Abstract
Direct-acting antivirals (DAAs) targeting proteins encoded by the hepatitis C virus (HCV) genome have great potential for the treatment of HCV infections. However, the efficacy of DAAs designed to target genotype 1 (G1) HCV against non-G1 viruses has not been characterized fully. In this study, we investigated the inhibitory activities of nonnucleoside inhibitors (NNIs) against the HCV RNA-dependent RNA polymerase (RdRp). We examined the ability of six NNIs to inhibit G1b, G2a, and G3a subgenomic replicons in cell culture, as well as in vitro transcription by G1b and G3a recombinant RdRps. Of the six G1 NNIs, only the palm II binder nesbuvir demonstrated activity against G1, G2, and G3 HCV, in both replicon and recombinant enzyme models. The thumb I binder JTK-109 also inhibited G1b and G3a replicons and recombinant enzymes but was 41-fold less active against the G2a replicon. The four other NNIs, which included a palm I binder (setrobuvir), two thumb II binders (lomibuvir and filibuvir), and a palm β-hairpin binder (tegobuvir), all showed at least 40-fold decreases in potency against G2a and G3a replicons and the G3a enzyme. This antiviral resistance was largely conferred by naturally occurring amino acid residues in the G2a and G3a RdRps that are associated with G1 resistance. Lomibuvir and filibuvir (thumb II binders) inhibited primer-dependent but not de novo activity of the G1b polymerase. Surprisingly, these compounds instead specifically enhanced the de novo activity at concentrations of ≥100 nM. These findings highlight a potential differential mode of RdRp inhibition for HCV NNIs, depending on their prospective binding pockets, and also demonstrate a surprising enhancement of de novo activity for thumb RdRp binders. These results also provide a better understanding of the antiviral coverage for these polymerase inhibitors, which will likely be used in future combinational interferon-free therapies.
INTRODUCTION
Nearly 3% of the world's population is infected with hepatitis C virus (HCV), a leading cause of chronic liver disease and hepatocellular carcinoma (1). A member of the Flaviviridae family, HCV is an enveloped virus which has a positive-sense, single-stranded RNA (ssRNA) genome of 9.6 kb. Upon infection, the genome is translated into a single polyprotein that is then processed into structural and nonstructural proteins. The genome shows substantial heterogeneity, and therefore HCV has been classified into six different genotypes (G1 to G6), which are approximately 35% divergent at the nucleotide level (2). Genotypes are further classified into subtypes (1a, 1b, 1c, etc.), with about 20% intersubtype nucleotide divergence (2).
Until recently, treatment of HCV infections involved a combination of pegylated interferon and ribavirin (PEG-IFN/RBV), a regimen that is lengthy and poorly tolerated and has various response rates among the HCV genotypes. Among patients infected with the most prevalent HCV genotype, G1, around 50% achieve a sustained virological response (SVR) with PEG-IFN/RBV therapy, compared to ∼80% of those infected with G2 or G3 viruses (3). For more than a decade, extensive efforts have been devoted to the development of direct-acting antivirals (DAAs), compounds which specifically inhibit HCV replication by targeting viral nonstructural proteins. Three protease inhibitors have so far been approved for treatment of HCV G1, in combination with PEG-IFN/RBV, and have increased SVR rates by nearly 30% compared to those with PEG-IFN/RBV therapy alone for that particular genotype (4–7). The first HCV nucleoside inhibitor (NI), sofosbuvir, was also recently approved for HCV treatment in combination with PEG-IFN/RBV, with SVR rates of around 90% in HCV patients, although the drug is less effective against G3a viruses in IFN-free regimens (8–10). These four approved HCV DAAs represent the forerunners of a group of around 30 DAAs in phase 2 or 3 clinical trials (11).
The HCV RNA-dependent RNA polymerase (RdRp) has long been a prime target for antiviral development because of its critical role in viral replication and the absence of a mammalian homologous enzyme. The RdRp has a “right-hand” structure with finger and thumb domains that encircle the active site, located in the palm domain (12–14). Current DAAs targeting the HCV RdRp are classified into two groups. Nucleoside inhibitors, such as sofosbuvir, are substrate analogues that cause termination during synthesis of new RNA molecules. In contrast, the binding of nonnucleoside inhibitors (NNIs) to the RdRp inhibits conformational changes essential for polymerase activity (15). HCV NNIs have been identified as encompassing a diverse range of chemical scaffolds (16). However, these have all been found to bind the RdRp at one of five NNI sites (reviewed in reference 17). Two binding sites lie within the thumb subdomain: thumb I (T1), to which compounds such as benzimidazole and indole derivatives (e.g., deleobuvir, BMS-791325, and TMC647055) bind, and thumb II (T2), which is the target site for thiophene-2-carboxylic acids (18), such as VX-222 (lomibuvir) and GS-9669, as well as dihydropyranones, such as PF-00868554 (filibuvir) (19). Two other NNI sites have been characterized within the palm domain, distinct from but in close proximity to the RdRp active site. The palm I site (P1) has been targeted by proline (20), benzodiazepine (21), and benzothiadiazine (22) derivatives, such as RG7790 (setrobuvir), ABT-333, and ABT-072, whereas compounds that bind to the palm II site (P2) include benzofurans, such as HCV-796 (nesbuvir) (23). Imidazopyridines, including the NNI GS-9190 (tegobuvir), are another class of compounds which bind to the palm site of the polymerase; however, unlike other palm binders, this binding uniquely involves an interaction with the β-hairpin which extends from the thumb into the palm domain (site P-β) (24, 25).
The lack of a cell culture model for HCV genotypes other than G1 and G2 (26) has significantly hindered the development of antivirals specifically targeting HCV genotypes G3 to G6. Studies to determine the efficacy of HCV G1-developed NNIs on G3 to G6 viruses have therefore largely involved recombinant enzymes made in Escherichia coli (27, 28) or the use of chimeric subgenomic replicons containing the NS5B coding region on a G1b/G2a backbone (29–32). However, chimeric replicons are often prone to replication fitness losses, and the contribution of the backbone RNA/proteins to the differential antiviral effects is poorly characterized (29). Recently, replicons of HCV G3a and G4a were described (33–35), therefore allowing a closer examination of the efficacies of HCV G1 DAAs against G2 to G6 viruses, which represent between 35% and 90% of all HCV infections in some countries (36–40).
The RdRp of HCV, like those of other members of the Flaviviridae family, can catalyze transcription through primed elongation as well as through a primer-independent (de novo) mechanism (41–43). De novo polymerase activity is believed to be the mechanism by which viral genome replication is initiated in vivo, and it is critical for the preservation of the terminal ends of the ssRNA genome during replication (42). The two modes of RdRp activity are believed to correspond to different conformations of the enzyme. A “closed” conformation, facilitated by the interactions between the finger and thumb domains, allows the de novo formation of a dinucleotide for replication initiation (44). The more relaxed “open” conformation of the HCV RdRp is formed by displacement of the β-hairpin loop as well as a C-terminal segment just upstream of the transmembrane domain, called the linker (45, 46). This open conformational change allows room for the nascent double-stranded RNA (dsRNA) and is thought to be responsible for the primer-extension activity of the RdRp (45, 46). Studies involving NNIs and recombinant RdRps have focused primarily on the ability of the NNIs to inhibit the primer-dependent activity of the HCV RdRp, and little is currently known on what effect NNIs have on de novo RdRp activity. Yi et al. (47) recently demonstrated that T2-binding HCV NNIs inhibited the primed activity but had little effect on the de novo activity of the HCV G1b RdRp. Conversely, compounds which bound to the P1 and P2 sites inhibited both modes of RdRp activity (47, 48). These compounds, however, were examined using only a narrow range of concentrations (0 to 200 nM), and the effects of NNI binding to the T1 and P-β sites remain to be examined (47). Furthermore, it is unclear how the various NNIs affect both modes of activity, i.e., de novo and primed elongation, across RdRps of different HCV genotypes.
In this study, RdRp inhibitors representing molecules that bind to all known sites on HCV polymerase were screened for the ability to inhibit G1b, G2a, and G3a replicons and G1b and G3a recombinant enzymes. Furthermore, the inhibitory profiles of HCV NNIs against de novo and primed RdRp activity were analyzed, revealing a novel mechanism of action for T2 and P-β binders.
