Abstract
Immunocompromised patients are predisposed to infections caused by influenza virus. Influenza virus may produce considerable morbidity, including protracted illness and prolonged viral shedding in these patients, thus prompting higher doses and prolonged courses of antiviral therapy. This approach may promote the emergence of resistant strains. Characterization of neuraminidase (NA) inhibitor (NAI)-resistant strains of influenza A virus is essential for documenting causes of resistance. In this study, using quantitative real-time PCR along with conventional Sanger sequencing, we identified an NAI-resistant strain of influenza A (H3N2) virus in an immunocompromised patient. In-depth analysis by deep gene sequencing revealed that various known markers of antiviral resistance, including transient R292K and Q136K substitutions and a sustained E119K (N2 numbering) substitution in the NA protein emerged during prolonged antiviral therapy. In addition, a combination of a 4-amino-acid deletion at residues 245 to 248 (Δ245-248) accompanied by the E119V substitution occurred, causing resistance to or reduced inhibition by NAIs (oseltamivir, zanamivir, and peramivir). Resistant variants within a pool of viral quasispecies arose during combined antiviral treatment. More research is needed to understand the interplay of drug resistance mutations, viral fitness, and transmission.
INTRODUCTION
Immunocompromised patients are predisposed to infections caused by influenza virus (1). In addition, immunocompromised patients are at risk of developing antiviral resistance and subsequent complications. In such patients, resistance may even emerge within a week of commencing therapy (1). Thus, it is not surprising that these patients can develop prolonged viral shedding for up to several months, whereas in immunocompetent individuals, viral shedding has been reported to occur for less than 10 days (2–5).
Two classes of anti-influenza A virus drugs are available, including the M2 channel blockers (adamantanes) and the neuraminidase (NA) inhibitors (NAIs) oseltamivir and zanamivir, with the last two agents also having activity against influenza B viruses (6).
NAI resistance is predominantly caused by specific substitutions in the eight catalytic sites or 11 framework residues of the sialic acid (SA) binding pocket of NA, with a subsequent decreased interaction between NAIs and the SA binding pocket occurring. Furthermore, the wide circulation of adamantane-resistant influenza A (H3N2) and influenza A (H1N1) 2009 pandemic (pH1N1) viruses as well as oseltamivir-resistant seasonal H1N1 viruses emerged between 2007 and 2009. This resistance could not be explained by antiviral use alone and may indicate the presence of other environmental pressures producing resistance through spontaneous mutations in NA.
Nevertheless, the relatively low incidence rate of NAI resistance among H3N2 and pH1N1 influenza viruses suggests that these mutants may have compromised viral fitness, producing low rates of direct transmission among humans. An experimental study in animal models suggests that an influenza A (H3N2) virus mutant with an R292K amino acid substitution and H1N1 (seasonal or pH1N1) virus with an H275Y (H274Y N2 numbering) amino acid substitution have lower replication rates and reduced transmissibility (7–10).
Data obtained from clinical studies or by using reverse genetics technology have demonstrated that single amino acid substitutions in catalytic or framework sites could result in subtype-specific NAI resistance or confer resistance across different NA types. NA substitutions, such as E119V, D151E, I222V, R224K, E276D, N249S, R292K, and R371K (N2 numbering) in H3N2 influenza virus and such as E119V, I222R/V, S247N, H275Y, and N294S in pH1N1 (N1 numbering) influenza virus have been documented to result in 50% inhibitory concentration (IC50) increases (≥10- to >100-fold) for NAIs. Although several substitutions, such as E119V, I222V, and N249S, may induce NAI resistance in both the pH1N1 and H3N2 subtypes, certain substitutions conferring NAI resistance seem to be more subtype specific among N2 (R292K, D151E, R371K, R224K, E276D, and R371K) and N1 (I223R/V, H275Y, and S247N) viruses (3, 11–13).
Differences in the chemical structures of oseltamivir (which contains an amino group and a hydrophobic side chain) and zanamivir (which contains a guanidino group and a glycerol side chain) seem to be key in the different susceptibilities of viruses to these drugs. It has been shown that in order to accommodate the side chain of oseltamivir, rotation and binding of E276 to R224 are necessary. Substitutions in NA, such as R292K, N294S, and H275Y, inhibit this rearrangement and therefore compromise oseltamivir affinity to the NAI binding site. Zanamivir has a higher structural homology to sialic acid, the natural substrate of neuraminidase, than oseltamivir. Therefore, reconfiguration of the NA binding site is not required for zanamivir activity. This may explain zanamivir's enhanced spectrum of activity compared to that of oseltamivir (14).
In addition to amino acid substitutions, NAI resistance in influenza A (H3N2) virus caused by a 4-amino-acid (aa) deletion (deletion of amino acids 245 to 248 [Δ245-248]) in NA has been observed clinically on two occasions and documented using a recombinant mutant NA protein (4, 5). This deletion has not been observed in other influenza virus types/subtypes. Recently, the co-occurrence of a deletion in neuraminidase (deletion of amino acids 247 to 250 [Δ247-250]) together with neuraminidase resistance substitutions, including E119V or R292K, in two immunocompromised patients treated with oseltamivir was reported (15). Other reports also indicate the loss of NA activity on passage in Madin-Darby canine kidney (MDCK) cells due to large internal deletions in the NA-coding RNA segment or a reduced dependence on NA following the accumulation of defective NA genes in the presence of NA inhibitors (16, 17).
One of the most frequent substitutions arising during or soon after oseltamivir administration has been reported to be E119V (a glutamic acid-to-valine substitution at position 119) (18–20). A novel Q136K substitution in NA confers clinical zanamivir resistance in influenza A (H3N2) virus and seasonal influenza A (H1N1) virus (21, 22). Although not observed clinically, in vitro studies have shown that Q136K arises in pH1N1 when exposed to zanamivir while undergoing serial passage in MDCK cells (23). A novel I223R (I222R in the NA2 numbering) mutation in pH1N1 resulted in oseltamivir, zanamivir, and peramivir resistance (24). Despite reduced susceptibility to zanamivir, Q136K alone was reported to have no effect on susceptibility to oseltamivir (22, 25). Of import, although there is a stable rate of oseltamivir and zanamivir resistance among circulating influenza A viruses, a higher prevalence of resistance has been documented in immunocompromised patients following antiviral administration (4, 5, 18, 26, 27, 49, 50).
In this analysis, we describe the molecular characteristics of an induced multidrug-resistant influenza A (H3N2) virus recovered from an immunocompromised patient containing several amino acid substitutions and Δ245-248.
MATERIALS AND METHODS
Patient history.
A 39-year-old female with a history of Philadelphia chromosome-positive acute B cell lymphoblastic leukemia (B-ALL) in complete remission underwent a matched, related allogeneic stem cell transplant in October 2012. She was maintained on cyclosporine and mycophenolate mofetil. In November 2012, she experienced grade 2 graft-versus-host disease (GVHD) of the skin and gastrointestinal tract. Methylprednisolone was initiated at 2 mg/kg of body weight intravenously (i.v.) once daily for 2 weeks, followed by a slowly tapering dose; she also received intermittent pulse doses of steroids. Additionally, she was administered alemtuzumab. Her hypogammaglobulinemia was treated with monthly intravenous immunoglobulin replacement.
