Significance
The normal function of the Huntingtin (HTT) protein is emerging. Here we report that selective autophagy requires an intact HTT protein in Drosophila and mouse CNS. We describe similarities in structure and binding activity between the C-terminal domain of HTT and the yeast autophagy scaffold protein Atg11, suggesting that HTT may normally function as a scaffold for various types of selective autophagy. Mice expressing an expanded repeat form of HTT also show deficits in protein clearance. Because autophagy is critical for clearance of cellular proteins, including mutant HTT, the impairment of normal HTT function by the polyQ expansion could suppress activity of the autophagy machinery. These results may have important implications when evaluating therapeutic strategies for HD.
Keywords: Huntington disease, selective autophagy, Huntingtin, polyglutamine, neurodegeneration
Abstract
Although dominant gain-of-function triplet repeat expansions in the Huntingtin (HTT) gene are the underlying cause of Huntington disease (HD), understanding the normal functions of nonmutant HTT protein has remained a challenge. We report here findings that suggest that HTT plays a significant role in selective autophagy. Loss of HTT function in Drosophila disrupts starvation-induced autophagy in larvae and conditional knockout of HTT in the mouse CNS causes characteristic cellular hallmarks of disrupted autophagy, including an accumulation of striatal p62/SQSTM1 over time. We observe that specific domains of HTT have structural similarities to yeast Atg proteins that function in selective autophagy, and in particular that the C-terminal domain of HTT shares structural similarity to yeast Atg11, an autophagic scaffold protein. To explore possible functional similarity between HTT and Atg11, we investigated whether the C-terminal domain of HTT interacts with mammalian counterparts of yeast Atg11-interacting proteins. Strikingly, this domain of HTT coimmunoprecipitates with several key Atg11 interactors, including the Atg1/Unc-51–like autophagy activating kinase 1 kinase complex, autophagic receptor proteins, and mammalian Atg8 homologs. Mutation of a phylogenetically conserved WXXL domain in a C-terminal HTT fragment reduces coprecipitation with mammalian Atg8 homolog GABARAPL1, suggesting a direct interaction. Collectively, these data support a possible central role for HTT as an Atg11-like scaffold protein. These findings have relevance to both mechanisms of disease pathogenesis and to therapeutic intervention strategies that reduce levels of both mutant and normal HTT.
Huntington disease (HD) is a fatal autosomal-dominant neurodegenerative disorder caused by an expansion of a CAG trinucleotide repeat encoding a polyglutamine (polyQ) tract near the N terminus of the 350-kD Huntingtin protein (HTT) (1). Identifying the normal biological function of the HTT protein is important in the effort to design and implement effective therapeutic interventions for HD, but has proved challenging.
In the mouse, loss of HTT leads to lethality during gastrulation at embryonic day 7 (2–4). Conditional inactivation of HTT in the mouse forebrain at postnatal or late embryonic stages causes a progressive neurodegenerative phenotype associated with neuronal degeneration, motor phenotypes, and early mortality (5). Loss of HTT in mouse cells reduces primary cilia formation, and deletion of HTT in ependymal cells leads to alteration of the cilia layer, suggesting a role for HTT in ciliogenesis (6). Mutant HTT expression and HTT knockdown have also been found to impair axonal trafficking of vesicles, mitochondria, and autophagosomes in neurons in vitro and in vivo (7–9). A clear molecular mechanism to relate these findings to the function of the HTT protein, however, has not yet emerged.
In contrast to the embryonic lethality observed in the mouse, Drosophila lacking the endogenous Htt gene develop normally. However, adult Drosophila HTT loss-of-function (LOF) flies show an accelerated neurodegenerative phenotype in a Drosophila model of HD, expressing human mutant exon 1 HTT protein (10). Loss of HTT function leads to defects in mobility and ultimately survival as the adult flies age, and to a mitotic spindle misorientation, which is also observed in the conditional mouse neuronal HTT knockout. Drosophila HTT expression in mouse cells treated with mouse Htt siRNA can compensate for the loss of mouse HTT, demonstrating functional conservation of the mitotic spindle and axonal transport roles of HTT from these widely divergent species (11, 12). In zebrafish, depletion of HTT produces symptoms of cellular iron deficiency, showing a requirement for HTT in cellular iron utilization (13), whereas knockout of HTT in Dictyostelium discoideum demonstrates it is needed for survival when nutrients are limiting (14).
In contrast to the sequence conservation among vertebrate and invertebrate HTT proteins, finding clear homologs of metazoan HTT in yeast has proven challenging. Bioinformatic studies suggest that the HTT protein shares a sequence relationship with three different components of the yeast autophagy system—Atg11, Atg23, and Vac8 (15)—leading us to propose that HTT carries out cellular functions analogous to those carried out by these yeast proteins. To test this hypothesis, we conducted functional studies showing that Drosophila and mouse knockouts for HTT have impaired autophagic processes. Molecular association studies using coimmunoprecipitation reveal that HTT has high affinity for mammalian components of the autophagy system that are known to interact with the relevant yeast autophagy proteins. These observations support the hypothesis that a normal function of the HTT protein is in the execution of autophagy pathways and that ablation of HTT function leads to defects in autophagy, which may be important to consider in evaluating therapeutic options for HD.
Results
HTT LOF in Drosophila Inhibits Autophagy.
