Abstract
Within ecosystems that are poor in carbohydrates, alternative substrates such as arginine may be of importance to coagulase-negative staphylococci (CNS). However, the versatility of arginine conversion in CNS remains largely uncharted. Therefore, a set of 86 strains belonging to 17 CNS species was screened for arginine deiminase (ADI), arginase, and nitric oxide synthase (NOS) activities, in view of their ecological relevance. In fermented meats, for instance, ADI could improve bacterial competitiveness, whereas NOS may serve as an alternative nitrosomyoglobin generator to nitrate and nitrite curing. About 80% of the strains were able to convert arginine, but considerable inter- and intraspecies heterogeneity regarding the extent and mechanism of conversion was found. Overall, ADI was the most commonly employed pathway, resulting in mixtures of ornithine and small amounts of citrulline. Under aerobic conditions, which are more relevant for skin-associated CNS communities, several strains shifted toward arginase activity, leading to the production of ornithine and urea. The obtained data indeed suggest that arginase occurs relatively more in CNS isolates from a dairy environment, whereas ADI seems to be more abundant in strains from a fermented meat background. With some exceptions, a reasonable match between phenotypic ADI and arginase activity and the presence of the encoding genes (arcA and arg) was found. With respect to the NOS pathway, however, only one strain (Staphylococcus haemolyticus G110) displayed phenotypic NOS-like activity under aerobic conditions, despite a wide prevalence of the NOS-encoding gene (nos) among CNS. Hence, the group of CNS displays a strain- and condition-dependent toolbox of arginine-converting mechanisms with potential implications for competitiveness and functionality.
INTRODUCTION
Spontaneously established communities of coagulase-negative staphylococci (CNS) occur in different mammal-based ecological niches, such as cheeses and fermented meats, as well as on human or animal skin (1, 2). In fermented meats, CNS are of technological interest, as they generate a desirable nitrosomyoglobin color through nitrate reductase activity, provide antioxidant protection through catalase activity, and contribute to aroma formation (3). Whereas the communities of lactic acid bacteria in fermented meats usually narrow down to a pronounced dominance by Lactobacillus sakei, the species diversity of CNS is wider (4, 5). Although Staphylococcus xylosus, Staphylococcus equorum, and Staphylococcus saprophyticus are the most frequently isolated CNS, many other species may be encountered, such as Staphylococcus carnosus, Staphylococcus cohnii, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus pasteuri, Staphylococcus sciuri, Staphylococcus succinus, and Staphylococcus warneri (2). The resulting CNS species diversity is not easy to predict, but it seems that variations in the raw materials play a role besides processing conditions, such as the effects of molding versus smoking (4, 5). On human and animal skin tissues, CNS are part of the natural microbiota, as is often seen for Staphylococcus chromogenes, Staphylococcus simulans, S. haemolyticus, S. xylosus, and S. epidermidis on bovine udders (1). The composition of udder-related CNS communities can vary considerably between herds and cows, which may play a role in the generation or prevention of infections, such as mastitis (1, 6).
Overall, metabolic heterogeneity exists within the Staphylococcus genus, also with respect to arginine metabolism (7). This potentially leads to differences in competitiveness in its specific ecological niches and to variable community compositions. Generally, carbohydrates are the primary energy source for staphylococci (7, 8). In fermented meats, for instance, carbohydrates are naturally scarce, whereas the fermentable sugars that are initially added to the sausage batter to boost fermentation are rapidly scavenged by the lactic acid bacteria. As a result, CNS must shift from common carbohydrates toward the use of alternative endogenous energy sources, such as nucleosides or arginine (9), a capacity that has also been demonstrated for meat-associated L. sakei strains (10–12).
In general, bacteria may use various strategies to catabolize arginine (13, 14). The arginine deiminase (ADI) pathway has been reported as a common route for arginine degradation in bacteria, including CNS (15), although variability on both species and strain levels exists (9). The ADI pathway leads to the production of extra ATP, improved survival under acid stress conditions via ammonia production, and the use of intermediate carbamoyl phosphate for pyrimidine biosynthesis. Three cytoplasmic enzymes are involved in this pathway, i.e., ADI, ornithine transcarbamylase (OTC), and carbamate kinase (CK) (15). Besides its omnipresence in the 45 finished genomes of S. aureus reported in the NCBI genome database, the ADI-encoding arcA gene has also been found in 10 out of 11 available genomes of CNS strains but not in that of S. saprophyticus ATCC 15305 (16). Depending on the pH, the intermediate citrulline can be partially excreted and subsequently converted into ornithine, likely involving a specific transporter, as shown for L. sakei (17).
As an alternative to the ADI pathway, arginine can be converted into ornithine and urea by the arginase enzyme (13). Arginase activity may be derepressed by oxygen, which at the same time represses the ADI pathway, as in Bacillus licheniformis (18). The arginase-encoding gene, arg, ubiquitous in the 45 finished S. aureus genomes in the NCBI genome database, has also been found in the finished genomes of S. saprophyticus (1 strain), Staphylococcus pseudintermedius (2 strains), and S. warneri (1 strain) but not in strains of S. epidermidis (2 strains), S. haemolyticus (1 strain), S. carnosus (1 strain), Staphylococcus lugdunensis (2 strains), and Staphylococcus pasteuri (1 strain) (16). Phenotypical arginase activity has been described for strains of S. saprophyticus (19), and proteomic data suggest that S. xylosus may also display it (20). Hypothetically, further bacterial conversion of ornithine into glutamate semialdehyde or glutamate may occur, or even into Δ1–pyrroline-5-carboxylate, proline, and α-ketoglutaric acid (13).
A third but hitherto poorly explored possibility of arginine conversion in staphylococci is provided by the action of nitric oxide synthase (NOS) (21). As such, arginine can be converted into citrulline and NO via the oxidation of the guanidinium group of arginine. This reaction consumes oxygen and NADPH-H+ as a cofactor. The presence of NOS in bacteria has been documented for members of Nocardia, Salmonella, and Bacillus (22–24). Although a presumed NOS gene has been found for all available reference sequences of the 56 finished staphylococcal genomes in the NCBI genome database (16), actual NOS activity has been persuasively described only for S. aureus (25, 26). From a technological point of view, the generation of NO by NOS-positive bacteria could be of interest for color formation in fermented meat products prepared without nitrate and nitrite (21, 27–31). Unfortunately, successful implementation of bacterial NOS activity in fermented meat products has thus far been insufficiently proven (14). Moreover, little is known on how widespread this potential for NOS activity is within the group of CNS from different ecological niches.
The aim of this study was to broaden the knowledge on the ability of CNS to use arginine as a substrate and to highlight the importance of the potential arginine-converting pathways involved. A particular focus was on the relevance of NOS activity, considering its unknown significance for CNS from different ecosystems and its potential applications for the meat industry.
MATERIALS AND METHODS
Microorganisms.
Eighty-six CNS strains isolated from different ecological niches (i.e., fermented sausages, meat starter cultures, milk, and bovine teat apex skin) were used during this study (Table 1). The strains originated from the laboratory collection of the Research Group of Industrial Microbiology and Food Biotechnology (Vrije Universiteit Brussel, Brussels, Belgium), with the exception of S. carnosus 833 (INRA, Clermont-Ferrand-Theix, France) (32), S. carnosus TM 300 (Department of Microbial Genetics, Universität Tübingen, Germany) (33), and S. epidermidis ATCC 12228 (Food and Drug Administration, Silver Spring, MD, USA) (34). In addition, S. carnosus 100M-5 (BioAgro, Chr. Hansen, Hørsholm, Denmark) and S. carnosus 10P1-3 and S. xylosus 10P1-1 (Danisco, Copenhagen, Denmark) were isolated from commercial meat starter cultures (35). The authenticity of all strains was confirmed at the species level through rpoB gene sequencing (36). All strains were stored at −80°C in brain heart infusion (BHI) medium (Oxoid, Basingstoke, Hampshire, United Kingdom), supplemented with 25% (vol/vol) glycerol as a cryoprotectant.
TABLE 1.
