Abstract
Bovine mastitis due to Mycoplasma californicum is often accompanied by huge economic losses, and the disease spreads very quickly. An appropriate molecular epidemiological analysis is needed to prevent and control infectious disease, but molecular epidemiological analysis methods for M. californicum have not yet been reported. Here we developed a combination of multiple-locus variable-number tandem repeat analysis (MLVA) and pulsed-field gel electrophoresis (PFGE) methods, which are common genotyping methods for various bacteria, for M. californicum. The MLVA is based on four interspersed repeat units that were found in the M. californicum genome data. The MLVA using these repeat units showed sufficient discriminatory power for a molecular epidemiological analysis; i.e., a Hunter-Gaston diversity index (HGDI) of 0.949, against M. californicum strains in Japan and M. californicum strain ATCC 33461. The PFGE for M. californicum also showed sufficient discriminatory power, with an HGDI of 0.985. Strain ATCC 33461 showed MLVA profiles and pulsotypes that differed greatly from those of strains from Japan. These results indicate that MLVA and PFGE are good tools for identifying M. californicum transmission events more accurately. Our combined MLVA and PFGE analysis suggests the persistence of M. californicum infection among herds in a specific area for a long period of time, as well as the movement of cows and heifers accompanying the expansion of M. californicum infection. Failure to identify asymptomatic infected cows is suspected as one of the central causes of the present M. californicum infection scenario in Japan.
INTRODUCTION
Mycoplasma californicum is a causal bacterium of bovine mastitis, arthritis, and pneumonia, as well as an indigenous bacterium of cattle (1, 2). In Japan, the causal agents of mycoplasmal mastitis were limited for many years to M. bovis and M. bovigenitalium (3), but the number of cases of mastitis caused by M. californicum has been increasing since 2005. As in the case of mastitis caused by M. bovis, M. californicum mastitis features strong infectivity, severe symptoms, and a poor response to treatment with antibiotics (2, 4, 5). Moreover, outbreaks of mycoplasmal mastitis are often accompanied by major economic losses because of the need to cull infected cows (2, 5).
An understanding of the genetic relatedness and clonal spread of mycoplasmal strains is necessary in order to control outbreaks (2). However, to the best of our knowledge, no molecular epidemiological analysis method for M. californicum has been reported. Multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) is a genotyping method that shows high discriminatory power, and it has been used as a marker for the strain typing of other mycoplasmas (6–8). Discrimination of the number of repeats at each locus produces a digital profile, providing a highly portable typing method that allows comparisons among laboratories. Pulsed-field gel electrophoresis (PFGE) is the gold standard of molecular epidemiological methods for many bacteria. Although PFGE is also a highly discriminatory typing method that is well suited to the investigation of microevolution and recent transmission events within a farm (9), it has several limitations, including difficulty in interpreting band patterns and evaluating correlations between band patterns in different image data among laboratories and limited reproducibility (10, 11).
All existing genotyping methods have virtues and weaknesses, and there is no one genotyping method that can meet all demands. High discriminatory power is one of the most essential factors in epidemiological analysis, and a proper combination of genotyping methods is needed to clarify the transmission events of a given pathogen. We attempted to develop a new useful MLVA and PFGE in order to obtain correct epidemiological information.
MATERIALS AND METHODS
M. californicum strains.
All of the M. californicum strains used in the present study were isolated from bovine milk obtained primarily from Hokkaido, Japan. The details of the 22 case farms of M. californicum infection are listed in Table 1 and Fig. 1. Milk samples were cultured in Hayflick's broth (12) for 2 to 7 days at 37°C, and 10 μl of cultured broth was then subcultured on Hayflick's agar containing 0.8% (wt/vol) Noble Agar (Difco, Sparks, MD, USA) at 37°C and 5% CO2 for 2 days. Hayflick's broth consists of PPLO broth (Difco) containing 0.002% (wt/vol) phenol red, 15% (vol/vol) horse serum, 2.5% (vol/vol) fresh yeast extract, 1,000 U/ml penicillin G (Banyu Pharmaceutical Co., Tokyo, Japan), 0.02% (wt/vol) thallium acetate, and 0.0024% (wt/vol) DNA from calf thymus and was adjusted to pH 7.5.
TABLE 1.
Details of the cases of M. californicum infection described in this report
| Case | Farm | Location | Mo/yr | Prevention and control procedure(s)a | No. of infected cows | No. of isolates testedb | Remarks |
|---|---|---|---|---|---|---|---|
| 1 | A | California | Aug 1972-May 1973 | T, S, C, Th | 50 | 1 (1/0) | Bovine mastitis outbreak, ATCC 33461 |
