Abstract
The important human pathogen Streptococcus pyogenes (group A Streptococcus [GAS]) produces a hyaluronic acid (HA) capsule that plays critical roles in immune evasion. Previous studies showed that the hasABC operon encoding the capsule biosynthesis enzymes is under the control of a single promoter, P1, which is negatively regulated by the two-component regulatory system CovR/S. In this work, we characterize the sequence upstream of P1 and identify a novel regulatory region controlling transcription of the capsule biosynthesis operon in the M1 serotype strain MGAS2221. This region consists of a promoter, P2, which initiates transcription of a novel small RNA, HasS, an intrinsic transcriptional terminator that inefficiently terminates HasS, permitting read-through transcription of hasABC, and a putative promoter which lies upstream of P2. Electrophoretic mobility shift assays, quantitative reverse transcription-PCR, and transcriptional reporter data identified CovR as a negative regulator of P2. We found that the P1 and P2 promoters are completely repressed by CovR, and capsule expression is regulated by the putative promoter upstream of P2. Deletion of hasS or of the terminator eliminates CovR-binding sequences, relieving repression and increasing read-through, hasA transcription, and capsule production. Sequence analysis of 44 GAS genomes revealed a high level of polymorphism in the HasS sequence region. Most of the HasS variations were located in the terminator sequences, suggesting that this region is under strong selective pressure. We discovered that the terminator deletion mutant is highly resistant to neutrophil-mediated killing and is significantly more virulent in a mouse model of GAS invasive disease than the wild-type strain. Together, these results are consistent with the naturally occurring mutations in this region modulating GAS virulence.
INTRODUCTION
Streptococcus pyogenes (group A streptococcus [GAS]) is a common human pathogen that causes a variety of diseases, including minor skin and throat infections, such as impetigo and pharyngitis, and life-threatening invasive infections, such as streptococcal toxic shock syndrome and necrotizing fasciitis. One of the most important virulence factors that aid GAS in evasion of the host immune system is the hyaluronic acid (HA) capsule. Highly encapsulated GAS strains are associated with both severe invasive infections and outbreaks of acute rheumatic fever (1). HA capsule is a high-molecular-mass linear polymer consisting of glucuronic acid and N-acetylglucosamine repeating units. As the structure of GAS capsular HA is identical to HA produced by mammalian tissues, this reduces the ability of the host immune system to detect and kill infecting GAS cells. Indeed, GAS capsule mutant strains have increased sensitivity to neutrophil-mediated killing and reduced virulence in murine models of GAS infection (2–6). The HA capsule not only enhances GAS immune evasion through molecular mimicry but also promotes GAS survival within neutrophil extracellular traps through the inhibition of the human antimicrobial peptide cathelicidin (7). Furthermore, in addition to its role in circumvention of the host immune response, the HA capsule also enhances GAS virulence by mediating adherence to CD44 expressed on the surface of human keratinocytes, thus promoting GAS colonization (8, 9).
The enzymes involved in HA capsule biosynthesis are encoded by the hasABC operon, which is present in all GAS serotypes thus far tested, except M4 and M22 (10). HA capsule production requires only the first gene of the operon, hasA, encoding hyaluronate synthase (11). The enzyme forms an HA polymer from UDP-activated sugar precursors. The operon also encodes two enzymes involved in biosynthesis of the activated sugars, UDP-glucose dehydrogenase (encoded by hasB) and UDP-glucose pyrophosphorylase (encoded by hasC) (12, 13). It has been shown that expression of the operon is under the control of a single promoter, P1, located upstream of hasA (14). Transcription of the hasABC operon is negatively regulated by the CovR/S two-component signal transduction system (also known as CsrR/S), which consists of the CovS sensor kinase and the CovR response regulator (15–19). It has been found that CovR recognizes and binds AT-rich DNA regions surrounding the −10 and −35 elements of the P1 promoter (15, 19, 20). Five short sites with consensus ATTARA have been proposed to act as CovR-binding motifs (19).
The CovR/S system is a global GAS regulator responsible for modulating the transcription of up to 10 to 15% of the genes in the genome, including important virulence determinants of GAS (21, 22). Spontaneous mutations in the covRS genes arise in vivo during infection, resulting in capsule overproduction and hypervirulence of GAS (22–24). Interestingly, some GAS isolates with a functioning CovR/S system also produce a large capsule, raising the possibility that additional layers of regulation of capsule expression exist (25).
In this study, we found a novel regulatory region upstream of P1 which controls transcription of the capsule operon. We demonstrated that deletion of this region has a positive effect on the has operon transcription. This novel region consists of two promoters and a transcriptional terminator that permits read-through transcription of hasABC. We showed that the CovR/S regulatory system negatively regulates P1 and one of the promoters, P2, upstream of P1. Moreover, our mouse studies with the terminator mutant indicate that this regulatory region significantly contributes to GAS virulence. Finally, we found that this region from multiple GAS strains displays a high level of sequence variation in the P2 promoter and the terminator sequences. Given the importance of this region in the regulation of capsule production, this observation suggests that the mutations might alter strain virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All plasmids, strains, and primers used in this study are listed in Tables S1, S2, and S3 in the supplemental material. S. pyogenes MGAS2221 and SF370, M1 serotype strains (26, 27), and the 2221ΔcovR mutant, the CovR deletion mutant of MGAS2221 (28), were used for most experiments and strain construction. The strains used for capsule assay are listed in Table S1 in the supplemental material. GAS cultures were grown in Todd-Hewitt broth supplemented with 0.2% yeast extract (THY) or on THY agar plates. GAS strains were cultured without aeration at 37°C. Escherichia coli strains were grown in Luria-Bertani (LB) medium or on LB agar plates at 37°C. When required, antibiotics were included at the following concentrations: ampicillin at 100 μg ml−1 for E. coli, chloramphenicol at 10 μg ml−1 for E. coli and 5 μg ml−1 for GAS, and spectinomycin at 200 μg ml−1 for E. coli and 100 μg ml−1 for GAS.
DNA techniques.
Plasmid DNA was isolated from E. coli by commercial kits (Qiagen) according to the manufacturer's instructions and used to transform E. coli and GAS strains. Plasmids were transformed into GAS by electroporation as described previously (29). Chromosomal DNA was purified from GAS as described previously (30). To construct single-base substitutions or deletion mutations, we used the QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Constructs containing mutations were identified by sequence analysis. Primers for site-directed mutagenesis are listed in Table S3 in the supplemental material. All constructs were confirmed by sequencing analysis (Eurofins MWG Operon).
Plasmid and strain construction. (i) Construction of isogenic mutant strains.
For construction of the mutants (2221ΔP2ΔhasS, 2221Δ63T, 2221Δ42T, 2221ΔP2, 2221P1−, 2221ΔP2P1−, 2221ΔcovRΔ42T, 2221ΔcovRΔP2ΔhasS, 2221ΔcovRP1−, 2221ΔcovRΔP2P1−, 2221ΔcovRΔ42TP1−, and SF370Δ63T mutants), either MGAS2221 or SF370 chromosomal DNA was used as the templates for amplification of a 1.4-kbp PCR product. The primer pair hasR-BamHI-f and hasR-XhoI-r (see Table S2 in the supplemental material) was used to amplify a DNA region flanking either side of the hasA upstream region. The PCR product was digested with BamHI and XhoI and ligated into the BglII/XhoI-digested pBBL740 plasmid (see Table S1 in the supplemental material). The integrational plasmid pBBL740 does not have a replication origin that is functional in GAS, so the plasmid can be maintained only by integrating into the GAS chromosome through homologous recombination. To create deletions in the hasA upstream sequence region, the resultant plasmid pBBL740hasS was employed for site-directed mutagenesis using the primer pair listed in Table S3 in the supplemental material. The resulting plasmids were transformed into the MGAS2221, 2221ΔcovR, and SF370 strains, and chloramphenicol-resistant colonies were selected on THY agar plates. Five randomly selected colonies that had the first crossover and wild-type (WT) capsule concentration were grown in liquid THY without chloramphenicol for ≥5 serial passages. Several potential double-crossover mutants were selected as previously described (31). The deletion in each mutant was confirmed by sequencing a PCR fragment, and the phenotypes of several mutant colonies were examined.
(ii) Reporter plasmids for luciferase expression.
For construction of transcriptional fusions of the P2 promoter with the firefly luciferase gene (luc), we employed the pKSM720 plasmid, which carries a promoterless luc with its ribosome-binding site (RBS) (32). To construct P2::luc fusion, MGAS2221 chromosomal DNA and the primer pair P2-BglII-f and P2-XhoI-r (see Table S2 in the supplemental material) were used to amplify a 141-bp PCR product. To construct a fusion of luc with the DNA fragment carrying P2, hasS, and the terminator (P2T::luc fusion), the primer pair P2-BglII-f and term-XhoI-r (see Table S2) was used to amplify a 184-bp PCR product from MGAS2221 DNA. The PCR products were then cloned into pKSM720 plasmid. The plasmids were designated pKSM720P2 and pKSM720P2T, respectively (see Table S1). The resulting plasmids were transformed into the MGAS2221 and 2221ΔcovR strains.
(iii) Plasmid for GAS expression of HasS.
For expression of HasS, a DNA fragment containing hasS with the P2 promoter region was amplified from MGAS2221 chromosomal DNA using primer pair hasS-f and hasS-r (see Table S2 in the supplemental material). The PCR product was then cloned into BglII/BamHI-digested pKSM720. The resultant plasmid, pKSMhasS, was transformed into MGAS2221 by electroporation, and transformants were selected on agar plates containing spectinomycin.
(iv) Plasmids for E. coli expression of covR.
For construction of a CovR expression plasmid, the primer pair covR-NcoI-f and covR-XhoI-r (see Table S2 in the supplemental material) was employed to amplify the covR gene from MGAS2221 chromosomal DNA. The PCR product was digested with NcoI and XhoI and ligated into NcoI/XbaI-digested pET21d. The resulting plasmid, pETCovR, contains covR fused at the C terminus with the sequence encoding a His tag. To mimic the phosphorylated state of CovR, the gene was mutated at aspartate 53 to glutamate (D53E mutation). The resultant pETCovRD53E plasmid was transferred into competent E. coli Rosetta(DE3) (Novagen) using the manufacturer's protocol.
Purification of the CovR D53E mutant.