Conversion of arginine (initial concentration of 17 mM) into citrulline, ornithine, and urea in MSM-A after 72 h under microaerobic and aerobic conditions, extended with a genotypic screening for the presence of arcA, arg, and nos genesa
| Staphylococcus species | Strain | Origin | Gene presenceb |
Concn (mean ± SD; mM)c |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Microaerobic |
Aerobic |
|||||||||
| arcA | arg | nos | Cit | Orn | Cit | Orn | Urea | |||
| S. arlettae | G130 | Teat apex skin | + | + | − | 2 ± 0 | 21 ± 1 | 6 ± 2 | 9 ± 5 | ND |
| G238 | Teat apex skin | − | + | − | ND | ND | ND | 1 ± 2 | 3 ± 2 | |
| S. auricularis | G162 | Teat apex skin | + | + | − | ND | 5 ± 1 | ND | 14 ± 1 | 16 ± 0 |
| S. capitis | G265 | Teat apex skin | − | − | − | ND | ND | ND | ND | ND |
| S. carnosus | 833 | Fermented sausage | + | − | 2d | 3 ± 1 | 17 ± 3 | 2 ± 1 | 12 ± 3 | ND |
| 10P1-3 | Starter culture | + | − | 2 | 3 ± 1 | 12 ± 0 | 3 ± 1 | 16 ± 4 | ND | |
| 100 M-5 | Starter culture | + | − | 2 | 3 ± 1 | 19 ± 2 | 2 ± 1 | 11 ± 3 | ND | |
| Carnimp | Starter culture | + | − | 2 | 3 ± 1 | 14 ± 3 | 3 ± 2 | 15 ± 5 | ND | |
| 318-5 | Starter culture | + | − | 2 | 3 ± 0 | 18 ± 1 | 3 ± 1 | 12 ± 3 | ND | |
| F4P1 | Fermented sausage | + | − | 2 | 4 ± 4 | 11 ± 2 | ND | 5 ± 2 | 5 ± 2 | |
| 1505-5 | Starter culture | + | − | 2d and 3d | 2 ± 2 | 17 ± 4 | 2 ± 1 | 15 ± 2 | ND | |
| SS3-4 | Fermented sausage | + | − | 2 | 3 ± 1 | 20 ± 2 | 3 ± 2 | 18 ± 3 | ND | |
| SS7-2 | Fermented sausage | + | − | 2 | 3 ± 0 | 18 ± 5 | 6 ± 0 | 11 ± 0 | ND | |
| 201-5 | Starter culture | + | − | 2 | 3 ± 0 | 21 ± 2 | ND | 15 ± 3 | ND | |
| 241-5 | Starter culture | + | − | 2 | 3 ± 0 | 15 ± 6 | 3 ± 1 | 12 ± 5 | ND | |
| TM300 | Fermented sausage | + | − | 2 | 5 ± 1 | 15 ± 6 | 4 ± 1 | 13 ± 4 | ND | |
| vk5 | Fermented sausage | + | − | 2 | 4 ± 1 | 17 ± 1 | 3 ± 1 | 6 ± 3 | ND | |
| S. chromogenes | G222 | Teat apex skin | + | + | 3d | ND | 15 ± 0 | ND | 12 ± 2 | 8 ± 3 |
| i160 | Teat apex skin | + | − | 3 | ND | ND | ND | ND | ND | |
| S. cohnii | 2.63 | Milk | − | + | − | ND | ND | ND | 12 ± 2 | 14 ± 1 |
| G166 | Teat apex skin | − | + | − | ND | ND | ND | 13 ± 1 | 9 ± 0 | |
| G207 | Teat apex skin | − | + | − | ND | ND | ND | 7 ± 4 | 15 ± 3 | |
| S. devriesei | G194 | Teat apex skin | + | − | 3d | ND | 5 ± 2 | ND | 5 ± 3 | ND |
| G246 | Teat apex skin | − | + | − | ND | ND | ND | 6 ± 2 | 5 ± 1 | |
| G267 | Teat apex skin | + | − | 3 | 7 ± 1 | 11 ± 9 | 3 ± 1 | 10 ± 5 | ND | |
| S. epidermidis | 2S7–4 | Fermented sausage | + | − | 1d and 3d | 2 ± 1 | 17 ± 0 | 2 ± 1 | 3 ± 2 | ND |
| ATCC 12228 | Unknown | + | − | 1 and 3 | 2 ± 1 | 20 ± 4 | 3 ± 1 | 16 ± 1 | ND | |
| G153 | Teat apex skin | + | − | 1 and 3 | 2 ± 1 | 14 ± 1 | 3 ± 1 | 13 ± 3 | ND | |
| G262 | Teat apex skin | + | +d | 1d and 2d | ND | 16 ± 4 | 3 ± 1 | 11 ± 5 | ND | |
| i162 | Teat apex skin | + | − | 1 and 2 | ND | 6 ± 4 | 2 ± 1 | 4 ± 2 | ND | |
| αSg3 | Fermented sausage | + | − | 1 and 3 | 3 ± 1 | 17 ± 4 | 4 ± 1 | 15 ± 2 | ND | |
| S. equorum | 2.55 | Milk | − | − | 1d | ND | ND | ND | ND | ND |
| 3FX-S13 | Fermented sausage | − | − | 1 | ND | 7 ± 0 | ND | ND | ND | |
| 4FX-S6 | Fermented sausage | − | + | 1,d 3,d and 4d | 4 ± 2 | 13 ± 1 | ND | 8 ± 1 | 6 ± 1 | |
| DBL-S8 | Fermented sausage | − | +d | 1 | 1 ± 0 | 7 ± 4 | ND | ND | ND | |
| DBL-S18 | Fermented sausage | − | + | 1 | ND | ND | ND | 4 ± 1 | 2 ± 3 | |
| DBX-S7 | Fermented sausage | − | + | 1 | 6 ± 1 | 15 ± 1 | ND | 10 ± 1 | 9 ± 0 | |
| DBX-S11 | Fermented sausage | − | +d | − | ND | ND | ND | ND | ND | |
| DBX-S21 | Fermented sausage | − | − | − | 5 ± 1 | 10 ± 2 | ND | 6 ± 0 | ND | |
| DFL-S9 | Fermented sausage | − | + | − | 6 ± 0 | 14 ± 2 | ND | 5 ± 2 | 8 ± 1 | |
| DFL-S19 | Fermented sausage | − | +d | 1 | ND | 17 ± 1 | ND | ND | ND | |
| DFL-S21 | Fermented sausage | − | − | 1 | 5 ± 1 | 9 ± 3 | ND | 3 ± 1 | ND | |
| DFL-S23 | Fermented sausage | − | + | 2d and 3d | ND | 6 ± 2 | ND | 7 ± 0 | 10 ± 1 | |
| DFX-S10 | Fermented sausage | − | + | 1 | ND | ND | ND | 7 ± 1 | 11 ± 1 | |
| G101 | Teat apex skin | − | + | 1 and 3 | ND | ND | ND | 4 ± 1 | 4 ± 1 | |
| S. fleurettii | 2.05 | Milk | − | − | − | ND | ND | ND | ND | ND |
| S. haemolyticus | 12S24-4 | Fermented sausage | + | − | 2d | 4 ± 0 | 7 ± 2 | 4 ± 1 | 7 ± 3 | ND |
| G110 | Teat apex skin | + | − | 3d | ND | ND | 9 ± 4 | ND | ND | |
| G191 | Teat apex skin | + | − | 3 | 3 ± 1 | 16 ± 2 | 4 ± 2 | 14 ± 6 | ND | |
| SB1–6 | Fermented sausage | + | +d | 3 | 11 ± 1 | 7 ± 1 | 2 ± 0 | 9 ± 2 | ND | |
| αS1–1 | Fermented sausage | + | + | 3 | 4 ± 1 | 17 ± 1 | 2 ± 2 | 13 ± 5 | ND | |
| S. pasteuri | DBL-S6 | Fermented sausage | − | − | 3 | ND | ND | ND | 3 ± 1 | 5 ± 0 |
| DFL-S3 | Fermented sausage | − | + | − | ND | ND | ND | 10 ± 3 | 10 ± 1 | |
| αS3-13 | Fermented sausage | − | +d | − | 7 ± 0 | 12 ± 1 | 5 ± 1 | 11 ± 5 | ND | |
| αS3-14 | Fermented sausage | + | − | − | 2 ± 2 | 13 ± 6 | 2 ± 2 | 16 ± 3 | ND | |
| S. saprophyticus | 3.02 | Milk | − | +d | 2,d 3,d and 4d | ND | ND | ND | ND | ND |
| DFL-12 | Fermented sausage | − | + | 3 and 4 | ND | ND | ND | ND | ND | |
| F2S11 | Fermented sausage | − | +d | 4 | ND | ND | ND | ND | ND | |
| FPP1 | Fermented sausage | + | − | 2 and 4 | 2 ± 0 | 19 ± 2 | 3 ± 1 | 12 ± 6 | ND | |
| FPS1 | Fermented sausage | − | + | 2,d 3,d and 4d | 2 ± 1 | 2 ± 0 | ND | 6 ± 4 | 4 ± 2 | |
| G089 | Teat apex skin | + | +d | 2, 3, and 4 | 5 ± 1 | 10 ± 1 | 3 ± 4 | 5 ± 1 | ND | |
| G131 | Teat apex skin | − | + | 2, 3, and 4 | ND | ND | ND | ND | ND | |
| G243 | Teat apex skin | − | +d | 2, 3, and 4 | ND | ND | ND | ND | ND | |
| αSG1 | Fermented sausage | − | +d | 3 and 4 | ND | 17 ± 3 | 2 ± 3 | 12 ± 8 | ND | |
| S. succinus | 3PA1 | Fermented sausage | − | − | − | ND | ND | ND | ND | ND |
| 4pb1 | Fermented sausage | − | − | − | ND | ND | ND | ND | ND | |
| SS0-7 | Fermented sausage | − | − | − | ND | ND | ND | ND | ND | |
| S. sciuri | G160 | Teat apex skin | + | + | − | ND | ND | ND | 3 ± 1 | 6 ± 1 |
| G173 | Teat apex skin | − | + | − | ND | ND | ND | 5 ± 1 | 2 ± 1 | |
| I20-1 | Fermented sausage | − | + | − | ND | ND | ND | 5 ± 1 | 8 ± 1 | |
| S. warneri | 2.06 | Milk | + | +d | − | ND | 17 ± 3 | 3 ± 2 | 15 ± 2 | ND |
| 2.25 | Milk | + | − | 3d | 2 ± 1 | 15 ± 3 | ND | 15 ± 2 | ND | |
| i184 | Teat apex skin | − | − | 1d | ND | ND | ND | 2 ± 2 | 2 ± 1 | |
| S. xylosus | 10P1-1 | Starter culture | − | +d | 1d and 3d | ND | ND | ND | ND | ND |
| 2.15 | Milk | − | + | 3 | ND | ND | ND | 8 ± 5 | 13 ± 1 | |
| 2.17 | Milk | − | + | 3 | ND | ND | ND | 4 ± 4 | 10 ± 1 | |
