Significance
Our results indicate that integration and excision of the toxin-linked cryptic satellite phage TLCϕ depend on a Xer recombination pathway different from the pathways so far described for other integrative mobile elements exploiting Xer (IMEXs) and for plasmid and chromosome dimer resolution. They also provide the most likely mechanism for the complete elimination of IMEX arrays that occurred in some Vibrio cholerae strains of the second wave of the current cholera pandemic.
Keywords: cholera, site-specific DNA recombination, lysogenic conversion, chromosome segregation, lateral gene transfer
Abstract
As in most bacteria, topological problems arising from the circularity of the two Vibrio cholerae chromosomes, chrI and chrII, are resolved by the addition of a crossover at a specific site of each chromosome, dif, by two tyrosine recombinases, XerC and XerD. The reaction is under the control of a cell division protein, FtsK, which activates the formation of a Holliday Junction (HJ) intermediate by XerD catalysis that is resolved into product by XerC catalysis. Many plasmids and phages exploit Xer recombination for dimer resolution and for integration, respectively. In all cases so far described, they rely on an alternative recombination pathway in which XerC catalyzes the formation of a HJ independently of FtsK. This is notably the case for CTXϕ, the cholera toxin phage. Here, we show that in contrast, integration of TLCϕ, a toxin-linked cryptic satellite phage that is almost always found integrated at the chrI dif site before CTXϕ, depends on the formation of a HJ by XerD catalysis, which is then resolved by XerC catalysis. The reaction nevertheless escapes the normal cellular control exerted by FtsK on XerD. In addition, we show that the same reaction promotes the excision of TLCϕ, along with any CTXϕ copy present between dif and its left attachment site, providing a plausible mechanism for how chrI CTXϕ copies can be eliminated, as occurred in the second wave of the current cholera pandemic.
The causative agent of the epidemic severe diarrheal disease cholera is the Vibrio cholerae bacterium. A major determinant of its pathogenicity, the cholera enterotoxin, is encoded in the genome of the filamentous cholera toxin phage, CTXϕ (1). Like many other V. cholerae filamentous phages, CTXϕ uses a host chromosomally encoded, site-specific recombination (Xer) machinery for lysogenic conversion (2–4). The Xer machinery normally serves to resolve chromosome dimers, which result from homologous recombination events between the two chromatids of circular chromosomes during or after replication. In V. cholerae, as in most bacteria, the Xer machinery consists of two tyrosine recombinases, XerC and XerD. They act at a unique specific chromosomal site, dif, on each of the two circular chromosomes, chrI and chrII, of the bacterium (5). Integrative mobile elements exploiting Xer (IMEXs) carry a dif-like site on their circular genome, attP (3, 4) (Fig. 1A). XerC and XerD promote their integration by catalyzing a recombination event between this site and a cognate chromosomal dif site (3, 4) (Fig. 1A). Based on the structure of their attP site, IMEXs can be grouped into at least three families (3, 4) (Fig. 1B). In all cases, however, a new functional dif site is restored after integration, which permits multiple successive integration events (Fig. 1A). Indeed, most clinical and environmental V. cholerae isolates harbor large IMEX arrays (6, 7).
Fig. 1.
Systems that use Xer. (A) Scheme depicting the sequential integration of IMEXs. Triangles represent attP and dif sites, pointing from the XerD binding site to the XerC binding site. Chromosomal DNA (black), TLCϕ DNA (blue), and CTXϕ DNA (magenta) are indicated. Dotted triangles represent nonfunctional CTXϕ sites. (B) Sequence alignment of dif1, attPCTX, attPVGJ, attPTLC, difA, and dif2. Bases differing from dif1 are indicated in color. Bases that do not fit the XerD binding site consensus are indicated in lowercase. XerC (●) and XerD (○) cleavage points are indicated. (C) Xer recombination pathways. XerC (light gray circles), XerD (dark gray circles), dif sites (red and black lines), and attPCTX and attPVGJ (magenta and green lines) are indicated. XerC and XerD catalysis-suitable conformations are depicted as horizontal and vertical synapses, respectively. Cleavage points are indicated as in B.
IMEX array formation participates in the continuous and rapid dissemination of new cholera toxin variants in at least three ways. First, CTXϕ integration is intrinsically irreversible because the active form of its attP site consists of the stem of a hairpin of its ssDNA genome, which is masked in the host dsDNA genome (8, 9) (Fig. 1 A and B). However, free CTXϕ genome copies can be produced by a process analogous to rolling circle replication after the integration of a second IMEX harboring the same integration/replication machinery, such as the RS1 satellite phage, which permits the production of new CTXϕ viral particles (10). Second, the V. cholerae Gillermo Javier filamentous phage (VGJϕ) belongs to a second category of IMEXs whose attP site permits cycles of integration and excision by Xer recombination (11). VGJϕ excision allows for the formation of hybrid molecules harboring the concatenated genomes of CTXϕ and VGJϕ, provided that VGJϕ integrated before CTXϕ (11). The hybrid molecules can be packaged into VGJϕ particles. VGJϕ particles have a different receptor than CTXϕ, which permits transduction of the cholera toxin genes to cells that do not express the receptor of CTXϕ (11–13). Finally, integration of the toxin-linked cryptic phage (TLCϕ), a satellite phage that defines a third category of IMEXs, seems to be a prerequisite to the toxigenic conversion of many V. cholerae strains (14, 15). IMEXs from this family are found integrated in the genome of many bacteria outside of the Vibrios, including human, animal, and plant pathogens, which sparked considerable interest in the understanding of how they exploit the Xer machinery at the molecular level (3, 4).
