Abstract
Francisella tularensis is a highly infectious bacterium that causes the potentially lethal disease tularemia. This extremely virulent bacterium is able to replicate in the cytosolic compartments of infected macrophages. To invade macrophages and to cope with their intracellular environment, Francisella requires multiple virulence factors, which are still being identified. Proteins containing tetratricopeptide repeat (TPR)-like domains seem to be promising targets to investigate, since these proteins have been reported to be directly involved in virulence-associated functions of bacterial pathogens. Here, we studied the role of the FTS_0201, FTS_0778, and FTS_1680 genes, which encode putative TPR-like proteins in Francisella tularensis subsp. holarctica FSC200. Mutants defective in protein expression were prepared by TargeTron insertion mutagenesis. We found that the locus FTS_1680 and its ortholog FTT_0166c in the highly virulent Francisella tularensis type A strain SchuS4 are required for proper intracellular replication, full virulence in mice, and heat stress tolerance. Additionally, the FTS_1680-encoded protein was identified as a membrane-associated protein required for full cytopathogenicity in macrophages. Our study thus identifies FTS_1680/FTT_0166c as a new virulence factor in Francisella tularensis.
INTRODUCTION
Francisella tularensis is a facultative intracellular bacterium that causes the potentially lethal disease tularemia. F. tularensis can infect a wide range of animal species, including humans. F. tularensis can be transmitted to humans through a number of routes; the most common is via the bite of an infected insect or another arthropod vector. The spectrum of human illness can range from the ulceroglandular form to the more serious pneumonic or typhoidal form of tularemia (1). The risk of serious human infection is associated mainly with two subspecies, the highly virulent F. tularensis subsp. tularensis (type A) and the less virulent F. tularensis subsp. holarctica (type B). Documented use of F. tularensis as a biological weapon in World War II and concerns over construction of antibiotic-resistant F. tularensis strains have led to an enhanced interest in unveiling mechanisms of virulence which may serve as promising targets for the development of treatments or effective prophylaxis in case of its misuse (2).
F. tularensis infects multiple cell types, including nonphagocytic and phagocytic cells (1, 2). Following entry into phagocytic host cells, Francisella is found in phagosomes that are characterized by the presence of early (EEA-1) and late (LAMP-1) endosomal markers (3). However, the bacterium subsequently modulates the fusion of the Francisella-containing phagosome with degradative lysosomes and escapes into the cytosol, where it replicates. Within 24 to 48 h after infection, Francisella reenters LAMP-1-positive endocytic compartments referred to as Francisella-containing autophagic vacuoles (3, 4).
Previous studies have identified a wide array of genes required by F. tularensis for adaptation to intracellular environments and/or evasion of phagocytic cell defense mechanisms. These include genes located in a 34-kb Francisella pathogenicity island (FPI), genes responsible for the presence of a noninflammatory lipopolysaccharide, protective capsule, and siderophores, and genes encoding proteins involved in resistance to various stress conditions (5–12).
Of the other candidates, tetratricopeptide repeat (TPR)- or SEL1-like (SLR) structural motif-containing proteins seem to be promising targets for more detailed studies. The TPR and SLR motifs share similar α-helical conformations but differ in consensus sequence, length, and superhelical topology (13). Despite this distinction, both motifs represent elegant modules for the assembly of various multiprotein complexes via mediating protein-protein interactions (13, 14); thus, such proteins are often involved in numerous cellular processes in both eukaryotic and prokaryotic organisms (14, 15). Several F. tularensis proteins with predicted TPR/SLR motifs have already been shown to be required for the fully expressed virulent phenotype. These proteins include the hypothetical SLR-containing protein DipA, the putative TPR-containing protein FTT_1244c from F. tularensis subsp. tularensis SchuS4 (4), and the putative TPR-containing proteins PilF and FTL_0205 from Francisella subsp. holarctica LVS (16, 17).
The goal of this study was to determine whether the three putative TPR-like proteins FTS_0201, FTS_0778, and FTS_1680 play a role in Francisella tularensis subsp. holarctica FSC200 (FSC200) virulence. Using functional genomics, in vitro and in vivo characterization, and proteomic studies, we discovered that the product of the FTS_1680 gene is a membrane-associated protein that contributes to the virulence mechanisms of Francisella. We found that FTS_1680 was required for intracellular replication, full virulence in mice, and heat stress tolerance. Additionally, we also tested whether inactivation of FTT_0166c, the ortholog of FTS_1680 in F. tularensis subsp. tularensis SchuS4, would result in a similar attenuated bacterial phenotype. We found that inactivation of FTT_0166c protein expression prolonged survival of mice and significantly decreased intracellular microbial replication within macrophages.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. F. tularensis subsp. holarctica FSC200 (18), acquired from the Francisella strain collection (FSC), was kindly provided by Åke Forsberg, Swedish Defense Research Agency, Umeå, Sweden. Wild-type FSC200 and the derived mutant strains were grown on McLeod agar supplemented with bovine hemoglobin (Becton Dickinson, Cockeysville, MD, USA) and IsoVitaleX (Becton Dickinson, Cockeysville, MD, USA) at 37°C with 5% CO2 or in liquid Chamberlain's medium (19) at 30°C, 37°C, or 42°C. Wild-type and mutant F. tularensis subsp. tularensis SchuS4 were grown on chocolate agar or in liquid brain heart infusion broth supplemented with 1% IsoVitaleX (Becton Dickinson, Cockeysville, MD, USA) at 30°C and 37°C. Escherichia coli strains were cultivated on Luria-Bertani (LB) agar and in LB broth at either 30°C or 37°C. When necessary, penicillin (100 U/ml), ampicillin (100 μg/ml), kanamycin (50 μg/ml for E. coli and 20 μg/ml for F. tularensis) or 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was used.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| α-Select Gold Efficiency | F− deoR endA1 recA1 relA1 gyrA96 hsdR17 (rK− mK+) supE44 thi-1 phoA Δ(lacZYA argF)U169 ϕ80lacZΔM15 λ− | Bioline |
| DHM1 | F′ cya-854 recA1 endA1 gyrA96 (Nalr) thi-1 hsdR17 spoT1 rfbD1 glnV44(AS) | BACTH system kit; Euromedex (46) |
| Francisella strains | ||
| FSC200 | F. tularensis subsp. holarctica, clinical isolate | Francisella strain collection (18) |
| SchuS4 | F. tularensis subsp. tularensis | USAMRIID strain collection |
| inFTS_0201 | FSC200 inFTS_0201 | This study |
| inFTS_0778 | FSC200 inFTS_0778 | This study |
| inFTS_1680 | FSC200 inFTS_1680 | This study |
| inFTS_1680 complemented | inFTS_1680 with pKK289FTS_1680 | This study |
| inFTT_0166c | SchuS4 inFTT_0166c | This study |
| Plasmids | ||
| pCR4-TOPO | Cloning vector, Ampr Kmr | Invitrogen |
| pKEK1140 | F. tularensis-specific TargeTron plasmid | 20 |
| pKEK1140/in0201 | pKEK1140 with FTS_0201 inserted | This study |
| pKEK1140/in0778 | pKEK1140 with FTS_0778 inserted | This study |
| pKEK1140/in1680 | pKEK1140 with FTS_1680 inserted | This study |
| pKEK1140/in0166c | pKEK1140 with FTT_0166c inserted | This study |
| pKK289gfp | Ft ori, p15a ori, Kmr, groES promoter | 21 |
| pKK289FTS_1680 | pKK289 with intact FTS_1680 | This study |
| pKNT25 | BACTH system plasmid | 46 |
| pUT18 | BACTH system plasmid | 47 |
| pUT18/1680 | pUT18 with intact FTS_1680 | This study |
| pKNT25/1670 | pKNT25 with intact FTS_1670 | This study |
| pKNT25/1167 | pKNT25 with intact FTS_1167 | This study |
| pKNT25/1166 | pKNT25 with intact FTS_1166 | This study |
| pKNT25/0277 | pKNT25 with intact FTS_0277 | This study |
| pKNT25/0263 | pKNT25 with intact FTS_0263 | This study |
TargeTron insertional mutagenesis.
The TargeTron gene knockout system was employed to mutate the FTS_0201, FTS_0784, FTS_1680, and FTT_0166c genes as previously described (20). Target sites for insertion and retargeting PCR primers (Table 2) were generated using the TargeTron gene knockout system (Sigma-Aldrich, Steinheim, Germany), and the resulting PCR product was digested (HindIII-BsrGI) and cloned into the Francisella targeting vector pKEK1140 (generously provided by Karl Klose, University of Texas at San Antonio, San Antonio, TX) (20). Prepared constructs were introduced into the FSC200 or SchuS4 strain by electroporation. The presence of the TargeTron insertion was determined using an intron-specific EBS universal primer combined with a gene-specific primer. Intron insertion of the targeted gene was determined using gene-specific primers that amplified across the insertion site. For the FSC200 mutants, positive clones were incubated in Chamberlain's medium at 37°C overnight and then streaked on McLeod agar (without kanamycin), with incubation at 37°C to remove the TargeTron temperature-sensitive plasmid. For the SchuS4 mutants, positive clones were incubated in brain heart infusion broth supplemented with 1% IsoVitaleX (Becton Dickinson, Cockeysville, MD, USA) overnight at 37°C and subsequently plated onto chocolate agar and maintained at 37°C. The insertion mutants were confirmed using PCR with the gene-specific primers.
TABLE 2.
Primers used in this study
Functional complementation.
To create the FTS_1680 complemented strain, functional complementation was performed in trans. A DNA fragment carrying the wild-type FTS_1680 gene was PCR amplified using FSC200 genomic DNA as a template, employing primers 1680_pKK289_F and 1680_pKK289_R (Table 2). The final PCR product was sequenced and then cloned downstream of the GroES promoter by replacing the green fluorescence protein-encoding gene in the shuttle vector pKK289gfp (21). The resulting construct, designated pKK289FTS_1680, was introduced into the mutant strain inFTS_1680 by electroporation.
Macrophage culture and infection.
