viral proteins containing ARMs. E4orf4 formed a trimolecular complex containing both Nup205 and PP2A. Furthermore, Nup205 complexed with E4orf4 was hypophosphorylated, suggesting that the protein is specifically targeted for dephosphorylation. An adenovirus mutant that does not express E4orf4 (Orf4−) displayed elevated early and reduced late gene expression relative to that of the wild type. We observed that knockdown of Nup205 resulted in the same phenotype as that of the Orf4− virus, suggesting that the proteins function as a complex to regulate viral gene expression. Furthermore, knockdown of Nup205 resulted in a more than a 4-fold reduction in the replication of wild-type adenovirus. Our data show for first time that Ad5 E4orf4 interacts with and modifies the NPC and that Nup205-E4orf4 binding is required for normal regulation of viral gene expression and viral replication.
IMPORTANCE Nuclear pore complexes (NPCs) are highly regulated conduits in the nuclear membrane that control transport of macromolecules between the nucleus and cytoplasm. Viruses that replicate in the nucleus must negotiate the NPC during nuclear entry, and viral DNA, mRNA, and proteins must then be exported from the nucleus. Several types of viruses restructure the NPC to facilitate replication, and the current study shows that adenovirus type 5 (Ad5) utilizes a novel mechanism to modify NPC function. We demonstrate that a subunit of the NPC, Nup205, is a phosphoprotein that is actively dephosphorylated by the Ad5-encoded protein E4orf4. Moreover, Nup205 is required by Ad5 to regulate viral gene expression and efficient viral replication. Nup205 is a nonstructural subunit that is responsible for the gating functions of the NPC, and this study suggests for the first time that the NPC is regulated by phosphorylation both during normal physiology and viral infection.
INTRODUCTION
Early region 4 (E4) of human adenovirus encodes seven polypeptides that regulate a variety of biological functions during viral replication, including transcription, nuclear-cytoplasmic transport of viral mRNAs, apoptosis, cell cycle, DNA repair, and host cell shutoff (1–5). E4 open reading frame 4 (E4orf4) is conserved among all eight species of human adenoviruses. In adenovirus type 2 (Ad2) and Ad5, E4orf4 encodes a 114-amino-acid protein with no homology to eukaryotic proteins other than an arginine-rich nucleolar targeting sequence (6, 7). E4orf4 has no intrinsic enzymatic activity and likely derives all of its reported functions through interaction with the cellular serine/threonine protein phosphatase 2A (PP2A) (6, 8–16) or other as-yet-unidentified polypeptides. E4orf4 interacts with the PP2A regulatory subunit, Bα, and recruits target phosphoproteins into complexes with PP2A, resulting in dephosphorylation of host and viral proteins.
Early studies on E4orf4 demonstrated that it suppresses transcriptional activity of the AP1 transcription factor by inducing its dephosphorylation (17). In the same study, E4orf4 was also shown to cause dephosphorylation of E1A and downregulate the E4 promoter activity in a negative-feedback mechanism (17). E4orf4 can also downregulate the transcription of c-Myc in a PP2A-dependent manner (12). E4orf4 has also been shown to downregulate E2 transcription by inhibiting E1A-mediated activation of the E2 promoter (18). A recent study identified the ACF chromatin-remodeling factor as interacting partner of E4orf4 that is also involved in downregulation of early viral gene expression (8).
In addition to transcriptional regulation, the E4orf4-PP2A complex also regulates posttranscriptional processes (1, 19, 20). E4orf4 specifically interacts with hyperphosphorylated forms of the cellular SR (serine/arginine) proteins SF2/ASF and SRp30c and promotes PP2A-dependent dephosphorylation of these proteins (21). During the early phase of infection, the alternative splicing of the late message L1-IIIa pre-mRNA is repressed by the SF2/ASF and SRp30c, which bind to an intronic repressor element at the IIIa 3′ splice site. E4orf4 enhances the alternative splicing and production of the late viral mRNA L1-IIIa, thereby shifting the viral gene expression from early to late phase (1).
Expressed by itself, E4orf4 also has the intriguing ability to induce p53-independent death selectively in tumor cells (22–24). This effect has been demonstrated in a wide range of cancer cells and is also dependent upon the activity of PP2A (13, 14, 24–26). In both mammalian cells and Saccharomyces cerevisiae, E4orf4-expressing cells become arrested or at least stalled in G2/M, and a significant amount of them become tetraploid and polyploid (11, 27). Although the precise mechanism of E4orf4-dependent killing remains to be identified, it is believed that E4orf4 exerts its toxicity by inducing mitotic catastrophe in infected cells (11). E4orf4 can interact with the cellular anaphase-promoting complex/cyclosome (APC/C), an important protein complex responsible for the mitotic progression; it is believed that by recruiting PP2A into complex with APC/C, E4orf4 can induce the premature activation of the APC/Ccdc20 form of the APC/C complex and arrest cells at the G2/M stage (11, 27, 28). E4orf4-induced toxicity has been shown to be dose dependent and is believed to result from the inhibition of PP2A activity (10, 16). As the levels of E4orf4 required to induce toxicity are considerably higher than those found in adenovirus-infected cells, this E4orf4 effect is not believed to play a role in furthering viral infection.
Although the primary function of E4orf4 during infection is to target specific cellular phosphoproteins to PP2A, only a small number of such host factors have been identified. In the current study, we used a proteomic approach to identify novel E4orf4 interacting proteins. In this manner we found a subunit of the nuclear pore complex (NPC), Nup205, which interacts with E4orf4 and becomes dephosphorylated. We observe that Nup205 activity is required for proper viral gene expression and replication.
MATERIALS AND METHODS
Cell lines and viruses.
H1299 (ATCC CRL-5803) and HeLa (ATCC CCL-2) cell lines were maintained in Dulbecco's modifed Eagle's medium (DMEM; Wisent Inc.), supplemented with 10% (vol/vol) fetal bovine serum (HyClone), 100 U/ml of penicillin, and 100 mg/ml of streptomycin (Wisent Inc.) at 37°C under 5% CO2.
Wild-type (wt) Ad5 (H5pg4100), Orf4− virus, and FLAG-tagged E4orf4 adenovirus (Ad-FLAG-E4orf4) have been previously described (6, 29, 30). LacZ adenovirus (Ad-LacZ) was previously described (31). All viruses were amplified in 293 cells and purified by ultracentrifugation on a cesium chloride gradient, and titers were determined using the fluorescence-forming units (FFU) method (29).
Plasmids and transfection.
To generate FLAG-tagged Nup205, the open reading frame (gift from Douglass Forbes, UCSD) was amplified using PCR with the addition of flanking NotI and BamHI sites. The resulting product was ligated into the plasmid p3XFlag-myc-CMV-26 (Sigma). Similarly, Nup205 truncation mutants were generated by PCR from the full-length template. HA-E4orf4 and all E4orf4 mutants have been described previously (14). The green fluorescent protein (GFP) fusion of E4orf4 has been described previously (7), as has been the GFP fusion with HIV Rev (32). Myc-PK was previously described in reference 33 and was a gift of Gideon Dreyfuss, and myc-PK-E4ARM was previously described in reference 7. FLAG-PP2A-B55α was previously described in reference 14. V5-PP2A-B55α was generated by amplifying the B55α open reading frame using PCR with the addition of flanking KpnI (5′) and EcoRI (3′) sites and subsequent ligation into pcDNA4/V5-His A (Invitrogen). The N-terminal hemagglutinin (HA)-tagged and C-terminal GFP-tagged large T antigen (LgT) constructs were made by amplifying the large T DNA sequence using PCR and ligating it into the vectors pCDNA3 HA (Invitrogen) and pEGFP-C1 (Clonetech), respectively. The source of the simian virus 40 (SV40) sequence was pBABE-zeo largeT (gift from Robert Weinberg). All constructs were confirmed by DNA sequencing.