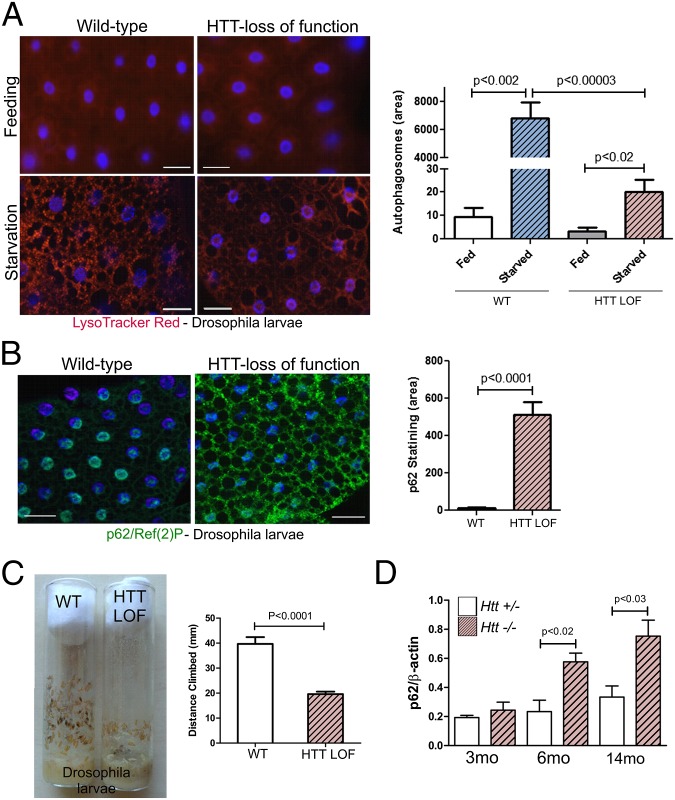
Given the similarities in amino acid sequence between human HTT and yeast Atg11, Atg23, and Vac8 (15), we asked whether normal Drosophila HTT plays a role in autophagy by assessing the ability of HTT LOF flies to mount a stress-activated autophagic response. Feeding larvae were immersed in 20% (wt/vol) sucrose for 3 h to induce a starvation response. Autophagy was monitored with both LysoTracker Red and by immunostaining with anti-p62/SQSTM1, a receptor protein for selective autophagy that is called Ref(2)P in Drosophila (16). In normal larvae, this starvation regimen induces the rapid appearance of many acidic LysoTracker Red-positive vesicles (Fig. 1A). In HTT LOF larvae, the appearance of acidic vesicles (primary lysosomes and autolysosomes) induced by starvation stress is dramatically suppressed. Consistent with a disruption of autophagy, accumulation of Ref(2)P increased markedly in the fat bodies of HTT LOF larvae (Fig. 1B). The data from Fig. 1 A and B was quantitated and found to be highly significant. These observations indicate that HTT-deficient Drosophila are unable to mount a stress-induced autophagic response. This phenotype is similar to that reported for LOF mutants of Drosophila RB1CC1/FIP200 and Atg7, proteins that are also essential for autophagy (17–19). In addition, we observed a climbing deficit in HTT LOF larvae, a behavioral defect typical of impaired autophagy. Late in the third larval stage (L3), a hormone signal prompts the initiation of a wandering behavior when larvae leave the food and climb the walls of the vial searching for an appropriate place to pupariate. HTT LOF larvae wander to pupriation sites significantly lower (approximately half) than controls (Fig. 1C). This behavior is similar to the motor deficit reported for RB1CC1/FIP200 and Atg7 LOF mutants (17, 18). These data demonstrate that loss of HTT in Drosophila results in an almost complete loss of starvation-induced autophagy.
Fig. 1.
Loss of HTT function in Drosophila larvae or conditional knockout of HTT in the mouse CNS inhibits autophagy. (A) Loss of Drosophila HTT function results in an inhibition of autophagy in starved larvae as shown by significant reduction in numbers of acidic vesicles monitored by LysoTracker Red in fat body cells. (Scale bars, 30 μm.) (B) Drosophila p62/SQSTM1, Ref(2)P, significantly accumulates in the cytoplasm of fat body cells with HTT LOF in starved larvae. (Scale bars, 30 μm.) (C) Just before pupariation, the locomotion of the HTT LOF larvae exhibit a statistically significant climbing deficit. (D) Httflox/−;nestin-cre-tg CNS conditional Htt knockout mice significantly accumulate p62/SQSTM1 in the striatal insoluble fraction with aging. n = 3 for each genotype.
Conditional Knockout of HTT in the Mouse CNS Supports a Role for HTT in Autophagy.
Because complete LOF of HTT in the mouse causes early embryonic lethality, we used the conditional inactivation of HTT in mouse forebrain to assess the impact of HTT LOF. HTT LOF at early postnatal or late embryonic stages causes a progressive neurodegenerative phenotype and early mortality (5), reminiscent of the phenotypes caused by loss of autophagy proteins, such as Atg5, Atg7, and RB1CC1/FIP200 in the CNS of mice. These conditional knockouts cause abnormal accumulation of ubiquitinated protein aggregates accompanied by increased apoptosis and neurodegeneration (20–22). We analyzed HTT conditional knockout mice (Httflox/−;nestin-cre-tg) for similar biochemical correlates and observed statistically significant increases in p62 in the pellet fraction from dissected striata by 6 mo of age compared with controls (Fig. 1D). This dysregulation is progressive, as by 14 mo accumulation of p62 in these fractions is even more pronounced (Fig. 1D and Fig. S1A). Dysregulation of autophagy is also suggested by a pronounced accumulation of lipofuscin and ubiquitin in thalamus from 7-mo-old conditional knockout mice (Fig. S1B), a brain region that shows relatively early lipofuscin accumulation in aging rodents and for which dysfunction is implicated in HD (23, 24). Lipofuscin and ubiquitin accumulation is similarly pronounced in striata from CAG140 HD mice (25, 26). Although most Htt−/− mutants die before 15 mo of age because of hydrocephalus, in one surviving 2-y-old mutant mouse with milder hydrocephalus, ubiquitin and p62 puncta and lipofuscin accumulation were also prevalent in the striatum, similar to what we observed in 2-y-old 140Q/+ mouse striatum (Fig. S1C). Taken together, these data are consistent with HTT playing a necessary essential role in autophagy that is impaired upon HTT knockout or mutation.