Xer recombination sites consist of 11-bp XerC and XerD binding arms, separated by an overlap region at the border of which recombination occurs (Fig. 1B). XerC and XerD each promote the exchange of a specific pair of strands (Fig. 1B). Recombination between dif sites is under the control of a cell division protein, FtsK, which restricts it temporally to the time of constriction and spatially to a specific zone within the terminus region of chromosomes (16–19). FtsK triggers the formation of a Holliday junction (HJ) by XerD catalysis, which is converted into product by XerC catalysis after isomerization (20, 21) (Fig. 1C). The intermediate HJ is stable enough to be converted into product by replication when XerC catalysis is impeded (5, 17) (Fig. 1C). The integration of IMEXs of the CTXϕ and VGJϕ families escapes FtsK control. The lack of homology in the overlap regions of their attP sites and the dif sites they target prevents any potential XerD-mediated strand exchange (Fig. 1B). CTXϕ and VGJϕ rely on the exchange of a single pair of strands by XerC catalysis for integration, with the resulting HJ being converted into product by replication (8, 9, 11) (Fig. 1C). In the case of CTXϕ, integration is facilitated by an additional host factor, EndoIII, which impedes futile cycles of XerC catalysis once the pseudo-HJ is formed (22) (Fig. 1C). In contrast, the overlap region of TLCϕ attP, attPTLC, is fully homologous to the overlaps of dif1 and difA, the two sites in which it was found to be integrated (Fig. 1B). Four integration pathways could thus be considered, depending on whether recombination is initiated by XerC or XerD catalysis, and whether it ends with a second pair of strand exchange or not. In addition, attPTLC lacks a consensus XerD binding site, which could affect the whole recombination process (Fig. 1B).
Here, we show that attPTLC is a poor XerD binding substrate. Nevertheless, we show that TLCϕ integration is initiated by XerD catalysis and that the resulting HJ is converted into product by XerC catalysis. We further show that TLCϕ integration is independent of FtsK. Finally, we demonstrate that the same reaction can lead to the excision of TLCϕ–CTXϕ arrays, providing a plausible mechanism for how all of the CTXϕ copies integrated on V. cholerae chrI can be eliminated in a single step, as occurred in ancestors of strains from the second wave of the current cholera pandemic (23–25).
Results
XerCD-Mediated dif1-Specific Integration of TLCϕ.
The complete genome of TLCϕ was obtained by PCR amplification using N16961 genomic DNA as a template (15). It was labeled with a cat resistance gene and was delivered to V. cholerae by conjugation. The presence of 1.8 kbp of additional DNA, including the resistance marker, did not impede TLCϕ-dependent replication and integration in V. cholerae (Figs. 2–4). To detect TLCϕ-integration events, we used a colorimetric screen based on functional Escherichia coli lacZ-dif fusions that were inserted in place of the dif site of one or the other of the two V. cholerae chromosomes, the endogenous V. cholerae lacZ gene and the dif site of the other chromosome having been deleted (7). IMEX integration disrupts the lacZ-dif ORF, which leads to the appearance of white colonies and/or white sectors (7, 9).
Fig. 2.
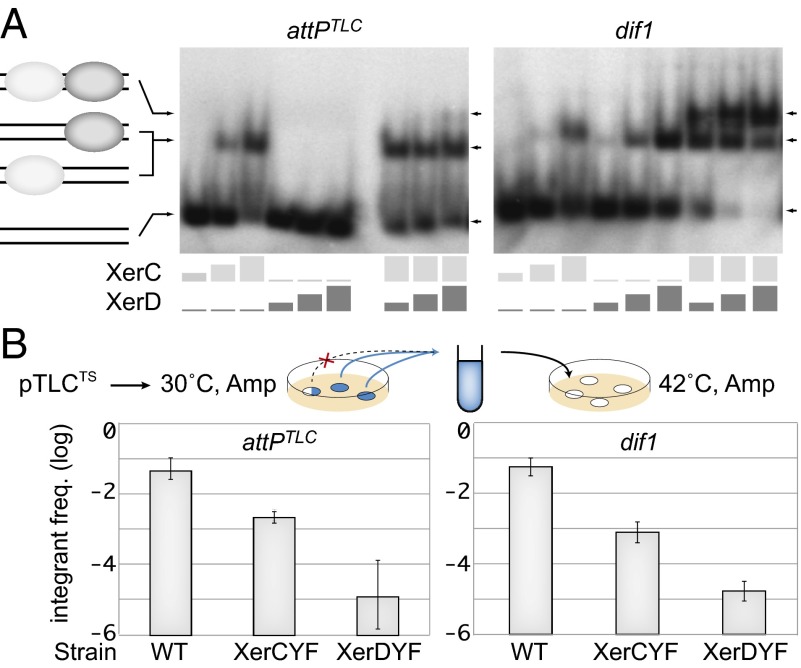
Integration of dif1 and TLCϕ TS plasmids. (A) In vitro XerCD binding assay on attPTLC and dif1. Light and dark gray rectangles indicate the respective concentrations of XerC (0 μg/mL, 15 μg/mL, 30 μg/mL, and 45 μg/mL) and XerD (0 μg/mL, 3.9 μg/mL, 7.9 μg/mL, and 11.8 μg/mL) in each lane. A scheme of the different products is indicated to the left of the gels. (B) Frequency (freq.) of integrants after overnight growth at the permissive temperature. A schematic of the assay is shown above the results. The frequency of integrants was estimated in pools of colonies without any visible integration after overnight growth (full blue circles). Colonies with visible integration were discarded (blue circles with white sectors). The results are shown on a logarithmic scale. Amp, ampicillin; WT, WT cells; XerCYF and XerDYF, strains harboring catalytically inactive XerC and XerD alleles, respectively.
