ABSTRACT
The helper-dependent adeno-associated virus type 2 (AAV-2) exhibits complex interactions with its helper adenovirus. Whereas AAV-2 is dependent on adenoviral functions for productive replication, it conversely inhibits adenoviral replication, both when its genome is present in trans after coinfection with both viruses and when it is present in cis, as in the production of recombinant adenovirus (rAd)/AAV-2 hybrid vectors. The notion that AAV-mediated inhibition of adenoviral replication is due predominantly to the expression of the AAV-2 Rep proteins was recently challenged by successful Rep78 expression in a rAd5 vector through recoding of the Rep open reading frame (ORF). We closely analyzed the relative contributions of AAV-2 nucleic acid elements and Rep protein expression to the inhibition of adenoviral replication in both of the above scenarios. When present in cis, a sequence element in the 3′ part of the rep gene, comprising only the AAV-2 p40 promoter and the AAV-2 intron sequence, which we termed the RIS-Ad, completely blocks adenoviral replication. p5/p19 promoter-driven Rep protein expression, on the other hand, only weakly inhibits rAd/AAV-2 vector propagation, and by inactivation of the RIS-Ad, it is feasible to generate first-generation rAd vectors expressing functional Rep proteins. The RIS-Ad plays no role in the inhibition of adenoviral replication in trans in a model closely mimicking AAV-2–Ad coinfection. In this case, expression of the Rep proteins is required, as well as the presence of an amplifiable inverted terminal repeat (ITR)-containing template. Thus, very different AAV-2 elements and mechanisms are involved in inhibition of adenoviral replication during rAd/AAV-2 vector propagation and after Ad-AAV coinfection.
IMPORTANCE This is the first study to systematically compare the contributions of AAV-2 protein expression and AAV-2 nucleic acid elements to the inhibition of adenoviral replication in rAd/AAV-2 hybrid vector generation and in AAV-2–adenovirus coinfection. This study shows that the two inhibitory processes are very different with regard to AAV-2 functions and the mechanisms involved. Whereas inhibition of rAd/AAV-2 hybrid vector propagation mostly involves a 3′ nucleic acid element in the rep gene, inhibition of an adenoviral genome in trans requires the Rep proteins and the AAV ITRs. These findings have important implications both for a basic understanding of the AAV replication cycle and for generation of rAd/AAV-2 hybrid vectors expressing the nonstructural and structural proteins of AAV-2.
INTRODUCTION
Adeno-associated virus (AAV) is a human parvovirus with a bipartite replication cycle. For productive replication, it depends upon coinfection or superinfection with a helper virus, such as adenovirus (1) or herpes simplex virus (HSV) (2). In the absence of a helper virus, it can establish a latent infection by integration into the host genome.
Both its apparent lack of pathogenicity and the possibility for site-specific integration in the presence of the large Rep proteins (3–5) have boosted the development of AAV as a vector for gene therapy applications. Recombinant AAV (rAAV) vectors contain a transgene with the corresponding control elements flanked by the AAV inverted terminal repeats (ITRs), which are the only AAV elements required in cis for amplification and packaging of the genome. Gene delivery approaches with rAAV vectors have allowed major improvements in the treatment of monogenic diseases, such as congenital blindness and hemophilia (6, 7). The rAAV vectors used in these studies generally do not contain the Rep gene and mainly persist in an episomal state. Therefore, additional efforts have been directed at achieving site-specific replacement of large transgenes by incorporation of the required AAV elements, Rep78/Rep68 and the AAV ITR-flanked transgene cassette, into large-capacity hybrid viruses derived from baculovirus (8), HSV type 1 (HSV-1) amplicons (9, 10), or adenovirus (11–14). A general observation made in the majority of these studies was that the presence of a complete rep gene strongly inhibits the replication of the corresponding hybrid viruses. To achieve propagation of recombinant adenovirus (rAd) vectors harboring a functional rep gene, complex strategies, such as assembly of full-length Rep proteins through recombination of two different rAd vectors (11), tightly regulated promoters driving Rep gene expression (13), and/or helper-dependent adenoviral vectors (13, 14), had to be applied. The inhibitory effects of the rep gene on adenoviral replication also hampered the generation of rAds expressing both the AAV Rep and structural (Cap) proteins, which would represent easily scalable systems for high-titer production of conventional rAAV vectors. Such rAd/AAVrep-cap vectors either could not be propagated at all or were genetically unstable (15). The observed inhibition has been attributed mainly to the expression of the large Rep proteins. In the AAV life cycle, Rep78 and Rep68, which are expressed from the p5 promoter, not only mediate site-specific integration of the AAV DNA in the absence of helper virus but also are essential for replication of the AAV genome in the presence of helper and for regulation of viral gene expression (16). The two former activities require binding of Rep78/Rep68 to a ribosome binding site (RBS) within the ITRs. However, the small Rep proteins Rep52 and Rep40, which are responsible for packaging of the newly single-stranded DNA (ssDNA) molecules into preformed capsids during a productive AAV infection (17), also seem to participate in inhibition of rAD propagation (12). To ablate the expression of Rep52 and Rep40, which present N-terminally truncated versions of Rep78 and Rep68 transcribed from the internally located p19 promoter (16), their start codon is mutated in the majority of rAd-Rep vectors (11, 13, 14). No inhibitory effects of the AAV cap gene on rAd replication have been described so far. The three structural proteins, VP1, VP2, and VP3, which share an open reading frame (ORF), are expressed from the p40 promoter, located in the 3′ part of the rep gene, through alternative splicing and the use of different start codons for VP2 and VP3 (16).
The contributions of different mechanisms to the inhibition of rAd/AAV hybrid vector propagation as well as to the long-described inhibition of wild-type adenovirus replication by increasing amounts of wild-type AAV (18, 19) remain to be clarified. Isolated Rep78 expressed under the control of heterologous promoters has been shown to inhibit early adenoviral gene expression and genome replication (20) as well as the maturation of adenoviral replication centers (21). Recently, however, synonymous codon pair recoding of Rep78 strongly challenged the solitary role of Rep protein expression in the rep gene-mediated inhibition of rAd replication by demonstrating the involvement of a cis inhibitory effect (22). In contrast to the wild-type AAV-2 sequence, the recoded nucleic acid sequence, which fully maintained the polypeptide sequence and the endonuclease properties of Rep78, could be propagated stably in the context of a first-generation adenoviral vector containing a tetracycline-inducible Rep expression cassette. Furthermore, recoding of only a 135-bp nucleic acid sequence in the 3′ part of the rep gene was sufficient for restoring adenoviral replication.
In the present study, we systematically studied the contributions of AAV-2 protein expression and AAV-2 sequence elements to the inhibition of adenoviral replication in rAd/AAV-2 hybrid vector generation and in AAV-2–adenovirus coinfections to determine whether the AAV-2 functions involved in both inhibitory processes are identical, as has been assumed implicitly for a long time.
MATERIALS AND METHODS
Cell culture and transfection.
HEK-293 (human embryonal kidney) cells were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 100 μg/ml of penicillin and streptomycin at 37°C with 5% CO2. Transfections were performed by the calcium phosphate precipitation technique as described previously (23), with 5 × 105 cells seeded the day before transfection, either in 25-cm2 flasks or in 6-cm dishes, using 3 ml medium.
Generation of rAds.
Generation of rAds was performed with the AdEasy system, largely as described previously (24, 25). Briefly, the sequences of interest were cloned into the pShuttle vector, and PmeI-linearized pShuttle constructs were electroporated into Escherichia coli BJ5183-AD-1 cells for homologous recombination with the E1/E3-deleted Ad5 genome, carried on the stably transformed pAdEasy plasmid. pAdEasy constructs were linearized with PacI, transfected into HEK-293 cells, and incubated for 12 to 14 days for the generation of the primary viral supernatant. The supernatant was prepared in 2 ml of medium by four freeze-thaw cycles in liquid nitrogen and a 37°C water bath. A sample of 0.5 ml of the supernatant was used for the first round of amplification in 10-cm dishes incubated for 3 to 6 days and then harvested in 2.5 ml of medium, again by four freeze-thaw cycles. Further rounds of amplification and rAd purification were performed as described previously (25). For each AdEasy construct, the generation of rAd was performed at least three times and with at least two different plasmid DNA preparations. The data are presented as means ± standard deviations.