On 4 December 2012, rhinorrhea, cough, and mild shortness of breath developed. Chest computed tomography (CT) scans over the next 2 weeks revealed worsening pulmonary infiltrates. A bronchoscopy was performed on 18 December, and bronchoalveolar lavage (BAL) fluid tested positive for influenza A (H3N2) virus. Oseltamivir therapy (75 mg twice daily orally [p.o.]) was administered. However, due to worsening pulmonary infiltrates on a repeat chest CT and a nasopharyngeal swab (NPS) specimen positive for influenza A (H3N2) virus on 27 December, the oseltamivir was increased to 150 mg p.o. twice daily. When a follow-up NPS specimen collected on 2 January 2013 was once again positive for influenza A (H3N2) virus, zanamivir was initiated at 10 mg twice daily by inhalation with minimal symptom improvement. A follow-up chest CT scan done on 7 January revealed further deterioration with increased ground glass opacities diffusely, and an NPS specimen collected on 8 January once again demonstrated influenza A (H3N2) virus. Inhaled zanamivir was switched to zanamivir at 600 mg i.v. every 12 h on 10 January and was continued for 10 days. Oseltamivir at 150 mg orally twice daily was added for the last 5 days, when an NPS specimen collected on 14 January revealed influenza A (H3N2) virus. Clinical as well as radiographic improvement was noted by 15 January. Thereafter, clinical deterioration characterized by a productive cough and shortness of breath as well as worsening of her chest CT findings on 23 January occurred. Oseltamivir was restarted at 150 mg every 12 h orally on 24 January (for 4 days), and a bronchoscopy performed on 25 January indicated the persistence of influenza A (H3N2) virus as well as Pseudomonas aeruginosa. Zanamivir (600 mg i.v. every 12 h) as well as piperacillin-tazobactam (4.5 g i.v. every 6 h) was reinitiated on 25 January for 5 days. Although follow-up NPS specimens collected on 31 January and 7 February revealed influenza A (H3N2) virus, no further antiviral therapy was administered due to clinical improvement (results represented asymptomatic shedding). Prior to her transfer to a rehabilitation facility on 7 March, an NPS specimen collected on 4 March was again positive for influenza A (H3N2) virus. Due to the persistence of influenza A (H3N2) virus in consecutive NPSs, testing for antiviral resistance was conducted on the NPS specimen collected on 4 March. Subsequently, other clinical specimens were sequenced retrospectively.
She was readmitted to the hospital on 14 March with a productive cough and shortness of breath. Bronchoscopy was performed and revealed Legionella pneumophila and influenza A (H3N2) virus. Oseltamivir at 150 mg p.o. twice daily was initiated on the same day with antibiotics. Both medications were given for 20 days. Following improvement, she was once more sent to a rehabilitation facility on 8 April.
Due to respiratory deterioration, she was admitted to her local hospital at the beginning of May. An NPS specimen collected on 3 May was negative for influenza A virus. She was transferred to the transplant center of Princess Margaret Cancer Center of the University Health Network on 7 May with severe respiratory distress, grade 4 hemorrhagic cystitis, and flare-up of her GVHD of the skin and gastrointestinal tract. A CT scan of the chest showed diffuse pulmonary consolidation. Due to respiratory decompensation, she required mechanical ventilation. Bronchoscopy performed on 8 May revealed influenza A (H3N2) virus, Pseudomonas aeruginosa, and cytomegalovirus. As there were other causes for her respiratory decompensation, no anti-influenza therapy was administered. Unfortunately, the patient's clinical condition deteriorated and she eventually succumbed to her GVHD and infections on 19 May 2013.
Ethics.
As the policy of the Research Ethics Board of the University Health Network does not require board approval for single patient case reports, no such approval was obtained.
Specimen collection, molecular detection, and sequencing.
All respiratory specimens (NPS or BAL fluid) collected from the patient were transferred to the Public Health Ontario Laboratory (PHL), Toronto, Ontario, Canada, for respiratory viral testing. Real-time reverse transcription-PCR (rRT-PCR) for influenza A and B viruses following the Centers for Disease Control and Prevention protocol as well as viral culture for respiratory viruses using rhesus monkey kidney cells (RMK-Diagnostic Hybrids, Inc., OH, USA) was performed (28).
Total nucleic acid extraction of all primary specimens and available culture material was performed using a NucliSens easyMAG extraction system instrument (bioMérieux Canada Inc., Québec, Canada), according to the manufacturer's instructions.
A SuperScript III One-Step reverse transcription-PCR (RT-PCR) system with Platinum Taq high-fidelity DNA polymerase (Invitrogen) and overlapping primers were used to amplify whole-genome segments of the NA, hemagglutinin (HA), nonstructural (NS), and matrix (M) proteins (29). Direct sequencing of the RT-PCR products was carried out with a BigDye Terminator cycle sequencing kit (version 3.1; Applied Biosystems) on an ABI 3730XL genetic analyzer (Applied Biosystems, CA, USA). Gene sequence assembly was performed using the Vector NTI ContigExpress program (Life Technologies, CA, USA). The BioEdit program was used for the raw sequence curation and also for the multiple alignments of the protein and nucleotide sequences (30).
To increase the NA sequence data resolution and identify the mutation and amino acid substitution frequency within a specimen and, subsequently, the presence of quasispecies, an Illumina MiSeq platform was utilized (Illumina, Inc., San Diego, CA, USA). Four fragments encompassing approximately 1 kb of the NA gene were amplified from all primary specimens. RT-PCR products were quantified and normalized. Fragments from each specimen were pooled, enzymatically fragmented, and ligated with Nextera-XT index pairs (Illumina, Inc., San Diego, CA, USA). The quality of the prepared library was checked by using an Agilent 2100 Hi-Sens DNA chip (Agilent Corporation, USA). A pooled library prepared using the Nextera-XT kit was run on an Illumina Miseq system using the 2× 150-bp paired-end reads option. Postrun fastq files were generated with onboard Miseq reporter software.
Data manipulation and mapping to the A/Victoria/361/2011 (H3N2) (the 2012/2013 vaccine strain) reference genome were done using programs available through Galaxy (31–33). In summary, the quality of the reads was checked by use of the fastQC:ReadQC tool, and Nextera adapters were removed using the Manipulate fastq program (34). Paired-end reads were mapped to the A/Victoria/361/2011 (H3N2) reference genome with the BWA software package using default parameters (35). Nucleotide substitutions were identified using the SAMtools utility (36). An indexed BAM file was created using SAMtools, the mapping results were visualized by use of the Integrative Genomics Viewer (IGV; version 2.3.25), and the presence of substitutions and deletions was visually verified (37).
Antigenic characterization.