| 2 | B | Okhotsk | Jun 2005 | Unknown | 2 | 3 (2/0) | First case of bovine mastitis in Japan |
| 3 | C | Kushiro | Jun 2006 | T, S, C | 7 | 7 (7/0) | Bovine mastitis outbreak after bronchitis in newly purchased cows |
| 4 | D | Nemuro | Jun 2007 | Unknown | 1 | 1 (1/0) | Bovine mastitis case |
| 5 | E | Kushiro | Aug-Sep 2007 | S, C | 14 | 15 (14/0) | Bovine mastitis outbreak after large purchase of cows for scale expansion; exchange of cows from farm F |
| 6 | F | Kushiro | Aug 2007 | S, C | 26 | 32 (26/1) | Bovine mastitis outbreak after large purchase of cows for scale expansion; exchange of cows from farm E |
| 7 | G | Kushiro | Feb-Apr 2008 | Unknown | 2 | 3 (2/1) | Bovine mastitis outbreak due to various mycoplasmas over a period of 1 yr |
| 8 | E | Kushiro | Mar 2008 | S, C | 16 | 16 (16/0) | Bovine mastitis outbreak after large purchase of cows for scale expansion; exchange of cows from farm F |
| 9 | F | Kushiro | Apr-Jun 2008 | S, C | 14 | 3 (3/0) | Bovine mastitis outbreak after large purchase of cows for scale expansion; exchange of cows from farm E |
| 10 | H | Okhotsk | Apr 2008 | Unknown | 20 | 25 (20/1) | Bovine mastitis outbreak after purchase of cows |
| 11 | I | Tokachi | Jul-Nov 2008 | Unknown | 33 | 31 (30/0) | Bovine mastitis outbreak in curing group and low milk production group for several months |
| 12 | J | Mie | Nov 2009 | Unknown | 4 | 1 (1/0) | First case of bovine mastitis outbreak in a district other than Hokkaido |
| 13 | K | Tokachi | May-Aug 2011 | Unknown | 3 | 5 (3/0) | Bovine mastitis |
| 14 | L | Tokachi | Nov-Dec 2011 | Unknown | Unknown | 10 (10/0) | Isolates from milk testing |
| 15 | M | Tokachi | Dec 2011-Nov 2012 | Unknown | 13 | 13 (13/0) | Bovine mastitis outbreak |
| 16 | N | Tokachi | Mar 2012 | Unknown | Unknown | 5 (4/0) | Isolates from milk testing |
| 17 | O | Tokachi | Sep 2012 | Unknown | Unknown | 2 (0/1) | Bulk tank milk culturing |
| 18 | P | Unknown | Unknown 2012 | Unknown | Unknown | 2 (1/0) | |
| 19 | Q | Tokachi | Dec 2012 | Unknown | Unknown | 5 (3/0) | Isolates from milk testing |
| 20 | R | Tokachi | Jan-Feb 2013 | Unknown | Unknown | 2 (2/0) | Isolates from milk testing |
| 21 | S | Tokachi | Feb-Mar 2013 | Unknown | Unknown | 2 (1/0) | Isolates from milk testing |
| 22 | T | Tokachi | Apr 2013 | Unknown | 1 | 1 (1/0) | Bovine mastitis case |
T, testing; S, segregation; C, culling; Th, therapy. All cases except 1 and 12 occurred in the Hokkaido district of Japan.
The values in parentheses are the number of cows sampled/number of bulk tanks sampled.
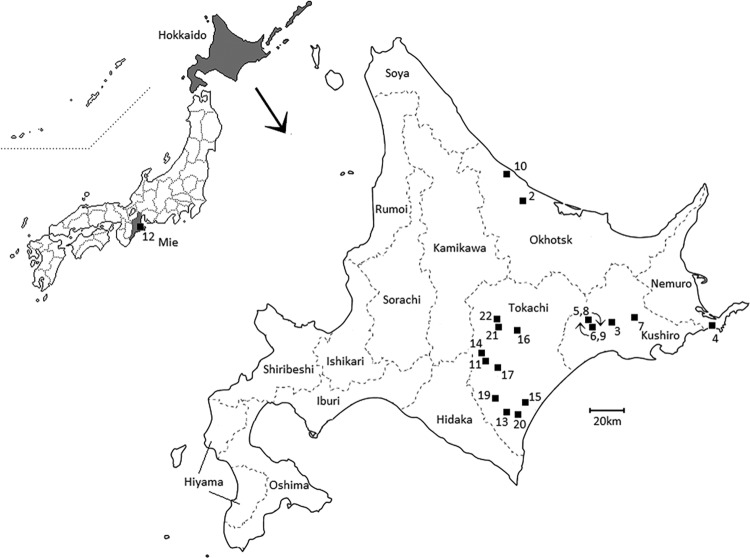
FIG 1.
Map of M. californicum infection cases. The numbers represent the cases of infection listed in Table 1. Movement of cows between farms is indicated by curved arrows.
One or two mycoplasma colonies were cloned from each sample. We made a preliminary identification of the species of mycoplasmal isolates by the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) pattern (13) and 16S rRNA gene (rrs) sequencing (14), and we made the final determination of bacterial species by growth inhibition (15) and metabolic inhibition tests (16). In the SDS-PAGE, mycoplasmal cells cultured for 2 to 4 days were harvested by centrifugation (13,000 × g for 6 min at 4°C), washed twice with 1 ml phosphate-buffered saline (PBS), and resuspended in 10 μl of PBS. The mycoplasmal cell suspension was mixed with an equal quantity of EzApply (ATTO, Tokyo, Japan) and then heated at 100°C for 1 min. Electrophoresis was performed for 85 min at 20 mA/gel with e-PAGEL (ATTO), which has a 12.5% gel concentration. A solution containing 24.8 mM Tris, 3.47 mM SDS, and 191.8 mM glycine was used for the SDS-PAGE buffer.
Mycoplasmal DNA was extracted from a 3-ml culture volume with InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA, USA). Rabbit antisera for use in the growth inhibition and metabolic inhibition tests were prepared as described by Senterfit (17). Mycoplasmal antigen for immunization was prepared from 500 to 1,000 ml of Hayflick's broth, and this broth was cultured for 3 to 4 days at 37°C. The broth culture was centrifuged (30,000 × g for 45 min at 4°C). The pellet was resuspended in 200 ml of PBS and then washed twice by centrifugation under the same conditions. The final pellet was resuspended in 4 to 8 ml of PBS. Four milliliters of antigen suspension was emulsified with an equal quantity of Freund's complete adjuvant. A total of 2 ml of the emulsified antigen was injected intramuscularly into the back of a rabbit and subcutaneously into its upper arms. On day 21, a serum sample was obtained and booster injections of a total of 2 ml into intramuscular sites were administered. The rabbit's serum was then collected on days 28, 35, and 42. We checked the antibody titer and cross-reactivity by conducting a growth inhibition test with-type strains of bovine mycoplasmas.
A total of 184 M. californicum strains were ultimately collected. We used these 184 strains and an additional strain of M. californicum, ATCC 33461, which is type strain ST-6 (18).
MLVA for M. californicum.
