ABSTRACT
Drosophila C virus (DCV) is a positive-sense RNA virus belonging to the Dicistroviridae family. This natural pathogen of the model organism Drosophila melanogaster is commonly used to investigate antiviral host defense in flies, which involves both RNA interference and inducible responses. Although lethality is used routinely as a readout for the efficiency of the antiviral immune response in these studies, virus-induced pathologies in flies still are poorly understood. Here, we characterize the pathogenesis associated with systemic DCV infection. Comparison of the transcriptome of flies infected with DCV or two other positive-sense RNA viruses, Flock House virus and Sindbis virus, reveals that DCV infection, unlike those of the other two viruses, represses the expression of a large number of genes. Several of these genes are expressed specifically in the midgut and also are repressed by starvation. We show that systemic DCV infection triggers a nutritional stress in Drosophila which results from intestinal obstruction with the accumulation of peritrophic matrix at the entry of the midgut and the accumulation of the food ingested in the crop, a blind muscular food storage organ. The related virus cricket paralysis virus (CrPV), which efficiently grows in Drosophila, does not trigger this pathology. We show that DCV, but not CrPV, infects the smooth muscles surrounding the crop, causing extensive cytopathology and strongly reducing the rate of contractions. We conclude that the pathogenesis associated with systemic DCV infection results from the tropism of the virus for an important organ within the foregut of dipteran insects, the crop.
IMPORTANCE DCV is one of the few identified natural viral pathogens affecting the model organism Drosophila melanogaster. As such, it is an important virus for the deciphering of host-virus interactions in insects. We characterize here the pathogenesis associated with DCV infection in flies and show that it results from the tropism of the virus for an essential but poorly characterized organ in the digestive tract, the crop. Our results may have relevance for other members of the Dicistroviridae, some of which are pathogenic to beneficial or pest insect species.
INTRODUCTION
The fruit fly Drosophila melanogaster has been used since the late 1980s as a model to decipher innate immunity. In particular, the study of host defense against bacterial and fungal infections in this organism has led to the discovery of an inducible humoral response in insects and the characterization of two evolutionarily conserved signaling pathways, Toll and IMD, regulating antimicrobial peptide gene expression through the induction of transcription factors of the NF-κB family (reviewed in references 1–3). Drosophila also has been used to characterize the role of hemocytes in the control of bacterial and parasitic infections (reviewed in reference 4), to study immune responses against intracellular pathogens (reviewed in reference 5), and to investigate host-microbe interactions at mucosal surfaces, e.g., the gut (reviewed in references 6–8).
More recently, several groups began to address antiviral immunity and virus-host interactions in Drosophila. A first arm of the antiviral immune system is based on RNA interference (RNAi) and involves three core components, Dicer-2, R2D2, and Argonaute 2 (AGO2). In association with the double-stranded RNA (dsRNA) binding protein R2D2, the RNase III Dicer-2 recognizes replicating viral RNAs and processes them into 21-nucleotide (nt)-long short interfering RNA (siRNA) duplexes. With the help of R2D2, these duplexes are loaded onto AGO2, an RNase H-like enzyme, where one of the two strands is cleaved and discarded, while the other (guide strand) is retained and used to target AGO2 toward viral RNA molecules. Genetic studies have shown that the siRNA pathway plays an important role in the control of many viruses with RNA or DNA genomes, several of which express suppressors of this host defense mechanism (viral suppressors of RNAi, or VSRs) (reviewed in references 9–11). A second layer of defense is composed of cellular mechanisms, such as apoptosis and autophagy, and the induced expression of genes contributing to the control of viral infection (reviewed in references 12–14). In contrast to RNAi, these responses appear to be virus specific, probably reflecting differences between type of genome, replication strategy, and/or tissue tropism. For example, using genome-wide microarrays, we previously reported that the dicistrovirus Drosophila C virus (DCV) induces a different set of genes than the nodavirus Flock House virus (FHV) and the alphavirus Sindbis virus (SINV). This difference in gene reprogramming profiles also is reflected by the fact that flies mutant for the Jak kinase Hopscotch are sensitive to infection by DCV and cricket paralysis virus (CrPV), another member of the Dicistroviridae family, but not to FHV or SINV (15). Beyond immunity, other studies revealed host cell factors required for viral infections (16–18) and host restriction factors (19, 20).
Lethality is used routinely as a readout for the efficiency of the antiviral immune response. However, virus-induced pathologies in flies still are poorly understood (21, 22). Here, we have investigated an aspect of the pathology caused by DCV, a natural pathogen of Drosophila frequently used to investigate antiviral immunity. We first noted that DCV infection, unlike those of FHV or SINV, leads to the downregulation of a large number of genes. Interestingly, many of these genes are expressed in the digestive tract. We further noted that DCV causes intestinal obstruction, and we tracked the cause of the defect to the infection by DCV of the visceral muscles surrounding the crop, a bilobed extensible sac found in the abdomen of Diptera that is used as a reservoir for nutrients.
MATERIALS AND METHODS
Drosophila strains and generation of transgenic lines.
Fly stocks were raised on standard cornmeal-agar medium at 25°C. Oregon-R, w*; P(GawB)Aug21/CyO, y1 w*; P(Akh-gal4.L)2/CyO, y+; P(Dot-GAL4.K)43A, y1 w*, w*; and P(UAS-GFP.S65T)Myo31DFT2 flies were obtained from the Bloomington Fly Stock Center (Bloomington, IN). wA5001, yw, and Oregon-R flies were used as wild-type controls. All flies were tested for Wolbachia infection and treated whenever necessary. For the construction of all reporter plasmids, PCR fragments of different sizes, amplified from the 5′-untranslated region (UTR) of Jon65Ai and Lysozyme E (LysE) genes, were inserted into pCasper expression vector containing the LacZ gene, the drosomycin poly(A) sequence, and an ampicillin resistance cassette. The PCR products were amplified by PCR (primer sequences are available upon request), digested with NotI and NheI restriction enzymes, and placed between the corresponding sites in pCasper transformation vector. The resulting constructs then were injected into Drosophila embryos (w− strain) to obtain transgenic lines. At least two independent lines were analyzed for each construct.
Viral infection.
Four- to 7-day-old flies were used in infection experiments. Infections were done by intrathoracic injection (Nanoject II apparatus; Drummond Scientific) of 4.6 nl of a viral suspension in 10 mM Tris-HCl, pH 7.5 (500 PFU/fly for DCV and FHV and 5 PFU/fly for CrPV). A lower inoculum was chosen for CrPV, which replicates rapidly in flies and is more virulent than DCV (23), to observe the development of symptoms over the same time window. Dose-response experiments ruled out the possibility that the difference observed between DCV and CrPV reflects the amount of virus injected. The injection of the same volume of 10 mM Tris-HCl, pH 7.5, was used as a control. All experiments were performed at 25°C.
RNA analysis.
Total RNA was extracted using Tri Reagent (Molecular Research Central, Inc.) according to the manufacturer's instructions and was analyzed by Northern blotting using standard procedures. The sequences of the primers used to generate the cDNA probes are available upon request.
β-Galactosidase reporter assay.
Four individual flies for each treatment were placed in the wells of a 96-well plate and homogenized in 100 μl of Chalfie buffer, pH 8 (10 mM Tris, pH 8.4, 100 mM NaCl, 1 mM MgCl2, 10 mM dithiothreitol [DTT]). 4-Methylumbelliferone β-d-galactopyranoside (MUG) (Sigma) was added to each well, and the kinetics of the appearance of a fluorescent product generated by the β-galactosidase was measured in a Fluoroscan Ascent plate reader (Labsystems). Fluorescence is indicated in arbitrary units, as previously described (24).
Antibody generation.