The CovR D53E mutant protein was expressed in E. coli Rosetta(DE3) harboring the pETCovRD53E plasmid. The protein was purified by Ni-nitrilotriacetic acid chromatography followed by size exclusion chromatography on Superdex 200. Based on the results obtained by analytical size exclusion chromatography, the CovR mutant protein forms stable dimers. Dimerization of CovR is considered necessary for transcription regulation of the has operon (33).
RNA extraction.
Total RNA was isolated from GAS grown to the mid-exponential phase (optical density at 600 nm [OD600] of 0.6) using the miRNeasy kit (Qiagen) according to the manufacturer's instructions. Contaminating genomic DNA was removed from eluted RNA samples with Turbo DNase (Applied Biosystems), with DNA removal being verified by PCR.
RT-PCR analysis.
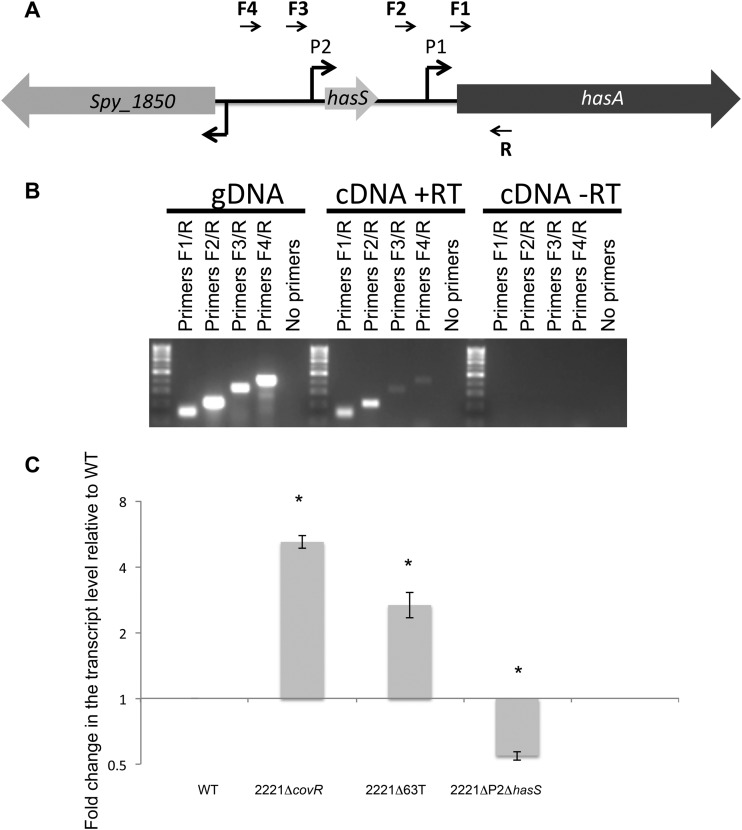
RNA isolated from MGAS2221 was used in two cDNA synthesis reactions, where one of the reaction mixtures did not have any reverse transcriptase added (−RT; to be used as a control against contaminating genomic DNA). Isolated genomic DNA (gDNA) from MGAS2221 and the +RT and −RT cDNA synthesis reactions were each used as the template in PCRs with primers R/F1, R/F2, R/F3, and R/F4 (see Table S2 in the supplemental material; see also Fig. 6A). Reaction products were separated by electrophoresis, and the gel was imaged.
FIG 6.
RT-PCR and qRT-PCR analyses of the hasA upstream region. (A) Schematic representation of the hasA upstream region. Shown is the approximate location of the promoters (bent arrows). Horizontal arrows indicate the positions of the primers used for RT-PCR. (B) RT-PCR confirming the transcription of the hasA upstream region. MGAS2221 gDNA and cDNA from reaction mixtures containing (+RT) or not containing (−RT) reverse transcriptase were used as the templates in four PCRs. The relative locations of the primers used (numbered R, F1, F2, F3, and F4) are shown in panel A. The samples without reverse transcriptase served as a control against gDNA contamination. (C) qRT-PCR analysis of the read-through transcription. Graph showing the mean values of fold change for the read-through mRNA levels in the mutants relative to those of MGAS2221. Both WT and mutant values were relative to those of the internal control gene, plr, with mutant values representing the fold change relative to that of WT, which was converted to 1. The experiments were performed in triplicate, and mean values ± SDs are shown. * indicates statistically significant differences relative to WT (P < 0.05, unpaired t test).
qRT-PCR.
To determine relative mRNA abundance of hasS, hasA, and the read-through product, we employed quantitative RT-PCR (qRT-PCR). The housekeeping gene plr was used as an internal standard for normalization. To obtain cDNA for qRT-PCR, 4 μg of total RNA was reverse transcribed with SuperScript III RT (Invitrogen) using both reverse target-specific primer and reverse plr-specific primer. qRT-PCRs were performed using an Mx3005P qPCR system (Agilent Technologies) under standard reaction conditions. The reaction mixture contained Power SYBR green PCR master mix (Applied Biosystems), cDNA, and both gene-specific primers (see Table S2 in the supplemental material). hasS was analyzed using the hasSRT-f and hasSRT-r primer pair. hasA was analyzed using the hasART-f and hasART-r primer pair. The read-through product was analyzed using the rtqRT-f and rtqRT-r primer pair. plr was analyzed in each cDNA reaction using the plr-f and plr-r primer pair. No-template and no-RT controls were included for each primer set and template. The expression levels of each target under each condition tested were normalized to the levels of plr transcript. Data are reported as the mean values of fold change for hasS, hasA, and the read-through mRNA levels in the mutants relative to those of the MGAS2221 WT strain. Each experiment was performed in triplicate, and mean values ± standard deviations (SDs) are shown. Error bars represent standard deviations from three independent experiments.
Expression and purification of core RNA polymerase from GAS.
Core RNA polymerase was purified from the GAS JRS4-PolHis strain (34) as previously described (35).
Expression and purification of σA of GAS from E. coli.
GAS σA was purified from E. coli Rosetta(DE3) carrying pEU7534 plasmid as previously described (36).
In vitro transcription and isolation of RNA products.
DNA template employed for the HasS transcriptional reaction was PCR amplified from MGAS2221 genomic DNA using the pair of primers 1 and RNA-has-r1 (see Table S2 in the supplemental material). DNA templates employed for hasA transcription were PCR amplified from MGAS2221 and 2221P1− genomic DNA using the P1-f and P1-r primer pair. In vitro transcription reaction was performed as previously described (37) with modifications. Briefly, purified GAS core RNA polymerase and σA, transcriptional buffer, SUPERase In RNase inhibitor (Ambion), and DNA template were mixed and incubated at room temperature for 10 min. The reaction was started by adding a mixture of ribonucleotides containing [α32P]UTP (PerkinElmer) and incubated at 37°C. For time course in vitro transcription, the sample aliquots were withdrawn at various time points. The reaction was terminated by adding ethanol for RNA precipitation. RNA products were resuspended in formamide loading dye (Thermo Scientific) and run on a 10% denaturing urea polyacrylamide gel. The gel was imaged using a Typhoon imaging system.
To determine 5′ and 3′ ends of RNA products, the in vitro transcription reaction mixture was incubated for 30 min and terminated by adding 2 units DNase I (Invitrogen) followed by incubation at 37°C for 15 min. DNase I was inactivated by adding EDTA to a final concentration of 5 mM. Products were ethanol precipitated, resuspended in formamide loading dye, and run on a 10% denaturing urea polyacrylamide gel. The radioactive bands were cut from a gel and incubated with probe elution buffer (RPA III kit; Invitrogen) overnight to extract RNA. RNA products were ethanol precipitated and used to identify 5′ and 3′ ends as described below.
RACE.
5′ rapid amplification of cDNA ends (5′-RACE) assays were carried out using the FirstChoice RLM-RACE kit (Invitrogen) as per the manufacturer's instructions with minor modifications. First, the dephosphorylation step was omitted from the protocol. Second, the products obtained by in vitro transcription were reverse transcribed using random decamers. The total cellular RNA isolated from MGAS2221 was reverse transcribed using the HasS-specific reverse primer hasSRT-r (see Table S2 in the supplemental material). 3′-RACE assays were carried out essentially as was described previously (38). PCR products were gel eluted and cloned in pCR2.1-TOPO using the TOPO TA cloning kit (Invitrogen). Eight to 10 clones were selected and sequenced to determine 5′ and 3′ ends of RNAs.
RPA.
RNase protection assay (RPA) was done by use of the RPA III RNase protection assay kit (Invitrogen). In brief, total RNA was isolated from GAS strains grown to the mid-exponential phase (OD600 of 0.6). An antisense RNA probe was prepared by PCR amplification using primer pair RPA-f and RPA-r. A 63-bp PCR product was subcloned into pCR2.1-TOPO (Invitrogen) so that the antisense RNA probe used in the RNase protection assay would be synthesized from the T7 promoter. Radiolabeled probe was prepared by transcription of SphI-digested plasmid by using a MAXIscript T7 kit (Invitrogen) and [α-32P]UTP (PerkinElmer). The probe was hybridized to 50 μg of total RNA overnight at 42°C. Protected hybrids were resolved on a 10% denaturing polyacrylamide sequencing gel.
Luciferase assay.
The luciferase assay was performed using a luciferase assay system (Promega). For assessment of luciferase reporter activity, the bacteria were grown in THY broth under spectinomycin selection to an OD600 of 0.6. Two-microliter aliquots of cell suspensions were pelleted and resuspended in various amounts of 1× lysis buffer to normalize to the optical density of the samples. The luciferase assay was read using a SpectraMax M5 multimode microplate reader (Molecular Devices). All experiments were performed in triplicate. For assessment of reporter activity during GAS growth, 2-ml aliquots of cell suspensions were withdrawn at various time points, and the OD600 was measured. The bacterial suspension was processed as described above.
Gel mobility shift assay.
The P2 promoter probe was generated by PCR amplification of MGAS2221 genomic DNA using the pair of primers 2 and RNA-has-r1 (see Table S2 in the supplemental material). The P2 promoter probe with a deleted HasS region was generated similarly by PCR amplification of 2221Δ42T genomic DNA. mga promoter probe was generated by PCR amplification of MGAS2221 genomic DNA using the mga-f and mga-r primer pair. PCR fragments were end labeled with [γ32P]ATP using T4 polynucleotide kinase (New England BioLabs). Electrophoretic mobility shift assay (EMSA) was performed essentially as described in the gel shift assay system (Promega). Briefly, a labeled promoter probe DNA (1.7 μM) was incubated with increasing concentrations of CovR (0.4 to 16.3 μM). Nonradioactive P2 (85 μM) and mga (100 μM) promoter DNA fragments were used in the reaction as specific and nonspecific competitors, respectively. As a nonspecific competitor, poly(dI-dC) · poly(dI-dC) (10 μg/ml) was used in the reaction. The reaction mixtures were run on a 10% native polyacrylamide gel. The gel was imaged using a Typhoon imaging system.