Cells were grown in 35-mm (for immunofluorescence) or 100-mm (for immunoprecipitation) dishes to about 70% confluence and transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols.
siRNA transfections.
Cells were grown in 24-well, 12-well, 35-mm or 100-mm dishes to about 50% confluence and transfected with Lipofectamine 2000 (Life Technologies) to deliver small interfering RNAs (siRNAs) according to the manufacturer's protocols. Experiments were conducted after 42 h posttransfection. siRNA duplexes used to knockdown Nup205 were as follows: Nup205 siRNA 1, 5′-CUGCGUCAGUUUAAAUUUCAA-3′ (Qiagen; SI02665257); Nup205 siRNA 2, 5′-CUGACAGGAAUUAUAAGUAAA-3′ (Qiagen; SI02665264); and nonsilencing control, 5′-AAUUCUCCGAACGUGUCACGU-3′ (Qiagen; 1022076).
Antibodies, immunoprecipitation, and Western blotting.
Affinity purification of E4orf4 binding proteins was performed exactly as previously described (34). Rabbit anti-FLAG followed and protein G beads (Sigma) were used to immunoprecipitate FLAG-tagged Nup205 in the Nup205-E4orf4-PP2A-Bα tri-molecular complex experiment. EZview Red anti-FLAG M2 affinity gel (Sigma; M2) was used to pull down FLAG-tagged proteins in all other immunoprecipitation (IP) experiments.
Whole-cell extracts were prepared by lysing cells in buffer X (50 mM Tris [pH 8.5], 250 mM NaCl, 1 mM EDTA, 1% NP-40, protease inhibitor minitablet [Roche]). Equal amounts of protein (lysate or immunoprecipitation samples) were separated by SDS-PAGE and transferred to 0.45-μm nitrocellulose membranes (Bio-Rad). Membranes were incubated with primary antibodies followed by appropriate horseradish peroxidase-coupled secondary antibodies (anti-rabbit or anti-mouse from Jackson ImmunoResearch or TrueBlot anti-mouse from eBioscience). Western Lightning Plus enhanced chemiluminescence substrate (PerkinElmer) was used to visualize proteins on autoradiography film.
E1A was detected with M73 monoclonal antibody (35) collected from ascites fluid. The anti-E4orf6 rabbit polyclonal antibody 1807 was described in reference 36, anti-E4orf4 rabbit polyclonal antibody 2419 was described in reference 24, and anti-Ad capsid polyclonal antibody L133 was described in reference 37 (gift from T. Dobner). Anti-Nup205 rabbit polyclonal was a gift from Douglass Forbes. Antitubulin monoclonal antibody was clone B-7 (Santa Cruz Biotechnology).
In vivo labeling.
Two million H1299 cells were seeded on 10-cm dishes 24 h prior to transfection. Cells were transfected with 6 μg of FLAG-tagged Nup205 and 4 μg of wt HA-E4orf4 together using Lipofectamine 2000 (Life Technologies) in 7.5 ml of Opti-MEM. The medium was changed 4 h after transfection and then again at 24 h posttransfection. Forty-eight hours posttransfection, the medium was changed to phosphate-free DMEM for 15 min. The medium then was changed for medium containing [32P]orthophosphate (PerkinElmer) at a concentration of 250 μCi/ml for 1 h at 37°C. The cells were then washed twice with cold phosphate-buffered saline (PBS), centrifuged, and then lysed in 1 ml of cold RIPA buffer (50 mm HEPES [pH 7.5], 150 mm NaCl, 1% Triton X-100, 0.1% SDS, and 0.2% sodium deoxycholate plus 1 complete miniprotease tablet and 1 PhosSTOP phosphatase inhibitor tablet per 10 ml [Roche]). The cells were lysed on ice for 30 min and then centrifuged for 15 min at 14,000 × g at 4°C. The lysate was then precleared with 10 μl of protein A agarose beads for 30 min. Finally, the lysate was incubated with 10 μl of anti-FLAG affinity agarose for 2 h on a rotating platform at 4°C. The beads were then washed 6 times with RIPA buffer. Bound proteins were eluted by boiling in 1× Laemmli buffer for 5 min. Half of the sample was resolved on a 6% SDS-PAGE gel and then dried and exposed to film.
Viral replication assays.
To measure the replication ability of adenovirus in H1299 cells with or without Nup205 knockdown, cells were infected with adenovirus at a multiplicity of infection (MOI) of 5 and harvested at 42 h postinfection (p.i.) by scraping. They were then lysed by three cycles of freeze-thaw, and the cell lysates were serially diluted in medium for infection of H1299 cells. Replication was measured by the virus yield as determined using a FFU assay at 40 h after infection (29, 38).
Fluorescence microscopy.
HeLa cells were cultured on glass coverslips in 6-well plates and were ∼60% confluent at the time of transfection with GFP fusion plasmids. Constructs were expressed for no more than 14 h and fixed with methanol (except for human immunodeficiency virus [HIV] Rev constructs). For GFP-Rev, fixation was performed with 3.2% formaldehyde for 5 min. Coverslips were counterstained with DAPI (4′,6-diamidino-2-phenylindole) and mounted on slides using DABCO (Sigma). Cells were then viewed using a Zeiss Axiovision 3.1 microscope equipped with Axiocam HR (Zeiss, Thornwood, NY) digital camera.
qPCR.
RNA was isolated from cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. DNase treatment and cDNA synthesis were performed with the QuantiTect reverse transcription kit (Qiagen) using 1 μg of RNA as a template according to the manufacturer's protocol. Quantitative real-time PCR (qPCR) was performed using a Realplex-2 Mastercycler (Eppendorf) and QuantiFast SYBR green master mix (Qiagen) supplemented with 1 μg of cDNA. Fold changes of viral transcripts were calculated using the threshold cycle (ΔΔCT) method relative to the housekeeping gene 18S rRNA. Primer sequences used for PCR were as follows: E1A forward primer, 5′-GTGCCCCATTAAACCAGTTG-3′; E1A reverse primer, 5′-GGCGTTTACAGCTCAAGTCC-3′; E4orf6 forward primer, 5′-GCTGGTTTAGGATGGTGGTG-3′; E4orf6 reverse primer, 5′-CCCTCATAAACACGCTGGAC-3′; tripartite leader forward primer, 5′-CGCTGTCTGCGAGGGCCAG-3′; tripartite leader reverse primer, 5′-GGCGGCGGAGTACCGTTCG-3′; 18S forward primer, 5′-CGGCTACCACATCCAAGGAA-3′; and 18S reverse primer, 5′-GCTGGAATTACCGCGGCT-3′.