Fig. 4.
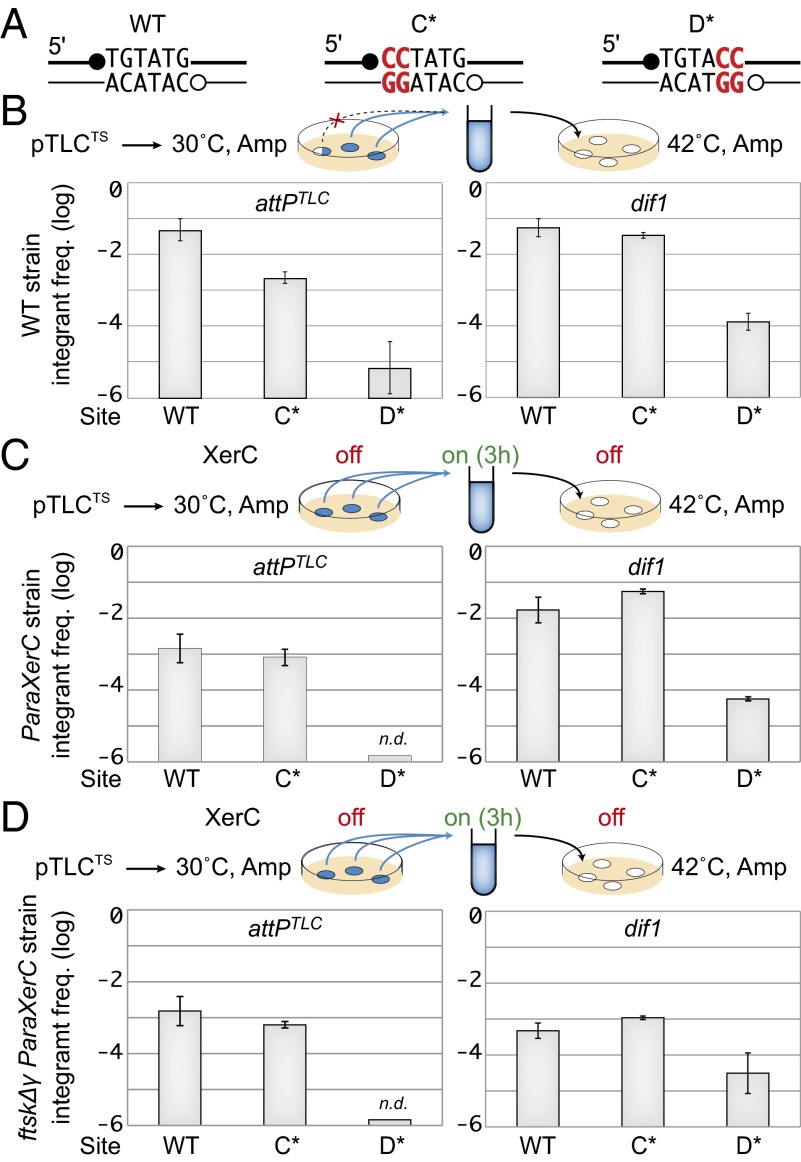
XerC promotes the resolution of the attPTLC/dif1 HJ recombination intermediate. (A) Frequency of integrants of dif1 and TLCϕ suicide vectors after 3 h of conjugation. WT, native promoter production of the recombinases; paraXerCD, production of the recombinases from a xerC-xerD operon under the pAra promoter integrated at the xerC locus; 0, no arabinose (ara). The gradual percentages of arabinose (black triangles; 0.002%, 0.02%, and 0.2%) are indicated. (B) Intramolecular recombination between plasmid-borne dif1 and attPTLC sites. (Left) Scheme of the substrate and of the recombination products. Arrows indicate the restriction sites used to differentiate fragments of the plasmid substrate (S1 and S2) from the HJ intermediate (HJ) and the full recombination products (P1 and P2). WT, C*, and D* indicate WT, C*, and D* versions of attPTLC, respectively. A, AlwNI; H, HpaI.
The overlap regions of the dif sites found in different V. cholerae strains are polymorphous. The chrII dif site of all V. cholerae strains characterized so far is dif2 (Fig. 1B). The most common chrI dif site of clinical isolates is dif1 (Fig. 1B). Several environmental strains carry a variant of dif1, difA, with the same overlap region (Fig. 1B). Delivery of TLCϕ to a strain harboring lacZ-dif1 or lacZ-difA reporters led to the appearance of white sectors in the resulting chloramphenicol-resistant colonies (Table 1). TLCϕ integration was below the detection limit in the absence of either XerC or XerD, suggesting that the presence of both recombinases was absolutely required for the process (Table 1). Not a single integration event was observed using a lacZ-dif2 reporter, demonstrating the specificity of the process (Table 1).
Table 1.
Integration rate of TLCϕ after 36 h of growth on plates
| Host | attB (chrI/chrII) | attP | Xer machinery | Integration, % |
| BS1 | dif1/− | TLC | 3.25 ± 0.59 | |
| BS47 | difA/− | TLC | 6.81 ± 0.25 | |
| BS3 | −/dif2 | TLC | <0.06 | |
| BS10 | dif1/− | TLC | ΔxerC | <0.06 |
| BS49 | dif1/− | TLC | ΔxerD | <0.06 |
| BS51 | dif1/− | TLC | XerCYF | 0.26 ± 0.01 |
| BS50 | dif1/− | TLC | XerDYF | <0.06 |
| BS1 | dif1/− | C* | 0.72 ± 0.18 | |
| BS1 | dif1/− | D* | <0.02 |
Results were obtained in at least three independent experiments. Over 1,000 colonies were screened for each condition. XerCYF and XerDYF are the catalytically inactive forms of the recombinases. C* and D* are modified sites inhibiting XerC and XerD strand exchanges, respectively.
attPTLC Lacks a Bona Fide XerD Binding Arm.
