Abstract
A novel nonheme chloroperoxidase (RhEst1), with promiscuous esterase activity for enantioselective hydrolysis of ethyl (S)-2,2-dimethylcyclopropanecarboxylate, was identified from a shotgun library of Rhodococcus sp. strain ECU1013. RhEst1 was overexpressed in Escherichia coli BL21(DE3), purified to homogeneity, and functionally characterized. Fingerprinting analysis revealed that RhEst1 prefers para-nitrophenyl (pNP) esters of short-chain acyl groups. pNP esters with a cyclic acyl moiety, especially that with a cyclobutanyl group, were also substrates for RhEst1. The Km values for methyl 2,2-dimethylcyclopropanecarboxylate (DmCpCm) and ethyl 2,2-dimethylcyclopropane carboxylate (DmCpCe) were 0.25 and 0.43 mM, respectively. RhEst1 could serve as an efficient hydrolase for the bioproduction of optically pure (S)-2,2-dimethyl cyclopropane carboxylic acid (DmCpCa), which is an important chiral building block for cilastatin. As much as 0.5 M DmCpCe was enantioselectively hydrolyzed into (S)-DmCpCa, with a molar yield of 47.8% and an enantiomeric excess (ee) of 97.5%, indicating an extremely high enantioselectivity (E = 240) of this novel and unique biocatalyst for green manufacturing of highly valuable chiral chemicals.
INTRODUCTION
Biological systems have evolved three classes of haloperoxidase enzymes capable of incorporating halogen atoms into molecules of organic substrates in the presence of hydrogen peroxide and halide ions (1). These classes are heme- and vanadium-dependent and nonheme haloperoxidases (2–4). High catalytic promiscuity has been found in haloperoxidases. For instance, sulfide oxidation (5), halogenation (6), phosphatase oxidation (7), epoxidation (8), indole oxidation (9), and amino oxidation (10) were catalyzed with heme- and vanadium-dependent haloperoxidases. The nonheme haloperoxidases were discovered to have significant differences from prosthetic group-dependent haloperoxidases (1, 11). Sequence and structure analysis showed that these enzymes belong to a larger group of esterase/lipase enzymes (12, 13).
Cilastatin, consisting of optically active (S)-(+)-2,2-dimethylcyclopropylformamide (or 2,2-dimethylcyclopropylcarboxylic acid) (DmCpCa), l-cysteine, and 7-chloro-2-oxoheptylic acid (Fig. 1), works as a renal dehydropeptidase inhibitor to prevent degradation of imipenem by renal dehydropeptidase in the kidney (14). The combination of cilastatin-imipenem (commercially known as Tienam) is the first choice in the clinic for the treatment of severe infections (15–17). The efficient preparation of (S)-DmCpCa, a key chiral precursor of cilastatin, draws much interest due to its vital role in saving people from bacterial infections.
FIG 1.
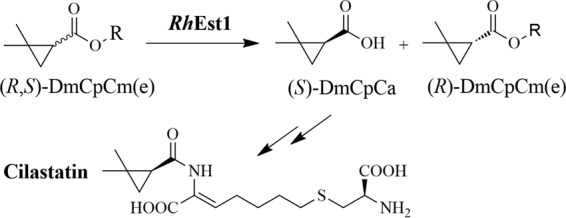
Enantioselective resolution of a racemic mixture of rac-DmCpCm(e) (that is, rac-DmCpCm or rac-DmCpCe) by RhEst1 for the production of a chiral cilastatin intermediate.
Highly efficient and enantioselective synthesis of optically pure organic acids, especially (S)-DmCpCa, represents a greater challenge than the synthesis of chiral alcohols by either chemical or enzymatic methods. Various strategies have been developed recently. With regard to chemical routes, the adoption of heavy metals or stoichiometric chiral resolution reagents, the use of precious chiral ligands, and also the relatively low enantioselectivity (E) value impede the practical applications (14, 18, 19). Enzymatic preparation of chiral synthons provides a direct, efficient, green, and highly chemoselective/enantioselective alternative, due to the excellent catalytic properties of diverse enzymes (20–22). Biocatalytic production of (S)-DmCpCa can be divided into three methods, depending on the starting substrates 2,2-dimethylcyclopropanecarbonitrile (23–25), 2,2-dimethylcyclopropanecarboxamide (26–28), and 2,2-dimethylcyclopropanecarboxylic esters (29, 30). The best record of enzymatic process was achieved by using recombinant (R)-selective nitrile hydratase and amidase from Comamonas acidovorans, which gave a 35% isolated yield and a 98.2% enantiomeric excess (ee) with 300 mM substrate loading (25). Zheng et al. previously isolated two (R)-amidase-producing strains, Delftia tsuruhatensis CCTCC 205114 and Brevibacterium epidermidis CCTCC 207076, which displayed a >99% ee but low substrate loading (10 mM) (26). In addition, the enantioselective resolution of 2,2-dimethylcyclopropanecarboxylic esters with hydrolases provides a much more efficient route. After testing several commercial lipases, Liang et al. found that only immobilized Candida antarctica lipase B (CALB) (Novozyme 435) showed suitable activity and enantioselectivity (30). In about 65 h, 65 mM ethyl 2,2-dimethylcyclopropanecarboxylate (DmCpCe) was enantioselectively resolved into (S)-DmCpCa with a 45.6% yield and a 99.2% ee. However, due mainly to the high dosage (16 g/liter), expensive price, and instable adsorption of CALB on its carrier, progress in industrial application has been very limited (31).
In order to discover a robust (S)-preferential DmCpCm hydrolase with industrial application potential, much research has been carried out in our laboratory. In our previous work, an active esterase-producing strain, Rhodococcus sp. strain ECU1013, was identified from soil samples by a target-reaction-oriented screening strategy (32). It was interesting to notice that there might be several isoenzymes in the strain with activities on either enantiomer of the racemic mixture (rac-DmCpCm). In this work, a shotgun gene library of the Rhodococcus sp. ECU1013 genome was constructed in order to identify the active (S)-preferential DmCpCm hydrolase (here designated RhEst1). After heterogeneous overexpression and purification of the recombinant esterase RhEst1, its biochemical properties and application potential for the production of a chiral cilastatin precursor were investigated in detail.
MATERIALS AND METHODS
Reagents.