Rabbit antiserum directed against the protease Jon65Ai was obtained after injection of the peptide NH2-IGNSVCENYYGSFSGDLICIPTPENK-OH by the company Biosynthesis. For Western blot experiments, the antiserum Jon65Ai BSYN5165 was used at the optimal dilution of 1:200. The specificity of the antiserum was ascertained by Western blotting on protein extracts prepared from wild-type or Jon65AiKG03203 null mutant flies (data are available upon request). Sucrose gradient-purified DCV or CrPV particles were injected intraperitoneally into 7-week-old BALB/c ByJ mice (3.E+08 per injection) three times, with an interval of 10 days. The first injection was performed in the presence of complete Freund's adjuvant. The second injection was performed using incomplete Freund's adjuvant, and the last was performed without Freund's adjuvant. Five days after the last virus injection, the serum of each mouse was harvested and stored at −20°C. The specificity of the antibodies was verified by immunofluorescence on cells infected by either DCV or CrPV (data are available upon request).
Microarray analysis.
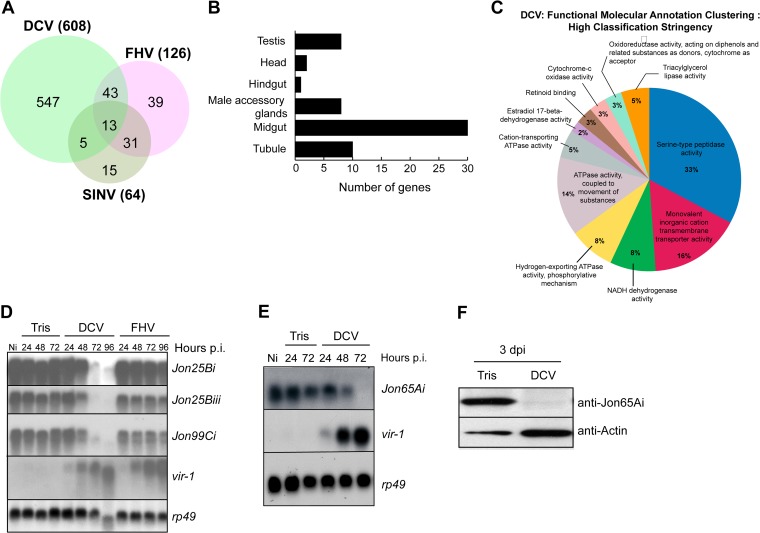
The Affymetrix DNA microarray data (accession number GSE31542; http://www.ncbi.nlm.nih.gov/geo/) and their primary analyses have been described (15, 25). The previously normalized data were used, taking advantage of new bioinformatic tools for functional annotation (26, 27). For this, we considered the genes repressed if they had a present flag, a P value of less than 0.05, and an at least 2-fold lower signal in the virus than Tris-injected samples (Fig. 1A). The DAVID functional annotation clustering website (http://david.abcc.ncifcrf.gov) was used with such a list with the GOTERM_MF_FAT molecular function annotation as gene ontology using the highest classification stringency (Fig. 1C). Drosophila gene expression by tissues was analyzed using the FlyAtlas website (http://flyatlas.org) (28). Genes for which the maximal value of expression in a specific tissue was higher than the means of expression in all tissues by a factor greater than 12 were considered tissue specific (Fig. 1B).
FIG 1.
Gene repression in DCV-infected flies. (A) Venn diagram showing the number of repressed genes in DCV-, FHV-, and SINV-infected flies. (B) DCV represses midgut-specific genes. The diagram shows the number of tissue-specific genes that are downregulated by a factor of at least two in DCV-infected flies. (C) Functional molecular annotation clustering of the DCV-repressed genes. Genes repressed by a factor of at least 2-fold were classified in different categories using the DAVID clustering tool. (D and E) Quantitative validation of the microarray analysis for four members of the Jonah family of serine proteases. vir-1 is a marker gene induced after viral infection, and rp49 is used as a loading control. Ni, noninfected. (F) Jon65Ai is repressed at the protein level. The Western blot is of proteins extracted from 10 dissected guts showing that the Jon65Ai protein is repressed 3 days after infection.
Oil red O staining.
Abdominal carcasses from virus-infected or control flies were dissected in 1× phosphate-buffered saline (PBS) 3 days postinjection (dpi) and fixed in 4% paraformaldehyde for 20 min. Samples then were incubated for 30 min in the Oil red O stain (6 ml of 0.1% Oil red O [Sigma] in isopropanol and 4 ml of MilliQ water, filtered through a 0.45-μm syringe filter) and quickly washed with MilliQ water (29). Samples then were mounted in 100% glycerol and observed under a phase-contrast microscope.
Immunostaining and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining.
For immunostaining experiments, fly guts were dissected in PBS containing 4% formaldehyde and fixed for 30 min. Samples were washed three times in PBS containing 0.2% Tween 20 and permeabilized and blocked for 1 h with PBS, 1% Tween 20, 2% bovine serum albumin (BSA). Samples were incubated overnight at 4°C with antibody against DCV or CrPV capsid (mouse polyclonal; 1:1,000) and anti-green fluorescent protein (GFP; rabbit polyclonal; 1:1,000; Molecular Probes) and were labeled for 4 h at 25°C with secondary antibodies (Alexa Fluor 546 donkey anti-mouse IgG and goat anti-rabbit IgG-fluorescein isothiocyanate [FITC]; 1:2,000; Molecular Probes). Slides were mounted in Vectashield medium containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories) and were examined by epifluorescence (Zeiss Axioskop 2) or confocal (Zeiss LSM700) microscopy.
For X-Gal staining, dissected tissues were placed in 1% glutaraldehyde in 1× PBS, fixed for 20 min, and then washed twice with 1× PBS. The staining was performed in a solution containing 5% X-Gal, 8.4 mM Na2HPO4, 1.6 mM NaH2PO4, 150 mM NaCl, 1 mM MgCl2, 3.5 mM K3Fe(CN)6, 3.5 mM K4Fe(CN)6. Before placing the samples between the slide and coverslip, colored samples were placed in two successive baths of 50% glycerol in 1× PBS. Samples next were mounted in 96% glycerol.
Colorimetric food ingestion and defecation rate assays.
Three groups of 10 flies were Tris or virus injected and placed on a tracking medium containing 0.1% bromophenol blue and 0.5% xylene cyanol. For food ingestion experiments, flies were collected 96 h postinjection, homogenized in 1× Tris-EDTA buffer containing 0.1% Triton X-100, and centrifuged at 14,000 rpm for 3 min. The optical density (OD) of the supernatants was measured as previously described (30). For the defecation assay, Tris- or virus-injected flies were kept on the tracking medium for 3 days and then transferred into empty vials for 5 h. The blue excretion spots, corresponding to the fly defecation rate, then were counted.
CAFE assay.
For the CAFE (capillary feeder) assay, virus-infected or control flies were transferred 72 h postinfection to a 24-well plate. Single flies were placed in individual wells filled with 500 μl of 1% agarose to provide moisture. One capillary, filled with 10 μl of 5% sucrose and 2% yeast extract (50× yeast extract ultrafiltrate; Y-4375; Sigma) solution, was provided per well (31). Flies were capillary fed for 24 h, and the volume of ingested food was measured for individual flies.
Crop contraction rate determination.
Crop contractions were measured as previously described (32). Briefly, 3 days postinjection, Tris, DCV, or CrPV Oregon-R flies were starved for 1 h at room temperature and cold anesthetized for 20 min. Animals were pinned dorsal side down and covered with physiological saline (5 mM HEPES, 128 mM NaCl, 36 mM sucrose, 4 mM MgCl2, 2 mM KCl, and 1.8 mM CaCl2, pH 7.1) and wings and legs were removed. The ventral epidermis at the junction of the thorax and the abdomen was carefully opened, exposing the crop. The preparation was allowed to equilibrate for 5 min in the saline solution. The number of crop contractions was counted at the base of the crop duct for 1 min, followed by a 1-min wait. The process was repeated 5 times to determine the crop contraction rate.