Hyaluronic acid capsule assay.
The amounts of hyaluronic acid capsule were determined in GAS cultured to mid-exponential phase of growth (OD600 of 0.5) as previously described (39) with minor modification. Bacteria from a 2-ml exponential-phase culture were washed twice with water and resuspended in 0.5 ml water. Capsule was released by shaking with 1 ml of chloroform for 1 h. Tubes were then recentrifuged at 14,000 × g for 10 min, and 0.2 ml of aqueous layer was removed and combined with 0.6 ml of a solution containing 20 mg of Stains-All (Sigma) and 60 μl acetic acid in 100 ml of 50% formamide. The hyaluronic acid content was determined by measuring absorbance at 640 nm. Absorbance values were compared with a standard curve generated with known concentrations of hyaluronic acid.
GAS survival in human blood.
Heparinized venous blood samples were collected from healthy donors in accordance with protocols approved by the Institutional Review Boards for Human Subjects at Montana State University. Bacteria were harvested at the mid-exponential phase of growth. One milliliter of heparinized human blood was inoculated with 1 × 105 GAS at 37°C for 1 h and 3 h with shaking. The survival of bacteria was determined by plating bacteria on THY agar plates and comparing CFU of each sample.
Polymorphonuclear leukocyte (PMN) isolation and PMN bactericidal assay.
Neutrophils were isolated under endotoxin-free conditions (<25.0 pg/ml) as described previously (40–42).
The ability of GAS to survive phagocytosis by neutrophils was assessed using synchronized phagocytosis as described previously (40) with the following modification: experiments were performed in 96-well plates. The percent survival of bacteria after incubation in the presence of PMNs was calculated using the formula (CFUn/CFU0) × 100, where CFU0 is the number of viable bacteria incubated with Hanks' balanced salt solution (HBSS) and CFUn is the number of surviving bacteria after incubation with PMNs.
Mouse infections.
All studies conformed to NIH guidelines and were approved by the Animal Care and Use Committee at Montana State University. Female BALB/c mice (Charles River Laboratories) were used for all mouse infection studies. Mice were inoculated via intraperitoneal injection with 100 μl of sterile saline containing 1 × 108 GAS CFU. To determine bacterial burdens, mice were sacrificed at 10 h postinfection and the peritoneal cavity was washed with 10 ml of HBSS using an 18-gauge needle and a 10-ml syringe. The washes were serially diluted in distilled water (dH2O) and plated on THY agar plates. To determine GAS burdens in tissues, hearts and kidneys were aseptically removed, washed in dH2O, and homogenized in dH2O. The number of CFU in the organs was quantified by plating serial dilutions of the homogenized material on THY agar plates.
Bioinformatics tools.
RNA secondary structures were predicted using the mfold program (43) (http://mfold.rna.albany.edu/?q=mfold). The prediction with the highest folding free-energy value was utilized.
Statistical data analysis.
All data sets were analyzed with GraphPad Prism, version 5.0 (GraphPad Software, San Diego, CA). All mouse data sets were analyzed with paired t tests on a log scale. Real-time PCR analysis and capsule and luciferase assays were performed a minimum of three times. Significant differences between two values were compared with a paired Student t test. Values were considered significantly different when P values were <0.05.
RESULTS
Identification of a small RNA (sRNA) upstream of the hasABC operon.
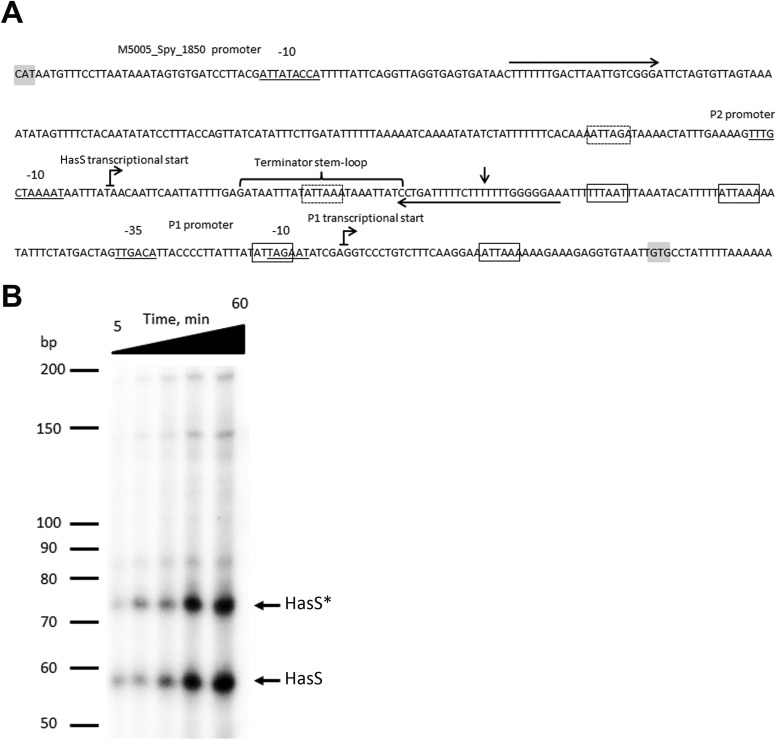
Our interest in the hasABC locus was driven in part by the importance of the HA capsule to GAS virulence and in part by the observation that the intergenic region upstream of hasA and downstream of M5005_Spy_1850 was 430 bp in size in M1 serotype strains. An intergenic region of 430 bp is unusual for GAS, where the average size is less than 100 bp, possibly indicating the existence of an uncharacterized gene or regulatory feature in this region. mfold analysis for RNA secondary structure (43) of the hasA upstream region in M1 serotype GAS shown in Fig. 1A predicted a potential stem-loop structure 25 bp long with a ΔG of −10.17 kcal/mol at 25°C. The stem-loop structure is followed by a string of U's and is predicted to act as a rho-independent terminator during transcription. This putative terminator is located approximately 50 bp upstream of the previously described P1 promoter (Fig. 1A) and does not overlap CovR-binding sites previously predicted upstream of the P1 promoter (19).
FIG 1.
The hasA upstream region encodes P1 and P2 promoters and a terminator. (A) Nucleotide sequence of the hasA upstream region in the GAS M1 serotype. −35, −10, and extended −10 promoter regions of P1 and P2 and M5005_Spy_1850 are underlined. Bent arrows denote the transcriptional start sites of hasA and hasS. The hasA transcriptional start site is according to the published results of primer extension analysis (63), and HasS was identified by 5′-RACE analysis. The vertical arrow depicts the termination site of HasS identified by 3′-RACE analysis of the in vitro transcriptional product. Horizontal arrows indicate the location of the primers, 1 and RNA-has-r1 (see Table S2 in the supplemental material), used for PCR amplification of a template employed for in vitro transcription of HasS. Gray shaded boxes indicate the location of start codons. The previously reported putative CovR-binding sites (19) in the hasA upstream region (open boxes) and putative CovR-binding sites in the P2 promoter and HasS regions (dotted boxes) are shown. Note that the fifth reported CovR-binding site lies within the hasA coding region and is not shown. (B) Time course of in vitro RNA synthesis from a PCR product carrying the P2 promoter and the terminator sequences. Shown is a 10% denaturing urea polyacrylamide gel containing in vitro transcribed RNA products using purified GAS RNA polymerase and σA (36). Lane 1, 5 min; lane 2, 10 min; lane 3, 20 min; lane 4, 30 min; lane 5, 1 h. The RNA ladder is labeled in bases. * denotes an HasS read-through product.
To test whether the terminator is associated with a transcript and identify the transcriptional start of the RNA, we employed 5′ rapid amplification of cDNA ends (5′-RACE) analysis. The transcriptional start was found upstream of the putative terminator in MGAS2221 (M1 serotype GAS strain). The 5′ terminus of the 5′-RACE product mapped to an adenine residue located 193 bp upstream of the hasA translation start codon (Fig. 1A). This site is preceded by a putative promoter, P2, which consists of a near-consensus −10 hexamer (5′-TAAAAT-3′) preceded by a −10 extension (5′-TTTG-3′) typical of σA-type promoters from Gram-positive bacteria (44–47). No consensus −35 sequence was present at a suitable distance from the −10 promoter sequence. It is noteworthy that the promoters with extended −10 sequence can function in the absence of a −35 site (44, 46). In addition to σA, GAS contains an alternative sigma factor, σX homolog, which recognizes a sequence called the “cin-box” (34). Examination of the hasA upstream sequence did not revealed any putative “cin-box” promoters, suggesting that σX does not control transcription in this region.
To examine the activity of the P2 promoter and investigate whether the predicted terminator is able to terminate efficiently, we assayed the transcription of the hasA upstream region in vitro with a GAS-specific transcription system which contains GAS core RNA polymerase and GAS housekeeping sigma factor σA. Previously, this system has been successfully employed to study transcription of GAS genes (33, 35, 36, 48, 49). RNA polymerase was purified from the GAS JRS4-PolHis strain, and recombinant GAS σA was purified from E. coli Rosetta(DE3) carrying pEU7534 plasmid (36). As a template for the in vitro reaction, we used a 240-bp PCR fragment carrying the P2 promoter, the terminator, and 82 bp downstream of the P2 promoter. We detected two major transcriptional products 58 and 73 nucleotides (nt) long (Fig. 1B). The transcriptional profiles were consistent with the products initiated from the P2 promoter. 5′-RACE analysis of the products validated our previously determined transcriptional start. 3′-RACE analysis revealed that the 58-nt product ends at the terminator and the 73-nt product is the result of read-through at the terminator.
In order to examine whether this region has additional promoters between P1 and P2, we analyzed in vitro transcription using a 300-bp PCR fragment carrying the P1 promoter and 110 bp upstream of the −35 box of P1 (see Fig. S1A in the supplemental material). We detected a 150-bp runoff transcript consistent with the product being initiated from the P1 promoter (see Fig. S1B). No transcript was observed when the −10 element of the P1 promoter was mutated from TAGAAT to TCTAGA.