Racemic ethyl 2,2-dimethylcyclopropanecarboxylate [(±)-DmCpCe] was kindly donated by Hisoar Pharmaceutical Co., Ltd. (Zhejiang, China). Racemic DmCpCa was prepared by chemical hydrolysis of racemic (±)-DmCpCe with 10% (wt/vol) NaOH and acidification with 20% (wt/vol) H2SO4 with a >90% yield and >98% purity. Racemic methyl 2,2-dimethylcyclopropanecarboxylate [(±)-DmCpCm] was prepared from (±)-DmCpCa and methanol by chemical esterification, which was verified by using nuclear magnetic resonance (NMR). Chromogenic p-nitrophenyl esters were previously prepared in our laboratory (32).
Construction and screening of the shotgun gene library.
Rhodococcus sp. strain ECU1013 was screened from soil samples and cultured in an enrichment medium (pH 7.0) containing the following components per liter of deionized water: 15 g glycerol, 5 g yeast extract, 5 g peptone, 5 g NaCl, 5 g KH2PO4, and 0.2 g MgSO4 (32). The genomic DNA of Rhodococcus sp. ECU1013 was extracted by employing the high-salt-concentration precipitation method (33). By strictly controlling the enzymatic digestion time, a shotgun gene library with a length of 2 to 6 kb was developed by digestion with Sau3AI and recovered with a DNA recovery kit (Tiangen, Shanghai, China). Based on the same cohesive terminus after digestion with Sau3AI and BamHI, the shotgun genes were ligated into BamHI/bacterial alkaline phosphatase (BAP)-digested plasmid pUC118. After transformation into Escherichia coli DH5α, a shotgun library expressing the proteins from Rhodococcus sp. ECU1013 with an endogenous promoter was constructed. A stepwise two-round screening strategy was adopted. Primary screening was carried out by inoculation onto LB agar plates supplemented with ampicillin (100 mg/liter) and tributyrin (0.25% [vol/vol]). After incubation at 37°C for 12 h, the potentially positive clones with visible halos produced by enzymatic hydrolysis of tributyrin were chosen and further cultured in LB liquid medium for secondary screening with DmCpCm as a real substrate. The ee and yield of the product were determined by using a gas chromatography (GC) instrument (GC-2014; Shimadzu) equipped with a flame ionization detector and a Chirasil-Dex CB capillary column (25 m by 0.25 mm; Varian Co., Palo Alto, CA, USA). N2 was used as the carrier gas. The temperatures of the detector and injector were both 280°C. The column temperature was kept at 80°C for 3 min, raised to 180°C at a rate of 10°C/min, and maintained for 5 min, as described previously (32). The retention times of (S)-DmCpCa and (R)-DmCpCa were 9.09 and 9.47 min, respectively.
Cloning, expression, and purification.
The positive clones with a significant product peak on GC chromatograms were sequenced. Open reading frames (ORFs) were analyzed with Omiga 2.0. Ten potential protein-coding genes with lengths of between 500 and 1,500 bp were identified. To achieve high expression levels, the positive genes were cloned into the pET28a vector under the T7 promoter. The coding regions of these genes were amplified from genomic DNA of Rhodococcus sp. ECU1013 by PCR (Bio-Rad, Hercules, CA, USA), and the amplified PCR fragments were ligated into the pGEM-T vector and transformed into E. coli DH5α. The resulting plasmids were sequence confirmed by Sunny Biotechnology Co., Ltd., Shanghai, China. The plasmids were then digested with the relevant restriction enzymes, and the restriction fragments were subcloned into the pET28a expression vector. The resulting expression plasmids were introduced into E. coli strain BL21(DE3). The positive transformants were cultivated in LB medium at 37°C at 180 rpm. When the optical density at 600 nm (OD600) reached 0.6 to 0.8, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 200 μM, and the cultivation temperature was decreased to 16°C for protein overexpression. After 12 h of cultivation, the cells were harvested by centrifugation at 5,000 × g for 10 min, and the pellets were washed twice with ice-chilled 100 mM potassium phosphate buffer (KPB) (pH 7.0). The cells were resuspended in the same buffer and disrupted by ultrasonication, and the cell lysates were centrifuged at 10,000 × g at 4°C for 20 min to remove the particulate fraction. To search for the (S)-preferential DmCpCm hydrolase (RhEst1), the activity of all the predicted proteins was examined with DmCpCm as the substrate. Purification of the corresponding enzyme RhEst1 was performed as described previously (34). The purity and molecular weight of RhEst1 were investigated by using SDS-PAGE and a gel exclusion chromatograph equipped with a TSK G2000 SWx1 column.
Enzyme activity and enantioselectivity assay.
The general activity assay was conducted spectrophotometrically by measuring the increase in the absorption of p-nitrophenol at 405 nm with p-nitrophenyl propionate (pNPPr) as the substrate. The reaction mixture, composed of 1.0 mM pNPPr and the appropriate amount of enzyme solution in KPB (pH 7.0; 100 mM), was incubated at 30°C for 5 min. One unit of activity was defined as the amount of enzyme required for the production of 1.0 μmol p-nitrophenol per minute under the assay conditions. The standard reaction for DmCpCm hydrolysis, with a mixture consisting of 20 mM DmCpCm in KPB, was performed at 30°C at 900 rpm in a Thermomixer (Eppendorf, Germany). After a certain time, the reaction was terminated by the addition of 20% H2SO4 to the mixture and extraction with an equal volume of ethyl acetate (containing 1 mM dodecane as an internal standard). The organic phase was transferred into a new Eppendorf tube, dried over anhydrous Na2SO4 for 12 h, and subjected to GC analysis. Enantioselectivity was measured and calculated by using a chiral GC instrument equipped with a CP-Chirasil-DEX CB column (Varian, USA), as reported previously (32).
Characterization of recombinant esterase RhEst1.
Kinetic constant analysis of the purified enzyme was performed by determining the initial rates with the substrate DmCpCm or DmCpCe at various concentrations (0.2 to 10 mM). The apparent Km and Vmax values of the purified enzyme were calculated from the Lineweaver-Burk plot (35). The pH profile of RhEst1 was investigated by using spectrophotometric assays with the following buffers (final concentration, 100 mM): sodium citrate (pH 3.0 to 6.0), sodium phosphate (pH 6.0 to 8.0), Tris-HCl (pH 8.0 to 9.0), and sodium carbonate (pH 9.0 to 10.0). The temperature profile of RhEst1 was determined at various temperatures (20°C to 70°C). The highest activity was regarded as 100%. Thermal stability was determined by preincubating purified RhEst1 (0.1 mg protein/ml) at the desired temperature (30°C, 40°C, or 50°C) in KPB (100 mM; pH 8.0) for a certain time period, followed by measurement of the residual activity. The activity at 0 h was regarded as 100%. Various chromogenic pNP ester substrates (substrate 1 [S1] to S18) with different acyl donors were adopted to examine the acyl group preference. The activities were spectrophotometrically assayed with 1 mM S1 to S18 by using standard protocols. All the presented results are average values of data from triplicate experiments.
