Abstract
We sequenced a novel conjugative blaKPC-2-harboring IncN plasmid, pYD626E, from an Escherichia coli sequence type 648 strain previously identified in Pittsburgh, Pennsylvania. pYD626E was 72,800 bp long and carried four β-lactamase genes, blaKPC-2, blaSHV-12, blaLAP-1, and blaTEM-1. In addition, it harbored qnrS1 (fluoroquinolone resistance) and dfrA14 (trimethoprim resistance). The plasmid profile and clinical history supported the in vivo transfer of this plasmid between Klebsiella pneumoniae and Escherichia coli.
TEXT
The rapid spread of Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae has become a major public health threat (1). The gene encoding KPC, blaKPC, is usually located on a plasmid and may transfer to other Enterobacteriaceae species. While KPC-producing Escherichia coli is still rare compared with KPC-producing K. pneumoniae, reports have emerged on E. coli strains acquiring blaKPC-carrying plasmids and becoming resistant to carbapenems (2–5). We previously reported a series of patients who had KPC-producing E. coli, and many of these patients also had a second species that produced KPC at approximately the same time (6). One of the patients in this report (patient 13) had KPC-2-producing E. coli (strain YD626E) and K. pneumoniae (strain YD626K) in the same bronchoalveolar lavage specimen. He was treated for ventilator-associated pneumonia with colistin but did not survive the hospitalization. The E. coli strain was unusual in that it also produced SHV-12, an extended-spectrum β-lactamase (ESBL) commonly seen in K. pneumoniae isolates. The plasmid in the E. coli transformant from this E. coli strain (pYD626E), obtained by electroporation, carried blaKPC-2 and blaSHV-12. We therefore conducted whole-plasmid sequencing to further characterize this unique plasmid.
The plasmid was extracted from the E. coli TOP10 transformant using the Qiagen plasmid maxi kit (Qiagen, Valencia, CA). Sequencing was performed on the PacBio RS II single-molecule real-time (SMRT) sequencing instrument (Pacific Biosciences, Menlo Park, CA) at the Yale Center for Genome Analysis. The average sequencing coverage was approximately 1,300× across the plasmid. The first-pass reads were assembled de novo using the hierarchical genome assembly process (HGAP) with the default settings of the SMRT Analysis v2.1 software package (Pacific Biosciences) (7). The single contig that represented the plasmid of interest was circularized and used as the reference for reassembling the first-pass reads within the default parameters of the consensus tool Quiver v1, also available in SMRT Analysis v2.1. This process was repeated until an internal consensus accuracy rate of 100% was achieved against the latest contig. No gap filling was required. The plasmid sequence was initially annotated with RAST (http://rast.nmpdr.org), and then it was further curated manually.
pYD626E is 72,800 bp long and has an IncN replicon with a GC content of 53.2%. It harbors 83 predicted open reading frames (ORFs) and is composed of a 39,230-bp core region and three distinct acquired regions (7,764 bp, 3,483 bp, and 22,323 bp) (Fig. 1). The core region includes the genes responsible for plasmid replication, horizontal transfer, and stability and maintenance functions, and it defines the plasmid backbone (8). pYD626E carries a core region that is similar to those of many other multidrug resistance IncN plasmids that have been identified in K. pneumoniae and E. coli from different countries, such as pBK31551, pECN580, pKo6, p9, p12, and pKOX105 (GenBank accession no. JX193301, KF914891, KC958437, FJ223607, FJ223605, and HM126016, respectively), and to the IncN prototype plasmid pR46 (AY046276), underscoring the high degree of plasticity of IncN plasmids and their ability to widely disseminate.
FIG 1.