The C-Terminal Domain of HTT Is Similar to Yeast Atg11.
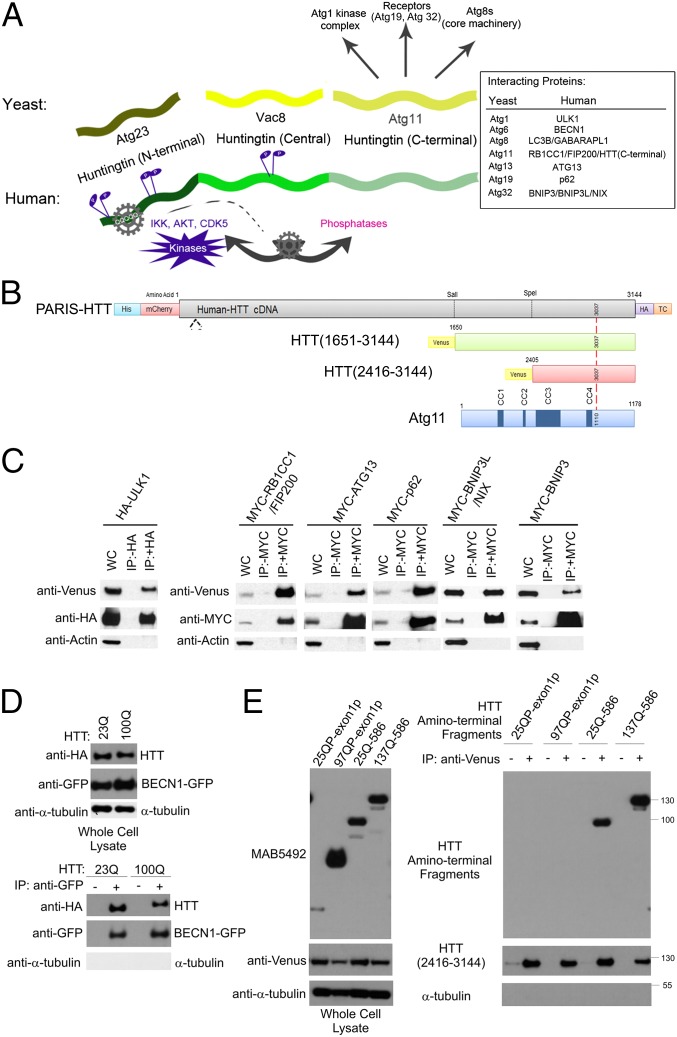
The in vivo data above support the hypothesis that HTT normally functions as a component of the autophagy system. Sequence analysis of HTT orthologs from mammals and invertebrates suggest that several regions of the HTT protein share similarity with Saccharomyces cerevisiae autophagic proteins (Fig. 2A and Figs. S2–S4) (15). The amino-terminal (N-terminal) domain of mammalian HTTs (human HTT amino acids 1–586) is similar to yeast Atg23 (Fig. S2), the central region of HTT (amino acids 745–1710) is similar to yeast Vac8 (Fig. S3), and the carboxyl-terminal (C-terminal) region of HTT (amino acids 1815–3144) is similar to yeast Atg11 (Fig. S4). This finding suggests the hypothesis that HTT is a protein in which the functionality of these three yeast proteins is combined into a single large polypeptide chain.
Fig. 2.
HTT copurifies with proteins required for regulation of autophagy. (A) HTT shares similarity with yeast proteins Atg23, Vac8, and Atg11 (15); its C-terminal Atg11-like domain may interact with mammalian homologs of yeast Atg11-binding proteins. A flexible hinge within the N-terminal domain of HTT may be modulated by HTT phosphorylation by IKK, AKT, and CDK5, activating the C-terminal Atg11-like domain of HTT to function in selective autophagy. (B) HTT constructs used in this study: Venus-HTT(2146–3144) (red) and Venus-HTT(1651–3144) (green). (C) Components of the Atg1 kinase complex and receptor proteins p62, BNIP3, and BNIP3L/NIX coimmunoprecipitate with HTT(2416–3144). HEK293T cells were cotransfected with Venus-HTT(2416–3144) and plasmids expressing HA-ULK1, MYC-RB1CC1/FIP200, MYC-ATG13, MYC-p62, MYC-BNIP3, and MYC-BNIP3L/NIX. Cell lysates were subjected to immunoprecipitation with (+) and without (−) antibodies against HA and MYC. (D) BECN-1 co-immunoprecipitation with full length HTT. HEK293T cells were cotransfected with pARIS-HTT (23Q or 100Q) and BECN1-GFP. Cell lysates were subjected to immunoprecipitation using antibodies against GFP. (E) 25QP and 97QP N-terminal HTT exon 1 fragments do not coimmunoprecipitate with Venus-HTT(2416–3144), but 586 amino acid HTT fragments do copurify. HEK293T cells were cotransfected with Venus-HTT(2416–3144) and with exon1 and 586 HTT fragment constructs. Cell lysates were subjected to immunoprecipitation using antibodies against Venus.