Optimization of rac-DmCpCe bioresolution.
Various organic solvents, including hexadecane, isooctane, n-heptane, n-hexane, cyclohexane, and toluene, were tested as organic phases of a biphasic reaction system. The effect of organic solvents on enzyme activity was examined with 5 ml enzyme solution (6.0 mg/ml) and 5 ml organic solvent. The presented results are average values of data from triplicate experiments with standard deviations. After incubation at 30°C at 180 rpm for 12 h, the reaction mixture was centrifuged (8,000 × g) to facilitate phase separation. The lower enzyme-containing phase was transferred into another tube for measurement of the residual activity by using standard protocols. RhEst1-mediated enantioselective resolution of racemic DmCpCe in the biphasic phase was performed by using 50% (vol/vol) organic solvent. The reaction mixture, consisting of 30 mg cell extract (lyophilized) of RhEst1 (18.8 U/mg using pNPPr as the substrate) in 5.0 ml KPB (pH 8.0; 100 mM) as well as 36 mM DmCpCe in 5.0 ml organic solvent, was carried out at 30°C at 180 rpm for 2 h. The reaction was terminated, and samples were extracted and analyzed as mentioned above. The effect of the substrate concentration on the conversion rate and enantioselectivity of RhEst1 was examined with a reaction mixture containing 20 ml KPB (pH 8.0; 100 mM), 200 mg or 500 mg RhEst1, and 100 to 500 mM DmCpCe at 30°C in a sealed flask with inner baffles. The reaction mixture was magnetically agitated until (S)-DmCpCe disappeared, and samples were intermittently removed and extracted to analyze the conversion rate. To maintain the pH at 8.0, the reaction mixture was titrated with 1.0 M NaOH.
Enantioselective synthesis of (S)-DmCpCa on a preparative scale.
A reaction mixture containing 100 ml KPB (pH 8.0; 100 mM), 2.5 g cell extract of RhEst1 (18.8 U/mg), and 51.9 mmol (7.42 g) DmCpCe was magnetically stirred at 30°C for 13 h. NaOH (1.0 M) was added by an autotitrator (785 DMP Titrino; Metrohm) to maintain the pH at 8.0. After the reaction was terminated, the reaction mixture was extracted with dichloromethane under alkaline conditions to remove the substrate, and 20% H2SO4 was then added to acidify the reaction mixture to pH <2.0. The mixture was then extracted with 100 ml dichloromethane three times. The organic layer was dried over anhydrous Na2SO4 and evaporated at 40°C at normal pressure.
Nucleotide sequence accession number.
The RhEst1 nucleotide sequence identified in this study has been deposited in the GenBank database under accession no. KJ179840.
RESULTS AND DISCUSSION
Discovery of RhEst1 from the shotgun gene library of Rhodococcus sp. ECU1013.
A shotgun gene library was successfully constructed by ligation of 2- to 6-kb fragments of the Sau3AI-digested Rhodococcus sp. ECU1013 genome into pUC118 (BamHI/BAP) and transformation into E. coli DH5α. After being transferred onto LB agar plates supplemented with ampicillin (100 mg/liter) and tributyrin (0.25% [vol/vol]), potentially positive clones, inserted with hydrolase expression fragments, produced transparent halos after incubation for <12 h at 37°C. The positive clones with apparent halos were then further cultured in LB liquid medium and subjected to secondary screening with DmCpCm as the substrate.
Around 20,000 clones were picked and transferred onto tributyrin agar plates. Among them, 225 transformants displayed apparent halos, with a positive rate of approximately 1%. However, upon further examination of the 225 clones with respect to their activity and enantioselectivity toward the hydrolysis of DmCpCm, only 12 clones showed an apparent product peak on their chiral GC chromatograms. Among them, 9 clones (75%) displayed an R preference, while the other 3 clones showed an S preference in the enantioselective resolution of (R,S)-DmCpCm. Because of the low expression level of foreign fragments in plasmid pUC118, the conversion rates of all 12 clones were <2%. All of the clones were sequenced, assembled, and predicted by using the NCBI online tool. Ten ORFs, predicted to be potential esterases or belonging to α/β hydrolases, were subcloned into the pET28a vector. Six of them were expressed as inclusion bodies. Fortunately, one of the remaining four subclones, which expressed the target enzyme in the soluble or partially soluble form, displayed obvious activity on our substrate and was designated RhEst1.
Sequence analysis indicated that RhEst1 consisted of 828 bp with a GC content of 65.6%, encoding a protein of 275 amino acids. A putative Shine-Dalgarno sequence (AGCGGAGGTGGGGATG [underlining indicating the Shine-Dalgarno sequence]) was located 5 bp upstream of the start codon ATG. The deduced protein was searched against the Protein Data Bank (PDB) (http://blast.ncbi.nlm.nih.gov/) by using the BLASTP program. The results showed that the majority of the retrieved proteins were haloperoxidases and esterases. The most closely related proteins include cofactor-free chloroperoxidase L (CPO-L) from Streptomyces lividans TK64 (GenBank accession no. AAA18642 and PDB accession no. 1A88) (33% identity) (36), nonheme bromoperoxidase A1 (BPO-A1) from Streptomyces aureofaciens ATCC 10762 (GenBank accession no. AAC43253 and PDB accession no. 1A8Q) (30% identity), and a stereoselective esterase from Pseudomonas putida IFO 12996 (GenBank accession no. ABA39859 and PDB accession no. 1ZOI) (30% identity). Based on amino acid sequences, a phylogenetic tree was constructed (Fig. 2) in order to verify the classification and evolutionary relationship of RhEst1 with other known haloperoxidases and lipases/esterases. Twenty-three lipases/esterases and haloperoxidases belonging to different families (families I to VIII of bacterial lipolytic enzymes) were selected and analyzed. As illustrated in Fig. 2, RhEst1 lies in the branch of family V and displays high sequence similarity with nonheme haloperoxidases, indicating that it is a potential member of the nonheme haloperoxidases. Sequence alignment of RhEst1 with six nonheme haloperoxidases was carried out (Fig. 3). Accordingly, the Gly-X-Ser-X-Gly and His-Gly motifs of nonheme haloperoxidases were detected in RhEst1, which is also the typical consensus for the serine-hydrolase family, including lipases/esterases (37). The distance between the two motifs is similar to the extent of ∼65 to 70 amino acids (38) in various lipases, N-acetylhydrolases, and esterases. Concerning the catalytic residues in nonheme haloperoxidases, Ser101, Asp225, and His253 were also conserved with other nonheme haloperoxidases and therefore were tentatively assigned as the catalytic triad of RhEst1.