Measurement of body weight, total glucose, triglycerides, and glycogen levels.
A group of 10 Tris- or virus-injected flies (5 males and 5 females) was weighted for body weight determination. Triglycerides (TG) and glucose (Glc) in Tris- or virus-infected flies were measured with Infinity triglycerides and glucose hexokinase kits (Thermo Scientific). Before extraction, the body weight of the three groups of 5 female flies was determined and used for normalization. For TG measurement, flies were homogenized in 1 ml of PBS containing 0.05% Tween 20. Samples were incubated for 5 min at 70°C. Five hundred μl of the supernatant was transferred into a new tube and centrifuged for 3 min at 14,000 rpm. Fifty μl of the supernatant was distributed per well of 96-well microtiter plates, to which were added 200 μl of Infinity TG kit. The plate was incubated for 5 min at 37°C, and the OD at 540 nm (OD540) was measured. For Glc titer determination, flies were homogenized in 200 μl of 1× Tris-EDTA buffer, supplemented with 0.1% Triton X-100, and centrifuged for 3 min at 14,000 rpm, and 100 μl of the supernatant was transferred into the wells of 96-well microtiter plates. One hundred μl of Infinity glucose hexokinase kit, supplemented with porcine kidney trehalase (T8778; Sigma), was added to each well, and the OD340 was measured. For both TG and Glc measurements, standard curves were generated according to the manufacturer's instructions. Glycogen levels of a group of 5 females were measured using the glycogen assay kit (700480; Cayman Chemical).
TEM analysis.
For transmission electron microscopy (TEM) analysis, fly digestive tracts were dissected in Schneider's medium and carefully placed into cellulose tubes (200-μm diameter; Leica Microsystems). Cellulose tubes containing the digestive tracts were placed in a fixative solution containing 2.5% glutaraldehyde and 4% paraformaldehyde. Samples were postfixed for 4 h in 1% osmium tetroxide at 4°C, rinsed, dehydrated through a graded ethanol series, and embedded in Epon resin. Ultrathin sections were contrasted with uranyl acetate and lead citrate. Sections were observed at 60 kV on a Hitachi 7500 or Philips CM120 transmission electron microscope.
Statistical analysis.
An unpaired two-tailed Student's t test was used to determine statistical significance. P values of less than 0.05 were considered significant (*, P < 0.05; **, P < 0.01; ***, P < 0.005). Data were analyzed using GraphPad Prism 4 for Macintosh (GraphPad Software Inc.).
RESULTS
Repressed gene expression in DCV-infected flies.
In a previous study, we reported that some 130 genes are upregulated 24 h and 48 h post-DCV infection (25). An analysis of the same expression data set indicates that an even larger set of genes, a total of 608, is downregulated by DCV (Fig. 1A). Two other RNA viruses, FHV and SINV, also repressed gene expression, but not to the same extent (see Table S1 in the supplemental material). The contrast is particularly striking between DCV, which represses more genes than it induces, and FHV, which shows the opposite pattern (600 genes induced versus 126 repressed), even though both viruses replicate rapidly to high titers in infected flies, causing lethality within 1 week at the doses used (Fig. 1A) (15, 33). In the following paragraphs, we focus on the comparison between DCV and FHV.
When we analyzed the pattern of tissue expression of the 608 genes repressed by DCV, we identified 62 tissue-specific genes (28). Half of these are expressed solely in the midgut (Fig. 1B), indicating that this tissue is strongly impacted by the infection. Genes uniquely expressed in the male genital tract (testis or accessory glands) or the Malpighian tubules also were repressed, suggesting that these tissues also are affected by DCV infection (see Table S2 in the supplemental material). Analysis of the molecular functions associated with the repressed genes revealed a strong enrichment for proteolytic activity, which represented one-third of the repressed genes (Fig. 1C). Many of these proteases are expressed in the digestive tract. Among these, the genes encoding the serine proteases of the Jonah family were spectacularly downregulated by DCV. Northern blot analysis confirmed the microarray data and revealed a dramatic reduction in the expression of several genes, including the Jonah family, following DCV infection. Importantly, these genes were not repressed following FHV infection (Fig. 1D and E). We raised antibodies against the Jon65Ai protease and confirmed that the protein was strongly repressed in the gut following DCV infection (Fig. 1F).
To gain insight into the mechanism of gene repression, we cloned the 5′ upstream region of two strongly repressed genes, Jon65Ai and LysE, upstream of the LacZ gene, and established transgenic flies. The expression of β-galactosidase was detected for both constructs in the midgut (Fig. 2A to D) and was strongly repressed in DCV-infected flies, indicating that repression occurs at the level of transcription. A short version of the Jon65Ai promoter composed of only 236 nucleotides of upstream sequences was sufficient to drive the expression of β-galactosidase in the midgut and repression by DCV (Fig. 2E to G). This repression does not reflect a global response to pathogenesis, because neither FHV nor bacteria significantly modified the expression of the reporters (Fig. 2B and D and data not shown). Altogether, these data suggest that gene repression following DCV infection reflects a virus-specific response or pathology which involves the midgut.
FIG 2.
Transcriptional repression of Jon65Ai and LysE during DCV infection. (A) X-Gal staining of dissected Drosophila tissues from Jon65Ai-LacZ flies. The β-galactosidase activity is detectable in the posterior midgut of Jon65Ai-LacZ flies. OV, ovaries; MT, Malpighian tubules; aMG, anterior midgut; pMG, posterior midgut; Cr, crop. (B) β-Galactosidase activity in extracts from Jon65Ai-LacZ flies significantly decreases 3 dpi in DCV- but not FHV- or CrPV-infected flies. The graph represents means ± standard deviations (SD) from three independent experiments. (C) X-Gal staining of dissected Drosophila tissues from LysE-LacZ transgenic flies. The β-galactosidase activity is detectable in the anterior midgut of LysE-LacZ flies. (D) β-Galactosidase activity significantly decreases 3 dpi in DCV- but not FHV-infected flies. The graph represents means ± SD from three independent experiments. ns, nonsignificant. (E to G) Analysis of Jon65Ai promoter region. Transgenic flies containing the LacZ gene under the control of truncated fragments of the Jon65Ai promoter (from bp −773 [E], −483 [F], or −236 [G]) were challenged with DCV, and β-galactosidase activity in single flies was measured 72 h later. Results correspond to the means ± SD from three independent experiments. Representative results for at least two independent transgenic lines are shown.
DCV infection triggers nutritional stress.