Antigenic characterization was performed at Canada's National Microbiology Laboratory, Public Health Agency of Canada, by hemagglutination inhibition (HI) assay. This was performed using 4 hemagglutination units of virus, 0.5% (vol/vol) turkey red blood cells, and postinfection ferret antisera against A/Victoria/361/2011-like H3N2 virus. HI titers were defined as the reciprocal of the highest dilution of serum that completely inhibited hemagglutination of 0.5% (vol/vol) turkey red blood cells.
NA inhibitors and NA inhibition assay.
Specimens were cultured in RMK cells, and those successfully grown (four isolates) were subjected to NAI testing for determination of susceptibility to oseltamivir carboxylate, peramivir, and zanamivir by chemiluminescent assay using an NA-Star kit (Applied Biosystems Ltd., Foster City, CA, USA). The NAI susceptibility of the influenza virus isolates was expressed as the 50% inhibitory concentration (IC50), as well as the fold change in the IC50 compared to that for the susceptible wild-type (WT) strain.
According to recently proposed WHO guidelines for antiviral susceptibility interpretation in influenza A viruses, we categorized isolates on the basis of the fold change of their IC50 relative to the reference IC50s. They were categorized as displaying normal inhibition or susceptibility (S; a <10-fold increase), reduced inhibition (RI; a 10- to 100-fold increase), and highly reduced inhibition (HRI; a >100-fold increase) (38).
Structural modeling.
The crystal structure of A/Tanzania/205/2010 (H3N2) NA in complex with oseltamivir (PDB accession no. 4GZP), which is 99% identical to reference strain A/Victoria/361/2011 (H3N2) NA, was superimposed with the crystal structure of NA from H2N2 in complex with zanamivir (PDB accession no. 3TIC), which is 92% similar to A/Victoria/361/2011 (H3N2) NA, and with the crystal structure of H5N1 NA (PDB accession no. 2HTU), which is 63% similar to A/Victoria/361/2011 (H3N2) NA. The NA protein chains were superimposed, and pairwise root mean square deviation (RMSD) values versus the NA with PDB accession number 4GZP (469 C-α atoms) were 0.22 Å over 353 C-α atoms and 0.77 Å over 344 C-α atoms, indicating good alignments (39, 40). Mutants were generated using the PyMOL system, with rotamers chosen to minimize clashes with protein atoms (41).
Viral load quantification.
A genomic influenza virus RNA control (AmpliRun influenza A virus H3 RNA control) was obtained from Vircell (Grenada, Spain), and the viral load was determined by real-time RT-PCR using a primer/probe targeting the matrix protein gene (CDC protocol).
Nucleotide sequence accession numbers.
The gene sequences of influenza A (H3N2) viruses obtained in this study have been submitted to the GenBank database under accession numbers KJ734739 to KJ734779.
RESULTS
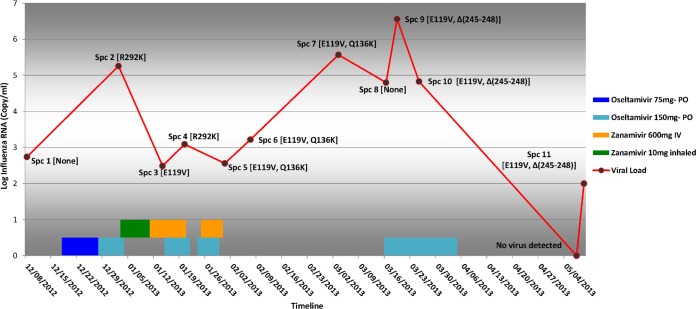
A total of 12 sequential specimens (9 NPS and 3 BAL fluid specimens) were submitted to the Public Health Ontario Laboratory over the course of 5 months (December 2012 to May 2013; Fig. 1).
FIG 1.
Viral load, antiviral therapy, and mutations conferring resistance to NAI in specimens obtained from an immunocompromised patient. The viral load in 12 consecutive specimens (Spc 1 to Spc 12) from the immunocompromised patient and the prescribed antiviral agent are shown as a red line and bars, respectively. The specimen and the dominant substitutions conferring resistance at each point are labeled on the line.
Influenza A (H3N2) virus was detected in 11 of the specimens and further characterized by genome sequencing. Despite the presence of quantifiable viral loads in all specimens, cell culture recovery of individual isolates was successful from only four specimens, and these isolates were further evaluated for NAI susceptibility using phenotypic methods. Sanger sequencing of the first isolate of influenza A (H3N2) virus (from 18 December 2012) and subsequent deep gene sequencing revealed no NAI resistance mutations. This was used as the background for comparative genome analysis.
Molecular analysis.
Complete HA, NA, M, and NS nucleotide sequences from the 11 influenza A (H3N2) virus-positive specimens were analyzed and compared with those of a reference strain, A/Victoria/361/2011 (H3N2) (Global Initiative on Sharing Avian Influenza Data accession no. EPI_ISL_134450); amino acid substitutions were identified in all four genes.
All four culture isolates recovered from the patient were characterized as A/Victoria/361/2011 (H3N2)-like virus by HI, and further HA genome sequencing analysis showed that they all belonged to an emerging subgroup in group 3C carrying HA substitutions Q33R and T128A (with the latter resulting in the loss of a potential glycosylation site), R142G and N145S (both of which are in antigenic site A), Q156S and V186G (both of which are in antigenic site B), Y219S (which is in antigenic site D), and N278K (which is in antigenic site C) within the circulating influenza A (H3N2) virus subtype (Table 1).
TABLE 1.
Nucleotide and amino acid homology to the A/Victoria/361/2011 reference strain and within intrahost varianta
| Protein | Homology to A/Victoria/361/2011 reference strain |
Homology within intrahost variantb |
||||
|---|---|---|---|---|---|---|
| % sequence homology |
Substitution(s)c | % sequence homology |
Substitution(s)d | |||
| Amino acid | Nucleotide | Amino acid | Nucleotide | |||
| NA | 98.4–99.7e | 98.4–99.7e | K258E and T329N | 98.7–99.5e | 98.4–99.7e | M51V, I77V, P81S, E119V, Q136K, R292K |
| HA | 98.2–98.5 | 98.8–99.1 | Q33R, T128A, R142G, N145S, Q156H, V186G, Y219S, N278K | 99.6–100 | 99.5–100 | L3I, L111I, N144K, P162S, N225D, M347V |
| M1 | 100 | 99.6–100f | None | 100 | 99.7–100f | None |
| M2 | 98.4–100 | None | 98.4–100 | E66G | ||
| NS1 | 97.8–98.6 | 99.1–99.4g | None | 99.1–100 | 99.7–100g | R37P, R118K, R193Q |
| NS2 | 99.1–100 | None | 99.1–100 | E36K | ||
NA, neuraminidase; HA, hemagglutinin; M1, matrix 1 protein; M2, matrix 2 protein; NS1, nonstructural protein 1; NS2, nonstructural protein 2.
Specimen 1, the earliest patient specimen sequenced, was considered a baseline to which all subsequent specimen sequences were compared.