Infection of HEK-293 cells with rAds.
Medium from 1 × 106 nontransfected HEK-293 cells or cells incubated for 16 h with the DNA-calcium phosphate precipitation mixture from a preceding transfection step was removed, and cells were infected with the rAd-containing freeze-thaw lysates for 1 h in a final volume of 500 μl for a 6-cm dish. Where possible, equal numbers of genomic particles were used for the infection step. However, for the first-round amplification step, generally 500 μl of the freeze-thaw supernatant of the primary transfection step was used, and if freeze-thaw lysates with low titers were obtained after the first amplification round, then the maximum volume of 500 μl was used, as indicated in the respective figure legends. After 1 h of incubation, medium was added to a final volume of 3 ml and was replaced by fresh medium after a further 3 h of incubation.
Plasmids.
A series of plasmids containing the complete AAV-2 genome, either in the wild-type form (pTAV2-0) or with different combinations of frameshift mutations in the Rep ORF (pTAV2-3 and pTAV2-8), generated by a 4-bp insertion at the unique BamHI site at AAV-2 position 1045, and in the Cap ORF (pTAV2-6 and pTAV2-8), generated by deletion of a 164-bp PfIMI restriction fragment between AAV-2 positions 3325 and 3489, has been described previously (26), as has the AAV-2 plasmid pdTR, which is devoid of the ITR sequences (27). The plasmid pTAV2-K340H was generated by introduction of a 1,650-bp SfiI/SwaI fragment from pKEX340H (28), carrying the K340H point mutation in the Rep ORF. Plasmids pTR-UF3 and pTR-UF5 (29), carrying a cytomegalovirus (CMV) enhancer/promoter-driven green fluorescent protein (GFP) cassette flanked by the AAV-2 ITRs, were kindly provided by Nick Muzyczka (University of Florida).
The transfer vector pShuttle-CMV-GFP, used as the positive control in the AdEasy system, was generated by subcloning of a KpnI/SalI fragment from pTR-UF5 into KpnI/SalI-digested pShuttle vector (Agilent). The plasmid pShuttle-AAV, which contains the complete AAV-2 genome except for the ITRs, was obtained by PCR amplification of AAV-2 nucleotides 154 to 4526 with primers generating BglII and XbaI sites for cloning into the pShuttle vector. For plasmid pShuttle-AAV-sRep, a 493-bp BstEII/SwaI restriction fragment from pMA-sRep was subcloned into pShuttle-AAV. pMA-sRep in turn was generated by gene synthesis (Geneart, Regensburg, Germany) and contains the AAV-2 genome from the BstEII site at nucleotide 1700 to the SwaI site at nucleotide 2193, with exclusive recoding of AAV nucleotides 1782 to 1916 according to the work of Sitaraman et al. (22). The K340H point mutation was introduced via the 1,650-bp SfiI/SwaI fragment from pKEX340H, as described above for pTAV2-0. All other pShuttle constructs were obtained by PCR amplification of the indicated AAV-2 nucleotides from pShuttle-AAV or pShuttle-AAV-sRep by use of primers generating corresponding restriction sites for subcloning into the pShuttle vector. All pAdEasy-AAV-2 constructs were generated from the corresponding pShuttle plasmids through homologous recombination of PmeI-linearized DNA in E. coli BJ5183-AD-1 cells (Agilent) containing the pAdEasy-1 plasmid (GenBank accession no. AY370909).
Extraction of viral DNA and Southern hybridization.
Extraction of viral DNA by a modified Hirt procedure was performed essentially as described previously (23).
For the quantitation of replicated adenoviral DNA, a 1-μg sample of the DNA was digested with an excess amount of DpnI (10 U of enzyme for 2 h), followed by phenol-chloroform extraction and precipitation with ethanol. The samples were dissolved in 10 μl of Tris-EDTA buffer, pH 7.6, and prediluted for quantitative real-time PCR as described below.
Southern blot analysis was performed as described previously (23), using 4 μg of Hirt-DNA digested with 40 U of DpnI for 2 h. After electrophoresis on a 0.8% agarose gel, DNAs were transferred by capillary blotting to a nylon membrane (Hybond-N; GE Healthcare) and hybridized with a biotin-11-dUTP-labeled 0.73-kb NotI GFP probe. This probe recognizes the replicative intermediates from recombinant plasmids pTR-UF5 and pTR-UF3, which contain AAV-2-ITR-flanked GFP transgene cassettes. The bound biotinylated probe was detected by use of a streptavidin-horseradish peroxidase (HRP) conjugate (high-sensitivity HRP conjugate; Thermo Scientific) followed by enhanced chemiluminescence (ECL).
Quantification of genomic rAd particles and adenoviral Hirt-DNA.
For determination of rAd genomic particles, 100-μl aliquots of freeze-thaw supernatants were treated with 500 U/ml benzonase (Merck) at 37°C for at least 1 h to degrade residual nonencapsidated DNA and subsequently treated with lysis buffer (1% [wt/vol] N-lauroylsarcosine, 25 mM Tris, pH 8.5, 10 mM EDTA, pH 8.0) containing proteinase K (Roth, Germany) at a final concentration of 400 μg/ml at 56°C for 2 h to digest adenoviral capsids. Adenoviral DNA was purified by phenol-chloroform extraction and dissolved in 20 μl of Tris-EDTA buffer, pH 7.6, after precipitation with ethanol. Both DNA extracted from adenoviral particles and DpnI-digested adenoviral Hirt-DNA, as described above, were prediluted 1:100 for determination of the copy numbers of adenoviral genomes, which was conducted on a Light-Cycler instrument, using a QuantiTect SYBR green PCR kit (Qiagen). Primers E4-Q1 (5′-TCA GCG TAA ATC CCA CAC TG-3′) and E4-Q2 (5′-CCA TTTGGCATG ACA CTA CG-3′) amplified a 202-bp sequence from the Ad5 E4 region, which contains 3 internal DpnI recognition sites.
Western blotting.
Immunoblot analysis of AAV-2 protein expression was performed with anti-Rep monoclonal antibody 303.9 diluted 1:10 or with anti-Cap monoclonal antibody B1 diluted 1:20 (both from Progen, Heidelberg, Germany), followed by incubation with horseradish peroxidase-conjugated secondary antibodies and ECL detection as described previously (23).
Flow cytometry analysis.
Transfected HEK-293 cells were trypsinized and resuspended in phosphate-buffered saline (PBS)–20% FCS for analysis in a FACSCalibur flow cytometer (BD Biosciences). Acquired data were analyzed with Cell Quest Pro software (BD Biosciences). Nontransfected HEK-293 cells served as negative controls to define the gates for separation of GFP-positive and -negative cells. A total of 10,000 cells were scored for each sample, and the total fluorescence of GFP-positive cells was calculated.
RESULTS
AAV-2-mediated inhibition of Ad5/AAV-2 hybrid vector replication in the absence of functional Rep protein expression requires only a small sequence element in the 3′ part of the rep gene.