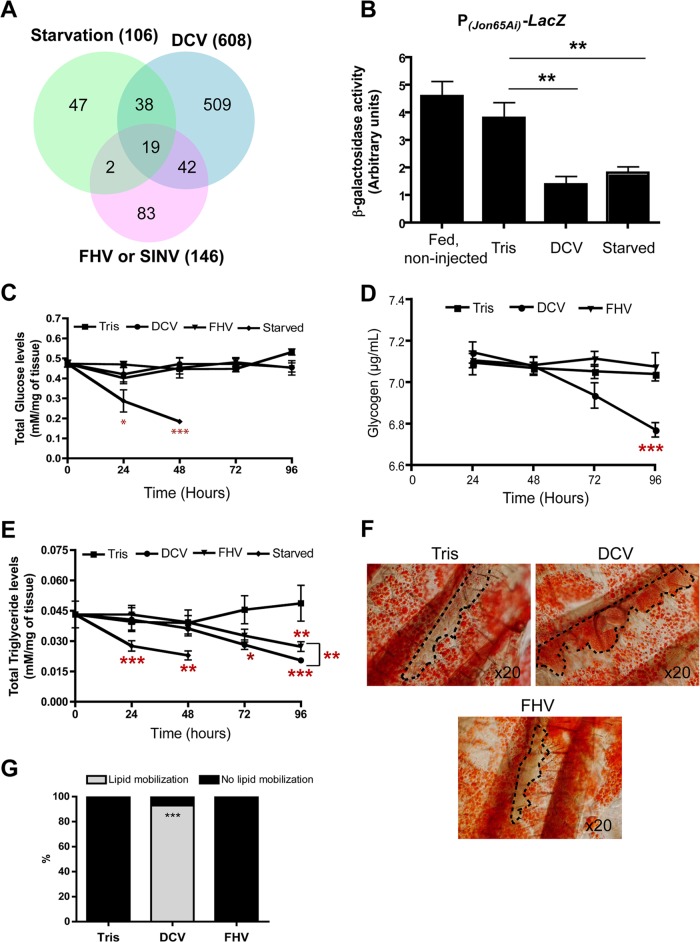
Starvation modulates gene expression in the digestive tract, and undernourishment is a common symptom associated with infection. We compared the transcriptome of starving flies (34) to that of virus-infected flies. Interestingly, 54% of the genes repressed by starvation also were repressed by DCV infection. This was not a global response to viral infection, as fewer than 20% of the genes repressed by starvation also were repressed by either FHV or SINV infection (Fig. 3A and data not shown). Genes strongly repressed by both starvation and DCV include members of the Jonah protease family, and we confirmed that the Jon65Ai (and also the LysE) reporter transgene was repressed by both starvation and DCV infection (Fig. 3B and data not shown).
FIG 3.
Infection by DCV triggers nutritional stress in Drosophila. (A) Venn diagram showing the overlap of downregulated genes between DCV-, FHV-, and SINV-infected and starved flies. (B) DCV infection and starvation downregulate the β-galactosidase activity in Jon65Ai-lacZ flies. The graph represents means ± SD from three independent experiments. (C) Quantification of glucose levels in virus-infected flies. The graph represents means ± SD from three independent experiments for each time point. (D) Glycogen levels decrease after DCV, but not FHV, infection. The graph represents means ± SD from three independent experiments for each time point. (E) Viral infection is accompanied by a decrease in triglyceride levels. The graph represents means ± SD from three independent experiments for each time point. (F) Lipid mobilization from fat body to oenocytes in Drosophila adults 3 days after infection with DCV but not FHV. Oenocytes are circled with dashed lines. (G) Quantification of lipid mobilization in infected flies. Tris mock-infected flies, n = 16; DCV, n = 14; FHV, n = 14.
We wondered whether the repression of protease expression in the midgut reflected a discontinuation of feeding in DCV-infected flies. We monitored glycemia in infected and control flies and did not observe significant differences (Fig. 3C). However, glycogen levels were significantly reduced in DCV-infected flies compared to those in Tris- or FHV-injected flies 4 days following infection, suggesting that DCV-infected flies mobilize their energy stores (Fig. 3D). We also observed a significant reduction in triglyceride levels in DCV-infected flies 3 and 4 days following infection compared with Tris-injected controls. Although triglyceride levels also decreased 4 days after infection with FHV, we found that the difference between the DCV- and FHV-infected flies was significant at this time point (Fig. 3E). In Drosophila larvae, specialized cells, the oenocytes, accumulate lipid droplets during starvation and participate in lipid catabolism (29). Interestingly, we observed a mobilization of triglycerides from the fat body to the oenocytes in DCV-injected but not in mock-injected or FHV-infected flies, providing further evidence for the induction of a nutrition stress in response to DCV infection (Fig. 3F and G).
DCV infection triggers intestinal obstruction.
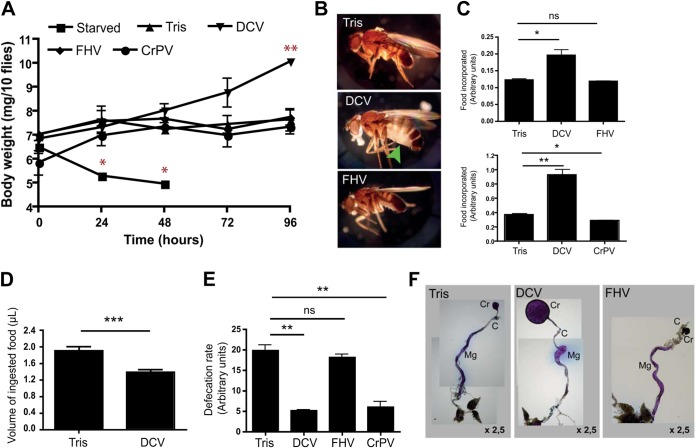
An additional readout for infection-induced anorexia is weight loss. Surprisingly, we observed that DCV-infected flies did not lose weight; on the contrary, they gained weight compared to control flies injected with Tris or an FHV suspension. In contrast, starved flies rapidly lost weight and died within 3 days (Fig. 4A). This observation is corroborated by the fact that DCV-infected flies can be recognized easily by their swollen abdomens (Fig. 4B). We measured food incorporation using the blue food colorimetric assay (30) and noted a significant increase in staining for DCV- but not Tris- or FHV-injected flies, suggesting that DCV infection increases food uptake (Fig. 4C). However, the precise quantification of the volume of food ingested using the CAFE assay revealed that the food uptake is reduced rather than increased in DCV-infected flies (Fig. 4D). Therefore, we monitored fly excretion and noted a strong decrease in defecation upon DCV infection (Fig. 4E), providing an explanation for the increase in food incorporation and body weight in response to DCV infection.
FIG 4.
DCV triggers an intestinal obstruction. (A) Increase in body weight is observed in DCV- but not FHV- or CrPV-infected flies. Groups of 10 flies (5 males and 5 females) were weighed at intervals of 24 h after starvation or injection with Tris (control), DCV, FHV, or CrPV. The graph represents means ± SD from three independent experiments. (B) The characteristic abdominal swelling observed in DCV-infected flies is indicated by the arrowhead. (C) Higher levels of incorporated food in DCV-infected flies. Groups of 10 flies (5 males and 5 females) were injected with Tris, DCV, FHV, or CrPV and kept on tracking medium for 4 days, and the incorporated food was measured by a colorimetric assay. Graphs represent means ± SD from three independent experiments, each performed in triplicate. (D) Measurement of the ingested volume of food in Tris-injected controls and DCV-infected flies using the CAFE assay. The graph represents means ± SD from two independent experiments. At least 15 individual flies were tested in each experiment. (E) Decreased defecation rates after DCV infection. Groups of 10 flies were injected with Tris, DCV, FHV, or CrPV and kept on tracking medium. Three days later, flies were transferred to empty vials, and blue spots corresponding to excretion were counted 5 h later. The graph represents means ± SD from three independent experiments, each performed in triplicate. (F) Dissected digestive tracts of Tris-, DCV-, and FHV-injected flies kept on tracking medium. The blue dye is equally distributed through the whole gut of Tris-injected controls and FHV-infected flies. The blue food accumulates preferentially in the crop and anterior part of the midgut in DCV-infected flies. C, cardia; Cr, crop; Mg, midgut.