MATERIALS AND METHODS
Compounds.
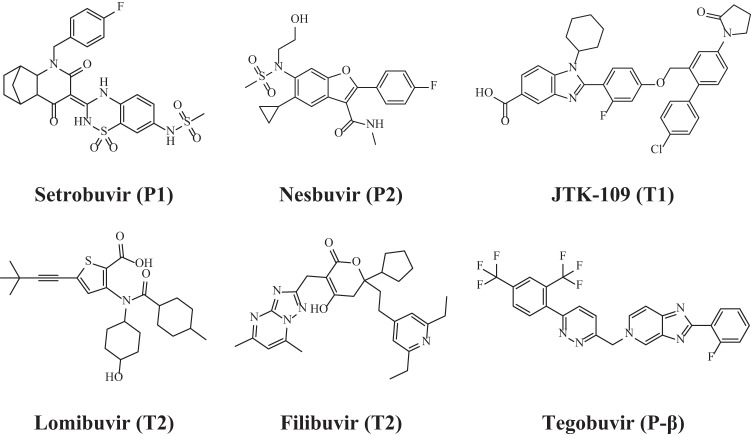
All compounds were purchased from commercial vendors, dissolved in 100% dimethyl sulfoxide (DMSO), and freshly diluted to the desired concentration on the day of the experiment. These compounds included 2′-C-methylcytidine (2CM) (Sigma-Aldrich, St. Louis, MO), JTK-109 (Dalton Pharma Services, Toronto, Canada), tegobuvir (GS-9190) and nesbuvir (HCV-796) (Haoyuan Chemexpress, Shanghai, China), filibuvir (PF-00868554) and setrobuvir (ANA-598) (Acme Biosciences, Palo Alto, CA), and lomibuvir (VX-222) (Selleckchem, Houston, TX). The structures of molecules examined in this study are shown in Fig. 1.
FIG 1.
Chemical structures of HCV NNIs examined in this study. Compounds were selected which bind to the palm (P1 and P2), thumb (T1 and T2), or palm β-hairpin interaction site (P-β) of RdRp. The binding site for each NNI is indicated in parentheses.
Quantitative RdRp assays.
Recombinant RdRps were expressed in E. coli and purified by affinity chromatography as described previously (49, 50). Recombinant RdRps used in this study were those from HCV G1b strain Con1 (GenBank accession number AJ238799) and G3a strain VRL69b (accession number EF189901) (50). The de novo activity of RdRps was measured by monitoring the formation of double-stranded RNA from a single-stranded homopolymeric template, poly(C), by using the fluorescent dye PicoGreen (Invitrogen, Carlsbad, CA) as described previously (49). Alternatively, radioactive-nucleotide incorporation assays (1 μCi [3H]rGTP/reaction mix) were also used to measure enzyme activity (50). Half-maximal inhibitory concentration (IC50) values were calculated for each compound by nonlinear regression in GraphPad Prism, version 6.02.
Gel-based RdRp assays.
Polyacrylamide gel-based assays were used to examine the two mechanisms of RdRp activity, i.e., primer extension and primer-independent (de novo) activities, using the method described by Yi et al. (47). The RNA template PE46 was designed to direct primer extension activity through a stable hairpin at the 3′ end, whereas the template LE19p can direct only de novo RdRp activity due to the addition of puromycin at the 3′ end of the RNA (47). Reactions were performed using either 1 μM PE46 or 0.5 μM LE19p. Each reaction mixture contained 240 nM RdRp, 0.2 mM rGTP, 0.1 mM (each) rCTP, rATP, and rUTP, 5 mM dithiothreitol (DTT), 2.5 mM MnCl2, and 20 mM Tris-HCl in a 25-μl final volume. Reaction mixtures were incubated for 5 h at 30°C in the presence of test compounds or the compound vehicle DMSO (0.5% [vol/vol]). PE46 products were run in 15% denaturing polyacrylamide gels containing 7 M urea (Bio-Rad, Hercules, CA), whereas LE19p products were run in urea-free 15% polyacrylamide gels (Bio-Rad). Gels were stained with SYBR green II (Invitrogen) and visualized on a Gel Doc molecular imager (Bio-Rad). RNA band intensities were quantified by densitometry using ImageJ (version 1.46r).
Cells, replicons, and plasmids.
A bicistronic HCV G1b replicon containing a luciferase reporter gene (51) was kindly provided by Ralf Bartenschlager (University of Heidelberg, Germany). The bicistronic G3a replicon S52/SG-Feo, containing a chimeric gene encoding firefly luciferase fused with neomycin phosphotransferase (33), was kindly provided by Charlie Rice (The Rockefeller University, New York). The plasmid for a tricistronic HCV G2a replicon (tri-JFH1) (52) was kindly provided by John McLauchlan (The University of Glasgow, Scotland). For the G2a replicon, RNA transcripts were generated and electroporated into Huh-7 cells for clonal selection. Cells were grown in the presence of 750 μg/ml of Geneticin for 3 weeks, and surviving colonies were isolated and characterized for replicon RNA levels and HCV NS5a expression as described previously (53).
HCV replicon assays.
The antiviral activity of HCV NNIs was examined using G1b, G2a, and G3a replicon-bearing cells and increasing concentrations of each test compound (0.01 nM to 100 μM). The nucleoside inhibitor 2CM was used as a positive control for replicon inhibition. Huh-7 (G1b and G2a) or Huh-7.5 cells (G3a) bearing the HCV subgenomic replicons were seeded into 96-well plates at a density of 5,000 cells/well in antibiotic-free Dulbecco's modified Eagle's medium (DMEM). On the next day, compounds were freshly diluted in complete medium, added to the cells, and incubated for 72 h. Untreated cells were incubated with 0.5% DMSO, the compound vehicle. Replication of the HCV replicons was determined by luciferase activity, measured using a luciferase assay system kit (Promega, Madison, WI) following the manufacturer's instructions. Luminescence was measured on a FLUOstar Optima microplate reader (BMG Labtech, Ortenberg, Germany). Replication of HCV replicons in treated cells was compared to that in untreated cells to calculate percent inhibition. Half-maximal effective concentration (EC50) values were determined by nonlinear regression in GraphPad Prism, version 6.02.
RESULTS
Inhibitory activity of NNIs against HCV replicons.
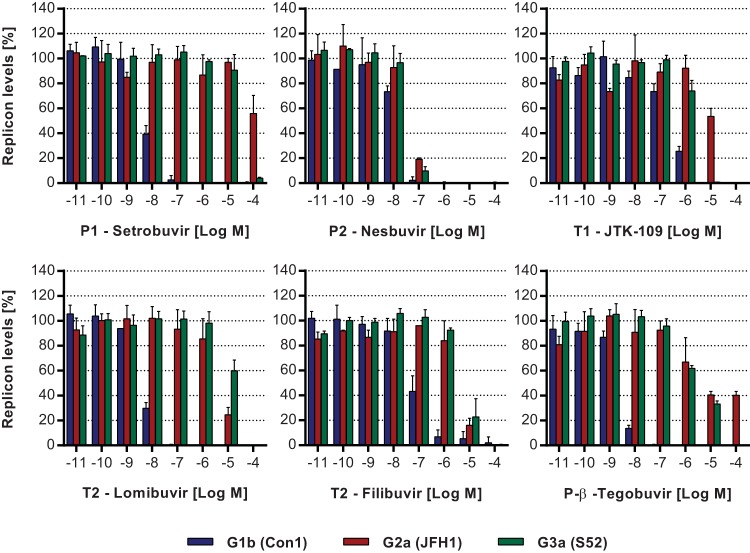
In order to examine the cross-genotype activity of HCV NNIs, replicons of HCV G1b, G2a, and G3a were used. Compounds were selected which bind to each of the five known NNI sites, and their ability to inhibit HCV replicon replication was assessed at increasing concentrations (0.01 nM to 100 μM). As a positive control, 2CM, an NI known to be an HCV cross-genotypic inhibitor, was also used. All three HCV replicons were susceptible to inhibition by 2CM. Specifically, the EC50 for the G1b replicon was 767.1 nM; however, 5-fold (3.8 μM) and 3-fold (2.2 μM) increases in EC50 were observed with the G2a and G3a replicons, respectively (Fig. 2; Table 1). In contrast, the five NNIs demonstrated differential inhibitory efficacies across replicons of genotypes 1b, 2a, and 3a. First, the P1 inhibitor setrobuvir demonstrated a significant loss of inhibitory activity against HCV G2a and G3a replicons in comparison to the EC50 of 8.1 nM for the G1b replicon. Only 44% ± 14.6% inhibition of G2a replicon replication was observed with the maximal concentration of 100 μM (>12,000-fold increase in EC50). Similarly, when examined using the G3a replicon, the EC50 was 26.1 μM, or >3,200-fold higher than that for the G1b replicon (Fig. 2; Table 1). The P2 inhibitor nesbuvir had an EC50 of 16.6 μM when examined using the G1b replicon, and it showed comparable activities against all three genotypes, with only 3-fold and 2-fold reductions in potency against G2a (EC50 = 43.3 μM) and G3a (EC50 = 39.9 μM) replicons, respectively, compared to the G1b replicon (Fig. 2; Table 1). Overall, these results show that the P2 binder nesbuvir is a better cross-genotype inhibitor than the NNI setrobuvir, which binds to the P1 site.