The XerD binding arm of attPTLC differs from the highly conserved XerD binding motif consensus of bacteria by 8 bp out of 11 bp (Fig. 1B). Correspondingly, XerD did not retard the electrophoretic migration of attPTLC in an acrylamide gel under conditions in which it efficiently retarded the migration of dif1 (Fig. 2A). In contrast, XerC bound as efficiently to attPTLC as it did to dif1 (Fig. 2A). Nevertheless, a faint band corresponding to the joint binding of XerC and XerD to attPTLC could be detected when the two recombinases were added together, suggesting that cooperative interactions between the recombinases partially compensated for the defective attPTLC XerD binding arm (Fig. 2A).
XerD Catalysis Is Necessary and Sufficient for TLCϕ Integration.
TLCϕ integration was not abolished in a strain in which the catalytic tyrosine of XerC was replaced by a phenylalanine (XerCYF) but went below the level of detection of the assay in a strain in which the catalytic tyrosine of XerD was replaced by a phenylalanine (XerDYF, Table 1). Catalytic mutations often affect substrate binding (26). We were therefore cautious about the TLCϕ integration results obtained in the XerDYF background because the lesser binding affinity of XerDYF could cumulate with the defective XerD binding arm of attPTLC (Fig. 2A). The exchange of strands promoted by tyrosine recombinases requires the stabilization of the invading strands by base-pairing interactions (9). Based on this rule, we introduced specific mutations in the recombination sites to block XerC or XerD strand exchanges, respectively (C* and D* mutations, Fig. 3A and Fig. S1). The presence of the C* mutation did not abolish TLCϕ integration, whereas no integrants were observed in the presence of the D* mutation (Table 1).
Fig. 3.
Integration of dif1 and TLCϕ TS plasmids harboring C* and D* mutated overlap regions. (A) Scheme of the WT, C*, and D* overlap regions. Mutations are indicated in red. Positions of XerC (●) and XerD (○) cleavage are indicated. (B) Frequency of integrants after overnight growth at the permissive temperature obtained with WT, C*, and D* versions of dif1 and attPTLC. The scheme legend is as in Fig. 2B. (C) Frequency of integrants after 3 h of growth at the permissive temperature in XerC-inducible cells. n.d., none detected; off, no XerC induction; on, XerC induction; ParaXerC, production of XerC from under the pAra promoter integrated at the xerC locus. The rest of the legend is as in Fig. 2B. (D) Frequency of integrants after 3 h of growth at the permissive temperature in ParaXerC ftsKΔγ cells. The legend is as in C.
An advantage of the colorimetric assay is that it can be used with replicative forms. However, it only provides qualitative results because the detection of white sectors depends on the size of the colonies, which is notably linked to the fitness of the strains. To obtain a quantitative view of the efficiency of integration, we used antibiotic resistance as a selection method for the integration of a replication-deficient form of TLCϕ. To this end, a portion of the TLCϕ genome lacking the whole of the cri nickase gene was cloned into a conjugative plasmid harboring a conditional thermosensitive (TS) origin of replication (Table S1). Disruption of the reporter dif-lacZ ORF was used to validate the specificity of the integration events. We monitored the frequency of integrants in fully blue colonies (i.e., in colonies in which integration was not yet visible), which were obtained after overnight growth at the permissive temperature under antibiotic selection (Fig. 2B). A TS suicide vector harboring dif1 was used as a control to validate the assay. For both dif1 and TLCϕ TS vectors, a 10-fold drop in integration was observed in the XerCYF background and a 100-fold drop was observed in the XerDYF background (Fig. 2B).
Results obtained with TS suicide vectors harboring C* and D* attPTLC were identical to the results obtained with WT attPTLC in XerCYF and XerDYF backgrounds, respectively (Fig. 3B). The frequencies of C* and D* dif1 integrants were 10-fold higher than the frequencies obtained with dif1 in the XerCYF and XerDYF backgrounds, respectively (Fig. 3B). However, D* dif1 integrants remained 100-fold less frequent than C* dif1 integrants (Fig. 3B).
attPTLC Integration Is Less Efficient Than dif1 Integration.
The frequency of integrants in colonies harboring a TS vector corresponds to the overnight growth integration/excision equilibrium. To compare the integration efficiencies of dif1 and TLCϕ TS vectors after a shorter time of growth, we used a strain in which the xerC gene was placed under the control of the arabinose promoter. The dif1 integrants were as frequent after 3 h of induction as after overnight growth, suggesting comparable excision and integration rates (Fig. 3C). This result fits well with the function of dif1, with the excision of the dif1 TS vector mimicking chromosome dimer resolution (19). Interestingly, a slightly higher integration frequency was observed with C* dif1 than with dif1, suggesting that completion of the dif1 recombination process by XerC catalysis might be more important for excision than for integration (Fig. 3C). In contrast, 10-fold fewer TLCϕ integrants were obtained after 3 h of growth compared with overnight growth, suggesting that attPTLC was less efficient than dif1 integration, which was compensated for during overnight growth by an even lower excision rate (Fig. 3C).
TLCϕ Integration Escapes FtsK Control.