RESULTS
Identification of novel E4orf4-interacting proteins.
To identify interacting partners of E4orf4, H1299 cells were infected with adenoviral vectors expressing FLAG-tagged E4orf4 (Ad-FLAG-E4orf4) (6) or control β-galactosidase (Ad-LacZ) (31). Immunoprecipitation (IP) using an anti-FLAG monoclonal antibody was used to purify E4orf4 complexes from cell extracts. Figure 1 shows typical silver-stained SDS-PAGE of immunoprecipitates obtained. Major bands on the gels were excised and submitted for analysis by mass spectrometry. In total, 10 proteins were identified in complexes with E4orf4. One of the proteins identified was serine/arginine splicing factor 1 (SRSF1, also known as ASF/SF2), which has been previously reported as an E4orf4-associated protein (20, 21). We also identified 9 other novel proteins interacting with E4orf4, including DNA-PK, Nup205, poly(A) binding protein, importin-β, iASPP, Hsp 70, p68 DEAD Box RNA helicase, ribophorin I, and α/β tubulin. The observation that E4orf4 interacts with Nup205, a subunit of the nuclear pore complex (NPC), was particularly intriguing considering that interactions with the NPC have been shown to regulate gene expression in many diverse types of virus (reviewed in reference 39). Further studies were therefore performed to characterize the E4orf4-Nup205 interaction and its potential role in viral replication.
FIG 1.
Identification of E4orf4 binding proteins. (A) Silver-stained SDS-PAGE (8% polyacrylamide) of immunoprecipitated FLAG-E4orf4 complexes from H1299 cells infected with Ad-E4orf4. Cells were infected with Ad-LacZ as a negative control. (B) 15% polyacrylamide silver-stained gel of the same samples as in panel A. The position of E4orf4 is indicated.
The ARM of E4orf4 is required for interaction with Nup205.
In order to determine the binding site for Nup205 on E4orf4, a series of previously characterized point mutants along the length of E4orf4 were utilized (14). Deletion mutations of E4or4 render the protein highly labile and necessitate the use of discrete point mutations for structure/function studies. In a previous study, these E4orf4 mutants were characterized for ability to bind to PP2A and activity for inducing cell death (14). Phenotypes of the E4orf4 mutants fell into two classes. Class I mutants were defective for interactions with PP2A and cell killing activity, and class II mutants interacted with PP2A but had disrupted killing activity. A subset of class I and II mutants were tested for interaction with Nup205 by cotransfecting wild-type or mutant HA-E4orf4 and FLAG-Nup205, followed by immunoprecipitation using anti-FLAG antibody. Figure 2A shows that only E4orf4 point mutations of the arginine-rich motif (ARM) between residues 69 and 75 (R69/70/72–75A) disrupted binding with Nup205. We then determined if the ARM of E4orf4 was sufficient for interaction with Nup205. Figure 2B shows that fusion of the ARM of E4orf4 to pyruvate kinase (PK) conferred Nup205 binding activity on the resulting fusion protein.
FIG 2.
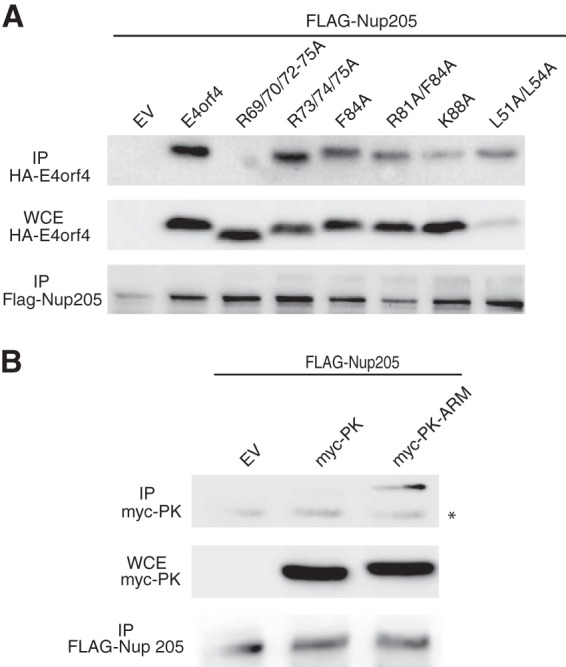
The arginine-rich motif (ARM) of E4orf4 is required for interaction with Nup205. (A) FLAG-Nup205 was cotransfected in H1299 cells with wild-type or the indicated point mutants of E4orf4. Immunoprecipitation (IP) from cell extracts was performed using anti-FLAG antibody. IP complexes were analyzed by Western blotting to detect E4orf4 (anti-HA) and Nup205 (anti-FLAG). Anti-HA Western blotting of whole-cell extract (WCE) was used to confirm expression of FLAG-Nup205 (middle portion). (B) H1299 cells were cotransfected with FLAG-Nup205 and empty vector (EV), myc-pyruvate kinase (myc-PK), or myc-PK fused to the E4orf4 ARM (myc-PK-ARM). IP from cell extracts was performed using anti-FLAG. IP complexes were analyzed by Western blotting to detect myc-PK and myc-PK-ARM (anti-myc). myc immunoblotting of WCE is used to confirm expression of PK fusions. The asterisks indicates a background band with anti-myc antibody.
The ARM is required for nuclear localization of E4orf4 and other nuclear viral proteins.
The observation that the ARM of E4orf4 was necessary and sufficient for E4orf4 interaction with Nup205 is intriguing considering that this sequence is also required to localize the protein to the nucleus (7). Several nuclear viral proteins also contain an ARM. For example, apoptin from chicken anemia virus (CAV Ap) and Rev from human immunodeficiency virus (HIV) are nuclear viral proteins that contain ARMs.
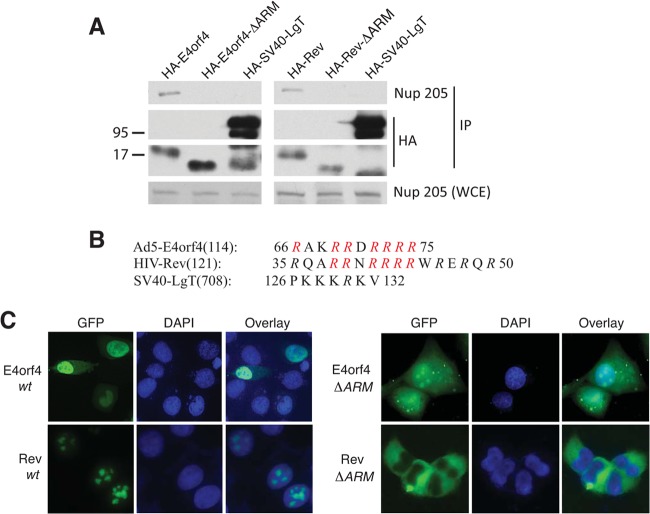
Figure 3A shows that HIV Rev is also able to interact with endogenous Nup205 and point mutation of the ARM (HA-Rev-ΔARM) abolished binding as observed with E4orf4. As a negative control, we tested SV40 large T antigen (LgT), a nuclear viral protein that utilizes a conventional nuclear localization signal (NLS), and showed that this protein does not bind Nup205. Point mutation of the ARM of both HIV Rev and E4orf4 abolished nuclear localization, indicating that interaction with Nup205 plays a role in nuclear import of these proteins (Fig. 3C).