FIG 2.
Inhibitory activities of HCV NNIs against subgenomic replicons. The activities of HCV NNIs were examined using subgenomic replicons of HCV G1b (blue bars), G2a (red bars), and G3a (green bars), which all contained luciferase reporter genes. Cells harboring HCV replicons were treated with the compounds, and luciferase activity was measured 72 h later. Results are shown as percentages of the mock (0.5% [vol/vol] DMSO) treatment level. Data are the averages and standard deviations of results from triplicate experiments.
TABLE 1.
Cross-genotype activities of HCV inhibitors against subgenomic replicons
| Compound | Binding site | G1b replicon |
G2a replicon |
G3a replicon |
|||||
|---|---|---|---|---|---|---|---|---|---|
| EC50 (nM) | 95% CIa | EC50 (nM) | 95% CIa | Fold changeb | EC50 (nM) | 95% CIa | Fold changeb | ||
| 2CM | NI | 767.1 | 520.5–1,130 | 3,765 | 1,548–9,156 | 5 | 2,249 | 1,437–3,517 | 3 |
| Setrobuvir | P1 | 8.1 | 6.1–10.6 | >100,000 | >12,000 | 26,060 | 17,690–38,390 | 3,217 | |
| Nesbuvir | P2 | 16.6 | 10.9–25.4 | 43.3 | 27.3–68.8 | 3 | 39.9 | 27.4–58.1 | 2 |
| JTK-109 | T1 | 257.1 | 153.6–430.2 | 10,620 | 6,434–17,520 | 41 | 1,508 | 944–2,409 | 6 |
| Lomibuvir | T2 | 5.9 | 4.4–7.9 | 3,998 | 2,406–6,643 | 678 | 12,030 | 8,744–16,540 | 2,039 |
| Filibuvir | T2 | 79.2 | 55.8–112.6 | 3,176 | 1,904–5,296 | 40 | 4,698 | 3,448–6,401 | 59 |
| Tegobuvir | P-β | 3.2 | 2.3–4.3 | 10,960 | 3,207–37,440 | 3,425 | 2,629 | 1,845–3,746 | 822 |
CI, confidence interval.
Fold change in EC50 compared to that obtained using the G1b replicon.
Among the thumb binders, JTK-109, which binds to the T1 site, inhibited the replication of the G1b replicon with an EC50 of 257.1 nM, while a 6-fold increase in EC50 was observed for the G3a replicon (EC50 = 1.5 μM) and a 41-fold increase in EC50 was observed for the G2a replicon (EC50 = 10.6 μM) (Fig. 2; Table 1). Both T2 binders, lomibuvir and filibuvir, inhibited the G1b replicon, with EC50s of 5.9 nM and 79.2 nM, respectively. These two T2 inhibitors appeared to be specific G1b inhibitors, with a significant loss of inhibitory activity observed with lomibuvir against the G2a and G3a replicons (678-fold and >2,000-fold increases, respectively, in the EC50), while filibuvir demonstrated 40-fold and 59-fold increases in the EC50 for the G2a and G3a replicons, respectively (Table 1). Lastly, tegobuvir, a P-β binder, inhibited the replication of the G1b replicon with an EC50 of 3.2 nM, but its inhibitory activity was significantly reduced against G2a (>3,400-fold) and G3a (822-fold) replicons compared to the G1b replicon (Fig. 2; Table 1).
Inhibitory activity of HCV NNIs against recombinant RdRps.
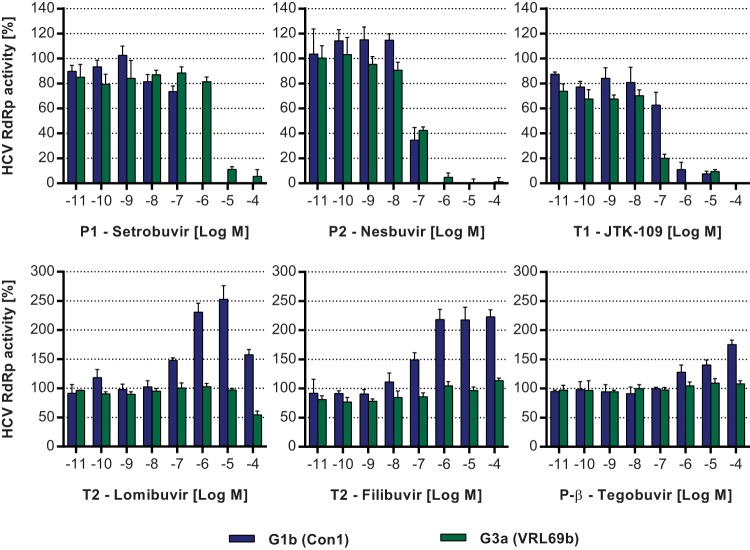
The effects of HCV NNIs on the de novo activity of G1b and G3a RdRps were analyzed using an RdRp fluorescence assay. Inhibition of recombinant RdRp activity was measured by the reduction of dsRNA produced from a poly(C) template, as determined by PicoGreen fluorescence, and compared to control reactions (49). Consistent with replicon results, the P1 binder setrobuvir inhibited the G1b RdRp with an IC50 of 157.5 nM and was 17-fold less potent when examined using the G3a RdRp (IC50 = 2.6 μM) (Fig. 3; Table 2). In contrast to setrobuvir, and also consistent with the replicon results, the P2 RdRp inhibitor nesbuvir demonstrated similar efficacies against both G1b and G3a RdRps, with IC50s of 76.7 nM and 75.0 nM, respectively (Fig. 3; Table 2). In a similar pattern, JTK-109 (T1 binder) showed comparable inhibitory activities against both G1b and G3a RdRps, with IC50s of 35.4 nM (G3a) and 107.6 nM (G1b) (Table 2).
FIG 3.
Effects of HCV NNIs on de novo activity of recombinant G1b and G3a RdRps. The inhibitory activities of HCV NNIs were examined using G1b (blue bars) and G3a (green bars) recombinant RdRps. Assays were performed using a homopolymeric poly(C) RNA template, and RdRp activity was measured via the quantitation of dsRNA formation using the fluorescent dye PicoGreen. RdRp activity is shown as a percentage of control reactions incubated with the compound vehicle DMSO (0.5% [vol/vol]). Data are the averages and standard deviations of results from triplicate experiments.
TABLE 2.
Inhibitory activities of HCV NNIs against recombinant RdRps
| Compound | Site | G1b RdRp |
G3a RdRp |
|||
|---|---|---|---|---|---|---|
| IC50 (nM) | 95% CIa | IC50 (nM) | 95% CIa | Fold changeb | ||
| Setrobuvir | P1 | 157.5 | 82.8–299.5 | 2,605.0 | 1,392–4,873 | 17 |
| Nesbuvir | P2 | 76.7 | 21.6–271.9 | 75.0 | 57.6–97.7 | 1 |
| JTK-109 | T1 | 107.6 | 45.6–253.8 | 35.4 | 15.0–83.6 | 0.3 |
| Lomibuvir | T2 | Enhanced | >100,000 | |||
| Filibuvir | T2 | Enhanced | >100,000 | |||
| Tegobuvir | P-β | Enhanced | >100,000 | |||
CI, confidence interval.