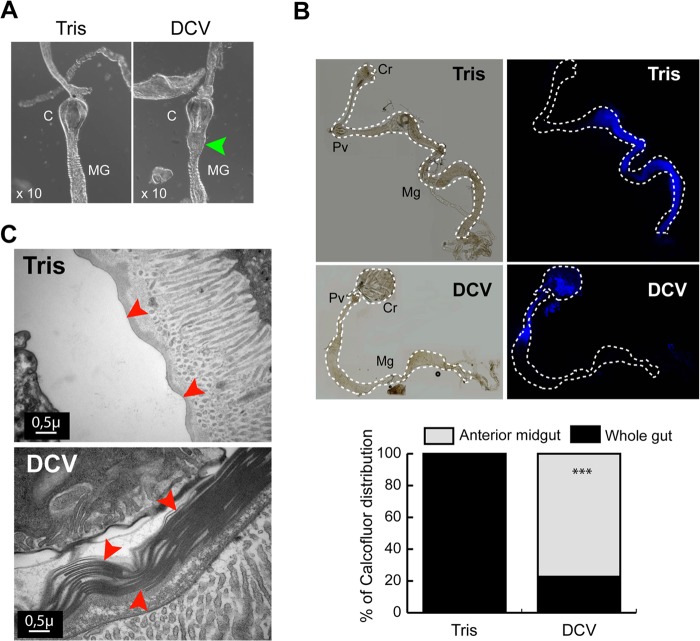
Dissection of DCV-infected flies following ingestion of colored food revealed a striking expansion of the crop, a storage contractile organ at the extremity of the foregut communicating with the entry of the midgut (35), where the blue-dyed food was found to concentrate. In contrast, the crop was not hypertrophied in FHV- or mock-infected flies, and the dyed food was observed all along the digestive tract (Fig. 4F). This suggested that the progression of the food bolus is blocked at the entry of the midgut in DCV-infected flies. The proventriculus (also called the cardia), a sphincter at the entry of the midgut, is a complex structure also involved in the production of the peritrophic matrix, a chitinous membrane isolating the ingested food and protecting the gut epithelium. A characteristic swelling of the posterior part of the cardia was observed in DCV-infected flies (Fig. 5A). Staining with the dye calcofluor, which binds chitin, revealed that the peritrophic matrix covered only the most anterior part of the midgut in DCV-infected flies (Fig. 5B). In addition, the ultrastructural examination of the cardia of DCV-infected flies revealed an accumulation of peritrophic matrix, suggesting that it was not unfolding properly in DCV infected flies (Fig. 5C). Altogether, our data indicate that DCV infection triggers intestinal obstruction in Drosophila.
FIG 5.
Alterations of the cardia and peritrophic matrix in DCV-infected flies. (A) Morphological changes of the Drosophila cardia during DCV infection. Digestive tracts were dissected 3 dpi from Tris- and DCV-infected flies. The posterior part of the cardia is swollen in DCV-infected flies (arrowhead). (B) Dissected digestive tracts from Tris- and DCV-injected flies, fed for 6 h on Calcofluor. The dye is equally distributed throughout the gut in Tris-injected controls. The position of midguts is indicated by dashed lines. Cr, crop; PV, proventriculus; Mg, midgut. The quantification of the intestinal obstruction of Tris-injected and DCV-infected flies is shown on the right. Tris, n = 19; DCV, n = 22. (C) Electron micrographs showing transversal sections of the proventriculus in Tris-injected and DCV-infected flies 3 days postinfection. Layers of peritrophic matrix (arrowheads) accumulate in the proventriculus of DCV-infected flies.
Intestinal obstruction reflects a specific feature of DCV infection rather than a response of the organism to viral infection.
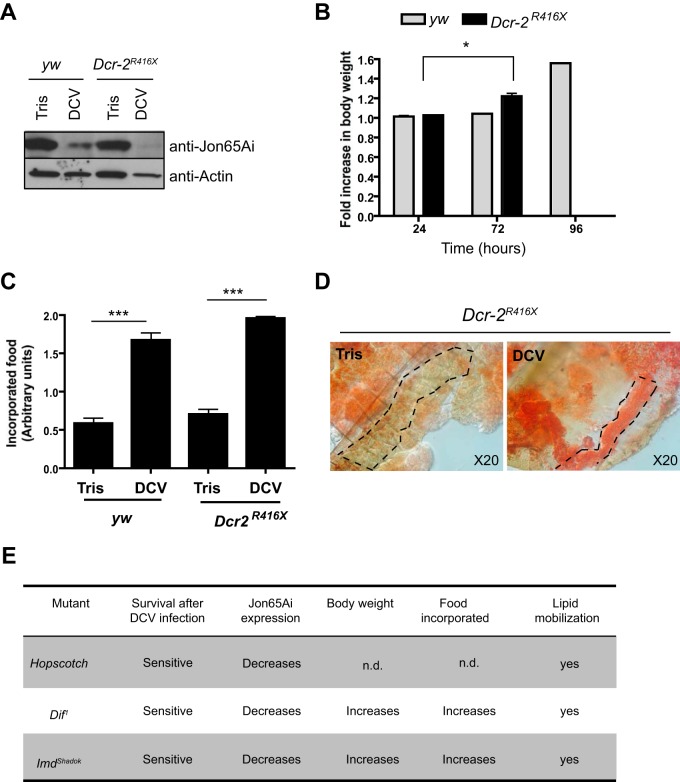
We next considered whether the effect of DCV on the digestive tract resulted from the host response to the infection. RNA interference, as well as three innate immune signaling pathways, Jak/STAT, Toll, and IMD, have been shown, or proposed, to participate in antiviral immunity in Drosophila (25, 33, 36–40). All of the symptoms (repression of Jon65Ai, increased weight and food incorporation, mobilization of triglycerides to oenocytes) were observed in Dcr-2 mutant flies, often more rapidly than in wild-type flies, indicating that they do not result from the activation of the siRNA pathway (Fig. 6A to D). Similar results were obtained for Hopscotch (Jak/STAT pathway), Dif (Toll pathway), and imd (IMD pathway) mutant flies (Fig. 6E), suggesting that the observed symptoms reflect a direct consequence of virus infection rather than a consequence of the host response.
FIG 6.
DCV-induced pathology does not result from activation of immune defense pathways. (A) Jon65Ai is repressed in the gut of Dcr-2 mutant flies. Western blot analysis of proteins extracted from the guts of Tris- and DCV-injected flies 72 h postinfection. (B) Increased body weight in Dcr-2 mutant flies after infection with DCV. The graph represents means ± SD from three independent experiments. All Dcr-2 mutants were dead at 96 h postinfection. (C) Increase of the incorporated food in Dcr-2 mutant flies after infection with DCV. The graph represents means ± SD from two independent experiments. (D) Lipids are mobilized from FB to oenocytes in Dcr-2 mutant flies, as shown by oil red O staining of dissected carcasses from Dcr-2 mutant flies. The dashed lines show the location of the oenocytes. (E) Table showing the development of DCV-induced symptoms in mutants for other antiviral immune pathways, Jak/STAT (Hopscotch), Toll (Dif), and IMD (imd).
We then investigated whether the pathology associated with DCV infection is specific to this virus or is common to other members of the Dicistroviridae family. DCV and CrPV are closely related, and their genomic RNAs share a high percentage of similarity. β-Galactosidase activity in Jon65Ai-lacZ transgenic flies was repressed after infection with DCV but not CrPV (Fig. 2B). In addition, CrPV infection did not lead to increased body weight 4 dpi (Fig. 4A) or to increased food incorporation (Fig. 4C). The only similarity that could be observed between CrPV- and DCV-infected flies was the decrease in defecation rates (Fig. 4E). We conclude that, with the exception of defecation, DCV triggers a specific set of symptoms, different from those for CrPV infection.
Different tropisms for DCV and CrPV in the Drosophila digestive tract.