VNTR markers, which are M. californicum tandem repeats (MCTRs), were identified previously in the sequenced genome of M. californicum strain HAZ 160_1 (accession no. AP013353) (19) with the Tandem Repeat Finder (http://tandem.bu.edu/trf/trf.html) (20). PCR primers for MLVA were designed as close as possible to the VNTR unit (21). We performed PCR amplification for selected MCTR loci with 10 strains from different mastitis cases as a preliminary selection. Each MCTR was amplified with the primers shown in Table 2 and the GeneAmp High Fidelity PCR System (Applied Biosystems, Carlsbad, CA, USA). The genome locations of MCTRs and gene prediction were analyzed with in silico Molecular Cloning Genomics Edition (IMCGE) software version 5.2.2 (In Silico Biology, Inc., Yokohama, Japan) with a BLASTP search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) as a guide.
TABLE 2.
Oligonucleotide primers used for the M. californicum MLVA
| Primera | Sequence (5′→3′) | Locationb | PCR product size (bp) |
|---|---|---|---|
| MCTR569_F | CACTAAACTCAGTAACAG | 568810–568827 | 107 + total size of repeat units |
| MCTR569_R | TGAAAAAGAGAAGACCG | 570262–570278 | |
| MCTR695_F | GATGATAAGTCTGCTTGTC | 695021–695039 | 346 + total size of repeat units |
| MCTR695_R | TTTTCATAGTCCGAACTGG | 696266–696284 | |
| MCTR698_F | TTTATCTCCGATTGCGTC | 697457–697474 | 256 + total size of repeat units |
| MCTR698_R | TGAAGCACTTTGTTGTCC | 698205–698222 | |
| MCTR701_F | CAACATTGCCGTTACTTGC | 701113–701131 | 347 + total size of repeat units |
| MCTR701_R | CTTAGGACCTTCCAATTCG | 702359–702377 |
F, forward primer; R, reverse primer.
Position in the M. californicum HAZ160_1 genome sequence (accession no. AP013353).
The size of each amplicon determined and the number of repeats calculated are listed in Tables 2 and 3. When the amplicon size was shorter than 1 kbp, the amplicon was purified with a LaboPass PCR purification kit (Cosmo Genetech, Seoul, South Korea) and sequenced on a 3130 genetic analyzer (Applied Biosystems) with a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) and the same primers in order to confirm the precise sizes and the repeat number of MCTRs. When the amplicon size was longer than 1 kbp, the amplicon size was estimated by sequence data and migration distance. The MLVA profiles are displayed in the order of the number of repeats of MCTR569, MCTR695, MCTR698, and MCTR701. If an amplicon was not produced, we expediently defined the number of repeats as zero for clustering of the dendrograms created.
TABLE 3.
Characteristics of MCTRs
| Name | Genome positiona (bp) | Locus | Repeat unit size (bp) | % identity between MCTRsb | Accession no. |
|---|---|---|---|---|---|
| MCTR569 | 568882 | MCAL160_0738d | 225c | 97 | AB916338 |
| MCTR695 | 695223 | MCAL160_0902d | 102 | 82 | AB916339 |
| MCTR698 | 697593 | MCAL160_0908d | 102 | 83 | AB916340 |
| MCTR701 | 701231 | MCAL160_0912d | 102 | 97 | AB916341 |
Position (5′ end) in the M. californicum HAZ160_1 genome sequence (accession no. AP013353).
Percentages of identity between MCTRs were calculated in MCTRs of M. californicum HAZ160_1.
An insertion or deletion of a 12-bp fragment in MCTR569 was confirmed in some strains.
Hypothetical protein.
PFGE for M. californicum.
Two-milliliter volumes of stationary-phase mycoplasmal cultures (optical density at 600 nm of approximately 0.3) were used for PFGE analysis. Mycoplasmal cells were harvested by centrifugation (13,000 × g for 6 min at 4°C), washed with 1 ml of PBS, and resuspended in 60 μl of PBS. Agarose plugs were made from a 1:1 mixture of 2% low-melting-point agarose (Bio-Rad) and the cell suspension. The plugs were incubated in 500 μl of lysis buffer (6 mM Tris-HCl, 1 mM NaCl, 100 mM EDTA, 0.5% [wt/vol] polyoxyethylene cetyl ether, 0.2% [wt/vol] deoxycholic acid sodium salt, 0.5% [wt/vol] N-lauroylsarcosine sodium salt, and 1.5 mg/ml [wt/vol] lysozyme, adjusted to pH 7.5) for 2 h at 37°C and then incubated in 500 μl of ESP buffer (0.5 M EDTA, 0.5% [wt/vol] N-lauroylsarcosine sodium salt, 1 mg/ml [wt/vol] proteinase K, adjusted to pH 7.5) for 12 h at 54°C.
The plugs were incubated twice in 500 μl of Tris-EDTA (TE) buffer (10 mM Tris HCl, 1 mM EDTA) with 1 mM phenylmethylsulfonyl fluoride for 30 min at room temperature and then washed three times with 1 ml of TE buffer for 30 min at room temperature. Restriction digestion of the plugs was performed in 100 μl of restriction enzyme solution containing 30 U of restriction enzymes (Wako, Osaka, Japan; TaKaRa Bio, Shiga, Japan) for 18 h at the optimal temperature. We tested 18 restriction enzymes (ApaI, BamHI, BglII, BlnI, ClaI, DraI, EcoRI, EcoRV, HindIII, KpnI, MluI, NarI, PstI, SacI, SalI, SmaI, XbaI, and XhoI) for the selection of a suitable restriction enzyme for the PFGE of M. californicum (see Fig. S1 in the supplemental material).
DNA fragments were separated by electrophoresis in 1% pulsed-field-certified agarose gels (Bio-Rad) in 0.5 × Tris-borate-EDTA (TBE) buffer with a CHEF-DRII system (Bio-Rad). The electrophoresis parameters were as follows: switch times of 4 to 5 s at 5.7 V/cm for 23.5 h and a buffer temperature of 10°C. Gels were stained with 0.5 μg/ml ethidium bromide for 1 h, destained in distilled water for 1 h, and photographed under UV light. We used a Low Range PFG Marker (New England BioLabs, Ipswich, MA, USA) for fragment size determination. To minimize the risk of false judgment in the image analysis, we first placed side-by-side samples that had the same or similar pulse patterns and electrophoresed them. A pulsotype is defined as a unique electrophoretic banding pattern evaluated by a combination of computer-aided and visual interpretations.
