Abstract
P1 bacteriophages lysogenize bacteria as independent plasmid-like elements. We describe here a P1-like bacteriophage, RCS47, carrying a blaSHV-2 gene, isolated from a clinical strain of Escherichia coli from phylogroup B1, and we report the prevalence of P1-like prophages in natural E. coli isolates. We found that 70% of the sequence of RCS47, a 115-kb circular molecule, was common to the reference P1 bacteriophage under GenBank accession no. AF234172.1, with the shared sequences being 99% identical. RCS47 had acquired two main foreign DNA fragments: a 9,636-bp fragment mobilized by two IS26 elements containing a blaSHV-2 gene, and an 8,544-bp fragment mobilized by two IS5 elements containing an operon encoding a dimethyl sulfoxide reductase. The reference P1 prophage plasmid replication gene belonged to the IncY incompatibility group, whereas that of RCS47 was from an unknown group. The lytic capacity of RCS47 and blaSHV-2 gene transduction, through the lysogenization of RCS47 in the recipient E. coli strains, were not demonstrated. The prevalence of P1-like prophages in various animal and human E. coli strain collections, as determined by the PCR detection of repL, the lytic replication gene, was 12.6%. No differences in the prevalences of these prophages were found between extended-spectrum β-lactamase (ESBL)-producing and non-ESBL-producing strains (P = 0.69), but this prevalence was lower in phylogroup B2 than in the other phylogroups (P = 0.008), suggesting epistatic interactions between P1 family phages and the genetic background of E. coli strains. P1-like phages are part of the mobile elements that carry antibiotic resistance. The high prevalence of P1-like prophages suggests their role may be underestimated.
INTRODUCTION
The spread of antibiotic-resistant pathogens is becoming an extremely serious clinical and public health problem worldwide (1). There is currently a worrying increase in the resistance to expanded-spectrum cephalosporins, which are regularly used for the empirical treatment of infections caused by Enterobacteriaceae. Acquired resistance to expanded-spectrum cephalosporins is mediated principally by extended-spectrum β-lactamases (ESBLs), the most prevalent of which are the ESBLs of the CTX-M, TEM, and SHV families (2). The rapidly increasing recognition of clinical isolates containing ESBLs worldwide has led to a growing interest in studies of the genetic elements responsible for their emergence and spread. The dissemination of ESBLs can be explained by two principal phenomena: the spread of plasmids bearing antibiotic resistance genes between bacterial strains (3), and the spread of epidemic clones bearing plasmids carrying resistance genes (4). Little is known about the contribution of other mobile genetic elements, such as bacteriophages, to the transfer of antibiotic resistance. Recent reports suggest that the horizontal transfer of antibiotic resistance genes by phages is much more prevalent than was previously believed (5, 6). For example, two β-lactamase genes (blaTEM and blaCTX-M) and one gene encoding a penicillin-binding protein (mecA) have been detected in the bacteriophage DNA fraction of sewage, river water, and fecal waste from pigs, poultry, and cattle, suggesting that phages are reservoirs of resistance genes in the environment (7, 8). A blaCTX-M-10 gene linked to phage-related elements has been described, suggesting the possible transfer of this gene from the chromosome of Kluyvera spp. to a plasmid via bacteriophage-mediated transduction (9). Phages from various lysogenic bacteria, such as Pseudomonas aeruginosa, Salmonella enterica serovar Typhimurium, Clostridium difficile, and group A streptococcus, have been reported to mediate transduction with antibiotic resistance genes (10–13). During their lysogenic cycle, these temperate phages replicate while integrated into the host chromosome. Alternatively, P1 and the closely related P7 bacteriophages, which infect and lysogenize Escherichia coli and several other enteric bacteria, replicate in their hosts as independent low-copy-number plasmid-like elements. In the early 1970s, Smith (14) isolated a P7 bacteriophage that conferred ampicillin resistance to a natural isolate of E. coli. This resistance was due to the acquisition of a blaTEM-1 gene located in a Tn3 transposon (GenBank accession no. AF503408.1). However, despite their capacity to acquire and transfer multidrug resistance genes due to their recombination systems (15, 16), no P1 family bacteriophages carrying resistance determinant genes, such as ESBL genes, have since been characterized in clinical isolates.
We report here the complete sequence of a P1-like bacteriophage carrying a blaSHV-2 gene isolated from a clinical strain of E. coli during a comparative genomics project focusing on plasmids carrying ESBL-encoding genes (RepliColScope [www.agence-nationale-recherche.fr/en/anr-funded-project/]). We also determined the prevalence of P1-like bacteriophages in natural E. coli isolates.
MATERIALS AND METHODS
Case study bacterial strains.
Strain 725, an E. coli strain from phylogroup B1 and O8:H19 serotype that produces an SHV-2 ESBL, was isolated in 2002 in Paris from an adult patient with a urinary tract infection (17). The mobile element encoding the SHV-2 enzyme, RCS47, was extracted from strain 725 by use of a Large-Construct kit (Qiagen, Courtaboeuf, France) and transferred by electroporation into E. coli strain DH10B, as previously described (18), to yield the E. coli TR725 strain (19).
Sequencing and annotation of phage RCS47.
Phage DNA was purified from the electroporant TR725 with the Qiagen Large-Construct kit and was sequenced on a 454 GS-FLX instrument, with titanium chemistry. A 3-kb-insert mate-pair library was constructed according to the Roche 454 protocol. The generated reads had a mean length of 350 bp (40× coverage). They were assembled into a single continuous contig using the Newbler software (20). The Magnifying Genomes (MaGe) software was used for gene annotation and comparative analysis, as described elsewhere (21). The automatic annotation was validated manually with the MaGe interface (www.cns.fr/agc/microscope).
DNA sequence analysis.
The nucleotide and protein sequences were compared through BLAST queries against the GenBank databases.
Induction and lytic capacity experiments of phage RCS47.