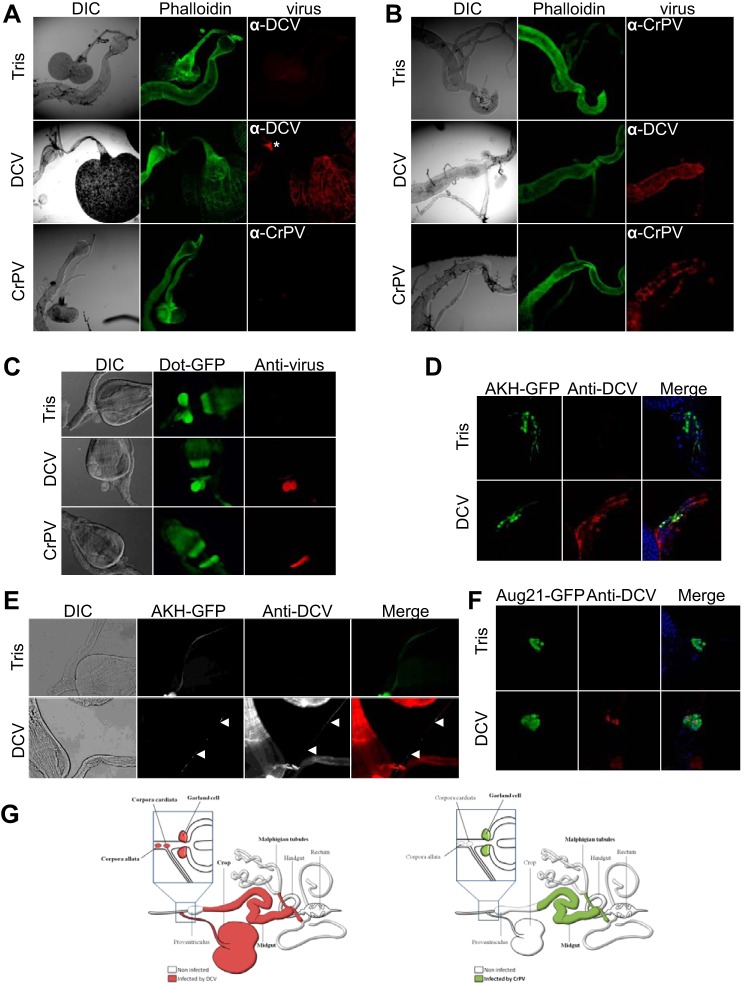
The above-described results suggest that the symptoms caused by DCV reflect the tissue tropism of the virus. To address this issue, we raised antibodies against DCV or CrPV and used them to identify the infected regions of the digestive tract. Both DCV and CrPV target the visceral muscles surrounding the midgut (from the posterior part of the cardia to the junction of the hindgut for DCV and from the one-third anterior part of the midgut until the junction of the hindgut for CrPV) (Fig. 7A and B). This probably explains the decreased defecation rate observed for both viruses. Interestingly, however, DCV, but not CrPV, was found in the muscles circling the lobes of the crop (Fig. 7A). Additional immunolabeling was observed for both viruses in small structures located close to the junction between the foregut and the proventriculus. A more detailed analysis using tissue-specific markers revealed that CrPV and DCV can be detected in the Garland cells (Fig. 7C), whereas DCV, but not CrPV, was found in the corpora allata and the corpora cardiaca (Fig. 7D to F). Importantly, genetic ablation experiments revealed that these small endocrine glands do not contribute to the pathogenesis described here (data not shown).
FIG 7.
DCV and CrPV have different tropisms for the Drosophila digestive tract. Immunostaining of the anterior (A) and posterior (B) parts of the midgut with anti-DCV and anti-CrPV antisera 3 dpi. Only DCV infects the muscles of the crop (A), whereas both DCV and CrPV are detectable in the visceral muscles surrounding the posterior midgut (B). The asterisk indicates the Drosophila ring gland. DIC, differential interference contrast. (C) Garland cells are infected by both DCV and CrPV. UAS-GFP expression was specifically triggered in garland cells with the Dot-Gal4 driver. (D) DCV infects corpora cardiacum (CC) cells. UAS-GFP expression was triggered in CC cells with the Akh-Gal4 driver. (E) Presence of DCV particles in a CC neuron innervating the crop muscle (arrowheads). (F) Corpora allatum (CA) cells are infected by DCV. UAS-GFP expression was triggered in CA cells with the Aug21-Gal4 driver. (G) Schematic summary representation of the tissue tropism of DCV and CrPV in the Drosophila digestive tract. Ten to 15 dissected flies were analyzed, and representative images are shown.
Electron microscopy confirmed the presence of particles of the same size as the virus in DCV-infected but not Tris-injected flies (Fig. 8A). Cellular alterations, such as vacuolization of the sarcoplasmic reticulum and swollen mitochondria, also were observed in DCV-infected cells, which suggested that the function of the crop muscle was affected by DCV infection (Fig. 8A). Indeed, crop contractions were strongly reduced in DCV-infected flies compared to Tris-injected controls. In contrast, infection by CrPV triggered an increase in crop contractions (Fig. 8B), possibly as a compensatory mechanism for the impaired function of the infected visceral muscle cells in the midgut. We conclude that DCV infection of the smooth muscle surrounding the crop affects the function of this organ, preventing correct processing of the food and affecting fly physiology.
FIG 8.
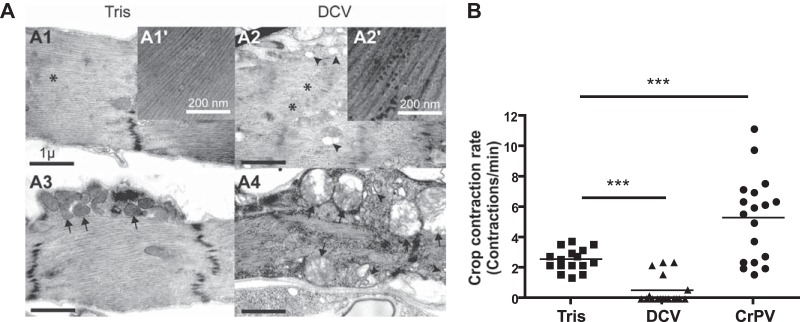
DCV infection of the crop triggers cytopathology and affects its function. (A) Electron micrographs showing the crop muscle of Tris-injected (A1 and A3) or DCV-infected (A2 and A4) Oregon-R flies 4 days postinfection. The presence of DCV particles is noticeable in the cytoplasm of the crop muscle cells (compare the magnified inset of A2′ to that of A1′). Disorganized myofibrils (asterisks), swollen mitochondria (arrows), and vacuolization of the sarcoplasmic reticulum (arrowheads) are present in the DCV-infected muscle cells surrounding the crop (compare A2 and A4 with A1 and A3). (B) Measurement of crop contraction rates in Tris-, DCV-, and CrPV-injected flies. The graph represents means ± SD from two independent experiments. At least 6 individual flies were tested per condition in each experiment.
DISCUSSION
In this study, we have investigated the pathogenesis triggered by DCV in Drosophila. We used intrathoracic injection to control accurately the amount of DCV inoculated. Importantly, crop infection and abdominal swelling also were observed in flies infected by a natural route (data not shown). For the past 25 years, the fruit fly has been used successfully as a model to decipher mechanisms of innate immunity using simple assays, such as induction of marker genes, determination of microbial load, and monitoring of survival. Although these assays proved valuable to dissect the basic molecular and cellular features of the Drosophila host defense system, they ignore the physiological events occurring in the infected flies between the inoculation of the infectious microorganism and the time when the induction of markers and microbial load are measured or when death occurs (41, 42). Studies on antiviral immunity so far make no exception and have focused on monitoring viral titer, survival, induction of marker genes, and profiling of small RNAs (15, 25, 37, 38, 40, 43–46). However, the description of the physiological consequences of infection is required to understand at the organism level both viral pathogenesis and host responses to infection. Our study addresses this important question for a natural viral pathogen of the model organism D. melanogaster.