Integration of a TS vector harboring dif1 was 100-fold less efficient when the chromosomal dif1 target site was displaced to the lacZ locus, which is outside of the normal FtsK loading region in V. cholerae (19) (Fig. S2). In contrast, a TS TLCϕ vector integrated as efficiently at the lacZ locus as at the dif1 locus, suggesting that it was not under the control of FtsK (Fig. S2). In agreement, no change was observed in the frequency of integrants of WT, C*, and D* TLCϕ TS vectors in FtsKΔγ cells compared with WT cells (Fig. 3D). As a point of comparison, note that a 10-fold decrease in the frequency of integrants of dif1 TS vectors was obtained in FtsKΔγ cells (Fig. 3D). The integration of C* dif1 TS vectors was similarly affected, confirming that dif1 integration relied on FtsK-dependent XerD catalysis (Fig. 3D). The residual integration observed with D* dif1 TS vectors was not affected, further indicating that it was linked to an FtsK-independent pathway (Fig. 3D).
Quantity of XerC and XerD Is a Limiting Factor of TLCϕ Integration.
Because of the low binding efficiency of XerD to attPTLC, we speculated that the concentration of the Xer recombinases within the cell could be a limiting factor for attPTLC/dif1 recombination. To test this hypothesis, we engineered a strain in which XerC and XerD production was placed under the control of the arabinose promoter. In the absence of arabinose, integration of a nonreplicative dif1 vector was over 10-fold less frequent than in a strain in which XerC and XerD were produced from their original promoters. Addition of arabinose increased dif1 integration up to WT levels (Fig. 4A). Integration of a nonreplicative TLCϕ vector was barely detectable in the absence of arabinose but reached a frequency almost 100-fold higher than the frequency obtained under normal XerCD production levels at 0.02% and 0.2% of arabinose (Fig. 4A). Western blot quantification of a His-tagged version of XerD suggested that at these two concentrations, the intracellular level of XerD was 100-fold higher than normal (Fig. S3).
attPTLC/dif1 HJs Are Resolved by XerC Catalysis.
Under conditions of expression of the Xer recombinases, TLCϕ integration was as frequent as dif1 integration, which suggested that attPTLC/dif1 recombination could be directly monitored on plasmids (Fig. 4A). Indeed, over 50% of a plasmid carrying dif1 and attPTLC in direct repeat was recombined after 3 h of arabinose induction (Fig. 4B). In addition, we observed a faint band migrating at the expected position of the HJ recombination intermediate (Fig. 4B). Two-dimensional gel analysis confirmed that this band corresponded to a four-way junction (Fig. S4). In similar experiments with tandem dif plasmids, dif/dif HJs were never detected, suggesting that they were more efficiently processed into product and/or back into substrate than attPTLC/dif1 HJs (5, 20).
Impeding XerD strand exchanges entirely abolished HJ and product formation, confirming that attPTLC/dif1 recombination was initiated by XerD catalysis (Fig. 4B). There is a perfect homology between the overlap regions of dif1 and attPTLC, which suggested that XerC catalysis might normally serve to convert attPTLC/dif1 HJs into product. In this case, impeding XerC catalysis would lead to HJ accumulation. In agreement with this idea, lesser recombination products and more HJs were detected when XerC-mediated strand exchanges were inhibited (Fig. 4B and Fig. S4). Taken together, these results demonstrate that attPTLC/dif1 recombination normally results from two successive pairs of strand exchange, with the first being promoted by XerD and the second by XerC.
XerCD Can Promote the Excision of an Integrated Copy of TLCϕ.
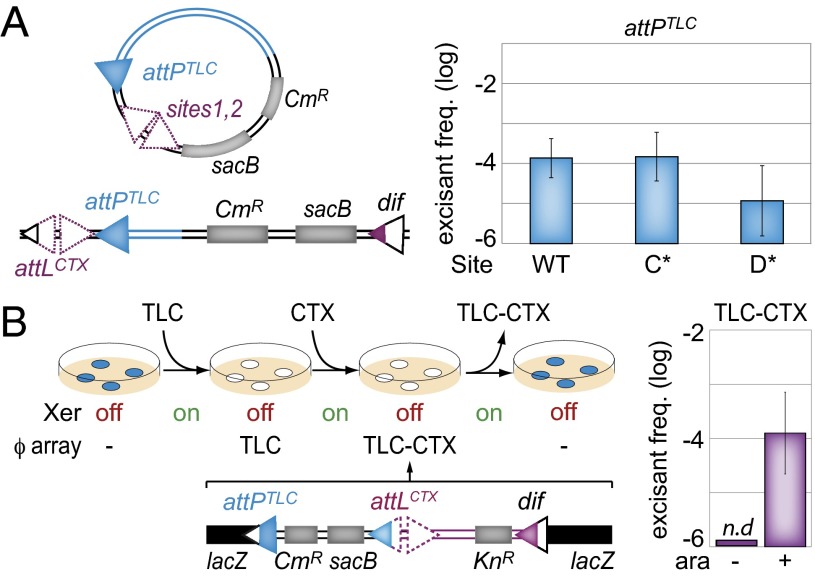
The attPTLC/dif1 intramolecular recombination reactions observed on plasmids mimic an excision reaction. Therefore, we decided to investigate the excision of chromosomal copies of TLCϕ harboring the sacB counterselection gene. We used a nonreplicative form of TLCϕ to ensure the rapid loss of any excised copy (Fig. 5A). WT, C*, and D* forms of the excision substrate were engineered by Xer site-specific recombination between dif1 and an extra CTXϕ attP (attPCTX) (Fig. 5A). Note that Xer-mediated excision of the sacB counterselection gene can only be due to a recombination event between attPTLC and dif1 because the active form of attPCTX is masked after integration. The correct arrangement of the different Xer recombination sites in the genome was checked by PCR.