Substitutions common to all 11 patient specimens sequenced shown.
Substitutions present in one or more of 10 specimens sequenced when compared to specimen 1 as a baseline.
Sequences with a gap (deletion of amino acids 245 to 248) were included in calculation of the data.
Homology percentage for complete M gene (M1 and M2).
Homology percentage for complete NS gene (NS1 and NS2).
Comparative sequence analysis of the 11 influenza A (H3N2) virus-positive specimens was undertaken for NA, HA, M, and NS. Intrahost nucleotide sequence divergence rates among the different specimens/isolates sequenced of 0 to 0.5%, 0.3 to 1.3%, 0 to 0.3%, and 0 to 0.3% were obtained for the HA, NA, M, and NS genome segments, respectively (Table 1).
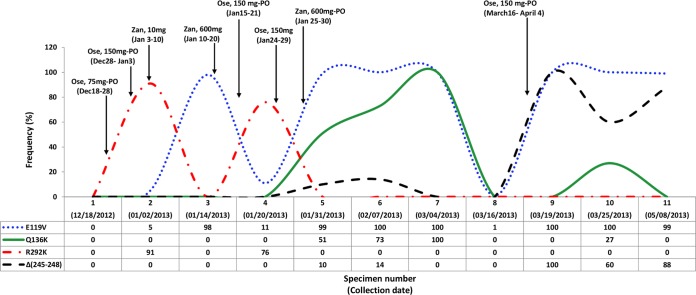
The highest and lowest rates of intrahost variability among nonsynonymous mutations were observed in the NA and M1 proteins, respectively. Sequence analysis of consecutive NA genes revealed the presence of R292K for the first time from specimen 2 (obtained after 14 days of initial oseltamivir administration). Next-generation sequencing (NGS) data suggested the presence of the R292K mutant and WT with relative percentages of 91% and 9%, respectively (Table 2). Although this mutant variant had reverted to the WT in the specimen obtained 2 weeks later (specimen 3), where the E119V variant emerged, the R292K variant reappeared in specimen 4 at a lower relative percentage of mutant and WT (76% and 24%, respectively). However, the exact time of emergence of the R292K substitutionwas not clear; it occurred sometime between 18 December 2012 and 2 January 2013, the collection dates of specimens 1 and 2, respectively. The E119V substitution emerged in specimen 3 (collected on 14 January) and persisted as a dual NAI resistance mutation accompanied by Q136K for more than 1 month (specimens 5, 6, and 7) from 31 January to 3 March 2013. Sequence chromatograms as well as deep gene sequencing showed the presence of a heterogeneous population of Q or K at amino acid position 136, with the proportion of the Q136K mutant increasing to 51%, 73%, and 100% in the three specimens, respectively (these specimens were obtained soon after a period of combination therapy with zanamivir at 600 mg i.v. every 12 h and high-dose oseltamivir at 150 mg p.o. twice daily). The initial specimen with quasispecies (specimen 5) containing the dual substitutions E119V (99%) and Q136K (51%) also revealed a low rate (10%) of a 4-aa (S245, A246, S247, and G248) deletion (Δ245-248) by deep gene sequencing (Fig. 2). This finding is in agreement with the phenotypic findings of highly reduced inhibition by oseltamivir, zanamivir, and peramivir with IC50s of 116.5, 114.31, and 36.5 nmol/liter, respectively (fold differences, 364, 285, and 180, respectively) (Table 3).
TABLE 2.
Comparison of NA mutations and amino acid substitutions obtained by NGS and the Sanger sequencing method in specimens recovered from an immunocompromised patient during the course of antiviral therapy
| Specimen no. | Date collected (mo/day/yr) | Results by deep gene sequencing |
Dominant genotype(s) by Sanger sequencing | |||||
|---|---|---|---|---|---|---|---|---|
| % virus with the following substitutions or deletion: |
Other mutations (% virus) | Dominant genotype(s) | ||||||
| E119V | Q136K | R292K | Δ245–248 | |||||
| 1 | 12/18/2012 | 0 | 0 | 0 | 0 | Y40H (22) | WT | WT |
| 2 | 01/02/2013 | 5 | 0 | 91 | 0 | R292K | R292K | |
| 3 | 01/14/2013 | 98 | 0 | 0 | 0 | E119V | E119V | |
| 4 | 01/20/2013 | 11 | 0 | 76 | 0 | R292K | R292K | |
| 5 | 01/31/2013 | 99 | 51 | 0 | 10 | E119V, Q136K | E119V, Q136Ka | |
| 6 | 02/07/2013 | 100 | 73 | 0 | 14 | V33A (30) | E119V, Q136K | E119V, Q136K |
| 7 | 03/04/2013 | 100 | 100 | 0 | 0 | E119V, Q136K | E119V, Q136K | |
| 8 | 03/16/2013 | 1 | 0 | 0 | 0 | M51V (98) | M51V | M51V |
| 9 | 03/19/2013 | 100 | 0 | 0 | 100 | S123A (44), S311N (40) | E119V, Δ245–248 | E119V, Δ245–248 |
| 10 | 03/25/2013 | 100 | 27 | 0 | 60 | E119V, Δ245–248 | E119V, Δ245–248, (V33Ab) | |
| 11 | 05/08/2013 | 99 | 0 | 0 | 88 | V122 M (9) | E119V, Δ245–248 | E119V, Δ245–248 |
Q136K mixed population.
Culture derived.
FIG 2.
Antiviral therapy time line and frequency of emerging mutations conferring resistance in a quasispecies population. Ose, oseltamivir; Zan, zanamivir.
TABLE 3.
Phenotypic characterization and viral load of consecutive influenza A/H3N2 viruses recovered from an immunocompromised patient during the course of antiviral therapya
| Virus strain or sample | Date collected (mo/day/yr) | NA mutation(s) by NGS (% virus) | Mean IC50 ± SD (nM)b |
Phenotype with O/Z/P treatment | Viral load (log10 no. of copies/ml) | ||
|---|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | Peramivir | |||||
| A/Washington/01/2007 | Not applicable | Not applicable | 0.32 ± 0.04 | 0.4 ± 0.22 | 0.2 ± 0.08 | S/S/S | Not applicable |
| A/Texas/12/2007 | Not applicable | Not applicable | 6.53 ± 1.0 (20) | 0.5 ± 0.24 | 0.2 ± 0.7 | RI/S/S | Not applicable |
| 1 | 12/18/2012 | None | ND | ND | ND | ND | 2.74 |
| 2 | 02/01/2013 | E119V (5), R292K (91) | ND | ND | ND | ND | 5.26 |
| 3 | 01/14/2013 | E119V (98) | ND | ND | ND | ND | 2.49 |
| 4 | 01/20/2013 | E119V (11), R292K (76) | ND | ND | ND | ND | 3.09 |
| 5 | 01/31/2013 | E119V (99), Q136K (51), Δ245–248 (10) | 116.5 ± 0.09 (364) | 114.31 ± 0.17 (285) | 36.5 ± 0.1 (180) | HRI/HRI/HRI | 2.56 |
| 6 | 07/02/2013 | Q136K (73), Δ245–248 (14) | ND | ND | ND | ND | 3.22 |
| 7 | 03/03/2013 | E119V (100), Q136K (100) | 104.21 ± 0.09 (325) | 88.14 ± 0.17 (220) | 20.19 ± 0.1 (101) | HRI/HRI/HRI | 5.57 |
| 8 | 03/16/2013 | E119V (1) | 0.1 ± 0.09 (1) | 0.32 ± 0.17 (1) | 0.12 ± 0.1 (1) | S/S/S | 4.8 |
| 9 | 03/19/2013 | E119V (100), Δ245–248 (100) | ND | ND | ND | ND | 6.56 |
| 10 | 03/25/2013 | E119V (100), Q136K (27), Δ245–248 (60) | 614.59 ± 0.09 (1,918) | 11.15 ± 0.17 (28) | 1.88 ± 0.1 (9) | HRI/RI/S | 4.83 |
| 11 | 07/05/2013 | E119V (99), Δ245–248 (88) | ND | ND | ND | ND | 3.32 |
S, susceptible (<10-fold increase in IC50); RI, reduced inhibition (10- to 100-fold increase in IC50); HRI, highly reduced inhibition (>100-fold increase in IC50); ND, not determined; O/Z/P, oseltamivir/zanamivir/peramivir.