Fold change in IC50 compared to that obtained with the G1b RdRp.
Surprisingly, when the T2 binders were examined using the RdRp fluorescence assay, an increase in the de novo activity of the HCV G1b RdRp was observed as measured by increased dsRNA formation (Fig. 3). Upon the addition of both lomibuvir and filibuvir, which inhibited the G1 replicon, the G1b RdRp de novo activity appeared to increase 50% at 100 nM and >100% at 1 μM (Fig. 3). This effect was not observed for the G3a RdRp, i.e., the de novo activity for the G3a enzyme was not induced by lomibuvir or filibuvir (Fig. 3). In fact, lomibuvir inhibited the G3a de novo RdRp activity, but only at higher concentrations (45.5% ± 6.6% inhibition at 100 μM), whereas filibuvir did not inhibit the G3a RdRp at any of the examined concentrations (Fig. 3). Induction of the de novo activity of the G1b RdRp was not limited to T2 binders, as tegobuvir, which binds to the P-β site, also specifically enhanced G1b RdRp activity, although this effect was not as potent as that of lomibuvir or filibuvir, with a 75.5% ± 7.5% increase in G1b RdRp activity at 100 μM (Fig. 3). Tegobuvir had no observable effect on the activity (enhancement or inhibition) of the G3a RdRp (Fig. 3).
In order to eliminate the possibility that the enhancement of G1b de novo RdRp activity by T2- and P-β-binding NNIs was an artifact of the PicoGreen assay system, an alternative radioactive-nucleotide incorporation assay was used, with lomibuvir as a representative of the three de novo activity-enhancing NNIs. A marked increase in the de novo activity of the G1b enzyme by lomibuvir was again seen with the radioactive assay; de novo activity increased by 39.1% ± 10.6% at 100 nM and by 115% ± 29.7% at 10 μM. Lomibuvir did not enhance the de novo activity of the G3a RdRp (data not shown). In the radioactive assay, lomibuvir also inhibited both G1b and G3a RdRps at very high concentrations, and it was more potent against the G3a RdRp (81% ± 1.5% inhibition) than against the G1b RdRp (41% ± 3.3% inhibition) at 100 μM (data not shown).
Gel-based examination of RdRp inhibition.
The mechanism of G1b RdRp inhibition by the different NNIs was further analyzed using two gel-based assays described by Yi et al. (47). Only the G1b RdRp was used for this analysis, as it is the target enzyme for all six NNIs used in this study. The PE46 template is designed to form a hairpin to initiate primed transcription (47), while the LE19p template has a puromycin modification of the 3′ terminus, allowing only de novo dsRNA synthesis (47). NNIs that bind to the palm and thumb sites (T1, T2, P1, and P2) inhibited the primer-dependent activity of the G1b RdRp, as assessed using the PE46 template (Fig. 4A). These inhibitors exerted inhibitory activity at concentrations between 10 nM and 100 nM, with >82% inhibition at 1 μM, with the exception of JTK-109, which required concentrations of 1 μM and 10 μM for 45% and 84% inhibitions of G1b RdRp activity, respectively (Fig. 4A).
FIG 4.
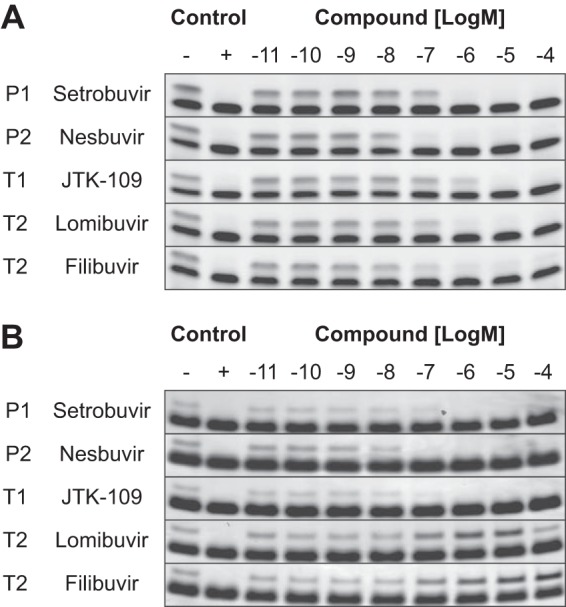
Effects of HCV G1 NNIs on primed and de novo RdRp activities. The effects of HCV NNIs on primed and de novo activities of the HCV G1b RdRp were analyzed using gel-based assays. Reaction mixtures were run in the absence of compound (negative control), in the absence of RdRp (positive control), and in the presence of increasing compound concentrations (0.01 nM to 100 μM). (A) Primer-dependent activity was examined using the PE46 RNA template, and reaction products were analyzed in a 15% denaturing gel. (B) Primer-independent or de novo RdRp activity was analyzed using LE19p, and products were run in a 15% nondenaturing gel. The RNA templates PE46 and LE19p are the bottom bands in both panels, and the RdRp products are the top bands. P1, P2, and T1 binders inhibited both modes of G1b RdRp activity at concentrations of >100 nM. At the same concentrations, however, T2 binders inhibited primed transcription (A) while unexpectedly enhancing de novo activity, by 80% (lomibuvir) and 27% (filibuvir) (B). The two T2 binders also enhanced G1b de novo RdRp activity at 1 μM, by 180% (lomibuvir) and 76% (filibuvir), as quantified by densitometry.
Gel-based analysis of the de novo G1b RdRp activity was performed to confirm observations of enhanced RdRp activity by T2- and P-β-binding NNIs in the quantitative fluorescence and radioactive in vitro assays (Fig. 3). In the gel-based assay, inhibitors which bound to the P1, P2, and T1 sites abolished the de novo activity of the HCV G1b RdRp when examined at concentrations of >100 nM (Fig. 4B). In contrast, an increase in de novo RdRp activity was again observed with the T2 inhibitors lomibuvir and filibuvir. The addition of lomibuvir resulted in an 80% increase in the de novo activity at 100 nM and a 180% increase at 1 μM (Fig. 4B), as quantified by densitometry (data not shown), but at a higher concentration (100 μM), a modest (4.5%) reduction in the activity was observed (Fig. 4B). In the gel-based assays, filibuvir also enhanced de novo transcription of the G1b RdRp, with a 27% increase at 100 nM, a 76% increase at 1 μM, a 144% increase at 10 μM, and a 240% increase at 100 μM, as quantified by densitometry (Fig. 4B). Overall, these results are consistent with the pattern observed using the fluorescence and radioactive RdRp assays, where T2 binders enhanced the de novo activity but inhibited primed elongation of the HCV G1b RdRp. The P-β binder tegobuvir had no observable effect on G1b RdRp activity when examined using either the primed or de novo gel-based assays (data not shown).
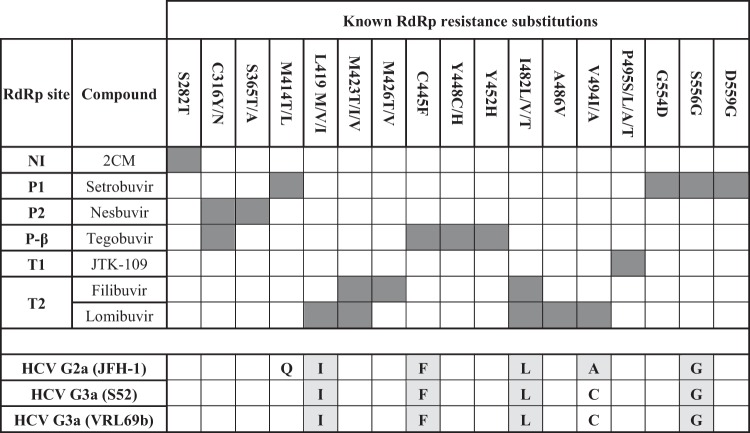
Analysis of resistance mutations in HCV RdRp sequences.