Taken together, our data indicate that the analyzed hasA upstream region encodes at least one additional promoter that initiates transcription of a small RNA as well as a larger RNA that includes the downstream hasA gene. We designated the sRNA HasS. Moreover, we further confirmed the existence of transcripts originating upstream of the P1 promoter region by RNase protection assay (RPA) using a radiolabeled antisense 58-nt RNA probe to HasS. The probe was used for hybridization with RNA isolated from three different strains: MGAS2221, a covR deletion mutant strain (2221ΔcovR), and MGAS2221 carrying a plasmid with hasS under the P2 promoter (2221/pKSMhasS). A weak signal corresponding to a 58-nt protected fragment was detected in the 2221ΔcovR and 2221/pKSMhasS strains but not in the wild-type (WT) strain (see Fig. S2 in the supplemental material). These data indicate that HasS is a low-abundance sRNA which is negatively regulated by CovR.
Deletion of the HasS region affects capsule operon transcription.
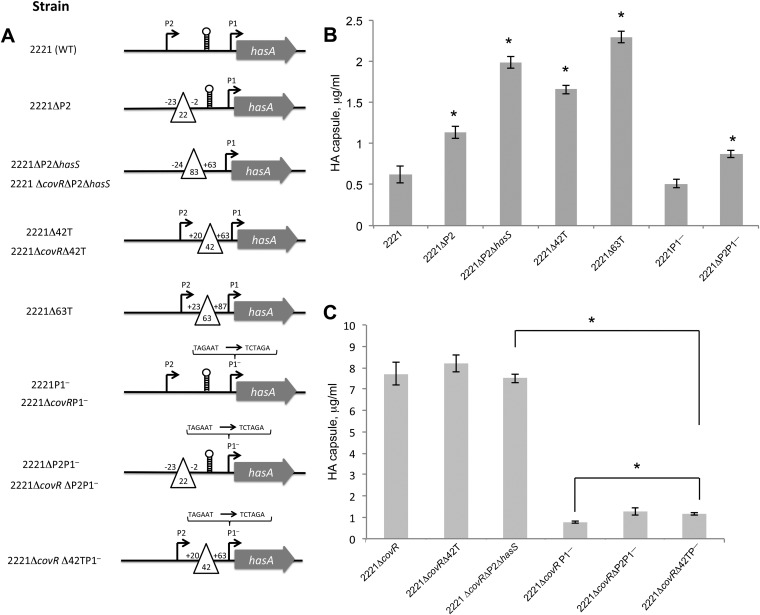
To facilitate investigation of what functions HasS and the terminator have in GAS, we constructed four mutants in the parental isolate MGAS2221: the 2221ΔP2, 2221ΔP2ΔhasS, 2221Δ42T, and 2221Δ63T mutants (Fig. 2A). In the 2221ΔP2 strain, we deleted the P2 promoter. In the 2221ΔP2ΔhasS strain, we deleted the P2 promoter and the entire hasS sequence, including the stem-loop terminator and the T-rich region located immediately downstream. In the 2221Δ42T strain, we created a 42-nt deletion which included the HasS stem-loop terminator sequence and the T-rich region located immediately downstream. In the 2221Δ63T strain, we created a 63-nt deletion which included the HasS stem-loop terminator sequence, the T-rich region, and 25 nt located immediately downstream. Since HasS is transcribed adjacent to the hasABC operon, we hypothesized that it regulates HA capsule production in GAS. To test this hypothesis, HA capsule concentration was measured from GAS cultures grown to the mid-exponential phase of growth. We found that the 2221ΔP2, 2221ΔP2ΔhasS, 2221Δ42T, and 2221Δ63T mutants had increased capsule expression relative to that of the WT strain (Fig. 2B).
FIG 2.
The HasS region regulates HA capsule production in GAS. (A) Schematic representations of the construction of the mutants used in this study. Bent arrows indicate the promoters in the hasA upstream region. The open triangles depict deletions that were constructed. The number of deleted nucleotides is indicated inside the triangles. The relative position of each deletion from the HasS transcriptional start site (+1) is shown. The terminator is shown as a stem-loop. A bracket denotes changes in the sequence of the −10 box of the P1 promoter. (B) Capsule amounts measured in the 2221 parental isolate and its mutants; (C) capsule amounts measured in the 2221ΔcovR strain and its mutants. The strains were harvested at the mid-exponential growth phase. The experiments were performed in triplicate, and mean values ± SDs are shown. * denotes statistically significant differences relative to the WT control, calculated using the Student test.
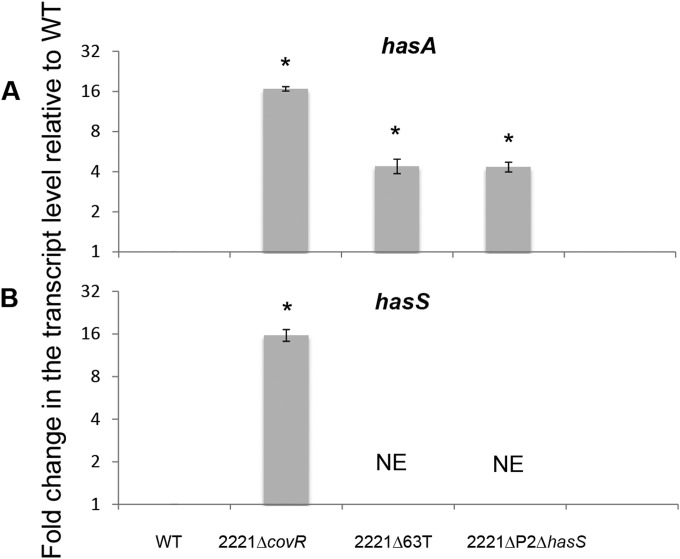
The 2221ΔP2ΔhasS and 2221Δ63T mutants demonstrated the most significant increase in capsule expression, producing 3 and 3.7 times more capsule, respectively, than the WT strain (Fig. 2B). To determine whether the elevated capsule amount resulted from upregulation of the has operon at a transcriptional level, we examined the transcription of hasA in the WT and the 2221ΔP2ΔhasS and 2221Δ63T mutants using quantitative RT-PCR (qRT-PCR) analysis. The mutants produced 4.5 times more transcript than the WT strain (Fig. 3A).
FIG 3.
qRT-PCR analysis of hasA and hasS mRNA transcripts. Graph showing the mean values of fold change for hasA (A) and hasS (B) mRNA levels in the mutants relative to those of MGAS2221. Both WT and mutant values were relative to those of the internal control gene plr, with mutant values representing the fold change relative to that of WT, which was converted to 1. The experiments were performed in triplicate, and mean values ± SDs are shown. NE denotes the lack of transcript, as the reverse primer for qRT-PCR is in the deleted region. * indicates statistically significant differences relative to WT (P < 0.05, unpaired t test).
To investigate whether upregulated has transcription is the result of deletion of previously unrecognized CovR-binding sites located in the HasS region, we constructed two mutants in the CovR deletion mutant background: 2221ΔcovRΔP2ΔhasS and 2221ΔcovRΔ42T (Fig. 2A). We found that the mutations had no effect on capsule production in this strain background. Taken together, our results indicate that the HasS region regulates the transcription of the capsule biosynthesis operon, and this regulation is most probably due to CovR binding.
CovR directly represses HasS transcription.
The CovR/S regulatory system negatively regulates transcription of the hasABC operon (15, 18, 19). To examine whether CovR also affects transcription of HasS, we employed several approaches. First, we examined transcription of hasS and hasA in the 2221ΔcovR mutant using qRT-PCR analysis. As expected, the 2221ΔcovR mutant has a 16-fold increase in hasA transcript relative to that of the WT strain, which is consistent with CovR being a negative regulator of the capsule operon (Fig. 3A). We found that the HasS transcription was about 16-fold higher in the 2221ΔcovR mutant than in the WT strain (Fig. 3B). These results indicate that CovR also represses the transcription of HasS.
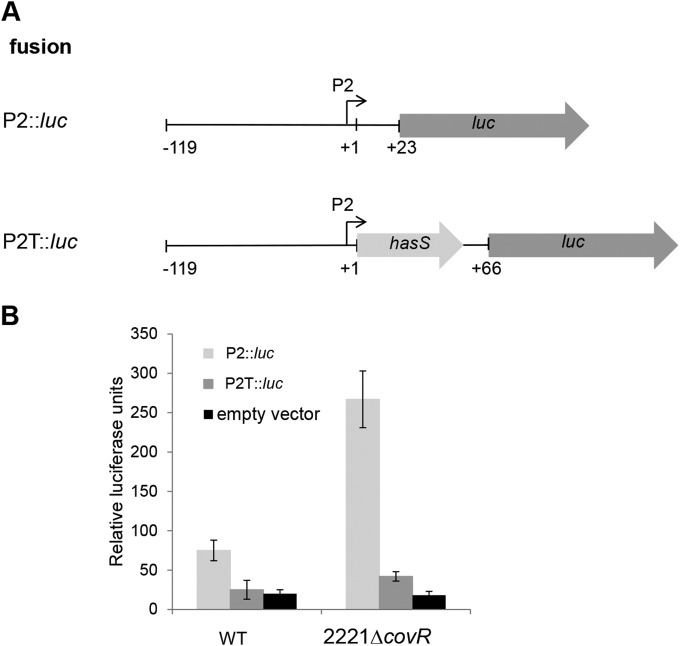
Second, we constructed transcriptional fusions of the P2 promoter with the luciferase (luc) reporter gene (Fig. 4A) and examined their activity in the WT and 2221ΔcovR mutant (Fig. 4B). The results showed that the transcriptional activity of the P2 promoter in the WT strain was 3 times higher than the activity obtained with the vector alone. The activity of the P2::luc fusion was 3.5-fold higher in the 2221ΔcovR strain than in the WT strain (Fig. 4B). The reporter activity slightly peaked in the late exponential phase and diminished rapidly during the stationary phase before largely disappearing in the late stationary phase in both strain backgrounds (see Fig. S3 in the supplemental material). Notably, a P2T::luc fusion, which consists of the P2 promoter followed by hasS and the terminator (Fig. 4A), produced the background level of luciferase activity in the WT, indicating that the predicted terminator terminates transcription under the conditions used in this experiment. The same construct was active in the 2221ΔcovR mutant (2-fold higher than the background level), suggesting that read-through transcription occurred in the absence of CovR (Fig. 4B). The luciferase assay data are consistent with CovR negatively regulating HasS transcription.