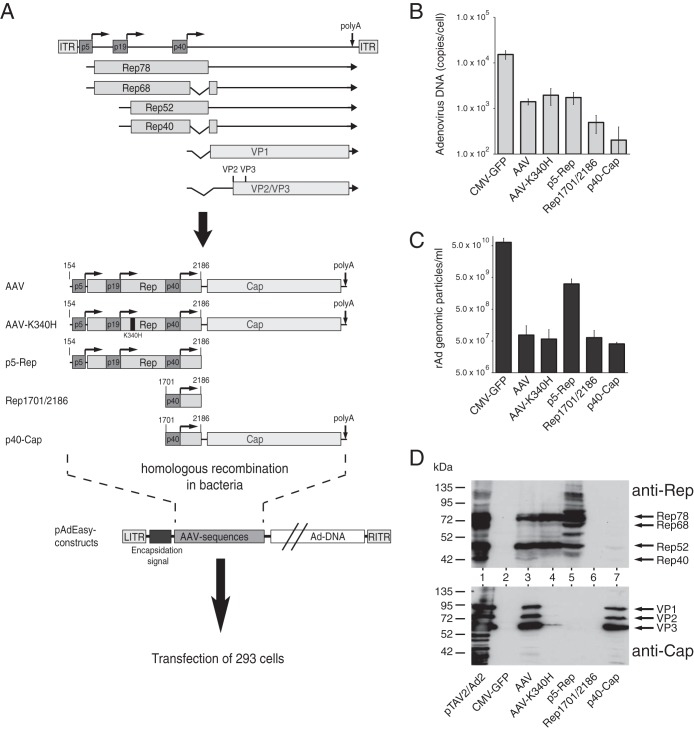
In initial experiments, we examined the replication properties of rAd/AAV-2 hybrid vectors carrying either the complete AAV-2 genome, except for the ITRs, or different AAV-2 subsequences, as depicted in Fig. 1A. These were introduced into the AdEasy system (24, 25), a well-established system for generation of recombinant adenoviral plasmids, through homologous recombination in bacteria followed by transfection into HEK-293 cells to produce the corresponding rAds (Fig. 1A, bottom panel). As a positive control, we transfected an AdEasy plasmid construct containing a human CMV (HCMV) promoter-driven GFP gene (CMV-GFP). In comparison to this positive control, the AdEasy construct containing the complete AAV genome except for the ITRs showed a decrease of >1 log in the amount of DpnI-resistant adenoviral replicative intermediates as scored at 12 days posttransfection (Fig. 1B, compare lanes CMV-GFP and AAV). For the AAV construct, amplification of freeze-thaw lysates of the primary transfection step cells for 6 days resulted in only about 5 × 107 genomic viral particles per ml (GP/ml) in the secondary supernatant, which was close to the background level of the assay. In contrast, the CMV-GFP control showed titers in the range of up to 1011 GP/ml, with early formation of fluorescent plaques and 100% GFP-positive cells after 2 to 3 days of the amplification step (Fig. 1C; note the logarithmic scales in Fig. 1B and C). The very low levels of genomic particles for the AAV construct corresponded to a lack of formation of infectious particles as scored by plaque assays (data not shown). AAV-mediated inhibition of adenoviral replication, however, obviously did not depend upon functional Rep protein expression, since an AAV genome encoding a well-characterized point mutation in the nucleotide binding site of the Rep proteins (AAV-K340H) (Fig. 1A), which has been demonstrated to abolish all biological activities of the Rep proteins, showed very similar effects (Fig. 1B and C). When the complete Rep78 coding sequence preceded by the physiological p5 promoter (p5-Rep) was examined, some residual rAd particle formation in the range of 5 × 109 GP/ml was scored in several independent experiments. Rather unexpectedly, AAV subsequences which harbored either the p40 promoter and the cap ORF (p40-cap) or only the region where the rep and cap genes overlap (Rep1701/2186 [named for AAV-2 nucleotides 1701 to 2186]) also led to a strongly reduced adenoviral DNA replication (Fig. 1B) and completely abrogated rAd particle formation (Fig. 1C). These results and the lack of infectious particle formation for the AAV wild-type, AAV-K340H, Rep1701/2186, and p40-Cap constructs were confirmed in repeated experiments using different plasmid preparations and also prolonged incubation times both for the primary rAd production by transfection and for the amplification step (data not shown). Western analysis of Rep and Cap protein expression levels after short-term transfection of the different pAdEasy-AAV constructs for 2 days showed the expected pattern of reactive protein bands (Fig. 1D), with no anti-Rep- or anti-Cap-responsive polypeptides obtained for the Rep1701/2186 construct. Thus, as proposed by the results of Sitaraman et al. (22), we could identify a sequence element in the 3′ part of the rep gene (AAV-2 nucleotides 1701 to 2186) which inhibits adenoviral replication in the absence of any further AAV-2 sequences. We denoted this sequence the Rep inhibition sequence for adenoviral replication (RIS-Ad).
FIG 1.
AAV elements involved in inhibition of adenoviral replication in the context of Ad5/AAV-2 hybrid vectors. (A) Genome organization of AAV-2 and schematic presentation of the AAV-2 sequences introduced into the AdEasy system for generation of recombinant adenoviruses. The viral genome is shown in the upper part of the figure, with the inverted terminal repeats (ITRs), the four Rep proteins (Rep78, Rep68, Rep52, and Rep40), the capsid proteins (VP1 to VP3), and the three promoters, at map units 5, 19, and 40, indicated by different shaded boxes. Right-angled arrows represent the transcription start sites of the promoters, and the vertical arrow indicates the common polyadenylation [poly(A)] site for all transcripts at map position 96. The lower part of the figure shows the AAV-2 sequences cloned into the pAdEasy plasmid by homologous recombination in bacteria. Characteristic nucleotide positions are indicated. (B) Amounts of newly replicated, DpnI-resistant adenoviral DNA obtained 12 days after transfection of PacI-linearized pAdEasy plasmids into HEK-293 cells, determined as numbers of genomic copies per cell by real-time PCR. (C) Amounts of recombinant adenoviral particles obtained in freeze-thaw cell supernatants after transfection of the PacI-linearized pAdEasy plasmids into HEK-293 cells for 12 days and amplification of the primary supernatants in HEK-293 cells for 6 days, determined as numbers of GP/ml of final cell supernatant by real-time PCR. (D) Western blot analysis of whole-cell extracts harvested 2 days after transfection of HEK-293 cells with the indicated pAdEasy-AAV-2 plasmids. Expression of Rep proteins was scored with monoclonal antibody 303.9 (upper panel), and that of capsid proteins was scored with monoclonal antibody B1 (lower panel). Arrows indicate the positions of the different Rep and capsid proteins. HEK-293 cells transfected with infectious AAV-2 plasmid pTAV2-0 and infected with adenovirus type 2 (multiplicity of infection [MOI] = 20) served as a positive control (lane pTAV2/Ad2).
Regarding the regulation of AAV gene expression, we made some interesting observations in these experiments. While the pAdEasy-p5-Rep construct does not contain the common polyadenylation site in the right part of the AAV-2 genome (Fig. 1A), Rep protein levels were comparable to those of the full-length AAV-2 construct (Fig. 1D, compare lanes AAV and p5-Rep). Furthermore, the p40-Cap construct, devoid of the p5/p19 promoter sequences shown to stimulate the p40 promoter in the presence of Rep proteins and adenovirus (30), showed levels of capsid protein expression similar to those of the complete AAV genome. In contrast, when the Rep proteins were rendered nonfunctional in the context of the full-length AAV genome (AAV-K340H), capsid expression levels were strongly reduced.
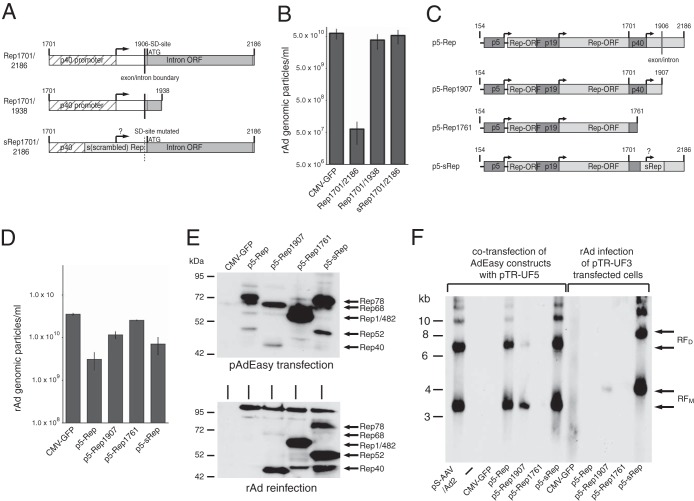
rAd-mediated constitutive expression of functional Rep proteins by inactivation of the RIS-Ad.