Evaluation of MLVA and PFGE.
We analyzed the MLVA profiles and pulsotypes with BioNumerics version 5.10 (Applied Maths, Sint-Martens-Latem, Belgium) according to criteria described previously (9, 22, 23). Briefly, a clustering dendrogram was created with the Euclidean coefficient for the MLVA and the Dice coefficient for the PFGE. The optimization and the position tolerance setting for the PFGE analysis were 1%, and pulsotypes that showed more than 80% similarity in this clustering dendrogram were considered to belong to the same cluster (23). Pulsotypes of the same cluster were assigned the same alphabetical name.
The discriminatory index of individual or combined MCTR loci and pulsotype was derived with the Hunter-Gaston diversity index (HGDI) (24). The HGDI is used to evaluate the discriminatory power of each typing method (24). As the discriminatory power becomes higher, this index nears 1.0, and an index of greater than 0.90 is desirable if the typing results are to be interpreted with confidence (24). The HGDI and 95% confidence intervals (Cls) were calculated by EpiCompare version 1.01 (Ridom GmbH, Würzburg, Germany). The correlation among typing methods or cases of M. californicum infection were also calculated with EpiCompare version 1.01, as were the adjusted Rand index (ARI) and Wallace coefficients (WCs). The ARI is used to evaluate the global congruence of two typing methods (25). It gives the overall concordance of two methods, taking into account the possibility that the agreement between partitions could arise by chance alone (25). The WC is also used to evaluate the global congruence of two typing methods (26). The WC can be more informative than the ARI by providing adirectional information about the partition relationship (26). As the congruence between two typing methods increases, the ARI and WC also near 1.0. We selected representative strains of combined genotypes of MLVA and PFGE in every case for the calculation of the discriminatory index and the correlation factor, and we used 85 strains in which the case and genotype do not overlap.
Prior to the molecular epidemiological examination, we carried out a preliminary examination by using four strains obtained from different cases of M. californicum infection: ATCC 33461 (case 1, MLVA 4-8-6-4, pulsotype G), HAZ160_1 (case 9, MLVA 6-9-5-9, pulsotype C4), HAZ175_1 (case 2, MLVA 5-6-5-2, pulsotype A18), and HAZ44_1 (case 6, MLVA 6-6-5-9, pulsotype A16). These four strains were used for the confirmation of MCTR, the selection of a suitable restriction enzyme for PFGE, and the in vitro stability study. In the in vitro stability study, each strain was passaged 20 times in Hayflick's broth and the identity of each MCTR locus and pulsotype was checked before and after the 20 passages. Additionally, a total of 14 M. californicum strains isolated from the milk of six mastitic cows at intervals of 14 to 25 days were used for the in vivo stability study (see Table S1 in the supplemental material).
RESULTS
The 184 strains used in the present study showed an SDS-PAGE pattern similar to that of M. californicum ATCC 33461, and their SDS-PAGE pattern was clearly different from those of the other bovine mycoplasmal species, i.e., M. arginini ATCC 10129, M. alkalescens ATCC 10135, M. canadense ATCC 29418, M. bovigenitalium ATCC 10122, M. bovis ATCC 10131, M. bovoculi ATCC 10141, M. bovirhinis ATCC 10118, M. dispar ATCC 27410 M. verecundum ATCC 27862, Acholeplasma laidlawii ATCC 10116, and A. axanthum ATCC 10138 (data not shown). Moreover, all of the strains showed over 99% similarity to rrs of M. californicum, and the growth inhibition and metabolic inhibition shown by rabbit antiserum against M. californicum ATCC 33461 were recognized in all of the strains (data not shown).
Evaluation of MLVA and PFGE.
We found four genomic sequences that consisted of tandem repeats in the sequenced M. californicum strain HAZ 160_1 genome, and we named them MCTR569, MCTR695, MCTR698, and MCTR701. We placed all of the MCTRs in hypothetical protein genes (Table 3). In all of the MCTRs, a part of the coding region of MCTR695 showed an amino acid sequence analogous to that of a putative multiple-banded antigen of Ureaplasma urealyticum (accession no. NC_011374 and U50459), and a part of the coding region of MCTR701 showed an amino acid sequence analogous to the putative phase-variable lipoprotein VsaA of M. pulmonis (accession no. U23947) (19, 27).
The numbers of variations in repeat numbers were 9 in MCTR569, 6 in MCTR695, 8 in MCTR698, and 10 in MCTR701 (Table 4). The size of the repeat units of MCTR695, MCTR698, and MCTR701 was 102 bp, and that of MCTR569 was 225 bp (Table 3). Therefore, the repeat numbers of all MCTRs can be easily estimated by simple electrophoresis with a 2% agarose gel (see Fig. S2 in the supplemental material). MCTR569 of some strains was confirmed to have an insertion or deletion of a 12-bp fragment. DNA sequence matching of repeat units was calculated by Tandem Repeat Finder. The percentages of identity between repeat units were 97% for MCTR569, 82% for MCTR695, 83% for MCTR698, and 97% for MCTR701 (Table 3). Moreover, the DNA sequences of the repeat units of MCTR695 and MCTR698 showed high similarity, with the highest percentage of similarity between them being 99%.
TABLE 4.
HGDIs and 95% CIs of the MLVA and PFGE by BamHI digestion used to characterize the present 85 M. californicum strains from bovine milk in Japan
| Item | No. of different types | HGDI | 95% CI |
|---|---|---|---|
| MCTR569 | 9 | 0.766 | 0.705–0.826 |
| MCTR695 | 6 | 0.674 | 0.616–0.732 |
| MCTR698 | 8 | 0.374 | 0.242–0.507 |
| MCTR701 | 10 | 0.75 | 0.675–0.825 |
| MLVA | 41 | 0.949 | 0.919–0.979 |
| PFGE clusters | 7 | 0.75 | 0.705–0.796 |
| Pulsotypes | 55 | 0.985 | 0.977–0.994 |
A total of 41 different MLVA profiles were identified in the present study (Table 4). The HGDIs of MCTR569, MCTR695, MCTR698 and MCTR701 were 0.766, 0.674, 0.374, and 0.75, respectively (Table 4). No MCTR with an extremely low HGDI was confirmed. Of the MCTRs, MCTR695 showed a comparatively high ARI and WC with PFGE clusters (0.554 and 0.784, respectively) (Tables 5 and 6). Moreover, MCTR695 showed a comparatively high WC with pulsotypes (0.942) (Table 6). MCTR698 also showed a comparatively high WC with PFGE clusters and pulsotypes (0.82 and 0.788, respectively) (Table 6). This MLVA targeted only four MCTRs, but the HGDI of the combined MCTRs was 0.949 (Table 4).