FIG 3.
Nup205 associates with HIV Rev via the ARM domain and is required for nuclear localization. (A) H1299 cells were transfected with HA-E4orf4 or HA-Rev or mutants that lack the ARM (ΔARM). HA-SV40-LgT is included as a negative control. Immunoprecipitation from cell extracts was performed using anti-HA. IP complexes were analyzed by Western blotting to detect endogenous Nup205 and HA-tagged viral proteins. Anti-Nup205 Western blotting of whole-cell extract was used to confirm expression of Nup205 (lower portion). (B) Schematic showing the ARMs of E4orf4 and HIV Rev. The nuclear localization sequence of SV40 is shown for comparison. Mutated residues in the ΔARM mutants are indicated in red. (C) Epifluorescence microscopy showing localization of EGFP-fused E4orf4, Rev, and ΔARM mutants.
A common region on Nup205 is required for interaction with viral ARMs.
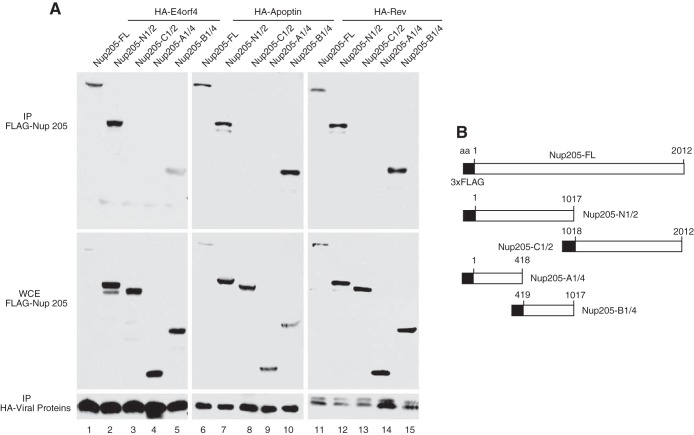
A series of deletion mutants of Nup205 was constructed to determine the binding site of E4orf4. A schematic of the FLAG-tagged Nup205 constructs is shown in Fig. 4B. Figure 4A shows that a central region from amino acids 419 to 1017 of Nup205 was minimally required to interact with E4orf4 (lane 5). Interestingly, the same region was also required to interact with the two other ARM-containing viral proteins, CAV apoptin (lane 10) and HIV Rev (lane 15). These data show that the E4orf4 and other ARM-containing proteins bind to Nup205 via the same region in the protein. Amino acid sequences rich in arginine are highly positively charged and can potentially interact with a suitably negatively charged region on Nup205. Analysis of the amino acid sequence of Nup205 between residues 419 and 1017 did not reveal any stretch of negatively charged amino acids, suggesting that interaction with the ARM may be a structural property of Nup205 rather than the primary amino acid sequence.
FIG 4.
ARM-containing viral proteins associated with the same domain on Nup205. (A) H1299 cells were cotransfected with wild-type or deletion mutants of FLAG-Nup205 and HA-tagged E4orf4, HA-apoptin, or HIV Rev. Immunoprecipitation from cell extracts was performed using anti-HA. IP complexes were analyzed by Western blotting to detect Nup205 and deletion mutants (anti-FLAG) and HA-tagged proteins (lower portion). Anti-FLAG Western blot of whole-cell extract (WCE) was used to confirm expression of FLAG-Nup205 and deletion mutants (middle portion). (B) Schematic of FLAG-Nup205 deletion constructs.
Nup205 is required for the nuclear localization of HIV Rev but not E4orf4.
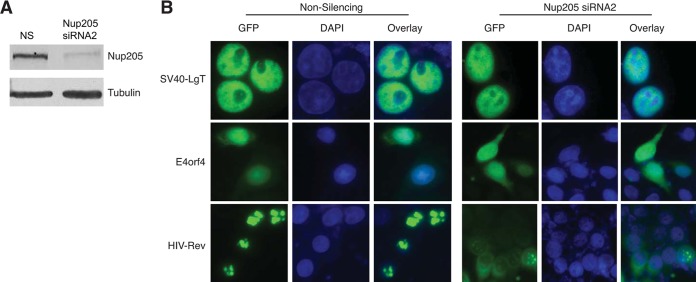
Having identified that the ARM is required for E4orf4 to interact with Nup205 and for E4orf4 to localize into the nucleus, we tested whether the nuclear localization was dependent upon Nup205. RNAi was used to knockdown expression of Nup205 in HeLa cells. The Western blot in Fig. 5A shows that siRNA targeting of Nup205 was able to silence protein expression very efficiently relative to a nonsilencing control siRNA. Enhanced GFP (EGFP)-tagged versions of E4orf4, HIV Rev, and LgT were then transfected into Nup205 knockdown and control cells. Figure 5B shows that knockdown of Nup205 resulted in only a slight increase in cytoplasmic levels of E4orf4. In contrast, nuclear localization of HIV Rev was mostly inhibited after depletion of Nup205. LgT, which includes a canonical NLS sequence, was not affected by Nup205 knockdown. These data suggest that although Nup205 may play a minor role in E4orf4 nuclear localization, other factors must be primarily involved, since only a modest localization defect was observed after Nup205 knockdown.
FIG 5.
Nup205 is required for nuclear localization of HIV Rev but not E4orf4. (A) Western blot demonstrating knockdown of Nup205 in HeLa cells. A tubulin blot is included as loading control. (B) Epifluorescence microscopy showing localization of EGFP-fused SV40-LgT, E4orf4, and HIV Rev in HeLa cells after transfection with nonsilencing or Nup205 siRNAs.
E4orf4 forms a trimolecular complex with PP2A-Bα and Nup 205.
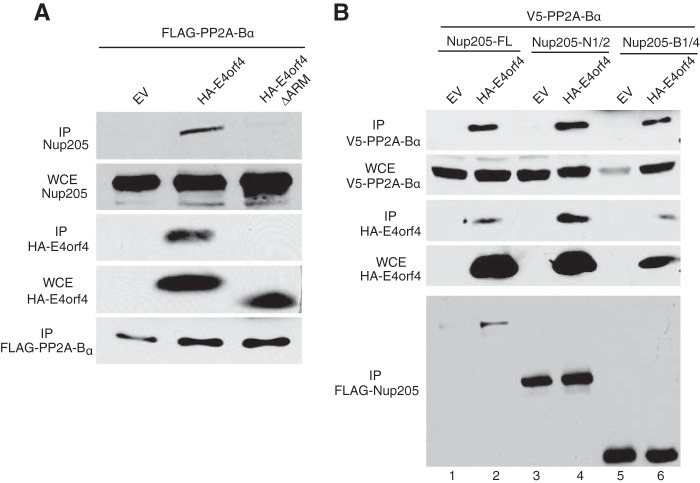
E4orf4 itself has no intrinsic enzymatic activity, and most of its functions are mediated through interaction with PP2A. E4orf4 can recruit target phosphoproteins into complexes with PP2A, resulting in dephosphorylation of host and viral proteins such as the SR splicing factors and the AP1 transcription factor (16, 21, 40). We therefore determined if E4orf4 could form a trimolecular complex with Nup205 and PP2A as was observed with other targets. A cDNA expressing a FLAG-tagged PP2A-Bα was cotransfected with those expressing HA-E4orf4 or a mutant form lacking the ARM (HA-E4orf4ΔARM). As shown in Fig. 6A, endogenous Nup205 coimmunoprecipitated with PP2A-Bα only in the presence of wt E4orf4, indicating that the three proteins exist as a trimolecular complex. Performing the inverse experiment, transfected FLAG-Nup205 also coimmunoprecipitated PP2A-Bα, but again, only in the presence of wt E4orf4 (Fig. 6B, lanes 1 and 2). As expected, even the deletion mutants of Nup205 that maintained binding to E4orf4 could also coimmunoprecipitate PP2A-Bα with E4orf4 (Fig. 6B, lanes 3 to 6). Taken together, these data show that E4orf4 is able to bring PP2A into a complex with the NPC through Nup205.