In order to gain insights into the differential efficacy of HCV inhibitors, the amino acid sequences for the G1, G2, and G3 RdRps were analyzed for known substitutions conferring resistance to all compounds tested in this study (reviewed in reference 54). A summary of amino acid substitutions associated with G1b resistance to the NNIs in this study is shown in Fig. 5. Residues are also shown for the RdRps from the G2a (JFH-1) and G3a (S52) replicon strains and for the G3a strain VRL69b. HCV G3a VRL69b was used for recombinant enzyme experiments in this study and differed from the S52 G3a RdRp by only 6 amino acids, none of which are known to confer resistance to HCV RdRp inhibitors (Fig. 5).
FIG 5.
Amino acid substitutions which confer resistance to HCV NNIs used in this study. Previously characterized substitutions in the HCV G1 RdRp which confer resistance to compounds analyzed in this study are shown. The corresponding amino acid residues for non-G1 RdRps analyzed in this study are shown along the bottom, and those that confer resistance to HCV NNIs are highlighted for G2a and G3a RdRps.
Examination of HCV G2a and G3a RdRp sequences revealed multiple naturally occurring resistance residues in both G2a and G3a RdRps, which likely confer resistance to HCV NNIs that bind to the P1, T2, and P-β sites but not to NNIs which bind to P2 or T1. Specifically, residues G556 and F445, implicated in G1 RdRp resistance to setrobuvir (P1) (55, 56) and tegobuvir (P-β) (25), respectively, were found in both G2a and G3a RdRp sequences (Fig. 5). Both G2a and G3a RdRps contained residues I419 and L482 (27, 29) (Fig. 5), which are associated with resistance to the T2 binders lomibuvir and filibuvir (47, 57, 58). Furthermore, the amino acid residue A494, also associated with T2 NNI resistance in G1b, was present in the G2a RdRp sequence but not in the G3a RdRp sequence (Fig. 5). The amino acid substitution V494A has been selected in vivo after treatment of G1 HCV-infected patients with lomibuvir (72), and also by us in vitro, using the G1b replicon (unpublished data). In contrast, none of the substitutions previously known to confer resistance to JTK-109 (T1) and nesbuvir (P2) were present in G2a or G3a RdRp (29) (Fig. 5).
DISCUSSION
The standard of care for treatment of HCV infection is currently undergoing significant changes, moving from traditional interferon-based regimens to shorter, interferon-free combination DAA regimens. Several dozen DAAs are in different stages of clinical trials (11), and four have so far been approved for use in combination with PEG-IFN/RBV. With all-oral, interferon-free combination therapies now on the horizon, a better understanding of the cross-genotypic specificities and modes of action for the different G1 NNIs is warranted, particularly as replicons are now available for other HCV genotypes. In this study, we examined six representative NNIs, which between them bind to all five RdRp binding pockets, for the ability to inhibit G1, G2, and G3 HCV replicons. Furthermore, the ability of the NNIs to inhibit both modes of transcription, i.e., de novo and primed elongation, was also assessed. Of the NNIs selected for this study, lomibuvir, setrobuvir, and tegobuvir are currently in phase II clinical trials, whereas clinical development has been halted for JTK-109, nesbuvir, and, more recently, filibuvir (59).
Examination of the cross-genotypic efficacy of the NNI setrobuvir (P1 binder) revealed that it had little effect on the replication of the G2a (EC50 > 100 μM) and G3a (EC50 = 26 μM) replicons compared to that of the G1b replicon (EC50 = 8.1 nM) (Table 1). Similarly, recombinant G3a RdRp was far less susceptible to inhibition by setrobuvir (IC50 = 2.6 μM) than G1b RdRp was (IC50 = 157.5 nM) (Table 2). The loss of activity of benzothiadiazines, such as setrobuvir, against G2a and G3a viruses is consistent with previous reports on chimeric replicons (29, 30, 32) and recombinant G2a and G3a RdRps (27, 28). Resistance to benzothiadiazines in G2a and G3a viruses appears to be due to the glycine residue at position 556 (55, 56) of the RdRp, an amino acid substitution which was detected in benzothiadiazine-resistant G1 replicons (60) and in G1 HCV-infected patients after treatment (61). Surprisingly, mutating G556 in the G3a RdRp to a serine, the residue naturally occurring in the G1b enzyme, did not confer susceptibility to benzothiadiazines, at least in recombinant RdRp studies (27). Thus, the precise mechanism of G3a resistance to this class of molecules remains to be resolved.
Of the six NNIs examined, only nesbuvir (site P2) demonstrated cross-genotypic inhibitory activity against G1b, G2a, and G3a replicons, with EC50s between 16.6 nM and 43.3 nM (Table 1). Nesbuvir also inhibited the de novo activity of the G3a RdRp, with an IC50 of 75 nM, compared to 76.7 nM for the G1b RdRp (Table 2). These findings are consistent with previous studies using HCV replicons and RdRps, where nesbuvir demonstrated equivalent potencies against all HCV genotypes (28–30, 32, 62), and correspond with the absence of naturally occurring substitutions in the P2 binding region of G2a and G3a RdRps.
In addition to inhibiting the G1b replicon (EC50 = 257.1 nM) and G1b RdRp (EC50 = 107.6 nM), the benzimidazole JTK-109 (site T1) demonstrated similar inhibitory activities against the G3a replicon (EC50 = 1.5 μM) and enzyme (IC50 = 35.4 nM). However, JTK-109 was 41-fold less potent against the G2a replicon (Fig. 2; Table 1). The reasons for the reduced efficacy of T1 binders, such as JTK-109, against G2a viruses are not fully understood (29). For instance, although substitutions at the P495 residue are known to confer resistance to JTK-109 in both G1b replicon and recombinant enzyme models (63), these residues are not observed in the G2a RdRp (Fig. 5). However, a naturally occurring alanine at position 494 in the G2a RdRp, which is not present in the susceptible G3a RdRp, may explain the reduced inhibitory activity of T1 binders against the G2a replicon (Fig. 5). Indeed, the V494A substitution has been associated with resistance of recombinant G1b RdRp to other thumb I binders, such as indole-N-acetamides (64), and further work is needed to confirm if this substitution confers JTK-109 resistance.
Resistance to the T2 inhibitors lomibuvir and filibuvir has been attributed to L419I and I482L substitutions in the HCV RdRp (47, 57, 58), both of which are present in G2a and G3a viruses (27, 29). This is consistent with our finding of a lack of activity for these drugs against G2a and G3a replicons (Fig. 5), against which T2 binders were much less potent than the case against the G1b replicon (Table 1). Tegobuvir (P-β binder) was less potent against the G2a (>3,400-fold) and G3a (822-fold) replicons than against the G1b replicon (Table 1). Resistance to tegobuvir in G2a and G3a replicons is likely due to the C445F substitution (Fig. 5), which lies in the β-hairpin and has been shown to confer resistance to this class of compounds in replicon studies (25). It is worth noting that the reduced potencies of lomibuvir, filibuvir, and tegobuvir against HCV G3a observed in this study are higher than those recently reported for a different G3a replicon based on the same HCV G3a strain, S52 (63-fold, 13-fold, and 22-fold, respectively) (62), but more consistent with levels reported previously using chimeric HCV replicons (29, 30, 32).
To our surprise, de novo but not primed RdRp activity of the recombinant G1b RdRp was enhanced approximately 2-fold by a 1 μM concentration of the T2 binders lomibuvir and filibuvir and, to a lesser degree, by the P-β binder tegobuvir (Fig. 3 and 4). The increased RdRp activity was detected using de novo RdRp assays with both fluorescent and radioactive outputs and was further confirmed by gel-based assays (Fig. 4), indicating that it was a true effect and not an artifact of the assay used. Furthermore, no increase in de novo activity was detected when these compounds were examined using the G3a RdRp. When tegobuvir was examined using the gel-based RdRp assay, no effect on G1b RdRp activity was detected, in contrast to the enhancement of G1b RdRp activity seen using the RdRp fluorescence assay. This may reflect a reduced assay sensitivity which meant that the lower level of RdRp enhancement seen with tegobuvir than with lomibuvir and filibuvir (Fig. 3) could not be detected in the gel-based assay.