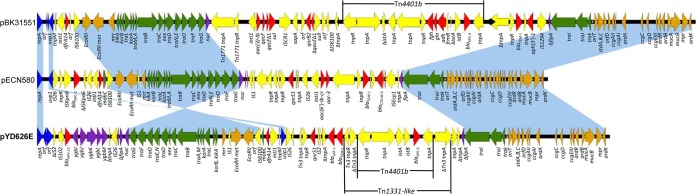
Comparative analysis of IncN plasmid pYD626E (GenBank accession no. KJ933392) with two other blaKPC-carrying IncN plasmids, pBK31551 (JX193301) and pECN580 (KF914891). Light-blue shading indicates shared backbone regions with a high degree of homology. ORFs are portrayed by arrows and are colored according to their putative functions. Dark-blue arrows indicate replication-associated genes. Green arrows indicate genes that are associated with plasmid conjugal transfers, and brown arrows indicate genes that are involved in plasmid stability. Red and yellow arrows indicate antimicrobial resistance genes and accessory genes of mobile elements, respectively. Dark-purple arrows indicate other backbone genes and inserted foreign genes. Bold type indicates the plasmid sequenced in this study (pYD626E).
Similar to other IncN plasmids, the core region in pYD626E encodes the IncN replicon, comprising the replication initiation protein gene repA and two modules involved in the conjugative transfer of plasmids (traGFOENDCBAML and traIJK). The first module is located downstream of the fipA (fertility inhibition protein, truncated) gene and, in addition to the tra genes, contains other backbone genes, including nuc (DNA degradation), eex (exclusion of DNA entry), korA and korB (regulation of transfer and replication), kikA (killing of Klebsiella oxytoca), and the EcoRII and EcoRII methyltransferase genes (possibly mediating lysis of cells which have lost the plasmid) (9). However, the first module in pYD626E is in an orientation inverse to that in most other IncN plasmids, except for the blaVIM-1-carrying plasmid pKOX105, which was found in a Klebsiella oxytoca strain isolated in Italy (GenBank accession no. HM126016) (10). The second module is located next to the second truncated copy of fipA, but it is upstream of the gene instead of downstream. Along with the tra genes, this second module carries the stbABC operon, which guarantees plasmid stability during conjugation, and it carries the ardAB and ardR systems, which regulate antirestriction functions. Furthermore, like most but not all other IncN plasmids, pYD626E possesses the ccgC and ccgD genes, which encode products that protect plasmid DNA from type I restriction enzymes (10, 11).
pYD626E has three distinct acquired regions that harbor a variety of antimicrobial resistance genes. The 3,483-bp resistance region is located upstream of the uvp1 gene next to the first core region, a previously described hot spot for integration within IncN plasmids (8), and this region has a class 1 integron. This integron harbors a trimethoprim resistance gene cassette, dfrA14, and has its 3′-conserved end truncated by the insertion of an IS6100 element, as has been observed in other blaKPC-carrying IncN plasmids (8, 10, 11). The 7,764-bp resistance region is located between the genes repA and fipA and is bracketed by the insertion elements IS102 and IS26, respectively. It carries blaSHV-12 and an incomplete operon involved in sugar metabolism (ygbI, ygbJ, ygbK, ygbL, and truncated ygbM). The blaSHV-12 gene together with the incomplete sugar metabolism operon exhibits 99% identity to fragments of several K. pneumoniae chromosomes, such as MGH 78578, KCTC 2242, and KPR0928 (GenBank accession no. CP000647, CP002910, and CP008831, respectively), and blaSHV-harboring plasmids, such as pTC2, pKEC-a3c, and p1658/97 (GenBank accession no. NC_019375.1, CP007558.1, and NC_004998.1, respectively). This shared identity supports the hypothesis that blaSHV originated from the K. pneumoniae chromosome (12). The 22,323-bp region located between uvp1 and the second copy of fipA is the primary cluster of resistance genes in pYD626E. It carries the β-lactamase genes blaKPC-2, blaTEM-1, and blaLAP-1 and the quinolone resistance gene qnrS1. qnrS1 is bracketed by a Tn3-like transposase gene and a truncated IS2 element. The qnrS1-containing region exhibits a high degree of identity (>99%) with several plasmids identified in Salmonella enterica serovar Typhimurium strains (GenBank accession no. HE652087, JN393220, EU715253, and AM746977). Between the regions containing qnrS1 and the blaKPC-2-carrying Tn4401 transposon reside the β-lactamase genes blaLAP-1 and blaTEM-1 in tandem. The blaKPC-2 gene in pYD626E is carried on a typical Tn4401b transposon. However, the Tn4401b transposon was inserted into the transposase gene of Tn1331, generating a 5-bp target duplication sequence (AGAAC) and forming a nested transposon (Tn1331-like). The nested Tn1331-like-Tn4401 transposon in pYD626E is highly similar to the Tn1331-Tn4401 transposon of the blaKPC-3-carrying plasmid pBK15692 (GenBank accession no. KC845573), which was previously identified in a K. pneumoniae strain from New York (13). However, the aminoglycoside-modifying enzyme genes aac(6′)-Ib and aadA1 and the β-lactamase genes blaOXA-9 and blaTEM-1 were missing in pYD626E, as were the inverted repeats (IRs). This indicates that the nested transposon in pYD626E might have originated from that of pBK15692 but that gene rearrangements occurred afterward and resulted in the loss of some resistance genes and IRs.