Bone marrow cells were isolated from femurs of 6- to 10-week-old BALB/c female mice and differentiated to bone marrow macrophages (BMMs) in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum, 10% L929 cell-conditioned medium (as a source of macrophage colony-stimulating factor [M-CSF]), 50 μg/ml streptomycin, and 50 U/ml penicillin for 6 to 7 days (22). Briefly, the BMMs were seeded at a concentration of 5 × 105 cells per well in 24-well tissue culture plates. F. tularensis bacteria were diluted into cell culture medium and used for infection of BMMs at a 50:1 (bacteria/cell) multiplicity of infection (MOI). Actual infection doses were determined by plating serial dilutions of the culture inoculum. Tissue culture plates were centrifuged at 400 × g for 5 min to start the infection and then incubated at 37°C for 30 min. Extracellular bacteria were then killed by gentamicin treatment (5 μg/ml) for 30 min. At 1, 6, 12, and 24 h postinfection, the infected BMMs were washed and then lysed with 0.1% sodium deoxycholate. Lysates were serially diluted and plated on McLeod agar to determine the number of CFU in each well.
The murine monocyte-macrophage cell line J774.2 (ECACC reference no. 85011428) was cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL) at 37°C with 5% CO2. Cells were seeded into 24-well plates at a density of approximately 3 × 105 cells/well. Cells were incubated overnight at 37°C with 5% CO2 and subsequently infected at an MOI of 500:1 with FSC200, inFTS_0201, inFTS_0784, or inFTS_1680. Infection and proliferation were performed as described above.
For the SchuS4 macrophage proliferation assay, J774.1 cells were infected at an MOI of 100:1 with SchuS4 or inFTT_0166c. After 2 h, cells were washed once with phosphate-buffered saline (PBS), and fresh medium containing 25 μg/ml gentamicin was added to kill the extracellular bacteria. Cells were harvested at 4 and 24 h postinfection, and the number of intracellular bacteria was enumerated by serial dilution and plating.
Mouse virulence studies.
For survival studies, groups of five 6- to 8-week-old female BALB/c mice were infected subcutaneously with 3 × 102 CFU/mouse of the wild-type FSC200 strain, the inFTS_1680 mutant, the inFTS_1680 complemented strain, the inFTS_0201 mutant, or the inFTS_0778 mutant. An additional group of mice (n = 5) was infected subcutaneously with 3 × 106 CFU/mouse of the inFTS_1680 mutant strain. Intraperitoneal challenges were carried out only with the inFTS_1680 mutant (using doses of 3 × 102 or 3 × 106 CFU/mouse), the wild-type FSC200 strain (3 × 102 CFU/mouse), and the inFTS_1680 complemented strain (3 × 102 CFU/mouse). Control groups of mice were inoculated with sterile saline only. The actual inoculation doses were confirmed by viable plate counting. Following infection, mice were observed for signs of illness or death daily for a total of 21 days.
Growth kinetic studies of the pathogen in mouse organs were performed using groups of three BALB/c mice infected intraperitoneally or subcutaneously with the wild-type strain FSC200, the inFTS_1680 complemented strain, or the inFTS_1680 mutant using a dose of 102 CFU/mouse of each strain. At the indicated times postinfection, livers, spleens, and lung tissues were recovered and homogenized in PBS, and the total bacterial burdens in each organ were determined by dilution plating onto McLeod agar plates.
For survival studies with the inFTT0166c mutant, groups of 6- to 8-week-old BALB/c mice (n = 10) were infected subcutaneously or intranasally with increasing doses of wild-type SchuS4 or inFTT0166c mutant bacteria. Bacteria for the infection were taken from a freshly grown chocolate agar plate incubated overnight at 37°C. The bacteria were resuspended in sterile PBS to an optical density at 600 nm (OD600) of 0.25 (approximately 1.5 × 109 CFU/ml) and diluted to the desired dose. Inoculum counts were verified by serial plating. Infected animals were monitored several times each day for signs of illness or death.
Colocalization of F. tularensis with LAMP-1 in BMMs.
To examine the ability of the inFTS_1680 mutant to escape from the host cell phagosomes (23), BMMs were infected as described above. At 1 h and 6 h postinfection, the BMMs were fixed with 3.8% paraformaldehyde for 30 min, followed by neutralization with 50 mM NH4Cl. After washing with PBS, macrophage membranes were permeabilized with 0.2% Triton X-100 for 15 min. For bacterial detection, purified rabbit polyclonal anti-F. tularensis serum was used at a concentration of 1:3,000 followed by detection with Alexa Fluor 488-labeled goat anti-rabbit IgG (Invitrogen, Molecular Probes, Eugene, OR, USA) at a dilution of 1:500. For the colocalization studies, BMMs were then labeled using rat monoclonal anti-mouse LAMP-1 antibody (1D4B; Santa Cruz Biotechnology, Santa Cruz, CA) at a concentration of 1:100, followed by detection with Alexa Fluor 594-labeled donkey anti-rat IgG (Invitrogen, Molecular Probes, Eugene, OR, USA) at a 1:500 dilution. Colocalization of bacteria with LAMP-1 was analyzed with a Nikon TE2000 confocal laser scanning microscope equipped with NIS Elements AR software.
Macrophage cytotoxicity assay.
For cytotoxicity experiments, BMMs were seeded in 96-well tissue culture plates at a concentration of 2 × 104 cells/well and allowed to adhere overnight at 37°C with 5% CO2. The next day, the BMMs were infected with bacterial cell suspensions at an MOI of 50:1. At 2 h, 24 h, and 48 h postinfection, the supernatant was collected and assayed for the presence of lactate dehydrogenase (LDH) according to manufacturer's instructions (cytotoxicity detection kit; Roche Diagnostics, Germany). Samples were measured with a Paradigm microplate reader (Beckman Coulter) at an absorbance of 490 nm. As a positive control (representing 100% cell lysis), uninfected BMMs were lysed with 0.1% natrium deoxycholate. Sample absorbance values were expressed as a percentage of the positive-control value.
Stress survival assay.
F. tularensis strains were grown overnight at 37°C in 2.5 ml of Chamberlain's medium supplemented with the appropriate antibiotic when applicable. The cultures were diluted in fresh Chamberlain's medium to an OD600 of 0.1. Five aliquots of each strain (0.25 ml each) were transferred into 96-well plates and subjected to one of the following stress conditions: pH 4.0, 4% NaCl, or iron depletion. Samples were then incubated at 37°C or 42°C (in the case of heat stress) for 24 h. Bacterial growth was determined by measuring the OD600 every 10 min for 24 h. CFU were calculated at the end of the experiment to verify the OD600 measurement. For oxidative stress experiments, stationary-phase bacteria from F. tularensis strain FSC200 or inFTS_1680 were diluted 1:10 into fresh Chamberlain's medium and exposed to 0.03% H2O2. Samples were harvested at 0, 20, and 40 min postexposure and viable bacteria enumerated by serial dilution on McLeod agar plates (5, 6).
For heat stress experiments with the inFTT_0166c mutant, stationary-phase bacteria from F. tularensis strain SchuS4 or inFTT0166c were diluted 1:10 into fresh Chamberlain's medium and exposed to heat stress (42°C) for 50 and 100 min. Viable bacteria were enumerated by serial dilution at the start of the experiment and at the indicated time points.
BACTH assay.
We performed the bacterial adenylate cyclase two-hybrid (BACTH) assay according to the manufacturer's instructions (BACTH system kit; Euromedex [reference no. EUK001]). Briefly, FTS_1680 protein was fused with the T18 adenylate cyclase subunit (N-terminal fusion to T18). Targeted proteins GroEL (FTS_1670), DnaK (FTS_1167), DnaJ2 (FTS_0277), GrpE (FTS_1166), and HtpG (FTS_0263) were fused to the T25 adenylate cyclase subunit (in-frame fusion at the N-terminal end of T25). The plasmids and primers used in this assay are listed in Tables 1 and 2, respectively. For each assay, E. coli DHM1 chemocompetent cells were transformed with 200 ng of each of the plasmids carrying the T25 and T18 fusions. The bacteria were then spread on LB plates containing 100 μg/ml ampicillin and 50 μg/ml kanamycin and incubated for 2 days at 30°C. Several clones were inoculated into 3 ml of LB containing 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 0.5 mM IPTG and incubated overnight at 30°C. The next day, 2 μl of each culture was dropped on MacConkey/maltose plates containing appropriate antibiotics and IPTG. The plates were then incubated for several days until a red coloration appeared. The empty pUT18 and pKNT25 plasmids were used as negative controls. For a positive controls, pUT18 and pKNT25 plasmids carrying the iglA and iglB genes, respectively, were used. The IglA-IglB protein/protein interaction has been previously demonstrated in F. tularensis (24).
Triton X-114 partitioning.
Cell lysis of wild-type FSC200 and inFTS_1680 mutant bacteria was performed using a French pressure cell (16,000 lb/in2). Fractions enriched in membrane proteins were collected by ultracentrifugation of the whole-cell lysate at 115,000 × g for 1 h at 4°C. The supernatant was discarded; the membrane pellet was resuspended in PBS and recollected by ultracentrifugation. The final membrane protein-containing pellet was resuspended in ice-cold PTX buffer (PBS containing 350 mM NaCl, 2% Triton X-114, and EDTA-free protease inhibitors) and incubated at 4°C with end-over-end rotation. After 1 h, the sample was centrifuged (12,000 rpm, 4°C, 30 min), and the supernatant was kept at 37°C for 10 min to initiate phase partitioning. Following centrifugation (14,000 rpm, 24°C, 10 min), the upper aqueous layer was removed. An equal volume of PBS containing 350 mM NaCl was added to the organic phase. The phase separation was repeated twice. The resulting detergent phase was resuspended in PBS, and the protein content was quantified by a bicinchoninic acid assay (Sigma-Aldrich, St. Louis, MO, USA).
2D gel electrophoresis.
Detergent-phase proteins were repeatedly precipitated with cold acetone prior to two-dimensional (2D) electrophoresis. The protein precipitate was resolubilized in a rehydration buffer containing 1% (wt/vol) ASB-14 surfactant. Isoelectric focusing, reduction, alkylation, and SDS-PAGE were performed as described previously (25).