Strains 725 and TR725 were cultured to the exponential growth phase at 37°C in LB (Luria-Bertani broth, also called lysogeny broth) (22). Bacterial suspensions were placed in a petri dish and irradiated with UV light for 25 s, with a 26-W germicidal UV light, as described by Kondo and Mitsuhashi (23). We then resuspended 3 ml of the induced culture in 30 ml of fresh LB and incubated the suspension for 2 h at 37°C with vigorous shaking. The cultures were centrifuged to remove bacterial cell debris, and the supernatants were passed through a filter with 0.22-μm pores and concentrated with a protein concentrator (100-kDa Amicon Ultra centrifugal filter units; Millipore, Bedford, MA) to yield a final volume of 0.3 ml (24). The same procedure was applied to the E. coli DH10B strain as a negative control. The phage suspensions obtained were stored at 4°C.
We checked that the suspension contained phage RCS47 DNA using PCR amplifications of the repL (the phage lytic replication gene), repA (the plasmid replication gene), and blaSHV-2 genes with the primers RepL-fw (5′-CCAATCAACCGTCGTTCGTG-3′) and RepL-rev (5′-TAAGCATATTTCCGCGCTGC-3′) for repL, RepA-RCS47-fw (5′-CAACGACGCAACCGAAGAA-3′) and RepA-RCS47-rev (5′-TACCTTGGCCGTCTCTTTGC-3′) for repA, and SHV-nestA (5′-TCGCCTGTGTATTATCTCCC-3′) and SHV-nestB (5′-GGTATCCCGCAGATAAATCA-3′) for blaSHV-2. We also checked that there was no bacterial chromosome present using PCR amplification of the TspE4C2.2 and yjaA chromosomal fragments present in the strains of phylogroups B1 (strain 725) and A1 (DH10B) of E. coli, respectively (25).
The lytic capacity of RCS47 was determined as described by Kutter (26), using E. coli strain K-12 (substrains MG1655 and MC1061) and five natural E. coli isolates of various phylogroups as sensitive strains. The cells in the exponential growth phase were embedded in LB top agar medium (0.7%) containing MC solution (10 mM MgCl2 and 5 mM CaCl2) overlaid on LB bottom agar. We spotted 10 μl of pure suspensions and 10-fold dilutions prepared from strains 725, TR725, and E. coli DH10B and 10 μl of a phage P1vir suspension, used as a positive control, on the surface of the plate. After overnight incubation at 37°C, the clear zones on the plate were considered to indicate the presence of a lytic phage.
To disprove that the suspension might contain colicins that killed the sensitive K-12 strains, the lytic capacity was also examined with suspensions treated with trypsin (250 μg/ml) for 20 min at 30°C (27).
Lysogenization assay.
Lysogenization was carried out as described by León et al. (28). Briefly, an overnight culture of the recipient strains (E. coli K-12 [MG1655, W1485, or MC1061], E. coli B, and as for the lytic capacity, five natural E. coli isolates of various phylogroups) in LB was centrifuged, and the cells were resuspended in an equivalent volume of MC solution. We then mixed 0.1 ml of the cell suspension with 0.1 ml of the RCS47 suspension obtained as described above. The mixture was then incubated for 30 min at 37°C and plated on LB agar supplemented with 1 μg/ml cefotaxime (CTX). We looked for CTX-resistant lysogenic colonies after 48 h of incubation at 37°C. We also assessed lysogenization by growing mixed cultures inoculated with strain 725 or TR725, both of which are tetracycline sensitive, and the tetracycline-resistant derivatives of E. coli W1485 or MC1061 in LB supplemented with CaCl2 (2.5 mM). The cultures were incubated overnight or for up to 48 h at room temperature. We then assessed growth directly on LB plates supplemented with CTX (1 µg/ml) and tetracycline (10 µg/ml).
Transmission electron microscopy.
Phage suspensions from both the 725 and TR725 strains prepared as described above were analyzed. The specimens were prepared for electron microscopy using the conventional negative-staining procedure. Drops of phage solutions (20 μl) were absorbed on Formvar carbon-coated grids for 5 min. The drops were then blotted and negatively stained with uranyl acetate (1%) for 1 min. The grids were then examined with a transmission electron microscope (Jeol JEM-1400) at 80 kV. Images were acquired using a digital camera (Gatan Orius) at ×40,000 magnification.
Prevalence of P1-like bacteriophages in E. coli populations.
We evaluated the prevalence of P1 bacteriophages in natural populations of E. coli by carrying out specific PCR assays to detect the presence of repL, the lytic replication gene of P1 family phages. These assays were carried out with consensus primers (see the primer sequences above) on 363 commensal and pathogenic strains from previously published and unpublished collections from various areas of France (17, 29–32). The 196 commensal strains originating from feces included 33 human and 163 animal strains: 147 of the animal strains were from mammals (107 wild animals, 36 farm animals, and 4 pets), and 16 were from birds (7 wild and 9 farmed). The 167 pathogenic strains, all of human origin, were isolated from patients with bacteremia (81 strains), urinary tract infections (68 strains), and other infections (18 strains), and they included 73 ESBL-producing strains. The type of repA, the plasmid replication gene, was determined in the strains positive for repL, with the repA primers described above and the IncY primers as described by Carattoli et al. (33). In addition, 18 strains of serogroup O8 and phylogroup B1 producing ESBL (7 SHV and 11 CTX-M) were screened (34, 35).
Statistical analysis.
For statistical analysis, we carried out Fisher's exact tests, and P values of <0.05 were considered significant.
Nucleotide sequence accession number.
The complete nucleotide sequence of RCS47 was deposited in GenBank under the accession no. FO818745.
RESULTS AND DISCUSSION
P1-like bacteriophage carrying an ESBL gene.