TABLE 5.
ARIs for the typing methods used to characterize the 85 M. californicum strains from bovine milk in Japan and cases of M. californicum infection
| Item | ARI |
||||||
|---|---|---|---|---|---|---|---|
| Cases | MCTR569 | MCTR695 | MCTR698 | MCTR701 | MLVA | PFGE clusters | |
| MCTR569 | 0.318 | ||||||
| MCTR695 | 0.267 | 0.183 | |||||
| MCTR698 | 0.06 | 0.042 | 0.158 | ||||
| MCTR701 | 0.136 | 0.109 | 0.11 | 0.219 | |||
| MLVA | 0.324 | 0.298 | 0.2 | 0.062 | 0.277 | ||
| PFGE clusters | 0.477 | 0.345 | 0.554 | 0.172 | 0.158 | 0.272 | |
| Pulsotypes | 0.117 | 0.034 | 0.054 | 0.008 | 0.028 | 0.064 | 0.085 |
TABLE 6.
WCs for the typing methods used to characterize the 85 M. californicum strains from bovine milk in Japan and cases of M. californicum infectiona
| Item | WC |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cases | MCTR569 | MCTR695 | MCTR698 | MCTR701 | MLVA | PFGE clusters | Pulsotypes | |
| Cases | 0.684 | 0.799 | 0.832 | 0.451 | 0.277 | 0.953 | 0.08 | |
| MCTR569 | 0.297 | 0.485 | 0.676 | 0.336 | 0.217 | 0.52 | 0.032 | |
| MCTR695 | 0.25 | 0.349 | 0.758 | 0.32 | 0.156 | 0.601 | 0.042 | |
| MCTR698 | 0.131 | 0.253 | 0.394 | 0.349 | 0.081 | 0.327 | 0.018 | |
| MCTR701 | 0.184 | 0.315 | 0.417 | 0.872 | 0.204 | 0.368 | 0.029 | |
| MLVA | 0.555 | 1.0 | 1.0 | 1.0 | 1.0 | 0.984 | 0.055 | |
| PFGE clusters | 0.389 | 0.488 | 0.784 | 0.82 | 0.369 | 0.201 | 0.058 | |
| Pulsotypes | 0.558 | 0.519 | 0.942 | 0.788 | 0.5 | 0.192 | 1.0 | |
Each value is the Wallace w1 coefficient of the item in the first column to the item in the column head and is also the Wallace w2 coefficient of the item in the column head to the item in the first column.
We sought a suitable restriction enzyme for the PFGE of M. californicum by using 18 restriction enzymes and four M. californicum strains. Pulse patterns produced by BamHI, BglII, BlnI, ClaI, EcoRI, EcoRV, HindIII, KpnI, MluI, PstI, SacI, SalI, XbaI, and XhoI showed more than 10 bands in each strain, but those produced by BglII, ClaI, EcoRI, EcoRV, HindIII, and XbaI showed too many bands of sizes too small for PFGE analysis (see Fig. S1 in the supplemental material). Moreover, the pulse patterns produced by restriction enzymes other than BamHI, BlnI, and MluI often contained indistinguishable bands. Among BamHI, BlnI, and MluI, the pulse patterns produced by BamHI showed especially clear bands (see Fig. S1). We therefore evaluated BamHI as a suitable restriction enzyme for the PFGE of M. californicum (see Fig. S1).
A total of 55 different pulsotypes were identified in the present study (Table 4). With respect to the PFGE, we recognized the obvious links between the PFGE (PFGE clusters and pulsotypes) and each case; i.e., multiple strains from each case showed identical or closely related pulsotypes (Fig. 2). On the other hand, the MLVA profiles also showed comparatively high WCs with the cases and PFGE clusters (0.555 and 0.984, respectively) (Table 6). The HGDI of the pulsotypes was 0.979, and thus the discriminatory power of PFGE was superior to that of MLVA typing.
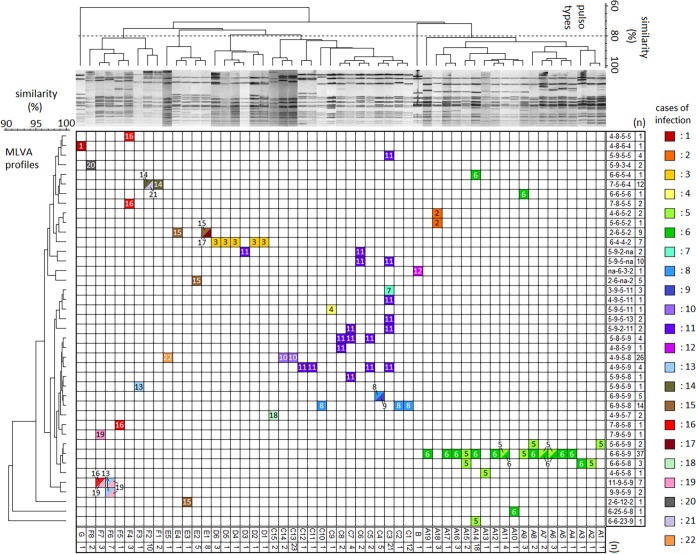
FIG 2.
Correlation between cases of infection and genotypes of M. californicum strains from bovine milk and bulk milk. A clustering dendrogram based on both MLVA typing and PFGE, in which the clustering of M. californicum strains is demonstrated, is shown. The boxes represent the cases of infection listed in Table 1, with their MLVA and PFGE profiles.