MS analysis and protein identification.
The colloidal blue-stained protein spots were excised from 2D gels and subjected to an in-gel tryptic digestion according to a recently described procedure (25). The digestion was stopped by acidifying the samples to a pH of 2 to 3 with trifluoroacetic acid. The in-gel-digested proteins were analyzed on a 4800 matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) mass spectrometer (MS) (AB Sciex, Foster City, CA). Acquisition of MS spectral data was performed in a mass range from m/z 800 to 4,000. Internal calibration of mass spectra was conducted utilizing tryptic autolytic peptides. Tandem mass spectra of the six most intense precursor ions having a minimum S/N ratio of 100 were acquired using a 1-kV MS/MS reflector in positive-ion mode. Data acquisition and processing were carried out using 4000 Series Explorer software v3.5 (AB Sciex). The mass spectral data obtained were analyzed using the F. tularensis OSU18 protein sequences database (NC_008369.1) using GPS Explorer Software v3.6 (AB Sciex) with the Mascot search algorithm v2.2. Trypsin was selected as the proteolytic enzyme, and one missed cleavage was allowed. Carbamidomethylation of cysteine residues and methionine oxidation were set as variable and fixed modifications, respectively. The mass tolerances of the precursor and fragment ions were 100 ppm and 0.25 Da, respectively. Proteins were considered identified with confidence when the GPS protein score confidence interval (%) was equal to 100% and a minimum of two peptide sequences per protein were identified.
Ethics statement.
All mouse experiments were performed in accordance with the guidelines of the Animal Care and Use Ethical Committee of the FMHS, University of Defence, Czech Republic. The research protocol was approved by this ethics committee under project no. 89–3/2013-3696. At USAMRIID, research was conducted under an IACUC-approved protocol in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
Statistical analysis.
For each strain and time in an experiment, the assay was performed in triplicate. Each experiment was independently repeated three times. All values were expressed as mean ± standard deviation (SD) and analyzed for significance using Student's two-tailed t test in the statistical software GraphPad Prism 6. Differences were considered statistically significant at a P value of <0.05. The 50% lethal dose (LD50) determinations were calculated using probit analysis (26). The mean time-to-death (TTD) comparisons were made using two-tailed t tests. Survival curve comparisons were made using Kaplan-Meier survival analysis with log rank comparisons. Survival rates were compared using Fisher's exact test with step-down Bonferroni correction. All statistical analyses were done using SAS software (SAS Institute, Cary, NC).
RESULTS
Selection of the targets for construction of the TargeTron insertion mutants.
To investigate the involvement of TPR-like-containing proteins in FSC200 pathogenesis, several genes encoding proteins from the TPR and SLR families were chosen as targets for inactivation. Selection was performed using the web-based prediction tool TPRpred (27). TPRpred is a profile-sequence comparison method for predicting TPRs and closely related solenoid structural motifs, pentatricopeptide repeats (PPRs), and SEL1-like repeats (SLRs). FTS_1680 (YP_007012402.1) contained five predicted TPRs at positions 24 to 57, 59 to 92, 94 to 127, 130 to 163, and 164 to 197 (99.83% probability); FTS_0201 (YP_007011286.1) had two predicted TPRs at positions 78 to 111 and 114 to 147 (99.95% probability); and FTS_0778 (YP_007011730) had four SLRs at positions 90 to 125, 126 to 161, 499 to 534, and 537 to 572 (100.00% probability) (Fig. 1). Disruption of FTS_0201 (TargeTron insertion site 347/348s; strain inFTS_0201), FTS_0778 (TargeTron insertion site 273/274s; strain inFTS_0778), and FTS_1680 (TargeTron insertion site 177/178s; strain inFTS_1680) (Fig. 1) was performed using retargeted mobile group II introns as described previously (20).
FIG 1.
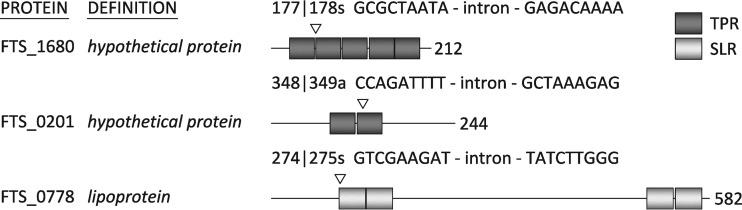
Schematic presentation of the domain positions (predicted by the web-based tool TPRpred [27]) in proteins originating from the TPR and SLR families with the TargeTron insertion sites. The TPR domains are depicted in dark gray, while the SEL1-like domains are highlighted in light gray. Accession numbers: FTS_1680 hypothetical protein, YP_007012402.1; FTS_0201 hypothetical protein, YP_007011286.1; FTS_0778 lipoprotein, YP_007011730. DNA sequences in which the intron is inserted are shown.
FTS_1680 is required for efficient intracellular proliferation in macrophage host cells.
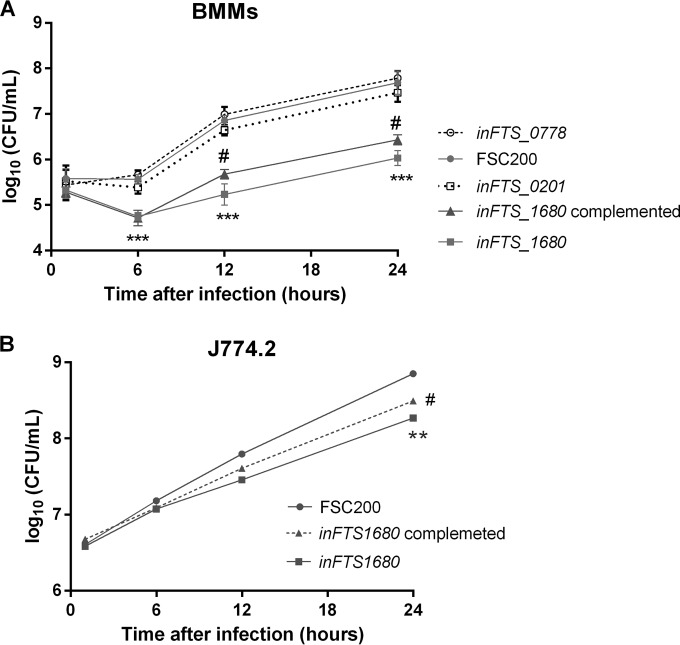
To explore the roles of the three TPR-like proteins FTS_0201, FTS_0778, and FTS_1680 during intracellular growth and virulence, we performed in vitro infection studies employing BMMs and macrophage-like cells (J774.2) as host cells. Cells were infected with the FSC200 wild-type strain or the inFTS_0201, inFTS_0778, or inFTS_1680 mutant strain, and numbers of intracellular bacteria were determined at 1, 6, 12, and 24 h after infection. The replication kinetics of the inFTS_0778 and inFTS_0201 mutant strains within BMMs (Fig. 2A) and J774.2 cells (data not shown) were indistinguishable from that of the wild-type strain. In contrast, the ability of the inFTS_1680 mutant to replicate within BMMs and J774.2 cells was significantly lower than that of the wild-type FSC200 strain (Fig. 2A and B). The difference in the intracellular growth between the inFTS_1680 and wild-type FSC200 strains was not due to an inherent growth defect, as both strains grew similarly in Chamberlain's chemically defined medium (see Fig. 8A). A statistically significant difference in intracellular replication was observed for the inFTS_1680 mutant at both 12 h and 24 h postinfection in BMMs and also at 24 h postinfection within J774.2 cells. Complementation of the inFTS_1680 strain resulted in partial restoration of the wild type phenotype, indicating that FTS_1680 contributes to F. tularensis intracellular replication. Importantly, insertional mutagenesis in genes (FTS_1681 and FTS_1682) from the predicted transcription unit containing FTS_1680 did not affect intracellular proliferation (data not shown), supporting the finding that the decreased proliferation observed with the inFTS_1680 mutant is solely a consequence of FTS_1680 disruption.
FIG 2.
FTS_1680 contributes to the intracellular survival in BMMs and J774.2 cells. The growth kinetics of the wild-type (filled circles), inFTS_1680 (filled squares), inFTS_1680 complemented (filled triangles), inFTS_0201 (empty squares), and inFTS_0778 (empty circles) strains inside murine bone marrow-derived macrophages (BMMs) (A) or J774.2 cells (B) are shown. The number of intracellular bacteria was determined at 1, 6, 12, and 24 h postinfection. Results are shown as the average log10 CFU per well ± SD for three independent experiments performed in triplicate (n = 9). Statistical significance was analyzed using an unpaired t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (comparing inFTS_1680 with the wild-type FSC200 strain). #, P < 0.05 (comparing the complemented strain with inFTS_1680).
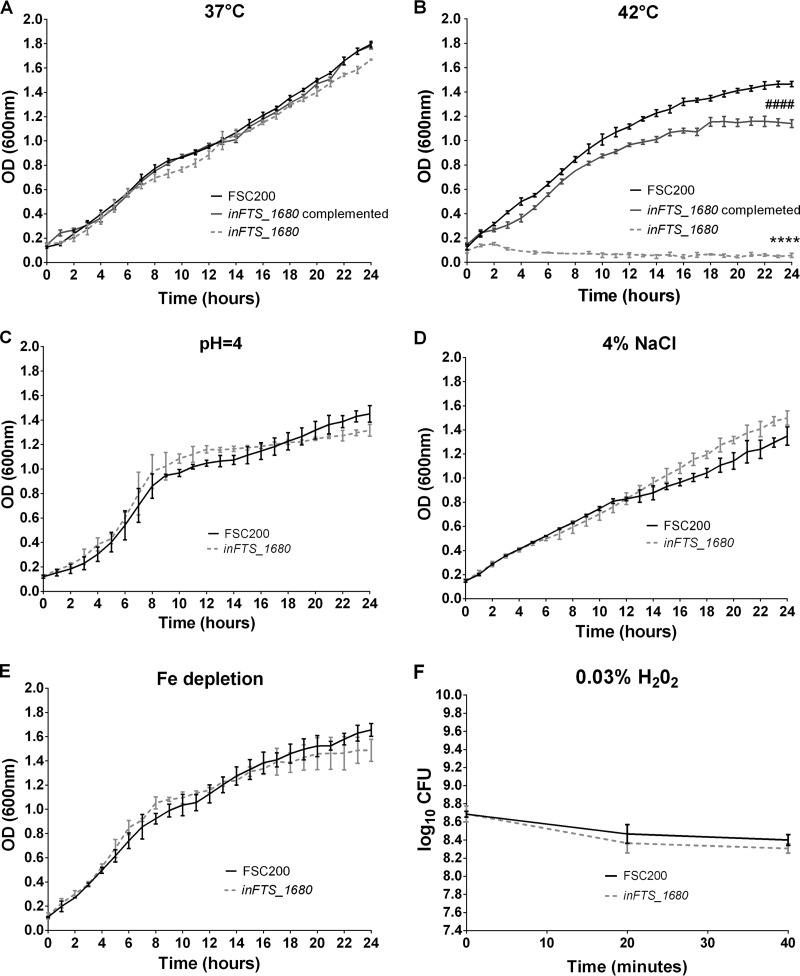
FIG 8.