FIG 6.
E4orf4 forms a trimolecular complex with PP2A-Bα and Nup205. (A) H1299 cells were cotransfected with FLAG-PP2A-Bα and wild-type HA-E4orf4, ΔARM mutant, or empty vector. Immunoprecipitation from cell extracts was performed using anti-FLAG to pull down FLAG-PP2A-Bα. IP complexes were analyzed by Western blotting to detect endogenous Nup205 (top), HA-E4orf4, and FLAG-PP2A-Bα. Western blotting of whole-cell extracts was used to confirm expression of endogenous Nup205 and HA-E4orf4. (B) H1299 cells were cotransfected with V5-PP2A-Bα, wild-type HA-E4orf4, or EV and FLAG-Nup205 or the indicated deletion mutants. IP from cell extracts was performed to using anti-FLAG to pull down FLAG-Nup205. IP complexes were analyzed by Western blotting to detect V5-PP2A-Bα (top), HA-E4orf4, and FLAG-Nup205 (bottom). Anti-HA Western blot of WCE is shown to confirm expression of E4orf4 and anti-V5 to confirm expression of PP2A-Bα.
E4orf4 modulates the phosphorylation state of Nup205.
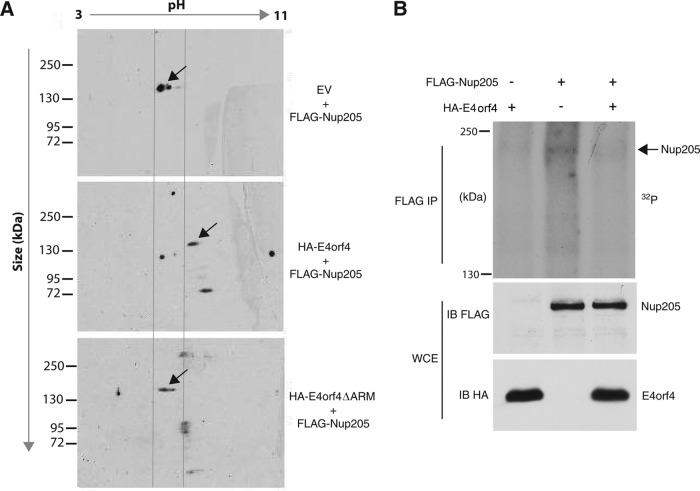
The above-described results show that E4orf4 may function by bringing PP2A into a complex with Nup205. This mechanism is similar to how E4orf4 modulates the splicing of viral late mRNAs by inducing PP2A-dependent dephosphorylation of the splicing factor ASF/SF2 (20). We therefore determined whether E4orf4 could similarly modulate the phosphorylation state of Nup205. There have been no studies examining phosphorylation of Nup205, although serine/threonine phosphorylation other NPC components has been shown to regulate nuclear envelope breakdown at mitosis (41). The bioinformatics tool NetPhos 2.0 predicts 48 serine and 16 threonine putative phosphorylation sites on Nup205. Two-dimensional gel electrophoresis was performed to analyze Nup205 in the presence or absence of E4orf4. FLAG-Nup205 was cotransfected in H1299 cells with either wt E4orf4, E4orf4ΔARM, or a vector control. Figure 7A shows that in the presence of wt E4orf4, the migration of Nup205 in the first dimension (pH) migrated with a higher isoelectric point (pI) relative to that of the vector control or the E4orf4ΔARM mutant. Since phosphorylated proteins generally have a lower pI value due to the negatively charged phosphate groups, a lower pI value suggests that the protein is hyperphosphorylated. Therefore, the increased pI value of Nup205 in the presence of wt E4orf4 suggests that the viral protein is mediating the dephosphorylation of Nup205.
FIG 7.
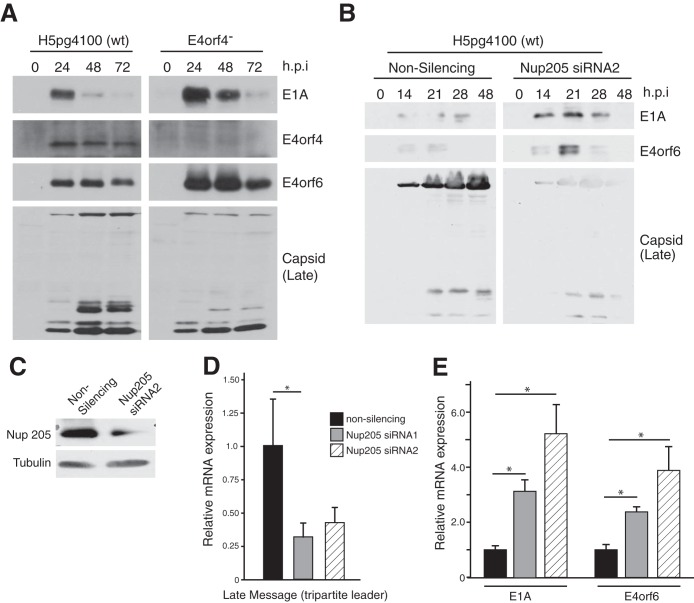
E4orf4 modulates the phosphorylation state of Nup205. (A) H1299 cells were cotransfected with FLAG-Nup205 and empty vector, HA-E4orf4, or HA-E4orf4ΔARM. Immunoprecipitation from cell extracts was performed using anti-FLAG. IP complexes were analyzed by Western blotting (anti-FLAG) after 2-dimensional gel electrophoresis to detect FLAG-Nup205. Isoelectric focusing was performed in the x axis on a pH gradient from 3 to 11, followed by size separation on the y axis. Arrows indicate the position of FLAG-Nup205, which migrates at approximately 200 kDa. (B) H1299 cells were transfected with HA-E4orf4, FLAG-Nup205, or both. Forty-eight hours posttransfection, cells were labeled with [32P]orthophosphate and immunoprecipitation was performed on cell extracts using anti-FLAG. Immunoprecipitates were resolved on SDS-PAGE to detect 32P-labeled proteins (top). Western blotting was performed on whole-cell extracts to confirm expression of Nup205 (anti-FLAG) and E4orf4 (anti-HA).