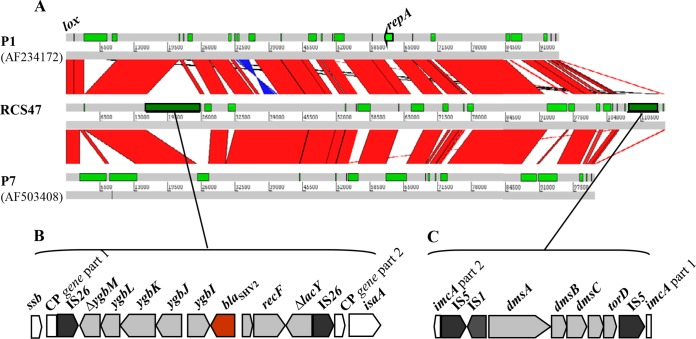
Plasmids are the main vectors of antimicrobial drug resistance. PCR-based typing methods are widely used for the identification and classification of plasmids, which is crucial for increasing our understanding of the epidemiology of antibiotic resistance emergence and dissemination (33). However, this technique excludes a significant fraction of plasmid groups. To obtain insight into the diversity of these plasmids, all the nontypeable plasmids of a well-characterized collection of strains (17, 19) were included in our project of comparative genomics of plasmids carrying ESBL-encoding genes, RepliColScope (www.agence-nationale-recherche.fr/en/anr-funded-project/). BLAST comparisons of the protein sequences against the GenBank databases showed that one of these plasmids, RCS47, displayed a high level of identity to the P1 bacteriophage genomes. The complete sequence of RCS47 was found to be 115,154 bp long and harbors 134 open reading frames, accounting for 88.2% of the genome (see Table S1 in the supplemental material). The G+C content of the RCS47 DNA is 49.35%. We found that 70% and 66% of the RCS47 sequence are common to the sequences of phage P1 c1-100 mod749::IS5 (GenBank accession no. AF234172.1) described by Lobocka et al. and Yarmolinsky (15, 16) and phage P7, a relative of P1 (GenBank accession no. AF503408.1), respectively. The shared sequences are 99% identical (Fig. 1A). P1 bacteriophages and their relatives lysogenize their hosts as autonomous plasmid-like elements. We therefore represented the RCS47 genome as a circle, with the site-specific recombination site lox assigned to the zero position (Fig. 2) (15).
FIG 1.
(A) Representation of the synteny between bacteriophage RCS47, P1 (GenBank accession no. AF234172.1), and P7 (GenBank accession no. AF503408.1), as determined with the Artemis comparison tool (ACT). The green rectangles represent specific genomic regions. Strand conservations are indicated in red, and strand inversions are indicated in blue. (B and C) Schematic representation of the inserted regions in bacteriophage RCS47: blaSHV-2 and its environment (B) and the dms operon (C). The gene names, derived from the closest BLAST homologs identified, are noted. The phage genes surrounding the inserted regions are represented in white, and the insertion sequences are represented in black. The blaSHV-2 resistance gene is shown in brown. Truncated genes are indicated by Δ. CP, conserved protein.
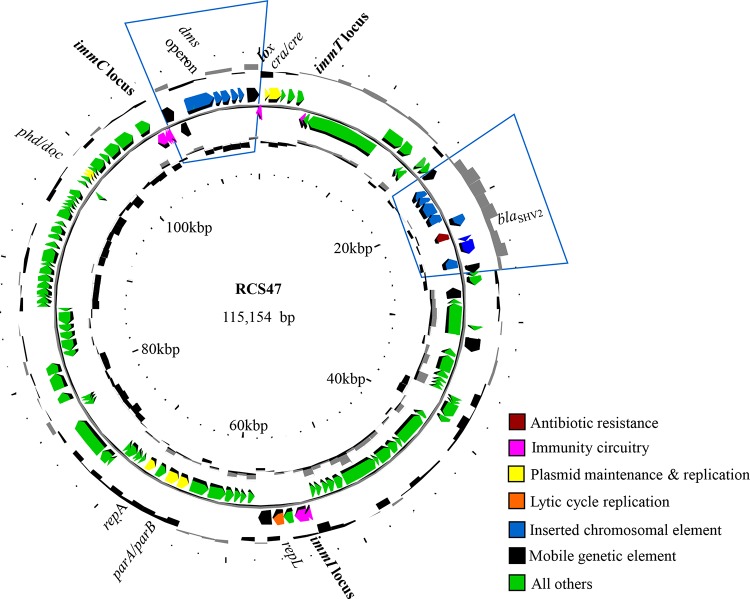
FIG 2.
Circular representation of the organization of the bacteriophage RCS47 genome. The circles display (from the outside) the (i) GC percent deviation (GC window − mean GC) in a 1,000-bp window, (ii) predicted coding sequences transcribed in a clockwise direction, (iii) predicted coding sequences transcribed in a counterclockwise direction, (iv) GC skew ([G + C]/[G − C]) in a 1,000-bp window, and (v) coordinates in kb pairs from the origin of replication. The color code for the various gene functions is shown below the map. The two segments inserted into the genome (Fig. 1B and C) are framed in blue. The imm loci are indicated in bold. Note that the dms operon has been inserted into the immC locus.
Phage RCS47 has acquired two main foreign elements of bacterial origin.
The sequences not present in the P1/P7 phages were essentially located in two regions. In the first region, a 9,636-bp DNA sequence flanked by two IS26 elements was inserted at nucleotide position 15948, interrupting a 684-bp gene of unknown function, downstream from a single-stranded DNA-binding protein-encoding gene, ssb. This segment contained a blaSHV-2 gene surrounded upstream by the ygb genes and the transcriptional regulator deoR gene and downstream by the recF gene and the truncated lacY gene (Fig. 1B). An identical blaSHV genetic environment has been found in several plasmids, such as p1648/97, an E. coli plasmid carrying blaSHV-5 (36), and pHNM1, an Enterobacter cloacae plasmid carrying blaSHV-5 (37). The only difference observed concerned the nucleotide sequence of blaSHV-5, which differs from that of blaSHV-2 by a single nucleotide mutation resulting in an amino acid substitution, E235K. No resistance gene other than the ESBL-encoding gene was detected in the sequence of phage RCS47. This region downstream from ssb seems to be an integration hot spot, as Iida et al. (38) reported the presence of an IS1 element involved in recombination (15, 39) in P1 phages, which is downstream from isaA (Fig. 1B).