FTS_1680 is required for heat stress tolerance. (A to E) Growth curves of F. tularensis strains FSC200 (black), inFTS_1680 (light gray), and complemented inFTS_1680 (dark gray) (supplemented with kanamycin at 20 μg/ml) in Chamberlain's medium incubated at 37°C (A) or 42°C (B) or under the following stress conditions: pH 4.0 (C), 4% NaCl (D), or iron depletion (E). Bacterial growth was determined by measuring the OD600 in pentaplicate every 10 min for 24 h. CFU were analyzed at the end of the experiment to verify OD600 measurement. (F) Oxidative stress experiments. Stationary-phase bacteria of F. tularensis strains FSC200 (black) or inFTS_1680 (light gray) were diluted 1:10 in fresh Chamberlain's medium and exposed to 0.03% H2O2. CFU were determined at the start of the experiment and after 20 and 40 min. The results shown are representative of three independent experiments. Statistical significance was analyzed at the 24-h time point using an unpaired t test, ****, P < 0.0001 (comparing inFTS_1680 with the wild-type FSC200 strain); ####, P < 0.0001 (comparing inFTS_1680 with the complemented strain).
FTS_1680 is important for virulence in a murine model of tularemia.
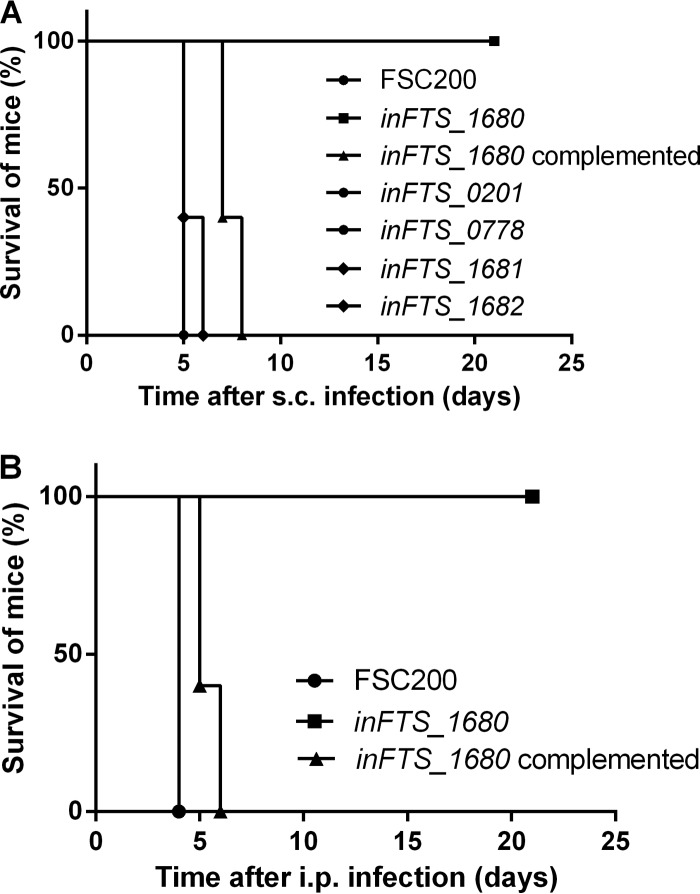
To determine whether the FTS_0201, FTS_0778, or FTS_1680 protein contributes to the ability of F. tularensis to cause disease in vivo, groups of five BALB/c mice were infected subcutaneously with the wild-type FSC200 strain, the inFTS_0201, inFTS_0778, or inFTS_1680 mutant strain, or the inFTS_1680 complemented mutant strain. Mice were observed for signs of illness for 21 days following infection. Mice infected with either the inFTS_0201 or the inFTS_0778 mutant strain at a dose of 3 × 102 CFU/mouse succumbed to disease 5 days after infection, comparable to the case for mice infected with a similar dose of the wild-type strain. In contrast, all mice infected with the inFTS_1680 mutant strain survived the infection with both 3 × 102 CFU/mouse (data not shown) and 3 × 106 CFU/mouse (Fig. 3A), although symptoms of illness were observed at both doses. To further verify the in vivo attenuation of the inFTS_1680 mutant strain, we also used an intraperitoneal model of infection. Similar to the case for the subcutaneous route, all mice infected with a dose of 3 × 106 CFU/mouse of the inFTS_1680 mutant strain survived, although symptoms of illness were observed as well (Fig. 3B). Additionally, the inFTS_1680 complemented strain exhibited a 2- to 3-day delay in TTD compared to the wild-type FSC200 strain. To exclude a possible polar effect on downstream genes surrounding FTS_1680, we also determined the virulence phenotypes of the inFTS_1681 and inFTS_1682 mutant strains. Both mutants showed a level of virulence comparable to that of wild-type FSC200 in mice (Fig. 3A). Altogether, these findings demonstrate the importance of the product of the FTS_1680 gene for in vivo virulence of F. tularensis.
FIG 3.
FTS_1680 is important for virulence in murine model of tularemia. Percent survival was determined from groups of five BALB/c mice infected subcutaneously (s.c.) with the inFTS_1680 mutant at a dose of 3 × 106 CFU/mouse, wild-type strain FSC200 (3 × 102 CFU), the inFTS_1680 complemented strain (3 × 102 CFU), the inFTS_0201 mutant (3 × 102 CFU), or the inFTS_0778 mutant (3 × 102 CFU) (A) or infected intraperitoneally (i.p.) with the inFTS_1680 mutant at a dose of 3 × 102 or 3 × 106 CFU/mouse, FSC200 (3 × 102 CFU), or the inFTS_1680 complemented strain (3 × 102 CFU) (B).
The inFTS_1680 mutant exhibits a growth defect in mouse organs.
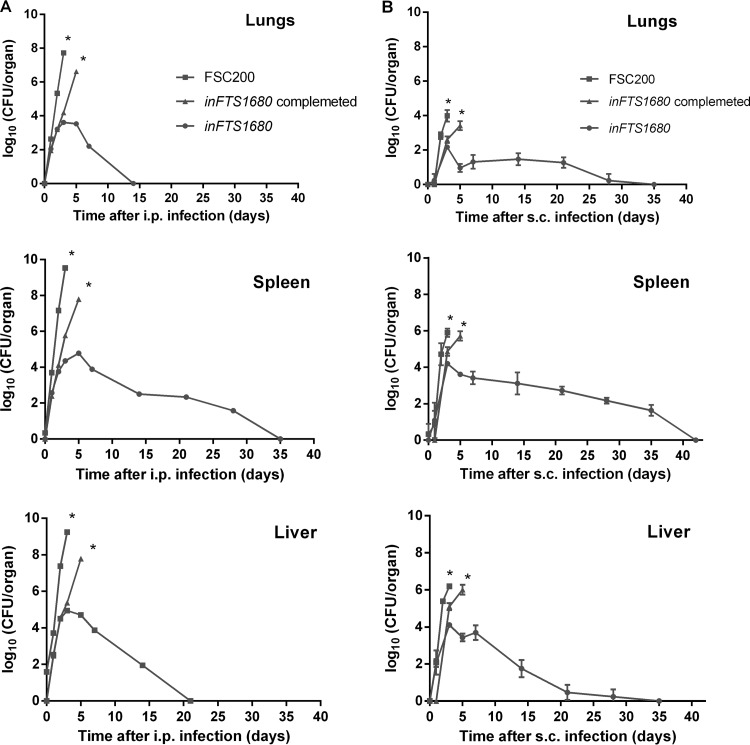
To investigate the ability of the inFTS_1680 mutant strain to persist and disseminate in host tissues, groups of three BALB/c mice were infected via the intraperitoneal route with a dose of 3 × 102 CFU/mouse of the wild-type strain or the inFTS_1680 mutant. Bacterial burdens were assayed in lung, spleen, and liver tissue homogenates at 2 h and at 1, 2, 3, 5, 7, 14, 21, 28, and 35 days postinfection (Fig. 4A). Our results showed that when the wild-type strain was administered via the intraperitoneal route, bacteria were detectable in the liver within 2 h after inoculation and in the spleen and lungs on day 1 after infection. At 3 days postinfection, the number of bacteria rapidly increased in all organs, reaching almost 1010 CFU in the liver, 109 CFU in the spleen, and 108 CFU within the lungs. None of the mice infected with the wild-type FSC200 strain survived longer than 5 days postinoculation, due to the rapid progression of disease. Although the inFTS_1680 mutant strain initially replicated in all organs studied, the bacterial loads began to decline after 3 days and were completely eliminated from the lungs (day 14), from the liver (day 21), and from the spleen (day 35). When the subcutaneous route was used (Fig. 4B), lower levels of bacteria were present in all examined organs but bacteria persisted in all organs for a longer time than with the intraperitoneal route of infection (Fig. 4A). These results demonstrate that the inFTS_1680 mutant is able to infect mice and to persist in infected organs, but it is unable to replicate effectively inside host tissues. We also infected mice with the inFTS_1680 complemented strain. On days 1, 2, 3, and 5 postinfection, we determined the number of bacteria in the lungs, liver, and spleen. Although the organ burdens in mice infected with the complemented strain did not reach the levels detected with the wild-type FSC200, they were significantly higher than with the inFTS_1680 mutant strain (Fig. 4A and B).