Fig. 5.
TLCϕ excision. (A) TLCϕ-sacB excision assay. (Left) Scheme of the assay. Sites 1 and 2 of attPCTX used to integrate the attPTLC-CmR-sacB insert at dif1 (purple triangles) and the sacB gene conferring sucrose sensitivity and the cat (CmR) gene conferring chloramphenicol resistance (gray rectangles) are indicated. (B) TLCϕ–CTXϕ excision assay. (Left) Scheme of the assay. ϕarray, integrated IMEXs; off, no arabinose induction of the recombinases; on, arabinose induction of the recombinases. (Right) In the absence of IMEXs, the lacZ gene is functional and colonies are blue on X-gal. After integration, colonies are white on X-gal. n.d., none detected.
Cells from colonies grown on chloramphenicol plates were diluted in fresh LB without antibiotic, grown until an OD600 of 0.2–0.3 was attained, and were spread on sucrose plates. Sucrose-resistant colonies were checked for the loss of cat. The precision of the recombination events was verified by PCR. We thus found that TLCϕ excision was possible but that it occurred much less frequently than its integration, in agreement with the results of Fig. 3 (Fig. 5A). Excision was affected by the presence of a D* mutation but not by a C* mutation, further suggesting that it relied on the same Xer recombination pathway as integration (Fig. 5A).
TLCϕ-Mediated Excision of CTXϕ.
The possibility of excising TLCϕ by Xer recombination prompted us to check if attPTLC/dif1 recombination could lead to the joint excision of other IMEXs, particularly CTXϕ. To this end, we engineered a TLCϕ–CTXϕ array at the lacZ-dif1 locus in a strain in which XerCD production was under the control of the arabinose promoter (Fig. 5B). The array included a cat resistance gene and a sacB counterselection gene in the TLCϕ prophage and a kanamycin resistance marker in place of the cholera toxin genes in the CTXϕ prophage (Fig. 5B). The correct arrangement of the integrated forms of TLCϕ and CTXϕ within the strain was checked by PCR. Recovery of the capacity to produce β-gal production was used to ascertain the precision of the excision events. Note that the replicative form of CTXϕ could be used because it is rapidly lost upon excision.
Induction of XerCD production led to the appearance of blue sucrose-resistant cells at a frequency of ∼3 × 10−4 (Fig. 5B). No blue sucrose-resistant colonies were obtained in the absence of XerCD induction (Fig. 5B). All (246 of 246) of these blue sucrose-resistant colonies proved to be both kanamycin- and chloramphenicol-sensitive, as expected for complete IMEX excision events (Table S2). The precision of the excision events was further checked by PCR amplification of the resulting junctions in 32 blue sucrose-resistant colonies. Thus, the presence of a TLCϕ copy on the attL side of an IMEX array can lead to its complete and precise excision by a single Xer-dependent recombination event. White sucrose-resistant colonies were also obtained. However, they were 10-fold less frequent than blue resistance colonies (Table S2). Their formation was independent of XerCD production (Fig. 5B). Only 71 of 108 of them were both kanamycin- and chloramphenicol-sensitive, with the others remaining resistant to one, the other, or both antibiotics (Table S2), and no specific pattern of rearrangement was observed by PCR. These results suggest that white sucrose-resistant colonies were due to partial deletion events and that no conservative recombination event other than Xer recombination can lead to the complete excision of IMEX arrays.
Discussion
Here, we characterized at the molecular level how TLCϕ, one of the numerous IMEXs that are integrated in the genome of toxigenic variants of V. cholerae, exploits the Xer recombination machinery of its host. We found that TLCϕ defines a third IMEX category with a strategy of exploitation of the Xer machinery different from the strategies so far described for other IMEXs and in chromosome and plasmid dimer resolution. In addition, we demonstrated that TLCϕ integration was reversible, which led to the joint elimination of any element that had integrated after it in a single Xer recombination step.
Paradoxical Strategy of Xer Recombination Exploitation.
attPTLC lacks a bona fide XerD binding site (Fig. 2A). Nevertheless, XerCD could efficiently recombine this degenerate half-site with dif1 or difA (Fig. 2B and Table 1). To our knowledge, the only other tyrosine recombinase for which efficient recombination between half-sites has been observed is Flp, the flippase of the 2-μm plasmid of Saccharomyces cerevisiae. Flp normally works as a homotetramer; however, in case of recombination between half-sites and full sites, the synapses imply three recombinases (27). In the case of Flp, trimer recombination is possibly related to the fact that active sites are assembled in trans (28). In contrast, Xer recombinases cleave DNA in cis. Using synthetic substrates designed specifically to impede the exchange of one or the other of the two strands, we further demonstrated that attPTLC/dif1 recombination was initiated by the exchange of the strands that were expected to be targeted by XerD (Figs. 2–5 and Table 1). Finally, we showed that recombination depended on XerD catalysis and that XerC catalysis was not necessary (Fig. 2 and Table 1). Taken together, these results suggested that recombination occurred within a heterotetramer containing two of each of the XerC and XerD molecules, with one of the two XerD molecules being bound to the degenerate half-site of attPTLC. However, fully efficient recombination required the overproduction of the Xer recombinases, probably because the absence of a bona fide XerD binding site in attPTLC impeded efficient synapse formation and/or destabilized synaptic complexes (Figs. 2 and 4).