The inhibitory sequence element in the 3′ part of the rep gene, which we termed the RIS-Ad, contains the p40 promoter and the AAV-2 intron (Fig. 2A, Rep1701/2186). To exclusively assess the contribution of Rep protein expression to the inhibition of replication of rAd/AAV-2 hybrid vectors, it was necessary to abolish the inhibitory effects of the RIS-Ad. Deletion of large portions of the 3′ part of the intron from the RIS-Ad (Fig. 2A, Rep1701/1938) completely abrogated its inhibitory activity (Fig. 2B). In addition, adenoviral replication could also be restored completely by recoding of the C-terminal Rep78/Rep52 amino acid coding sequence from AAV-2 nucleotides 1782 to 1916 (Fig. 2A and B, sRep1701/2186 [for scrambled Rep]), as described by Sitaraman et al. (22). Neither the Rep78/Rep52 ORF nor a second ORF of 82 amino acids contained completely within the intron should be affected by the sRep mutations. For both Rep1701/1938 and sRep1701/2186, primary transfection and the amplification step were associated with plaque formation that was easily visible by light microscopy. Based on these results, AdEasy-p5-Rep constructs containing either a Rep78 sequence recoded in the region from AAV-2 nt 1782 to 1916 (Fig. 2C, p5-sRep) or a protein similar to Rep68 (Fig. 2C, p5-Rep1907 [identical to Rep68 except for the 7 C-terminal Rep68-specific amino acids]) were evaluated for formation of rAd vectors capable of expressing functional Rep proteins under the control of the AAV-2 p5 promoter. For both constructs, we obtained genomic particle titers slightly higher than that with the wild-type Rep78 sequence after transfection and first-round amplification, but still 5- to 10-fold lower than that for the positive control (Fig. 2D, p5-Rep1907 and p5-sRep). Compared to the Rep expression pattern 2 days after transfection of the pAdEasy constructs (Fig. 2E, upper panel), the pattern obtained after reinfection with the freeze-thaw lysates from the first amplification round was shifted toward the small Rep proteins (Fig. 2E, lower panel). However, in contrast to the wild-type Rep78 construct, both the protein similar to Rep68 and the partially recoded Rep78 protein could be expressed in clearly detectable amounts from the corresponding virus preparations. The rAd–wild-type Rep78 preparations either did not express any anti-Rep-reactive bands at all (as in Fig. 1E) or expressed only bands clearly smaller than Rep78 and Rep52 (data not shown), which indicates a loss of Rep sequences in the rAd preparations. As a control for further inhibitory nucleic elements possibly located in the Rep sequence, we used a construct expressing Rep amino acids 1 to 482 (Fig. 2C, p5-Rep1761), which expresses Rep proteins lacking the nuclear localization signal. The corresponding rAd could be amplified to titers similar to that of the control (Fig. 2D) and expressed the expected bands after both transfection and reinfection (Fig. 2E, upper and lower panels). To assay the functional properties of the protein similar to Rep68 and the recoded Rep78 protein expressed by the recombinant adenoviruses, HEK-293 cells transfected with recombinant AAV plasmids containing the AAV-2 ITRs were either cotransfected with the pAd-Easy-p5-Rep constructs as positive controls (Fig. 2F, left panel) or infected with the corresponding rAd preparations (Fig. 2F, right panel). AAV ITR-dependent DNA replication with the typical monomeric and dimeric replication intermediates could be scored after reinfection with the rAD-p5-sRep preparation and, to a lesser degree, with the rAD-p5-Rep1907 preparation (Fig. 2F, right panel). Thus, although expression of the large Rep proteins under the control of their physiological p5 promoter seems to inhibit the replication of the corresponding adenoviral vectors to some degree, generation of rAd vectors expressing these proteins can be achieved by standard procedures, without transient suppression of Rep protein expression.
FIG 2.
Constitutive expression of functional large Rep proteins from recombinant adenoviruses by inactivation of the 3′ rep inhibitory sequence (RIS-Ad). (A) Schematic presentation of the 3′ rep inhibitory sequence (Rep1701/2186) and the 3′-deleted (Rep1701/1938) and partially recoded (sRep1701/2186) sequences contained within the pAdEasy constructs. The p40 transcription start site at AAV-2 nucleotide 1853 is represented by a right-angled arrow, and the splice donor (SD) site at nucleotide 1906 is represented by a vertical line. (B) Amounts of recombinant adenoviral particles (numbers of GP/ml) in freeze-thaw supernatants obtained after reinfection of HEK-293 cells for 6 days with primary freeze-thaw lysates harvested 12 days after transfection of HEK-293 cells with the indicated pAdEasy plasmids. (C) Schematic presentation of pAdEasy-p5-Rep constructs. The p5, p19, and p40 promoters, the Rep78 ORF, and the recoded rep sequence (sRep) are indicated by differentially shaded boxes. The p5, p19, and p40 transcription start sites are represented by right-angled arrows, and the splice donor (SD) site at position 1906 is indicated by a vertical line. (D) Amounts of recombinant adenoviral particles (numbers of GP/ml) obtained after transfection and first-round amplification of the indicated pAdEasy-p5-Rep plasmids as described for panel B. (E) Western blot analysis of whole-cell extracts harvested either 2 days after transfection of the indicated pAdEasy-p5-Rep plasmids (upper panel) or 3 days after infection with the corresponding rAd supernatants (MOI of 1,000 GP/cell) from the first-round amplification (lower panel). (F) Southern blot detection of DpnI-resistant replicative rAAV intermediates in viral Hirt-DNA preparations obtained either 2 days after cotransfection of HEK-293 cells with the pAdEasy-p5-Rep constructs and pTR-UF5 (left lanes) or 3 days after infection of pTR-UF3-transfected HEK-293 cells with the corresponding rAd preparations from the first-round amplification (MOI of 1,000 GP/cell) (right lanes). pTR-UF5 and pTR-UF3 both contain an AAV-2 ITR-flanked GFP expression cassette, but they harbor differences in the transgene cassette leading to monomeric and dimeric replicative intermediates of different sizes (RFM and RFD). These were detected with a 0.7-kb GFP hybridization probe containing the complete GFP ORF and are indicated by arrows.
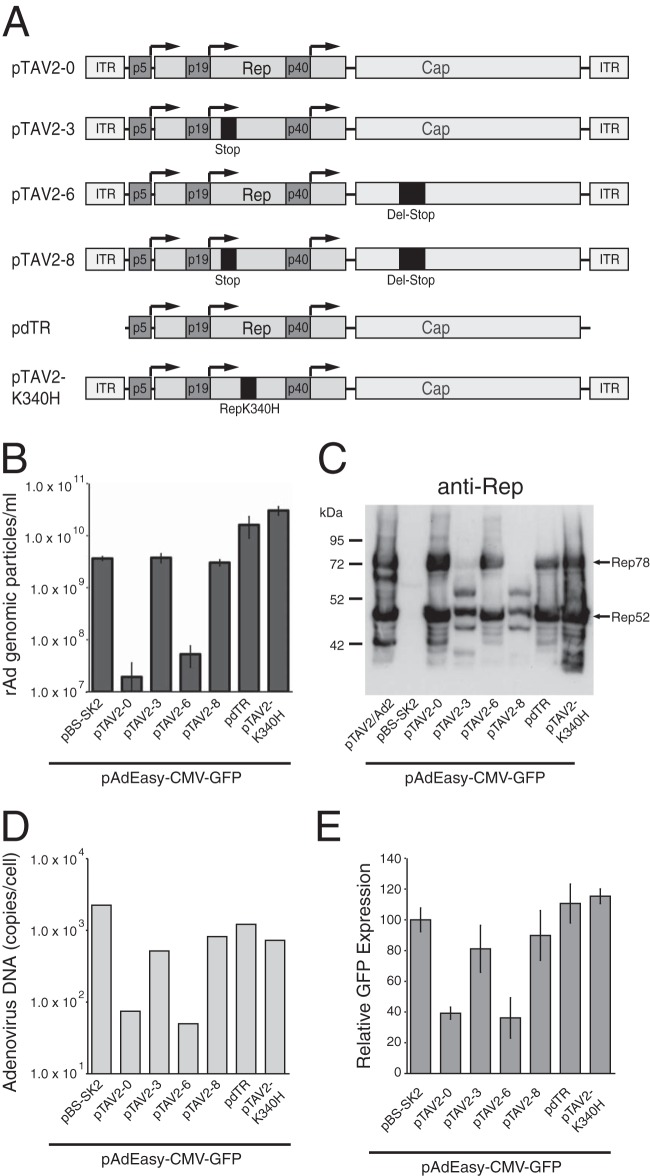
Rep protein expression strongly inhibits adenoviral replication in trans, in an AAV ITR-dependent manner.