Most studies to date that have analyzed NNI RdRp inhibition have used primer-dependent assays, so it is likely that the enhancement of de novo RdRp activity has gone unnoticed. In fact, both thiophene-2-carboxylic acids (18) and dihydropyranones (19, 65), the scaffolds from which lomibuvir and filibuvir, respectively, were developed, were identified from high-throughput screening studies using primed RdRp assays, and further evaluation was achieved using the replicon model. In a recent study, Yi et al. reported no observable effect on de novo activity of the G1b RdRp by lomibuvir or filibuvir (47). However, lomibuvir and filibuvir were examined over a relatively narrow range of concentrations (50 to 200 nM), and indeed, a small increase in RdRp activity can be observed in Fig. 3 from that paper for lomibuvir and filibuvir at a concentration of 200 nM (47). In contrast to lomibuvir and filibuvir, tegobuvir was identified using HCV replicon and infectious (JFH-1) cell culture systems (24). Interestingly, in another recent study, tegobuvir was shown to increase the G1b primed RdRp activity by 50% at a concentration of 3.7 μM but resulted in a 40% inhibition at 100 μM, an observation that the authors were unable to explain (66). Although the RdRp assay used in the latter study uses a self-primed RNA template, no measures were taken to block the de novo RdRp activity; therefore, both modes of activity may be in operation in that assay system. The effect was not observed with the T2 binders lomibuvir and filibuvir in the same study (66). In summary, the enhancement of the HCV RdRp activity by T2 and P-β NNIs reported in this study can be observed in two previous reports (47, 66); however, the effect has so far gone unnoticed or unexplained.
The mechanism of action for tegobuvir and T2 inhibitors is poorly understood. Tegobuvir requires intracellular activation to form a covalent inhibitor of the HCV RdRp (67) and is not thought to interact with the enzyme before activation (66), although, interestingly, no activation was needed to enhance de novo RNA synthesis in the present study. In contrast, recent evidence suggests that filibuvir and lomibuvir reduce RdRp binding to RNA but do not block the interaction completely (66). Structural analysis of the RdRp in complex with thiophene-2-carboxylic acids indicated that these molecules bind to the closed conformation of the enzyme (68) and induce conformational changes that may interfere with enzymatic activity (68, 69). Although the authors of that study suggested that this resulted in an initiation-incompetent enzyme, our results indicate that filibuvir and lomibuvir in fact increase the initiation (de novo) but inhibit the elongation activity of the G1b RdRp. Recently, recombinant HCV RdRp was proposed to exist as a mixture of conformations which are in dynamic equilibrium, with stabilization of one conformation occurring at the expense of the other in solution (45). Given that formation of the first few phosphodiester bonds and the transition to elongation are two known rate-limiting steps of the HCV RdRp reaction (44, 70, 71), it is likely that T2 and P-β enhance the initiation efficiency (de novo) of the HCV RdRp by stabilizing the RdRp conformations required for these rate-limiting steps. However, further studies are needed to validate such effects and to detail the mechanism of action for the thumb-interacting subset of HCV NNIs.
In summary, we have analyzed the inhibitory activities of six representative HCV inhibitors that bind to the five NNI sites across three genotypes: G1b, G2a, and G3a. Our data indicate that only the P2 inhibitor nesbuvir is cross-genotypic, as naturally occurring amino acid residues in non-G1 RdRps largely confer resistance against other NNIs. We also report a previously uncharacterized enhancement of de novo RdRp activity by the T2 binders lomibuvir and filibuvir, as well as the imidazopyridine tegobuvir (P-β), which improves our understanding of the mechanisms by which these compounds exert antiviral activity against HCV.
ACKNOWLEDGMENTS
We thank Ralf Bartenschlager, Charlie Rice, John McLauchlan, and Mark Harris for reagents.
A.A.E. and P.A.W. conceived and designed the experiments; A.A.E. and E.T. performed the experiments; A.A.E., M.W.D., and P.A.W. analyzed the data; M.W.D. contributed reagents, materials, and analysis tools; P.A.W. obtained funding; and A.A.E. and P.A.W. wrote the paper.
We declare that we have no conflicts of interest.
This work was partially funded by a National Health and Medical Research Council project grant (APP1010327) and an Australian Research Council Discovery grant (DP120104073).
Footnotes
Published ahead of print 22 September 2014
REFERENCES
- 1.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342. 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 2.Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973. 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 3.Manns MP, Wedemeyer H, Cornberg M. 2006. Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55:1350–1359. 10.1136/gut.2005.076646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364:2405–2416. 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 5.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206. 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarrazin C, Hezode C, Zeuzem S, Pawlotsky JM. 2012. Antiviral strategies in hepatitis C virus infection. J. Hepatol. 56(Suppl 1):S88–S100. 10.1016/S0168-8278(12)60010-5. [DOI] [PubMed] [Google Scholar]
- 7.Jacobson I, Dore G, Foster G, Fried M, Radu M, Rafalskiy V, Moroz L, Craxi A, Peeters M, Lenz O. 2013. Simeprevir (TMC435) with peginterferon/ribavirin for chronic HCV genotype-1 infection in treatment-naïve patients: results from QUEST-1, a phase III trial. J. Hepatol. 58:S574. [Google Scholar]
- 8.Gane EJ, Stedman CA, Hyland RH, Ding X, Svarovskaia E, Symonds WT, Hindes RG, Berrey MM. 2013. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N. Engl. J. Med. 368:34–44. 10.1056/NEJMoa1208953. [DOI] [PubMed] [Google Scholar]
- 9.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR, Jacobson IM, Kowdley KV, Nyberg L, Subramanian GM, Hyland RH, Arterburn S, Jiang D, McNally J, Brainard D, Symonds WT, McHutchison JG, Sheikh AM, Younossi Z, Gane EJ. 2013. Sofosbuvir for previously untreated chronic hepatitis C infection. N. Engl. J. Med. 368:1878–1887. 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 10.Lawitz E, Lalezari JP, Hassanein T, Kowdley KV, Poordad FF, Sheikh AM, Afdhal NH, Bernstein DE, DeJesus E, Freilich B, Nelson DR, Dieterich DT, Jacobson IM, Jensen D, Abrams GA, Darling JM, Rodriguez-Torres M, Reddy KR, Sulkowski MS, Bzowej NH, Hyland RH, Mo H, Lin M, Mader M, Hindes R, Albanis E, Symonds WT, Berrey MM, Muir A. 2013. Sofosbuvir in combination with peginterferon alfa-2a and ribavirin for non-cirrhotic, treatment-naive patients with genotypes 1, 2, and 3 hepatitis C infection: a randomised, double-blind, phase 2 trial. Lancet Infect. Dis. 13:401–408. 10.1016/S1473-3099(13)70033-1. [DOI] [PubMed] [Google Scholar]
- 11.Scheel TK, Rice CM. 2013. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 19:837–849. 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 96:13034–13039. 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937–943. 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 14.Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417–1426. 10.1016/S0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 15.Caillet-Saguy C, Simister PC, Bressanelli S. 2011. An objective assessment of conformational variability in complexes of hepatitis C virus polymerase with non-nucleoside inhibitors. J. Mol. Biol. 414:370–384. 10.1016/j.jmb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Haudecoeur R, Peuchmaur M, Ahmed-Belkacem A, Pawlotsky JM, Boumendjel A. 2013. Structure-activity relationships in the development of allosteric hepatitis C virus RNA-dependent RNA polymerase inhibitors: ten years of research. Med. Res. Rev. 33:934–984. 10.1002/med.21271. [DOI] [PubMed] [Google Scholar]