FIG 2.
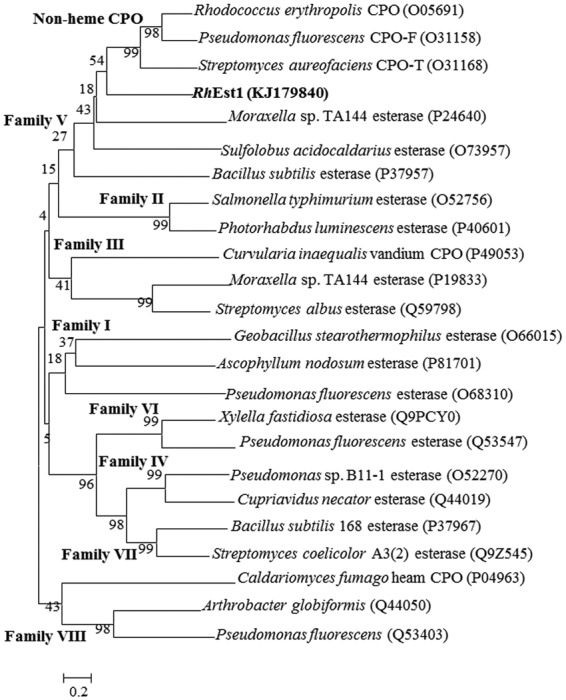
Phylogenetic analysis of RhEst1 and closely related proteins. The phylogenetic tree was constructed with MEGA 6.0 software by the neighbor-joining method. The bootstrap values were based on 10,000 replicates. All protein sequences, except for RhEst1, were retrieved from Swiss-Prot (http://www.uniprot.org/). Numbers in parentheses are SwissProt accession numbers of published sequences. The value near each node is the bootstrap value.
FIG 3.
Sequence alignment of RhEst1 and nonheme haloperoxidase. The proteins are as follows: BPO-A2 (Streptomyces aureofaciens; SwissProt accession no. P29715), CPO-T (Streptomyces aureofaciens; accession no. O31168), CPO (Serratia marcescens; accession no. L7ZQX6), CPO-F (Pseudomonas fluorescens; accession no. O31158), CPO-P (Pseudomonas pyrrocinia; accession no. P25026), CPO-L (Streptomyces lividans; accession no. P49323). The template of secondary structure is referred to as BPO-A2 (PDB accession no. 1A8Q). Boxes indicate amino acids with high identity. Consensus amino acids are displayed with a dark gray background. Amino acids of the catalytic triad are marked by triangles.
Biochemical properties and kinetic parameters of RhEst1.
RhEst1 with an N-terminal His tag was purified to electrophoretic homogeneity by nickel affinity chromatography (see Fig. S1 in the supplemental material). The purified enzyme migrated as a single band with a size of about 35.0 kDa by SDS-PAGE, in agreement with its theoretical value. Additionally, the purified protein was eluted from the TSK G2000 SWx1 column as a single peak with an elution volume corresponding to an apparent molecular mass of 69.8 kDa, indicating that RhEst1 is a homodimeric enzyme consisting of two identical subunits.
The activity of purified RhEst1 was measured at various pH values ranging from 3.0 to 10.0 by using standard protocols. The maximum activity was observed at pH ∼8.5, and >90% of the maximum activity was retained at between pH 8.0 and 9.0 (Fig. 4A). The effect of temperature on RhEst1 activity was investigated at temperatures from 20°C to 70°C. As shown in Fig. 4B, the optimum temperature was found to be 60°C. Furthermore, the thermostability of RhEst1 was examined at temperatures of 30°C, 40°C, and 50°C. According to thermal inactivation curves in Fig. 4C, the enzyme had half-lives (t1/2) of 112, 22, and 0.24 h at 30°C, 40°C, and 50°C, respectively. This finding suggests that although higher temperatures resulted in a rapid loss of activity, RhEst1 was indeed quite stable under mild reaction conditions (30°C to 40°C). The properties of RhEst1 and chloroperoxidase from Serratia marcescens (SmCPO) were also compared. The optimum pNP phosphate hydrolysis activity of SmCPO was found to be between pH 5.2 and pH 6.1. The enzymatic reactions can proceed at comparatively high temperatures, and the activity decrease was not observed until 65°C (7). The optimum reaction temperature of SmCPO was slightly higher than that of RhEst1. This difference between RhEst1 and SmCPO also confirms the novelty of RhEst1.
FIG 4.