We checked the in vitro stability of the MLVA and PFGE after 20 passages in Hayflick's broth. All of the MLVA profiles and pulsotypes were identical after 20 passages. With respect to the in vivo stability study, resampled isolates from four of the six mastitic cows showed MLVA profiles and pulsotypes identical to those of the first isolates (see Table S1 in the supplemental material). The MLVA profiles of the resampled isolates from the remaining two mastitic cows were identical to those of the first isolates, but their pulsotypes changed slightly (see Table S1).
MLVA typing was able to distinguish between strains with identical pulsotypes as it had for other bacteria, and vice versa (Fig. 2) (28). Although PFGE is the primary method of analysis in molecular epidemiological genotyping, MLVA typing is considered to be complementary to PFGE.
Epidemiological analysis of cases of M. californicum infection.
We examined a total of 185 M. californicum strains isolated from 22 cases of infection for their epidemiological backgrounds by using MLVA typing and PFGE (Fig. 1 and 2). Of these, 16 cases were confirmed as clinical mastitis due to M. californicum (cases 1 to 13, 15, and 22) but the M. californicum strains in the remaining 6 cases were isolated from bulk tank milk or milk from cows without clinical symptoms.
The case 1 strain, ATCC 33461, showed genotypes that are greatly different from those of other strains from Hokkaido, Japan. Moreover, the case 12 strain, which was isolated in Mie Prefecture (which is not in Hokkaido; Fig. 1), also showed genotypes that are greatly different from those of the Hokkaido strains. The strains from Hokkaido were broadly distinguished into five clusters (A, C, D, E, and F) by PFGE. Strains assigned to cluster A were isolated from cases 2, 5, and 6. Of these, some of the strains from cases 5 and 6 had identical MLVA profiles and pulsotypes (Fig. 2). In fact, exchanges of a large number of cows took place between farms E and F before the outbreak, and the transportation of cows was suspected to have contributed to the dissemination of M. californicum (Table 1; Fig. 1). Moreover, the strains from these cases showed various closely related MLVA profiles and pulsotypes (Fig. 2).
The strains assigned to cluster C were isolated from cases 4, 7, 8, 9, 10, 11, and 18. Of these, some of the strains from cases 8 and 9 had identical MLVA profiles and pulsotypes (Fig. 2). Again, exchanges of cows between farms E and F before the outbreak are thought to have contributed to the spread of the infection (Table 1; Fig. 1). Strains from case 11 also showed various MLVA profiles and pulsotypes (Fig. 2), whereas those from case 10 showed mostly the same MLVA profile and pulsotype (MLVA 4-9-5-8, pulsotype C13) (Fig. 2). Although the strains from case 7 and some of those from case 11 showed the same pulsotype (C3), their MLVA profiles differed (Fig. 2).
Most of the strains assigned to cluster D were isolated from case 3, and all of the strains from case 3 showed identical MLVA profiles. Strains assigned to cluster E were isolated from cases 15, 17, and 22. Of these, some strains from case 15 and all of the strains from case 17 had identical MLVA profiles and pulsotypes (Fig. 2). These cases were identified in different years but were located in the Tokachi area (Table 1; Fig. 1). Strains assigned to cluster F were isolated from cases 13, 14, 16, 19, 20, and 21. Of these, some strains from case 14 and all of the strains from case 21 shared identical MLVA profiles and pulsotypes (Fig. 2). Again, these cases were identified in different years but were located in the Tokachi area, as were cases 15 and 17 (Table 1; Fig. 1). Some of the strains from cases 13, 16, and 19 also had identical MLVA profiles and pulsotypes (Fig. 2). These three cases were also identified in the Tokachi area but at different times (Table 1; Fig. 1).
DISCUSSION
Molecular genotyping methods are generally anticipated to show higher discriminatory ability than phenotyping methods. Many of these techniques are based on a diversified genomic region; i.e., direct repeat, mutable genes, the variety of foreign genes, etc. Of these, a genotyping method reflecting a genomic background is often used as the clustering technique for multiple strains, and the resulting data are often used to make phylogenetic trees. Many genotyping methods have been developed and used effectively in a wide range of studies of many bacterial species.
Regarding bovine mycoplasma species, genotyping methods have been developed for M. mycoides subsp. mycoides SC and M. bovis, the genome information of which was shown relatively early, i.e., PFGE (29, 30), MLVA (7, 8), multilocus sequence typing (31, 32), PCR and restriction fragment length polymorphism (33), and more, but such methods have not been developed yet for many other mycoplasmal species. Reports of survey research of bovine mycoplasmosis species other than M. mycoides subsp. mycoides SC and M. bovis are thus scarce.
Although the MLVA we developed for M. californicum consisted of only four MCTRs, the HGDI of this MLVA typing is higher than the HGDIs of MLVA typing for other mycoplasmas (6, 8), and the new MLVA indicates sufficient discriminatory ability for genotyping (24). An MCTR that showed extremely few variations in the repeat number among the four loci was not identified by this MLVA, unlike the MLVA results obtained for Staphylococcus aureus (28), M. pneumoniae (6), M. mycoides subsp. mycoides SC (7), and M. bovis (8). Thus, all of the MCTR loci contributed to the discriminatory power of this method, with variations in the repeat number occurring at all loci. Although MCTR569 of some strains was confirmed to have an insertion or a deletion of a 12-bp fragment, it was not difficult to carry out the MLVA. Nadon et al. (21) advocated avoiding repeat units with insertions and/or deletions in order to facilitate consistent sizing, but the insertion and deletion in MCTR569 is far shorter than that of the repeat units, and the number of insertions and deletions in MCTR569 occurred at only one location, and thus, the number of repeat units of MCTR569 could be easily estimated by simple electrophoresis. Moreover, the results of both the present in vivo and in vitro stability studies demonstrated the high stability of this MLVA typing method. These results suggest that our MLVA typing method is both easy to use and reliable.