- 17.Bartenschlager R, Lohmann V, Penin F. 2013. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 11:482–496. 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 18.Chan L, Das SK, Reddy TJ, Poisson C, Proulx M, Pereira O, Courchesne M, Roy C, Wang W, Siddiqui A. 2004. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. 1. Sulfonamides. Bioorg. Med. Chem. Lett. 14:793–796. 10.1016/j.bmcl.2003.10.067. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Tatlock J, Linton A, Gonzalez J, Jewell T, Patel L, Ludlum S, Drowns M, Rahavendran SV, Skor H. 2009. Discovery of (R)-6-cyclopentyl-6-(2-(2,6-diethylpyridin-4-yl) ethyl)-3-((5,7-dimethyl-[1,2,4] triazolo [1,5-a] pyrimidin-2-yl) methyl)-4-hydroxy-5,6-dihydropyran-2-one (PF-00868554) as a potent and orally available hepatitis C virus polymerase inhibitor. J. Med. Chem. 52:1255–1258. 10.1021/jm8014537. [DOI] [PubMed] [Google Scholar]
- 20.Gopalsamy A, Chopra R, Lim K, Ciszewski G, Shi M, Curran KJ, Sukits SF, Svenson K, Bard J, Ellingboe JW. 2006. Discovery of proline sulfonamides as potent and selective hepatitis C virus NS5b polymerase inhibitors. Evidence for a new NS5b polymerase binding site. J. Med. Chem. 49:3052–3055. 10.1021/jm060168g. [DOI] [PubMed] [Google Scholar]
- 21.Nyanguile O, Pauwels F, Van den Broeck W, Boutton CW, Quirynen L, Ivens T, van der Helm L, Vandercruyssen G, Mostmans W, Delouvroy F, Dehertogh P, Cummings MD, Bonfanti JF, Simmen KA, Raboisson P. 2008. 1,5-Benzodiazepines, a novel class of hepatitis C virus polymerase nonnucleoside inhibitors. Antimicrob. Agents Chemother. 52:4420–4431. 10.1128/AAC.00669-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhanak D, Duffy KJ, Johnston VK, Lin-Goerke J, Darcy M, Shaw AN, Gu B, Silverman C, Gates AT, Nonnemacher MR, Earnshaw DL, Casper DJ, Kaura A, Baker A, Greenwood C, Gutshall LL, Maley D, DelVecchio A, Macarron R, Hofmann GA, Alnoah Z, Cheng HY, Chan G, Khandekar S, Keenan RM, Sarisky RT. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322–38327. 10.1074/jbc.M205566200. [DOI] [PubMed] [Google Scholar]
- 23.Kneteman NM, Howe AY, Gao T, Lewis J, Pevear D, Lund G, Douglas D, Mercer DF, Tyrrell DLJ, Immermann F. 2009. HCV796: a selective nonstructural protein 5B polymerase inhibitor with potent anti-hepatitis C virus activity in vitro, in mice with chimeric human livers, and in humans infected with hepatitis C virus. Hepatology 49:745–752. 10.1002/hep.22717. [DOI] [PubMed] [Google Scholar]
- 24.Vliegen I, Paeshuyse J, De Burghgraeve T, Lehman LS, Paulson M, Shih IH, Mabery E, Boddeker N, De Clercq E, Reiser H, Oare D, Lee WA, Zhong W, Bondy S, Purstinger G, Neyts J. 2009. Substituted imidazopyridines as potent inhibitors of HCV replication. J. Hepatol. 50:999–1009. 10.1016/j.jhep.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih IH, Vliegen I, Peng B, Yang H, Hebner C, Paeshuyse J, Purstinger G, Fenaux M, Tian Y, Mabery E, Qi X, Bahador G, Paulson M, Lehman LS, Bondy S, Tse W, Reiser H, Lee WA, Schmitz U, Neyts J, Zhong W. 2011. Mechanistic characterization of GS-9190 (tegobuvir), a novel nonnucleoside inhibitor of hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 55:4196–4203. 10.1128/AAC.00307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann V, Bartenschlager R. 2014. On the history of hepatitis C virus cell culture systems. J. Med. Chem. 57:1627–1642. 10.1021/jm401401n. [DOI] [PubMed] [Google Scholar]
- 27.Pauwels F, Mostmans W, Quirynen LM, van der Helm L, Boutton CW, Rueff AS, Cleiren E, Raboisson P, Surleraux D, Nyanguile O, Simmen KA. 2007. Binding-site identification and genotypic profiling of hepatitis C virus polymerase inhibitors. J. Virol. 81:6909–6919. 10.1128/JVI.01543-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.May MM, Lorengel H, Kreuter J, Zimmermann H, Ruebsamen-Schaeff H, Urban A. 2011. RNA-dependent RNA polymerases from different hepatitis C virus genotypes reveal distinct biochemical properties and drug susceptibilities. Biochim. Biophys. Acta 1814:1325–1332. 10.1016/j.bbapap.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Herlihy KJ, Graham JP, Kumpf R, Patick AK, Duggal R, Shi ST. 2008. Development of intergenotypic chimeric replicons to determine the broad-spectrum antiviral activities of hepatitis C virus polymerase inhibitors. Antimicrob. Agents Chemother. 52:3523–3531. 10.1128/AAC.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam AM, Espiritu C, Bansal S, Micolochick Steuer HM, Niu C, Zennou V, Keilman M, Zhu Y, Lan S, Otto MJ, Furman PA. 2012. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob. Agents Chemother. 56:3359–3368. 10.1128/AAC.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenaux M, Eng S, Leavitt SA, Lee Y-J, Mabery EM, Tian Y, Byun D, Canales E, Clarke MO, Doerffler E. 2013. Preclinical characterization of GS-9669, a thumb site II inhibitor of the hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 57:804–810. 10.1128/AAC.02052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong KA, Xu S, Martin R, Miller MD, Mo H. 2012. Tegobuvir (GS-9190) potency against HCV chimeric replicons derived from consensus NS5B sequences from genotypes 2b, 3a, 4a, 5a, and 6a. Virology 429:57–62. 10.1016/j.virol.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 33.Saeed M, Scheel TK, Gottwein JM, Marukian S, Dustin LB, Bukh J, Rice CM. 2012. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob. Agents Chemother. 56:5365–5373. 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu M, Corsa AC, Xu S, Peng B, Gong R, Lee YJ, Chan K, Mo H, Delaney WT, Cheng G. 2013. In vitro efficacy of approved and experimental antivirals against novel genotype 3 hepatitis C virus subgenomic replicons. Antiviral Res. 100:439–445. 10.1016/j.antiviral.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 35.Saeed M, Gondeau C, Hmwe S, Yokokawa H, Date T, Suzuki T, Kato T, Maurel P, Wakita T. 2013. Replication of hepatitis C virus genotype 3a in cultured cells. Gastroenterology 144:56.e7–58.e7. 10.1053/j.gastro.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 36.Frank C, Mohamed MK, Strickland GT, Lavanchy D, Arthur RR, Magder LS, El Khoby T, Abdel-Wahab Y, Aly Ohn ES, Anwar W, Sallam I. 2000. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet 355:887–891. 10.1016/S0140-6736(99)06527-7. [DOI] [PubMed] [Google Scholar]
- 37.Ray SC, Arthur RR, Carella A, Bukh J, Thomas DL. 2000. Genetic epidemiology of hepatitis C virus throughout Egypt. J. Infect. Dis. 182:698–707. 10.1086/315786. [DOI] [PubMed] [Google Scholar]
- 38.McOmish F, Yap PL, Dow BC, Follett EA, Seed C, Keller AJ, Cobain TJ, Krusius T, Kolho E, Naukkarinen R. 1994. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J. Clin. Microbiol. 32:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chowdhury A, Santra A, Chaudhuri S, Dhali GK, Maity SG, Naik TN, Bhattacharya SK, Mazumder DN. 2003. Hepatitis C virus infection in the general population: a community-based study in West Bengal, India. Hepatology 37:802–809. 10.1053/jhep.2003.50157. [DOI] [PubMed] [Google Scholar]