| 2S7-2 | Fermented sausage | − | − | − | 3 ± 1 | 10 ± 3 | ND | 10 ± 2 | ND | |
| 3PA3 | Fermented sausage | + | +d | 1 and 3 | ND | ND | 5 ± 4 | 8 ± 2 | ND | |
| 3PA6 | Fermented sausage | + | + | 3 | ND | ND | ND | 5 ± 2 | 4 ± 0 | |
| fns1 | Fermented sausage | − | + | 3 | ND | ND | ND | 2 ± 2 | 6 ± 1 | |
| FPS2 | Fermented sausage | − | +d | 3 | ND | ND | ND | ND | ND | |
| G211 | Teat apex skin | + | + | 1 and 3 | 2 ± 0 | 18 ± 1 | ND | 10 ± 5 | 10 ± 5 | |
| G223 | Teat apex skin | − | + | 3 | ND | ND | ND | 4 ± 0 | 7 ± 2 | |
| p20g1 | Fermented sausage | − | +d | 3 | ND | ND | ND | ND | ND | |
| TP4 | Fermented sausage | − | + | 3 | ND | ND | ND | 5 ± 0 | 6 ± 5 | |
| W1–1 | Fermented sausage | − | + | 3 | 3 ± 1 | 6 ± 1 | ND | 8 ± 7 | 15 ± 0 | |
No urea was found under microaerobic conditions.
+, present; −, absent. Number indicates the primer set(s) (see Table 2) giving a positive signal.
Cit, citrulline; Orn, ornithine; ND, not detected.
Confirmed by sequencing the PCR products (sequence identity between the PCR products and the corresponding targeted gene for the Staphylococcus species with the best hit ranged between 90% and 100%).
Genotypic screening for arginine conversion potential.
Genomic DNA of all 86 CNS strains was isolated from respective overnight cultures in BHI medium at 30°C. Cell pellets were obtained by centrifugation of 1 ml of these cultures (21,036 × g for 20 min) and washed with TES buffer (50 mM Tris base, 1 mM EDTA, 6.7% [mass/vol] sucrose, pH 8.0). Cell pellets were mixed with a lysis solution (20 mM Tris, 2 mM EDTA, 1% [vol/vol] Triton X-100, 50 μg ml−1 lysozyme, 0.5 U μl−1 mutanolysin [Sigma-Aldrich, St. Louis, MO, USA]) and incubated at 37°C for 60 min to release DNA. Subsequently, the NucleoSpin 96 tissue kit (Macherey-Nagel, Düren, Germany) was used to purify the DNA by following the manufacturer's instructions. The DNA concentration was quantified using a NanoDrop 2000 apparatus (Thermo Scientific, Wilmington, NC, USA).
Several primer pairs were designed to screen for the presence of genes potentially involved in arginine catabolism pathways (Table 2), in addition to previously designed primers targeting the arcA gene of the ADI pathway (9). The primer pair targeting the arginase-encoding gene (arg) was based on its corresponding sequences in strains of Staphylococcus caprae, Staphylococcus capitis, S. epidermidis, S. saprophyticus, Staphylococcus simiae, S. warneri, and S. xylosus. Four primer pairs targeting the NOS-encoding gene (nos) were designed based on annotated sequences, covering species heterogeneity. Primer pair NOS-1 was based on all available annotated NOS sequences in strains of S. capitis, S. epidermidis, S. equorum, and S. warneri; primer pair NOS-2 was based on all available annotated NOS sequences in strains of S. carnosus and S. aureus; primer pair NOS-3 was based on the annotated NOS sequence in S. haemolyticus JCSC1435; and primer pair NOS-4 was based on the annotated NOS sequences in S. saprophyticus subsp. saprophyticus ATCC 15305 and S. saprophyticus subsp. saprophyticus KACC 16562. Finally, three primer pairs, based on the gene sequence of otc (encoding OTC of the ADI pathway) of S. haemolyticus JCSC1435 (37), were designed using SNPbox (38) (Table 2), resulting in overlapping PCR amplicons and covering the whole otc gene of S. haemolyticus JCSC1435. All primer pairs were synthesized by Integrated DNA Technologies (Leuven, Belgium).
TABLE 2.
Primer pairs used to screen for the presence of the arg, nos, and otc genes in Staphylococcus strains
| PCR target | Primer pair | Nucleotide sequences (forward and reverse, respectively) (5′ to 3′) | Amplicon size (bp) | Species | ENA accession no. of the entries used for primer design |
|---|---|---|---|---|---|
| arg | ARG-F and ARG-R | THGGDATGAGRGATTTRGA and GCAAARTGRCTTTCHCKATA | 247 | S. caprae, S. capitis, S. epidermidis, S. saprophyticus, S. simiae, S. warneri, S. xylosus | EFS16894, BAE17866, ABF13437, EEE48798, EES42258, EEQ79567, EHJ09071, EGG96003, EGS40237 |
| nos | NOS-1-F and NOS-1-R | CAAAAATGGCATGGAGAAAC and CCATCCATTAAAAGGTGCTG | 523 | S. capitis, S. epidermidis, S. equorum, S. warneri | EEE48709, EEQ78851, EFA89185, EFV88123, EGG97234, EGG63344, EGG71090 EGG71063, EGS40499, EGS80208, EGS78236, EGS80733, EHM67228, EHQ72179, EHQ76423, EHQ75636, EHR79954, EHR81536, EHR82566, EHR90879, EHR89224, EHR94024, EHR95252, EHR98881, EHS00755, EHQ81520, EEE48709, EEQ78851, EFA89185, EFV88123, EGG97234, EGG63344, EGG71090, EGG71063, EGS40499, EGS80208, EGS78236, EGS80733, EHM67228, EHQ72179, EHQ76423, EHQ75636, EHR79954, EHR81536, EHR82566, EHR90879, EHR89224, EHR94024, EHR95252, EHR98881, EHS00755, EHQ81520, EJE31238, EJE36529, EJE38416, EJE39628, EJE43873, EJE45333, CCI58543 |
| NOS-2-F and NOS-2-R | GGNGATCCTGCTGAAAAAGA and GCAGCGGTNAAATGATCNAC | 490 | S. aureus, S. carnosus | CAL28392, AAW36946 | |
| NOS-3-F and NOS-3-R | TGGACGCTTATTTTGGGATT and GGGGCTAACCAAGACCATTT | 768 | S. haemolyticus | BAE04347 | |
| NOS-4-F and NOS-4-R | TTGGATGGCAAGGTCAGTAT and ACCAATGTGGGAGACAAAGA | 541 | S. saprophyticus subsp. saprophyticus | BAE18022, EHY92989 | |
| otc | OTC-1-F and OTC-1-R | AAACATGTTATTGTAAACATTTTGTC and CACACAGGCACACCTGAATA | 594 | S. haemolyticus | AP006716 |
| OTC-2-F and OTC-2-R | ACGCCGGTATAGAACAGCAAAAA and CGTCAGGTTCACCCATTGATACC | 618 | S. haemolyticus | AP006716 | |
| OTC-3-F and OTC-3-R | CCCTTAAAGAATTAAATCCAACAGAT and GATTACAACTTCATAACCTTTTTGG | 586 | S. haemolyticus | AP006716 |
PCR assays were performed using a T300 thermocycler (Biometra GmbH, Göttingen, Germany), containing 200 ng of genomic DNA, 200 μM each deoxynucleoside triphosphate (dNTP), 20 pmol of each primer, 5 μl of 10× PCR buffer, 1.25 U of Taq DNA polymerase (Roche, Basel, Switzerland), and sterile ultrapure water in a final total volume of 50 μl. For the PCR assay targeting the arcA gene, the conditions were as previously described (9). For the other genotypic screening experiments, the following conditions were used for the PCR assays with the different primer pairs: initial denaturation at 94°C for 120 s, 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C (primer pair NOS-2), 50°C (primer pairs NOS-1 and NOS-4), or 45°C (primer pair NOS-3 and primer pairs for the genes encoding arginase and OTC) for 60 s, and extension at 72°C for 60 s, with a final extension at 72°C for 420 s. The optimum annealing temperatures for the PCR conditions were determined by performing a gradient PCR with a TProfessional basic gradient thermocycler (Biometra GmbH).