Next, we compared the blaKPC-2-carrying plasmids from YD626E (pYD626E) and YD626K (pYD626K). pYD626E and pYD626K rendered comparable susceptibility profiles in the E. coli TOP10 transformants (Table 1). The two plasmids were conjugated to E. coli strain J53Azi by broth mating at a relatively high frequency (approximately 1 × 10−4/donor). PCR and subsequent sequencing were performed to screen resistance genes on pYD626K. In addition to blaKPC-2, pYD626K harbored all other resistance genes present on pYD626E, including blaSHV-12, blaTEM-1, blaLAP-1, dfrA14, and qnrS1. In addition, pYD626K fell into the same incompatibility group as the IncN plasmid pYD626E (14). Plasmid fingerprint analysis showed highly related but not identical restriction patterns between pYD626E and pYD626K (Fig. 2). These findings suggest that E. coli YD626E likely acquired the KPC-encoding plasmid from K. pneumoniae YD626K in this patient or vice versa. In either case, the IncN plasmid appears to have undergone minor structural rearrangements upon interspecies transfer that did not affect the complement of resistance genes.
TABLE 1.
MICs for E. coli YD626E, K. pneumoniae YD626K, and the corresponding blaKPC-positive E. coli TOP10 transformants
| Strain | MIC (μg/ml) ofa: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IPM | ETP | LVX | GEN | AMK | TET | SXT | NAL | CST | TGC | |
| E. coli YD626E | 3 | 6 | >32 | 1 | 2 | 8 | 1 | >256 | 0.094 | 0.38 |
| E. coli TOP10 (pYD626E)b | 0.75 | 0.19 | 0.38 | 0.38 | 1 | 2 | 1.5 | 3 | 0.047 | 0.25 |
| K. pneumoniae YD626K | 4 | 8 | >32 | 0.75 | 3 | 6 | 0.75 | >256 | 0.094 | 0.5 |
| E. coli TOP10 (pYD626K)c | 1 | 0.19 | 0.25 | 0.38 | 1 | 2 | 1 | 4 | 0.047 | 0.25 |
MICs were determined by Etest. IPM, imipenem; ETP, ertapenem; LVX, levofloxacin; GEN, gentamicin; AMK, amikacin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; NAL, nalidixic acid; CST, colistin; TGC, tigecycline.
E. coli TOP10 transformant of YD626E.
E. coli TOP10 transformant of YD626K.
FIG 2.