To evaluate whether the C-terminal region of HTT was functionally related to Atg11 in yeast, we asked whether it would coimmunoprecipitate with human orthologs of known yeast Atg11-interacting proteins. In yeast, the Atg1/Unc-51–like autophagy activating kinase 1 (ULK1) kinase complex is a key regulator of autophagic activation and inhibition and is comprised of several Atg proteins, including Atg1, Atg11, Atg13, and Vac8 (27, 28). In mammalian cells this complex includes ULK1, RB1CC1/FIP200, and ATG13 (Fig. 2A). To test for interaction with members of this complex, a minimal fragment of the C-terminal domain of HTT (amino acids 2416–3144) that contains the Atg11 CC3- and CC4-like domains involved in interactions with autophagy proteins (28, 29), was fused in frame to an N-terminal Venus tag (Fig. 2B). When epitope-tagged and overexpressed in HEK293T cells, we found that these three mammalian proteins coimmunoprecipitated with the C-terminal HTT fragment. In addition, this fragment also copurified with autophagic receptor protein p62/SQSTM1, an ortholog of yeast Atg19 that is known to interact with Atg11 (27, 30) (Fig. 2C).
The mammalian homolog of Atg6, BECN1/Beclin-1 is part of the phosphatidylinositol 3-kinase complex required for induction of autophagy and is phosphorylated and activated by ULK1 (31). Because Atg6 has a genetic interaction with Vac8 (32), we investigated the potential interaction between BECN1 and HTT, because the central domain of HTT resembles Vac8 (Fig. S3) (15). BECN1 coimmunoprecipitated with full-length WT (23Q) and mutant (100Q) HTT in cells (Fig. 2D). These data are consistent with HTT functioning like Atg11 and Vac8 in metazoans.
The N- and C-Terminal Domains of HTT Interact.
In yeast, Atg23 and Atg11 work together to regulate autophagosome formation and Atg9 trafficking (15). In HTT, the N-terminal domain resembles the yeast Atg23 protein (Fig. S2), whereas the C-terminal domain is similar to Atg11 (Fig. S4). If these structural similarities are functionally relevant, one would predict that the N- and C-terminal domains of HTT might interact with each other. Consistent with this hypothesis, the N-terminal 586 amino acid-HTT fragment coimmunoprecipitated with the C-terminal region tested above (amino acids 2416–3144) and expansion of the polyQ repeat did not affect the interaction (Fig. 2E). The HTT fragment encoded by 90 amino acid exon 1 was not sufficient for this interaction, suggesting that HTT residues within the 91–586 amino acid domain are required.
HTT May Play a Role in Mitophagy.
In yeast, selective autophagic clearance of mitochondria, mitophagy, requires an interaction between Atg11 and the protein Atg32, a receptor protein anchored to the outer mitochondrial membrane (33). The mammalian counterparts of Atg32 are the integral membrane receptors BNIP3 (BCL2/adenovirus E1B 19 kd-interacting protein 3) and BNIP3L/NIX (34, 35). Immunoprecipitation of MYC-tagged BNIP3 or BNIP3L/NIX after cotranfection with Venus-tagged HTT(2416–3144) followed by Western blotting (Fig. 2C) resulted in robust staining with anti-Venus antibody but not with anti-actin, demonstrating that the C-terminal HTT(2416–3144) interacts with the mammalian counterparts of yeast Atg32 that bind Atg11 during mitophagy.
BNIP3 and BNIP3L/NIX were both originally isolated as proteins that, when overexpressed in cultured mammalian cells (e.g., MCF-7, HeLa, and rat fibroblasts), cause apoptotic cell death, suggesting that destabilizing mitophagic balance may have severe consequences for the cell (36). Furthermore, overexpression of Atg11 from a multicopy plasmid in yeast results in cell death (37). Consistent with a disruption in cellular homeostasis, transiently transfecting C-terminal HTT fragments into rat primary cortical neurons (Fig. S5) caused cell death, both when expressing either the minimal CC3 and CC4-like region (amino acids 2416–3144) or a longer fragment that comprises the full Atg11-like protein (amino acids 1651–3144).
HTT and Atg11 Have WXXL Domains.
Atg8-family proteins are evolutionarily conserved proteins that are components of the core autophagic machinery and play a crucial role in the formation of autophagosomes. Yeast Atg8 is highly similar to the mammalian Atg8 family, which is comprised of at least three MAP1 light-chain 3 proteins (LC3A, LC3B, LC3C), and four GABA receptor-associated protein (GABARAP) and GABARAP-like proteins (GABARAPL1/Atg8L/GEC-1, GABARAPL2/Gate-16, and GABARAPL3) (38). Atg8s are ubiquitin-like modifiers that become conjugated to phosphatidylethanolamine by a process similar to ubiquitination. Lipidated Atg8s localize to phagophores (autophagosome precursors) and autophagosomes and are involved in the recruitment of the selective cargo to the forming autophagosome. Atg11 has recently been shown to directly interact with Atg8 (39). To examine whether the C-terminal domain of HTT interacts with the Atg8-family of proteins in cells, the two different C-terminal HTT fragments were tested in coimmunoprecipitation experiments with mammalian Atg8 homologs LC3B and GABARAPL1.