DNA isolated from S. carnosus TM 300 was used as a positive control for PCR assays targeting arcA, whereas DNA isolated from S. epidermidis ATCC 12228, S. carnosus TM 300, and S. saprophyticus ATCC 15305 was used as a positive control for the PCR assays with the NOS-1, NOS-2, and NOS-4 primer pairs, respectively. In addition, DNA isolated from S. saprophyticus ATCC 15305 and S. carnosus TM 300 was used as a positive control for the PCR assays targeting arg and otc, respectively.
Following amplification, the presence of PCR products and their sizes were controlled on a 1.0% (mass/vol) agarose gel (Life Technologies, Carlsbad, CA, USA). A selected number of PCR products (indicated in Table 1) were purified using the Wizard SV gel and PCR cleanup system (Promega, Madison, WI, USA) and submitted to Sanger sequencing (VIB Genetic Service Facility, Antwerp, Belgium). To assess whether the intended gene fragment was amplified, the sequences were aligned to the NCBI nonredundant protein sequence database (nr) using the blastx tool.
The genotypic screening for the presence of arcA, arg, and nos genes was performed for all 86 CNS strains, whereas the screening for the otc gene was limited to S. haemolyticus G110.
Media for phenotypic investigation and conversion studies.
BHI medium was used for inoculum buildup. For the small-scale screening (10 ml and 100 ml) and large-scale kinetic experiments (10 liters), a meat simulation medium (MSM) was used (39), which contained (per liter) 8.8 g Lab Lemco powder (Oxoid), 11.0 g bacteriological peptone (Oxoid), 2.2 g granulated yeast extract (VWR International, Darmstadt, Germany), 1 ml Tween 80 (VWR International), 0.038 g MnSO4·4H2O (VWR International), and 40 g NaCl (VWR International). The MSM was supplemented in all cases with 17 mM arginine (Sigma-Aldrich), further referred to as MSM-A.
For a subset of experiments dealing with attempts to stimulate NOS activity, modifications of the MSM-A included the removal of NaCl or the addition of glucose (5 g liter−1), FeSO4·7H2O (0.5 g liter−1), MgSO4·0.7H2O (0.2 g liter−1), CaCO3 (0.2 g liter−1), methanol (0.125% [vol/vol]), tetrahydrobiopterin (H4B) (0.005 g liter−1), hemin (0.002 g liter−1), or filter-sterilized myoglobin extract from beef muscle (50 ml). Myoglobin extract was prepared by extraction of 100 g of beef muscle with 50 ml of phosphate buffer (1 M; pH 6.0), blending for 5 min, and centrifuging at 7,200 rpm for 30 min.
All components were dissolved in deionized water and, prior to sterilization, the medium was adjusted to pH 6.5 for the screening experiments (a favorable pH condition for growth) or to pH 5.8 for the large-scale fermentations (mimicking the initial pH value of meat fermentations). MSM-A agar was prepared by adding 12 g liter−1 of granulated agar (Oxoid) prior to medium sterilization.
Mannitol salt phenol red agar (MSA; VWR International) was used for enumeration of the CFU to determine the cell counts of the CNS cultures (in CFU ml−1).
Phenotypic screening for arginine conversion under microaerobic and aerobic conditions.
The inocula for the small-scale screening experiments were prepared by subculturing the CNS strains in BHI medium at 30°C for 15 h. Both microaerobic and aerobic conditions were imposed to take into account ecological variation in oxygen exposure. For the screening experiments under microaerobic conditions, 0.1 ml of a subculture was inoculated into 10 ml of MSM-A, which was subsequently incubated at 30°C. For the screening experiments under aerobic conditions, 1.0 ml of a subculture was inoculated into 100 ml of MSM-A, which was subsequently incubated at 30°C in a shaking incubator at 160 rpm (Certomat BS-1; Sartorius Stedim Systems, Melsungen, Germany). All screening experiments were performed in triplicate.
Samples were collected after 72 h; arginine, citrulline, and ornithine concentrations were determined using a Waters 2695 liquid chromatograph coupled to a Quattro Micro mass spectrometer (Waters Corp., Milford, MA, USA), as described previously (11). Due to matrix interference, all quantifications were carried out through standard addition; the original sample concentrations with corresponding errors were calculated as described previously (40). Urea samples were prepared with 500 μl of cell-free culture supernatant and 500 μl of acetonitrile and were measured through high-performance liquid chromatography with UV detection at 190 nm (Waters 600S; Waters Corp.), equipped with an Inertsil Amide 5020-07815 column (GLSciences, Tokyo, Japan). The eluents used were 95% acetonitrile and 5% ultrapure water. Quantification was performed via an external standard curve with standards prepared in the same way as the samples. The detection limit was established above 1 mM for all three components.
For the samples related to the screening of S. haemolyticus strains under aerobic conditions, ammonia was measured in triplicate via a colorimetric kit based on the action of the enzyme l-glutamate dehydrogenase (ammonia assay kit; Sigma-Aldrich).
Conversion kinetics of arginine by Staphylococcus haemolyticus G110.
Large-scale experiments (10 l) were performed in MSM-A with S. haemolyticus G110, a strain selected based on its atypical phenotypic behavior (i.e., the only strain producing citrulline without ornithine under aerobic conditions; see Results). The inocula were prepared through three subcultures. The first subculture was obtained after inoculation into 10 ml of BHI medium (pH 7.4) and subsequent incubation at 30°C for 10 h. Next, 0.1 ml of the first subculture was added to 10 ml of MSM-A (pH 6.5). After incubation at 30°C for 20 h, 1 ml was transferred into 100 ml of MSM-A (pH 6.5). After a final incubation at 30°C for 15 h, this third subculture was used as a final inoculum.
The large-scale experiments with S. haemolyticus G110 were performed under aerobic and microaerobic conditions to determine the effect of atmospheric oxygen on growth and arginine conversion. Aerobic conditions were applied with an airflow of 4.0 liters/min, whereas microaerobic conditions were carried out without airflow. The experiments were carried out in 15-liter Biostat C fermentors (Sartorius Stedim Biotech, Göttingen, Germany), containing a working volume of 10 liters. The fermentor was sterilized in situ at 121°C and a pressure of 210 kPa for 20 min. The fermentation temperature was kept at 30°C. During fermentation, the pH of the medium was kept constant at pH 5.8 through automatic addition of a 5 M NaOH and a 5 M HCl solution. The stirring speed was fixed at 100 rpm under microaerobic conditions and at 300 rpm when air was pumped into the fermentor. In the case of aeration, 10 ml of antifoam (Sigma-Aldrich) was added to the medium. Temperature, pH, and agitation were controlled online (Micro MFCS for Windows NT software; Sartorius AG). After each sampling, the sampling valve was sterilized during 20 min through an external steam generator (J. Strobel & Söhne GmbH & Co., Munich, Germany).
The concentrations of arginine, citrulline, and ornithine were determined as described above. Calibration was performed with external standards, and the samples were analyzed in triplicate.
Additional screening for phenotypic NOS activity with Staphylococcus carnosus strains under different conditions.