FIG 4.
Transcriptional analysis of the P2 promoter. (A) The schematic representations of the transcriptional fusions to the luc reporter. The fusion names, the fragment from the hasA upstream sequence that is carried by each plasmid (thin line), the promoters (bent arrows), hasS (light-gray arrow), and the luc gene (dark-gray arrow) are shown. Numbers indicate nucleotide positions relative to the HasS transcriptional start (+1). (B) Relative promoter activity determined by luciferase assay at mid-exponential growth phase for P2::luc and P2T::luc reporter plasmids in the WT and 2221ΔcovR strains. Luciferase activity expressed by each strain was determined as described in Materials and Methods. Each data point represents the average, and the error bars denote the standard deviations derived from at least 3 experiments from independent clones done in triplicate.
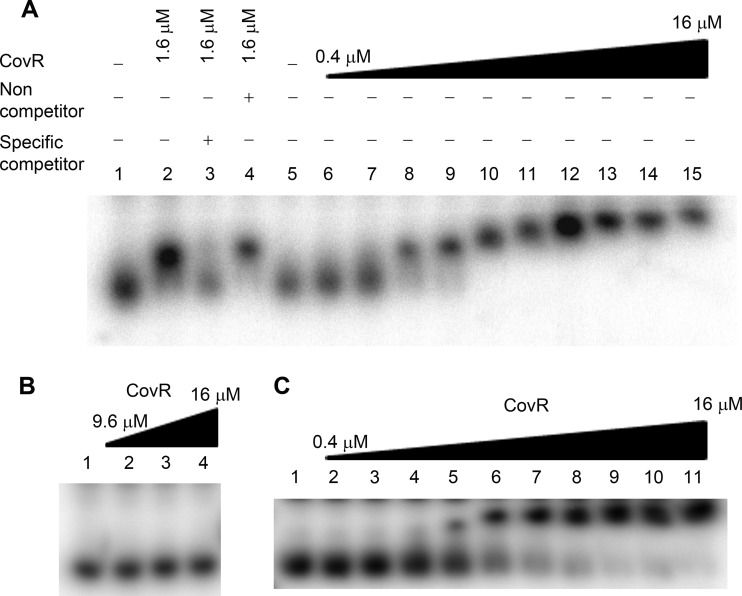
Lastly, we studied CovR protein binding to the P2 promoter region using an electrophoretic mobility shift assay (EMSA). We found that CovR binds to a 270-bp DNA region which carries the P2 promoter, and binding is dependent upon the concentration of CovR (Fig. 5A). As seen in Fig. 5A, the binding was lost when CovR was preincubated with the specific competitor. Furthermore, the binding was specific, since CovR failed to bind a 160-bp DNA fragment amplified from an unrelated mga promoter region (Fig. 5B). Similar to the hasS genomic locus, the mga promoter region is also AT rich.
FIG 5.
CovR binds specifically to the P2 promoter region. Gel mobility shift assay of increasing concentration of CovR binding to labeled DNA fragments (1.4 μM) from P2 (A), mga (B), and P2Δ42T (C) promoter regions. (A) The gel mobility shift assay was carried out in the presence (+) or absence (–) of specific competitor (85 μM P2 promoter DNA fragment) or nonspecific competitor (100 μM mga promoter DNA fragment). Lanes 1 and 5, probe alone; lanes 2 to 4 and 9, 1.6 μM CovR; lane 6, 0.4 μM CovR; lane 7, 0.8 μM CovR; lane 8, 1.2 μM CovR; lane 10, 3.2 μM CovR; lane 11, 4.8 μM CovR; lane 12, 6.4 μM CovR; lane 13, 9.6 μM CovR; lane 14, 12.8 μM CovR; lane 15, 16 μM CovR. (B) Lane 1, probe alone; lane 2, 9.6 μM CovR; lane 3, 12.8 μM CovR; lane 4, 16 μM. (C) Lane 1, probe alone; lane 2, 0.4 μM CovR; lane 3, 0.8 μM CovR; lane 4, 1.2 μM CovR; lane 5, 1.6 μM CovR; lane 6, 3.2 μM CovR; lane 7, 4.8 μM CovR; lane 8, 6.4 μM CovR; lane 9, 9.6 μM CovR; lane 10, 12.8 μM CovR; lane 11, 16 μM CovR.
As it has been suggested by Federle and Scott (19), CovR binds to the ATTARA consensus sequence. Examination of the P2 promoter region shows two putative consensus CovR-binding sequences. The first site is located 17 bp upstream of the extended −10 box of P2, and the second site is present in the HasS terminator (Fig. 1A). To investigate whether the HasS terminator region contains CovR-binding sequences, we introduced a deletion (Δ42T) in the P2 promoter DNA fragment. As seen in Fig. 5C, binding of CovR to this probe requires a higher concentration of the protein relative to binding to the WT probe, suggesting that the affinity of CovR for the WT probe is higher. This observation implies that the terminator has the CovR-binding sequences. Taken together, our data indicate that CovR binds the P2 promoter and represses HasS transcription.
Detection of read-through transcription from HasS into hasA.
We analyzed whether there could be read-through from the P2 promoter into the downstream hasA gene. This was tested using RT-PCR using a reverse primer (primer R in Fig. 6A) embedded within hasA and a series of four forward primers located at increasing distances from the hasA promoter (primers F1, F2, F3, and F4 in Fig. 6A), where F1 is located downstream of the P1 promoter, F2 is located between the P2 and P1 promoters, and F3 and F4 are located upstream of the P2 promoter. Surprisingly, PCR products of the expected sizes were obtained for all four PCRs using cDNA synthesized from MGAS2221 RNA as the template (Fig. 6B). No PCR products were observed in reactions that used template generated from control reactions with no reverse transcriptase (−RT). Thus, the data indicate that (i) there is an additional promoter upstream of the P2 and (ii) at least some of the HasS transcripts are being cotranscribed with hasA.
The results of analysis of transcriptional reporters prompted us to test whether CovR effects the read-through transcription. We determined transcription of the read-through region in the WT strain and the 2221ΔcovR mutant using qRT-PCR analysis. The 2221ΔcovR mutant has a 5-fold increase in read-through transcript relative to the WT strain (Fig. 6C). This result is in agreement with the enhanced activity of the P2 promoter in this mutant.
Next, we investigated whether elevated capsule production in the 2221ΔP2ΔhasS, 2221Δ42T, and 2221Δ63T strains is due to increased read-through transcription to the downstream hasA or rather results from deletion of CovR-binding sequences present in the terminator region. To test this hypothesis, we determined the read-through transcription in the WT, 2221ΔP2ΔhasS, and 2221Δ63T strains (Fig. 6C). We found that the 2221Δ63T mutant produced 2.5 times more read-through transcript than the WT, which is consistent with a lack of the terminator in this mutant. In contrast, 2221ΔP2ΔhasS produced 2 times less read-through transcript than the WT, indicating that the deletion of the P2 promoter decreases read-through transcription. Detection of read-through transcription in the 2221ΔP2ΔhasS mutant further confirms the presence of an additional promoter upstream of the P2 promoter. Our data also indicate that the phenotype of this mutant is not due to a positive effect of terminator deletion on the read-through transcription. The results suggest that the deleted region has regulatory elements, CovR-binding sequences, that contribute to capsule transcription.
The P1 promoter is not active in strain MGAS2221.
To investigate the contribution of the P1 promoter to transcription of the hasABC operon, we constructed two mutants. In the MGAS2221 parental isolate and the 2221ΔcovR mutant, we mutated the −10 box of the P1 promoter, creating the 2221P1− and 2221ΔcovRP1− mutants (Fig. 2A). The mutation changes the −10 sequence from TAGAAT to TCTAGA. As we demonstrated above, this mutation completely abrogates transcription from the P1 promoter (see Fig. S1B in the supplemental material). Remarkably, we found that the 2221P1− mutant had no significant defect in capsule production (Fig. 2B), and the 2221ΔcovRP1− mutant showed a significant decrease in capsule expression, producing 10 times less capsule than the 2221ΔcovR mutant (Fig. 2C). This result indicates that in the MGAS2221 strain, the P1 promoter is not active, probably due to negative regulation of the CovR/S system, and the genes in the operon are transcribed from P2 and the additional putative promoter upstream of P2. Moreover, comparison of capsule levels produced by the mutants revealed that the 2221ΔcovRP1− mutant expressed 2 times more capsule than the 2221P1− mutant (see Fig. S4 in the supplemental material), which is in agreement with increased transcription from the P2 promoter in the absence of CovR.
Capsule expression is controlled by the putative promoter upstream of the P2 promoter in MGAS2221.
To examine the contribution of the P2 promoter to transcription of the hasABC operon, we deleted the P2 promoter in two strain backgrounds, 2221P1− and 2221ΔcovRP1−, creating the 2221ΔP2P1− and 2221ΔcovRΔP2P1− mutants (Fig. 2A). Surprisingly, we found that both mutants had a slight increase (1.7-fold) in capsule expression relative to that of the 2221P1− and 2221ΔcovRP1− strains (Fig. 2B and C; see also Fig. S4 in the supplemental material), indicating a minor role of the P2 promoter in the hasABC operon expression. Together, our data suggest that the putative promoter upstream of P2 controls capsule expression in the MGAS2221 strain. The increase in capsule expression in both mutants might indicate that the RNA product from the HasS region represses capsule expression.
To further investigate the role of the terminator in blocking transcription from the upstream promoters, we deleted the terminator in the 2221ΔcovRP1− strain, creating the 2221ΔcovRΔ42TP1− mutant (Fig. 2A). The 2221ΔcovRP1− strain was selected for mutagenesis, because in this genetic background, the terminator function in regulation of capsule operon transcription is not masked by repressive activity of CovR and transcription from the active P1 promoter. We observed a slight increase (1.5-fold) in capsule expression relative to that of the parent strain (Fig. 2B; see also Fig. S4 in the supplemental material). Thus, our data indicate that in the MGAS2221 strain, the terminator is not effective in vivo.
Genetic diversity in the HasS region.