FIG 4.
The inFTS_1680 mutant exhibits reduced bacterial organ burdens. Bacterial burdens in the spleens, lungs, and livers of BALB/c mice infected intraperitoneally (i.p.) (A) or subcutaneously (s.c.) (B) with 3 × 102 CFU/mouse of wild-type FSC200 (filled squares), inFTS_1680 (filled circles) or, inFTS_1680 complemented strain (filled triangles) are shown. The asterisks (*) indicate that all mice infected with the wild-type FSC200 or inFTS_1680 complemented strain died after day 5. Results are shown as the average log10 CFU per organ ± SD at the indicated time points of infection.
Description of the genomic locus surrounding FTS_1680.
The genomic region surrounding FTS_1680 is highly conserved between the various Francisella strains, including 3 consecutive genes in the same orientation (FTS_1680, FTS_1681, and FTS_1682) (Fig. 5). FTS_1680 encodes a hypothetical protein (212 amino acids), the FTS_1681 product is annotated as the outer membrane protein assembly factor BamB (456 amino acids), and FTS_1682 encodes a drug:H+ antiporter-1 (DHA1) family protein (393 amino acids). Protein BLAST searches indicated that FTS_1680 and FTS_1681 share 28% identity (query cover 93%; E value = 2e−15) with YfgM (which contains TPR motifs) of E. coli K-12 and 26% identity (query cover 94%; E value = 6e−48) with YfgL of E. coli K-12. Proteins YfgM and YfgL are linked to temperature tolerance (28), and very recently YfgM has been described as a protein involved in the SecYEG system of E. coli (29). To exclude possible polar effects on downstream genes, we evaluated levels of the mRNA transcripts of the genes surrounding FTS_1680. Quantitative reverse transcription-PCR (qRT-PCR) did not reveal any significant effect of insertional inactivation of FTS_1680 on expression of FTS_1681 and FTS_1682 (data not shown).
FIG 5.
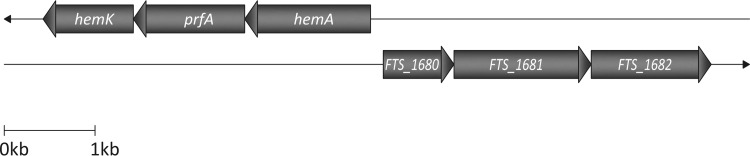
Schematic size-scaled diagram of the organization of the genomic region surrounding FTS_1680 in FSC200. The genomic region surrounding FTS_1680 is highly conserved between the various Francisella strains, including 3 consecutive genes in the same orientation (FTS_1680, FTS_1681, and FTS_1682). FTS_1680 encodes a hypothetical protein (212 amino acids), FTS_1681 is annotated to encode outer membrane protein assembly factor BamB (456 amino acids), and FTS_1682 encodes the drug:H+ antiporter-1 (DHA1) family protein (393 amino acids) belonging to the subclass of the major facilitator superfamily (MFS).
The inFTS_1680 mutant escapes into the host cell cytosol to a lesser extent than the wild-type FSC200 strain.
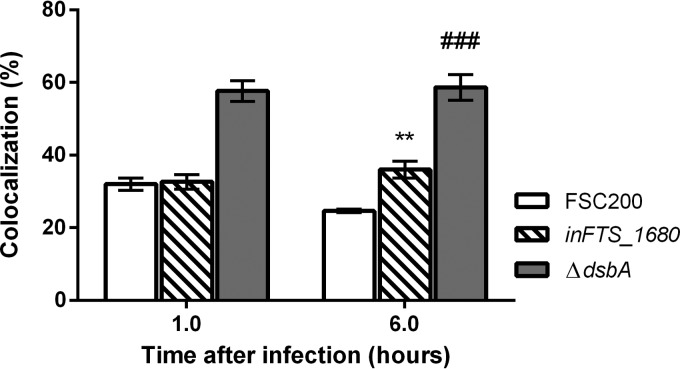
The typical Francisella intramacrophage infectious cycle includes successful phagosomal escape and active multiplication in the host cell cytosol. Based on the intracellular growth defect of the inFTS_1680 mutant in BMMs, we analyzed the ability of the inFTS_1680 mutant to escape from the host cell phagosome using a colocalization study with LAMP-1 as a marker of late endosomes. As a positive control, we used the ΔdsbA mutant, which has been previously shown to be unable to escape into the host cytosol (30). The inFTS_1680 mutant exhibited almost the same level of colocalization with LAMP-1 as the wild-type strain. The percentage of colocalization of the mutant with LAMP-1 was 31% at 1 h and 35.3% at 6 h after infection. In comparison, the colocalization of the wild-type FSC200 strain with LAMP-1 was 31.3% at 1 h and significantly declined to 23.5% at 6 h after infection (Fig. 6). However, LAMP-1 colocalization of the wild-type strain or the inFTS_1680 mutant did not reach the value detected for the ΔdsbA mutant strain (Fig. 6). Despite the observed statistical difference in the number of wild-type versus mutant bacteria in the phagosome at 6 h, it seems likely that the inability of inFTS_1680 to fully proliferate in the cell cytosol is responsible for attenuation rather than defects in phagosomal escape.
FIG 6.
The inFTS_1680 mutant is able to escape from the phagosomes of BMMs. Quantification of colocalization of LAMP-1 with wild-type (open bars), inFTS_1680 mutant (striped bars) or ΔdsbA mutant (gray bars) bacteria is shown. At each time point, 100 infected cells were examined from three different coverslips. Results are presented as the mean of triplicate samples ± SD, and the results shown are representative of three independent experiments. Statistical significance was analyzed using an unpaired t test. **, P < 0.01 (comparing inFTS_1680 with the wild-type FSC200 strain); ###, P < 0.001 (comparing inFTS_1680 with the ΔdsbA mutant).
Role of FTS_1680 in F. tularensis-induced BMM cell cytotoxicity.
To investigate the role of the FTS_1680 protein in cellular cytopathogenic effects associated with F. tularensis, we infected BMMs with the wild-type FSC200 strain or the inFTS_1680 mutant and measured the release of lactate dehydrogenase (LDH) into the cell supernatant. After 2 h of infection, the LDH level released from cells infected with the inFTS_1680 mutant (approximately 4%) was comparable to the LDH release detected in cells infected with the wild-type FSC200 strain (approximately 6%) (Fig. 7). After 24 h of infection, the LDH levels increased to 5% and 11% for the inFTS_1680 and parental FSC200 strains, respectively (Fig. 7). At 48 h postinfection, the level of LDH release was 37% for the inFTS_1680 mutant strain, which was significantly lower than the LDH release detected for the wild-type FSC200 strain (54%) (Fig. 7). The LDH assay showed that the inFTS_1680 mutant strain induces a time-dependent loss of host cell membrane integrity at levels significantly lower than those of the wild-type strain.
FIG 7.
Induction of cytotoxicity in BMMs. BMMs were infected with the wild-type strain (gray bars) or the inFTS_1680 mutant strain (open bars). At 2 h, 24 h, and 48 h after infection, culture supernatants were collected and assayed for LDH activity using the LDH cytotoxicity detection kit (Roche). LDH activity is expressed as a percentage of the level for noninfected lysed cells (positive lysis control). Results are presented as the means of quadruplicate samples ± SD, and the results shown are representatives of three separate experiments. Statistical significance was analyzed using an unpaired t test, **, P < 0.01.
FTS_1680 is involved in heat stress tolerance.
Growth of the inFTS_1680 mutant strain and the inFTS_1680 complemented strain in broth at 37°C was comparable to that of the wild-type FSC200 strain (Fig. 8A). To examine a possible involvement of the FTS_1680 protein in stress tolerance, we monitored the growth of the inFTS_1680, inFTS_1680 complemented, and wild-type FSC200 strains under stress-inducing culture conditions. Under heat stress conditions, the inFTS_1680 mutant strain grew substantially more slowly than the wild-type strain (Fig. 8B). Complementation of inFTS_1680 restored the growth of the mutant to a level similar to that of the parental strain (Fig. 8B). Conversely, the insertion mutants inFTS_1681 and inFTS_1682 grew similarly to FSC200 at 42°C (data not shown). This demonstrates that the observed lower resistance to heat stress of the inFTS_1680 mutant strain is solely due to the inactivation of the FTS_1680 gene. Sensitivities of the inFTS_1680 mutant to low pH (Fig. 8C), osmotic (Fig. 8D), iron depletion (Fig. 8E), and oxidative (Fig. 8F) stresses were comparable to those of the wild-type strain, indicating that FTS_1680 is not required for tolerance to these stress conditions.
Protein-protein interaction analysis of FTS_1680 with representatives of the heat shock proteins.
Involvement of FTS_1680 in heat stress tolerance and the predicted presence of TPR motifs suggest that FTS_1680 may interact with heat shock proteins found in F. tularensis. Using the BACTH assay, we tested FTS_1680 interaction with GroEL (FTS_1670), DnaK (FTS_1167), DnaJ2 (FTS_0277), GrpE (FTS_1166), or HtpG (FTS_0263). Under our experimental conditions, we were unable to detect an interaction between FTS_1680 and any of the selected proteins.
FTS_1680 is detected in a lipoprotein-enriched membrane fraction.