A set of 12 S. carnosus strains that yielded a positive signal for the genotypic NOS screening (see Results) were further investigated for phenotypic NOS activity under different conditions. This particular species was chosen as a case study, not only because it displayed a clear presence of the NOS-encoding gene and an absence of the potentially competing arg gene (both favored under aerobic conditions; see Results) but also because of its widespread use as starter culture for meat fermentation. A wide range of different conditions was used, as to take into account the variability occurring in different ecosystems (e.g., fermented meats versus animal skin). The screening experiments were performed in MSM-A at different initial pH values (pH 5.8, pH 7.0, and pH 7.5) and a constant temperature of 30°C, as well as at different temperatures (12°C, 20°C, and 40°C) and an initial pH of 6.5. In addition, screening experiments were performed at 30°C in modified MSM-A medium at an initial pH of 6.5 (see “Phenotypic screening for arginine conversion under microaerobic and aerobic conditions” above). In all cases, aerobic conditions were applied using a shaking incubator at 160 rpm for 72 h, except for the experiments at 12°C, for which incubation took place during 7 days. Finally, a screening experiment was performed on MSM-A agar, both in the presence and absence of myoglobin extract, to investigate the potential effect of growth of the investigated S. carnosus strains on solid surfaces. In all cases, concentrations of arginine, citrulline, and ornithine in the media (or in a stomacher-generated extract of the agar) were determined as described above.
RESULTS
Genotypic screening for arginine conversion potential.
Based on the primer pairs used, the arcA and arg genes were present in 39 and 47 of the strains tested, respectively (Table 1). A minority of 12 strains possessed both genes. Strikingly, S. carnosus systematically possessed the arcA gene but never the arg gene. In contrast, the arg gene in the absence of the arcA gene was found in all three strains of S. cohnii. In addition to the arcA and arg genes, the nos gene was also widely distributed among CNS, as it could be detected in 63 of the 86 strains tested (Table 1), based on the primer pairs used (Table 2). However, it was not found in any of the strains investigated belonging to the species Staphylococcus arlettae, Staphylococcus auricularis, S. capitis, S. cohnii, S. succinus, and S. sciuri. Of specific interest was the nos gene found in S. haemolyticus G110, the only strain displaying an NOS-like phenotypic behavior (see below). In the same strain, no arg gene was found, but the arcA gene was present. To verify if citrulline production without ornithine could be due to a truncated ADI pathway lacking OTC, a screening for the genetic elements needed for the three subunits of OTC (otc) in this particular strain was also performed. Three PCR amplicons of the expected size were obtained, confirming that the genetic underpinning for OTC was available.
Phenotypic screening for arginine conversion under microaerobic and aerobic conditions.
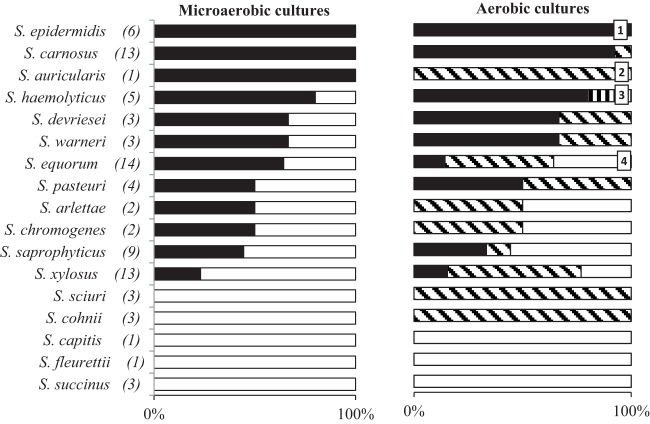
Conversion of arginine into citrulline, ornithine, and/or urea was very common among CNS strains (Table 1; Fig. 1). About 80% of all strains tested were able to at least partially degrade arginine within the investigated time frame of 72 h. However, substantial heterogeneity was found, ranging from no detectable conversion to exhaustion of arginine and resulting in an array of concentrations of citrulline, ornithine, and/or urea. Interspecies heterogeneity was apparent, but variability was situated on strain level as well. The latter was, for instance, the case within the S. saprophyticus species, where both strong conversion and failure to convert arginine were found among the strains investigated. Whereas S. saprophyticus FPP1 was an avid convertor, arginine conversion by strain FPS1 was much weaker, and a majority of five out of nine strains showed no arginine conversion at all.
FIG 1.
Distribution of the phenotypic patterns of arginine conversion by 86 coagulase-negative staphylococci in MSM-A after 72 h of incubation. 1, arginine deiminase (ADI) activity; 2, arginase (ARG) activity; 3, nitric oxide synthase (NOS) activity; 4, none.
Besides differences in the extent of arginine conversion, the mechanisms involved also displayed heterogeneity (Fig. 1). Under microaerobic conditions, all arginine-converting strains (57% of the total) displayed ADI activity, which was indicated by the conversion of arginine into ornithine and, in several cases, some extracellular citrulline (Table 1). Under aerobic conditions, however, the number of strains displaying ADI activity fell to 42%. Also, 34% of the strains exerted arginase activity, leading to equimolar combinations of ornithine and urea. NOS-like activity, indicated by the production of citrulline (9 mM) without ornithine under aerobic conditions, was also found, albeit only for one strain, i.e., S. haemolyticus G110 (Table 1). To confirm that the latter was not due to an incomplete ADI pathway due to a dysfunctional OTC, the production levels of ammonia were compared between the different investigated strains of the species S. haemolyticus. Increases in ammonia concentrations were found after 72 h of incubation for all strains displaying ADI activity, ending up with 19.78 ± 2.46, 22.66 ± 0.06, 22.66 ± 0.59, and 23.13 ± 1.03 mM ammonia for S. haemolyticus 12s24-4, SB1-6, αs1-1, and G191, respectively. In contrast, the ammonia level for S. haemolyticus G110 (3.93 ± 2.52 mM) was in the order of magnitude of the background ammonia concentration in the medium (3.00 ± 0.06 mM), indicating the absence of ADI activity.
Some indicative patterns of phenotypic idiosyncrasy were found at the species level, despite a few exceptions (Fig. 1, Table 1). Strains of S. carnosus and S. epidermidis, for instance, generally displayed strong ADI activity, independently of whether aeration was applied. In contrast, ADI activity was absent or uncommon in strains of, for instance, S. cohnii, S. sciuri, S. succinus, and S. xylosus. For some species, such as S. equorum, several strains lost their ability of ADI activity or shifted to arginase activity under aerobic conditions. Other strains that did not display any arginine conversion microaerobically displayed arginase activity under aerobic conditions, as was the case within the groups of S. cohnii and S. sciuri. For the three investigated strains of S. succinus, no arginine conversion could be found regardless of the aerobicity.
In general, no clear correlation between the origin of the CNS strains and the type of arginine-converting mechanisms was present. Still, ADI activity was found in 71% of the strains from meat origin (fermented sausage and starter cultures) and only in 39% of the strains from dairy-related origin (teat apex skin and milk) under microaerobic conditions. Aerobically, these numbers were 48% and 30%, respectively, whereas arginase activity accounted for 27% and 42% of the strains, respectively.
In contrast to the NOS pathway, for which the gene was often present but activity was limited to one strain, phenotypic and genotypic results concurred rather well for the ADI and arginase pathways. However, some inconsistencies were found (Table 1). In CNS strains where neither the arcA nor the arg gene was found with the primer pairs used, no substantial arginine conversion was seen, except for S. xylosus 2S7-2 and some S. equorum strains (i.e., 3FX-S13, DBX-S21, and DFL-S21). The latter exceptions were indicative of incomplete coverage by the primer pairs used. Coverage of the arcA gene in the genus S. equorum was particularly low, as the gene was never detected, although 79% of the strains from this species displayed ADI activity. In addition, the presence of the arcA or arg gene was not always a marker for phenotypic activity. In S. saprophyticus, for instance, the arg gene was often found, although no arginase activity was retrieved, except for one strain (S. saprophyticus FPS1). As another example, S. sciuri G160 was positive for the arcA gene but did not display ADI activity, suggesting a problem with primer specificity or a dysfunctional ADI pathway.
Overall, to verify that the above-mentioned discrepancies were not due to the amplification of unrelated gene fragments, a wide selection of PCR products were sequenced and compared to the NCBI nonredundant protein sequence database. Results indicated that the PCR products corresponded to the targeted genes with high identities (Table 1).
Conversion kinetics of arginine by Staphylococcus haemolyticus G110.
When S. haemolyticus G110 was grown in MSM-A at a constant pH of 5.8 (Table 3), maximum cell concentrations were obtained after 24 h under aerobic and microaerobic conditions, corresponding with 9.1 and 7.3 log CFU ml−1, respectively, decreasing to 5.5 log CFU ml−1 after 5 days for both conditions.
TABLE 3.