Proteins that dock onto Atg8s have a “WXXL” interaction surface with a consensus sequence W/F/Y-X-X-L/I/V/F. In mammals, this domain is also be referred to as a “LIR,” for LC3-interacting region (38, 40), and is found in many proteins, including p62, OPTN/optineurin, and BNIP3L/NIX, which are also Atg8 family-interacting proteins (Fig. S6A). The C-terminal domains of both Atg11 and HTT contain a highly conserved region (Figs. S4 and S6B) with a consensus LIR, coinciding with tryptophan (W) 3037 in human HTT. Using a web resource for prediction of Atg8-family interacting proteins, iLIR (38), we find that human full-length HTT has nine such xLIR motifs (LIR motifs containing flanking residues common to other Atg8 family-interacting proteins) including the phylogenetically conserved motif at W3037, which has the highest position-specific scoring matrix score (Figs. S6C and S7). An additional 45 WXXL motifs were also identified within full-length HTT (Fig. S8), again similar to yeast Atg11, which has 4 xLIRs and 23 WXXL domains predicted by the iLIR web resource (Fig. S9).
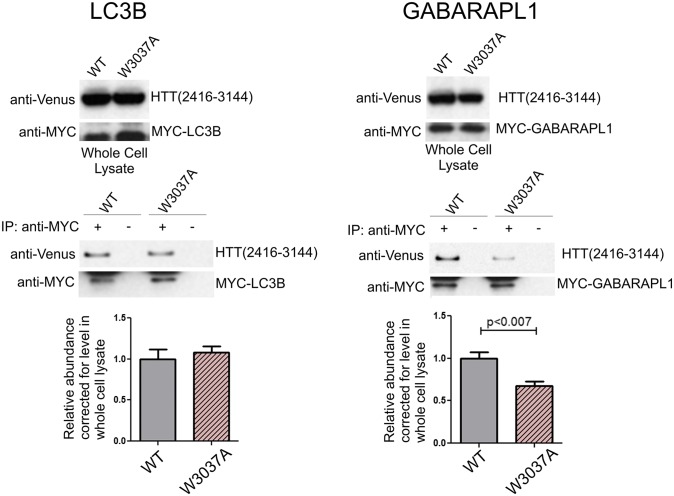
The p62 and OPTN proteins each contain only one predicted xLIR, and mutation of the p62 xLIR at W338A or the OPTN xLIR at F178A blocks their interaction with LC3 (41, 42). We found that Venus-HTT(2416–3144) and Venus-HTT(1651–3144) specifically coimmunoprecipitated with both MYC-LC3B and MYC-GABARAPL1. To test whether the interaction of HTT with either GABARAPL1 or LC3B was dependent on the presence of the conserved W3037 motif, a W-to-A mutation was introduced (W3037A) into both C-terminal HTT polypeptides. This mutation did not affect the interaction with the longer construct (Fig. S6D), but significantly reduced the interaction of GABARAPL1 with the shorter amino acid 2416–3144 HTT fragment (Fig. 3). The interaction with LC3B was not affected by the presence of the W3037A mutation. These data suggest that the higher number of combined xLIRS and WXXL domains within the longer HTT fragment (26 vs. 11) may compensate for the mutation, but that this highly conserved motif at HTT W3037 is likely a functional LIR, as evidenced by reduced binding to GABARAPL1. The domain of mammalian Atg8s that interacts directly with WXXL domains, as shown by crystal structure, is negatively charged for GABARAPL1 but positively charged for LC3B (42). Therefore, it is possible that the positively charged HTT residue R3035 adjacent to the WXXL at W3037 may directly interact with GABARAPL1 E7 and D8 negatively charged residues, but not with LC3B R10 and R11 positively charged residues. There may therefore be a structural basis for the loss of GABARAPL1 interaction with HTT W3037A; this will be evaluated in future studies.
Fig. 3.
The C-terminal domain of HTT interacts with GABARAPL1 and LC3B. Venus-HTT(2416–3144) coimmunoprecipitates with mammalian Atg8s. HEK293T cells were cotransfected with Venus-HTT(2416–3144) and MYC-LC3B or MYC-GABARAPL1. Cell lysates were subjected to immunoprecipitation (IP) using anti-MYC. The resulting precipitates were examined by immunoblot analysis with the indicated antibodies. W3037A mutation significantly reduces the coimmunoprecipitation of Venus-HTT(2416–3144) with MYC-GABARAPL1 but not with MYC-LC3B.
Discussion
Autophagy is an evolutionarily conserved lysosomal degradation pathway. Here we show that the HTT protein in Drosophila and mice is essential for normal selective autophagy and we propose that the HTT proteins of metazoans encapsulate the function of autophagy proteins Atg11, Atg23, and Vac8 in yeast. Specifically, the protein Atg11 is known to regulate selective autophagy and act as a scaffold/adaptor protein that brings the autophagic core machinery into contact with targets for degradation. Vac8 is part of the Atg1 complex and plays a role in nucleophagy in the formation of nucleus/vacuole junctions, whereas Atg23 and Atg11 work together to regulate autophagosome formation and Atg9 trafficking in yeast (15). We find that the C-terminal domain of HTT, similar in structure to yeast Atg11, can interact with the Atg1/ULK1 kinase complex, receptor proteins and the Atg8 family of proteins, suggesting that HTT may have activity as a scaffold for selective forms of macroautophagy and microautophagy, in essence functioning as a mammalian Atg11.