As for the correlation between MCTRs and various factors, MCTR695 showed comparatively high ARI and WC values with PFGE clusters. MCTR695 also showed a comparatively high WC with pulsotypes. These findings indicate that the analysis of the results of MCTR695 accords well with that of PFGE (25, 26). Although MCTR695 was identified as part of a putative highly variable protein gene, this marker seems to be unambiguous and reflects the genomic similarities of M. californicum strains. MCTR698 also showed a comparatively high WC with PFGE clusters and pulsotypes, and thus it had the same tendency as MCTR695 (25, 26).
As for the restriction enzyme used for PFGE of M. californicum, our preliminary examination suggested that BamHI was suitable. BamHI could provide a distinguishable pulse pattern and was cost-effective; the cost of BamHI is approximately $0.25 per sample. Moreover, PFGE with BamHI showed higher discriminatory power than MLVA and showed a comparatively high correlation to MLVA. We thus concluded that PFGE is the primary method of analysis in molecular epidemiological genotyping and that MLVA typing is complementary to PFGE.
Genotyping information is helpful in developing an understanding of the invasion and spread of M. californicum in each case of infection. In the present study, MLVA and PFGE objectively showed the dissemination of M. californicum infection with the transportation of cows, as well as the persistence of M. californicum infection for a long period of time among herds in specific areas. In mastitis cases due to M. bovis, asymptomatic carrier cows were a significant factor (34). Additionally, M. californicum has been isolated from swabbing solution samples from the vulvovaginal tracts of cows (34). Some strains of M. californicum in the present study were isolated from the milk of cows that did not show clinical symptoms of bovine mastitis. The previous study (34) and the present data may indicate that asymptomatic infected cows play an important role in the dissemination of M. californicum, as well as in that of M. bovis, and that failure to identify these cows is a significant hindrance factor in the prevention of mycoplasmal bovine diseases. The roles of asymptomatic carriers, contaminated shedders (from milk, nasal and genital secretions, etc.), and contaminated environmental materials (e.g., bedding, feeding stuff, water, biofilm) that may be involved in the persistence of M. californicum within herds are relevant issues that should be clarified in the future.
The genotypical situation varied from case to case, and we found both cases with many genotypes (cases 5, 6, and 11) and a case whose genotypes were mostly identical (case 10). If many genotypes are confirmed, this suggests that many cows are the source of infection at the beginning of the outbreak. On the other hand, if most strains show identical genotypes, only a few cows may be the source of infection, or many cows carrying strains that show the same genotype under specific conditions may be the source of infection. Genotyping information is fundamental in developing an understanding of each outbreak and in determining the appropriate measures to take to prevent recurrence.
In Hokkaido, Japan, milking cows and heifers are frequently moved from farm to farm for scale expansion, replacement, and rearing. Although this management style is unfavorable for animal hygiene, it is also becoming established in many areas other than Hokkaido. It is thus essential to obtain epidemiological information that can identify mycoplasma carrier animals and the farms at the center of any mycoplasmal disease outbreak in order to prevent the spread of disease. The present genotyping method is a useful means of obtaining various types of epidemiological information.
A more precise understanding of the transmission of M. californicum is necessary to control the spread of disease. In the present study, MLVA typing and PFGE for M. californicum showed high discriminatory ability. An analysis that combines these two methods will provide a more precise picture of transmission events than either analysis alone. Ultimately, it is hoped that these techniques will contribute to a marked reduction in mycoplasmal bovine diseases such as mastitis.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the Livestock Hygiene Service Center and the Federation of Hokkaido Agricultural Mutual Aid Associations for their help with the collection of bovine mycoplasmal mastitic milk samples.
This work was supported in part by grants from the National Agriculture and Food Research Organization (NARO) of Japan.
We have no conflicts of interest.
Footnotes
Published ahead of print 3 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02488-14.
REFERENCES
- 1.Hewicker-Trautwein M, Feldmann M, Kehler W, Schmidt R, Thiede S, Seeliger F, Wohlsein P, Ball HJ, Buchenau I, Spergser J, Rosengarten R. 2002. Outbreak of pneumonia and arthritis in beef calves associated with Mycoplasma bovis and Mycoplasma californicum. Vet. Rec. 151:699–703. [PubMed] [Google Scholar]
- 2.Jasper DE. 1982. The role of Mycoplasma in bovine mastitis. J. Am. Vet. Med. Assoc. 181:158-162. [PubMed] [Google Scholar]
- 3.Eguchi M. 1994. Bovine mycoplasmal mastitis. J. Vet. Clin. 373:3–10 (In Japanese.) [Google Scholar]
- 4.Ball HJ, Campbell JN. 1989. Antibiotic treatment of experimental Mycoplasma californicum mastitis. Vet. Rec. 125:377–378. 10.1136/vr.125.14.377. [DOI] [PubMed] [Google Scholar]
- 5.Mackie DP, Ball HJ, Logan EF. 1986. Mycoplasma californicum mastitis in the dry dairy cow. Vet. Rec. 119:350–351. 10.1136/vr.119.14.350. [DOI] [PubMed] [Google Scholar]
- 6.Dégrange S, Cazanave C, Charron A, Renaudin H, Bébéar C, Bébéar CM. 2009. Development of multiple-locus variable-number tandem-repeat analysis for molecular typing of Mycoplasma pneumoniae. J. Clin. Microbiol. 47:914–923. 10.1128/JCM.01935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAuliffe L, Ayling RD, Nicholas RA. 2007. Identification and characterization of variable-number tandem-repeat markers for the molecular epidemiological analysis of Mycoplasma mycoides subspecies mycoides SC. FEMS Microbiol. Lett. 276:181–188. 10.1111/j.1574-6968.2007.00936.x. [DOI] [PubMed] [Google Scholar]