Conversion of arginine (initial concentration of 17 mM) into citrulline and ornithine as well as growth in MSM-A by Staphylococcus haemolyticus G110 under different aeration conditions
| Condition | Conversion parameter | Concn (mean ± SD) or growth by indicated time (h)a |
||||
|---|---|---|---|---|---|---|
| 0 | 12 | 24 | 48 | 120 | ||
| Aerobic | Arg (mM) | 18 ± 2 | 19 ± 1 | 18 ± 1 | 11 ± 0 | 3 ± 1 |
| Orn (mM) | ND | ND | ND | ND | ND | |
| Cit (mM) | ND | ND | 4 ± 1 | 7 ± 0 | 15 ± 1 | |
| Log (CFU ml−1) | 5.6 ± 0.2 | 8.1 ± 0.1 | 9.1 ± 0.1 | 7.4 ± 0.1 | 5.5 ± 0.1 | |
| Microaerobic | Arg (mM) | 18 ± 2 | 19 ± 2 | 17 ± 1 | 17 ± 2 | 8 ± 2 |
| Orn (mM) | ND | ND | ND | ND | 3 ± 2 | |
| Cit (mM) | ND | ND | ND | ND | 2 ± 1 | |
| Log (CFU ml−1) | 6.2 ± 0.1 | 6.1 ± 0.1 | 7.3 ± 0.1 | 7.2 ± 0.1 | 5.5 ± 0.1 | |
ND, not determined.
Under aerobic conditions, arginine conversion started after 24 h. Arginine was never completely depleted during the duration of the experiments, leading to a residual concentration of 3 mM. Ornithine was never found, but citrulline production started upon arginine decrease and reached a level of 15 mM at the end of the experiments. Under microaerobic conditions, arginine levels remained elevated, and these levels did not decrease below 8 mM. Minor concentrations of ornithine (3 mM) and citrulline (2 mM) were found in the medium after 120 h. This suggests that a certain amount of ADI activity was present, provided that enough time was given for arginine metabolism.
Additional screening for phenotypic NOS activity with Staphylococcus carnosus strains under different conditions.
No shifts in arginine conversion patterns were found under any of the conditions tested for the investigated nos-positive S. carnosus strains, compared to the initial screening (data not shown). Although pH and temperature slightly interfered with the overall kinetics and extent of arginine conversion through effects on bacterial growth, mixtures of citrulline and ornithine were obtained in all cases. ADI pathway activity also took place when other components (ions, H4B, hemin, myoglobin extract, and methanol) were added to the MSM-A, as well as when NaCl was omitted. Likewise, ADI pathway activity was found when the strains were grown on arginine-containing MSM-A agar, even when supplemented with myoglobin extract.
DISCUSSION
Certain ecosystems, such as mammal skin and spontaneously fermented meats, may contain a variety of CNS species (1, 2). The selection for specific staphylococcal communities depends on differences in adaptive behavior of the naturally occurring CNS strains. The capacity to utilize specific substrates other than carbohydrates may partially explain differences in competitiveness between CNS species, possibly even on the strain level. Arginine forms an interesting candidate substrate, due to its relative prevalence in meat and its potential to yield ATP and improve acid stress tolerance through the ADI pathway (9, 41). From the present study, it is clear that ADI activity is very common in CNS, but not in all species, and even displays heterogeneity at the strain level. Although not all-explanatory, a functional ADI pathway may nevertheless represent a competitive benefit to certain CNS species. This may, for instance, be the case in acidified meat matrices, as suggested for S. carnosus (9). In addition to the native arc operon, competitiveness can also be affected by the presence of a second gene cluster encoding a complete ADI pathway, such as the arginine catabolic mobile element (ACME). The presence of an ACME has been observed among CNS, including S. epidermidis, S. haemolyticus, S. capitis, S. xylosus, and S. chromogenes, but is mostly restricted to clinical isolates (9, 42–45). In this context, it is important to notice that biofilm formation by S. epidermidis may benefit from the ammonia released by ACME-derived ADI activity (41).
Besides its dependency on the presence of an arc operon, consumption of arginine by CNS also depends on the environmental conditions, in particular the atmospheric status. As demonstrated in the present study, aerobicity often causes a shift of ADI activity toward arginase activity, which could not be found under microaerobic conditions. Such effect has been described for other bacteria, too, including B. licheniformis (18). As a consequence, the arginase pathway is probably less relevant in fermented sausages, where oxygen levels are low, especially toward the end of the fermentation and in the core of sausages with a large caliber. Data from the present study suggest that arginase occurs relatively more in isolates from a dairy environment, whereas ADI seems to be more abundant in strains from a fermented meat background. Nevertheless, some meat-associated CNS possess arginase activity while lacking ADI machinery, as found for several sausage isolates of S. xylosus and S. equorum in the present study. It has yet to be established whether this arginase potential is obsolete or not during meat fermentation, for instance, through gene expression studies. In addition, the relevance of arginase for skin-associated CNS should be further investigated, as this may have important clinical implications. Some CNS are not only important opportunistic pathogens in humans; they may also affect mastitis in dairy herds (1, 6, 36). It has been suggested that the arginase activity of Helicobacter pylori (46) and Bacillus anthracis (47) plays an in vivo role in host-pathogen interactions, via inhibition of host nitric oxide production and T cell proliferation (48). A second role could be to regulate levels of arginine and ornithine within the cell for protein synthesis and polyamine formation (19, 49, 50).
An alternative arginine conversion mechanism is provided by the NOS pathway, also requiring oxygen. The biological role of the NOS pathway in bacteria and its dependency on environmental conditions are yet to be clarified (51). The available hypotheses state that the NOS enzyme may protect bacteria against oxidative stress (52, 53) and antimicrobials, such as aminoglycosides (54, 55). Theoretically, arginine can also be converted into citrulline without ornithine formation through argininosuccinate lyase (ASL) and synthase (ASS) activities (56, 57), although these enzymes have not been described in staphylococci and are generally described as favored toward arginine formation rather than conversion (56). Despite its unclear ecological role, NOS activity by CNS strains may have a potential industrial application as a natural alternative to nitrite for color formation in fermented meats. This is of concern not only because of the demand for clean-label food products but also because of the suspected contribution of cured meats to colon cancer development (58, 59). Until present, attempts to find NOS-producing starter cultures for meat fermentation have not been very persuasive. The use of NOS-positive lactic acid bacteria does not seem likely, despite preliminary reports (21, 27–29, 60, 61); only two of the 57 finished genomes of Lactobacillus species report a flavodoxin/nitric oxide synthase protein (16). In meat-associated CNS, NOS activity has not been convincingly confirmed either, although there have been some preliminary and controversial attempts with respect to S. xylosus (31, 62). First, in vitro generation of NO may be due to the conversion of nitrate traces in the medium (i.e., about 0.6 mg nitrate liter−1 in the applied broth models) rather than to actual NOS activity (63). Second, it remains unclear to which degree the activity of the NOS enzyme can be modulated by environmental conditions and process factors. For instance, NOS enzyme activity obtained from S. aureus ATCC 6538P is optimal at pH 6.5 and 47.5°C and is induced by the presence of methanol in the culture medium (25). Nevertheless, optimal conditions for NOS functionality in meat-related CNS are still to be known, as demonstrated in the present study. A third problem is related to the fact that the NOS pathway is oxygen dependent, so questions remain about its relevance for meat fermentations. As a result, the sausage caliber might play a role, since this will determine overall oxygen gradients. Finally, it needs to be evaluated if candidate strains do not produce enterotoxins or biogenic amines. Overall, NOS-producing starter cultures for meat fermentation would mean a technological breakthrough only when these serious bottlenecks are overcome.
Conclusions.
Coagulase-negative staphylococci possess a toolbox of different arginine-converting mechanisms with potential implications for competitiveness and functionality. The use of the ADI pathway is relatively common among CNS but not universal. Further research is needed to evaluate whether the presence of ADI activity indeed promotes the fitness of CNS during meat fermentations, especially at the strain level, as this may be of importance for starter culture development. The use of the arginase and NOS pathways is restricted to growth under aerobic conditions and thus probably less applicable during meat fermentation. Nevertheless, their relevance for other ecological niches needs to be considered, in particular with respect to clinical contexts in humans and animals. In addition, further research is required to investigate why the genetic potential for NOS is not usually expressed in CNS and to discover which conditions could trigger NOS activity.
ACKNOWLEDGMENTS
We acknowledge the financial support of the Research Council of the Vrije Universiteit Brussel (OZR, SRP, IRP, HOA, and IOF projects), the Hercules Foundation (project UABR 09/004), the Research Foundation-Flanders (project FWOAL632), and Flanders' FOOD (project NITRILOW).
We thank Ralf Rosenstein (Universität Tübingen, Germany) for kindly providing Staphylococcus carnosus strain TM 300, as well as Maarten Janssens, Tom Balzarini, and Wim Borremans (Vrije Universiteit Brussel, Belgium) for their technical assistance. Luc De Vuyst is acknowledged for his critical reading of the manuscript.