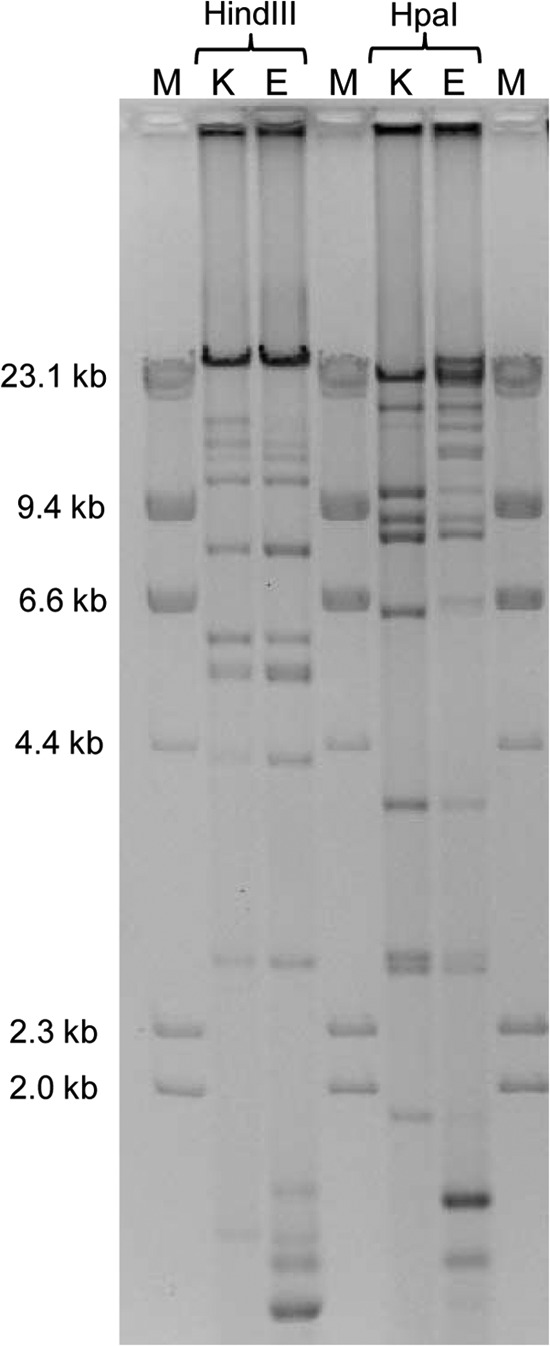
Plasmid profiles of blaKPC-carrying plasmids of YD626E (pYD626E) and YD626K (pYD626K) using the restriction enzymes HindIII and HpaI. M, marker; K, plasmid pYD626K; E, plasmid pYD626E.
Here, we present the complete sequence of a unique blaKPC-2-harboring plasmid from an E. coli sequence type 648 (ST648) strain. Its structure was characterized by the incorporation of a blaSHV-12-containing region similar to certain K. pneumoniae chromosomes and a qnrS1-containing region similar to several S. enterica plasmids into the IncN backbone. The coexistence of blaKPC and blaSHV on a single plasmid is rare and has been reported in only an IncFIIK plasmid (coharboring blaKPC-2 and blaSHV-11 [encoding a non-ESBL resistance pattern]) and an IncX plasmid (coharboring blaKPC-2 and blaSHV-12), both of which were from K. pneumoniae isolates (15, 16). In addition, ST648 has been associated with various β-lactamases, including ESBLs, NDM, and KPCs (6, 17), and is a predominant multidrug-resistant clone observed worldwide in humans, companion animals, livestock, and wild birds (18–20). The emergence of the conjugative plasmid encoding KPCs, ESBLs, and qnr genes in E. coli ST648 highlights the potential of blaKPC to effectively spread to high-risk clones utilizing multidrug resistance plasmids as the vehicles.
Plasmid sequence accession number.
The plasmid sequence reported in this article has been submitted to GenBank under accession no. KJ933392.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health research grants R21AI107302 and R01AI104895 (to Y.D.).