In the second region, an 8,544-bp DNA sequence flanked by two IS5 elements was inserted at nucleotide position 106327, interrupting the imcA gene, which, together with imcB, is part of ImmC (Fig. 1C). ImmC is a component of the tripartite immunity circuitry involved in the two life strategies of P1 phages, the lytic and lysogenic strategies (16). This segment contains an IS1 element and a dms operon encoding a dimethyl sulfoxide (DMSO) reductase enzyme allowing bacteria, such as E. coli, to grow anaerobically, using DMSO as a respiratory oxidant (40).
Phage RCS47 displays many genetic similarities to the P1 and P7 genomes in terms of the immunity circuitry, the host specificity DNA inversion system, and the lytic cycle replication protein.
Phage P1 lysogens are extremely stable and are not easily induced. C1 is the major repressor protein of P1 lytic functions. Derivatives of phage P1 with a temperature-sensitive C1 variant encoded by an allele of c1 (c1-100) have been used to overcome the maintenance of lysogeny (15). With respect to the wild-type c1 sequences described by Osborne, Stovall, and Baumstark (41), P1 c1-100 Tn9 contains two amino acid substitutions, L190P and G193C. As P7, which encodes a thermostable protein C1, carries only the first substitution, Lobocka et al. (15) suggested that the G193C substitution probably confers thermosensitivity on the C1 protein. The phage RCS47 C1 amino acid sequence, like that of P7, has the L190P substitution but not the G193C substitution. We also observed an A34T substitution. The ImmI region of phage RCS47, which contains a C1 control operon (c4, icd, and ant1/2), is similar to that of P7. The C segment inversion system (tail fiber genes) expressed by the phage determines the specificity of P1 adsorption to different hosts (15, 42). The nucleotide sequences of the C segments of phages RCS47 and P7 are 99% identical (seven amino acid substitutions) and showed an inversion with respect to P1 c1-100 Tn9 (Fig. 1A). In RCS47, we observed a frameshift (insertion of an adenine residue after codon 135) in the recombinase cin gene, which controls the inversion of the C segment. This mutation may affect the host specificity of RCS47, decreasing its host spectrum (42, 43).
The lytic initiator protein of replication, RepL, of phage RCS47 was compared with those of P1 c1-100 Tn9 and P7, as well as with 21 RepL protein sequences extracted from GenBank. Most of these sequences were obtained during the whole-genome sequencing of strains of Enterobacteriaceae. Phylogenetic analysis by unweighted-pair group method using average linkages (UPGMA) showed this protein to be strongly conserved, with <1.5% divergence between the sequences (see Fig. S1 in the supplemental material).
Phage RCS47 differs from P1 and P7 in terms of its plasmid replication and maintenance module and restriction modification systems.
The RepA proteins of P1 c1-100 Tn9 and P7 belong to the IncY plasmid incompatibility group (44), whereas RCS47 RepA, which displays only 42% identity to the P1 c1-100 Tn9 and P7 sequences, belongs to a different Inc group. We found no similarities between this sequence and those of any other replication proteins from known Inc group plasmids (33, 45). GenBank analysis showed that RCS47 RepA is present in only the genomes of phages, such as pO111_2 (46) and p12579_1 (47), with which it displayed 99% and 100% identity, respectively, at the protein level. The IncY replicon mobile elements associated with strains producing ESBL genes have been described in several epidemiologic studies (19, 48, 49) but, unlike RCS47 RepA, the IncY replicon is found on both phages and plasmids (10; T. Billard-Pomares and C. Branger, unpublished data). Only the sequencing of the mobile element can distinguish between these two sites.
Surprisingly, comparisons of the partitioning proteins of RCS47, ParA and ParB, which are encoded by genes in the same region as repA (Fig. 2), showed them to be identical to those of P7 but distantly related to those of P1 (50). In terms of segregation, the RCS47 prophage is therefore incompatible with P7 but compatible with P1 (51). These data suggest that unlike the lytic replication module, which is strongly conserved between P1 family phages, the plasmid replication and partitioning operons are polymorphic, with various combinations of modules from different origins. This combinatory diversity probably makes it possible to avoid incompatibility with other mobile elements.
Some genes present in the P1 and P7 genomes were found to be absent from RCS47: the genes of the restriction modification system, mod and res, which may protect the lysogen against the entry of foreign DNA, and the superinfection exclusion simABC operon, which protects against infection with another P1 or homologous phage (15).
Absence of lytic capacity and transduction of phage RCS47.
We then investigated whether the RCS47 phage (i) could be induced, (ii) had lytic capacities, and (iii) could transduce the ESBL gene. Following UV irradiation, we were able to obtain phage suspensions containing the ESBL gene from the natural isolate 725 and the DH10B electroporant TR725, both of which gave positive PCR results for repL, repA, and blaSHV-2. The PCRs amplifying E. coli chromosome fragments gave negative results, confirming that the phage PCR amplifications did not result from bacterial strain contamination. The lytic capacity of RCS47 was then assessed by spotting the suspensions onto agar overlays of several E. coli strains widely used for P1 assays and several natural isolates. After 24 h of incubation at 37°C, unlike the P1 (Fig. 3D) and P7 phages, which provide turbid plaques (52), the concentrated suspensions of both 725 and TR725 induced a complete clearing of the spots (Fig. 3A and B) of the E. coli test strains, which was not observed with the dilutions. Similar clear spots were obtained after trypsin treatment of the suspensions (colicin degraded, phage unaffected) (53) (data not shown), and no clearing of spots was observed with the suspension obtained with the DH10B strain without phage RCS47 (Fig. 3C), ruling out the possibility of bacteriocins causing lysis. These data were in favor of a phenomenon called “lysis from without,” which is partly understood (54). Lysis from without is observed at high densities and not at lower densities of phages, probably due to phage adsorption above a threshold value on the cell wall inducing cell destruction (54, 55).
FIG 3.