Few studies have investigated the physiopathology of viral infections in insects so far. A striking example is the case of baculoviruses, which express cathepsins and chitinases that cooperate to break down the cuticle, resulting in the “melting” of the larvae and the release into the environment of large quantities of occluded virions. Other viral gene products control the physiology and behavior of infected insects, highlighting the importance of viral genes in the pathogenesis of baculovirus infection (reviewed in reference 47). In other cases, the cell or organ tropism of the virus plays a major role in the pathogenesis. For example, the infection of tracheal cells by the densovirus Junonia coenia densovirus correlates with decreased oxygen consumption by larvae (48). Along the same lines, the neurotropism of some dicistroviruses (e.g., CrPV, aphid lethal paralysis virus, and acute bee paralysis virus) triggers paralysis (reviewed in reference 49). We show here that DCV infection of the crop muscles participates in the pathogenesis caused by this virus, and that this specific tissue tropism explains the different pathogenesis triggered by the related viruses DCV and CrPV. These results support a recent study that reported that DCV infection triggers a significant increase in fresh mass and a decreased metabolic rate and suggested that the infection interferes with the processing of the food after it has been ingested (21).
The difference of tropism between DCV and CrPV is intriguing, as these viruses are closely related, especially in their capsid proteins, sharing 56% and 66% identity between ORF1 and ORF2, respectively (50). Also intriguing is the fact that CrPV does not infect the crop muscles, although this virus has a wide host and tissue tropism, infecting members from several orders of the Insecta class (Hemiptera, Orthoptera, Lepidoptera, Hymenoptera, and Diptera) and several tissues besides the nervous system (e.g., fat body, tracheae, and muscles). Many dicistroviruses infect primarily gut tissues (reviewed in reference 49), and it will be interesting to see if some of them share with DCV the ability to infect the crop muscles. Interestingly, we noted other differences in the tropism of DCV and CrPV along the digestive tract. For example, DCV appears to preferentially infect the longitudinal muscles in the midgut, whereas CrPV is found primarily in the circular muscles. These two types of muscles have different embryonic origins (51) and may express, at their surface, different proteins serving as receptors for DCV or CrPV. Another difference between the two viruses is the infection by DCV, but not CrPV, of cells from the corpora allata and corpora cardiaca, two small endocrine glands producing juvenile hormone and adipokinetic hormone (AKH), respectively (Fig. 7G). Interestingly, AKH neurons originating from the posterior part of the corpora cardiaca project onto the crop lobes, suggesting that DCV spreads from the crop muscles to this gland through a neural pathway. Interestingly, the list of DCV-repressed genes goes beyond the digestive tract (Fig. 1B) and the response to starvation (Fig. 3A). In some tissues, gene repression may reflect the DCV replication strategy (e.g., internal ribosome entry site [IRES] virus), a hypothesis that could be tested by analyzing global gene expression in CrPV-infected flies.
The pathology triggered by DCV described here suggests several hypotheses regarding the cause of death for the infected flies. First, it is tempting to speculate that starvation contributes to the lethality, as food accumulates in the crop and does not access the site where it is normally digested, the midgut. Interestingly, mutations in the gene dropdead (drd) have a gut phenotype very similar to that of DCV-infected flies: drd mutant flies have a bloated appearance and enlarged crop (52), resulting from impaired movement of the ingested food to the midgut (53). drd mutant flies also exhibit widespread neurodegeneration in the brain, suggesting that the crop expansion results from the defective opening of the stomodeal valve, which controls entry into the midgut (35). Whatever the exact mechanism, drd mutant flies have significantly reduced levels of triglycerides and glycogen stores, like DCV-infected flies, and malnutrition has been proposed to contribute to their premature death (53, 54). An alternative cause of death is the production or accumulation of toxic compounds. For example, in mammals, the mobilization of lipid stores during fasting leads to the production of ketone bodies, which can decrease the pH of the blood and have toxic effects (ketoacidosis). However, we did not detect signs of acidification of the hemolymph in DCV-infected flies, even though the pH of the crop itself was very acidic (data not shown). The latter observation probably reflects the yeast fermentation in the crop, which may contribute to the release of toxic metabolites. Finally, a third possible cause of death is dehydration. Indeed, water absorption occurs in the midgut and hindgut and could be affected by the retention of ingested liquids in the crop. We noticed that DCV-infected flies spontaneously gather around drops of water when placed in petri dishes, whereas FHV- or CrPV-infected flies do so only occasionally. Along the same lines, DCV-infected flies spend most of their time attached to the extremity of the liquid-filled capillaries during the CAFE assay (data not shown). Interestingly, two of the water channels expressed in Malpighian tubules, CG4019 and CG17664 (55), are downregulated following infection by DCV but not FHV or SINV, suggesting that flies are trying to save water.
Whatever the exact cause of death, we have in this study uncovered an important aspect of the pathogenesis caused by systemic DCV infection in the fruit fly. We further show that the major symptoms result from the tissue tropism of DCV for the crop muscles. These results complement our previous study demonstrating that the sensitivity of flies defective for KATP channels to FHV, but not DCV, infection results from the tropism of FHV for the cardiomyocytes in the dorsal vessel (22). Altogether, these studies reveal that two small nonenveloped positive-sense RNA viruses sharing the ability to replicate rapidly in flies, killing them within a week or so (25), trigger different pathophysiological events. In light of these results, we note that very little is known of the interaction between important human-pathogenic arthropod-borne viruses (i.e., arboviruses), such as dengue virus, West Nile virus, or chikungunya virus, and their insect vectors. Our data from the model insect Drosophila suggest that the identification of the tissues or organs playing a critical role in the multiplication of the virus, or the resistance/endurance to the infection, holds promise for the development of new prophylactic strategies aimed at decreasing the transmission of arboviruses to vertebrate hosts.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Centre National de la Recherche Scientifique (CNRS) and grants from the NIH (PO1 AI070167) and the Investissement d'Avenir Program Laboratoire d'Excellence (NetRNA ANR-10-LABX-36 and I2MC ANR-11-EQPX-0022). S.C. was supported by fellowships from MENRT and the Fondation pour la Recherche Medicale.
We thank Johan Arnold, Gaël Schmitt, Estelle Santiago, and Adrien Franchet for providing technical and scientific assistance at different stages of the project; the staff members of the TEM facilities of the Institut de Neurosciences Cognitives et Integratives, the IGBMC (Strasbourg), and the University of Wisconsin (Madison) for electron microscopy analysis; Bader Al-Anzi for discussion and advice; and Dominique Ferrandon for critical reading of the manuscript and discussion.
Footnotes
Published ahead of print 24 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02320-14.