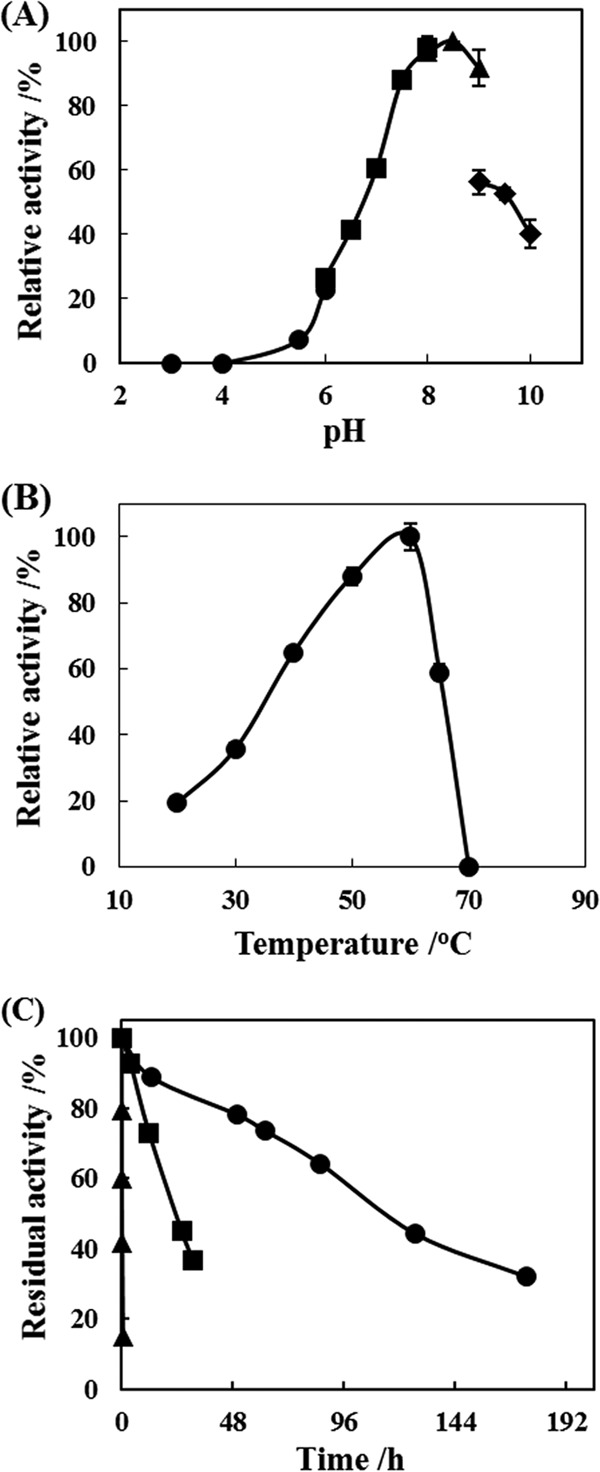
Effects of temperature and pH on activity of purified RhEst1. (A) The pH profile was calculated according to standard protocols by using the following buffers (100 mM): sodium citrate (●), sodium phosphate (◼), Tris-HCl (▲), and sodium carbonate (◆). The enzyme activity of 198 μmol/min/mg protein was defined as 100%. (B) Temperature profile determined at various temperatures (20°C to 70°C) in KPB (pH 8.0; 100 mM). The enzyme activity of 522 μmol/min/mg protein was defined as 100%. (C) Thermostability was estimated at 30°C (●), 40°C (◼), and 50°C (▲). The specific activity of the enzyme at 100% was 193 U/mg.
The kinetic constants of purified RhEst1 were calculated from a Lineweaver-Burk double-reciprocal plot. The Km values of the esterase toward DmCpCm and DmCpCe were 0.25 and 0.43 mM, respectively. Considering that the solubility (2.6 g/liter; ca. 18.3 mM) of DmCpCe was 43-fold higher than the Km (0.43 mM), the Km values of the enzyme are consistent with its high activity at a relatively low concentration of the dissolved substrate and are beneficial for the enantioselective preparation of optically pure (S)-DmCpCa.
Acyl preference of RhEst1.
Eighteen chromogenic pNP esters with different acyl groups (Fig. 5), including aliphatic, cyclic, and aromatic acyls, were chosen from the chromogenic pNP ester library developed recently in our laboratory for rapid fingerprinting of lipolytic/esterolytic enzymes (39).
FIG 5.
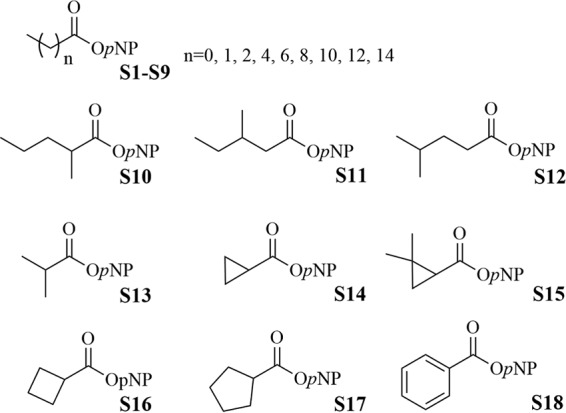
Various chromogenic pNP esters with different acyl groups employed for characterization of the substrate (acyl) specificity of RhEst1.
The substrate profile of RhEst1 (Fig. 6) revealed a preference for short-chain aliphatic esters. Among all the linear aliphatic pNP esters (S1 to S9), the catalytic activity decreased with increasing fatty acid chain length, except for para-nitrophenyl acetate (pNPA) (S1). Propionyl (S2) seems to be the best acyl moiety of the pNP esters, with a specific activity of 141 U · mg−1. No activity was detected for the long-chain aliphatic esters S7, S8, and S9. Compared to straight-chain aliphatic esters, branch-chained aliphatic esters were relatively poorer substrates to be hydrolyzed by RhEst1 (S10, S11, and S12 versus S4). However, the specific activity of branched pNP-isobutyrate (iBu) (S13) was higher than that of linear pNP-normal butyrate (nBu) (S3), perhaps due to the more compact size of iso-butanoyl. For esters with a cyclic side chain, the highest activity was observed on the ester with a cyclobutyl side chain (S16). In contrast, cyclopropanecarboxylic acid esters (S14 and S15), especially the pharmaceutically relevant 2,2-dimethylcyclopropanecarboxylic acid esters (S15, an analog of DmCpCm or DmCpCe), were much more difficult to hydrolyze. RhEst1 did not display any hydrolysis activity toward benzoyl ester (S18). Compared with other esterases reported previously, RhEst1 has a relatively high activity toward pNP propionate (39).
FIG 6.
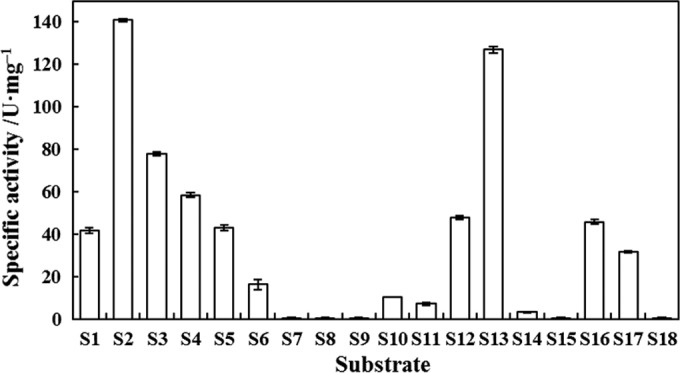
Activity fingerprint of RhEst1 with an array of chromogenic esters made of p-nitrophenol and various carboxylic acids.
Organic solvent tolerance of RhEst1 for biphasic resolution of rac-DmCpCe.