- 8.Pinho L, Thompson G, Rosenbusch R, Carvalheira J. 2012. Genotyping of Mycoplasma bovis isolates using multiple-locus variable-number tandem-repeat analysis. J. Microbiol. Methods 88:377–385. 10.1016/j.mimet.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulvey MR, Chui L, Ismail J, Louie L, Murphy C, Chang N, Alfa M, Canadian Committee for the Standardization of Molecular Methods 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:3481–3485. 10.1128/JCM.39.10.3481-3485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, Fussing V, Salmenlinna S, Vuopio-Varkila J, El Solh N, Cuny C, Witte W, Tassios PT, Legakis N, van Leeuwen W, van Belkum A, Vindel A, Laconcha I, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Coombes G, Cookson B. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574–1585. 10.1128/JCM.41.4.1574-1585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freundt EA. 1983. Culture media for classic mycoplasmas, p 127–135 In Razin S, Tully JG. (ed), Methods in mycoplasmology, vol 1 Mycoplasma characterization. Academic Press, New York, NY. [Google Scholar]
- 13.McNulty MS, Gowans EJ, Houston MJ, Fraser G. 1975. Neuraminidase content of strains of Newcastle disease virus which differ in virulence. J. Gen. Virol. 27:399–402. 10.1099/0022-1317-27-3-399. [DOI] [PubMed] [Google Scholar]
- 14.Pettersson B, Uhlén M, Johansson KE. 1996. Phylogeny of some mycoplasmas from ruminants based on 16S rRNA sequences and definition of a new cluster within the hominis group. Int. J. Syst. Bacteriol. 46:1093–1098. 10.1099/00207713-46-4-1093. [DOI] [PubMed] [Google Scholar]
- 15.Clyde WA., Jr 1964. Mycoplasma species identification based upon growth inhibition by specific antisera. J. Immunol. 92:958–965. [PubMed] [Google Scholar]
- 16.Senterfit LB, Jensen KE. 1966. Antimetabolic antibodies to Mycoplasma pneumoniae measured by tetrazolium reduction inhibition. Proc. Soc. Exp. Biol. Med. 122:786–790. 10.3181/00379727-122-31252. [DOI] [PubMed] [Google Scholar]
- 17.Senterfit LB. 1983. Preparation of antigens and antisera, p 401–404 In Razin S, Tully JG. (ed), Methods in mycoplasmology, vol 1 Mycoplasma characterization. Academic Press, New York, NY. [Google Scholar]
- 18.Jasper DE. 1977. Mycoplasma and mycoplasma mastitis. J. Am. Vet. Med. Assoc. 170(10 Pt 2):1167–1172. [PubMed] [Google Scholar]
- 19.Hata E, Murakami K. 2014. Complete genome sequence of Mycoplasma californicum strain HAZ160_1 from bovine mastitic milk in Japan. Genome Announc. 2:e00684–14. 10.1128/genomeA.00684-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580. 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nadon CA, Trees E, Ng LK, Møller Nielsen E, Reimer A, Maxwell N, Kubota KA, Gerner-Smidt P, MLVA Harmonization Working Group 2013. Development and application of MLVA methods as a tool for inter-laboratory surveillance. Euro Surveill. 18:20565 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy KJ, Oppenheim BA, Gossain S, Gao F, Hawkey PM. 2006. Use of variations in staphylococcal interspersed repeat units for molecular typing of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 44:271–273. 10.1128/JCM.44.1.271-273.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mørk T, Tollersrud T, Kvitle B, Jørgensen HJ, Waage S. 2005. Comparison of Staphylococcus aureus genotypes recovered from cases of bovine, ovine, and caprine mastitis. J. Clin. Microbiol. 43:3979–3984. 10.1128/JCM.43.8.3979-3984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubert L, Arabie P. 1985. Comparing partitions. J. Classif. 2:193–218. 10.1007/BF01908075. [DOI] [Google Scholar]
- 26.Wallace DL. 1983. A method for comparing two hierarchical clusterings: comment. J. Am. Stat. Assoc. 78:569–576. [Google Scholar]
- 27.Shen X, Gumulak J, Yu H, French CT, Zou N, Dybvig K. 2000. Gene rearrangements in the vsa locus of Mycoplasma pulmonis. J. Bacteriol. 182:2900–2908. 10.1128/JB.182.10.2900-2908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hata E, Katsuda K, Kobayashi H, Uchida I, Tanaka K, Eguchi M. 2010. Genetic variation among Staphylococcus aureus strains from bovine milk and their relevance to methicillin-resistant isolates from humans. J. Clin. Microbiol. 48:2130–2139. 10.1128/JCM.01940-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusiluka LJ, Ojeniyi B, Friis NF, Kokotovic B, Ahrens P. 2001. Molecular analysis of field strains of Mycoplasma capricolum subspecies capripneumoniae and Mycoplasma mycoides subspecies mycoides, small colony type isolated from goats in Tanzania. Vet. Microbiol. 82:27–37. 10.1016/S0378-1135(01)00352-2. [DOI] [PubMed] [Google Scholar]
- 30.McAuliffe L, Kokotovic B, Ayling RD, Nicholas RA. 2004. Molecular epidemiological analysis of Mycoplasma bovis isolates from the United Kingdom shows two genetically distinct clusters. J. Clin. Microbiol. 42:4556–4565. 10.1128/JCM.42.10.4556-4565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer A, Shapiro B, Muriuki C, Heller M, Schnee C, Bongcam-Rudloff E, Vilei EM, Frey J, Jores J. 2012. The origin of the ‘Mycoplasma mycoides cluster' coincides with domestication of ruminants. PLoS One 7(4):e36150. 10.1371/journal.pone.0036150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manso-Silván L, Dupuy V, Lysnyansky I, Ozdemir U, Thiaucourt F. 2012. Phylogeny and molecular typing of Mycoplasma agalactiae and Mycoplasma bovis by multilocus sequencing. Vet. Microbiol. 161:104–112. 10.1016/j.vetmic.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 33.Bashiruddin JB. 1998. PCR and RFLP methods for the specific detection and identification of Mycoplasma mycoides subsp. mycoides SC. Methods Mol. Biol. 104:167–178. [DOI] [PubMed] [Google Scholar]
- 34.Punyapornwithaya V, Fox LK, Hancock DD, Gay JM, Alldredge JR. 2010. Association between an outbreak strain causing mycoplasma bovis mastitis and its asymptomatic carriage in the herd: a case study from Idaho, USA. Prev. Vet. Med. 93:66–70. 10.1016/j.prevetmed.2009.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.