Footnotes
Published ahead of print 3 October 2014
REFERENCES
- 1.Vanderhaeghen W, Piepers S, Leroy F, Van Coillie E, Haesebrouck F, De Vliegher S. 2014. Invited review: effect, persistence, and virulence of coagulase-negative Staphylococcus species associated with ruminant udder health. J. Dairy Sci. 97:5275–5293. 10.3168/jds.2013-7775. [DOI] [PubMed] [Google Scholar]
- 2.Ravyts F, De Vuyst L, Leroy F. 2012. Bacterial diversity and functionalities in food fermentations. Eng. Life Sci. 12:356–367. 10.1002/elsc.201100119. [DOI] [Google Scholar]
- 3.Leroy F, Verluyten J, De Vuyst L. 2006. Functional meat starter cultures for improved sausage fermentation. Int. J. Food Microbiol. 106:270–285. 10.1016/j.ijfoodmicro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 4.Janssens M, Myter N, De Vuyst L, Leroy F. 2012. Species diversity and metabolic impact of the microbiota are low in spontaneously acidified Belgian sausages with an added starter culture of Staphylococcus carnosus. Food Microbiol. 29:167–177. 10.1016/j.fm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Janssens M, Myter N, De Vuyst L, Leroy F. 2013. Community dynamics of coagulase-negative staphylococci during spontaneous artisan-type meat fermentations differ between smoking and moulding treatments. Int. J. Food Microbiol. 166:168–175. 10.1016/j.ijfoodmicro.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 6.Braem G, Stijlemans B, Haken W, Vliegher S, Vuyst L, Leroy F. 2014. Antibacterial activities of coagulase-negative staphylococci from bovine teat apex skin and their inhibitory effect on mastitis-related pathogens. J. Appl. Microbiol. 116:1084–1093. 10.1111/jam.12447. [DOI] [PubMed] [Google Scholar]
- 7.Götz F, Bannerman T, Schleifer KH. 2006. The genera Staphylococcus and Macrococcus, p 5–75 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: a handbook on the biology of bacteria, 3rd ed, vol 4 Springer Science and Business Media, LLC, New York, NY. [Google Scholar]
- 8.Rosenstein R, Nerz C, Biswas L, Resch A, Raddatz G, Schuster SC, Götz F. 2009. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 75:811–822. 10.1128/AEM.01982-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janssens M, Van der Mijnsbrugge A, Sánchez Mainar M, Balzarini T, De Vuyst L, Leroy F. 2014. The use of nucleosides and arginine as alternative energy sources by coagulase-negative staphylococci in view of meat fermentation. Food Microbiol. 39:53–60. 10.1016/j.fm.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Rimaux T, Rivière A, Illeghems K, Weckx S, De Vuyst L, Leroy F. 2012. Expression of the arginine deiminase pathway genes in Lactobacillus sakei is strain-dependent and is affected by the environmental pH. Appl. Environ. Microbiol. 78:4874–4883. 10.1128/AEM.07724-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rimaux T, Vrancken G, Pothakos V, Maes D, De Vuyst L, Leroy F. 2011. The kinetics of the arginine deiminase pathway in the meat starter culture Lactobacillus sakei CTC 494 are pH-dependent. Food Microbiol. 28:597–604. 10.1016/j.fm.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 12.Rimaux T, Vrancken G, Vuylsteke B, De Vuyst L, Leroy F. 2011. The pentose moiety of adenosine and inosine is an important energy source for the fermented-meat starter culture Lactobacillus sakei CTC 494. Appl. Environ. Microbiol. 77:6539–6550. 10.1128/AEM.00498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunin R, Glansdorff N, Piérard A, Stalon V. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudhamsu J, Crane BR. 2009. Bacterial nitric oxide synthases: what are they good for? Trends Microbiol. 17:212–218. 10.1016/j.tim.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Zúñiga M, Pérez G, González-Candelas F. 2002. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 25:429–444. 10.1016/S1055-7903(02)00277-4. [DOI] [PubMed] [Google Scholar]
- 16.NCBI Resource Coordinators. 2014. Database resources of the National Center for Biotechnology Information. 42:D7–D17. 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rimaux T, Rivière A, Hebert EM, Mozzi F, Weckx S, De Vuyst L, Leroy F. 2013. A putative transport protein is involved in citrulline excretion and re-uptake during arginine deiminase pathway activity by Lactobacillus sakei. Res. Microbiol. 164:216–225. 10.1016/j.resmic.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Broman K, Lauwers N, Stalon V, Wiame JM. 1978. Oxygen and nitrate in utilization by Bacillus licheniformis of arginase and arginine deiminase routes of arginine catabolism and other factors affecting their syntheses. J. Bacteriol. 135:920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deutch CE. 2011. l-Proline nutrition and catabolism in Staphylococcus saprophyticus. Antonie Van Leeuwenhoek 99:781–793. 10.1007/s10482-011-9552-7. [DOI] [PubMed] [Google Scholar]
- 20.Planchon S, Desvaux M, Chafsey I, Chambon C, Leroy S, Hébraud M, Talon R. 2009. Comparative subproteome analyses of planktonic and sessile Staphylococcus xylosus C2a: new insight in cell physiology of a coagulase-negative staphylococcus in biofilm. J. Proteome Res. 8:1797–1809. 10.1021/pr8004056. [DOI] [PubMed] [Google Scholar]
- 21.Morita H, Yoshikawa H, Sakata R, Nagata Y, Tanaka H. 1997. Synthesis of nitric oxide from the two equivalent guanidino nitrogens of l-arginine by Lactobacillus fermentum. J. Bacteriol. 179:7812–7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YJ, Rosazza JPN. 1995. Purification and characterization of nitric oxide synthase (NOSNoc) from a Nocardia species. J. Bacteriol. 177:5122–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi DW, Oh HY, Hong SY, Han JW, Lee HW. 2000. Identification and characterization of nitric oxide synthase in Salmonella typhimurium. Arch. Pharm. Res. 23:407–412. 10.1007/BF02975456. [DOI] [PubMed] [Google Scholar]
- 24.Pant K, Bilwes AM, Adak S, Stuehr DJ, Crane BR. 2002. Structure of a nitric oxide synthase heme protein from Bacillus subtilis. Biochemistry 41:11071–11079. 10.1021/bi0263715. [DOI] [PubMed] [Google Scholar]
- 25.Choi WS, Chang MS, Han JW, Hong SY, Lee HW. 1997. Identification of nitric oxide synthase in Staphylococcus aureus. Arch. Pharm. Res. 237:554–558. [DOI] [PubMed] [Google Scholar]
- 26.Hong IS, Kim YK, Choi WS, Seo DW, Yoon JW, Han JW, Lee HY, Lee HW. 2003. Purification and characterization of nitric oxide synthase from Staphylococcus aureus. FEMS Microbiol. Lett. 222:177–182. 10.1016/S0378-1097(03)00254-4. [DOI] [PubMed] [Google Scholar]
- 27.Arihara K, Kushida H, Kondo Y, Itoh M, Luchansky JB, Cassens RG. 1993. Conversion of metmyoglobin to bright red myoglobin derivatives by Chromobacterium violaceum, Kurthia sp. and Lactobacillus fermentum JCM1173. J. Food Sci. 58:38–42. 10.1111/j.1365-2621.1993.tb03205.x. [DOI] [Google Scholar]
- 28.Møller JKS, Jensen JS, Skibsted LH, Knöchel S. 2003. Microbial formation of nitrite-cured pigment, nitrosylmyoglobin, from metmyoglobin in model systems and smoked fermented sausages by Lactobacillus fermentum strains and a commercial starter culture. Eur. Food Res. Technol. 216:463–469. 10.1007/s00217-003-0681-8. [DOI] [Google Scholar]
- 29.Gündoğdu AK, Karahan AG, Ca̧kmakç ML. 2006. Production of nitric oxide (NO) by lactic acid bacteria isolated from fermented products. Eur. Food Res. Technol. 223:35–38. 10.1007/s00217-005-0097-8. [DOI] [Google Scholar]
- 30.Götterup J, Olsen K, Knöchel S, Tjener K, Stahnke LH, Møller JKS. 2007. Relationship between nitrate/nitrite reductase activities in meat associated staphylococci and nitrosylmyoglobin formation in a cured meat model system. Int. J. Food Microbiol. 120:303–310. 10.1016/j.ijfoodmicro.2007.08.034. [DOI] [PubMed] [Google Scholar]