Recent whole-genome analysis of GAS (10) reported that serotypes of M4 and M22 strains, including two M4 serotype genomes available in the NCBI database, MGAS10750 and GA40634 (accession numbers CP000262.1 and AURU01000008, respectively), lack a 3,980-bp fragment carrying the hasABC operon. The alignment of the DNA regions flanking the 3,980-bp locus indicates that the genomic arrangements of two reference M4-serotype strains are similar to those of four sequenced genomes of Streptococcus dysgalactiae subsp. equisimilis (accession numbers HE858529.1, CP002215.1, AP010935.1, and AP011114.1), the closest known genetic relatives of GAS. Interestingly, the flanking regions of M4 serotype strains are more homologous to GAS genomes, with an average identity of 99% for upstream and downstream flanks, than to S. dysgalactiae genomes, with average identities of 79% and 97% (see Fig. S5 in the supplemental material). These observations suggest that a 3,980-bp fragment carrying hasS and the hasABC operon has been acquired by GAS during its separate evolution and has become widely distributed among the strains.
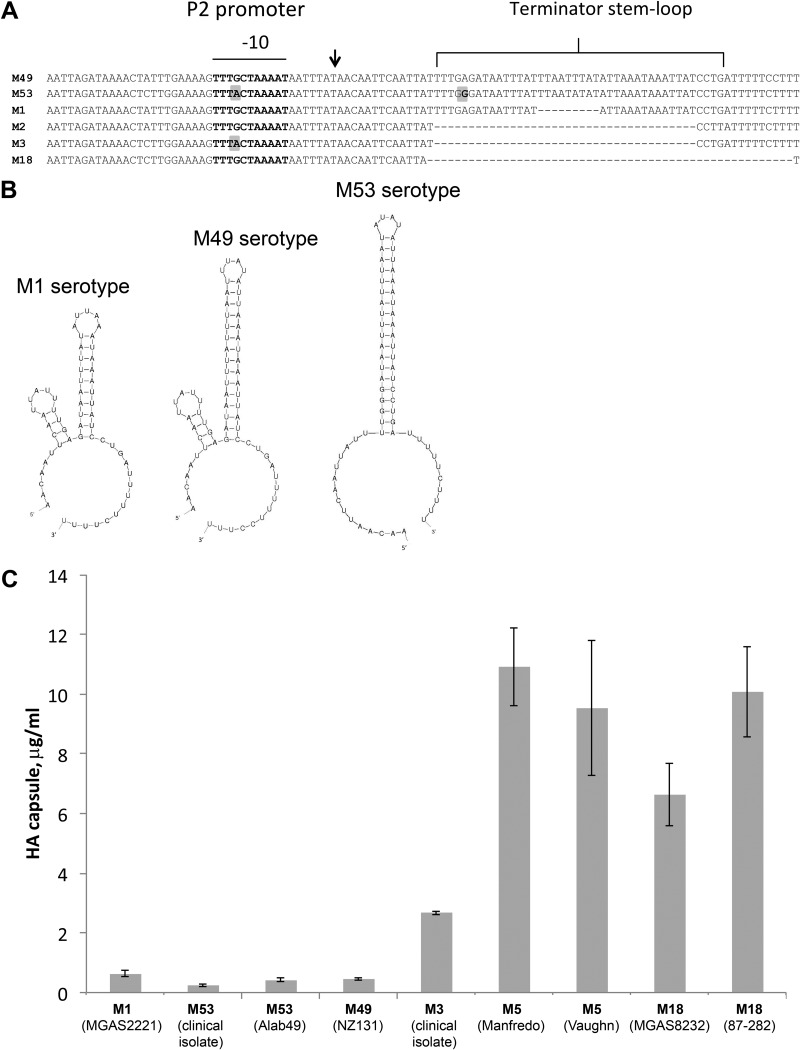
Sequence analysis of the hasA upstream region in 44 GAS genomes available in the NCBI database revealed a high level of polymorphism in the hasS sequence (see Table S4 in the supplemental material). We found that 12 GAS strains display a nucleotide change in a TG motif of the extended −10 sequence of the P2 promoter (Fig. 7A). The mutation changes TTTGCTAAAAT to TTTACTAAAAT and might result in the differential expression of HasS between strains. Furthermore, we observed that the stem-loop structure of the HasS intrinsic terminator is 34 bp long in 20 GAS genomes (M12, M24, M28, M49, M59, and M66 serotypes) (Fig. 7A and B; see also Table S4 in the supplemental material). Seven GAS strains (M6, M53, and M41.2 serotypes) have an SNP which would result in a more stable terminator stem-loop of 42 bp long (Fig. 7A and B; see also Table S4 in the supplemental material). In all M1 serotype strains, the stem-loop is 25 bp long due to a deletion of 9 nt (Fig. 7A and B). Remarkably, in 10 strains (M2, M3, M5, M12, M14, and M18 serotypes), the stem-loop is completely deleted (Fig. 7A; see also Table S4 in the supplemental material). Four strains (M5 and M6 serotypes) contain an IS1239 insertion element located immediately downstream of the HasS terminator sequence and upstream of the P1 promoter (see Table S4 in the supplemental material). Finally, in three M6 serotype strains, the hasS sequence contains three 29-nt tandem repeats (see Fig. S6 in the supplemental material).
FIG 7.
Genetic diversity in the terminator region correlates with HA capsule expression. (A) Sequence alignment of the hasS region from serotypes M1, M2, M3, M18, M49, and M53. The extended −10 promoter region is marked with a horizontal line. The terminator hairpin is highlighted by a bracket. Vertical arrow denotes the HasS transcriptional start site. Gray boxes indicate nucleotide changes in the promoter and terminator sequence regions. (B) The secondary structures of HasS from serotype M1, M49, and M53 were predicted using mfold analysis (43). (C) Production of HA capsule by different GAS serotypes. Capsule amount measured in the strains harvested at the mid-exponential growth phase. The experiments were performed in triplicate, and mean values ± SDs are shown.
Genetic diversity in the terminator region correlates with capsule expression.
It has been previously reported that M3 serotype strains produce greater capsule levels than M1 isolates (25) and M18 serotype strains display highly encapsulated phenotypes (50, 51). To assess whether genetic variability found in the terminator region correlates with capsule expression, we compared HA capsule levels in the GAS isolates harboring deleted terminator (M3, M5, and M18 isolates), with the GAS strains harboring the weak terminator (M1 strain) and the strong terminator (M49 and M53 strains). Only strains carrying WT covRS were analyzed to rule out a confounding influence from the transcriptional regulator on the data. We found that M3, M5, and M18 isolates produced greater capsule concentration than M1, M49, and M53 strains (Fig. 7C). In contrast, capsule expression in M49 and M53 strains was lower than in the M1 strain (Fig. 7C). Hence, the differences in capsule concentration produced by GAS might be associated with sequence variation in the HasS terminator region.
Deletion of the HasS terminator affects virulence of GAS.
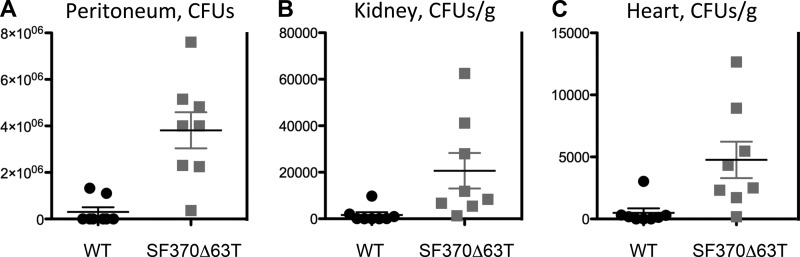
Since the HasS genomic regions from different GAS strains show variability in the terminator sequence, we analyzed the importance of this region for virulence. The experiments were conducted with the SF370 strain, which displays relatively low virulence in mice and is also an M1 serotype (52). Similar to the 2221Δ63T strain, the SF370Δ63T mutant produced 4-fold increases in capsule expression relative to that of the SF370 strain. We used GAS intraperitoneal injections to compare dissemination of the mutant in kidney, heart, and peritoneum with that of the SF370 strain in a mouse model of invasive infection (53). Ten hours after inoculation, the mutant-infected mice showed a significantly higher bacterial count than the SF370 strain-infected mice in all analyzed organs (Fig. 8).
FIG 8.
Viable bacteria recovered from organs of BALB/c mice at 10 h postinfection. Organs from which bacteria were recovered include the peritoneum (A), kidney (B), and heart (C). Mice were infected intravenously with either the WT (strain SF370) or the SF370Δ63T mutant. Black circles represent WT-infected mice (n = 8), and gray boxes represent SF370Δ63T mutant-infected mice (n = 8). The horizontal bar is the mean of the eight mice per treatment group; error bars represent standard errors of the means. P values were determined using an unpaired t test: P = 0.0006 for peritoneum, P = 0.027 for kidney, and P = 0.014 for heart.
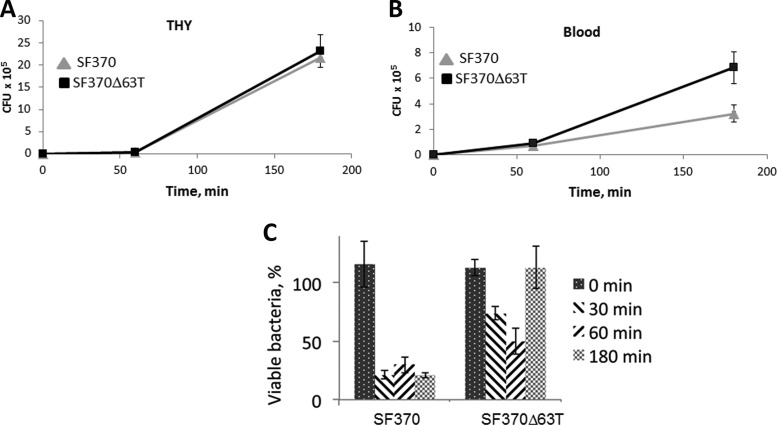
In order to better understand the mechanism behind enhanced virulence of the mutant, we compared the mutant and the SF370 strain growth in human blood. We found that the deletion of the terminator does not affect bacterial growth in THY broth (Fig. 9A). In contrast, the mutant grew more rapidly in human blood than the SF370 strain (Fig. 9B). To determine whether the fast growth of the mutant in blood was due to less efficient killing of the mutant by PMNs, we analyzed survival of the SF370 strain and the mutant in a human neutrophil bactericidal assay. We discovered that deletion of the terminator sequence makes the bacteria significantly more resistant to neutrophil killing (Fig. 9C). In summary, our experiments demonstrate that the HasS terminator region negatively regulates GAS virulence.