The mean IC50s for A/Washington/01/2007, A/Texas/12/2007, and clinical isolates were derived from 26, 23 and 4 replicates, respectively. Data in parentheses indicate the fold difference in IC50 compared to the reference susceptible strain, A/Washington/01/2007.
Even though the virus in specimen 7 was resistant to oseltamivir and had an IC50 similar to that of the virus in specimen 5, specimen 7, which contained 100% E119V and Q136K dual mutations in NA, had a lower zanamivir IC50 (88.14 nmol/liter; 220-fold that of the WT) and peramivir IC50 (20.19 nmol/liter; 101-fold that of the WT) than specimen 5. There was no evidence of Δ245-248 in this specimen (Table 3).
The NA mutant viruses disappeared and were replaced by WT virus in specimen 8, collected on 16 March 2013. However, specimen 9, collected on 19 March (90 and 60 days after the first administration of oseltamivir and zanamivir, respectively), showed a 100% prevalence of the 4-aa deletion (Δ245-248), a return to 100% E119V variant, and subpopulations of viruses with the S123A (44%) and S311N (40%) substitutions in the NA gene. Despite the appearance of the combination of E119V and a low rate of Δ245-248 during therapy (specimens 5 and 6) and the subsequent disappearance of them both (specimen 8, collected on 16 March), a variant containing both E119V (100%) and Δ245-248 (100%) reappeared (specimen 9, collected on 19 March) and was sustained in various proportions in the two subsequent specimens obtained.
Specimens 5, 6, and 7 carried Q136K in NA along with the E119V and N144K substitutions in the NA and HA genes, respectively. However, the N144K substitution was identified in only one of the specimens containing E119V and Δ245-248 (specimen 11). Interestingly, an E36K substitution in the NS2 protein was identified only in specimens with the E119V variant and a high level of virus with Δ245-248 (specimens 9, 10, and 11). Sequencing of the M2 protein showed the presence of the S31N resistance marker in the M2 protein, suggesting that all specimens were resistant to M2 inhibitors (adamantine). Comparison of the sequences obtained from primary specimens and isolates cultured from specimens 5, 7, 8, and 10 showed the presence of V33A in NA and showed that it was the only culture-driven substitution, which was detected in specimen 10, although the presence of a subpopulation (comprising 30% of the viruses) containing the same substitution was detected through deep gene sequencing of specimen 6.
Structural modeling analysis.
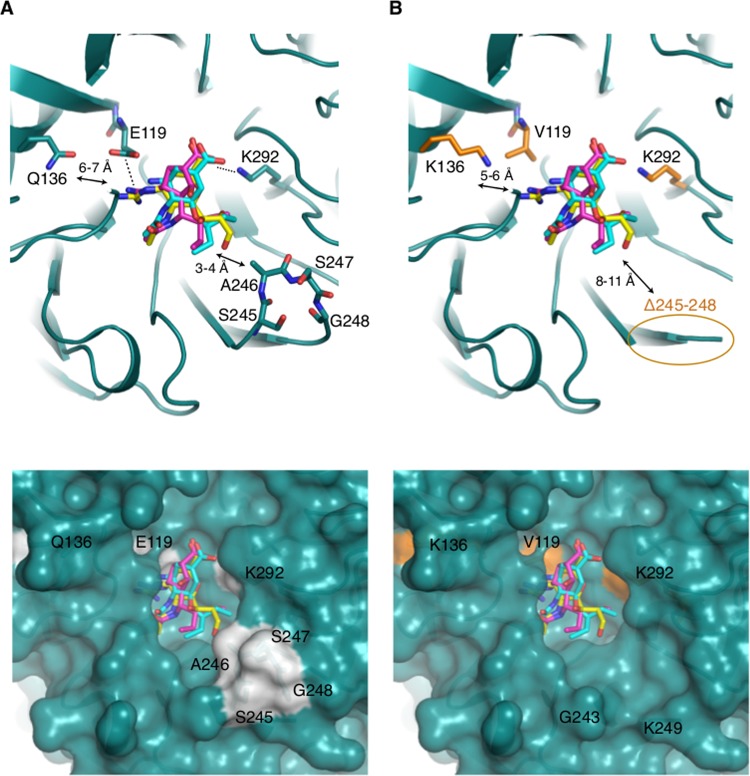
Molecular modeling suggests that although Q136 does not directly interact with oseltamivir due to its relative distance from the drug (6 to 7 Å), it is positioned in a constricted area of the active site near other residues (R156 and D151) involved in the interaction with the inhibitors. Q136K introduces repulsive charges to the guanidino group of both zanamivir and peramivir.
Our analysis also revealed that within amino acids 245 to 248, A246 (along with R224, I222, and E276) forms a hydrophobic side pocket adjacent to the NA cavity (within 3 to 4 Å) (Fig. 3). This side pocket appears to accommodate the glycerol side chain of oseltamivir and zanamivir.
FIG 3.
Structural analysis of modes of NAI binding to A/Victoria/361/2011 (H3N2) NA. (A) (Top) Overlay of crystal structure of NA from H3N2 (PDB accession no. 4GZP) (blue-green) in complex with oseltamivir (cyan), docked zanamivir (yellow), and docked peramivir (pink). Dashes indicate electrostatic interactions. Arrows indicate the distance range between Q136 or A246 and the NAIs. (Bottom) Surface structure of the NAI binding pocket in complex with oseltamivir and zanamivir. The surface structure of residues 245 to 248 is highlighted in gray. (B) (Top) Overlay of NA from H3N2 (PDB accession no. 4GZP) with NAIs, as described in the legend to panel A, with substitutions introduced and colored in brown. Δ245-248 is circled. (Bottom) Conformational changes on the surface structure around the NAI pocket following the deletion of residues 245 to 248 (Δ245-248).