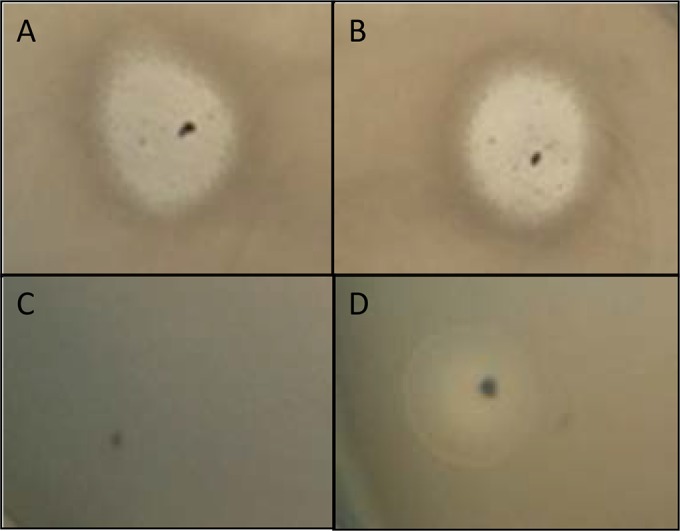
Spotting of 4 sets of bacteriophages onto a layer of E. coli K-12 MG1655. (A) Undiluted bacteriophage suspension from strain 725. (B) Undiluted bacteriophage suspension from the electroporant TR725 strain. (C) Negative control (E. coli DH10B strain). (D) Bacteriophage P1vir (positive control).
The successful in vitro transduction of antibiotic markers, such as chloramphenicol, mediated by the P1 phage in E. coli strains or Salmonella enterica serovar Typhi strains has been reported (23, 56). We assessed the ability of RCS47 to lysogenize E. coli strains and, therefore, to transfer the ESBL gene. Using the lysogenization procedure described above, we obtained no colonies resistant to CTX with the E. coli strains tested.
To better understand the lysis phenotype obtained above, phage suspensions prepared from the 725 and TR725 strains were then examined by electron microscopy. They showed the presence of icosahedral DNA-containing heads of about 75 nm and structures that resemble tail tubes, reminiscent of what has been observed with P1 lysates (57). However, in our samples, the potential tail tubes were never found to be attached to the heads, and we did not observe sheath structures (Fig. 4). This is in agreement with the positive repL and repA PCRs obtained with the RCS47 phage suspension. The RCS47 phage is probably defective in sheath subunit production, as suggested by Walker and Walker (57). Gene 22, which has been proposed to encode the sheath protein gp22 (15, 57), was present on the phage genome. However, gp22 contains one amino acid substitution (L307I) that is not found in P1 and P7 gp22. Further studies are required to explore the potential role of this mutation. According to Lobocka et al. (15), another unidentified protein may be involved in the maturation of the sheath. All the genes on the operons coding for the tail assembly were present, with the exception of pmgF, a gene coding for a protein with a putative morphogenic function (15). The interruption by an IS5 of the imcA gene, which is part of ImmC involved in the two life strategies of P1 phages, did not affect the lytic cycle, as DNA-containing heads were observed by electron microscopy. Together, these results confirm the fact that we observed a lysis from without but no turbid plaques. In conclusion, the RCS47 phage, without a sheath, supposedly cannot inject its DNA into the E. coli strain (58), and it is thereby is a defective phage no longer able to lysogenize E. coli hosts.
FIG 4.
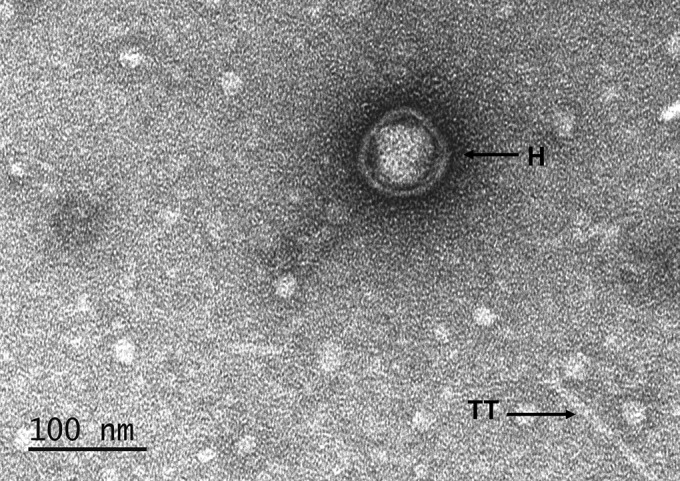
Electron microscopy of phage RCS47 suspension obtained from the 725 E. coli strain showing DNA-containing head (H) and possibly a tail tube (TT). Note the absence of sheath.
P1 family phages are frequent in natural populations of E. coli.
Despite our lack of success in establishing the lysogeny of phage RCS47 carrying blaSHV-2 in recipient strains, P1-related phages may nevertheless be potential vectors of antibiotic resistance genes. In the course of our comparative genomics project, we found another P1-like phage of the same repA type coelectroporated with a plasmid carrying a CTX-M-15-encoding gene (T. Billard-Pomares and C. Branger, unpublished data). We thus assessed the prevalence of P1-related prophages in natural populations of E. coli by PCR amplification of the lytic replication gene, repL, as this gene seems to be well conserved and representative of the P1 family. We detected repL in 46 of the 363 (12.6%) strains tested, demonstrating that P1-related prophages are common in E. coli populations. The prevalence of repL differed between the phylogroups (Table 1). This prevalence was lower in phylogroup B2 than in all the other groups (4/86 [4.6%] versus 42/277 [15.1%]; P = 0.008). No difference in prevalence was found between the ESBL-producing and non-ESBL-producing clinical isolates (8/73 [12.3%] isolates versus 15/290 [13.1%] isolates; P = 0.69). The prevalence of repL was significantly lower in the wild-animal commensal strains (4/114 [3.5%]) than in the domestic-animal commensal strains (13/49 [25.5%]), human commensal strains (9/33 [27.2%]), and human-pathogenic strains (20/169 [11.8%]) (P = 0.00004, 0.00002, and 0.015, respectively). The prevalence of repL in human-pathogenic strains was lower than that in domestic-animal commensal strains and human commensal strains (P = 0.021 and 0.03, respectively) (see Fig. S2 in the supplemental material). The low prevalence of repL in the wild-animal commensal strains may be accounted for by the high proportion of phylogroup B2 E. coli strains in this population (59) and the low prevalence of repL in the phylogroup B2 strains. Similarly, the prevalence of repL in the human-pathogenic strains was lower than that in the domestic-animal commensal and human commensal strains, as phylogroup B2 strains are predominant among the E. coli strains responsible for extraintestinal infections (31). We also determined the prevalences of IncRCS47 and IncY, the incompatibility groups of the plasmid replication gene repA of RCS47 and the reference P1 phage, respectively (15, 44), in the 46 strains testing positive for repL. IncY was found in 14 of the 46 (30.4%) strains, and IncRCS47 was found in 11 of the 46 (23.9%) strains, indicating a low diversity of repA types in P1-like phages.