The loss of HTT function in a number of different organisms and tissue types has led to a plethora of phenotypic consequences (43). The possibility that HTT plays a role as an autophagic scaffold has the potential to unite a diverse set of observations regarding HTT function and has relevance to therapeutic interventions in HD. In Fig. S10, we outline a range of cellular processes which have been shown to involve selective autophagy and to require the specific functional components we suggest are attributable to HTT; many of these have been reported to be disrupted in HTT knockouts. For example, in zebrafish loss of HTT function is associated with a defect in iron metabolism, which at present is mechanistically unclear. A defect in selective autophagic clearance of the iron-binding protein ferritin (ferritinophagy) with loss of HTT function in HD patients and mice may therefore reflect an impairment in ferritinophagy (13, 44, 45). Both mutant HTT expression and HTT knockdown are found to impair axonal trafficking of vesicles, mitochondria, and autophagosomes in neurons in vitro and in vivo (7–9, 12), consistent with a loss of Atg11/Atg23-like membrane trafficking.
Autophagy proteins can regulate cell cycle progression, mitosis, and selective midbody ring autophagic clearance (46–48); therefore, HTT activity at the mitotic spindle may reflect its function as part of the autophagic machinery. We also find that HTT knockout in the mouse CNS causes accumulation of p62- and ubiquitin-containing aggregates, which may reflect a loss of aggrephagy, the selective autophagic disposal of protein oligomers and aggregates (49). Atg11 is a scaffold protein required for selective targeting of oligomers to the yeast vacuole in the Cvt pathway. Two Atg11-interacting proteins required for the Cvt pathway, Atg20 and Atg24, are similar in amino acid sequence to HTT-interacting proteins OPTN and HAP1, respectively (15), further supporting the similarity in aggrephagy function between HTT and Atg11.
Loss of HTT function in mouse cells reduces cilia formation, and ciliogenesis is altered in HD. The association of HTT with OPTN directly links it to the RAB8 protein, important for ciliogenesis (6, 41, 50, 51). These data support a role for HTT in ciliogenesis/ciliophagy (52). A loss of WT HTT function may similarly cause dysregulation of RNA-mediated gene silencing because HTT associates with Argonaute and P bodies/stress granules, both shown to be cleared by selective autophagy/granulophagy (53–55). These associations suggest that many of the phenotypic consequences caused by loss of HTT function can be understood in terms of disrupted selective autophagy.
HD is a progressive neurodegenerative disease associated with protein accumulation and dysfunction and a growing number of studies suggest that fundamental defects in autophagy may underlie pathogenesis of this disease. In HD, autophagosomes form normally and are eliminated by lysosomes, but fail to efficiently trap cytosolic cargo in their lumen. The number of lipid droplets increases in HD cells, paralleling their reduced association with autophagosomes, which may reflect a loss of selective autophagic degradation of fats and lipids, lipophagy. Indeed, the accumulation of lipofuscin, which represents reduced lipid degradation and is a hallmark of aging and neurodegeneration, is observed in postmortem HD brain (56, 57). Relevant to the proposal that HTT is a critical component of the autophagy machinery, when autophagy core proteins are reduced through knockdown (20–22), neurodegeneration is observed that is similar to that observed in HD models containing an expanded polyglutamine repeat within HTT (1). In Drosophila, HTT LOF animals are viable with no obvious developmental defects; however, removing endogenous Drosophila HTT leads to accelerated neurodegeneration in flies challenged with expanded human HTT exon 1 (Q93) and to loss of autophagy in starvation-stressed larvae (Fig. 1) (10). The loss of stress-induced autophagy can account for the accelerated degeneration when challenged with mutant HTT, consistent with a role for WT HTT in autophagy in flies.
Inactivation of HTT in mouse brain results in progressive neurodegeneration and in an accumulation of aggregates (Fig. S1 B and C) (5). Conditional neuronal knockdown of autophagy core proteins Atg5 and Atg7 has previously been shown to cause neurodegeneration with corresponding accumulation of aggregates (20, 21). Knockdown of the ULK1 kinase complex protein RB1CC1/FIP200, a protein with sequence similar to yeast Atg11 (39) and the C-terminal domain of HTT (Fig. S4), causes cerebellar neurodegeneration and aggregate accumulation (22), whereas knockdown of HTT results in striatal cell loss (1), although at a much slower rate of progression, possibly because of redundancy in Atg11-like protein function. Thus, differential expression and use of mammalian Atg11-like proteins in brain may contribute to the selective neuronal cell death observed in neurodegenerative diseases. BECN1 can regulate autophagic clearance of full-length and fragmented mutant HTT in cell culture and colocalizes with mutant HTT in inclusions in the transgenic R6/2 HD mouse brain (58), raising the possibility that sequestration of BECN1 in mutant HTT aggregates can contribute to disease by disrupting autophagy. Here we find that BECN1 associates with full-length WT and mutant HTT. Taken together, these findings have significance for therapeutics that involve reduction of normal HTT function if HTT is indeed a critical scaffold protein involved in selective autophagy.
The central domain of HTT shares similarity with Vac8 (Fig. S3), a protein that functions together with Atg11 in yeast nucleophagy, suggesting that the central and C-terminal domains of HTT may play a role in mammalian nucleophagy (15). Impairment of nucleophagic transcription factor clearance caused by a loss in WT HTT function may contribute to HD pathogenesis, because transcriptional dysregulation has been demonstrated in HD and autophagic clearance of transcription factors has been recently shown to be progressively dysregulated with aging and neurodegeneration (43, 59). In addition, we observe that the C-terminal domain of HTT coimmunoprecipitates with mitophagy receptor proteins p62, BNIP3, and BNIP3L/NIX. A loss of mitophagic function may contribute to the known abnormalities in mitochondrial function and bioenergetics that occur in HD (60).