Viral load.
The viral load was significantly higher after initial administration of oseltamivir and the emergence of a heterogeneous population of R and K amino acids at residue 292, with the R292K mutant being dominant (specimen 2). Although the viral load dropped to levels lower than those documented on admission while the patient was on combination therapy with oseltamivir and zanamivir, the E119V mutant emerged and a remnant of the R292K mutant remained.
DISCUSSION
Influenza virus infection is associated with greater morbidity and mortality as well as a higher rate of resistance in immunocompromised patients than in other individuals (42–44).
The dynamics of emerging mutations conferring resistance to NAIs have not been fully elucidated. The presence of viral quasispecies in conjunction with selective pressure caused by antivirals, immunosuppressive agents, host immune response, and the adaptation rate of the influenza virus, which is influenced by fitness costs, affects the complex interaction of influenza virus with its host. The interplay of such factors can result in an unpredictable effectiveness of antiviral agents, including the emergence of resistant strains, in immunocompromised patients.
In this study, we were able to retrospectively detect the emergence of three different influenza A (H3N2) virus amino acid substitutions (R292K, E119V, Q136K) and one deletion (Δ245-248) in the NA protein arising simultaneously or consecutively during NAI therapy and resulting in NAI resistance in an immunocompromised patient. Although zanamivir resistance was documented in specimen 5 (the first culture-positive specimen), this was most likely due to the Q136K substitution and not the E119V substitution that was present in the same specimen. E119V has previously been associated with oseltamivir resistance but not zanamivir resistance, whereas Q136K has been associated with zanamivir resistance. Q136K has been reported in two influenza A (H3N2) virus-positive specimens from Myanmar (one each in 2007 and 2008) (22). Zanamivir-resistant seasonal influenza A (H1N1) viruses with the Q136K substitution have also been described (21). E119V and R292K are the most frequent substitutions causing clinical resistance to oseltamivir in influenza A (H3N2) virus, and previous reports indicated an association of R292K with the in vitro inhibition of zanamivir (12, 13, 45). There did not appear to be a direct correlation between the prevalence of each mutation and the particular NAI being administered to the patient. E119V first emerged during oseltamivir therapy (5% prevalence in specimen 2), yet it became much more prevalent (98% prevalence in specimen 3) after 11 days of zanamivir therapy, even though E119V does not cause zanamivir resistance. R292K disappeared between specimen 2 (91% prevalence) and specimen 3, despite continued exposure to zanamivir, which this mutation confers resistance to. This may be explained by the reduced fitness of viruses carrying the R292K substitution, which outweighed the ability of R292K to permit growth in the presence of zanamivir (8, 46).
Using deep gene sequencing, a quasispecies population of NAI-resistant influenza virus containing the dominant R292K substitution (specimen 2, 91%) and a minor population of virus with the E119V substitution (specimen 2, 5%) was identified during the patient's initial 2-week course of oseltamivir administration. This scenario was subsequently replaced by one with E119V (98%) during zanamivir administration (specimen 3). Thereafter, a lower proportion of virus with R292K (76%) dominated along with a minor population of virus with E119V (11%) during a second course of oseltamivir therapy in the absence of zanamivir (Fig. 2). Such a transition may have resulted from the biological behavior of the virus and the preferred selection of a mutant under multiple pressures, such as immunosuppressive therapy, antiviral medications, and host immune response.
This finding is in agreement with previous reports of the emergence of the R292K or E119V substitution in oseltamivir-treated immunocompromised patients infected with influenza A (H3N2) virus (18, 20, 26).
In contrast to previously reported cases of the emergence of the E119V substitution following oseltamivir therapy, deep gene sequencing revealed the presence of quasispecies, with the E119V variant being found at a very low rate (5%) in the specimen obtained after the first course of oseltamivir therapy. However, this rate increased to 98% in specimen 3, collected while the patient was on zanamivir therapy. A reduced prevalence of the E119V variant to 11% during the second course of oseltamivir therapy (specimen 4) was followed by its presence at a high rate (99%) in the subsequent specimen collected immediately after combination therapy with oseltamivir and zanamivir. We are not able to confirm if selective pressure solely caused by zanamivir is responsible for the magnitude of the E119V variant or not.
Interestingly, following 5 days of administration of zanamivir and oseltamivir in combination with antibiotics (piperacillin-tazobactam), E119V (99%) and Q136K (51%) viruses, along with a minor population of virus with Δ245-248 (10%), were detected (specimen 5). The domination of the E119V variant with the complete disappearance of the R292K variant may suggest possible selection of the E119V substitution by zanamivir, even though the E119V substitution by itself does not cause zanamivir resistance in H3N2 influenza viruses (45).
Despite no further antiviral administration, we observed an increase in the relative proportion of the Q136K variant in 3 consecutive specimens, specimens 5, 6, and 7 (51%, 73%, and 100%, respectively; Table 2). Surprisingly, the virus in specimen 7 showed a lower zanamivir IC50 (88.14 nmol/liter, 220-fold increase, HRI) than specimen 5 (114.31 nmol/liter, 285-fold increase, HRI). Another difference between the two specimens was the presence of Δ245-248 in specimen 5 but not specimen 7. Δ245-248 (first detected in specimen 5) most likely emerged as a result of repeated courses of NAI therapy. It subsequently flourished during oseltamivir therapy, when it was present in 100% of sequences obtained on 19 March (specimen 9).
Despite the presence of few mutations, the high level of intrahost homology (99.5 to 100%) among the HA, M, and NS genes may be supporting evidence for a common genetic background of the virus populations (Table 1). However, due to the presence of some mutation variation within specimens, we cannot rule out the possibility of reinfection with contemporary circulating H3N2 influenza viruses specifically between times of patient discharge and readmission (7 to 14 March 2013). The mutations documented in genes besides NA may be compensatory in nature, serving to maintaining viral fitness.
Although the mechanism by which substitutions at the framework residue 119 alter the susceptibility to NAIs has not been completely elucidated, the root cause may be the loss of an electrostatic interaction between the amino (NH3+) groups of oseltamivir and the carboxylate (COO−) groups of glutamic acid at amino acid 119. Our analysis shows that the charged interaction between E119 and the guanidino group of zanamivir and peramivir could be interrupted by an E119K substitution. Therefore, the lack of an increase in the zanamivir and peramivir IC50s could possibly imply that the activities of these NAIs are not affected by the interaction of the larger guanidino groups with E119, as these groups have more hydrogen bond donor groups to compensate for the loss of the interaction with E119.
The Q136K substitution, which introduces a larger side chain at this position, would be expected to affect the binding of NAIs due to conformational changes in the enzyme-inhibitor complex. This is reflected in resistance to zanamivir, and the structural modeling suggests that the charge repulsion between the guanidino group of the NAI and lysine at position 136 may have a complementary effect. Likewise, the higher IC50 of peramivir could be the result of the larger guanidino groups that this NAI possesses, which prevent it from fitting into the altered active-site configuration. In contrast, oseltamivir lacks a functional group approaching lysine at position 136, and therefore, there was no change in its IC50.