TABLE 1.
Distribution of the repL gene between phylogroups, according to strain origin
| Phylogroup | No. (%) of strains of the indicated type: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All types |
Human pathogenic |
Human commensal |
Domestic-animal commensal |
Wild-animal commensal |
||||||
| Total | repL present | Total | repL present | Total | repL present | Total | repL present | Total | repL present | |
| A | 65 (17) | 10 (16.3) | 30 (16.1) | 3 (10) | 14 (42.4) | 4 (28.5) | 18 (36.7) | 3 (16.6) | 3 (2.6) | 0 |
| B1 | 82 (22.5) | 10 (12.1) | 16 (9.58) | 2 (12.5) | 9 (27.2) | 0 | 18 (36.7) | 6 (33.3) | 39 (34.2) | 2 (5.1) |
| B2 | 86 (24.2) | 4 (4.6) | 54 (33.5) | 1 (1.8) | 4 (12.1) | 2 | 2 (4) | 0 | 26 (22.8) | 1 (3.8) |
| C | 39 (12.3) | 6 (15.3) | 35 (22.1) | 6 (17.1) | 1 (3) | 0 | 3 (6.1) | 0 | 0 | 0 |
| D | 51 (14.3) | 8 (15.6) | 22 (13.7) | 3 (13.6) | 5 (15.1) | 3 | 5 (10.2) | 2 | 19 (16.6) | 0 |
| E, F, Escherichia clades, ungrouped | 40 (10.4) | 8 (20) | 10 (4.8) | 5 | 0 | 0 | 3 (6.1) | 2 | 27 (23.6) | 1 (3.7) |
| Total | 363 | 46 (12.6) | 167 | 20 (19.7) | 33 | 9 (27.2) | 49 | 13 (26.5) | 114 | 4 (3.5) |
Lastly, we searched for P1-like bacteriophages in 18 ESBL-producing strains of the same B1 phylogroup/O8 serogroup as the 725 strain. Four of them were positive (22%) for repL, of which one and three are of IncRCS47 and IncY, respectively, indicating that the strains were not more prone to be lysogenized by the phage than were the other strains of the species.
These data indicate that P1-like bacteriophages are widespread in natural E. coli isolates, including ESBL-producing isolates, suggesting a high potential for gene transfer. However, the presence of these phages is not linked to ESBL but seems to be related to phylogroup. Such associations between mobile elements and the genetic background have been described (17, 18) and probably correspond to epistatic interactions between P1 family phages and the genetic background of E. coli strains.
In conclusion, we describe here a temperate phage related to P1, isolated from a clinical strain of E. coli carrying an ESBL-encoding gene. This phage was one of the nontypeable plasmids from our collection (19). Our sequencing project allowed us to reveal that P1-like phages, which lysogenize bacteria as autonomous plasmid-like elements, are part of the mobile elements that acquire and have the capacity to transfer antibiotic resistance. Yarmolinsky (16) stressed that despite their ability to acquire and transfer multidrug resistance genes, P1 phages do not seem to compromise antibiotic treatment. The high prevalence of P1-like phages in E. coli populations and our identification of a P1-like phage carrying an ESBL gene suggest that the role of P1-like phages in disseminating antibiotic resistance may be underestimated. The transfer of resistance genes by temperate bacteriophages integrating into the bacterial chromosome has been well described (10–12). In contrast, the different ways in which P1-like phages potentially transfer resistance genes have to be explored more thoroughly. Indeed, besides the fact that P1-like phages can insert resistance genes through their recombination systems (15), they can transfer resistance plasmids via transduction (60). P1-like phages carrying resistance genes can also be transferred to bacteria through conjugation with a conjugative plasmid (26, 39, 56, 60).
Our findings indicate that more plasmid sequencing efforts are required, particularly for nontypeable plasmids, to identify the various vectors involved in the transmission of antibiotic resistance markers.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Agence Nationale pour la Recherche (grant ANR-10-GENM-0012) and the Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia, and the European Regional Development Fund (ERDF) (grant CN2012/303).
For electron microscopy, we thank Maya Salhi for technical assistance and the Multiscale Electron Imaging platform (METi) of the Bio-Research Federation of Toulouse, CNRS FR3451, Bat. IBCG, Toulouse, France.
Footnotes
Published ahead of print 18 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03183-14.