REFERENCES
- 1.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. 2007. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7:862–874. 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 2.Lindsay SA, Wasserman SA. 2014. Conventional and non-conventional Drosophila Toll signaling. Dev. Comp. Immunol. 42:16–24. 10.1016/j.dci.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleino A, Silverman N. 2014. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 42:25–35. 10.1016/j.dci.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honti V, Csordas G, Kurucz E, Markus R, Ando I. 2014. The cell-mediated immunity of Drosophila melanogaster: hemocyte lineages, immune compartments, microanatomy and regulation. Dev. Comp. Immunol. 42:47–56. 10.1016/j.dci.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Pean CB, Dionne MS. 2014. Intracellular infections in Drosophila melanogaster: host defense and mechanisms of pathogenesis. Dev. Comp. Immunol. 42:57–66. 10.1016/j.dci.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Lee WJ, Brey PT. 2013. How microbiomes influence metazoan development: insights from history and Drosophila modeling of gut-microbe interactions. Annu. Rev. Cell Dev. Biol. 29:571–592. 10.1146/annurev-cellbio-101512-122333. [DOI] [PubMed] [Google Scholar]
- 7.Buchon N, Broderick NA, Lemaitre B. 2013. Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat. Rev. Microbiol. 11:615–626. 10.1038/nrmicro3074. [DOI] [PubMed] [Google Scholar]
- 8.Ferrandon D. 2013. The complementary facets of epithelial host defenses in the genetic model organism Drosophila melanogaster: from resistance to resilience. Curr. Opin. Immunol. 25:59–70. 10.1016/j.coi.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, Wang X, Ding SW. 2010. Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe 8:12–15. 10.1016/j.chom.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nayak A, Tassetto M, Kunitomi M, Andino R. 2013. RNA interference-mediated intrinsic antiviral immunity in invertebrates. Curr. Top. Microbiol. Immunol. 371:183–200. 10.1007/978-3-642-37765-5_7. [DOI] [PubMed] [Google Scholar]
- 11.Karlikow M, Goic B, Saleh MC. 2014. RNAi and antiviral defense in Drosophila: setting up a systemic immune response. Dev. Comp. Immunol. 42:85–92. 10.1016/j.dci.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Lamiable O, Imler JL. 2014. Induced antiviral innate immunity in Drosophila. Curr. Opin. Microbiol. 20C:62–68. 10.1016/j.mib.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Cherry S. 2014. Viruses and antiviral immunity in Drosophila. Dev. Comp. Immunol. 42:67–84. 10.1016/j.dci.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kingsolver MB, Huang Z, Hardy RW. 2013. Insect antiviral innate immunity: pathways, effectors, and connections. J. Mol. Biol. 425:4921–4936. 10.1016/j.jmb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, Pfeffer S, Hoffmann JA, Imler JL. 2013. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 190:650–658. 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins NE, Faria VG, Nolte V, Schlotterer C, Teixeira L, Sucena E, Magalhaes S. 2014. Host adaptation to viruses relies on few genes with different cross-resistance properties. Proc. Natl. Acad. Sci. U. S. A. 111:5938–5943. 10.1073/pnas.1400378111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. 2006. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2:e102. 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherry S, Perrimon N. 2004. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat. Immunol. 5:81–87. 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magwire MM, Bayer F, Webster CL, Cao C, Jiggins FM. 2011. Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a Duplication. PLoS Genet. 7:e1002337. 10.1371/journal.pgen.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magwire MM, Fabian DK, Schweyen H, Cao C, Longdon B, Bayer F, Jiggins FM. 2012. Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet. 8:e1003057. 10.1371/journal.pgen.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnold PA, Johnson KN, White CR. 2013. Physiological and metabolic consequences of viral infection in Drosophila melanogaster. J. Exp. Biol. 216:3350–3357. 10.1242/jeb.088138. [DOI] [PubMed] [Google Scholar]
- 22.Eleftherianos I, Won S, Chtarbanova S, Squiban B, Ocorr K, Bodmer R, Beutler B, Hoffmann JA, Imler JL. 2011. ATP-sensitive potassium channel (K[ATP])-dependent regulation of cardiotropic viral infections. Proc. Natl. Acad. Sci. U. S. A. 108:12024–12029. 10.1073/pnas.1108926108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Krutchinsky A, Gross J, Antoniewski C, Andino R. 2010. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 17:547–554. 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung AC, Criqui MC, Rutschmann S, Hoffmann JA, Ferrandon D. 2001. Microfluorometer assay to measure the expression of beta-galactosidase and green fluorescent protein reporter genes in single Drosophila flies. Biotechniques 30:594–600. [DOI] [PubMed] [Google Scholar]
- 25.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 6:946–953. 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 26.Huang Da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chintapalli VR, Wang J, Dow JA. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39:715–720. 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez E, Wiggins D, Fielding B, Gould AP. 2007. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445:275–280. 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- 30.Ayres JS, Schneider DS. 2009. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 7:e1000150. 10.1371/journal.pbio.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. 2007. Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. U. S. A. 104:8253–8256. 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duttlinger A, Berry K, Nichols R. 2002. The different effects of three Drosophila melanogaster dFMRFamide-containing peptides on crop contractions suggest these structurally related peptides do not play redundant functions in gut. Peptides 23:1953–1957. 10.1016/S0196-9781[02]00179-1. [DOI] [PubMed] [Google Scholar]
- 33.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 7:590–597. 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 34.Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. 2005. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1:323–330. 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Stoffolano JG, Jr, Haselton AT. 2013. The adult Dipteran crop: a unique and overlooked organ. Annu. Rev. Entomol. 58:205–225. 10.1146/annurev-ento-120811-153653. [DOI] [PubMed] [Google Scholar]
- 36.Zambon RA, Nandakumar M, Vakharia VN, Wu LP. 2005. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 102:7257–7262. 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312:452–454. 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. 2006. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20:2985–2995. 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costa A, Jan E, Sarnow P, Schneider D. 2009. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One 4:e7436. 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. 2009. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 5:e1000582. 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirasu-Hiza MM, Schneider DS. 2007. Confronting physiology: how do infected flies die? Cell Microbiol. 9:2775–2783. 10.1111/j.1462-5822.2007.01042.x. [DOI] [PubMed] [Google Scholar]
- 42.Ayres JS, Schneider DS. 2012. Tolerance of infections. Annu. Rev. Immunol. 30:271–294. 10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- 43.Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. 2008. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 4:387–397. 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueller S, Gausson V, Vodovar N, Deddouche S, Troxler L, Perot J, Pfeffer S, Hoffmann JA, Saleh MC, Imler JL. 2010. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 107:19390–19395. 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han YH, Luo YJ, Wu Q, Jovel J, Wang XH, Aliyari R, Han C, Li WX, Ding SW. 2011. RNA-based immunity terminates viral infection in adult Drosophila in the absence of viral suppression of RNA interference: characterization of viral small interfering RNA populations in wild-type and mutant flies. J. Virol. 85:13153–13163. 10.1128/JVI.05518-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabin LR, Zheng Q, Thekkat P, Yang J, Hannon GJ, Gregory BD, Tudor M, Cherry S. 2013. Dicer-2 processes diverse viral RNA species. PLoS One 8:e55458. 10.1371/journal.pone.0055458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friesen PD, Miller LK. 2007. Insect viruses. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 48.Mutuel D, Ravallec M, Chabi B, Multeau C, Salmon JM, Fournier P, Ogliastro M. 2010. Pathogenesis of Junonia coenia densovirus in Spodoptera frugiperda: a route of infection that leads to hypoxia. Virology 403:137–144. 10.1016/j.virol.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 49.Bonning BC, Miller WA. 2010. Dicistroviruses. Annu. Rev. Entomol. 55:129–150. 10.1146/annurev-ento-112408-085457. [DOI] [PubMed] [Google Scholar]
- 50.Huszar T, Imler JL. 2008. Drosophila viruses and the study of antiviral host-defense. Adv. Virus Res. 72:227–265. 10.1016/S0065-3527(08)00406-5. [DOI] [PubMed] [Google Scholar]
- 51.Klapper R. 2000. The longitudinal visceral musculature of Drosophila melanogaster persists through metamorphosis. Mech. Dev. 95:47–54. 10.1016/S0925-4773(00)00328-2. [DOI] [PubMed] [Google Scholar]
- 52.Blumenthal EM. 2008. Cloning of the neurodegeneration gene drop-dead and characterization of additional phenotypes of its mutation. Fly 2:180–188. 10.4161/fly.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peller CR, Bacon EM, Bucheger JA, Blumenthal EM. 2009. Defective gut function in drop-dead mutant Drosophila. J. Insect Physiol. 55:834–839. 10.1016/j.jinsphys.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sansone CL, Blumenthal EM. 2012. Developmental expression of drop-dead is required for early adult survival and normal body mass in Drosophila melanogaster. Insect Biochem. Mol. Biol. 42:690–698. 10.1016/j.ibmb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beyenbach KW, Skaer H, Dow JA. 2010. The developmental, molecular, and transport biology of Malpighian tubules. Annu. Rev. Entomol. 55:351–374. 10.1146/annurev-ento-112408-085512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.