Since A246 (along with R224 and I222) is directly involved in forming the hydrophobic pocket adjacent to the NA cavity, conformational changes at the surface level caused by Δ245-248 could disturb the hydrophobic side pocket (Fig. 3). Since this pocket accommodates the glycerol side chain of sialic acid and zanamivir, it may potentially alter NAI binding (40). However, more experiments involving reverse genetics are needed to confirm its effect.
Due to the lack of a sufficient resolution for the detection of minor viral mutant subpopulations (<20% to 25%), conventional sequencing methods, such as Sanger sequencing, might not be an optimal resource for the identification of mutations conferring antiviral resistance in quasispecies populations (47). However, the recent technology of deep gene sequencing, which involves the sequencing of numerous copies of a gene, is a useful tool in the identification of resistant or other quasispecies variants present in clinical specimens at a low frequency. The clinical significance of these quasispecies is not fully understood at this time. The combination of highly error-prone RNA-dependent RNA polymerase (RdRP), which has an average mutation rate of >7.3 × 10−5 nucleotide per replication cycle among the influenza viruses (48), and selective pressures, such as the host immune response, which may be influenced by chemotherapy and/or antiviral treatment, could facilitate the emergence of a dominant mutant with a selective advantage.
In this retrospective investigation, we identified 3 specimens (specimens 5, 7, and 10) with various proportions of viruses with the E119V and Q136K substitutions and Δ245-248 and peramivir IC50s ranging from just below RI to HRI. Due to the simultaneous presence of several mutations conferring resistance in our study, the NAI phenotypic results could not be attributed to individual mutations or Δ245-248. A synergistic effect of the dual substitutions E119V and Q136K and Δ245-248 cannot be ruled out. Future testing of rescued mutants through site-directed mutagenesis and Δ245-248 in an identical genetic background will be more insightful and help provide a better understanding of the role of each amino acid substitution or deletion alone or in combination with the other substitutions/deletions documented in this study. Previous studies with clinical specimens and in vitro using reverse genetics have confirmed that Δ245-248 causes NAI resistance but no change in fitness or transmissibility in ferret experiments (4, 5).
These observations support the need for further research that elucidates the evolutionary changes which arise in influenza viruses as a result of selective pressure from antiviral exposure and documents the fitness cost to H3N2 mutants carrying antiviral resistance mutations. Such work may assist in developing more effective influenza treatment strategies for immunocompromised patients.
Footnotes
Published ahead of print 22 September 2014
REFERENCES
- 1.Kunisaki KM, Janoff EN. 2009. Influenza in immunosuppressed populations: a review of infection frequency, morbidity, mortality, and vaccine responses. Lancet Infect. Dis. 9:493–504. 10.1016/S1473-3099(09)70175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubareva LV, Matrosovich MN, Brenner MK, Bethell RC, Webster RG. 1998. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 178:1257–1262. 10.1086/314440. [DOI] [PubMed] [Google Scholar]
- 3.Weinstock DM, Gubareva LV, Zuccotti G. 2003. Prolonged shedding of multidrug-resistant influenza A virus in an immunocompromised patient. N. Engl. J. Med. 348:867–868. 10.1056/NEJM200302273480923. [DOI] [PubMed] [Google Scholar]
- 4.Abed Y, Baz M, Boivin G. 2009. A novel neuraminidase deletion mutation conferring resistance to oseltamivir in clinical influenza A/H3N2 virus. J. Infect. Dis. 199:180–183. 10.1086/595736. [DOI] [PubMed] [Google Scholar]
- 5.Memoli MJ, Hrabal RJ, Hassantoufighi A, Jagger BW, Sheng Z, Eichelberger MC, Taubenberger JK. 2010. Rapid selection of a transmissible multidrug-resistant influenza A/H3N2 virus in an immunocompromised host. J. Infect. Dis. 201:1397. 10.1086/651610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC. Antiviral drug-resistance among influenza viruses. Guidance on the use of influenza antiviral agents. CDC; Atlanta, GA: http://www.cdc.gov/flu/professionals/antivirals/antiviral-drug-resistance.htm Accessed 20 September 2013. [Google Scholar]
- 7.Herlocher ML, Carr J, Ives J, Elias S, Truscon R, Roberts N, Monto AS. 2002. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res. 54:99–111. 10.1016/S0166-3542(01)00214-5. [DOI] [PubMed] [Google Scholar]
- 8.Yen HL, Herlocher LM, Hoffmann E, Matrosovich MN, Monto AS, Webster RG, Govorkova EA. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob. Agents Chemother. 49:4075–4084. 10.1128/AAC.49.10.4075-4084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan S, Boltz DA, Seiler P, Li J, Bragstad K, Nielsen LP, Webby RJ, Webster RG, Govorkova EA. 2010. Oseltamivir-resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathog. 6:e1001022. 10.1371/journal.ppat.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ives JA, Carr JA, Mendel DB, Tai CY, Lambkin R, Kelly L, Oxford JS, Hayden FG, Roberts NA. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 55:307–317. 10.1016/S0166-3542(02)00053-0. [DOI] [PubMed] [Google Scholar]
- 11.Boivin G. 2013. Detection and management of antiviral resistance for influenza viruses. Influenza Other Respir. Viruses 7(Suppl 3):S18–S23. 10.1111/irv.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abed Y, Baz M, Boivin G. 2006. Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir. Ther. 11:971–976. [PubMed] [Google Scholar]
- 13.Whitley RJ, Hayden FG, Reisinger KS, Young N, Dutkowski R, Ipe D, Mills RG, Ward P. 2001. Oral oseltamivir treatment of influenza in children. Pediatr. Infect. Dis. J. 20:127–133. 10.1097/00006454-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Samson M, Pizzorno A, Abed Y, Boivin G. 2013. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res. 98:174–185. 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Piralla A, Gozalo-Margüello M, Fiorina L, Rovida F, Muzzi A, Colombo AA, Alessandrino PE, Baldanti F. 2013. Different drug-resistant influenza A(H3N2) variants in two immunocompromised patients treated with oseltamivir during the 2011-2012 influenza season in Italy. J. Clin. Virol. 58:132–137. 10.1016/j.jcv.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Gulati S, Smith DF, Air GM. 2009. Deletions of neuraminidase and resistance to oseltamivir may be a consequence of restricted receptor specificity in recent H3N2 influenza viruses. Virol. J. 6:22. 10.1186/1743-422X-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nedyalkova MS, Hayden FG, Webster RG, Gubareva LV. 2002. Accumulation of defective neuraminidase (NA) genes by influenza A viruses in the presence of NA inhibitors as a marker of reduced dependence on NA. J. Infect. Dis. 185:591–598. 10.1086/339358. [DOI] [PubMed] [Google Scholar]