To analyze the contribution of the RIS-Ad and Rep protein expression to the well-documented inhibition of adenoviral replication after AAV-Ad coinfection, we performed cotransfections of the pAdEasy-CMV-GFP plasmid with different AAV-2 constructs as depicted in Fig. 3A and used the formation of rAd-CMV-GFP viral particles after a 13-day transfection step and amplification of the primary freeze-thaw lysates for 5 days as the readout. This assay system should closely mimic AAV-Ad coinfections, since both viral genomes are present in the same cells, with the minor restriction that the AAV sequences are already present in a double-stranded form as opposed to the single-stranded form present after AAV infection. Cotransfection of the complete AAV-2 genome, including the ITRs (Fig. 3A, pTAV2-0), led to a strong inhibition of adenoviral replication, with only background amounts of genomic particles (Fig. 3B, pTAV2-0). With a series of AAV mutants (Fig. 3A) carrying frameshift mutations in either the rep gene (pTAV2-3), the cap gene (pTAV2-6), or both (pTAV2-8), we could clearly demonstrate that inhibition in trans requires Rep protein expression and no capsid protein expression (Fig. 3B). Importantly, inhibition could be abrogated completely by deletion of the AAV ITRs (Fig. 3A and B, pdTR). Since the presence of the ITRs did not lead to a marked increase in Rep protein levels (Fig. 3C, compare pTAV2-0 and pdTR lanes), ITR-dependent amplification of the AAV DNA genome per se seems to be required for inhibition. Furthermore, the mere presence of the ITRs was not sufficient for inhibition, as demonstrated with an AAV genome carrying a point mutation in the nucleotide binding site of the Rep proteins (pTAV-K340H). In short-term cotransfection assay mixtures harvested after 3 days, the inhibitory effect of the Rep proteins and the ITRs was associated with a >10-fold decrease in adenoviral genome replication (Fig. 3D) and a clearly reduced GFP expression (Fig. 3E).
FIG 3.
AAV-mediated inhibition of adenoviral replication in trans requires functional Rep protein expression and the AAV ITRs. (A) Schematic presentation of AAV-2 constructs cotransfected with the pAdEasy-CMV-GFP plasmid. The three promoter sequences, the Rep and Cap ORFs, and the ITRs are indicated by differentially shaded boxes. A frameshift mutation in the Rep ORF (Stop), a point mutation in the Rep nucleotide binding site (RepK340H), and a deletion-frameshift mutation in the Cap ORF (Del-Stop) are indicated by darker shaded boxes. (B) The AAV plasmids were cotransfected into HEK-293 cells with equal amounts of PacI-linearized pAdEasy-CMV-GFP DNA, and the rAd-CMV-GFP titers obtained after 13 days of transfection and amplification of the primary freeze-thaw lysates for 5 days were assayed as numbers of GP/ml. (C) Western blot analysis of Rep expression levels in whole-cell extracts harvested 2 days after cotransfection of HEK-293 cells as described for panel B. Anti-Rep-responsive bands for the Rep frameshift mutants (pTAV2-3 and pTAV2-8) were probably due to translation initiation at cryptic start sites. HEK-293 cells transfected with infectious AAV-2 plasmid pTAV2-0 and infected with adenovirus type 2 (MOI = 20) served as a positive control (lane pTAV2/Ad2). Positions of Rep78 and Rep52 proteins are indicated. (D) Amounts of newly replicated, DpnI-resistant adenoviral DNA obtained 3 days after cotransfection of HEK-293 cells as described for panel B, represented as numbers of genomic copies/cell. (E) Relative GFP expression levels, scored by fluorescence-activated cell sorter analysis 3 days after cotransfection of HEK-293 cells as described for panel B.
The RIS-Ad does not inhibit adenoviral replication in trans.
The above findings were further refined with constructs containing only the left part of the AAV-2 genome, with the three promoters and the Rep open reading frame (Fig. 4A). Whereas the p5/p19-driven Rep78 and Rep52 proteins (Fig. 4B), either in the wild-type form (p5Rep) or in the partially recoded form (p5-sRep), showed only a minor inhibition, and the Rep68/Rep40-like proteins (p5Rep1907) showed no inhibition of adenoviral replication in the cotransfection assay (Fig. 4C), production of rAd-CMV-GFP particles dropped to background levels after cotransfection of the corresponding constructs now flanked by the ITRs (Fig. 4B, pTR-Rep, pTR-Rep1907, and pTR-sRep). Again, Rep protein levels were very similar for both kinds of constructs (Fig. 4B), so the effect of the ITRs was not mediated indirectly by enhanced Rep expression.
FIG 4.
No involvement of the RIS-Ad in AAV-mediated inhibition of adenoviral replication in trans. (A) Schematic presentation of the AAV-2 Rep sequences cotransfected with the pAdEasy-CMV-GFP plasmid. The three promoter sequences, the Rep ORF, and the ITRs are indicated by differentially shaded boxes. For the Rep1701/2186 constructs, the p40 promoter is indicated by a hatched box and the intron by a gray shaded box. (B) Western blot analysis of Rep protein levels in whole-cell extracts harvested 2 days after cotransfection of HEK-293 cells with equal amounts of PacI-linearized pAdEasy-CMV-GFP DNA and the p5-Rep expression plasmids. (C) rAd-CMV-GFP titers obtained after 13 days of cotransfection analogous to that described for panel B and amplification of the primary freeze-thaw lysates for 6 days, assayed as numbers of GP/ml. (D) rAd-CMV-GFP titers (numbers of GP/ml) after 13 days of primary cotransfection with equal amounts of PacI-linearized pAdEasy-CMV-GFP DNA and the constructs harboring the 3′ rep sequence from AAV-2 nucleotides 1701 to 2186, in either the wild-type or recoded (sRep) form, both flanked by the AAV ITRs, followed by amplification of the primary freeze-thaw lysates for 6 days. Where indicated, the construct for the p5-driven Rep68-like protein (p5-Rep1907) was additionally cotransfected.
In line with the finding that AAV-mediated inhibition of adenoviral replication in trans required functional Rep protein expression, the RIS-Ad, which had completely abolished replication when present in cis in the context of the rAd/AAV hybrid vectors, had no effect in the cotransfection assay in either the absence (data not shown) or presence of the ITRs (Fig. 4A and D, pTR-Rep1701/2186). The ITR-flanked construct, however, could complement the inhibition mediated by the Rep68/Rep40-like proteins (Fig. 4D, pTR-Rep1701/2186 + p5-Rep1907), which demonstrates that the ITR template necessary for this inhibition can also be provided from a second plasmid.
Inactivation of the RIS-Ad is not sufficient to obtain rAd/AAV hybrid vectors containing the complete AAV genome.
As shown above, functional Rep78 protein under the control of the authentic p5 promoter could be expressed from rAd vectors by inactivation of the RIS-Ad through recoding of a small portion of the Rep78/52 ORF in the 3′ part of the rep gene (sRep; AAV nucleotides 1782 to 1916). We therefore asked whether the introduction of the sRep sequence could also restore the replicative properties of rAd vectors containing the complete AAV genome except for the ITRs (Fig. 5A, AAV-sRep). However, in contrast to the case with the corresponding Rep78 construct (compare Fig. 2C to F, p5-sRep), no adenoviral particles could be obtained after transfection of the recoded AdEasy/AAV plasmid (Fig. 5B). Furthermore, additional inactivation of functional Rep protein expression by introduction of the K340H point mutation also did not result in the formation of rAd particles (Fig. 5A and B, AAV-sRep-K340H). The same negative result was obtained with an AAV-sRep construct containing a frameshift mutation for Rep protein inactivation (data not shown). These findings point to the presence of further inhibitory elements in the AAV-2 genome, in addition to the RIS-Ad and the Rep proteins. In short-term transfection assays, the pAdEasy-AAV constructs showed the expected pattern of AAV-2 protein expression, with the sRep construct expressing much smaller amounts of capsid proteins than the wild-type construct and a further reduction in capsid protein levels observed after introduction of the Rep K340H point mutation (Fig. 5C).