As a more direct demonstration that E4orf4 can modulate the phosphorylation state of Nup205, we performed metabolic labeling using [32P]orthophosphate in H1299 cells cotransfected with E4orf4 or an empty vector control. Figure 7B, lane 2, shows that FLAG-Nup205 immunoprecipitated from 32P-labeled cell extracts migrated on SDS-PAGE as a major band at approximately 200 kDa, the correct molecular mass of Nup205. Furthermore, in the presence of E4orf4 the intensity of the band is reduced considerably (Fig. 6B, lane 2). These data show for the first time that Nup205 exists as a phosphoprotein and that E4orf4 can reduce the phosphorylation of Nup205.
E4orf4 and Nup205 regulate viral gene expression.
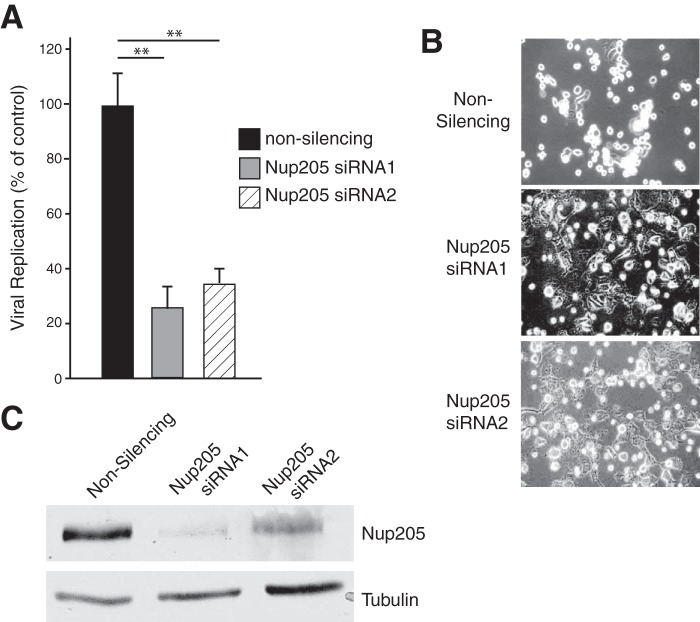
Previous studies have demonstrated that E4orf4 regulates viral gene expression during infection primarily by downregulation of early promoters. Figure 8A shows that a viral mutant that lacks expression of E4orf4 displays elevated expression of early protein production (E1A and E4orf6), consistent with previous studies (8, 17, 18). In contrast, we observe that late protein production as measured by Western blot detection of capsid proteins is markedly reduced in the Orf4− virus relative to the wt. In order to determine if Nup205 plays a role in E4or4 gene regulation, Nup205 mRNA expression was knocked down in H1299 cells and subsequently infected with wt virus. Figure 8B shows that cells with Nup205 knockdown displayed a pattern of gene expression similar to that of the E4orf4− virus, with elevated early gene expression and low late gene expression. The efficiency of Nup205 knockdown is shown in Fig. 8C. In order to determine if the alterations in viral gene expression were at the level of mRNA, real-time RT-PCR analysis was performed on E1A and E4orf6 at 7.5 h postinfection. Late viral gene expression was assessed at 23 h postinfection by analyzing the abundance of the tripartite leader containing mRNAs. We observed that late mRNAs were reduced relative to the nonsilencing control (Fig. 8D), whereas early viral mRNAs were elevated (Fig. 8E), which is in agreement with results obtained by Western blot analysis. The similar phenotypes of E4orf4− virus and cells with knockdown of Nup205 suggest that these proteins act as a complex to regulate gene viral expression at the mRNA level.
FIG 8.
E4orf4 and Nup205 regulate viral gene expression. (A) Western blot showing expression of indicated viral proteins at various hours postinfection (h.p.i) in H1299 cells infected with wild-type (H5pg400) Ad5 or the E4orf4− virus. (B) Western blot showing expression of viral proteins at indicated h.p.i. in H1299 cells transfected with nonsilencing or siRNA targeting Nup205. (C) Western blot demonstrating knockdown of Nup205 in H1299 cells. A tubulin blot is included as loading control. (D) Quantitative RT-PCR of late viral gene expression (mRNAs with tripartite leader) at 23 hours postinfection in H1299 cells transfected with nonsilencing or siRNAs targeting Nup205. (E) qRT-PCR of early gene expression (E1A and E4orf6) at 7.5 hours postinfection.
Knockdown of Nup205 impairs viral replication and delays the cytopathic effect.
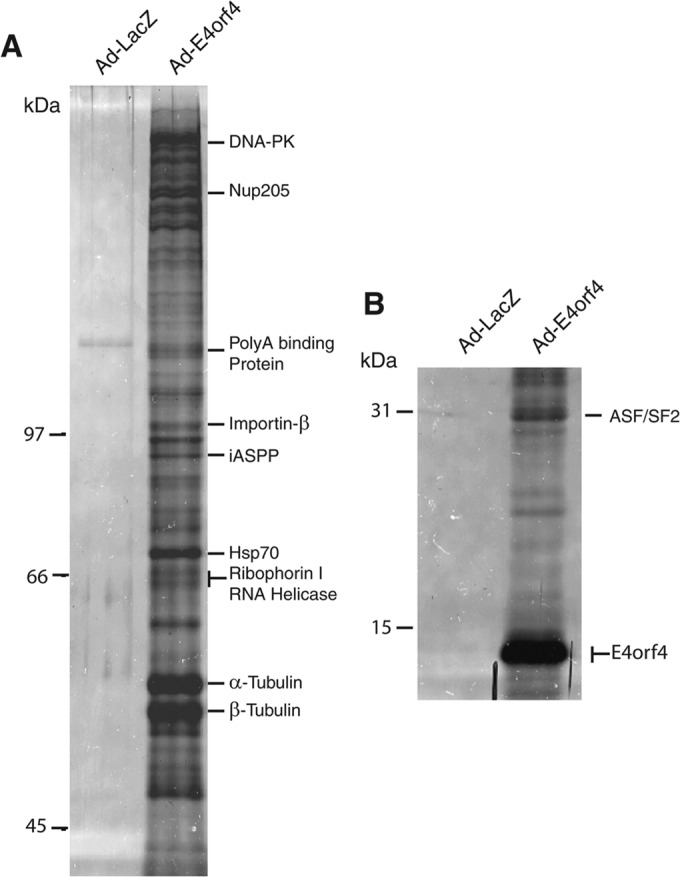
Since Nup205 was required for proper viral gene expression, we then determined if it is also required for effective viral replication. Replication of wt Ad5 was measured in H1299 cells following siRNA knockdown of Nup205 or a nonsilencing control. Virus yields were quantitatively determined using the FFU assay (29, 38). H1299 cells with knockdown of Nup205 did not exhibit major deleterious effects and appeared similar to nonsilencing control cells during the course of the experiment (data not shown). However, analysis of viral replication in Fig. 9A showed that knockdown of Nup205 using two different siRNA duplexes resulted in replication of 30% and 35% relative to the nonsilencing control. In addition, 30 h postinfection, most of the H1299 cells treated with nonsilencing siRNA displayed a rounded and swollen appearance characteristic of viral cytopathic effect (CPE), whereas in cells treated with the two Nup205 siRNAs, only half had such morphology (Fig. 9B). The efficiency of Nup205 knockdown is shown in Fig. 9C. Taken together, these data show that adenovirus replication is significantly compromised in cells with reduced levels of Nup205.
FIG 9.