REFERENCES
- 1.Coque TM, Baquero F, Canton R. 2008. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 13:pii=19044 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19044. [PubMed] [Google Scholar]
- 2.Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686. 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238. 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028. 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 5.Muniesa M, Colomer-Lluch M, Jofre J. 2013. Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Future Microbiol. 8:739–751. 10.2217/fmb.13.32. [DOI] [PubMed] [Google Scholar]
- 6.Brabban AD, Hite E, Callaway TR. 2005. Evolution of foodborne pathogens via temperate bacteriophage-mediated gene transfer. Foodborne Pathog. Dis. 2:287–303. 10.1089/fpd.2005.2.287. [DOI] [PubMed] [Google Scholar]
- 7.Colomer-Lluch M, Imamovic L, Jofre J, Muniesa M. 2011. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 55:4908–4911. 10.1128/AAC.00535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colomer-Lluch M, Jofre J, Muniesa M. 2011. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One 6:e17549. 10.1371/journal.pone.0017549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver A, Coque TM, Alonso D, Valverde A, Baquero F, Cantón R. 2005. CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrob. Agents Chemother. 49:1567–1571. 10.1128/AAC.49.4.1567-1571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banks DJ, Porcella SF, Barbian KD, Martin JM, Musser JM. 2003. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A streptococcus. J. Infect. Dis. 188:1898–1908. 10.1086/379897. [DOI] [PubMed] [Google Scholar]
- 11.Blahová J, Králiková K, Krcméry V, Mlynarcík D, Trupl J. 1998. Transduction of imipenem resistance by wild-type bacteriophages carried by three strains of Pseudomonas aeruginosa isolated from a single source. J. Antimicrob. Chemother. 41:660–662. 10.1093/jac/41.6.660. [DOI] [PubMed] [Google Scholar]
- 12.Goh S, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P. 2013. Phage φC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio 4(6):e00840-13. 10.1128/mBio.00840-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmieger H, Schicklmaier P. 1999. Transduction of multiple drug resistance of Salmonella enterica serovar Typhimurium DT104. FEMS Microbiol. Lett. 170:251–256. 10.1111/j.1574-6968.1999.tb13381.x. [DOI] [PubMed] [Google Scholar]
- 14.Smith HW. 1972. Ampicillin resistance in Escherichia coli by phage infection. Nat. New Biol. 238:205–206. 10.1038/newbio238205a0. [DOI] [PubMed] [Google Scholar]
- 15.Lobocka MB, Rose DJ, Plunkett G, III, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. 2004. Genome of bacteriophage P1. J. Bacteriol. 186:7032–7068. 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarmolinsky MB. 2004. Bacteriophage P1 in retrospect and in prospect. J. Bacteriol. 186:7025–7028. 10.1128/JB.186.21.7025-7028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branger C, Zamfir O, Geoffroy S, Laurans G, Arlet G, Thien HV, Gouriou S, Picard B, Denamur E. 2005. Genetic background of Escherichia coli and extended-spectrum beta-lactamase type. Emerg. Infect. Dis. 11:54–61. 10.3201/eid1101.040257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deschamps C, Clermont O, Hipeaux MC, Arlet G, Denamur E, Branger C. 2009. Multiple acquisitions of CTX-M plasmids in the rare D2 genotype of Escherichia coli provide evidence for convergent evolution. Microbiology 155:1656–1668. 10.1099/mic.0.023234-0. [DOI] [PubMed] [Google Scholar]
- 19.Marcadé G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, Denamur E, Arlet G. 2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 63:67–71. 10.1093/jac/dkn428. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Blaxter ML. 2010. Comparing de novo assemblers for 454 transcriptome data. BMC Genomics 11:571. 10.1186/1471-2164-11-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Médigue C. 2006. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 34:53–65. 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595–600. 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo E, Mitsuhashi S. 1964. Drug resistance of enteric bacteria. IV. Active transducing bacteriophage P1 CM produced by the combination of R factor with bacteriophage P1. J. Bacteriol. 88:1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allué-Guardia A, García-Aljaro C, Muniesa M. 2011. Bacteriophage-encoding cytolethal distending toxin type V gene induced from nonclinical Escherichia coli isolates. Infect. Immun. 79:3262–3272. 10.1128/IAI.05071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5:58–65. 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 26.Kutter E. 2009. Phage host range and efficiency of plating. Methods Mol. Biol. 501:141–149. 10.1007/978-1-60327-164-6_14. [DOI] [PubMed] [Google Scholar]
- 27.Schaller K, Krauel A, Braun V. 1981. Temperature-sensitive, colicin M-tolerant mutant of Escherichia coli. J. Bacteriol. 147:135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.León M, Santander J, Curtiss R, III, Robeson J. 2012. Natural lysogenization and transduction in Salmonella enterica serovar Choleraesuis by bacteriophage P1. Res. Microbiol. 164:1–5. 10.1016/j.resmic.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skurnik D, Le Menac'h A, Zurakowski D, Mazel D, Courvalin P, Denamur E, Andremont A, Ruimy R. 2005. Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob. Agents Chemother. 49:3062–3065. 10.1128/AAC.49.7.3062-3065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skurnik D, Ruimy R, Andremont A, Amorin C, Rouquet P, Picard B, Denamur E. 2006. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57:1215–1219. 10.1093/jac/dkl122. [DOI] [PubMed] [Google Scholar]
- 31.Lefort A, Panhard X, Clermont O, Woerther PL, Branger C, Mentré F, Fantin B, Wolff M, Denamur E, COLIBAFI Group 2011. Host factors and portal of entry outweigh bacterial determinants to predict the severity of Escherichia coli bacteremia. J. Clin. Microbiol. 49:777–783. 10.1128/JCM.01902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228. 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 34.Blanco M, Alonso MP, Nicolas-Chanoine MH, Dahbi G, Mora A, Blanco JE, López C, Cortés P, Llagostera M, Leflon-Guibout V, Puentes B, Mamani R, Herrera A, Coira MA, García-Garrote F, Pita JM, Blanco J. 2009. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 63:1135–1141. 10.1093/jac/dkp122. [DOI] [PubMed] [Google Scholar]