- 40.White PA, Zhai X, Carter I, Zhao Y, Rawlinson WD. 2000. Simplified hepatitis C virus genotyping by heteroduplex mobility analysis. J. Clin. Microbiol. 38:477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851–863. 10.1128/JVI.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong W, Uss AS, Ferrari E, Lau JY, Hong Z. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017–2022. 10.1128/JVI.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kao CC, Singh P, Ecker DJ. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251–260. 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- 44.Harrus D, Ahmed-El-Sayed N, Simister PC, Miller S, Triconnet M, Hagedorn CH, Mahias K, Rey FA, Astier-Gin T, Bressanelli S. 2010. Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis. J. Biol. Chem. 285:32906–32918. 10.1074/jbc.M110.151316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scrima N, Caillet-Saguy C, Ventura M, Harrus D, Astier-Gin T, Bressanelli S. 2012. Two crucial early steps in RNA synthesis by the hepatitis C virus polymerase involve a dual role of residue 405. J. Virol. 86:7107–7117. 10.1128/JVI.00459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosley RT, Edwards TE, Murakami E, Lam AM, Grice RL, Du J, Sofia MJ, Furman PA, Otto MJ. 2012. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 86:6503–6511. 10.1128/JVI.00386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi G, Deval J, Fan B, Cai H, Soulard C, Ranjith-Kumar CT, Smith DB, Blatt L, Beigelman L, Kao CC. 2012. Biochemical study of the comparative inhibition of hepatitis C virus RNA polymerase by VX-222 and filibuvir. Antimicrob. Agents Chemother. 56:830–837. 10.1128/AAC.05438-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomei L, Altamura S, Bartholomew L, Bisbocci M, Bailey C, Bosserman M, Cellucci A, Forte E, Incitti I, Orsatti L. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938–946. 10.1128/JVI.78.2.938-946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eltahla AA, Lackovic K, Marquis C, Eden JS, White PA. 2013. A fluorescence-based high-throughput screen to identify small compound inhibitors of the genotype 3a hepatitis C virus RNA polymerase. J. Biomol. Screen. 18:1027–1034. 10.1177/1087057113489883. [DOI] [PubMed] [Google Scholar]
- 50.Jones LA, Clancy LE, Rawlinson WD, White PA. 2006. High-affinity aptamers to subtype 3a hepatitis C virus polymerase display genotypic specificity. Antimicrob. Agents Chemother. 50:3019–3027. 10.1128/AAC.01603-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vrolijk JM, Kaul A, Hansen BE, Lohmann V, Haagmans BL, Schalm SW, Bartenschlager R. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201–209. 10.1016/S0166-0934(03)00134-4. [DOI] [PubMed] [Google Scholar]
- 52.Jones DM, Domingues P, Targett-Adams P, McLauchlan J. 2010. Comparison of U2OS and Huh-7 cells for identifying host factors that affect hepatitis C virus RNA replication. J. Gen. Virol. 91:2238–2248. 10.1099/vir.0.022210-0. [DOI] [PubMed] [Google Scholar]
- 53.Street A, Macdonald A, McCormick C, Harris M. 2005. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular β-catenin and stimulation of β-catenin-responsive transcription. J. Virol. 79:5006–5016. 10.1128/JVI.79.8.5006-5016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Legrand-Abravanel F, Nicot F, Izopet J. 2010. New NS5B polymerase inhibitors for hepatitis C. Expert Opin. Invest. Drugs 19:963–975. 10.1517/13543784.2010.500285. [DOI] [PubMed] [Google Scholar]
- 55.Thompson PA, Patel R, Showalter RE, Li C, Applemon JR, Steffy K. 2008. In vitro studies demonstrate that combinations of antiviral agents that include HCV polymerase inhibitor ANA598 have the potential to overcome viral resistance. Hepatology 48:1164A. [Google Scholar]
- 56.Le Pogam S, Seshaadri A, Kosaka A, Chiu S, Kang H, Hu S, Rajyaguru S, Symons J, Cammack N, Nájera I. 2008. Existence of hepatitis C virus NS5B variants naturally resistant to non-nucleoside, but not to nucleoside, polymerase inhibitors among untreated patients. J. Antimicrob. Chemother. 61:1205–1216. 10.1093/jac/dkn085. [DOI] [PubMed] [Google Scholar]
- 57.Le Pogam S, Kang H, Harris SF, Leveque V, Giannetti AM, Ali S, Jiang WR, Rajyaguru S, Tavares G, Oshiro C, Hendricks T, Klumpp K, Symons J, Browner MF, Cammack N, Najera I. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146–6154. 10.1128/JVI.02628-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi ST, Herlihy KJ, Graham JP, Nonomiya J, Rahavendran SV, Skor H, Irvine R, Binford S, Tatlock J, Li H. 2009. Preclinical characterization of PF-00868554, a potent nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 53:2544–2552. 10.1128/AAC.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahn J, Flamm SL. 2014. Frontiers in the treatment of hepatitis C virus infection. Gastroenterol. Hepatol. 10:90–100. [PMC free article] [PubMed] [Google Scholar]
- 60.Delang L, Vliegen I, Froeyen M, Neyts J. 2011. Comparative study of the genetic barriers and pathways towards resistance of selective inhibitors of hepatitis C virus replication. Antimicrob. Agents Chemother. 55:4103–4113. 10.1128/AAC.00294-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Torres M, Lawitz E, Cohen D, Larsen LM, Menon R, Collins C, Marsh T, Gibbs S, Bernstein B. 2009. Treatment-naïve, HCV genotype 1-infected subjects show significantly greater HCV RNA decreases when treated with 28 days of ABT-333 plus peginterferon and ribavirin compared to peginterferon and ribavirin alone. Hepatology 50:5A. [Google Scholar]
- 62.Yu M, Corsa AC, Xu S, Peng B, Gong R, Lee Y-J, Chan K, Mo H, Delaney W, IV, Cheng G. 2013. In vitro efficacy of approved and experimental antivirals against novel genotype 3 hepatitis C virus subgenomic replicons. Antiviral Res. 100:439–445. 10.1016/j.antiviral.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 63.Tomei L, Altamura S, Bartholomew L, Biroccio A, Ceccacci A, Pacini L, Narjes F, Gennari N, Bisbocci M, Incitti I, Orsatti L, Harper S, Stansfield I, Rowley M, De Francesco R, Migliaccio G. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225–13231. 10.1128/JVI.77.24.13225-13231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rydberg EH, Cellucci A, Bartholomew L, Mattu M, Barbato G, Ludmerer SW, Graham DJ, Altamura S, Paonessa G, De Francesco R. 2009. Structural basis for resistance of the genotype 2b hepatitis C virus NS5B polymerase to site A non-nucleoside inhibitors. J. Mol. Biol. 390:1048–1059. 10.1016/j.jmb.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 65.Love RA, Parge HE, Yu X, Hickey MJ, Diehl W, Gao J, Wriggers H, Ekker A, Wang L, Thomson JA. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575–7581. 10.1128/JVI.77.13.7575-7581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winquist J, Abdurakhmanov E, Baraznenok V, Henderson I, Vrang L, Danielson UH. 2013. Resolution of the interaction mechanisms and characteristics of non-nucleoside inhibitors of hepatitis C virus polymerase. Antiviral Res. 97:356–368. 10.1016/j.antiviral.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 67.Hebner CM, Han B, Brendza KM, Nash M, Sulfab M, Tian Y, Hung M, Fung W, Vivian RW, Trenkle J, Taylor J, Bjornson K, Bondy S, Liu X, Link J, Neyts J, Sakowicz R, Zhong W, Tang H, Schmitz U. 2012. The HCV non-nucleoside inhibitor tegobuvir utilizes a novel mechanism of action to inhibit NS5B polymerase function. PLoS One 7:e39163. 10.1371/journal.pone.0039163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Biswal BK, Cherney MM, Wang M, Chan L, Yannopoulos CG, Bilimoria D, Nicolas O, Bedard J, James MNG. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202–18210. 10.1074/jbc.M413410200. [DOI] [PubMed] [Google Scholar]
- 69.Biswal BK, Wang M, Cherney MM, Chan L, Yannopoulos CG, Bilimoria D, Bedard J, James MN. 2006. Non-nucleoside inhibitors binding to hepatitis C virus NS5B polymerase reveal a novel mechanism of inhibition. J. Mol. Biol. 361:33–45. 10.1016/j.jmb.2006.05.074. [DOI] [PubMed] [Google Scholar]
- 70.Shim JH, Larson G, Wu JZ, Hong Z. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J. Virol. 76:7030–7039. 10.1128/JVI.76.14.7030-7039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferrari E, He Z, Palermo RE, Huang H-C. 2008. Hepatitis C virus NS5B polymerase exhibits distinct nucleotide requirements for initiation and elongation. J. Biol. Chem. 283:33893–33901. 10.1074/jbc.M803094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang M, Zhang EZ, Ardzinski A, Tigges A, Davis A, Sullivan JC, Nelson M, Spanks J, Dorrian J, Nicolas O, Bartels DJ, Rao BG, Rijnbrand R, Kieffer TL. 2014. Genotypic and phenotypic analyses of hepatitis C virus variants observed in clinical studies of VX-222, a nonnucleoside NS5B polymerase inhibitor. Antimicrob. Agents Chemother. 58:5456–5465. 10.1128/AAC.03052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]