FIG 5.
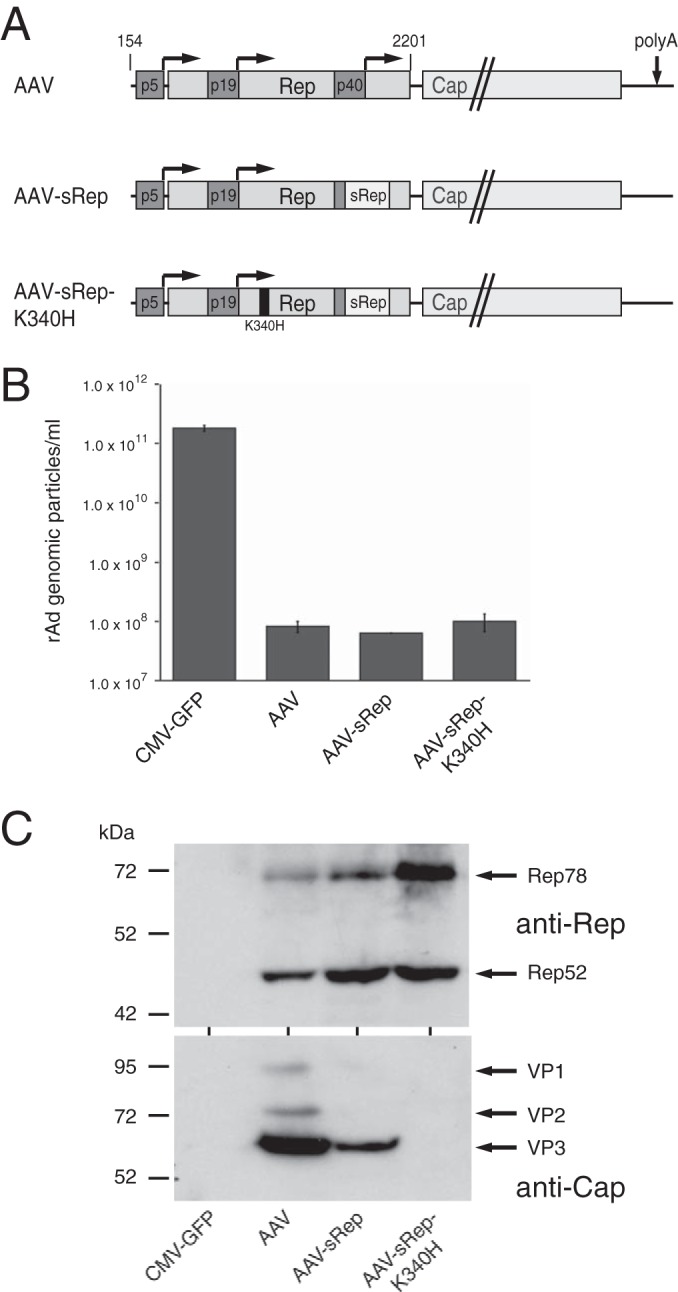
Inactivation of the RIS-Ad does not relieve suppression of adenoviral replication in the context of the complete AAV genome. (A) Schematic presentation of the AAV-2 sequences of the pAdEasy constructs transfected into HEK-293 cells for production of recombinant adenoviruses. The three AAV-2 promoter sequences and the Rep and Cap ORFs are indicated by differentially shaded boxes, the sRep sequence in the 3′ part of the rep gene by a white box, and the K340H point mutation by a black line. (B) Titers of recombinant adenoviruses (numbers of GP/ml) obtained after transfection of the indicated PacI-linearized pAdEasy-AAV constructs for 13 days and amplification of the primary freeze-thaw lysates for 6 days. (C) Western blot analysis of Rep and Cap expression levels in whole-cell extracts after transfection of the pAdEasy-AAV constructs for 2 days.
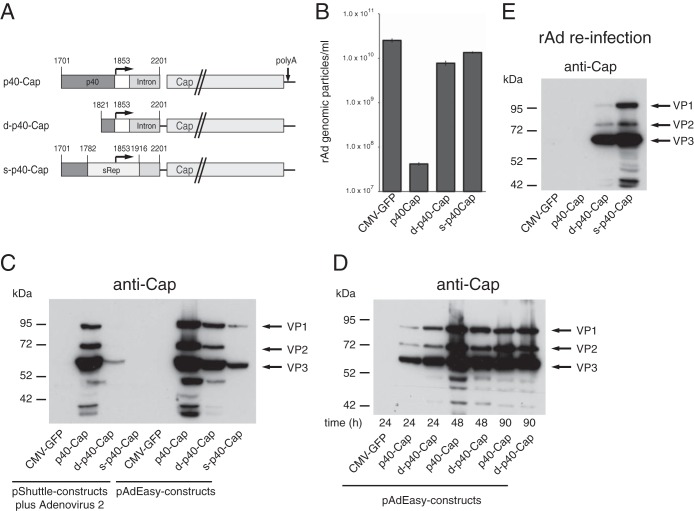
rAd-mediated constitutive expression of Cap proteins by inactivation of the RIS-Ad.
To address the existence of additional inhibitory sequence elements located in the cap gene and to also examine the possibility of expressing the AAV-2 Cap proteins from recombinant rAd vectors, the pAdEasy construct containing the isolated p40 promoter in front of the Cap ORF was subjected to further modifications. The initial construct had shown Cap expression levels similar to those with the complete AAV genome in short-term transfection assays (Fig. 1D) but had not given rise to rAd particles (Fig. 1C). By introduction of the sRep sequence or by deletion of the p40 promoter sequences, except for the TATA box (Fig. 6A, s-p40-Cap and d-p40-Cap, respectively), we were able to restore the replication levels of the corresponding rAd vectors to those of the CMV-GFP control construct (Fig. 6B). In the context of the pShuttle constructs used for the generation of the AdEasy constructs by homologous recombination, d-p40-Cap and s-p40-Cap showed very low VP expression levels in the presence of adenovirus superinfection compared to the wild-type construct (Fig. 6C, left part). Transfection of the d-p40-Cap and s-p40-Cap AdEasy constructs, however, led to clearly detectable levels of all three capsid proteins (Fig. 6C, right part). At some time points posttransfection, the expression rates of the d-p40-Cap construct even exceeded those of the wild-type construct (Fig. 6D). Thus, a lower p40 promoter activity may be compensated by adenoviral sequences in the AdEasy plasmid or by template replication. Most importantly, for both d-p40-Cap and s-p40-Cap, substantial levels of capsid protein expression were obtained after reinfection with the rAd preparation from the first amplification round (Fig. 6E), and the preparations could be amplified further in subsequent reinfection rounds, with titers similar to those of the rAd-CMV-GFP control virus (data not shown). Thus, changing sequences within the p40 promoter allows for the expression of the capsid proteins by rAd vectors, which argues against the presence of isolated further elements in the right side of the AAV-2 genome that are inhibitory for adenoviral replication in cis.