Footnotes
Published ahead of print 2 September 2014
REFERENCES
- 1.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13:785–796. 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai JC, Zhang R, Hu YY, Zhou HW, Chen GX. 2014. Emergence of Escherichia coli sequence type 131 isolates producing KPC-2 carbapenemase in China. Antimicrob. Agents Chemother. 58:1146–1152. 10.1128/AAC.00912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mavroidi A, Miriagou V, Malli E, Stefos A, Dalekos GN, Tzouvelekis LS, Petinaki E. 2012. Emergence of Escherichia coli sequence type 410 (ST410) with KPC-2 β-lactamase. Int. J. Antimicrob. Agents 39:247–250. 10.1016/j.ijantimicag.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Robledo IE, Aquino EE, Vazquez GJ. 2011. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob. Agents Chemother. 55:2968–2970. 10.1128/AAC.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goren MG, Navon-Venezia S, Chmelnitsky I, Carmeli Y. 2010. Carbapenem-resistant KPC-2-producing Escherichia coli in a Tel Aviv medical center, 2005 to 2008. Antimicrob. Agents Chemother. 54:2687–2691. 10.1128/AAC.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YA, Qureshi ZA, Adams-Haduch JM, Park YS, Shutt KA, Doi Y. 2012. Features of infections due to Klebsiella pneumoniae carbapenemase-producing Escherichia coli: emergence of sequence type 131. Clin. Infect. Dis. 55:224–231. 10.1093/cid/cis387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10:563–569. 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 8.Carattoli A, Aschbacher R, March A, Larcher C, Livermore DM, Woodford N. 2010. Complete nucleotide sequence of the IncN plasmid pKOX105 encoding VIM-1, QnrS1 and SHV-12 proteins in Enterobacteriaceae from Bolzano, Italy compared with IncN plasmids encoding KPC enzymes in the USA. J. Antimicrob. Chemother. 65:2070–2075. 10.1093/jac/dkq269. [DOI] [PubMed] [Google Scholar]
- 9.Bhagwat AS, Johnson B, Weule K, Roberts RJ. 1990. Primary sequence of the EcoRII endonuclease and properties of its fusions with β-galactosidase. J. Biol. Chem. 265:767–773. [PubMed] [Google Scholar]
- 10.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob. Agents Chemother. 57:269–276. 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Hu H, Chavda KD, Zhao S, Liu R, Liang H, Zhang W, Wang X, Jacobs MR, Bonomo RA, Kreiswirth BN. 2014. Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob. Agents Chemother. 58:2422–2425. 10.1128/AAC.02587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drieux L, Decre D, Frangeul L, Arlet G, Jarlier V, Sougakoff W. 2013. Complete nucleotide sequence of the large conjugative pTC2 multireplicon plasmid encoding the VIM-1 metallo-β-lactamase. J. Antimicrob. Chemother. 68:97–100. 10.1093/jac/dks367. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 57:5019–5025. 10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228. 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, Siu LK. 2014. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J. Antimicrob. Chemother. 69:628–631. 10.1093/jac/dkt409. [DOI] [PubMed] [Google Scholar]
- 16.Kassis-Chikhani N, Frangeul L, Drieux L, Sengelin C, Jarlier V, Brisse S, Arlet G, Decre D. 2013. Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57:618–620. 10.1128/AAC.01712-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 66:2002–2005. 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 18.Guenther S, Grobbel M, Beutlich J, Bethe A, Friedrich ND, Goedecke A, Lubke-Becker A, Guerra B, Wieler LH, Ewers C. 2010. CTX-M-15-type extended-spectrum β-lactamases-producing Escherichia coli from wild birds in Germany. Environ. Microbiol. Rep. 2:641–645. 10.1111/j.1758-2229.2010.00148.x. [DOI] [PubMed] [Google Scholar]
- 19.Ewers C, Bethe A, Stamm I, Grobbel M, Kopp PA, Guerra B, Stubbe M, Doi Y, Zong Z, Kola A, Schaufler K, Semmler T, Fruth A, Wieler LH, Guenther S. 2014. CTX-M-15-d-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J. Antimicrob. Chemother. 69:1224-1230. 10.1093/jac/dkt516. [DOI] [PubMed] [Google Scholar]
- 20.Cortés P, Blanc V, Mora A, Dahbi G, Blanco JE, Blanco M, Lopez C, Andreu A, Navarro F, Alonso MP, Bou G, Blanco J, Llagostera M. 2010. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76:2799–2805. 10.1128/AEM.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