FIG 9.
Deletion of the HasS terminator affects growth of bacteria in human blood and phagocytosis by PMNs. (A) Growth of the WT (strain SF370) and the SF370Δ63T mutant in THY broth; (B) growth of the WT (strain SF370) and the SF370Δ63T mutant in human blood; (C) percentage of GAS survival following 0, 30, 60, and 180 min of incubation with PMNs relative to untreated bacteria incubated with HBSS. The percent survival was calculated using the formula (CFUn/CFU0) × 100, where CFU0 is the number of viable bacteria incubated with HBSS and CFUn is the number of surviving bacteria after incubation with PMNs. The experiments with whole blood/PMNs from three separate blood donors were performed. Panels A, B, and C show the results of one representative donor (means ± SDs of triple measurements from two independently growing clones).
DISCUSSION
Invasive bacterial infections represent an important public health problem, because they are associated with high morbidity and mortality in humans. It is widely accepted that most GAS strains causing invasive infections arise from pharyngeal and other superficial infections (54). Invasive GAS infections are often associated with spontaneous mutations in the genes encoding the CovR/S system, resulting in massive HA capsule production and hypervirulence of GAS (7, 22, 55). Although GAS requires the capsule for protection against the host immune response during infection, capsule biosynthesis also represents a great metabolic burden on the cell, potentially slowing cell growth and division. Thus, capsule expression requires effective and sensitive control mechanisms to avoid excessive capsule overproduction. In this work, we analyzed the upstream region of the capsule operon promoter and identified and characterized a transcriptionally active region comprised of a novel noncoding sRNA, HasS. HasS is present in all available genomes of GAS except the M4 serotype, which does not have the hasABC operon also. Although other streptococcal species like Streptococcus equi, Streptococcus uberis, Streptococcus parauberis, and Streptococcus iniae harbor the capsule biosynthesis operon, they lack HasS, indicating that this sRNA is GAS specific.
Recent studies employing computation prediction methods and intergenic tiling array approaches discovered multiple candidate sRNAs in the GAS genome (56–58). However, HasS was missed from these studies, probably due to its low expression in the bacteria. This implies that more sensitive methods of genome-wide analysis are required for a comprehensive identification of sRNAs in GAS. While the numerous sRNAs were predicted in GAS genomes, only some of them were experimentally validated, and a very limited number were functionally studied. The best-characterized GAS sRNA is FasX, which is involved in the regulation of two important virulence factors, the collagen-binding pilus (59) and streptokinase (60, 61). The regulation of key virulence factors by FasX demonstrates that sRNAs play a prominent role in GAS pathogenesis.
Previously, it has been shown that CovR represses hasABC transcription by binding to an extending region comprising five AT-rich sequences located upstream and downstream of the P1 promoter (19). The close location of hasS to hasA in the majority of GAS strains suggests that they might share the same regulatory region, which coordinates their expression in response to environmental signals. Federle and Scott (19) did not search for CovR-binding sites in the HasS genomic region, possibly due to the fact that they used the type M6 GAS strain JRS4, which contains the IS1239 transposase inserted between hasS and the P1 promoter. We demonstrated that HasS transcription initiated from the P2 promoter is also negatively regulated by CovR. The gel shift assay using the DNA fragment from the HasS genomic region indicates the presence of CovR-binding sequences within the terminator region of HasS. These findings show that CovR regulation of the P2 promoter is direct.
We found that in the absence of the P1 promoter, the MGAS2221 strain produces the normal level of capsule, indicating that this promoter is fully repressed by CovR and does not contribute to capsule operon transcription, under the conditions tested. This finding is in line with our observation that the HasS terminator does not work efficiently, allowing read-through transcription into hasA. Furthermore, our RT-PCR experiments and analyses of read-through transcription in the P2 deletion mutant and capsule production by the mutants with inactive P1 and P2 promoters indicate that there is another promoter upstream of the P2 promoter. In the MGAS2221 strain, this putative promoter controls the hasA transcription, allowing the pathogen to maintain a low basal level of capsule in the presence of active CovR. Consistent with this notion, deletion of the terminator in the MGAS2221 strain leads to release of CovR repression, enhanced read-through, and hasABC transcription and capsule production.
Intriguingly, deletion of the P2 promoter also has a positive effect on capsule expression, suggesting that the RNA product from this region, HasS may negatively control capsule expression. It is possible that HasS acts as a trans-acting regulatory sRNA by base pairing with the hasA transcript, leading to its degradation. Another hypothesis is that HasS may regulate capsule expression indirectly by affecting other targets. It has been recently reported that the insertion of a transposon at 9 nt upstream of the P2 −10 promoter element resulted in the increased capsule production at 37°C in the CovR deletion mutant of GAS HSC5 (M14 serotype) (62). This report further suggests activity of the HasS region at the RNA level. As the HasS region contains CovR-binding sites, the phenotype of our P2 promoter deletion mutants could also be a result of elevated P1 promoter activity caused by decreased binding of CovR to this DNA locus. Thus, the regulatory region of the capsule operon is much more complex than previously envisioned, and it remains to be determined what the exact roles of CovR, HasS, the terminator, and the additional promoter upstream of P2 are in the regulation of capsule biosynthesis.
Current evidence is in favor of a hypothesis that GAS invasive strains arise during minor skin or throat infections as a result of the accumulation of spontaneous mutations in critical regulatory regions. A striking and unexpected feature of this novel regulatory region is a remarkable variability among GAS clinical isolates. Our analysis of the sequences of GAS strains of different serotypes identified single nucleotide polymorphisms, deletions, and insertions clustered in the HasS region. These polymorphisms are likely to affect the expression and biological function of HasS. Notably, a high degree of divergence was detected in the terminator region, suggesting that this regulatory element is under strong selective pressure in GAS. The terminator sequence is found frequently weakened by mutations/deletions or completely deleted. Moreover, among six analyzed M12 serotype strains, four strains have lost the terminator, suggesting that this deletion is a recent evolutionarily event. In this study, we compared virulence of the serotype M1 SF370 strain with that of the mutant lacking the terminator. We observed that the mutant has increased growth in human blood, is highly resistant to neutrophil killing, and is considerably more virulent in the mouse model of GAS invasive disease than the WT strain. Thus, our data indicate that this regulatory region significantly contributes to GAS virulence.
The observed genetic diversity in the HasS region might in part explain the wide phenotypic variation in encapsulation and virulence among GAS clinical isolates. Our data and published observations (25) suggest that deletion of the terminator sequences correlates with enhanced capsule expression in GAS isolates. Thus, our results suggest that naturally occurring mutations in the terminator region enhance GAS virulence and predispose the pathogen to switch from superficial to invasive infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R21AI113253 from the NIAID and P20 RR020171 from the National Center for Research Resources (to N.K.), by NIH grant R01 AI087747 from the NIAID (to P.S.), and by Postdoctoral Fellowship 13POST16820024 from the American Heart Association (to M.F.). We also acknowledge the University of Kentucky Viral Production Core, which is partially supported by grant P20GM103486 from the National Institute of General Medical Sciences.
We thank Mark McAlpine for the technical assistance with mouse infections. We thank Kevin S. McIver of the University of Maryland for providing pKSM720 and pEU7534 vectors and the JRS4-PolHis strain, Michael R. Wessels of Harvard Medical School for providing the 87-282 and Vaughn strains, Michael McShan of the University of Oklahoma for providing the Manfredo strain, Debra E. Bessen of New York Medical College for providing the Alab49 strain, and Benfang Lei of Montana State University for providing the pBBL740 plasmid. We thank Daniel Nelson of the University of Maryland for the kind gift of PlyC lysin.
Footnotes
Published ahead of print 6 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02035-14.