FIG 6.
rAd-mediated expression of AAV-2 capsid proteins after inactivation of the RIS-Ad. (A) Schematic presentation of the AAV-2 portion of the pAdEasy-p40-Cap constructs transfected into HEK-293 cells for production of recombinant adenoviruses. The p40 promoter, the intron, the Cap ORF, and the sRep sequence are indicated by differentially shaded boxes. (B) Titers of recombinant adenoviruses (numbers of GP/ml) obtained after transfection of the PacI-linearized pAdEasy-p40-Cap plasmids for 13 days and amplification of the primary freeze-thaw lysates for 6 days. (C) Western blot analysis of Cap protein expression levels in whole-cell extracts after transfection of HEK-293 cells with the pAdEasy-p40-Cap constructs (right) or of adenovirus type 2-infected cells (MOI = 20) with the corresponding pShuttle plasmids (left) for 2 days. (D) Time course of Cap expression after transfection of the pAdEasy-p40-Cap construct in the wild-type form (p40-Cap) and the p40 promoter-deleted form (d-p40-Cap) for the indicated periods. (E) Western blot analysis of whole-cell extracts harvested 3 days after infection (MOI of 1,000 GP/ml, except for p40-Cap [MOI of 10 GP/ml]) with the rAd preparations from the first-round amplification.
DISCUSSION
Almost since its initial identification as a contaminant of adenoviral preparations, it has become evident that AAV not only relies on helpers, such as adenovirus or different members of the herpesvirus family, for a productive replication cycle but also inhibits the replication of the respective helper, which is especially evident in the case of adenovirus (18, 19). The AAV-mediated inhibition of adenoviral replication has been attributed to the expression of the Rep proteins (20, 21, 31), and based on these findings, the difficulties encountered in various attempts to incorporate the rep gene region into the context of replication-competent first-generation adenoviral vectors (11, 12, 32) were also assigned to Rep protein expression. A recent study by Sitaraman et al. (22) strongly changed this prevailing picture of the rep gene-mediated inhibition of Ad/AAV hybrid vector propagation by showing the involvement of cis inhibitory effects. Ours is the first study to compare side by side the contributions of AAV-2 protein expression and cis elements to the inhibition of adenoviral replication in rAd/AAV hybrid vector generation and in AAV-2–Ad coinfection.
AAV-2–Ad coinfection was analyzed by determining the replication of a recombinant adenoviral vector genome cotransfected with different AAV-2 constructs. Whereas cotransfection of AAV-2 sequences differs from AAV-2 infection in that the AAV-2 genome is already present in a double-stranded, transcription-competent form, the kinetics of Rep expression should not be enhanced dramatically in the former case, since the onset of Rep protein expression was also found as early as 10 to 12 h after AAV-2–Ad coinfection (33). Transfection of the AAV-2 sequences offered the opportunity to manipulate genetic elements otherwise essential for the formation of infectious AAV-2 particles. This led to the important finding that strong inhibition of adenoviral replication in trans requires not only Rep protein expression but also AAV ITR-dependent DNA amplification. When expressed under the control of the authentic promoters in the absence of ITR sequences, only Rep78 and/or Rep52 slightly inhibits adenoviral replication. This inhibition is likely due to the C-terminal domain of Rep78/Rep52, absent in Rep68/Rep40, which was shown to downregulate adenoviral replication by inhibiting cyclic AMP-dependent protein kinase (PKA) through direct protein interaction (31). The dependence of Rep-mediated inhibition on the presence of an amplifiable DNA template also explains the rather weak inhibitory capacity of the Rep proteins under the control of the strong heterologous CMV promoter observed in earlier studies (20). Regarding the mechanism of inhibition of adenoviral propagation in trans by the combination of Rep proteins and ITR sequences, we hypothesize that Rep-dependent amplification of the ITR-flanked genome in the nuclear adenoviral replication compartments (21) inhibits adenoviral DNA amplification, most likely by direct competition for cellular replication and, possibly, transcription factors. Whereas direct Rep-mediated inhibition of early adenoviral gene expression could also contribute to the reduced propagation, this inhibition represents a consequence rather than a prerequisite for inhibition of adenoviral DNA replication (20).
In contrast to the AAV-mediated inhibition of adenoviral replication in trans, the lack of production of progeny virus with first-generation Ad/AAV hybrid vectors is largely due to a sequence element in the 3′ part of the rep gene, comprising the p40 promoter and the AAV-2 intron. This element, denoted the RIS-Ad, is not able to inhibit adenoviral replication in trans. This finding strongly suggests that it interferes with adenoviral DNA replication directly at the nucleic acid level. The identification of the RIS-Ad is in line with the recent findings of Sitaraman et al. (22), who investigated propagation of first-generation Ad5 vectors carrying the AAV-2 rep gene but used a tetracycline-inducible promoter instead of the physiological p5 promoter for expression of the large Rep proteins and omitted the Cap coding sequences. They found that recoding of AAV-2 nucleotides 1782 to 1916 was sufficient for propagation of rAd vectors after silencing of Rep protein expression, and we confirmed this finding by introduction of their recoded sequence (denoted sRep) into the RIS-Ad. Another possibility for ablating the inhibitory potential of the RIS-Ad is the deletion of large parts of the p40 promoter sequences. The sRep mutations affect several potential transcription factor binding sites in the p40 promoter, such the GGT element, the SP1 and AP1 sites, and the TATA box, as well as the p40 transcription start site, and lead to strongly reduced capsid expression. These findings suggest that at least part of the inhibition mediated by the RIS-Ad may be due to p40 promoter activity.
Based on the assumption that primarily Rep protein expression is responsible for the failure to generate rAd vectors containing a functional rep gene, many investigators have tried to minimize Rep78/Rep68 protein expression by using either promoters exhibiting very low expression in HEK-293 cells or inducible systems and have additionally ablated Rep52/Rep40 expression by mutation of the corresponding translation initiation codon (11–14, 22). Whereas these approaches were successful with helper-dependent (HD) adenoviral vectors propagated in the presence of a second adenoviral vector, they still failed with first-generation Ad vectors. Our results propose that the inhibition of rAd vector propagation mediated by the RIS-Ad may be lost in HD-Ad vector production, because the helper virus, which provides the adenoviral functions in trans, is not inhibited by the RIS-Ad.
By inactivation of the RIS-Ad, we assessed the contribution of functional Rep protein expression to the limited propagation of rAd/Rep vectors. We demonstrated that functional large Rep proteins can be expressed in the context of adenoviral vectors, even from the p5 promoter constitutively active in the HEK-293 cell line. Only a minor reduction of vector amplification was observed compared to the case for the control vector. Whereas we did not differentiate between the effects of the large and small Rep proteins, the shift toward preferential expression of the small Rep proteins after infection with the adenoviral preparation compared to transfection of the original adenoviral plasmids suggests that the large Rep proteins inhibit adenoviral replication. To eliminate the minor inhibition of adenoviral vector propagation mediated by the Rep proteins, future approaches for rAD-mediated expression of the Rep proteins based on inactivation of the RIS-Ad may rather use inducible promoters silenced during the propagation step to achieve higher titers. Minimization of Rep protein expression may also limit the selective loss of Rep sequences, as encountered in the sole study so far reporting the successful incorporation of the complete AAV-2 genome into a first-generation adenoviral vector (15). Such a loss of Rep sequences, maybe preferentially over the RIS-Ad element, probably also accounts for the small size or complete lack of Rep-reactive bands we observed after infections with the rAd/p5-Rep wild-type preparations originating from the adenoviral plasmids carrying the RIS-Ad.
Whereas inactivation of the RIS-Ad allows rAd-mediated, p5/p19-driven expression of the large and small Rep proteins, it unexpectedly does not restore propagation of rAd vectors containing the complete Rep and Cap coding regions. Furthermore, additional inactivation of functional Rep protein expression by introduction of a point mutation in the Rep nucleotide binding site (Fig. 5) or a frameshift mutation also did not result in rAd vector production. These results point to the possible presence of additional inhibitory elements in the cap gene. However, inactivation of the RIS-Ad in the rAd-p40-Cap vectors allows for highly efficient rAd vector propagation with titers comparable to those of the control virus. Thus, in addition to Rep protein expression and the RIS-Ad, further inhibitory effects of the AAV genome on rAd vector propagation exist, which may be mediated by complex interactions of elements present in the rep and cap regions.