Due to the low solubility (2.6 g/liter) and relatively high volatility (400 Pa at 25°C) of rac-DmCpCe, a biphasic reaction system was attempted. Six organic solvents, which are frequently used as an organic phase, were introduced into the RhEst1-catalyzed kinetic resolution of rac-DmCpCe. After incubation with an equal volume of organic solvent at 30°C at 180 rpm for 12 h, the residual activity of RhEst1 located in the aqueous phase was determined. As shown in Table 1, RhEst1 displayed >60% of the initial activity in all the tested solvents. At the same time, all the eep (enantiomeric excess of product) values observed for all the tested solvent systems were >97%. With increased logP (logarithm of the partition coefficient of the solvent in a standard mixture of 1-octanol and water) values, the residual activity and conversion rate increased. The highest activity and conversion rate were found with hexadecane. However, in the neat aqueous system (as a control), the residual activity and conversion rate were the best, 89.1% and 46.5%, respectively. Although extension of the reaction time in the biphasic system further improved the conversion rate, it still could not exceed the result of the aqueous monophase system (data not shown). The inhibition of enzyme activity by organic solvents and the long reaction time might account for the loss (evaporation) of the substrate and the lower conversion rate. In spite of the high conversion rate, the extremely high boiling point of hexadecane, which is higher than those of DmCpCe (substrate) and DmCpCa (product), might be disadvantageous for the isolation of the target product (S)-DmCpCa. Isooctane would be a good choice of organic phase for composing a biphasic system, if necessary.
TABLE 1.
Effect of organic solvents on bioresolution of rac-DmCpCe
| Organic solvent | LogP | Mean residual activity (%) ± SD | Conversion rate (%) | eep (%) |
|---|---|---|---|---|
| Control | 89.1 ± 2.3 | 46.5 | 99.3 | |
| Hexadecane | 8.9 | 83.5 ± 0.7 | 38.6 | 98.6 |
| Isooctane | 4.5 | 76.0 ± 3.3 | 33.9 | 98.4 |
| n-Heptane | 4.0 | 77.0 ± 0.9 | 22.6 | 97.7 |
| n-Hexane | 3.5 | 73.7 ± 0.9 | 17.8 | 97.3 |
| Cyclohexane | 3.2 | 63.8 ± 0.1 | 18.3 | 98.0 |
| Toluene | 2.5 | 75.3 ± 1.4 | 13.1 | 98.0 |
Performance of RhEst1 in preparative synthesis of (S)-DmCpCa.
The effect of substrate loading on the enantioselective resolution of rac-DmCpCe in a neat aqueous system was investigated by using tightly closed anaerobic bottles with inner plugs to prevent the evaporation of the substrate. The racemic ester with various loads of 100, 200, and 500 mM was hydrolyzed by the same dosage of RhEst1 (10 mg/ml). Within 6 h, approximately 47.8 mM (S)-DmCpCa with an eep of ∼97.0% was produced from 100 mM rac-DmCpCe. The reaction was then terminated, since no (S)-substrate was detected. In the case of 200 mM substrate loading, the production of (S)-DmCpCa was increased up to 96.2 mM after an ∼10-h reaction, with a molar yield of 48.1% and an eep of 96.3%. However, when the substrate loading concentration was further increased to 500 mM, only a 115.5 mM product was detected, affording a 98.0% eep but merely a 23.1% yield (Fig. 7). Furthermore, some protein-like precipitates were observed in the reaction mixture, indicating that the enzyme had lost its activity. Therefore, another batch of reaction mixtures with 500 mM substrate was made, with an increased dosage (25 mg/ml) of RhEst1, as illustrated in Fig. 7. Under such conditions, (S)-ester was efficiently hydrolyzed into (S)-acid. At about 21 h, the production of (S)-DmCpCa reached 239.2 mM, giving a 47.8% yield and a 97.5% eep (E = 240), and no (S)-ester was detected in the reaction mixture. Similarly, preparative synthesis of (S)-DmCpCa was performed in 100 ml KPB (pH 8.0; 100 mM) containing 2.5 g RhEst1 and 51.9 mmol DmCpCe (ca. 7.42 g). After enzymatic resolution for ∼13 h, 1.82 g (30.8% yield and 98.0% ee) of (S)-DmCpCa was recovered after a simple workup.
FIG 7.
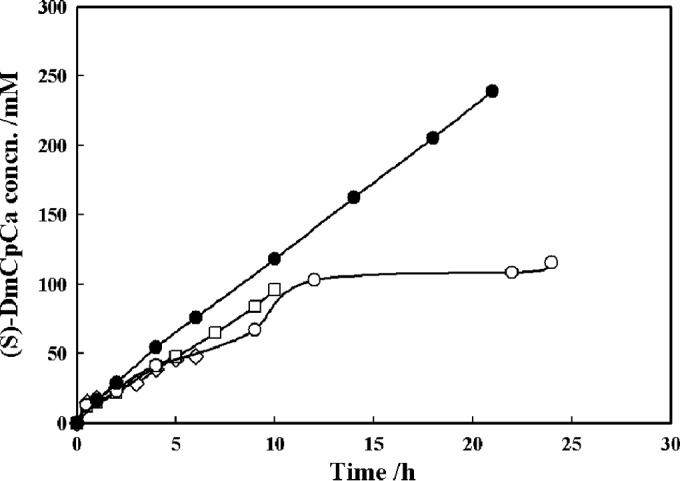
Progress curves of the RhEst1-catalyzed kinetic resolution of rac-DmCpCe with different substrate loads. ◇, 100 mM; □, 200 mM; ○, 500 mM with 10 g/liter RhEst1 and with pH control; ● 500 mM with 25 g/liter RhEst1 and with pH control.
In fact, various lipases (mostly from commercial sources) have been tested for the enantioselective hydrolysis of rac-DmCpCe under their respective appropriate reaction conditions (29). As listed in Table 2, five lipases were reported to have activity and enantioselectivity. At a fixed substrate loading concentration of 65 mM, lipase PS-D, lipase F-AP15, and lipase L-1754 showed high activities in spite of the undesired opposite enantiopreference. Novozyme 435 from Candida antarctica showed the desired enantioselectivity but moderate activity. At 30°C, after a 64-h reaction, the conversion reached only 25.6% at a 99.0% eep (E = 278) with a 16-g/liter catalyst loading concentration. Compared with the commercial lipase Novozyme 435, the esterase RhEst1 showed high activity and substrate tolerance. As much as 500 mM rac-DmCpCe could be enantioselectively hydrolyzed into (S)-DmCpCa. Thus, RhEst1 was demonstrated to be a promising biocatalyst for potential application in biomanufacturing of optically active chiral acids, especially (S)-DmCpCa, a key building block for that production of cilastatin with a large market. Most importantly, RhEst1 displayed lower sequence similarity (<20%) with commercial lipases listed in Table 2 and could be used as a much more economic and efficient alternative to CALB for the preparation of key chiral synthons for cilastatins.
TABLE 2.