The default states of XerC and XerD within most recombination synapses are active and inactive, respectively (17, 20, 21). In agreement with this idea, the Xer reactions exploited by all other mobile elements so far characterized, whether IMEXs or plasmids, are initiated by XerC catalysis. In this respect, the attPTLC/dif1 recombination pathway is similar to the chromosome dimer resolution pathway. However, in the case of chromosome dimer resolution, activation of XerD catalysis requires a direct interaction with the FtsK cell division protein, which restricts it spatially (19). In contrast, attPTLC/dif1 recombination is independent of FtsK (Fig. 3) and is not spatially restricted (Fig. S2). Finally, in all of the other IMEXs so far characterized, HJ intermediates cannot be processed into products by Xer catalysis. Instead, we found that XerC-mediated strand exchanges could resolve attPTLC/dif1 HJ intermediates into products (Fig. 4B). More HJs were observed upon inhibition of XerC-mediated strand exchanges, further suggesting that HJ resolution by a second pair of strand exchanges is more efficient than resolution by replication (Fig. 4B).
Putative Regulation of attPTLC/dif1 Recombination.
All of the experiments presented in this study were performed using the whole or a large piece of the TLCϕ genome. Indeed, we failed to detect any recombination events with attPTLC alone. This observation suggests that attPTLC/dif1 recombination depends on factors additional to XerC and XerD. These factors could be implicated in synapse formation and/or activation of XerD catalysis. They could also help favor integration instead of excision. Future work will aim at identifying putative attPTLC/dif1 accessory elements and at characterizing their mechanism of action.
Contribution of TLCϕ to the Evolution of Toxigenic V. cholerae Variants.
Most clinical and environmental V. cholerae isolates harbor large IMEX arrays on chrI (4, 6, 7). Their history of formation can be traced based on the relative position of the different IMEXs they harbor. It does not reflect the phylogenetic lineage of the isolates, which suggests that chrI IMEXs are constantly eliminated and rapidly reacquired (3, 4). In particular, elimination of entire chrI IMEX arrays has occurred in the ancestors of some strains from the second wave of the current cholera pandemic, such as the B33 Mozambique V. cholerae strain (23). This scenario was experimentally reproduced under laboratory conditions (24, 25). Paradoxically, however, CTXϕ integration is intrinsically irreversible (Fig. 1A). Here, we showed how TLCϕ excision could lead to the precise elimination of entire chrI IMEX arrays (Fig. 5 and Table S2). Two other mechanisms have been proposed for the elimination of IMEX arrays. First, CTXϕ copies can be eliminated in a stepwise fashion by homologous recombination events between any CTXϕ and RS1 copies within an array (24, 25). However, entire elimination of the array implies a final recombination event between two regions of less than <18 nt of homology. Our results indicate that such recombination events are 10-fold less frequent than the single-step Xer-dependent excision of TLCϕ–CTXϕ arrays (Fig. 5). In addition, they are highly imprecise, with most of them leading to partial deletions (Table S2). Second, we previously reported that Xer-dependent excision was a key aspect of the life cycle of VGJϕ, which led to CTXϕ excision, provided that VGJϕ integrated first (11). However, IMEXs of the VGJϕ family are only very rarely found integrated in front of CTXϕ in the genome of clinical and environmental strains (4, 6, 7). In contrast, TLCϕ is almost invariably the first integrated element of chrI IMEX arrays (4, 6, 7), probably because its integration is a prerequisite to CTXϕ integration on this chromosome (14, 15). Taken together, these results suggest that Xer recombination between attPTLC and dif1 is the most likely mechanism for the elimination of entire chrI IMEX arrays. Thus, TLCϕ is a major contributor to the ecological cycle that allows for the constant and rapid acquisition of new cholera toxin gene variants via the continuous assembly and elimination of large IMEX arrays in clinical strains.
Materials and Methods
Strains and Plasmids.
Relevant strains, plasmids, and oligonucleotides are described in Tables S1, S3, and S4, respectively. All V. cholerae reporter strains were constructed by natural transformation or by double-crossover integration/excision methods. Engineered strains were confirmed by PCR and sequencing.
EMSA Experiments.
Five-nanomolar 32P-labeled synthetic DNA probes obtained by the annealing of purified oligonucleotides were incubated with purified XerCD recombinases in a buffer containing 0.1 μg/mL BSA, 100 mM NaCl, 40% (vol/vol) glycerol, 1 mM EDTA, and 25 mM Tris⋅HCl at pH 7.5. The different DNA/protein complexes were resolved by migration through a 5% (vol/vol) 29:1 acrylamide/bisacrylamide gel in 0.5× Tris/borate/EDTA for 2 h at 150 V at 4 °C.
Integration Assays.
E. coli β2163 donors and V. cholerae recipients were grown to an OD600 of 0.3, mixed at a 1:10 ratio, and incubated for 3 h. Conjugants were selected for the plasmid antibiotic resistance and meso-diaminopimelic acid autotrophy. The specificity of the integration events of suicide vectors was checked using X-gal. TS vector conjugants were recovered at 30 °C on X-gal plates to select for fully blue colonies. Serial dilutions were then plated at 42 °C and at 30 °C to determine the overnight frequency of integrants. In the case of Xer-inducible strains, Xer production was induced for 3 h in liquid culture with an arabinose concentration of 0.2% before plating.
Excision Assays.
Integrants harboring the sacB and cat genes were grown to an OD600 of 0.2–0.3. Serial dilutions were plated on LB agar plates supplemented with 18% sucrose at 25 °C.
Intraplasmid Recombination Assay.
Plasmids were electroporated into V. cholerae XerCD-inducible cells. Transformed fresh bacteria were grown to an OD600 of 0.5 and induced with 0.2% arabinose for 3 h.