Knockdown of Nup205 impairs viral replication and delays the cytopathic effect. (A) Replication of wild-type Ad5 (H5pg400) in H1299 cells transfected with nonsilencing or two separate siRNAs targeting Nup205. (B) Light micrographs showing appearance of cells infected with wild-type Ad4 under conditions of Nup205 knockdown or control. (C) Western blot demonstrating knockdown of Nup205 in H1299 cells. A tubulin blot is included as a loading control.
DISCUSSION
Many types of viruses, including adenoviruses, interact with the NPC in order to facilitate entry into the nucleus during initial infection (reviewed in reference 39). Our study has demonstrated for the first time a mechanism by which adenoviral proteins interact with the nuclear pore complex during viral replication to regulate viral gene expression. We propose a mechanism by which the E4orf4 protein recruits the PP2A phosphatase into a trimolecular complex with Nup205 and thereby causes dephosphorylation of the protein and alters its function. The NPC is an enormous complex (approximately 60 MDa) composed of about 30 distinct nucleoporins (Nups) known to be extensively regulated by phosphorylation. For example, phosphorylation of NPC components during mitosis mediates the disassembly of the NPC (41).
The function of Nup205 within the NPC remains poorly understood. The Nups can be broadly categorized as being components of the outer structural skeleton of the NPC or the central pore that mediates import/export of cargoes across the nuclear membrane. Nup205 is one of the components of the central pore that is responsible for the gating functions of the NPC and is not required for formation or integrity of the NPC (42). Interestingly, knockdown of Nup205 in Caenorhabditis elegans was shown to increase the size limit of nonnuclear macromolecules that could passively enter the nucleus (43). These changes occurred without affecting the active transport of proteins into the nucleus. Therefore, E4orf4 interactions with Nup205 may alter the dynamics of viral and/or host cell proteins across the nuclear membrane as a means to enhance replication. In late stages of adenovirus infection, export of viral mRNAs from the nucleus is enhanced, whereas host mRNAs are blocked through an unknown mechanism (reviewed in references 3 and 44). Since we observed that Nup205 knockdown caused reduced late gene expression but appeared to enhance early gene expression, E4orf4 targeting of Nup205 may provide a molecular explanation for the pattern of adenovirus gene expression during late phases.
E4orf4 stimulated the dephosphorylation of Nup205 as it does other cellular targets, such as SR splicing factors, which suggests that the activity of Nup205 is itself regulated by phosphorylation under normal physiological conditions. However, no study to date has mapped phosphorylation sites on Nup205 or determined how these modifications may affect activities such as transcription and mRNA transport. Judging from the large number of potential phosphorylation sites that are present on Nup205, it is likely that the protein is dynamically regulated by signaling and/or cell cycle-dependent kinases. Further studies into the phosphorylation of Nup205 may therefore provide additional insights into the regulation of gene expression in normal physiology as well as during viral infection. In addition to inducing dephosphorylation of Nup205, another possible function for the E4orf4-Nup205 interaction may be to localize PP2A to the nuclear pore, where it may, in turn, dephosphorylate host factors as they are transported. For example, the phosphorylation states of E1A, SR proteins, myc, and AP1 have all been shown to be reduced in the presence of E4orf4 (8, 17, 21). E4orf4 complexes associated with Nup205 may therefore dephosphorylate such proteins as they are transported through the NPC.
Our results suggest that activity of Nup205 is required for adenoviral replication. Interestingly, Nup205 may be required for replication of other viruses. For example, a recent study describing a genome-wide screen for host factors required for replication of influenza virus also identified Nup205 (45). In addition, our data also suggest that nuclear localization of HIV Rev is dependent on Nup205. It would be interesting to determine if Nup205 knockdown also has inhibitory effects on HIV replication as it does with adenovirus and influenza virus. In addition to adenoviruses, several other virus types have been shown to alter the properties of the NPC during the course of replication (reviewed in reference 39). For example, herpesvirus (46), picornavirus (47–49), and HIV (50) have all been shown to induce reorganization of NPC subunits without affecting the overall integrity of the pore. Although these viral interactions with the NPC differ greatly in their specific mechanisms, they may act as conserved strategies to control the transport of macromolecules during replication and regulate both viral and cellular gene expression.
ACKNOWLEDGMENTS
We thank Sabrina Schreiner for insightful discussions and Bob Weinberg, Thomas Dobner, and Douglass Forbes for reagents.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR), MOP-179122 (J.G.T.), MOP-93753 (P.E.B. and J.G.T.), and the Natural Science and Engineering Research Council of Canada (J.G.T.). T.J.K. was supported by a doctoral studentship from the CIHR. Y.L. was supported by a studentship from the FRSQ.
Footnotes
Published ahead of print 10 September 2014
REFERENCES
- 1.Täuber B, Dobner T. 2001. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene 278:1–23. 10.1016/S0378-1119(01)00722-3. [DOI] [PubMed] [Google Scholar]
- 2.Imperiale MJ, Akusjnarvi G, Leppard KN. 1995. Post-transcriptional control of adenovirus gene expression. Curr. Top. Microbiol. Immunol. 199(Part 2):139–171. [DOI] [PubMed] [Google Scholar]
- 3.Dobner T, Kzhyshkowska J. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25–54. 10.1007/978-3-642-56597-7_2. [DOI] [PubMed] [Google Scholar]
- 4.Leppard KN. 1997. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J. Gen. Virol. 78(Part 9):2131–2138. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg DH, Ketner G. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miron MJ, Blanchette P, Groitl P, Dallaire F, Teodoro JG, Li S, Dobner T, Branton PE. 2009. Localization and importance of the adenovirus E4orf4 protein during lytic infection. J. Virol. 83:1689–1699. 10.1128/JVI.01703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miron MJ, Gallouzi IE, Lavoie JN, Branton PE. 2004. Nuclear localization of the adenovirus E4orf4 protein is mediated through an arginine-rich motif and correlates with cell death. Oncogene 23:7458–7468. 10.1038/sj.onc.1207919. [DOI] [PubMed] [Google Scholar]
- 8.Brestovitsky A, Sharf R, Mittelman K, Kleinberger T. 2011. The adenovirus E4orf4 protein targets PP2A to the ACF chromatin-remodeling factor and induces cell death through regulation of SNF2h-containing complexes. Nucleic Acids Res. 39:6414–6427. 10.1093/nar/gkr231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Mui MZ, Chan F, Roopchand DE, Marcellus RC, Blanchette P, Li S, Berghuis AM, Branton PE. 2011. Genetic analysis of B55alpha/Cdc55 protein phosphatase 2A subunits: association with the adenovirus E4orf4 protein. J. Virol. 85:286–295. 10.1128/JVI.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Brignole C, Marcellus R, Thirlwell S, Binda O, McQuoid MJ, Ashby D, Chan H, Zhang Z, Miron MJ, Pallas DC, Branton PE. 2009. The adenovirus E4orf4 protein induces G2/M arrest and cell death by blocking protein phosphatase 2A activity regulated by the B55 subunit. J. Virol. 83:8340–8352. 10.1128/JVI.00711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Szymborski A, Miron MJ, Marcellus R, Binda O, Lavoie JN, Branton PE. 2009. The adenovirus E4orf4 protein induces growth arrest and mitotic catastrophe in H1299 human lung carcinoma cells. Oncogene 28:390–400. 10.1038/onc.2008.393. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Israel H, Sharf R, Rechavi G, Kleinberger T. 2008. Adenovirus E4orf4 protein downregulates MYC expression through interaction with the PP2A-B55 subunit. J. Virol. 82:9381–9388. 10.1128/JVI.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shtrichman R, Sharf R, Kleinberger T. 2000. Adenovirus E4orf4 protein interacts with both Balpha and B' subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Balpha. Oncogene 19:3757–3765. 10.1038/sj.onc.1203705. [DOI] [PubMed] [Google Scholar]