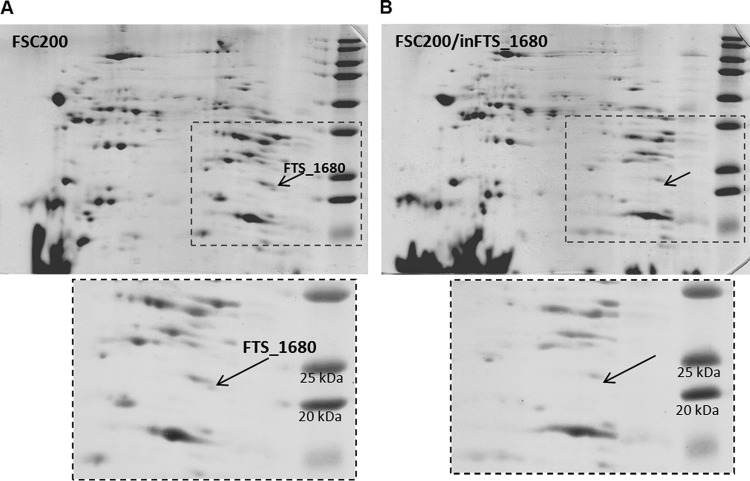
The LipPred tool for prediction of lipoprotein signal sequences (31) identified the protein FTS_1680 as a lipoprotein with a prediction confidence of 0.99, based on the lipobox [LVI][ASTVI][GAS]C sequence variations. We verified this prediction using a Triton X-114 phase partitioning (32) followed by a separation of the detergent-phase proteins by 2D SDS-PAGE. The quality of fractionation was first verified by the immunodetection of proteins with a known localization (outer membrane protein FopA) and affiliation to the lipoprotein class of membrane proteins (lipoprotein DsbA) in both aqueous and detergent phases (data not shown). The protein FTS_1680 was repeatedly identified among the Triton X-114 phase partitioning of the membrane protein-enriched fraction of FSC200 (Fig. 9A). The comparison of protein patterns of the detergent-enriched fractions collected from the parental FSC200 and inFTS_1680 mutant strains also confirmed the absence of the protein FTS_1680 in the mutant strain (Fig. 9A and B). This finding confirms membrane association of the FTS_1680 protein and also supports its predicted acylation.
FIG 9.
(A) Representative 2D SDS-PAGE separation of a lipoprotein-enriched fraction obtained by Triton X-114 phase partitioning of F. tularensis subsp. holarctica FSC200. The protein encoded by FTS_1680 was detected in a basic region of the broad nonlinear pH range of 3 to 10 and among the masses 25 and 20 kDa (in accordance with its theoretical molecular mass and pI of 23.8 kDa and 9.39, respectively) and identified by mass spectrometry. (B) Representative 2D SDS-PAGE separation of the lipoprotein-enriched fraction which was obtained by Triton X-114 phase partitioning of the mutant strain inFTS_1680. The protein encoded by FTS_1680 was not detected or identified in the neighboring spots.
Effect of inactivation of the FTT_0166c gene, the FTS_1680 ortholog in the type A strain, on the phenotype of Francisella tularensis strain SchuS4.
FTT_0166c is a predicted TPR-containing protein within the fully virulent F. tularensis type A strain SchuS4. It shares 98% identity to FTS_1680 at the amino acid level. We hypothesized that, similar to the case for FTS_1680, inactivation of FTT_0166c may lead to an attenuated phenotype in the SchuS4 background. Using TargeTron mutagenesis, we created strain inFTT_0166c. This mutant strain carries an intron insertion at the same position in the coding sequence as in inFTS_1680, 177/178s. To investigate virulence of strain inFTT_0166c in an animal model of tularemia, we infected mice subcutaneously with increasing doses of the wild-type SchuS4 or inFTT_0166c mutant bacteria and compared LD50s, survival rates, mean TTDs, and survival curves. As shown in Table 3, we found no significant difference between the wild-type and mutant strains in the calculated LD50 (approximately 1 CFU) or survival rate. However, we did observe that animals infected with the mutant strain survived longer than wild-type SchuS4-infected mice. This observation was supported by a statistically significant increase in the TTD for mutant infected animals across all doses (Table 3). Additionally, comparison of survival curves yielded a larger survival estimate for the inFTT_0166c-infected mice than for SchuS4-infected mice (P < 0.0001). To examine whether the observed difference was independent of route of administration, we repeated the experiment, but this time animals were challenged by the intranasal route. Similar to the case for the subcutaneous route, animals infected intranasally with the inFTT_0166c mutant had a statistically significantly increased TTD and survival estimate compared to the SchuS4-infected animals; however, we failed to observe a statistically significant difference in LD50 or survival rates (Table 3).
TABLE 3.
Comparisons of survival and time to death between mice challenged with SchuS4 (wild type) or with the inFTT_0166c mutant strain
| Inoculation | SchuS4a |
inFTT_0166cb |
TTD comparison (P value) | ||||
|---|---|---|---|---|---|---|---|
| CFU | % Survivors | Mean TTD (days) ± SD | CFUa | % Survivors | Mean TTD (days) ± SD | ||
| Subcutaneous | 0.1 | 70 | 5.7 ± 0.47 | 0.1 | 80 | 7.5 ± 0.50 | 0.0020 |
| 1 | 20 | 6.4 ± 0.48 | 1 | 50 | 8.0 ± 0.63 | <0.0001 | |
| 8 | 0 | 5.0 ± 0.00 | 11 | 0 | 6.8 ± 0.98 | <0.0001 | |
| 76 | 0 | 5.0 ± 0.00 | 107 | 0 | 6.4 ± 0.66 | <0.0001 | |
| 756 | 0 | 4.5 ± 0.50 | 1067 | 0 | 5.2 ± 0.40 | 0.0092 | |
| Intranasal | 0.1 | 90 | 6.0 | 0.1 | 100 | NCc | NC |
| 1 | 70 | 6.0 ± 0.00 | 1 | 90 | 6.0 | NC | |
| 7 | 0 | 6.0 ± 0.00 | 9 | 30 | 6.6 ± 0.50 | 0.0398 | |
| 74 | 0 | 5.0 ± 0.00 | 89 | 10 | 6.0 ± 0.00 | 0.0016 | |
| 741 | 0 | 5.0 ± 0.00 | 889 | 0 | 6.0 ± 0.00 | 0.0016 | |
The LD50 was ∼1 CFU for both inoculation routes.
The LD50s were ∼1 CFU and ∼6 CFU for the subcutaneous and intranasal inoculation routes, respectively.
NC, not calculated.
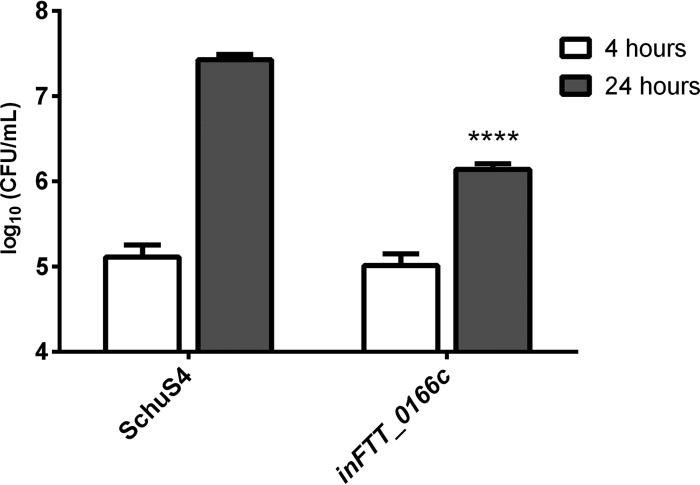
Further we hypothesized that, similar to the case for FTS_1680, the inactivation of FTT_0166c in the fully virulent SchuS4 strain may lead to a decreased ability to proliferate in macrophages. Mouse macrophage-like J774.1 cells were infected with the wild-type SchuS4 strain or the inFTT_0166c mutant strain and assayed for intracellular replication at 4 and 24 h postinfection. No significant difference was observed between CFU counts of SchuS4 and inFTT_0166c at 4 h (Fig. 10). However, at 24 h, the inFTT_0166c mutant exhibited a significant reduction in CFU compared to SchuS4 (Fig. 10). These data demonstrated that FTT_0166c is important for intracellular replication of the fully virulent SchuS4 strain and corroborated the findings observed with the inFTS_1680 mutant of FSC200 (Fig. 2A and B).
FIG 10.
The FTT_0166c gene is required for optimal intracellular replication within J774.1 cells. The number of intracellular bacteria was determined at 4 h (open bars) and 24 h (gray bars) postinfection. Data represent the average from two independent experiments run in triplicate (n = 6). Error bars indicate SD. Statistical significance was analyzed using an unpaired t test, ****, P < 0.0001.
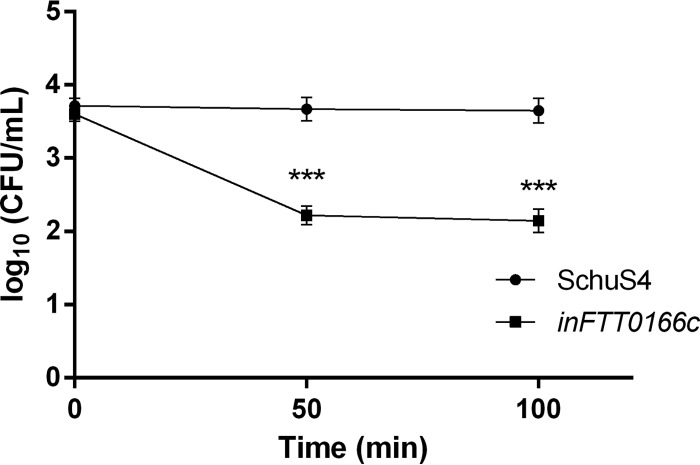
Resistance of the inFTT_0166c mutant to heat stress (42°) was also tested, and we observed an average 1.5-log decrease in viable numbers of mutant bacteria after 50 and 100 min compared to those of the parental SchuS4 strain (Fig. 11). Taken together, these data demonstrated that FTT_0166c is required for survival of SchuS4 under heat stress conditions and correlated with the findings observed with the inFTS_1680 mutant of FSC200 (Fig. 8B).
FIG 11.