We find that the HTT N-terminal Atg23-like domain interacts with its Atg11-like C-terminal domain. We postulate that posttranslational modifications within the N-terminal domain of HTT, which regulate WT and mutant HTT abundance and mutant HTT-mediated toxicity, may serve as potential modulators of this intramolecular interaction and, hence, regulation of autophagy. These modifications include phosphorylation of serines 13, 16, and 421 and acetylation of lysines 9 and 444 (15, 61). Phosphorylation of HTT serines 13 and 16 may change the conformation of a flexible hinge in the N-terminal domain of HTT to reduce the ability of HTT to fold back on itself (62), thereby potentially allowing the autophagy activity of the Atg11 C-terminal domain to be activated. Indeed, mimicking this modification reduces mutant HTT-mediated toxicity in vivo (63), consistent with a central role for this phosphorylation event.
We propose here that HTT may function as a scaffold for selective autophagy, based on structural similarities with Atg11, Atg23, and Vac8, interactions with Atg11 partners, the presence of a motif that can regulate at least one of these interactions, and findings that loss of HTT function recapitulates phenotypes in common with loss of core autophagy proteins. A loss of neuronal selective autophagy with age may contribute to the accumulation of protein aggregates, disruption of mitochondrial function, and inflammation that are hallmarks of HD and other polyQ diseases, as well as diseases such as Alzheimer’s and Parkinson’s diseases, frontotemporal dementia, and amyotropic lateral sclerosis. Given that autophagy has been implicated in many of these diseases, targeting HTT itself may provide an attractive therapeutic target for intervention in these disorders, suggesting that these findings may have broad relevance.
Materials and Methods
Reagents.
Plasmids and antibodies used in this study are described in SI Materials and Methods.
Cell Culture and Transfection, Immunoprecipitation, and Western Blot.
HEK293T cells were cultured, lysed, and sonicated. For immunoprecipitation, 500 μg of HEK293T cell lysate was incubated with anti-HA, anti-MYC or anti-Living Colors antibody and Dynabeads (Invitrogen) and incubated on a rotator overnight at 4 °C. Immunoprecipitates were washed three times and analyzed by Western blot, and statistics are described in SI Materials and Methods.
Primary Cortical Neuron Survival Assay.
Rat (Rattus norvegicus) colonies were maintained in accordance with the University of California, San Francisco Assurance of Compliance with PHS Policy on Humane Care and Use of Laboratory Animals by Awardee Institutions number A3400-01. The animals were euthanized using CO2, resulting in rapid and painless death, consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. Rat primary cortical neurons were dissected, plated and transfected, imaged and survival analysis performed as described in SI Materials and Methods.
Nestin-cre Conditional Htt Knockout Mouse.
All procedures using mice were approved by the Institutional Animal Care and Use Committee of the University of Virginia, and were in accordance with NIH guidelines. For conditional knockout of Htt expression in neuronal progenitors, we used a nestin-cre transgenic line. Httflox/−;nestin-cre-tg mice were obtained from crosses between Htt+/−; nestin-cre-tg males and Httflox/flox females. Brains were frozen and subjected to analysis as described in SI Materials and Methods. p62/SQSTM1 was detected by Western analysis in the striatal insoluble pellet fraction and quantitated as previously described (26).
Drosophila HTT LOF Experiments.
Autophagy was analyzed in HTT LOF Drosophila larvae (10) as described in SI Materials and Methods. Briefly, early third-instar larvae synchronized at 88 h after egg laying were starved through flotation in sucrose for 3 h at room temperature. Fat bodies were dissected and processed with LysoTracker Red or fixed and subjected to immunohistochemistry with anti-p62/Ref(2)p. Climbing behavior was analyzed by averaging the distance from the food surface of the 5 highest pupae in each of 10 vials.
Supplementary Material
Acknowledgments
We thank Drs. Asa Gustafsson (University of California, San Diego) for the BNIP3 plasmid; K. C. Walls and Frank LaFerla (University of California, Irvine) for the Beclin-1 plasmid; Do-Hyung Kim (University of Minnesota) for ULK1, ATG13/mAtg13, and RB1CC1/FIP200 plasmids; Jorge Moscat (Sanford-Burnham Medical Research Institute) for the p62 plasmid; Dr. Gabor Juhasz (Eötvös Loránd University) for the Drosophila p62/Ref(2)P antibody; Dr. Sheng Zhang (University of Texas) for the Huntingtin loss-of-function Drosophila; and Drs. Ana Maria Cuervo, Erich Wanker, Alexander Osmand, Pinar Coskun, Jorge Busiglio, and Pablo Helguera for helpful discussion. This work was supported in part by the Hereditary Disease Foundation, CHDI Foundation Inc., the Fox Family Foundation, and NIH Grants NS072453 (to J.S.S.), NS052789 (to L.M.T.), NS043466 (to S.O.Z.), NS045283 (to J.L.M.), and GM053396 (to D.J.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420103111/-/DCSupplemental.