Comparison of different enzymes for preparation of (S)- or (R)-DmCpCa
| Enzyme | Substrate load (mM) | Time (h) | Temp (°C) | ees (%)b | eep (%) (R or S preference) | Conversion rate (%) | E value | Reference |
|---|---|---|---|---|---|---|---|---|
| Novozyme 435 | 65 | 64 | 30 | 34.0 | 99.0 (S) | 25.6 | 278 | 34 |
| Lipozyme IM | 65 | 20 | 30 | 1.9 | 65.9 (S) | 2.8 | 5 | 34 |
| Lipase PS-D | 65 | 20 | 50 | 97.8 | 85.0 (R) | 53.5 | 55 | 34 |
| Lipase F-AP15 | 65 | 20 | 40 | 98.3 | 85.0 (R) | 53.6 | 58 | 34 |
| Lipase L-1754 | 65 | 20 | 40 | 95.0 | 79.0 (R) | 54.7 | 31 | 34 |
| RhEst1 | 500 | 21 | 30 | 97.7 | 97.5 (S) | 47.8a | 240 | This work |
Molar isolation yield.
ees, enantiomeric excess of substrate.
In summary, this study not only provides useful guidance for further application of RhEst1 to biochemical production of valuable chiral chemicals but also establishes an efficient process for the direct synthesis of (S)-DmCpCa with excellent enantiopurity. Since RhEst1 displays the highest sequence and structure similarity with nonheme haloperoxidase, its halogenation activity and molecular mechanism will be further studied in detail.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (grant no. 21276082); the Ministry of Science and Technology, People's Republic of China (grant no. 2011CB710800 and 2011AA02A210); and the Shanghai Commission of Science and Technology (grant no. 11431921600).
Footnotes
Published ahead of print 19 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01597-14.
REFERENCES
- 1.Littlechild J. 1999. Haloperoxidases and their role in biotransformation reactions. Curr. Opin. Chem. Biol. 3:28–34. 10.1016/S1367-5931(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 2.Pfeifer O, Pelletier I, Altenbuchner J, von Pée KH. 1992. Molecular cloning and sequencing of a non-heme bromoperoxidase gene from Streptomyces aureofaciens ATCC 10762. J. Gen. Microbiol. 138:1123–1131. 10.1099/00221287-138-6-1123. [DOI] [PubMed] [Google Scholar]
- 3.Hemrika W, Renirie R, Dekker HL, Barnett P, Wever R. 1997. From phosphatases to vanadium peroxidases: a similar architecture of the active site. Proc. Natl. Acad. Sci. U. S. A. 94:2145–2149. 10.1073/pnas.94.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franzen S, Roach MP, Chen YP, Dyer RB, Woodruff WH, Dawson JH. 1998. The unusual reactivities of Amphrite ornata dehaloperoxidase and Notomastus lobatus chloroperoxidase do not arise from a histidine imidazole proximal heme iron ligand. J. Am. Chem. Soc. 120:4658–4661. 10.1021/ja973212d. [DOI] [Google Scholar]
- 5.Colonna S, Gaggero N, Manfredi A, Casella L, Gullotti M, Carrea G, Pasta P. 1990. Enantioselective oxidations of sulfides catalyzed by chloroperoxidases. Biochemistry 29:10465–10468. 10.1021/bi00498a006. [DOI] [PubMed] [Google Scholar]
- 6.Dembitsky VM. 2003. Oxidation, epoxidation and sulfoxidation reactions catalyzed by haloperoxidases. Tetrahedron 59:4701–4720. 10.1016/S0040-4020(03)00701-4. [DOI] [Google Scholar]
- 7.Preobrazhenskaya YV, Voskovoev AI, Burd VN. 2003. Phosphatase activity of non-heme chloroperoxidase from the bacterium Serratia marcescens. FEBS Lett. 536:41–44. 10.1016/S0014-5793(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 8.Allain EJ, Hager LP, Deng L, Jacobsen EN. 1993. Highly enantioselective epoxidation of disubstituted alkene with hydrogen-peroxide catalyzed by chloroperoxidase. J. Am. Chem. Soc. 115:4415–4416. 10.1021/ja00063a091. [DOI] [Google Scholar]
- 9.van Deurzen MPJ, van Rantwijk F, Sheldon RA. 1996. Synthesis of substituted oxindoles by chloroperoxidase catalyzed oxidation of indole. J. Mol. Catal. B Enzym. 2:33–42. 10.1016/1381-1177(96)00008-2. [DOI] [Google Scholar]
- 10.Itoh N, Izumi Y, Yamada H. 1987. Haloperoxidase-catalyzed halogenation of nitrogen-containing aromatic heterocycles represented by nucleic bases. Biochemistry 26:282–289. 10.1021/bi00375a039. [DOI] [Google Scholar]
- 11.Itoh N, Izumi Y, Yamada H. 1986. Characterization of non-heme type bromoperoxidase in Corallina pilulifera. J. Biol. Chem. 261:5194–5200. [PubMed] [Google Scholar]
- 12.Wiesner W, von Pée KH, Lingens F. 1988. Purification and characterization of a novel bacterial non-heme chloroperoxidase from Pseudomonas pyrrocinia. J. Biol. Chem. 263:13725–13732. [PubMed] [Google Scholar]
- 13.Pelletier I, Pfeifer O, Altenbuchner J, von Pée KH. 1994. Cloning of a second non-heme bromoperoxidase gene from Streptomyces aureofaciens ATCC 10762: sequence analysis, expression in Streptomyces lividans and enzyme purification. Microbiology 140:509–516. 10.1099/00221287-140-3-509. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Yang F, Du H, Mahmum Hossain M, Bennett D, Grubisha DS. 1998. The synthesis of S-(+)-2,2-dimethylcyclopropane carboxylic acid: a precursor for cilastatin. Tetrahedron Asymmetry 9:3971–3977. 10.1016/S0957-4166(98)00421-2. [DOI] [Google Scholar]
- 15.Kahan FM, Kropp H, Sundelof JG, Birnbaum J. 1983. Thienamycin: development of imipenen-cilastatin. J. Antimicrob. Chemother. 12:1–35. 10.1093/jac/12.suppl_D.1. [DOI] [PubMed] [Google Scholar]
- 16.Clissold SP, Todd PA, Campoli-Richards DM. 1987. Imipenem/cilastatin. A review of its antibacterial activity, pharmacokinetic properties and therapeutic efficacy. Drugs 33:183–241. [DOI] [PubMed] [Google Scholar]