FTT_0166c is required for growth under heat stress conditions. Stationary-phase bacteria of F. tularensis strains SchuS4 (circles) or inFTT_0166c (squares) were diluted 1:10 in fresh Chamberlain's medium and then exposed to heat stress (42°C). The bacteria were spread on chocolate agar plates at 50 and 100 min, and viable bacteria were counted. Results are presented as the average CFU/ml for three independent experiments. Statistical significance was analyzed using an unpaired t test. ***, P < 0.001.
DISCUSSION
In recent years, there have been several reports indicating that TPR-containing proteins of bacterial pathogens are directly linked to virulence-associated functions. The TPRs within class II chaperones of Yersinia, Pseudomonas, Shigella, Salmonella enterica, enteropathogenic Escherichia coli, and Chlamydia muridarum (LcrH, PcrH, IpgC, SicA, CesD, and SycD, respectively) have been shown to be key elements for binding the cognate translocators of type III secretion systems (TTSS) (33). Moreover, TPR domains seem to be of high importance for another TPR-containing protein, PilF, which participates in biogenesis of Tfp in Pseudomonas (reviewed in reference 15). SLR-containing proteins such as HcpA from Helicobacter pylori, EnhC and LpnE from Legionella pneumophila, AlgK from Pseudomonas aeruginosa, and ExoR from both Sinorhizobium meliloti and Rhizobium leguminosarum, MotX from Vibrio parahaemolyticus, and MerG from Pseudomonas strain K-62 have also been shown to play an important role in bacterial pathogenesis (reviewed in reference 14).
F. tularensis is able to successfully survive and cause disease in a host due to its unique strategy of intracellular survival. The mechanisms of cell entry, rapid phagosomal escape, active cytosolic multiplication, and dissemination following cell lysis are still under investigation. Since TPR-like-containing proteins are ubiquitous and fulfill many biological functions, we hypothesized that TPR-like-containing proteins might also play a role in Francisella virulence. Therefore, we created mutant strains with intron insertions in genes FTS_0201, FTS_0778, and FTS_1680, which encode TPR/SLR-containing proteins in F. tularensis FSC200, and also in the gene FTT_0166c in F. tularensis SchuS4 (FTS_1680 ortholog). We subsequently investigated the effects of these mutations on F. tularensis virulence. First, we analyzed the abilities of the inFTS_0201, inFTS_0778, and inFTS_1680 mutants to replicate within mouse macrophages. Only the inFTS_1680 mutant exhibited a significant intracellular growth defect in primary BMMs (Fig. 2A) and slightly restricted replication in the J774.2 macrophage cell line (Fig. 2B). Although we observed differences in the levels of intracellular replication between these two cell lines, the inFTS_1680 mutant showed significantly reduced replication in both cell lines, implying its importance for optimal intracellular replication. Phenotypic differences in intracellular proliferation within differing phagocytic cell lines have been observed with other attenuated Francisella mutants, as previously reported (5, 6, 34).
Based on the inFTS_1680 intracellular growth defect in mouse macrophages, we also assayed intracellular growth of the inFTT_0166c mutant in the J774 macrophage cell line. Similar to the case for the inFTS_1680 mutant, intracellular proliferation of the inFTT_0166c mutant was also significantly reduced in these cells. These results suggest that the proteins encoded by FTS_1680 and FTT_0166c are required for optimal intracellular proliferation in both the type B and type A strains of Francisella, respectively.
TTD studies in BALB/c mice showed that the inFTS_0201 and inFTS_0778 mutant strains were as virulent as the wild-type strain, suggesting that both FTS_0201 and FTS_0778 are not important for virulence in the subcutaneous murine model of tularemia (Fig. 3A). In contrast to our studies with inFTS_0201, it has previously been reported that intraperitoneal infection of BALB/c mice with a disrupted FTL_0205 gene (FTS_0201 ortholog) in the F. tularensis LVS strain revealed a strong attenuation (6). A possible explanation for the discrepancy in the reported virulence between the inFTS_0201 and inFTL_0205 mutant strains could be the variable number of NKD repeats existing in these orthologs, the clear differences in the challenge model, or simply that within the FSC200 strain other virulence factors mitigate the effect of FTS_0201 disruption, similarly to data published by Meibom et al. (35).
TTD studies with the inFTS_1680 mutant showed an attenuated phenotype. Importantly, 100% of mice infected subcutaneously with as much as 3 × 106 CFU of the inFTS_1680 mutant all survived infection. Importantly, all mice infected with either the wild-type strain or inFTS_1680 complemented strain with a dose of 3 × 102 CFU became moribund at 5 to 7 days after infection. Since virulence can vary significantly between subcutaneous and intraperitoneal models of infection (36), we also tested the inFTS_1680 and the inFTS_1680 complemented strains by the intraperitoneal route of infection. Similar to our observations with mice infected subcutaneously, all mice infected with the inFTS_1680 mutant survived infection, whereas all mice infected with the wild-type FSC200 or inFTS_1680 complemented strain succumbed to infection. These results demonstrate that the observed attenuation of the inFTS_1680 mutant occurs independent of the route of exposure. Likewise, the SchuS4-derived inFTT_0166c mutant also exhibited a route-independent attenuated phenotype. However, compared to that of its FSC200 ortholog, the attenuation observed was weaker. Nevertheless, the fact that a delay in TTD was observed across all doses for both routes supports the conclusion that this gene plays a role in virulence of the highly virulent type A strain. It is likely that the observed difference in attenuation can be attributed to the significant difference in virulence between the FSC200 and SchuS4 strains (35). Taken together, our findings support the hypothesis that FTS_1680 and its ortholog are needed for the full virulence of both type A and type B strains.
To understand the in vivo attenuation of inFTS_1680, we evaluated the ability of this mutant to persist and disseminate within selected host organs. The inFTS_1680 mutant was able to replicate at low levels in the liver, spleen, and lungs during the first days postinfection; however, the viable number of mutant bacteria declined with time. In contrast, the wild-type and complemented inFTS_1680 mutant strains replicated to high numbers in all these organs during the first days, after which no mice survived the infection (Fig. 4). It appears in some cases that proliferation in nonmacrophage cell types is sufficient to sustain F. tularensis pathogenesis (34). The rapid elimination of the inFTS_1680 mutant from the organs tested suggests that this strain may be defective in replication within nonmacrophage cells as well, which may also contribute to its attenuation.
Further, we investigated the intracellular fate of the inFTS_1680 mutant inside BMMs by employing a LAMP-1 marker as an indicator of late endosomes. Our results showed that the inFTS_1680 mutant is able to escape from the phagosome, albeit at a frequency slightly lower than that of the wild-type FSC200 (Fig. 6). However, it seems likely that the predominant role of FTS_1680 might be to help Francisella to multiply to high numbers within the cytosol. The inFTS_1680 mutant was able to replicate within the cytosolic compartment; however, it was unable to replicate to the high number observed for wild-type bacteria. This suggests that FTS_1680 is required to maintain or support high-level replication within the cytosol. In this respect, FTS_1680 resembles virulence factors playing a specific role during multiplication in the cytosol after phagosomal escape (reviewed in reference 37), such as the Francisella SEL1-like protein DipA (4). The DipA mutant does not proliferate within the cells, despite the ability to escape from the phagosome, and is completely attenuated in in vivo infection models (4). Additionally, we found that the FTS_1680 protein also contributes to FSC200 cytopathogenicity (Fig. 7). As shown previously (38, 39), the inability to induce full cytotoxicity might be associated with the observed defect in cytosolic replication.
A common characteristic of TPR-like repeats is that they create multiprotein complexes and mediate protein-protein interactions, often functioning as cochaperones in particular of the heat shock proteins (15). In the present work, the inFTS_1680 mutant showed a loss of ability to resist heat stress. Therefore, we assayed protein-protein interactions of FTS_1680 with some typical representatives of the heat shock proteins by employing the BACTH system. Unfortunately, we were unable to detect any interaction between FTS_1680 and the targeted proteins. Although the BACTH system is presented as useful for detection of protein interactions outside the cytoplasm (40), the results may be false negative since transient interactions may not be detected. Another possibility could be a localization of the interaction in the periplasmic space. This is a known obstacle for another bacterial two-hybrid system (41), and it is not clear if it also is a complication for analysis when employing the BACTH assay. We attempted to explore a chaperone activity, but we were unable to purify the native recombinant protein.
Interestingly, FTS_1680 also shows high similarity to YfgM from E. coli, that was recently shown to interact with the SecYEG translocon (29). YfgM might function in a periplasmic chaperone network while playing a role in the δ(E)-dependent envelope stress response (29). Direct interaction with SecA and SecE has not been observed to date; however, it cannot be ruled out. It is worthwhile to note that both of these proteins influence replication of Francisella in host organs (42), which parallels our in vivo observations.
In order to characterize FTS_1680 in more detail, we performed proteomic characterization. FTS_1680 is predicted to be a membrane protein, and to confirm this prediction, we exploited Triton X-114 phase partitioning for isolation of Francisella membrane-associated proteins. Based on our results, we can conclude that FTS_1680 is a membrane protein. Taking LipPred tool prediction into account, it can be speculated that FTS_1680 might be modified with an acyl moiety that could simply function as an anchor to the membrane, thus allowing FTS_1680 to assemble membrane-associated protein complexes. From this point of view, it is interesting that DipA was found to be on the surface of the SchuS4 strain, where it forms a membrane-associated complex with the outer membrane protein FopA during intramacrophage growth (43).
We should note that for both our in vitro and in vivo experiments, we were unable to fully complement the observed phenotype by supply the native gene in trans. Our approach of creating insertion mutants by employing the TargeTron gene knockout system for F. tularensis, which utilizes retargeted mobile group II introns, raises the question of potential polar effects influencing the observed phenotype. However, we feel that for these studies this possibility is remote. In our study, we used trans-complementation with a plasmid where the coding sequence is cloned downstream of the GroES promoter (21). Lack of full complementation may be attributable to expression differences between the native and GroES promoters or other limitations intrinsic to trans-complementation (44, 45). However, the wild-type phenotype was almost fully restored in in vivo experiments and when assessing heat stress tolerance. Moreover, other experimental steps were performed in order to rule out potential polar effects. qRT-PCR did not show any changes in FTL_1681, FTL_1682, and FTL_1683 expression in the inFTS_1680 mutant. Mutagenesis of FTL_1681 and FTL_1682 had no effect on the proliferation, virulence phenotype, and ability to resist heat stress. These observations support the conclusion that downstream genes, which are most likely to exhibit polar effects, do not influence the virulent phenotype. Thus, our observations might be the sole result of inactivation of FTS_1680.