References
- 1.Ross CA, Tabrizi SJ. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10(1):83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 2.Duyao MP, et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995;269(5222):407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 3.Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet. 1995;11(2):155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 4.Nasir J, et al. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81(5):811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 5.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26(3):300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 6.Keryer G, et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Invest. 2011;121(11):4372–4382. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong YC & Holzbaur EL (2014) The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci 34(4):1293-1305. [DOI] [PMC free article] [PubMed]
- 8.Trushina E, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24(18):8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118(1):127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S, Feany MB, Saraswati S, Littleton JT, Perrimon N. Inactivation of Drosophila Huntingtin affects long-term adult functioning and the pathogenesis of a Huntington’s disease model. Dis Model Mech. 2009;2(5-6):247–266. doi: 10.1242/dmm.000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godin JD, et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67(3):392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Zala D, Hinckelmann MV, Saudou F. Huntingtin’s function in axonal transport is conserved in Drosophila melanogaster. PLoS ONE. 2013;8(3):e60162. doi: 10.1371/journal.pone.0060162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lumsden AL, Henshall TL, Dayan S, Lardelli MT, Richards RI. Huntingtin-deficient zebrafish exhibit defects in iron utilization and development. Hum Mol Genet. 2007;16(16):1905–1920. doi: 10.1093/hmg/ddm138. [DOI] [PubMed] [Google Scholar]
- 14.Myre MA, et al. Deficiency of huntingtin has pleiotropic effects in the social amoeba Dictyostelium discoideum. PLoS Genet. 2011;7(4):e1002052. doi: 10.1371/journal.pgen.1002052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steffan JS. Does Huntingtin play a role in selective macroautophagy? Cell Cycle. 2010;9(17):3401–3413. doi: 10.4161/cc.9.17.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nezis IP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180(6):1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M, et al. Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy. 2013;9(8):1201–1213. doi: 10.4161/auto.24811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juhász G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21(23):3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy P, et al. Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy. 2014;10(3):453–467. doi: 10.4161/auto.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 21.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 22.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285(5):3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassubek J, Juengling FD, Ecker D, Landwehrmeyer GB. Thalamic atrophy in Huntington’s disease co-varies with cognitive performance: a morphometric MRI analysis. Cereb Cortex. 2005;15(6):846–853. doi: 10.1093/cercor/bhh185. [DOI] [PubMed] [Google Scholar]
- 24.Nakano M, Oenzil F, Mizuno T, Gotoh S. Age-related changes in the lipofuscin accumulation of brain and heart. Gerontology. 1995;41(Suppl 2):69–79. doi: 10.1159/000213726. [DOI] [PubMed] [Google Scholar]
- 25.Zheng S, Ghitani N, Blackburn JS, Liu JP, Zeitlin SO. A series of N-terminal epitope tagged Hdh knock-in alleles expressing normal and mutant huntingtin: Their application to understanding the effect of increasing the length of normal Huntingtin’s polyglutamine stretch on CAG140 mouse model pathogenesis. Mol Brain. 2012;5:28. doi: 10.1186/1756-6606-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng S, et al. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010;6(2):e1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26(1):9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16(4):1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aoki Y, et al. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell. 2011;22(17):3206–3217. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraft C, Peter M, Hofmann K. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12(9):836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 31.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoppins S, et al. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195(2):323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki K. Selective autophagy in budding yeast. Cell Death Differ. 2013;20(1):43–48. doi: 10.1038/cdd.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanna RA, et al. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287(23):19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parzych KR, Klionsky DJ. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20(3):460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16(7):939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geng J, Baba M, Nair U, Klionsky DJ. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol. 2008;182(1):129–140. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalvari I, et al. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy. 2014;10(5):913–925. doi: 10.4161/auto.28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li F, Chung T, Vierstra RD. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014;26(2):788–807. doi: 10.1105/tpc.113.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farré JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013;14(5):441–449. doi: 10.1038/embor.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333(6039):228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noda NN, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13(12):1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 43.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90(3):905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 44.Simmons DA, et al. Ferritin accumulation in dystrophic microglia is an early event in the development of Huntington’s disease. Glia. 2007;55(10):1074–1084. doi: 10.1002/glia.20526. [DOI] [PubMed] [Google Scholar]
- 45.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maskey D, et al. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun. 2013;4:2130. doi: 10.1038/ncomms3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dotiwala F, et al. DNA damage checkpoint triggers autophagy to regulate the initiation of anaphase. Proc Natl Acad Sci USA. 2013;110(1):E41–E49. doi: 10.1073/pnas.1218065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat Cell Biol. 2009;11(1):65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- 49.Lamark T, Johansen T. Aggrephagy: Selective disposal of protein aggregates by macroautophagy. Int J Cell Biol. 2012;2012:736905. doi: 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hattula K, Peranen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr Biol. 2000;10(24):1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- 51.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129(6):1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 52.Wrighton KH. Cytoskeleton: Autophagy and ciliogenesis come together. Nat Rev Mol Cell Biol. 2013;14(11):687. doi: 10.1038/nrm3686. [DOI] [PubMed] [Google Scholar]
- 53.Savas JN, et al. Huntington’s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci USA. 2008;105(31):10820–10825. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gibbings D, et al. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14(12):1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153(7):1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13(5):567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cortes CJ, La Spada AR. The many faces of autophagy dysfunction in Huntington’s disease: From mechanism to therapy. Drug Discov Today. 2014;19(7):963–971. doi: 10.1016/j.drudis.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shibata M, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281(20):14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 59.Lu T, et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507(7493):448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenstock TR, Duarte AI, Rego AC. Mitochondrial-associated metabolic changes and neurodegeneration in Huntington’s disease—From clinical features to the bench. Curr Drug Targets. 2010;11(10):1218–1236. doi: 10.2174/1389450111007011218. [DOI] [PubMed] [Google Scholar]
- 61.Thompson LM, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187(7):1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caron NS, Desmond CR, Xia J, Truant R. Polyglutamine domain flexibility mediates the proximity between flanking sequences in huntingtin. Proc Natl Acad Sci USA. 2013;110(36):14610–14615. doi: 10.1073/pnas.1301342110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu X, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64(6):828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.