- 17.Edwards JR, Betts MJ. 2000. Carbapenems: the pinnacle of the β-lactam antibiotics or room for improvement? J. Antimicrob. Chemother. 45:1–4. 10.1093/jac/45.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Mori A, Arai I, Yamamoto H, Nakai H, Arai Y. 1986. Asymmetric Simmons-Smith reactions using homochiral protecting groups. Tetrahedron 42:6447–6458. 10.1016/S0040-4020(01)88107-2. [DOI] [Google Scholar]
- 19.Kang J, Lim GJ, Yoon SK, Kim MY. 1995. Asymmetric cyclopropanation using new chiral auxiliaries derived from D-fructose. J. Org. Chem. 60:564–577. 10.1021/jo00108a018. [DOI] [Google Scholar]
- 20.Loughlin WA. 2000. Biotransformations in organic synthesis. Bioresour. Technol. 74:49–62. 10.1016/S0960-8524(99)00145-5. [DOI] [Google Scholar]
- 21.Straathof AJJ, Panke S, Schmid A. 2002. The preparation of fine chemicals by biotransformations. Curr. Opin. Biotechnol. 13:548–556. 10.1016/S0958-1669(02)00360-9. [DOI] [PubMed] [Google Scholar]
- 22.Panke S, Held M, Wubbolts M. 2004. Trends and innovations in industrial biocatalysis for the production of fine chemicals. Curr. Opin. Biotechnol. 15:272–279. 10.1016/j.copbio.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Wang MX, Feng GQ. 2002. Enzymatic synthesis of optically active 2-methyl- and 2,2-dimethylcyclopropanecarboxylic acids and their derivatives. J. Mol. Catal. B Enzym. 18:267–272. 10.1016/S1381-1177(02)00105-4. [DOI] [Google Scholar]
- 24.Yeom SJ, Kim HJ, Oh DK. 2007. Enantioselective production of 2,2-dimethylcyclopropane carbonitrile using the nitrile hydratase and amidase of Rhodococcus erythropolis ATCC 25544. Enzyme Microb. Technol. 41:842–848. 10.1016/j.enzmictec.2007.07.007. [DOI] [Google Scholar]
- 25.Shaw NM, Robins KT, Kiener A. 2003. Lonza: 20 years of biotransformations. Adv. Synth. Catal. 345:425–435. 10.1002/adsc.200390049. [DOI] [Google Scholar]
- 26.Zheng RC, Wang YS, Zheng YG. 2010. Enantioselective hydrolysis of (R)-2,2-dimethylcyclopropane carboxamide by immobilized cells of an R-amidase-producing bacterium, Delftia tsuruhatensis CCTCC M 205114, on an alginate capsule carrier. J. Ind. Microbiol. Biotechnol. 37:503–510. 10.1007/s10295-010-0696-7. [DOI] [PubMed] [Google Scholar]
- 27.Jin SJ, Zheng RC, Zheng YG, Shen YC. 2008. (R)-enantioselective hydrolysis of 2,2-dimethylcyclopropanecarboxamide by amidase from a newly isolated strain Brevibacterium epidermidis ZJB-07021. J. Appl. Microbiol. 105:1150–1157. 10.1111/j.1365-2672.2008.03841.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang ZY, Ni Y, Lu ZY, Liao XR, Zheng YG, Sun ZH. 2011. Industrial production of S-2,2-dimethylcyclopropanecarboxamide with a novel recombinant R-amidase from Delftia tsuruhatensis. Process Biochem. 46:182–187. 10.1016/j.procbio.2010.08.005. [DOI] [Google Scholar]
- 29.Wang P, Zhu JA, He JY. 2010. Enantioselective synthesis of S-(+)-2,2-dimethylcyclopropanecarboxylic acid from ethyl-2,2-dimethylcyclopropane carboxylate catalyzed by lipase Novozyme 435. Chin. J. Catal. 31:651–655. 10.3724/SP.J.1088.2010.91101. [DOI] [Google Scholar]
- 30.Liang FY, Huang J, He JY, Wang P. 2012. Improved enantioselective hydrolysis of racemic ethyl-2,2-dimethylcyclopropanecarboxylate catalyzed by modified Novozyme 435. Biotechnol. Bioprocess Eng. 17:952–958. 10.1007/s12257-012-0135-x. [DOI] [Google Scholar]
- 31.Royon D, Daz M, Ellenrieder G, Locatelli S. 2007. Enzymatic production of biodiesel from cotton seed oil using t-butanol as a solvent. Bioresour. Technol. 98:648–653. 10.1016/j.biortech.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 32.Liu CH, Pan J, Ye Q, Xu JH. 2013. Enzymatic production of cilastatin intermediate via highly enantioselective hydrolysis of methyl (±)-2,2-dimethylcyclopropane carboxylate using newly isolated Rhodococcus sp. ECU1013. Appl. Microbiol. Biotechnol. 97:7659–7667. 10.1007/s00253-013-5038-z. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 34.Xu GC, Yu HL, Zhang XY, Xu JH. 2012. Access to optically active aryl halohydrins using a substrate-tolerant carbonyl reductase discovered from Kluyveromyces thermotolerans. ACS Catal. 2:2566–2571. 10.1021/cs300430g. [DOI] [Google Scholar]
- 35.Lineweaver H, Burk D. 1934. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56:658–666. 10.1021/ja01318a036. [DOI] [Google Scholar]
- 36.Hofmann B, Tolzer S, Pelletier I, Altenbuchner J, von Pée KH, Hecht HJ. 1998. Structural investigation of the cofactor free chloroperoxidase. J. Mol. Biol. 279:889–900. 10.1006/jmbi.1998.1802. [DOI] [PubMed] [Google Scholar]
- 37.Brenner S. 1988. The molecular evolution of genes and proteins: a tale of two serines. Nature 334:528–530. 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- 38.Lee JH, Boyapati G, Song KB, Rhee SK, Kim CH. 2000. Cloning and sequence analysis of the estA gene encoding enzyme for producing (R)-β-acetylmercaptoisobutyric acid from Pseudomonas aeruginosa 1001. J. Biosci. Bioeng. 90:684–687. 10.1016/S1389-1723(00)90019-7. [DOI] [PubMed] [Google Scholar]
- 39.Qian L, Liu JY, Liu JY, Yu HL, Li CX, Xu JH. 2011. Fingerprint lipolytic enzymes with chromogenic p-nitrophenyl esters of structurally diverse carboxylic acids. J. Mol. Catal. B Enzym. 73:22–26. 10.1016/j.molcatb.2011.07.010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