- 35.Coelho A, Mora A, Mamani R, López C, González-López JJ, Larrosa MN, Quintero-Zarate JN, Dahbi G, Herrera A, Blanco JE, Blanco M, Alonso MP, Prats G, Blanco J. 2011. Spread of Escherichia coli O25b:H4-B2-ST131 producing CTX-M-15 and SHV-12 with high virulence gene content in Barcelona (Spain). J. Antimicrob. Chemother. 66:517–526. 10.1093/jac/dkq491. [DOI] [PubMed] [Google Scholar]
- 36.Zienkiewicz M, Kern-Zdanowicz I, Gołebiewski M, Zyliñska J, Mieczkowski P, Gniadkowski M, Bardowski J, Cegłowski P. 2007. Mosaic structure of p1658/97, a 125-kilobase plasmid harboring an active amplicon with the extended-spectrum beta-lactamase gene blaSHV-5. Antimicrob. Agents Chemother. 51:1164–1171. 10.1128/AAC.00772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garza-Ramos U, Davila G, Gonzalez V, Alpuche-Aranda C, López-Collada VR, Alcantar-Curiel D, Newton O, Silva-Sanchez J. 2009. The blaSHV-5 gene is encoded in a compound transposon duplicated in tandem in Enterobacter cloacae. Clin. Microbiol. Infect. 15:878–880. 10.1111/j.1469-0691.2009.02790.x. [DOI] [PubMed] [Google Scholar]
- 38.Iida S, Meyer J, Bächi B, Stålhammar-Carlemalm M, Schrickel S, Bickle TA, Arber W. 1983. DNA restriction–modification genes of phage P1 and plasmid p15B. Structure and in vitro transcription. J. Mol. Biol. 165:1–18. [DOI] [PubMed] [Google Scholar]
- 39.Iida S. 1980. A cointegrate of the bacteriophage P1 genome and the conjugative R plasmid R100. Plasmid 3:278–290. 10.1016/0147-619X(80)90041-4. [DOI] [PubMed] [Google Scholar]
- 40.Sambasivarao D, Weiner JH. 1991. Dimethyl sulfoxide reductase of Escherichia coli: an investigation of function and assembly by use of in vivo complementation. J. Bacteriol. 173:5935–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osborne FA, Stovall SR, Baumstark BR. 1989. The c1 genes of P1 and P7. Nucleic Acids Res. 17:7671–7680. 10.1093/nar/17.19.7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iida S, Meyer J, Kennedy KE, Arber W. 1982. A site-specific, conservative recombination system carried by bacteriophage P1. Mapping the recombinase gene cin and the cross-over sites cix for the inversion of the C segment. EMBO J. 1:1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehnherr H. 2006. Bacteriophage P1, p 350–364 In Calendar R. (ed), The bacteriophages, 2nd ed. Oxford University Press, New York, NY. [Google Scholar]
- 44.Jaffé-Brachet A, Briaux-Gerbaud S. 1981. Curing of P1 prophage from Escherichia coli K-12 recA(P1) lysogens superinfected with P1 bacteriophage. J. Virol. 37:854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. 2010. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54:4168–4177. 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944. 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kyle JL, Cummings CA, Parker CT, Quiñones B, Vatta P, Newton E, Huynh S, Swimley M, Degoricija L, Barker M, Fontanoz S, Nguyen K, Patel R, Fang R, Tebbs R, Petrauskene O, Furtado M, Mandrell RE. 2012. Escherichia coli serotype O55:H7 diversity supports parallel acquisition of bacteriophage at Shiga toxin phage insertion sites during evolution of the O157:H7 lineage. J. Bacteriol. 194:1885–1896. 10.1128/JB.00120-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novais A, Cantón R, Valverde A, Machado E, Galán JC, Peixe L, Carattoli A, Baquero F, Coque TM. 2006. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-alpha, and IncFI groups. Antimicrob. Agents Chemother. 50:2741–2750. 10.1128/AAC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hrabák J, Empel J, Bergerová T, Fajfrlík K, Urbásková P, Kern-Zdanowicz I, Hryniewicz W, Gniadkowski M. 2009. International clones of Klebsiella pneumoniae and Escherichia coli with extended-spectrum beta-lactamases in a Czech hospital. J. Clin. Microbiol. 47:3353–3357. 10.1128/JCM.00901-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayes F, Austin SJ. 1993. Specificity determinants of the P1 and P7 plasmid centromere analogs. Proc. Natl. Acad. Sci. U. S. A. 90:9228–9232. 10.1073/pnas.90.19.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouet JY, Nordstrom K, Lane D. 2007. Plasmid partition and incompatibility–the focus shifts. Mol. Microbiol. 65:1405–1414. 10.1111/j.1365-2958.2007.05882.x. [DOI] [PubMed] [Google Scholar]
- 52.Walker DH, Jr, Walker JT. 1976. Genetic studies of coliphage P1. II. Relatedness to P7. J. Virol. 19:271–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordon DM, O'Brien CL. 2006. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 152:3239–3244. 10.1099/mic.0.28690-0. [DOI] [PubMed] [Google Scholar]
- 54.Abedon ST. 2011. Lysis from without. Bacteriophage 1:46–49. 10.4161/bact.1.1.13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delbrück M. 1940. The growth of bacteriophage and lysis of the host. J. Gen. Physiol. 23:643–660. 10.1085/jgp.23.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kondo E, Mitsuhashi S. 1966. Drug resistance of enteric bacteria. VI. Introduction of bacteriophage P1CM into Salmonella Typhi and formation of PldCM and F-CM elements. J. Bacteriol. 91:1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker JT, Walker DH., Jr 1983. Coliphage P1 morphogenesis: analysis of mutants by electron microscopy. J. Virol. 45:1118–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J, Chen CY, Shiomi D, Niki H, Margolin W. 2011. Visualization of bacteriophage P1 infection by cryo-electron tomography of tiny Escherichia coli. Virology 417:304–311. 10.1016/j.virol.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lescat M, Clermont O, Woerther PL, Glodt J, Dion S, Skurnik D, Djossou F, Dupont C, Perroz G, Picard B, Catzeflis F, Andremont A, Denamur E. 2013. Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host-dependent population structure. Environ. Microbiol. Rep. 5:49–57. 10.1111/j.1758-2229.2012.00374.x. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe T, Furuse C, Sakaizumi S. 1968. Transduction of various R factors by phage P1 in Escherichia coli and by phage P22 in Salmonella Typhimurium. J. Bacteriol. 96:1791–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.