- 14.Marcellus RC, Chan H, Paquette D, Thirlwell S, Boivin D, Branton PE. 2000. Induction of p53-independent apoptosis by the adenovirus E4orf4 protein requires binding to the Balpha subunit of protein phosphatase 2A. J. Virol. 74:7869–7877. 10.1128/JVI.74.17.7869-7877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinberger T, Shenk T. 1993. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J. Virol. 67:7556–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mui MZ, Kucharski M, Miron MJ, Hur WS, Berghuis AM, Blanchette P, Branton PE. 2013. Identification of the adenovirus E4orf4 protein binding site on the B55alpha and Cdc55 regulatory subunits of PP2A: Implications for PP2A function, tumor cell killing and viral replication. PLoS Pathog. 9:e1003742. 10.1371/journal.ppat.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller U, Kleinberger T, Shenk T. 1992. Adenovirus E4orf4 protein reduces phosphorylation of c-Fos and E1A proteins while simultaneously reducing the level of AP-1. J. Virol. 66:5867–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannervik M, Fan S, Strom AC, Helin K, Akusjarvi G. 1999. Adenovirus E4 open reading frame 4-induced dephosphorylation inhibits E1A activation of the E2 promoter and E2F-1-mediated transactivation independently of the retinoblastoma tumor suppressor protein. Virology 256:313–321. 10.1006/viro.1999.9663. [DOI] [PubMed] [Google Scholar]
- 19.Kanopka A, Muhlemann O, Akusjarvi G. 1996. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381:535–538. 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 20.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185–187. 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 21.Estmer Nilsson C, Petersen-Mahrt S, Durot C, Shtrichman R, Krainer AR, Kleinberger T, Akusjarvi G. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20:864–871. 10.1093/emboj/20.4.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcellus RC, Teodoro JG, Wu T, Brough DE, Ketner G, Shore GC, Branton PE. 1996. Adenovirus type 5 early region 4 is responsible for E1A-induced p53-independent apoptosis. J. Virol. 70:6207–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shtrichman R, Kleinberger T. 1998. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J. Virol. 72:2975–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavoie JN, Nguyen M, Marcellus RC, Branton PE, Shore GC. 1998. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J. Cell Biol. 140:637–645. 10.1083/jcb.140.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shtrichman R, Sharf R, Barr H, Dobner T, Kleinberger T. 1999. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc. Natl. Acad. Sci. U. S. A. 96:10080–10085. 10.1073/pnas.96.18.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branton PE, Roopchand DE. 2001. The role of adenovirus E4orf4 protein in viral replication and cell killing. Oncogene 20:7855–7865. 10.1038/sj.onc.1204862. [DOI] [PubMed] [Google Scholar]
- 27.Kornitzer D, Sharf R, Kleinberger T. 2001. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in Saccharomyces cerevisiae and interacts with the anaphase-promoting complex/cyclosome. J. Cell Biol. 154:331–344. 10.1083/jcb.200104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mui MZ, Roopchand DE, Gentry MS, Hallberg RL, Vogel J, Branton PE. 2010. Adenovirus protein E4orf4 induces premature APCCdc20 activation in Saccharomyces cerevisiae by a protein phosphatase 2A-dependent mechanism. J. Virol. 84:4798–4809. 10.1128/JVI.02434-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groitl P, Dobner T. 2007. Construction of adenovirus type 5 early region 1 and 4 virus mutants. Methods Mol. Med. 130:29–39. 10.1385/1-59745-166-5:29. [DOI] [PubMed] [Google Scholar]
- 30.Tollefson AE, Kuppuswamy M, Shashkova EV, Doronin K, Wold WS. 2007. Preparation and titration of CsCl-banded adenovirus stocks. Methods in molecular medicine. 130:223–235. 10.1385/1-59745-166-5:223. [DOI] [PubMed] [Google Scholar]
- 31.Bacchetti S, Graham F. 1993. Inhibition of cell-proliferation by an adenovirus vector expressing the human wild type-p53 protein. Int. J. Oncol. 3:781–788. [DOI] [PubMed] [Google Scholar]
- 32.Heilman DW, Teodoro JG, Green MR. 2006. Apoptin nucleocytoplasmic shuttling is required for cell type-specific localization, apoptosis, and recruitment of the anaphase-promoting complex/cyclosome to PML bodies. J. Virol. 80:7535–7545. 10.1128/JVI.02741-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siomi H, Dreyfuss G. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551–560. 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teodoro JG, Heilman DW, Parker AE, Green MR. 2004. The viral protein Apoptin associates with the anaphase-promoting complex to induce G2/M arrest and apoptosis in the absence of p53. Genes Dev. 18:1952–1957. 10.1101/gad.1198404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harlow E, Franza BR, Jr, Schley C. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boivin D, Morrison MR, Marcellus RC, Querido E, Branton PE. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 73:1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kindsmüller K, Groitl P, Hartl B, Blanchette P, Hauber J, Dobner T. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. U. S. A. 104:6684–6689. 10.1073/pnas.0702158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philipson L. 1961. Adenovirus assay by the fluorescent cell-counting procedure. Virology 15:263–268. 10.1016/0042-6822(61)90357-9. [DOI] [PubMed] [Google Scholar]
- 39.Le Sage V, Mouland AJ. 2013. Viral subversion of the nuclear pore complex. Viruses 5:2019–2042. 10.3390/v5082019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller U, Roberts MP, Engel DA, Doerfler W, Shenk T. 1989. Induction of transcription factor AP-1 by adenovirus E1A protein and cAMP. Genes Dev. 3:1991–2002. 10.1101/gad.3.12a.1991. [DOI] [PubMed] [Google Scholar]
- 41.Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144:539–550. 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. 2010. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J. Cell Biol. 189:1129–1142. 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galy V, Mattaj IW, Askjaer P. 2003. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol. Biol. Cell 14:5104–5115. 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flint SJ, Gonzalez RA. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272:287–330. 10.1007/978-3-662-05597-7_10. [DOI] [PubMed] [Google Scholar]
- 45.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 46.Copeland AM, Newcomb WW, Brown JC. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660–1668. 10.1128/JVI.01139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gustin KE, Sarnow P. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240–249. 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gustin KE, Sarnow P. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 76:8787–8796. 10.1128/JVI.76.17.8787-8796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park N, Katikaneni P, Skern T, Gustin KE. 2008. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J. Virol. 82:1647–1655. 10.1128/JVI.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monette A, Pante N, Mouland AJ. 2011. HIV-1 remodels the nuclear pore complex. J. Cell Biol. 193:619–631. 10.1083/jcb.201008064. [DOI] [PMC free article] [PubMed] [Google Scholar]