REFERENCES
- 1.Stollerman GH, Dale JB. 2008. The importance of the group a Streptococcus capsule in the pathogenesis of human infections: a historical perspective. Clin. Infect. Dis. 46:1038–1045. 10.1086/529194. [DOI] [PubMed] [Google Scholar]
- 2.Moses AE, Wessels MR, Zalcman K, Alberti S, Natanson-Yaron S, Menes T, Hanski E. 1997. Relative contributions of hyaluronic acid capsule and M protein to virulence in a mucoid strain of the group A Streptococcus. Infect. Immun. 65:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wessels MR, Bronze MS. 1994. Critical role of the group A streptococcal capsule in pharyngeal colonization and infection in mice. Proc. Natl. Acad. Sci. U. S. A. 91:12238–12242. 10.1073/pnas.91.25.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wessels MR, Goldberg JB, Moses AE, DiCesare TJ. 1994. Effects on virulence of mutations in a locus essential for hyaluronic acid capsule expression in group A streptococci. Infect. Immun. 62:433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. 1991. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc. Natl. Acad. Sci. U. S. A. 88:8317–8321. 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashbaugh CD, Warren HB, Carey VJ, Wessels MR. 1998. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Invest. 102:550–560. 10.1172/JCI3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole JN, Pence MA, von Kockritz-Blickwede M, Hollands A, Gallo RL, Walker MJ, Nizet V. 2010. M protein and hyaluronic acid capsule are essential for in vivo selection of covRS mutations characteristic of invasive serotype M1T1 group A Streptococcus. mBio 1(4):e00191-10. 10.1128/mBio.00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cywes C, Stamenkovic I, Wessels MR. 2000. CD44 as a receptor for colonization of the pharynx by group A Streptococcus. J. Clin. Invest. 106:995–1002. 10.1172/JCI10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schrager HM, Alberti S, Cywes C, Dougherty GJ, Wessels MR. 1998. Hyaluronic acid capsule modulates M protein-mediated adherence and acts as a ligand for attachment of group A Streptococcus to CD44 on human keratinocytes. J. Clin. Invest. 101:1708–1716. 10.1172/JCI2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores AR, Jewell BE, Fittipaldi N, Beres SB, Musser JM. 2012. Human disease isolates of serotype m4 and m22 group a Streptococcus lack genes required for hyaluronic acid capsule biosynthesis. mBio 3(6):e00413–12. 10.1128/mBio.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeAngelis PL, Papaconstantinou J, Weigel PH. 1993. Molecular cloning, identification, and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J. Biol. Chem. 268:19181–19184. [PubMed] [Google Scholar]
- 12.Ashbaugh CD, Alberti S, Wessels MR. 1998. Molecular analysis of the capsule gene region of group A Streptococcus: the hasAB genes are sufficient for capsule expression. J. Bacteriol. 180:4955–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JN, Aziz RK, Kuipers K, Timmer AM, Nizet V, van Sorge NM. 2012. A conserved UDP-glucose dehydrogenase encoded outside the hasABC operon contributes to capsule biogenesis in group A Streptococcus. J. Bacteriol. 194:6154–6161. 10.1128/JB.01317-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dougherty BA, van de Rijn I. 1994. Molecular characterization of hasA from an operon required for hyaluronic acid synthesis in group A streptococci. J. Biol. Chem. 269:169–175. [PubMed] [Google Scholar]
- 15.Bernish B, van de Rijn I. 1999. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J. Biol. Chem. 274:4786–4793. 10.1074/jbc.274.8.4786. [DOI] [PubMed] [Google Scholar]
- 16.Federle MJ, McIver KS, Scott JR. 1999. A response regulator that represses transcription of several virulence operons in the group A Streptococcus. J. Bacteriol. 181:3649–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath A, DiRita VJ, Barg NL, Engleberg NC. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin JC, Wessels MR. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol. Microbiol. 30:209–219. 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 19.Federle MJ, Scott JR. 2002. Identification of binding sites for the group A streptococcal global regulator CovR. Mol. Microbiol. 43:1161–1172. 10.1046/j.1365-2958.2002.02810.x. [DOI] [PubMed] [Google Scholar]
- 20.Miller AA, Engleberg NC, DiRita VJ. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol. Microbiol. 40:976–990. 10.1046/j.1365-2958.2001.02441.x. [DOI] [PubMed] [Google Scholar]
- 21.Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. U. S. A. 99:13855–13860. 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2:e5. 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia AF, Abe LM, Erdem G, Cortez CL, Kurahara D, Yamaga K. 2010. An insert in the covS gene distinguishes a pharyngeal and a blood isolate of Streptococcus pyogenes found in the same individual. Microbiology 156:3085–3095. 10.1099/mic.0.042614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyoshi-Akiyama T, Ikebe T, Watanabe H, Uchiyama T, Kirikae T, Kawamura Y. 2006. Use of DNA arrays to identify a mutation in the negative regulator, csrR, responsible for the high virulence of a naturally occurring type M3 group A streptococcus clinical isolate. J. Infect. Dis. 193:1677–1684. 10.1086/504263. [DOI] [PubMed] [Google Scholar]
- 25.Cao TN, Liu Z, Cao TH, Pflughoeft KJ, Trevino J, Danger JL, Beres SB, Musser JM, Sumby P. 2014. Natural disruption of two regulatory networks in serotype M3 group a streptococcus isolates contributes to the virulence factor profile of this hypervirulent serotype. Infect. Immun. 82:1744–1754. 10.1128/IAI.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658–4663. 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771–782. 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 28.Trevino J, Perez N, Ramirez-Pena E, Liu Z, Shelburne SA, III, Musser JM, Sumby P. 2009. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect. Immun. 77:3141–3149. 10.1128/IAI.01560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoff JS, DeWald M, Moseley SL, Collins CM, Voyich JM. 2011. SpyA, a C3-like ADP-ribosyltransferase, contributes to virulence in a mouse subcutaneous model of Streptococcus pyogenes infection. Infect. Immun. 79:2404–2411. 10.1128/IAI.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caparon MG, Scott JR. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556–586. 10.1016/0076-6879(91)04028-M. [DOI] [PubMed] [Google Scholar]
- 31.Trevino J, Liu Z, Cao TN, Ramirez-Pena E, Sumby P. 2013. RivR is a negative regulator of virulence factor expression in group A Streptococcus. Infect. Immun. 81:364–372. 10.1128/IAI.00703-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinkel TL, McIver KS. 2008. CcpA-mediated repression of streptolysin S expression and virulence in the group A streptococcus. Infect. Immun. 76:3451–3463. 10.1128/IAI.00343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gusa AA, Gao J, Stringer V, Churchward G, Scott JR. 2006. Phosphorylation of the group A streptococcal CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. J. Bacteriol. 188:4620–4626. 10.1128/JB.00198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Opdyke JA, Scott JR, Moran CP., Jr 2001. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol. Microbiol. 42:495–502. 10.1046/j.1365-2958.2001.02657.x. [DOI] [PubMed] [Google Scholar]
- 35.Almengor AC, Walters MS, McIver KS. 2006. Mga is sufficient to activate transcription in vitro of sof-sfbX and other Mga-regulated virulence genes in the group A Streptococcus. J. Bacteriol. 188:2038–2047. 10.1128/JB.188.6.2038-2047.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gusa AA, Scott JR. 2005. The CovR response regulator of group A streptococcus (GAS) acts directly to repress its own promoter. Mol. Microbiol. 56:1195–1207. 10.1111/j.1365-2958.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- 37.Wade KH, Schyns G, Opdyke JA, Moran CP., Jr 1999. A region of sigmaK involved in promoter activation by GerE in Bacillus subtilis. J. Bacteriol. 181:4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941–950. 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 39.Schrager HM, Rheinwald JG, Wessels MR. 1996. Hyaluronic acid capsule and the role of streptococcal entry into keratinocytes in invasive skin infection. J. Clin. Invest. 98:1954–1958. 10.1172/JCI118998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyum A. 1968. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Invest. Suppl. 97:77–89. [PubMed] [Google Scholar]
- 41.Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, Long RD, Dorward DW, Gardner DJ, Lina G, Kreiswirth BN, DeLeo FR. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761–1770. 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 42.Voyich JM, Sturdevant DE, Braughton KR, Kobayashi SD, Lei B, Virtaneva K, Dorward DW, Musser JM, DeLeo FR. 2003. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. U. S. A. 100:1996–2001. 10.1073/pnas.0337370100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415. 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goupil-Feuillerat N, Corthier G, Godon JJ, Ehrlich SD, Renault P. 2000. Transcriptional and translational regulation of alpha-acetolactate decarboxylase of Lactococcus lactis subsp. lactis. J. Bacteriol. 182:5399–5408. 10.1128/JB.182.19.5399-5408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helmann JD. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351–2360. 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabelnikov AG, Greenberg B, Lacks SA. 1995. An extended -10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J. Mol. Biol. 250:144–155. 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 47.Voskuil MI, Chambliss GH. 1998. The −16 region of Bacillus subtilis and other Gram-positive bacterial promoters. Nucleic Acids Res. 26:3584–3590. 10.1093/nar/26.15.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Churchward G, Bates C, Gusa AA, Stringer V, Scott JR. 2009. Regulation of streptokinase expression by CovR/S in Streptococcus pyogenes: CovR acts through a single high-affinity binding site. Microbiology 155:566–575. 10.1099/mic.0.024620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts SA, Churchward GG, Scott JR. 2007. Unraveling the regulatory network in Streptococcus pyogenes: the global response regulator CovR represses rivR directly. J. Bacteriol. 189:1459–1463. 10.1128/JB.01026-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, Sylva GL, Sturdevant DE, Ricklefs SM, Porcella SF, Parkins LD, Beres SB, Campbell DS, Smith TM, Zhang Q, Kapur V, Daly JA, Veasy LG, Musser JM. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. U. S. A. 99:4668–4673. 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smoot JC, Korgenski EK, Daly JA, Veasy LG, Musser JM. 2002. Molecular analysis of group A Streptococcus type emm18 isolates temporally associated with acute rheumatic fever outbreaks in Salt Lake City, Utah. J. Clin. Microbiol. 40:1805–1810. 10.1128/JCM.40.5.1805-1810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venturini C, Ong CL, Gillen CM, Ben-Zakour NL, Maamary PG, Nizet V, Beatson SA, Walker MJ. 2013. Acquisition of the Sda1-encoding bacteriophage does not enhance virulence of the serotype M1 Streptococcus pyogenes strain SF370. Infect. Immun. 81:2062–2069. 10.1128/IAI.00192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voyich JM, Braughton KR, Sturdevant DE, Vuong C, Kobayashi SD, Porcella SF, Otto M, Musser JM, DeLeo FR. 2004. Engagement of the pathogen survival response used by group A Streptococcus to avert destruction by innate host defense. J. Immunol. 173:1194–1201. 10.4049/jimmunol.173.2.1194. [DOI] [PubMed] [Google Scholar]
- 54.Shea PR, Beres SB, Flores AR, Ewbank AL, Gonzalez-Lugo JH, Martagon-Rosado AJ, Martinez-Gutierrez JC, Rehman HA, Serrano-Gonzalez M, Fittipaldi N, Ayers SD, Webb P, Willey BM, Low DE, Musser JM. 2011. Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 108:5039–5044. 10.1073/pnas.1016282108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engleberg NC, Heath A, Miller A, Rivera C, DiRita VJ. 2001. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J. Infect. Dis. 183:1043–1054. 10.1086/319291. [DOI] [PubMed] [Google Scholar]
- 56.Patenge N, Billion A, Raasch P, Normann J, Wisniewska-Kucper A, Retey J, Boisguerin V, Hartsch T, Hain T, Kreikemeyer B. 2012. Identification of novel growth phase- and media-dependent small noncoding RNAs in Streptococcus pyogenes M49 using intergenic tiling arrays. BMC Genomics 13:550. 10.1186/1471-2164-13-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez N, Trevino J, Liu Z, Ho SC, Babitzke P, Sumby P. 2009. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLoS One 4:e7668. 10.1371/journal.pone.0007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tesorero RA, Yu N, Wright JO, Svencionis JP, Cheng Q, Kim JH, Cho KH. 2013. Novel regulatory small RNAs in Streptococcus pyogenes. PLoS One 8:e64021. 10.1371/journal.pone.0064021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z, Trevino J, Ramirez-Pena E, Sumby P. 2012. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus. Mol. Microbiol. 86:140–154. 10.1111/j.1365-2958.2012.08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392–406. 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 61.Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P. 2010. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol. Microbiol. 78:1332–1347. 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang SO, Wright JO, Tesorero RA, Lee H, Beall B, Cho KH. 2012. Thermoregulation of capsule production by Streptococcus pyogenes. PLoS One 7:e37367. 10.1371/journal.pone.0037367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crater DL, van de Rijn I. 1995. Hyaluronic acid synthesis operon (has) expression in group A streptococci. J. Biol. Chem. 270:18452–18458. 10.1074/jbc.270.31.18452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.