Supplementary Material
Acknowledgments
We thank M. Blokesh and J. Bischerour for V. cholerae chromosomal engineering tools. This study received financial support from the Agence Nationale pour la Recherche [ANR-11-BLAN-O2401] and Fondation Bettencourt Schueller (2012 Coup d’Elan Award). C.M. was the recipient of a fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.J.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404047111/-/DCSupplemental.
References
- 1.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272(5270):1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 2.Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417(6889):656–659. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- 3.Das B, Bischerour J, Barre FX. Molecular mechanism of acquisition of the cholera toxin genes. Indian J Med Res. 2011;133:195–200. [PMC free article] [PubMed] [Google Scholar]
- 4.Das B, Martínez E, Midonet C, Barre F-X. Integrative mobile elements exploiting Xer recombination. Trends Microbiol. 2013;21(1):23–30. doi: 10.1016/j.tim.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Val M-E, et al. FtsK-dependent dimer resolution on multiple chromosomes in the pathogen Vibrio cholerae. PLoS Genet. 2008;4(9):e1000201. doi: 10.1371/journal.pgen.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477(7365):462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun J, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA. 2009;106(36):15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Val M-E, et al. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol Cell. 2005;19(4):559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Das B, Bischerour J, Val M-E, Barre F-X. Molecular keys of the tropism of integration of the cholera toxin phage. Proc Natl Acad Sci USA. 2010;107(9):4377–4382. doi: 10.1073/pnas.0910212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moyer KE, Kimsey HH, Waldor MK. Evidence for a rolling-circle mechanism of phage DNA synthesis from both replicative and integrated forms of CTXphi. Mol Microbiol. 2001;41(2):311–323. doi: 10.1046/j.1365-2958.2001.02517.x. [DOI] [PubMed] [Google Scholar]
- 11.Das B, Bischerour J, Barre F-X. VGJphi integration and excision mechanisms contribute to the genetic diversity of Vibrio cholerae epidemic strains. Proc Natl Acad Sci USA. 2011;108(6):2516–2521. doi: 10.1073/pnas.1017061108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campos J, et al. Novel type of specialized transduction for CTX phi or its satellite phage RS1 mediated by filamentous phage VGJ phi in Vibrio cholerae. J Bacteriol. 2003;185(24):7231–7240. doi: 10.1128/JB.185.24.7231-7240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos J, Martínez E, Izquierdo Y, Fando R. VEJphi, a novel filamentous phage of Vibrio cholerae able to transduce the cholera toxin genes. Microbiology. 2010;156(Pt 1):108–115. doi: 10.1099/mic.0.032235-0. [DOI] [PubMed] [Google Scholar]
- 14.Rubin EJ, Lin W, Mekalanos JJ, Waldor MK. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol Microbiol. 1998;28(6):1247–1254. doi: 10.1046/j.1365-2958.1998.00889.x. [DOI] [PubMed] [Google Scholar]
- 15.Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM. Satellite phage TLCφ enables toxigenic conversion by CTX phage through dif site alteration. Nature. 2010;467(7318):982–985. doi: 10.1038/nature09469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornet F, Louarn J, Patte J, Louarn JM. Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes Dev. 1996;10(9):1152–1161. doi: 10.1101/gad.10.9.1152. [DOI] [PubMed] [Google Scholar]
- 17.Barre FX, et al. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000;14(23):2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy SP, Chevalier F, Barre FX. Delayed activation of Xer recombination at dif by FtsK during septum assembly in Escherichia coli. Mol Microbiol. 2008;68(4):1018–1028. doi: 10.1111/j.1365-2958.2008.06212.x. [DOI] [PubMed] [Google Scholar]
- 19.Demarre G, et al. Differential management of the replication terminus regions of the two Vibrio cholerae chromosomes during cell division. PLoS Genet. 2014;10(9):e1004557. doi: 10.1371/journal.pgen.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aussel L, et al. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108(2):195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 21.Zawadzki P, et al. Conformational transitions during FtsK translocase activation of individual XerCD-dif recombination complexes. Proc Natl Acad Sci USA. 2013;110(43):17302–17307. doi: 10.1073/pnas.1311065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bischerour J, Spangenberg C, Barre F-X. Holliday junction affinity of the base excision repair factor Endo III contributes to cholera toxin phage integration. EMBO J. 2012;31(18):3757–3767. doi: 10.1038/emboj.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faruque SM, et al. Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc Natl Acad Sci USA. 2007;104(12):5151–5156. doi: 10.1073/pnas.0700365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh K, Guo F, Van Duyne GD. Synapsis of loxP sites by Cre recombinase. J Biol Chem. 2007;282(33):24004–24016. doi: 10.1074/jbc.M703283200. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Jayaram M. Role of partner homology in DNA recombination. Complementary base pairing orients the 5′-hydroxyl for strand joining during Flp site-specific recombination. J Biol Chem. 1995;270(8):4042–4052. doi: 10.1074/jbc.270.8.4042. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Narendra U, Iype LE, Cox MM, Rice PA. Crystal structure of a Flp recombinase-Holliday junction complex: Assembly of an active oligomer by helix swapping. Mol Cell. 2000;6(4):885–897. [PubMed] [Google Scholar]
- 27.Kamruzzaman M, et al. RS1 satellite phage promotes diversity of toxigenic Vibrio cholerae by driving CTX prophage loss and elimination of lysogenic immunity. Infect Immun. 2014;82(9):3636–3643. doi: 10.1128/IAI.01699-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim EJ, et al. Molecular Insights Into the Evolutionary Pathway of Vibrio cholerae O1 Atypical El Tor Variants. PLoS Pathog. 2014;10(9):e1004384. doi: 10.1371/journal.ppat.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.