- 31.Li PJ, Kong BH, Chen Q, Zheng DM, Liu N. 2013. Formation and identification of nitrosylmyoglobin by Staphylococcus xylosus in raw meat batters: a potential solution for nitrite substitution in meat products. Meat Sci. 93:67–72. 10.1016/j.meatsci.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Montel MC, Talon R, Cantonnet M, Fournaud J. 1992. Identification of Staphylococcus from French dry sausage. Lett. Appl. Microbiol. 15:73–77. 10.1111/j.1472-765X.1992.tb00728.x. [DOI] [Google Scholar]
- 33.Wagner E, Doskar J, Götz F. 1998. Physical and genetic map of the genome of Staphylococcus carnosus TM300. Microbiology 144:509–517. 10.1099/00221287-144-2-509. [DOI] [PubMed] [Google Scholar]
- 34.Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577–1593. 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 35.Ravyts F, Steen L, Goemaere O, Paelinck H, De Vuyst L, Leroy F. 2010. The application of staphylococci with flavour-generating potential is affected by acidification in fermented dry sausages. Food Microbiol. 27:945–954. 10.1016/j.fm.2010.05.030. [DOI] [PubMed] [Google Scholar]
- 36.Braem G, De Vliegher S, Verbist B, Piessens V, Van Coillie E, De Vuyst L, Leroy F. 2013. Unraveling the microbiota of teat apices of clinically healthy lactating dairy cows, with special emphasis on coagulase-negative staphylococci. J. Dairy Sci. 96:1499–1510. 10.3168/jds.2012-5493. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui LZ, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292–7308. 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weckx S, De Rijk P, Van Broeckhoven C, Del-Favero J. 2004. SNPbox: Web-based high-throughput primer design from gene to genome. Nucleic Acids Res. 32:170–172. 10.1093/nar/gkh369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leroy F, De Vuyst L. 2005. Simulation of the effect of sausage ingredients and technology on the functionality of the bacteriocin-producing Lactobacillus sakei CTC 494 strain. Int. J. Food Microbiol. 100:141–152. 10.1016/j.ijfoodmicro.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Vrancken G, Rimaux T, De Vuyst L, Leroy F. 2008. Kinetic analysis of growth and sugar consumption by Lactobacillus fermentum IMDO 130101 reveals adaptation to the acidic sourdough ecosystem. Int. J. Food Microbiol. 128:58–66. 10.1016/j.ijfoodmicro.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Lindgren JK, Thomas VC, Olson ME, Chaudhari SS, Nuxoll AS, Schaeffer CR, Lindgren KE, Jones J, Zimmerman MC, Dunman PM, Bayles KW, Fey PD. 2014. Arginine deiminase in Staphylococcus epidermidis functions to augment biofilm maturation through pH homeostasis. J. Bacteriol. 196:2277–2289. 10.1128/JB.00051-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, Higashi J, Sullam PM, Lin J, Wong KI, King KA, Otto M, Sensabaugh GF, Diep BA. 2009. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One 4:1–9. 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pi BR, Yu MH, Chen YG, Yu YS, Li LJ. 2009. Distribution of the ACME-arcA gene among meticillin-resistant Staphylococcus haemolyticus and identification of a novel ccr allotype in ACME-arcA-positive isolates. J. Med. Microbiol. 58:731–736. 10.1099/jmm.0.007351-0. [DOI] [PubMed] [Google Scholar]
- 44.Barbier F, Lebeaux D, Hernandez D, Delannoy AS, Caro V, Francois P, Schrenzel J, Ruppe E, Gaillard K, Wolff M, Brisse S, Andremont A, Ruimy R. 2011. High prevalence of the arginine catabolic mobile element in carriage isolates of methicillin-resistant Staphylococcus epidermidis. J. Antimicrob. Chemother. 66:29–36. 10.1093/jac/dkq410. [DOI] [PubMed] [Google Scholar]
- 45.Shore AC, Rossney AS, Brennan OM, Kinnevey PM, Humphreys H, Sullivan DJ, Goering RV, Ehricht R, Monecke S, Coleman DC. 2011. Characterization of a novel arginine catabolic mobile element (ACME) and staphylococcal chromosomal cassette mec composite island with significant homology to Staphylococcus epidermidis ACME type II in methicillin-resistant Staphylococcus aureus genotype ST22-MRSA-IV. Antimicrob. Agents Chemother. 55:1896–1905. 10.1128/AAC.01756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGee DJ, Zabaleta J, Viator RJ, Testerman TL, Ochoa AC, Mendz GL. 2004. Purification and characterization of Helicobacter pylori arginase, RocF: unique features among the arginase superfamily. Eur. J. Biochem. 271:1952–1962. 10.1111/j.1432-1033.2004.04105.x. [DOI] [PubMed] [Google Scholar]
- 47.Viator RJ, Rest RF, Hildebrandt E, McGee DJ. 2008. Characterization of Bacillus anthracis arginase: effects of pH, temperature, and cell viability on metal preference. BMC Biochem. 9:15. 10.1186/1471-2091-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zabaleta J, McGee DJ, Zea AH, Hernandez CP, Rodriguez PC, Sierra RA, Correa P, Ochoa AC. 2004. Helicobacter pylori arginase inhibits T cell proliferation and reduces the expression of the TCR ζ-chain (CD3ζ). J. Immunol. 173:586–593. 10.4049/jimmunol.173.1.586. [DOI] [PubMed] [Google Scholar]
- 49.Townsend DE, Kaenjak A, Jayaswal RK, Wilkinson BJ. 1996. Proline is biosynthesized from arginine in Staphylococcus aureus. Microbiology 142:1491–1497. 10.1099/13500872-142-6-1491. [DOI] [PubMed] [Google Scholar]
- 50.Bewley MC, Jeffrey PD, Patchett ML, Kanyo ZF, Baker EN. 1999. Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily. Struct. Fold Des. 7:435–448. 10.1016/S0969-2126(99)80056-2. [DOI] [PubMed] [Google Scholar]
- 51.Crane BR, Sudhamsu J, Patel BA. 2010. Bacterial nitric oxide synthases. Annu. Rev. Biochem. 79:445–470. 10.1146/annurev-biochem-062608-103436. [DOI] [PubMed] [Google Scholar]
- 52.Gusarov I, Nudler E. 2005. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc. Natl. Acad. Sci. U. S. A. 102:13855–13860. 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel BA, Crane BR. 2010. When it comes to antibiotics, bacteria show some NO-how. J. Mol. Cell Biol. 2:234–236. 10.1093/jmcb/mjp044. [DOI] [PubMed] [Google Scholar]
- 54.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. 2009. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325:1380–1384. 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCollister BD, Hoffman M, Husain M, Vázquez-Torres A. 2011. Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob. Agents Chemother. 55:2189–2196. 10.1128/AAC.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu G, Morris SM. 1998. Arginine metabolism: nitric oxide and beyond. Biochem. J. 336:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Husson A, Brasse-Lagnel C, Fairand A, Renouf S, Lavoinne A. 2003. Argininosuccinate synthetase from the urea cycle to the citrulline-NO cycle. Eur. J. Biochem. 270:1887–1899. 10.1046/j.1432-1033.2003.03559.x. [DOI] [PubMed] [Google Scholar]
- 58.Cross AJ, Ferrucci LM, Risch A, Graubard BI, Ward MH, Park Y, Hollenbeck AR, Schatzkin A, Sinha R. 2010. A large prospective study of meat consumption and colorectal cancer risk: an investigation of potential mechanisms underlying this association. Cancer Res. 70:2406–2414. 10.1158/0008-5472.CAN-09-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sindelar JJ, Milkowski AL. 2012. Human safety controversies surrounding nitrate and nitrite in the diet. Nitric Oxide 26:259–266. 10.1016/j.niox.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 60.Adawi D, Kasravi FB, Molin G, Jeppsson B. 1997. Effect of Lactobacillus supplementation with and without arginine on liver damage and bacterial translocation in an acute liver injury model in the rat. Hepatology 25:642–647. 10.1002/hep.510250325. [DOI] [PubMed] [Google Scholar]
- 61.Yarullina DR, Il'inskaia ON, Aganov AV, Silkin NI, Zverev DG. 2006. Alternative pathways of nitric oxide formation in lactobacilli: evidence for nitric oxide synthase activity by EPR. Microbiology 75:731–736. 10.1134/S0026261706060026. [DOI] [PubMed] [Google Scholar]
- 62.Morita H, Sakata R, Nagata Y. 1998. Nitric oxide complex of iron(II) myoglobin converted from metmyoglobin by Staphylococcus xylosus. J. Food Sci. 63:352–355. [Google Scholar]
- 63.Xu J, Verstraete W. 2001. Evaluation of nitric oxide production by lactobacilli. Appl. Microbiol. Biotechnol. 56:504–507. 10.1007/s002530100616. [DOI] [PubMed] [Google Scholar]