In conclusion, the present study investigating the involvement of TPR-like proteins in the pathogenesis of F. tularensis identified the locus FTS_1680/FTT_0166c as a novel virulence factor that is required for proper intracellular replication of the microbe, heat stress tolerance, and in vivo virulence. Furthermore, the FTS_1680-encoded protein was identified as a membrane-associated protein necessary for fully expressed cytopathogenicity. Thus, FTS_1680/FTT_0166c, in addition to PilF, FTL_205, and DipA, represents another protein from the TPR-like family that is important for Francisella virulence. Identification of FTS_1680/FTT_0166c protein interactions and elucidation of FTS_1680/FTT_0166c functions could contribute to a deeper understanding of the unique mechanisms behind F. tularensis intracellular survival.
ACKNOWLEDGMENTS
We thank Karl Klose and Stephen A. Rodriguez (UTSA, San Antonio, TX) for generously providing the pKEK1140 targeting vector and Maria Safarova for technical assistance with the mouse infection studies. Additionally, we thank Steve Kerns (USAMRIID) for help with the statistical analysis for SchuS4 mouse experiments.
This work was supported by grants 160/50/15011 from the Grant Agency of Charles University, Prague, Czech Republic (SVV260065, DTRA project D-CZ-10-00001, and DTRA project CB3387 PA D-CZ-11-0001).
The opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
The authors declare no conflicts of interest.
Footnotes
Published ahead of print 22 September 2014
REFERENCES
- 1.Ellis J, Oyston PC, Green M, Titball RW. 2002. Tularemia. Clin. Microbiol. Rev. 15:631–646. 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oyston PC, Sjostedt A, Titball RW. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2:967–978. 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 3.Santic M, Al-Khodor S, Abu Kwaik Y. 2010. Cell biology and molecular ecology of Francisella tularensis. Cell. Microbiol. 12:129–139. 10.1111/j.1462-5822.2009.01400.x. [DOI] [PubMed] [Google Scholar]
- 4.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U. S. A. 103:14578–14583. 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meibom KL, Dubail I, Dupuis M, Barel M, Lenco J, Stulik J, Golovliov I, Sjostedt A, Charbit A. 2008. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol. Microbiol. 67:1384–1401. 10.1111/j.1365-2958.2008.06139.x. [DOI] [PubMed] [Google Scholar]
- 6.Dieppedale J, Sobral D, Dupuis M, Dubail I, Klimentova J, Stulik J, Postic G, Frapy E, Meibom KL, Barel M, Charbit A. 2011. Identification of a putative chaperone involved in stress resistance and virulence in Francisella tularensis. Infect. Immun. 79:1428–1439. 10.1128/IAI.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell BL, Mohapatra NP, Gunn JS. 2010. Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect. Immun. 78:2189–2198. 10.1128/IAI.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohapatra NP, Soni S, Bell BL, Warren R, Ernst RK, Muszynski A, Carlson RW, Gunn JS. 2007. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect. Immun. 75:3305–3314. 10.1128/IAI.00351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanistanon D, Powell DA, Hajjar AM, Pelletier MR, Cohen IE, Way SS, Skerrett SJ, Wang X, Raetz CR, Ernst RK. 2012. Role of Francisella lipid A phosphate modification in virulence and long-term protective immune responses. Infect. Immun. 80:943–951. 10.1128/IAI.06109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen B, Meeker A, Ramakrishnan G. 2010. The fslE homolog, FTL_0439 (fupA/B), mediates siderophore-dependent iron uptake in Francisella tularensis LVS. Infect. Immun. 78:4276–4285. 10.1128/IAI.00503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llewellyn AC, Jones CL, Napier BA, Bina JE, Weiss DS. 2011. Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS One 6:e24201. 10.1371/journal.pone.0024201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenco J, Pavkova I, Hubalek M, Stulik J. 2005. Insights into the oxidative stress response in Francisella tularensis LVS and its mutant DeltaiglC1+2 by proteomics analysis. FEMS Microbiol. Lett. 246:47–54. 10.1016/j.femsle.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 13.Allan RK, Ratajczak T. 2011. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16:353–367. 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittl PR, Schneider-Brachert W. 2007. Sel1-like repeat proteins in signal transduction. Cell Signal. 19:20–31. 10.1016/j.cellsig.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 15.Cerveny L, Straskova A, Dankova V, Hartlova A, Ceckova M, Staud F, Stulik J. 2013. Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect. Immun. 81:629–635. 10.1128/IAI.01035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asare R, Abu Kwaik Y. 2010. Molecular complexity orchestrates modulation of phagosome biogenesis and escape to the cytosol of macrophages by Francisella tularensis. Environ. Microbiol. 12:2559–2586. 10.1111/j.1462-2920.2010.02229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin A, Mann BJ. 2006. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6:69. 10.1186/1471-2180-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson A, Berglund L, Eriksson U, Goransson I, Wollin R, Forsman M, Tarnvik A, Sjostedt A. 2000. Comparative analysis of PCR versus culture for diagnosis of ulceroglandular tularemia. J. Clin. Microbiol. 38:22–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chamberlain RE. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13:232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez SA, Davis G, Klose KE. 2009. Targeted gene disruption in Francisella tularensis by group II introns. Methods 49:270–274. 10.1016/j.ymeth.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonquist L, Lindgren H, Golovliov I, Guina T, Sjostedt A. 2008. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 76:3502–3510. 10.1128/IAI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, Celli J. 2008. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76:5488–5499. 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Straskova A, Cerveny L, Spidlova P, Dankova V, Belcic D, Santic M, Stulik J. 2012. Deletion of IglH in virulent Francisella tularensis subsp. holarctica FSC200 strain results in attenuation and provides protection against the challenge with the parental strain. Microbes Infect. 14:177–187. 10.1016/j.micinf.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Broms JE, Lavander M, Sjostedt A. 2009. A conserved alpha-helix essential for a type VI secretion-like system of Francisella tularensis. J. Bacteriol. 191:2431–2446. 10.1128/JB.01759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balonova L, Hernychova L, Mann BF, Link M, Bilkova Z, Novotny MV, Stulik J. 2010. Multimethodological approach to identification of glycoproteins from the proteome of Francisella tularensis, an intracellular microorganism. J. Proteome Res. 9:1995–2005. 10.1021/pr9011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finney DJ. 1971. Statisical logic in the monitoring of reactions to therapeutic drugs. Methods Infect. Med. 10:237–245. [PubMed] [Google Scholar]
- 27.Karpenahalli MR, Lupas AN, Soding J. 2007. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics 8:2. 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang M, Sullivan SM, Walker AK, Strahler JR, Andrews PC, Maddock JR. 2007. Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques. J. Bacteriol. 189:3434–3444. 10.1128/JB.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotzke H, Palombo I, Muheim C, Perrody E, Genevaux P, Kudva R, Muller M, Daley DO. 2014. YfgM is an Ancillary Subunit of the SecYEG Translocon in Escherichia coli. J. Biol. Chem. 289:19089–19097. 10.1074/jbc.M113.541672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin A, Scott DW, Thompson JA, Mann BJ. 2009. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect. Immun. 77:152–161. 10.1128/IAI.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor PD, Toseland CP, Attwood TK, Flower DR. 2006. LIPPRED: a web server for accurate prediction of lipoprotein signal sequences and cleavage sites. Bioinformation 1:176–179. 10.6026/97320630001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bordier C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604–1607. [PubMed] [Google Scholar]
- 33.Pallen MJ, Chaudhuri RR, Henderson IR. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6:519–527. 10.1016/j.mib.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Horzempa J, O'Dee DM, Shanks RM, Nau GJ. 2010. Francisella tularensis DeltapyrF mutants show that replication in nonmacrophages is sufficient for pathogenesis in vivo. Infect. Immun. 78:2607–2619. 10.1128/IAI.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meibom KL, Forslund AL, Kuoppa K, Alkhuder K, Dubail I, Dupuis M, Forsberg A, Charbit A. 2009. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect. Immun. 77:1866–1880. 10.1128/IAI.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U. S. A. 104:6037–6042. 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meibom KL, Charbit A. 2010. The unraveling panoply of Francisella tularensis virulence attributes. Curr. Opin. Microbiol. 13:11–17. 10.1016/j.mib.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Maier TM, Casey MS, Becker RH, Dorsey CW, Glass EM, Maltsev N, Zahrt TC, Frank DW. 2007. Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect. Immun. 75:5376–5389. 10.1128/IAI.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pechous R, Celli J, Penoske R, Hayes SF, Frank DW, Zahrt TC. 2006. Construction and characterization of an attenuated purine auxotroph in a Francisella tularensis live vaccine strain. Infect. Immun. 74:4452–4461. 10.1128/IAI.00666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Battesti A, Bouveret E. 2012. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58:325–334. 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 41.Karimova G, Ladant D, Ullmann A. 2002. Two-hybrid systems and their usage in infection biology. Int. J. Med. Microbiol. 292:17–25. 10.1078/1438-4221-00182. [DOI] [PubMed] [Google Scholar]
- 42.Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. 2007. Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75:3089–3101. 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chong A, Child R, Wehrly TD, Rockx-Brouwer D, Qin A, Mann BJ, Celli J. 2013. Structure-function analysis of DipA, a virulence factor required for intracellular replication. PLoS One 8:e67965. 10.1371/journal.pone.0067965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robertson GT, Child R, Ingle C, Celli J, Norgard MV. 2013. IglE is an outer membrane-associated lipoprotein essential for intracellular survival and murine virulence of type A Francisella tularensis. Infect. Immun. 81:4026–4040. 10.1128/IAI.00595-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, Monack DM. 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 74:6642–6655. 10.1128/IAI.01250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233–2243. 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756. 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]