- 18.Baz M, Abed Y, McDonald J, Boivin G. 2006. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin. Infect. Dis. 43:1555–1561. 10.1086/508777. [DOI] [PubMed] [Google Scholar]
- 19.Sheu TG, Deyde VM, Okomo-Adhiambo M, Garten RJ, Xu X, Bright RA, Butler EN, Wallis TR, Klimov AI, Gubareva LV. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 52:3284–3292. 10.1128/aac.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiso M, Mitamura K, Sakai-Tagawa Y, Shiraishi K, Kawakami C, Kimura K, Hayden FG, Sugaya N, Kawaoka Y. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759–765. 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- 21.Hurt AC, Holien J, Parker M, Kelso A, Barr I. 2009. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J. Virol. 83:10366–10373. 10.1128/JVI.01200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dapat C, Suzuki Y, Saito R, Kyaw Y, Myint YY, Lin N, Oo HN, Oo KY, Win N, Naito M, Hasegawa G, Dapat IC, Zaraket H, Baranovich T, Nishikawa M, Saito T, Suzuki H. 2010. Rare influenza A (H3N2) variants with reduced sensitivity to antiviral drugs. Emerg. Infect. Dis. 16:493–496. 10.3201/eid1603.091321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaminski MM, Ohnemus A, Staeheli P, Rubbenstroth D. 2013. Pandemic H1N1 influenza A virus carrying a Q136K mutation in the neuraminidase gene is resistant to zanamivir but exhibits reduced fitness in the guinea pig transmission model. J. Virol. 87:1912–1915. 10.1128/JVI.02507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eshaghi A, Patel SN, Sarabia A, Higgins RR, Savchenko A, Stojios PJ, Li Y, Bastien N, Alexander DC, Low DE, Gubbay JB. 2011. Multidrug-resistant pandemic (H1N1) infection in immunocompetent child. Emerg. Infect. Dis. 17:1472–1474. 10.3201/eid1708.102004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okomo-Adhiambo M, Nguyen HT, Sleeman K, Sheu TG, Deyde VM, Garten RJ, Xu X, Shaw MW, Klimov AI, Gubareva LV. 2010. Host cell selection of influenza neuraminidase variants: implications for drug resistance monitoring in A(H1N1) viruses. Antiviral Res. 85:381–388. 10.1016/j.antiviral.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Ison MG, Gubareva LV, Atmar RL, Treanor J, Hayden FG. 2006. Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J. Infect. Dis. 193:760–764. 10.1086/500465. [DOI] [PubMed] [Google Scholar]
- 27.Couturier BA, Bender JM, Schwarz MA, Pavia AT, Hanson KE, She RC. 2010. Oseltamivir-resistant influenza A H1N1 virus in immunocompromised patients. Influenza Other Respir. Viruses 4:199–204. 10.1111/j.1750-2659.2010.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization. 2009. CDC protocol of real-time RT-PCR for influenza A (H1N1). World Health Organization Collaborating Centre for Influenza at CDC, Atlanta, GA: http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf Accessed 29 September 2013. [Google Scholar]
- 29.Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ, Sitz J, Koo H, Bolotov P, Dernovoy D, Tatusova T, Bao Y, St George K, Taylor J, Lipman DJ, Fraser CM, Taubenberger JK, Salzberg SL. 2005. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 437:1162–1166. 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- 30.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98. [Google Scholar]
- 31.Goecks J, Nekrutenko A, Taylor J, The Galaxy Team 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86. 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19:Unit 19.10.1–19.10.21. 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15:1451–1455. 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A, Galaxy Team 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26:1783–1785. 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization. 2012. Meetings of the WHO Working Group on Surveillance of Influenza Antiviral Susceptibility—Geneva, November 2011 and June 2012. Wkly. Epidemiol. Rec. 87:369–380. [PubMed] [Google Scholar]
- 39.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varghese JN, McKimm-Breschkin JL, Caldwell JB, Kortt AA, Colman PM. 1992. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins 14:327–332. 10.1002/prot.340140302. [DOI] [PubMed] [Google Scholar]
- 41.Schrödinger LLC. 2010. The PyMOL molecular graphics system, version 1.5.0.4. Schrödinger, LLC, New York, NY. [Google Scholar]
- 42.Boivin G, Goyette N, Bernatchez H. 2002. Prolonged excretion of amantadine-resistant influenza A virus quasi species after cessation of antiviral therapy in an immunocompromised patient. Clin. Infect. Dis. 34:E23–E25. 10.1086/338870. [DOI] [PubMed] [Google Scholar]
- 43.Gooskens J, Jonges M, Claas ECJ, Meijer A, van den Broek PJ, Kroes ACM. 2009. Morbidity and mortality associated with nosocomial transmission of oseltamivir-resistant influenza A(H1N1) virus. J. Am. Med. Assoc. 301:1042–1046. 10.1001/jama.2009.297. [DOI] [PubMed] [Google Scholar]
- 44.Hurt AC, Chotpitayasunondh T, Cox NJ, Daniels R, Fry AM, Gubareva LV, Hayden FG, Hui DS, Hungnes O, Lackenby A, Lim W, Meijer A, Penn C, Tashiro M, Uyeki TM, Zambon M. 2012. Antiviral resistance during the influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect. Dis. 12:240–248. 10.1016/S1473-3099(11)70318-8. [DOI] [PubMed] [Google Scholar]
- 45.McKimm-Breschkin JL. 2013. Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza Other Respir. Viruses 7:25–36. 10.1111/irv.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sleeman K, Guo Z, Barnes J, Shaw M, Stevens J, Gubareva LV. 2013. R292K substitution and drug susceptibility of influenza A(H7N9) viruses. Emerg. Infect. Dis. 19:1521–1524. 10.3201/eid1909.130724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghedin E, Holmes EC, DePasse JV, Pinilla LT, Fitch A, Hamelin ME, Papenburg J, Boivin G. 2012. Presence of oseltamivir resistant pandemic A/H1N1 minor variants before drug therapy with subsequent selection and transmission. J. Infect. Dis. 206:1504–1511. 10.1093/infdis/jis571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drake JW. 1993. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 90:4171–4175. 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antón A, López-Iglesias AA, Tórtola T, Ruiz-Camps I, Abrisqueta P, Llopart L, Marcos MÁ, Martínez MJ, Tudó G, Bosch F, Pahissa A, de Anta MT, Pumarola T. 2010. Selection and viral load kinetics of an oseltamivir-resistant pandemic influenza A (H1N1) virus in an immunocompromised patient during treatment with neuraminidase inhibitors. Diagn. Microbiol. Infect. Dis. 68:214–219. 10.1016/j.diagmicrobio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz-Carrascoso G, Casas I, Pozo F, González-Vincent M, Pérez-Breña P. 2010. Prolonged shedding of amantadine- and oseltamivir-resistant influenza A(H3N2) virus with dual mutations in an immunocompromised infant. Antivir. Ther. 15:1059–1063. 10.3851/IMP1657. [DOI] [PubMed] [Google Scholar]