With regard to the long-pursued goal of using rAD/AAV vectors to provide all the helper functions needed in trans for the simplified high-titer production of recombinant AAVs (rAAVs) by an infection step (15), both efficient Rep and Cap protein expression is required. Since inactivation of the RIS-Ad by introduction of the recoded sRep sequence seems to inhibit p40 activity and also affects the splice donor required for generation of the spliced transcripts encoding the Cap proteins, no major Cap expression levels can be expected from the corresponding adenoviral constructs. The initial experiments with the constructs containing the complete AAV-2 coding region confirmed these caveats. Somewhat surprisingly, however, substantial levels of Cap protein expression could be achieved with the AdEasy constructs containing only the p40-Cap cassette, both after introduction of the sRep sequence and after deletion of most of the p40 sequences upstream of the TATA box, and these also produced high levels of Cap proteins after reinfection with the rAd preparations. Thus, the amplification of the adenoviral genome, and maybe transcription factor sites located in the vicinity of the p40-cap cassette, can compensate for the reduction in gene expression required for inactivation of the RIS-Ad. Based on our findings, high-titer generation of rAAVs with the help of recombinant adenoviral vectors may be feasible by expression of the Rep and Cap proteins from two different adenoviral vectors.
In summary, we have set the framework for a better understanding of the complex interactions between AAV-2 and its helper adenovirus, with important implications for the future construction of adenoviral vectors expressing the AAV-2 Rep and Cap proteins.
ACKNOWLEDGMENT
We thank Nick Muzyczka (University of Florida) for kindly providing plasmids pTR-UF3 and pTR-UF5.
Footnotes
Published ahead of print 1 October 2014
REFERENCES
- 1.Atchison RW, Casto BC, Hammon WM. 1965. Adenovirus-associated defective virus particles. Science 149:754–756. 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 2.Buller RM, Janik JE, Sebring ED, Rose JA. 1981. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 40:241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotin RM, Siniscalco M, Samulski RJ, Zhu XD, Hunter L, Laughlin CA, McLaughlin S, Muzyczka N, Rocchi M, Berns KI. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U. S. A. 87:2211–2215. 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linden RM, Ward P, Giraud C, Winocour E, Berns KI. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U. S. A. 93:11288–11294. 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samulski RJ, Zhu X, Xiao X, Brook JD, Housman DE, Epstein N, Hunter LA. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941–3950 (Erratum, 11:1228, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maguire AM, High KA, Auricchio A, Wright JF, Pierce EA, Testa F, Mingozzi F, Bennicelli JL, Ying GS, Rossi S, Fulton A, Marshall KA, Banfi S, Chung DC, Morgan JI, Hauck B, Zelenaia O, Zhu X, Raffini L, Coppieters F, De Baere E, Shindler KS, Volpe NJ, Surace EM, Acerra C, Lyubarsky A, Redmond TM, Stone E, Sun J, McDonnell JW, Leroy BP, Simonelli F, Bennett J. 2009. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet 374:1597–1605. 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mingozzi F, High KA. 2011. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 12:341–355. 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 8.Palombo F, Monciotti A, Recchia A, Cortese R, Ciliberto G, La Monica N. 1998. Site-specific integration in mammalian cells mediated by a new hybrid baculovirus-adeno-associated virus vector. J. Virol. 72:5025–5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heister T, Heid I, Ackermann M, Fraefel C. 2002. Herpes simplex virus type 1/adeno-associated virus hybrid vectors mediate site-specific integration at the adeno-associated virus preintegration site, AAVS1, on human chromosome 19. J. Virol. 76:7163–7173. 10.1128/JVI.76.14.7163-7173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Mukherjee S, Fraefel C, Breakefield XO, Allen PD. 2002. Herpes simplex virus type 1 amplicon vector-mediated gene transfer to muscle. Hum. Gene Ther. 13:261–273. 10.1089/10430340252769789. [DOI] [PubMed] [Google Scholar]
- 11.Carlson CA, Shayakhmetov DM, Lieber A. 2002. An adenoviral expression system for AAV rep78 using homologous recombination. Mol. Ther. 6:91–98. 10.1006/mthe.2002.0634. [DOI] [PubMed] [Google Scholar]
- 12.Recchia A, Parks RJ, Lamartina S, Toniatti C, Pieroni L, Palombo F, Ciliberto G, Graham FL, Cortese R, La Monica N, Colloca S. 1999. Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc. Natl. Acad. Sci. U. S. A. 96:2615–2620. 10.1073/pnas.96.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Recchia A, Perani L, Sartori D, Olgiati C, Mavilio F. 2004. Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors. Mol. Ther. 10:660–670. 10.1016/j.ymthe.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Lieber A. 2006. A helper-dependent capsid-modified adenovirus vector expressing adeno-associated virus rep78 mediates site-specific integration of a 27-kilobase transgene cassette. J. Virol. 80:11699–11709. 10.1128/JVI.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang HG, Wang YM, Xie JF, Liang X, Hsu HC, Zhang X, Douglas J, Curiel DT, Mountz JD. 2001. Recombinant adenovirus expressing adeno-associated virus cap and rep proteins supports production of high-titer recombinant adeno-associated virus. Gene Ther. 8:704–712. 10.1038/sj.gt.3301454. [DOI] [PubMed] [Google Scholar]
- 16.Berns KI, Giraud C. 1996. Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218:1–23. [DOI] [PubMed] [Google Scholar]
- 17.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282–3291. 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casto BC, Armstrong JA, Atchison RW, Hammon WM. 1967. Studies on the relationship between adeno-associated virus type 1 (AAV-1) and adenoviruses. II. Inhibition of adenovirus plaques by AAV; its nature and specificity. Virology 33:452–458. [DOI] [PubMed] [Google Scholar]
- 19.Casto BC, Atchison RW, Hammon WM. 1967. Studies on the relationship between adeno-associated virus type I (AAV-1) and adenoviruses. I. Replication of AAV-1 in certain cell cultures and its effect on helper adenovirus. Virology 32:52–59. [DOI] [PubMed] [Google Scholar]
- 20.Timpe JM, Verrill KC, Trempe JP. 2006. Effects of adeno-associated virus on adenovirus replication and gene expression during coinfection. J. Virol. 80:7807–7815. 10.1128/JVI.00198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weitzman MD, Fisher KJ, Wilson JM. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sitaraman V, Hearing P, Ward CB, Gnatenko DV, Wimmer E, Mueller S, Skiena S, Bahou WF. 2011. Computationally designed adeno-associated virus (AAV) Rep78 is efficiently maintained within an adenovirus vector. Proc. Natl. Acad. Sci. U. S. A. 108:14294–14299. 10.1073/pnas.1102883108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winter K, von Kietzell K, Heilbronn R, Pozzuto T, Fechner H, Weger S. 2012. Roles of E4orf6 and VA I RNA in adenovirus-mediated stimulation of human parvovirus B19 DNA replication and structural gene expression. J. Virol. 86:5099–5109. 10.1128/JVI.06991-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 95:2509–2514. 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, Vogelstein B, He TC. 2007. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2:1236–1247. 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 26.Heilbronn R, Burkle A, Stephan S, zur Hausen H. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weger S, Wistuba A, Grimm D, Kleinschmidt JA. 1997. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J. Virol. 71:8437–8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horer M, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt JA. 1995. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J. Virol. 69:5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zolotukhin S, Potter M, Hauswirth WW, Guy J, Muzyczka N. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70:4646–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarty DM, Christensen M, Muzyczka N. 1991. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J. Virol. 65:2936–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Pasquale G, Chiorini JA. 2003. PKA/PrKX activity is a modulator of AAV/adenovirus interaction. EMBO J. 22:1716–1724. 10.1093/emboj/cdg153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher KJ, Kelley WM, Burda JF, Wilson JM. 1996. A novel adenovirus-adeno-associated virus hybrid vector that displays efficient rescue and delivery of the AAV genome. Hum. Gene Ther. 7:2079–2087. 10.1089/hum.1996.7.17-2079. [DOI] [PubMed] [Google Scholar]
- 33.Redemann BE, Mendelson E, Carter BJ. 1989. Adeno-associated virus rep protein synthesis during productive infection. J. Virol. 63:873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]